Abstract
Lyme borreliosis (LB) is a tick-borne spirochetal infection caused by three Borrelia species: Borrelia afzelii, B. garinii, and B. burgdorferi sensu stricto. LB evolves in two stages: a skin lesion called erythema migrans and later, different disseminated forms (articular, neurological, cardiac…). Previous research based on analysis of ospC sequences allowed the definition of 58 groups (divergence of <2% within a group and >8% between groups). Only 10 of these groups include all of the strains isolated from disseminated forms that are considered invasive. The aim of this study was to determine whether or not invasive strains belong to restricted ospC groups by testing human clinical strains isolated from disseminated forms. To screen for ospC genetic diversity, we used single-strand conformation polymorphism (SSCP) analysis. Previously known ospC sequences from 44 different strains were first tested, revealing that each ospC group had a characteristic SSCP pattern. Therefore, we studied 80 disseminated-form isolates whose ospC sequences were unknown. Of these, 28 (35%) belonged to previously known invasive groups. Moreover, new invasive groups were identified: six of B. afzelii, seven of B. garinii, and one of B. burgdorferi sensu stricto. This study confirmed that invasive strains are not distributed among all 69 ospC groups but belong to only 24 groups. This suggests that OspC may be involved in the invasiveness of B. burgdorferi.
Lyme borreliosis (LB) is the most commonly reported tick-borne infection in Europe and North America. The etiological agent belongs to the complex Borrelia burgdorferi sensu lato, which comprises at least 11 species, of which only 3 are pathogenic for humans: B. burgdorferi sensu stricto, B. afzelii, and B. garinii (2, 4). LB evolves in two stages that may occur independently. The first is a primary skin lesion that spreads from the site of the tick bite and is called erythema migrans (EM). Later, bacteria may disseminate to different organs and induce different clinical manifestations, i.e., neurological (neuroborreliosis [NB]), articular (arthritis), cardiac, cutaneous (multiple EM [MEM]), acrodermatitis chronica atrophicans (ACA), and lymphocytoma benigna cutis (LBC). Each of these manifestations is associated with a distinct pathogenic species, and more frequently a given patient experiences only one of these delayed manifestations (1, 5, 22, 23). The expression patterns of outer surface protein A (OspA) and OspC play an important role in the infection cycle. In unfed ticks, spirochetes express OspA but not OspC. During tick feeding, OspA synthesis is repressed whereas OspC synthesis is induced. The switch is associated with increased temperature and with exposure to tick hemolymph factor (10, 16, 19). Thus, OspC is the major outer surface protein expressed in early infection. Moreover, Masuzawa et al. (14) have shown that OspC expression is associated with infectivity. The ospC gene is highly variable (9, 12, 21, 25, 26). On the basis of ospC sequence analysis, Wang et al. (24) defined major ospC groups (divergence of <2% within a group and >8% between groups). Furthermore, Seinost et al. (17) and Baranton et al. (3) found that, among 149 sequences from data banks, 58 ospC groups can be defined within the three pathogenic species. Only 10 of these groups contain all of the strains (76 sequences) isolated from different clinical samples in disseminated forms (referred to secondary sites). These strains are hereafter called invasive. Two groups are individualized within B. afzelii, four within B. garinii, and four within B. burgdorferi sensu stricto. These groups are defined as invasive. Other ospC groups comprise strains isolated either from primary skin lesions or from ticks and are defined as noninvasive or of unknown invasiveness. In this study, we extended the investigation to 80 clinical strains isolated from secondary sites and of which the ospC sequence was unknown. We used single-strand conformation polymorphism (SSCP) analysis to study the genetic variability of the ospC gene. We determined the distribution among defined invasive groups. Our results confirm that invasive strains belong to given ospC groups. This suggests that the ospC gene could be one of the determinants involved in the invasiveness of strains leading to disseminated forms of the disease.
MATERIALS AND METHODS
Borrelia strains.
One hundred twenty-four B. burgdorferi strains were tested in this study. Forty-four strains whose ospC sequences are known were chosen, as indicated in Table 1. Eighty human-invasive strains were selected, including 61 Slovenian isolates. Fifty strains were isolated from different clinical samples (skin, blood, and cerebrospinal fluid [CSF]) in disseminated forms (ACA, MEM, LBC, and NB). Thirty strains isolated in the early stage were also included in this study. As they were isolated from the blood of patients with EM, they were considered to be invasive. Strains were cultured at 33°C in BSK-H medium (Sigma). The cultures were routinely monitored by dark-field microscopy for growth and contamination.
