The t(4;14) translocation leads to the simultaneous overexpression of two genes, FGFR3 and MMSET, in myeloma plasma cells. The findings of this study suggest that MMSET is implicated in the pathogenesis of the t(4;14) translocation.
Keywords: MMSET, WHSC1, myeloma
Abstract
Background
The recurrent immunoglobulin translocation, t(4;14)(p16;q32) occurs in 15% of multiple myeloma patients and is associated with poor prognosis, through an unknown mechanism. The t(4;14) up-regulates fibroblast growth factor receptor 3 (FGFR3) and multiple myeloma SET domain (MMSET) genes. The involvement of MMSET in the pathogenesis of t(4;14) multiple myeloma and the mechanism or genes deregulated by MMSET upregulation are still unclear.
Design and Methods
The expression of MMSET was analyzed using a novel antibody. The involvement of MMSET in t(4;14) myelomagenesis was assessed by small interfering RNA mediated knockdown combined with several biological assays. In addition, the differential gene expression of MMSET-induced knockdown was analyzed with expression microarrays. MMSET gene targets in primary patient material was analyzed by expression microarrays.
Results
We found that MMSET isoforms are expressed in multiple myeloma cell lines, being exclusively up-regulated in t(4;14)-positive cells. Suppression of MMSET expression affected cell proliferation by both decreasing cell viability and cell cycle progression of cells with the t(4;14) translocation. These findings were associated with reduced expression of genes involved in the regulation of cell cycle progression (e.g. CCND2, CCNG1, BRCA1, AURKA and CHEK1), apoptosis (CASP1, CASP4 and FOXO3A) and cell adhesion (ADAM9 and DSG2). Furthermore, we identified genes involved in the latter processes that were differentially expressed in t(4;14) multiple myeloma patient samples.
Conclusions
In conclusion, dysregulation of MMSET affects the expression of several genes involved in the regulation of cell cycle progression, cell adhesion and survival.
Introduction
Multiple myeloma (MM) is characterized by the clonal expansion of plasma cells in the bone marrow. Recent advances in autologous stem cell transplantation and chemotherapeutic treatments have improved the long-term survival of patients with MM. However, despite such advances, MM remains an incurable disease for which there is a need to identify novel therapeutic targets. There is a well recognized range of clinical outcomes in MM reflecting the heterogeneous nature of the disease which is characterized by multiple genetic abnormalities. In the past two decades our understanding of the molecular pathology of MM has increased significantly. This increase in knowledge has led to the identification of a wealth of chromosomal and genetic abnormalities, amongst which recurrent translocations targeting the immunoglobulin heavy chain locus (IGH) at the 14q32 locus are the best characterized. These translocations occur in up to 60% of MM patients and deregulate a number of oncogenes including cyclins D1 (11q13) and D3 (6p21), C-MAF (16q23) together with FGFR3 and MMSET (4p16).1 These molecular events are thought to occur early in the natural history of the disease promoting myelomagenesis and defining the clinical outcome. The most important of these translocations clinically is the t(4;14), which is present in 15–20% of MM cases and is associated with poor prognosis.1
The t(4;14) translocation leads to the simultaneous overexpression of two genes, FGFR3 and MMSET.2,3 While the role and mechanism by which FGFR3, a receptor tyrosine kinase promotes myelomagenesis has been shown in various in vitro and in vivo studies, the involvement of MMSET upregulation in t(4;14) myeloma is still poorly explored.4–10 Moreover, about 30% of MM tumors with the t(4;14) translocation have recently been reported to lack expression of FGFR3 due to the loss of der(14).11 Interestingly, the poor prognosis associated with the t(4;14) in such patients lacking FGFR3 expression remains unchanged, lending further support to a potential role for MMSET.5, 11 MMSET has recently been shown to have histone methyltransferase activity and knockdown studies have demonstrated that MMSET upregulation contributes to cellular adhesion, clonogenic growth and tumorigenicity.12 Despite these studies, the genes that are directly or indirectly regulated by MMSET and the mechanisms by which MMSET promotes t(4;14) MM remain unknown.
In this study, using RNAi mediated knockdown and overexpression of the MMSET isoform, REIIBP combined with global expression arrays on myeloma cell lines and patient samples, we have identified genes involved in cell cycle progression, cell adhesion and regulation of apoptosis that are differentially expressed by MMSET in t(4;14) MM. This study significantly sheds light on our better understanding of the molecular mechanisms by which MMSET promotes t(4;14) myelomagenesis.