TABLE 1.
Strains previously identified as invasivea (A1 to G4) or noninvasive on the basis of their sequences
| Strain(s) | Vector(s) | Source (condition) | Origin(s) | ospC group | Seinost ospC group designation (17) |
|---|---|---|---|---|---|
| B. afzelii | |||||
| ACA1 | Skin (ACA) | Sweden | A1 | ||
| DK3, DK8 | Skin (ACA) | Denmark | A2 | ||
| PKo, DK26 | Skin (EM) | Germany, Denmark | A2 | ||
| E61 | Skin (EM) | Austria | NIb | ||
| Simon | Skin (EM) | Austria | NI | ||
| H9 | Skin (EM) | Austria | NI | ||
| J1 | Ixodes persulcatus | Japan | NI | ||
| VS461 | I. ricinus | Switzerland | NI | ||
| B. burgdorferi sensu stricto | |||||
| 28354 | I. scapularis | United States | B1 | K | |
| 297 | CSF (NB) | United States | B1 | K | |
| DK7 | Skin (ACA) | Denmark | B2 | B | |
| 61BV3 | Skin (EM) | Germany | B2 | B | |
| ZS7 | I. ricinus | Switzerland | B2 | B | |
| HB19 | Blood (arthritis) | United States | B3 | I | |
| Pka | I. ricinus | Germany | B4 | A | |
| IP1, IP2, IP3 | CSF (NB) | France | B4 | A | |
| HII | Blood (arthritis) | Italy | B4 | A | |
| P1F | Synovia (arthritis) | Switzerland | B4 | A | |
| B31, 26816 | I. scapularis | United States | B4 | A | |
| Mil | I. ricinus | Slovakia | NI | J | |
| 20006 | I. ricinus | France | NI | P | |
| 212 | I. ricinus | France | NI | Q | |
| ESP1, Ne-56 | I. ricinus | Spain, Switzerland | NI | R | |
| Z136 | I. ricinus | Germany | NI | S | |
| Son188 | I. pacificus | United States | NI | F | |
| B. garinii | |||||
| N34, Far03 | I. ricinus; I. uriae | Germany, Sweden | G1 | ||
| W | CSF (NB) | Austria | G2 | ||
| VSDA | CSF (NB) | Switzerland | G3 | ||
| PBi | CSF (NB) | Germany | G4 | ||
| DK6 | CSF (NB) | Denmark | G4 | ||
| KL11 | I. ricinus | Czech Republic | G4 | ||
| NBS16 | I. ricinus | Sweden | NI | ||
| NBS23 | I. ricinus | Sweden | NI | ||
| 153 | I. ricinus | France | NI | ||
| 20047 | I. ricinus | France | NI | ||
| T25 | I. ricinus | Germany | NI | ||
| BITS | I. ricinus | Italy | NI |
A strain was considered invasive if it was isolated from secondary sites or when it belonged to a group comprising strains isolated from secondary sites.
NI, noninvasive group.
DNA isolation.
Bacterial cultures were harvested by centrifugation (10,000 × g; 10 min). The bacterial pellet was washed in phosphate-buffered saline, resuspended in water, heated at 100°C for 10 min, and then stored at −20°C.
PCR.
A 277-bp fragment of the variable central part of ospC, suitable in size for SSCP analysis, was amplified by using forward primer SC3 (5′-AAAGCTATTGGTAAAGTAAT-3′; bp 226 to 245; Genset) and reverse primer OspC92 (5′-GTTTTTAAAATAGCTTTTTTTG-3′; bp 491 to 470; Eurogentec), which are based on consensus sequences for the three pathogenic species.