Design and Methods
Patients and sample preparation
This study was performed on 231 bone marrow (BM) samples with informed consent obtained in accordance with the Declaration of Helsinki received by the Leukaemia Research Fund UK Myeloma Forum Cytogenetic Database between March 2001 and December 2005. Plasma cells were isolated from newly diagnosed myeloma patients to a purity of more than 90% using CD138 microbeads and magnetic assisted cell sorting (Miltenyi Biotech, Bergisch Gladbach, Germany). Selected cells were used for FISH and RNA extraction as described.13, 14
Genome-wide expression arrays
For expression arrays, 100 ng of total tumor RNA was amplified using the 2-cycle target labeling kit (Affymetrix) and as specified by the manufacturer’s instructions. Amplified cRNA (15μg) was hybridized to the Human genome U133 Plus 2.0 arrays as described.13,14 All samples were analyzed with dCHIP 2006 (www.dchip.org).15 In addition, functional clustering analysis was performed using DAVID Bioinformatics Resource 2007 (http://david.abcc.ncifcrf.gov).16
Comparison of expression array samples
MMSET knockdown samples were compared to control samples using dCHIP, for each cell line. In addition, patient samples (n=231) were used to compare those with and without a t(4;14) translocation (Online Supplementary Appendix).
Multiple myeloma cell lines and cell culture
All cell lines were acquired from either ATCC or DSZM, with the exception of KMS-11, which was kindly provided by Dr. Otsuki (Kawasaki Medical School, Japan) (Online Supplementary Appendix).
Nuclear-cytosol extraction and Western blotting analysis
The cytosolic and nuclear fractions of MM cell lines were isolated and Western blotting analysis performed (Online Supplementary Appendix).
MMSET knockdown and REIIBP cloning
MM cell lines were permeabilized to siRNAs using streptolysin-O (SLO) as described previously17–19 (Online Supplementary Appendix).
Cell proliferation assay
Cell proliferation was assessed using the CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega, UK) (Online Supplementary Appendix).
Statistical analysis
Statistical significance was determined by the two-way ANOVA test and Pearson’s correlation using GraphPad Prism version 4.00 for Windows (GraphPad Software, California, US). The minimal level of significance was p<0.05.
Results
MMSET isoforms are up-regulated at the protein level in multiple myeloma cell lines with the t(4;14) translocation
MMSET isoforms up-regulated in MM cell lines and patient samples with the t(4;14) have been studied using quantitative real time PCR (qRT-PCR).5,20 However, due to the complex nature of the alternative splicing of MMSET, the use of any region of MMSET mRNA amplifies or detects multiple transcript isoforms, making the data difficult to interpret accurately (Figure 1A). More recently, antibodies have been used to determine the expression of MMSET isoforms in cell lines.12,21 However, these studies have used a limited number of cell lines carrying the t(4;14) translocation and are, therefore, unable to show the extent of the expression of the different MMSET isoforms in t(4;14) MM. In order to characterize the expression patterns of MMSET isoforms we have studied a panel of MM cell lines carrying the t(4;14) translocation using a novel antibody developed to recognize a C-terminal epitope (Figure 1B). In this analysis we found that, MMSET II, MB4-2II, MB4-3II and REII-BP isoforms were present and are up-regulated in t(4;14)-positive MM cell lines (Figure 1C). We also noted that some MMSET isoforms are expressed in RPMI8226 and U266 cell lines which lack the t(4;14) translocation; however, the level of expression is considerably lower than that seen in cell lines carrying the t(4;14) translocation.
Figure 1.
MMSET gene diagram and predicted expressed variants. (A) MMSET gene diagram depicting the location of MB4-1, MB4-2 and MB4-3 t(4;14) breakpoints including the breakpoints of some of the cell lines used in this study. Exons and introns are represented by boxes and spaces between boxes respectively. Transcription start codons are represented by solid/dotted arrows. The location of the siRNAs used in this study are represented by the thin black lines. (B) MMSET predicted protein variants.5 Shaded boxes represent the protein domains predicted by SMART and the solid dark line represents the epitope recognized by the antibody used in this study. PWWP-proline-tryptophan-tryptophan-proline domain; HMG-High mobility group domain and PHD-Plant homeodomain. (C) Western blotting analysis of the MMSET isoforms expressed in myeloma cell lines using the 5306 antibody. (D) Cellular localization of MMSET isoforms in t(4;14)-positive myeloma cell lines. The nuclear (N) and cytosol (C) fractions are depicted.