Amplification was processed in 25 μl of a solution containing 0.2 μM each primer, 0.2 mM each deoxynucleoside triphosphate, 0.625 U of Taq polymerase (Q. Bio gene), and 1× Taq buffer (1.5 mM MgCl2). The amplification reaction was carried out in a DNA thermal cycler (Touch Down Hybaid) under the following conditions: initial denaturation at 93°C for 1 min, followed by 35 cycles of denaturation at 93°C for 1 min, annealing at 48°C for 1 min, and extension at 72°C for 30 s. Negative controls were included to check for contamination. Amplification was checked by agarose gel electrophoresis. A 5-μl volume of each sample was loaded onto a 1% TBE 1X (Tris-borate-EDTA) agarose gel and revealed by ethidium bromide staining.
SSCP.
A 3-μl volume of the PCR product was added to 3 μl of denaturation solution (94% formamide, 0.05% xylene cyanol) and heated at 95°C for 5 min. Samples were loaded on a nondenaturing polyacrylamide gel (GeneGel Excel 12,5/24; Amersham Pharmacia Biotech). Electrophoresis was performed in a temperature-controlled electrophoresis system (GenePhor; Amersham Pharmacia Biotech) at 6°C with a first run at 600 V, 25 mA, and 15 W for 10 min and then at 600 V, 37 mA, and 21 W for 2 h 30 min.
Gels were revealed by silver staining (Plus One DNA Silver Staining Kit; Amersham Pharmacia Biotech) in accordance with the manufacturer's instructions.
DNA sequencing.
The partial ospC gene (sizes ranged from 534 to 601 bp) was sequenced (n = 25) as previously described (13), by Genome Express, Montreuil, France.
Phylogenetic analysis.
ospC gene sequences recorded from the GenBank database and new sequences (sizes ranged from 445 to 460 bp) were aligned manually by using VSM software and analyzed by the unweighted pair group method with mathematic averages (UPGMA) (18). Phylogenic trees were drawn with Mega software (11).
RESULTS AND DISCUSSION
SSCP is a screening method based on the secondary structure of a single-stranded DNA fragment. Different single-stranded DNA sequences result in different conformational foldings. These conformational polymorphisms can be discriminated by their electrophoretic mobilities on polyacrylamide gels (8, 15). This method is widely used for mutation analysis (20). Theoretically, three bands are detectable on the gel (two single DNA strands and one double DNA strand), but heteroduplex formation causes several conformations that coexist in the gel. Under our conditions, different electrophoretic mobilities were indicative of sequence heterogeneities, irrespective of the genetic distance between sequences and of their mutation rates. This method was previously used (17, 24) to provide evidence for different groups of ospC mobility classes within B. burgdorferi sensu stricto. The authors also demonstrated that each mobility class had a unique sequence. Our first objective was to confirm that such data are also applicable to B. garinii and B. afzelii. The analysis was initiated by selecting strains from our collection (n = 44) that belong to the three pathogenic species and whose ospC sequence is available in data banks. Therefore, these strains could be classified into either invasive (n = 26) or noninvasive (n = 18) groups on the basis of their sequences (Table 1). Within each ospC group, the sequences were very similar, with less than 2% nucleotide differences. SSCP patterns were determined for all of the strains. Moreover, SSCP reproducibility was checked, revealing that the PCR product obtained in repeated experiments from a given DNA always yielded the same SSCP pattern and one PCR product always yielded the same SSCP pattern in different migrations.
Within the 26 invasive strains, we identified 11 distinct patterns, each corresponding to 1 of the 10 invasive groups previously described (A1 and -2, B1 to -4, and G1 to -4; Table 1) (3, 17). However, within invasive group G4, two different SSCP patterns were observed. From the 18 noninvasive strains, 17 different SSCP patterns were recorded, in complete accordance with the sequence analysis (data not shown). These results confirmed that each ospC group should correspond to a specific SSCP pattern, in agreement with previous studies (17, 24). Therefore, SSCP patterns could be used as references for the assignment of clinical strains to invasive or noninvasive groups.
Further, since SSCP methodology has been validated as a powerful and reproducible screening tool, we tested 80 clinical invasive European isolates from different secondary sites (Table 2). Additionally, some ospC sequences were determined in order to confirm the assignment of strains to a given invasive group.
TABLE 2.