MMSET isoforms localize to different cellular compartments
To gain further insight into the biological role of MMSET isoforms in t(4;14) MM, the cellular location of MMSET isoforms was evaluated by isolating the cytosolic and nuclear fractions of t(4;14)–positive cell lines followed by Western blot analysis. The MMSET II, MB4-2II and MB4-3II isoforms were primarily present in the nuclear fraction, whereas the REIIBP was present in both cellular fractions in MM cell lines carrying the t(4;14) translocation (Figure 1D). Furthermore, the level of REIIBP was variable in both cytosolic and nuclear fractions across the cell lines with the t(4;14) translocation, with JIM-1, KMS-11 and LP-1 having higher levels of REIIBP in the nucleus in relation to the cytoplasm. In contrast, REIIBP was present at much higher levels in the cytoplasm compared to the nucleus in the JIM-3 and H929 cell lines. The expression of PARP-1 (control for nuclear fraction) and α-tubulin (control for cytosolic fraction) for each cell line tested indicate that the two fractions were successfully purified with minimal cross-contamination (Figure 1D). These data clearly demonstrate that different MMSET isoforms localize to different cellular compartments. In addition, these data suggest that the MMSET II, MB4-2II, MB4-3II and REIIBP, despite sharing the C-terminal end, may have different biological roles in t(4;14) myeloma.
MMSET knockdown affects cell proliferation, cell cycle and apoptosis
In order to demonstrate the involvement of MMSET upregulation in the pathogenesis of t(4;14) MM we knocked down MMSET expression using specific siRNAs. MMSET knockdown was initially optimized in three myeloma cell lines, RPMI8226, JIM-3 and H929 using the streptolysin-O method.19 We found that two siRNAs, 273 and 490, designed within exons 23 and 24 of MMSET respectively reduced the expression of MB4-2II and MB4-3II isoforms by greater than 90% in the t(4;14)-positive cell lines JIM-3 and H929 in relation to the negative control siRNA (Figure 2A). The expression of REIIBP was not affected by the knockdown, this finding was expected as REIIBP has an extended half-life of 65 hours compared to four hours for MMSET II, MB4-2II and MB4-3II isoforms (data not shown).
Figure 2.
(A) Knockdown of MMSET expression in JIM-3 and H929 human MM cell lines. MMSET was knocked down by transfecting 273 and 490 siRNAs using SLO-permeabilization. MMSET knockdown was assessed at 48h post-transfection by Western blotting analysis. Detection of α-tubulin expression was used to ensure equal loading. (B) Knockdown of MMSET affects proliferation in cells with the t(4;14) translocation. The effect of MMSET on JIM-3 and H929 cells, both of which carry the t(4;14) translocation and RPMI8226 proliferation was assessed by exposing cells to SLO only, SLO with Neg CTL siRNA, 273 siRNA and 490 siRNA. Growth was assessed at 0, 24, 48 and 72 hours by CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay. Values represent mean ± SD of three independent experiments. ** represents p<0.05, two way ANOVA test. (C) Knockdown of MMSET affects cell cycle of human MM cells carrying the t(4;14) translocation. The effect of MMSET knockdown on JIM-3, H929 and RPMI8226 cell cycle was assessed by exposing cells to SLO only, SLO with Neg. CTL siRNA and 273 siRNA for 48 hours. Cell cycle profile was evaluated by flow cytometry using propidium iodide. The data is a representation of three independent experiments.
In addition, we evaluated the effect of MMSET knockdown on cell proliferation and cell cycle progression by MTS assay and propidium iodide staining respectively. MMSET knockdown significantly impaired the proliferation of JIM-3 and H929 cell lines (p<0.05, two-way ANOVA). In contrast, the proliferation of the t(4;14)-negative cell line RPMI8226 was unaffected (Figure 2B). The knockdown of MMSET expression using either siRNA 273 or 490 had a similar effect on the proliferation of MM cell lines, comparable with our observations that both siRNAs efficiently knockdown MMSET expression. Furthermore, MMSET knockdown led to a reduction of JIM-3 and H929 cells in the Go-G1, S and G2-M phases of the cell cycle associated with a substantial increase in the sub-G1 phase of the cell cycle and an increase in cells undergoing apoptosis (Figure 2C and 3A). In addition, MMSET knockdown induced the cleavage of caspase 9 (marker of the intrinsic pathway) and a significant reduction of pro-caspase 3 (Figure 3A). Lastly, the cell cycle status of the t(4;14) negative cell line RPMI8226 was unaffected.
Figure 3.
(A) The effect of MMSET knockdown on JIM-3 and H929 apoptosis was assessed by exposing cells to SLO only, SLO with Neg. CTL siRNA, 273 siRNA and 490 siRNA for 48 h. Apoptotic cells were identified by annexin-V by flow cytometry. Data represents three independent experiments. MMSET knockdown induced apoptosis via the intrinsic pathway. (B) Genes affected by the MMSET deregulation. Venn diagram showing the probesets that overlap among MMSET-suppressed JIM-3 and H929 cells plus those in RPMI8226-REIIBP cells and of t(4;14)Pos tumor samples. The genes corresponding to the probe sets are in brackets. (C) Overall survival comparison of t(4;14)Pos patients with and without FGFR3 expression. (D) Correlation of MMSET with DSG2 and ADAM9 levels in patient samples (Pearson’s correlation, p<0.05). (E) Correlation of MMSET with DSG2 levels using the GSE2658 series deposited at the Gene Expression Omnibus (Pearsons’s correlation, p<0.05).