Clinical strains isolated from secondary sites with unknown ospC sequences
| Strain(s) | Clinical data | Origin(s) | ospC group | Accession no. of ospC sequences determined in this study |
|---|---|---|---|---|
| B. afzelii | ||||
| Spet1793/01, Sbri | Skin (ACA) | Slovania | A1 | |
| Svod, Srej | Blood (EM) | Slovenia | A1 | |
| ACA2a | Skin (ACA) | Sweden | New invasive group A3 | AY150206 |
| P/stoa | Skin (ACA) | Germany | New invasive group A4 | AY150205 |
| Skoz | Blood (EM) | Slovenia | A4 | |
| 5 isolates (Skol,a Ssima) | Blood (MEM) | Slovenia | New invasive group A5 | AY150203, AY150202 |
| Shri | Blood (meningitis) | Slovenia | A5 | |
| 22 isolates | Blood (EM) | Slovenia | A5 | |
| Spria | Blood (MEM) | Slovenia | New invasive group A6 | AY150204 |
| Sspe, Sdol | Blood (EM) | Slovenia | A6 | |
| Sobla | Skin (ACA) | Slovenia | New invasive group A7 | AY150201 |
| Shrva | Skin (ACA) | Slovenia | New invasive group A8 | AY150200 |
| Srav, Svirb | CSF (NB) | Slovania | A1 | |
| GR4135b | CSF (NB) | Austria | A1 | |
| Sjam, Specb | CSF (NB) | Slovania | A5 | |
| Sabub | CSF (NB) | Slovania | A6 | |
| B. garinii | ||||
| 60, Schenk, PATO3 | CSF (NB) | Austria | G1 | |
| 387a | CSF (NB) | Germany | G1 | AY150188 |
| Skos,a Sspaa | CSF (NB) | Slovenia | G1 | AY150194, AY150197 |
| 57, GR4229 | CSF (NB) | Austria | G2 | |
| Sdom | CSF (NB) | Slovenia | G2 | |
| Silc | Blood (EM) | Slovenia | G2 | |
| 239, VSBP | CSF (NB) | Austria, Switzerland | G4 | |
| Scar, Stama | CSF (NB) | Slovenia | G4 | AY150187 |
| Skle, Ssko | Blood (EM) | Slovenia | G4 | |
| VSBMa | CSF (NB) | Switzerland | New invasive group G5 | AY150185 |
| PBra | CSF (NB) | Germany | New invasive group G6 | AY150186 |
| GrLil | CSF (NB) | Austria | New invasive group G7 | |
| Sbosa | CSF (NB) | Slovenia | New invasive group G8 | AY150193 |
| IBS-8,a Smrza | CSF (NB) | France, Slovenia | New invasive group G10 | AY150189, AY150196 |
| Smara | CSF (NB) | Slovenia | New invasive group G11 | AY150195 |
| IBS-9a,b | Skin (LBC) | France | G1 | AY150190 |
| Skotb | Skin (ACA) | Slovenia | G4 | |
| Spet 114/95a,b | Skin (ACA) | Slovenia | G5 | AY150192 |
| Sles,a,b Ssoba,b | Skin (ACA) | Slovenia | G7 | AY150199, AY150198 |
| Spet 1058/01a,b | Skin (ACA) | Slovenia | New invasive group G9 | AY150191 |
| B. burgdorferi sensu stricto | ||||
| Lenz, Holzer | Cardiac muscle, blood | Austria | B4 (A)c | |
| Shaja,b | Skin (ACA) | Slovenia | B2 (B)c | AY150208 |
| Szid,a,b BRE-13a,b | Skin (ACA), CSF (NB) | Slovenia, France | New invasive group B5 (Q)c | AY150209, AY150207 |
Strain whose ospC sequence was sequenced in this study.
Strain isolated in a unusual clinical manifestation.
Seinost ospC group designation (17).
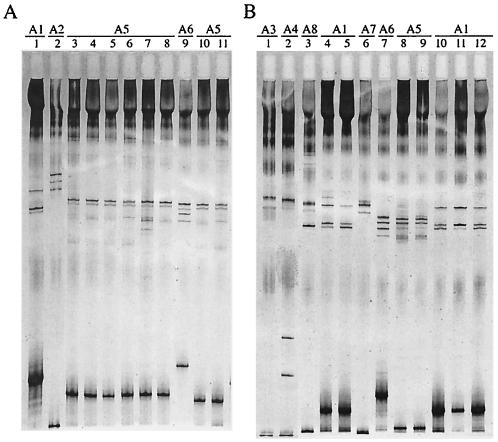
Forty-six invasive B. afzelii strains were analyzed by SSCP. Seven distinct SSCP patterns were recorded (Fig. 1), corresponding to seven different ospC groups in accordance with the phylogenetic analysis of sequences (Fig. 2). Seven strains exhibited the typical pattern of invasive group A1. No pattern corresponding to previously described group A2 was observed. The 39 remaining strains belonged to six new distinct invasive groups designated A3 to A8 (Table 2 and Fig. 2). Strains E61 and Simon, which have been previously designated noninvasive (3), had the same SSCP pattern as invasive strains ACA2 and P/sto, respectively (Table 1 and data not shown). ospC sequencing and phylogenetic analysis confirmed that these strains should now be included in new invasive groups A3 and A4 (Table 2 and Fig. 2). Invasive group A5 comprised isolate Orth, which was not previously considered to be invasive on the basis of the sequence data (Fig. 2). Although invasive strains Spri, Sspe, and Sdol exhibited identical SSCP patterns that were different from that of noninvasive strain H9, sequencing data allowed them to be assigned to new invasive group A6 (data not shown; Table 2 and Fig. 2). Therefore, group A6 exhibited two different patterns.
FIG. 1.
ospC SSCP patterns of 21 invasive B. afzelii isolates. A 277-bp fragment of the central variable part of the ospC gene was amplified by PCR, denatured by heating and formamide, and electrophoresed in a nondenaturing polyacrylamide gel. (A) Lanes: 1, B. afzelii ACA1; 2, B. afzelii DK8; 3, B. afzelii Shri; 4, B. afzelii Skol; 5: B. afzelii Szaj (a strain of the five Slovenian isolates from blood [MEM]); 6: B. afzelii Savb (a strain of the five Slovenian isolates from blood [MEM]); 7, B. afzelii Ssim; 8, B. afzelii Svre (a strain of the five Slovenian isolates from blood [MEM]); 9, B. afzelii Spri; 10, B. afzelii Sper (a strain of the 22 Slovenian isolates from blood [EM] [Table 2]), 11, B. afzelii Slet (a strain of the 22 Slovenian isolates from blood [EM] [Table 2]). (B) Lanes: 1, B. afzelii ACA2; 2, B. afzelii P/sto; 3, B. afzelii Shrv; 4, B. afzelii Spet 1793/01; 5, B. afzelii Sbri; 6, B. afzelii Sobl; 7, B. afzelii Sabu; 8, B. afzelii Sjam; 9, B. afzelii Spec; 10, B. afzelii Srav; 11 B. afzelii Svir; 12, B. afzelii GR4135.
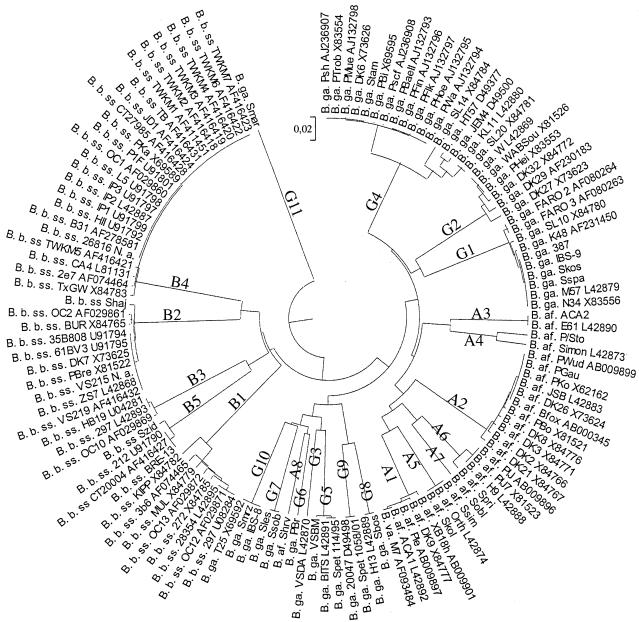
FIG. 2.