MMSET knockdown differentially regulates genes involved in cell cycle progression, apoptosis and cell adhesion
We demonstrate above that MMSET knockdown affects key pathways essential for cell proliferation, cell cycle progression and cell survival; however, the important genes mediating these effects remain unclear. In order to address this question we determined the genes that are regulated by MMSET using an approach combining siRNA mediated Knockdown with global expression arrays. In this experiment two myeloma cell lines, JIM-3 and H929 were transiently transfected with 273 and negative control siRNAs in three separate experiments. Forty-eight hours post-transfection, total RNA was isolated and expression patterns determined using Human Genome Plus U133 2.0 arrays. To exclude those genes whose expression is regulated by MMSET in a cell line specific fashion, we determined the genes that were deregulated in both t(4;14)-positive cell lines (average fold difference greater than 1.2; p<0.05). As a result of this analysis we identified 389 genes from which 288 were down-regulated and 101 genes were up-regulated (Online Supplementary Table S1). Importantly, transcript levels of genes associated with interferon responses such as OAS genes, interferon genes or STAT1 were not affected by the MMSET knockdown (data not shown). Of particular interest, the knockdown affected the expression of 29 genes associated with the regulation of cell cycle progression (Table 1). These genes have been implicated in cell cycle arrest in a wide range of cells, consistent with the cell cycle arrest induced by the 273 siRNA. Furthermore, 18 genes implicated in the regulation of programmed cell death were also affected by the MMSET knockdown (Table 2). These data suggest that these genes are involved in the apoptosis induced by MMSET suppression. Lastly, the expression level of genes involved in cell adhesion (DSG2 and ADAM9) were decreased by the suppression of MMSET expression (Online Supplementary Table S1). It is worth mentioning that FGFR3, which is also up-regulated by the t(4;14) translocation was not highlighted as one of the genes regulated by MMSET siRNA mediated knockdown, suggesting that our analysis selectively excluded FGFR3 and FGFR3 regulated genes.
Table 1.
List of genes associated with the regulation of cell cycle progression, whose expression changes with MMSET knockdown.
Table 2.
List of genes associated with apoptosis deregulated by the MMSET knockdown.
Genes differentially expressed by MMSET in cell lines are differentially expressed in patients with t(4;14) translocation
In addition to studying cell lines it is critical to understand the impact of MMSET in primary patient material. We have done this by studying the clinical impact of the presence of the t(4;14) within a series of uniformly treated patients. This case series is representative of other case series in the literature clearly showing the adverse impact on overall survival of the t(4;14) both with and without the deletion of FGFR3 (Figure 3B).
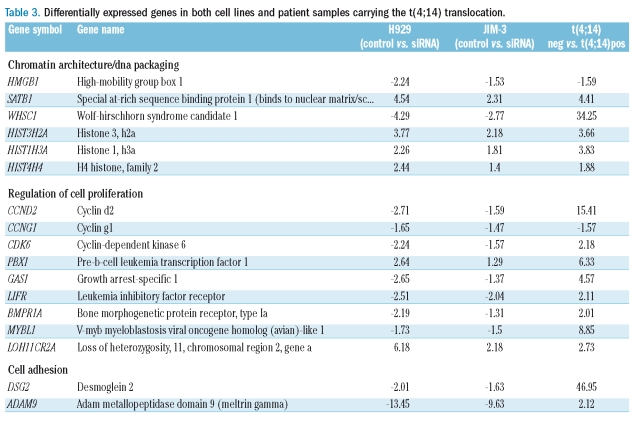
In order to determine potential MMSET targets and understand their relevance in patient samples, we identified genes that were significantly deregulated in both the cell lines and in patient samples carrying the t(4;14) translocation (average fold difference greater than 1.2; p<0.05) (Figure 3C). Total RNA was extracted from CD138+ selected cells from 37 t(4;14)-positive myeloma samples, as well as from 194 CD138+ selected t(4;14)-negative MM samples. In addition, as part of this analysis, REIIBP isoform was cloned and over-expressed in the t(4;14) negative cell line RPMI8226 (Online Supplementary Figure S1), followed by total RNA extraction. As a consequence of this analysis we identified 73 genes that were differentially expressed in both the t(4;14)-positive series and by MMSET knockdown and REIIBP upregulation in cell lines (Online Supplementary Table S2). Of these, we identified genes involved in cell cycle regulation (CCND2, CCNG1, GAS1 and LOH11CR2A), cell proliferation (MYBL1, LIFR and PBX1), cell adhesion (ADAM9 and DSG2) chromatin structure/DNA packaging (HMGB1, SATB1, HIST3H2A, HIST1H3A, HIST1H4A and WHSC1) (Table 3). Interestingly, from the 389 genes previously identified as being regulated by the MMSET knockdown, only 68 (17.5%) were found to be differentially expressed in t(4;14)-positive patient tumor samples. These data suggest that at least 82% of the genes differentially expressed by MMSET suppression may be either indirect targets of MMSET or are regulated as a consequence of the suppression of proliferation, survival and cell cycle progression. Surprisingly, of the 186 genes identified to be differentially expressed by the REIIBP upregulation only 5 genes (CD9, MYO1E, HNRPA3, ZNF677 and C3orf14) were found to be differentially expressed in t(4;14) positive tumor samples (Online Supplementary Tables S2 and S3).
Table 3.
Differentially expressed genes in both cell lines and patient samples carrying the t(4;14) translocation.
MMSET upregulation affects the adhesion profile of myeloma plasma cells
The expression changes identified in this study show a distinct adhesion molecule profile characteristic of t(4;14) myeloma. This observation is particularly interesting because of the potential of these genes to modulate cell adhesion mediated drug resistance (CAMDR), which could explain the clinical behavior of t(4;14) myeloma. The two genes ADAM9 and DSG2 were consistently deregulated at 2-fold and 46.9-fold respectively. In order to further validate these data we examined the correlation of DSG2 and ADAM9 expression with MMSET expression. This analysis demonstrated a significant correlation of both DSG2 and ADAM9 expression with MMSET expression (Pearson’s correlation, p<0.05) (Figure 3D). We validated this observation further and show that DSG2 expression was correlated with MMSET levels in an additional data set [GSE2658 deposited at the Gene Expression Omnibus (GEO)],22 confirming our initial result (Figure 3E). Thus, we demonstrate that knockdown of MMSET is associated with the differential expression of DSG2 and that their expression is also correlated in primary patient samples.
Discussion
The t(4;14) translocation leads to the direct upregulation of both FGFR3 and MMSET, defining them both as candidate oncogenes relevant to the pathogenesis of t(4;14) myeloma.1,2,20,23–25 Recent studies have shown that MMSET has histone methyltransferase activity and interacts with co-repressors such as HDACs, suggesting that it may have a role in regulating gene transcription. However, the mechanism by which MMSET is involved and regulates the pathogenesis of t(4;14) MM is unclear. In this present study, we have assessed the involvement of MMSET in the pathogenesis of t(4;14) MM using siRNA mediated knockdown. In addition, we determined the genes that are globally regulated by MMSET in cell lines and in patient samples carrying the t(4;14). This study unveils important clues about the mechanisms by which MMSET promotes the pathogenesis of t(4;14) MM through its effects on cell cycle progression apoptosis and adhesion.
Using a novel antibody that recognizes an epitope at the C-terminal end of MMSET, we found that MMSET II, MB4-2II, MB4-3II and REIIBP isoforms are expressed and up-regulated in t(4;14)-positive human cell lines. Unexpectedly, MMSET II and REIIBP were found to be expressed in the RPMI8226 cell line, which does not carry the t(4;14) translocation; however they were expressed at a much lower level when compared with cells harboring the translocation. This finding suggests that MMSET isoforms are expressed in myeloma cells and are up-regulated as a consequence of the t(4;14) translocation. In addition, we demonstrate that MMSET isoforms expressed in t(4;14)-positive MM cell lines are predominantly nuclear, a finding that is in concordance with recent reports.12 We suppressed the expression of MMSET using siRNAs in t(4;14)-positive cell lines and demonstrate that cell proliferation, cell cycle progression and apoptosis were affected. These data suggest that MMSET upregulation as a result of t(4;14) translocation is an important event in pathogenesis of t(4;14) MM, which is consistent with two recent reports.12,21 A key observation is that transfection of MMSET into the t(4;14)-negative RPMI8226 cell line did not effect cell proliferation, cell cycle or apoptosis, demonstrating that the low expression level of MMSET in such a cell line has little or no involvement in key cellular processes.
In order to gain further insight into the pathogenic mechanisms underlying MMSET deregulation we determined the global transcriptional downstream effect of MMSET knockdown in t(4;14)-positive cell lines, as well as by comparing these results with over-expression of REIIBP in t(4;14)-positive cells. In consonance with the cell biology outcome occurring as a result of MMSET knockdown, we identified key genes differentially expressed by the MMSET suppression involved in cell cycle regulation, apoptosis and cell adhesion processes.
Among the cell cycle genes, CCND2 was identified as being up-regulated in the t(4;14) subset and found to be down-regulated by the MMSET knockdown. In addition, among the genes that regulate cell cycle progression, several key players involved in cell cycle surveillance (CHEK1, CDC25A, CCNG1, LOH11CR2A and BRCA1), centriole biogenesis (PLK4) and chromosome segregation (AURKA) were identified and shown to have decreased expression levels upon MMSET siRNA treatment. AURKA siRNA targeting was recently shown to induce apoptotic cell death in myeloma cell lines.26 This report suggests that the decreased expression of AURKA may contribute to the enhanced apoptosis induced by MMSET knockdown.
The role of CHEK1, CDC25A, CCNG1, BRCA1, LOH11CR2A, PLK4 and AURKA in cell cycle regulation combined with CCND2, a G1-S transition regulator, and their decreased transcript levels are consistent with the effects of MMSET knockdown on cell cycle progression of t(4;14)-positive cells.
In addition to the affect on cell cycle progression, MMSET knockdown induced apoptosis in t(4;14)-positive cells. Our expression array data demonstrate that MMSET knockdown up-regulated the expression of several genes, such as CASP1, CASP4 and FOXO3A, whose overexpression triggers programmed cell death in human cells.27, 28 In addition, FOXO3A responds to cellular stress by inducing cell cycle arrest, repair of damaged DNA and apoptosis.29–31 In myeloma cells, FOXO3A is a direct target of the antiapoptotic AKT of the PI3K pathway that plays an important role in the survival of myeloma cells by preventing the nuclear translocation and activation of pro-apoptotic proteins such as Bim and Fas ligands.32
Further to the effect of MMSET suppression on cell cycle progression and apoptosis, cell adhesion was also shown to be affected.12 We observed that expression of cell adhesion genes decreased with MMSET knockdown including ADAM9 and DSG2. ADAM9 is expressed in myeloma cells and has been shown to promote cell adhesion by interacting with αvβ5 integrin on osteoblasts stimulating IL6 production.33,34 Whereas the cadherin family member, desmoglein 2 (DSG2), a component of cell-cell adhesion in desmosome formation is associated with cell proliferation, activation of PI3K/AKT and NF-κB signaling pathways in keratinocytes.35 DSG2 has been previously reported to be differentially expressed in patient samples with t(4;14).22 The downregulation of ADAM9 and DSG2 by the MMSET knockdown suggests that upregulation of MMSET promotes cell-stroma and cell-osteoblast interaction, possibly leading to the activation of several paracrine and signaling pathways enhancing cell survival and cell adhesion mediated drug resistance (CAMDR) to conventional therapy, therefore affecting the clinical outcome.
Further to identify genes deregulated in cell lines we have also identified genes that are differentially expressed by MMSET in patient samples. Of the 389 genes shown to be differentially expressed by the MMSET knockdown in myeloma cell lines, 17.5% were expressed in t(4;14) tumor samples, suggesting that these may be direct targets of MMSET. From these we have identified genes consistently deregulated affecting cell cycle (CCND2, CCNG1, GAS1 and LOH11CR2A), cell adhesion (ADAM9 and DSG2) and cell proliferation (MYBL1 and LIFR), which are inversely expressed in tumor samples when compared to MMSET knockdown cell lines, making them excellent targets for future analysis. Upregulation of LIFR and MYBL1 in t(4;14) tumors and their decreased expression by the MMSET knockdown suggest that MMSET is involved in the IL6 signaling pathway and proliferation of t(4;14) myeloma cells. MYBL1 is a key regulator of gene transcription, cell growth and differentiation that is expressed in germinal center B cells and its expression is suppressed towards either memory or plasma cells.36 Whereas, LIFR is an important component of the IL6RκJAK/STATκRAS/MAPK signaling pathway regulating cell cycle progression in myeloma cells.37,38
Our data as a whole suggest that MMSET is implicated in t(4;14) pathogenesis, by promoting cell cycle progression through the regulation of key cell cycle regulators cell adhesion. Cell adhesion enhanced by MMSET upregulation can activate paracrine and signaling pathways enhancing t(4;14) myeloma cell survival and CAMDR to conventional therapy, providing a possible answer for why the myeloma patients with t(4;14) have a poor clinical outcome.
Acknowledgments
we would like to thank Bud Flanagan and Kay Kendall leukemia funds for funding this work.
Footnotes
Authorship and Disclosures
JLRB: designed, performed, collected, analyzed the data and wrote the paper; BW, NJMB: analyzed the data; FMR: collected the data. NJD, MJ, AA, DG, FED: wrote the paper; GJM: designed and wrote the paper. The authors reported no potential conflicts of interest.
References
- 1.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev Cancer. 2002;2:175–87. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 2.Bergsagel PL, Nardini E, Brents L, Chesi M, Kuehl WM. IgH translocations in multiple myeloma: a nearly universal event that rarely involves c-myc. Curr Top Microbiol Immunol. 1997;224:283–7. doi: 10.1007/978-3-642-60801-8_30. [DOI] [PubMed] [Google Scholar]
- 3.Richelda R, Ronchetti D, Baldini L, Cro L, Viggiano L, Marzella R, et al. A novel chromosomal translocation t(4; 14)(p16.3; q32) in multiple myeloma involves the fibroblast growth-factor receptor 3 gene. Blood. 1997;90:4062–70. [PubMed] [Google Scholar]
- 4.Chesi M, Brents LA, Ely SA, Bais C, Robbiani DF, Mesri EA, et al. Activated fibroblast growth factor receptor 3 is an oncogene that contributes to tumor progression in multiple myeloma. Blood. 2001;97:729–36. doi: 10.1182/blood.v97.3.729. [DOI] [PubMed] [Google Scholar]
- 5.Keats JJ, Maxwell CA, Taylor BJ, Hendzel MJ, Chesi M, Bergsagel PL, et al. Overexpression of transcripts originating from the MMSET locus characterizes all t(4;14)(p16;q32)-positive multiple myeloma patients. Blood. 2005;105:4060–9. doi: 10.1182/blood-2004-09-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plowright EE, Li Z, Bergsagel PL, Chesi M, Barber DL, Branch DR, et al. Ectopic expression of fibroblast growth factor receptor 3 promotes myeloma cell proliferation and prevents apoptosis. Blood. 2000;95:992–8. [PubMed] [Google Scholar]
- 7.Ronchetti D, Greco A, Compasso S, Colombo G, Dell’Era P, Otsuki T, et al. Deregulated FGFR3 mutants in multiple myeloma cell lines with t(4;14): comparative analysis of Y373C, K650E and the novel G384D mutations. Oncogene. 2001;20:3553–62. doi: 10.1038/sj.onc.1204465. [DOI] [PubMed] [Google Scholar]
- 8.Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–6. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 9.Sibley K, Fenton JA, Dring AM, Ashcroft AJ, Rawstron AC, Morgan GJ. A molecular study of the t(4;14) in multiple myeloma. Br J Haematol. 2002;118:514–20. doi: 10.1046/j.1365-2141.2002.03618.x. [DOI] [PubMed] [Google Scholar]
- 10.Trudel S, Ely S, Farooqi Y, Affer M, Robbiani DF, Chesi M, et al. Inhibition of fibroblast growth factor receptor 3 induces differentiation and apoptosis in t(4;14) myeloma. Blood. 2004;103:3521–8. doi: 10.1182/blood-2003-10-3650. [DOI] [PubMed] [Google Scholar]
- 11.Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–9. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 12.Lauring J, Abukhdeir AM, Konishi H, Garay JP, Gustin JP, Wang Q, et al. The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood. 2008;111:856–64. doi: 10.1182/blood-2007-05-088674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 14.Walker BA, Leone PE, Jenner MW, Li C, Gonzalez D, Johnson DC, et al. Integration of global SNP-based mapping and expression arrays reveals key regions, mechanisms, and genes important in the pathogenesis of multiple myeloma. Blood. 2006;108:1733–43. doi: 10.1182/blood-2006-02-005496. [DOI] [PubMed] [Google Scholar]
- 15.Zhong S, Li C, Wong WH. ChipInfo: Software for extracting gene annotation and gene ontology information for microarray analysis. Nucleic Acids Res. 2003;31:3483–6. doi: 10.1093/nar/gkg598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35(Web Server issue):W169–75. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giles RV, Spiller DG, Clark RE, Tidd DM. Antisense morpholino oligonucleotide analog induces missplicing of C-myc mRNA. Antisense Nucleic Acid Drug Dev. 1999;9:213–20. doi: 10.1089/oli.1.1999.9.213. [DOI] [PubMed] [Google Scholar]
- 18.Spiller DG, Tidd DM. Nuclear delivery of antisense oligodeoxynucleotides through reversible permeabilization of human leukemia cells with streptolysin O. Antisense Res Dev. 1995 Spring;5:13–21. doi: 10.1089/ard.1995.5.13. [DOI] [PubMed] [Google Scholar]
- 19.Brito JL, Davies FE, Gonzalez D, Morgan GJ. Streptolysin-O reversible permeabilisation is an effective method to transfect siRNAs into myeloma cells. J Immunol Methods. 2008;333:147–55. doi: 10.1016/j.jim.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 20.Stewart JP, Thompson A, Santra M, Barlogie B, Lappin TR, Shaughnessy J., Jr Correlation of TACC3, FGFR3, MMSET and p21 expression with the t(4;14)(p16.3;q32) in multiple myeloma. Br J Haematol. 2004;126:72–6. doi: 10.1111/j.1365-2141.2004.04996.x. [DOI] [PubMed] [Google Scholar]
- 21.Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood. 2008;111:3145–54. doi: 10.1182/blood-2007-06-092122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–8. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chesi M, Bergsagel PL, Kuehl WM. The enigma of ectopic expression of FGFR3 in multiple myeloma: a critical initiating event or just a target for mutational activation during tumor progression. Curr Opin Hematol. 2002;9:288–93. doi: 10.1097/00062752-200207000-00005. [DOI] [PubMed] [Google Scholar]
- 24.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–34. [PubMed] [Google Scholar]
- 25.Malgeri U, Baldini L, Perfetti V, Fabris S, Vignarelli MC, Colombo G, et al. Detection of t(4;14)(p16.3;q32) chromosomal translocation in multiple myeloma by reverse transcription-polymerase chain reaction analysis of IGH-MMSET fusion transcripts. Cancer Res. 2000;60:4058–61. [PubMed] [Google Scholar]
- 26.Evans R, Naber C, Steffler T, Checkland T, Keats J, Maxwell C, et al. Aurora A kinase RNAi and small molecule inhibition of Aurora kinases with VE-465 induce apoptotic death in multiple myeloma cells. Leuk Lymphoma. 2008;49:559–69. doi: 10.1080/10428190701824544. [DOI] [PubMed] [Google Scholar]
- 27.Feng Q, Li P, Leung PC, Auersperg N. Caspase-1zeta, a new splice variant of the caspase-1 gene. Genomics. 2004;84:587–91. doi: 10.1016/j.ygeno.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Kamens J, Paskind M, Hugunin M, Talanian RV, Allen H, Banach D, et al. Identification and characterization of ICH-2, a novel member of the interleukin-1 beta-converting enzyme family of cysteine proteases. J Biol Chem. 1995;270:15250–6. doi: 10.1074/jbc.270.25.15250. [DOI] [PubMed] [Google Scholar]
- 29.Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, et al. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–68. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 30.Martinez-Gac L, Alvarez B, Garcia Z, Marques M, Arrizabalaga M, Carrera AC. Phosphoinositide 3-kinase and Forkhead, a switch for cell division. Biochem Soc Trans. 2004;32(Pt 2):360–1. doi: 10.1042/bst0320360. [DOI] [PubMed] [Google Scholar]
- 31.Tran NL, Adams DG, Vaillancourt RR, Heimark RL. Signal transduction from N-cadherin increases Bcl-2. Regulation of the phosphatidylinositol 3-kinase/Akt pathway by homophilic adhesion and actin cytoskeletal organization. J Biol Chem. 2002;277:32905–14. doi: 10.1074/jbc.M200300200. [DOI] [PubMed] [Google Scholar]
- 32.Younes H, Leleu X, Hatjiharissi E, Moreau AS, Hideshima T, Richardson P, et al. Targeting the phosphatidylinositol 3-kinase pathway in multiple myeloma. Clin Cancer Res. 2007;13:3771–5. doi: 10.1158/1078-0432.CCR-06-2921. [DOI] [PubMed] [Google Scholar]
- 33.Karadag A, Zhou M, Croucher PI. ADAM-9 (MDC-9/meltrin-gamma), a member of the a disintegrin and metalloproteinase family, regulates myeloma-cell-induced interleukin-6 production in osteoblasts by direct interaction with the alpha(v)beta5 integrin. Blood. 2006;107:3271–8. doi: 10.1182/blood-2005-09-3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou M, Graham R, Russell G, Croucher PI. MDC-9 (ADAM-9/Meltrin gamma) functions as an adhesion molecule by binding the alpha(v)beta(5) integrin. Biochem Biophys Res Commun. 2001;280:574–80. doi: 10.1006/bbrc.2000.4155. [DOI] [PubMed] [Google Scholar]
- 35.Brennan D, Hu Y, Joubeh S, Choi YW, Whitaker-Menezes D, O’Brien T, et al. Suprabasal Dsg2 expression in transgenic mouse skin confers a hyperproliferative and apoptosis-resistant phenotype to keratinocytes. J Cell Sci. 2007;120:758–71. doi: 10.1242/jcs.03392. [DOI] [PubMed] [Google Scholar]
- 36.Golay J, Broccoli V, Lamorte G, Bifulco C, Parravicini C, Pizzey A, et al. The A-Myb transcription factor is a marker of centroblasts in vivo. J Immunol. 1998;160:2786–93. [PubMed] [Google Scholar]
- 37.Gado K, Domjan G, Hegyesi H, Falus A. Role of INTERLEUKIN-6 in the pathogenesis of multiple myeloma. Cell Biol Int. 2000;24:195–209. doi: 10.1006/cbir.2000.0497. [DOI] [PubMed] [Google Scholar]
- 38.Gu ZJ, Costes V, Lu ZY, Zhang XG, Pitard V, Moreau JF, et al. Interleukin-10 is a growth factor for human myeloma cells by induction of an oncostatin M autocrine loop. Blood. 1996;88:3972–86. [PubMed] [Google Scholar]