UPGMA-based tree of invasive ospC sequences from 100 isolates available in data banks (indicated by their accession numbers) and the 25 ospC sequences determined in this study. B. b. ss, B. burgdorferi sensu stricto; B. ga., B. garinii; B. af., B. afzelii; B. va., B. valaisiana.
Twenty-nine invasive B. garinii strains were also tested by SSCP. Eighteen strains exhibited a pattern previously assigned to an invasive group; seven patterns referred to invasive group G1, four patterns referred to G2, and seven patterns referred to G4. Seven distinct SSCP patterns were identified among the 11 remaining strains (data not shown). ospC sequencing confirmed that these B. garinii strains belonged to seven new distinct invasive groups designated G5 to G11 (Table 2 and Fig. 2). The SSCP patterns of groups G5 (strains VSBM, Spet 114/95, and BITS), G9 (strains Spet 1058/01 and 20047), and G10 (strains IBS-8, Smrz, and T25), which have been previously designated distinct noninvasive groups (3), should be now assigned to invasive groups (data not shown; Table 1 and Fig. 2). A sequence homologous to that of group G8 was also found in data banks (Fig. 2). The three remaining invasive groups (G6, G7, and G11) constituted new ospC groups (Table 2 and Fig 2).
Finally, we tested five B. burgdorferi sensu stricto isolates. Two SSCP patterns referred to invasive group B4, and one pattern referred to B2. The two remaining strains, which had the same SSCP pattern as strain 212 and were previously identified as noninvasive, should now be included in new invasive group B5 (data not shown; Table 1 and Fig. 2). Wang et al. (24) and Seinost et al. (17) have screened a large number of ospC genes from North American human and tick B. burgdorferi sensu stricto isolates. This study allowed us to list all of the ospC groups found in North America. As additional invasive group B5 found in this study comprised only strains from Europe, it had not been identified in those previous studies. Genetic diversity studies based on the whole genome (7) or on the ospC gene (13) have shown that North American B. burgdorferi sensu stricto isolates are more heterogeneous than European ones and that some groups are restricted to either North America or Europe. Analysis of all of the B. burgdorferi sensu stricto ospC sequences found in data banks revealed that invasive groups B1 to -4 are all found in North America, whereas only groups B2, B4, and B5 are found in Europe, suggesting that groups B1 and B3 are restricted to North America and B5 is restricted to Europe.
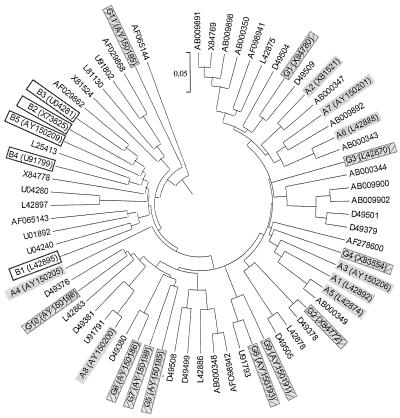
Previous studies based on ospC sequences (3, 17) allowed the definition of 10 invasive groups within 58 groups. Within the 80 strains tested, 28 (35%) belonged to previously defined invasive groups and the 52 remaining strains (65%) belonged to the 14 new invasive ospC groups identified in this study. Nine of these 14 groups comprise isolates that, on the basis of their ospC sequences, were previously classified as noninvasive, increasing the proportion of invasive groups. However, invasive ospC groups A7, A8, G6, G7, and G11 constituted previously unobserved ospC groups. At the same time, new ospC groups were also identified and added to data banks, increasing the global ospC diversity to 69 ospC groups. Thus, ospC sequence analysis allowed us to define 69 groups among 204 sequences, including 24 invasive groups (125 sequences) (Fig. 3 [a table listing nucleotide identities for the newly described ospC groups is available]). All of these results confirmed that invasive strains were not distributed within all ospC groups but belonged to restricted ospC groups (24 [35%] of 69) (Fig. 3). Moreover, the strains additionally studied in this work were selected for their invasiveness, thus increasing the probability of finding only new invasive groups.
FIG. 3.
ospC genetic diversity tree (UPGMA method) drawn from one representative sequence, indicated by accession numbers, from each of 69 ospC groups of the three pathogenic species (204 sequences). Invasive groups are identified by their coding abbreviations (A1 to G11). B. afzelii invasive group, plain gray shading; B. burgdorferi sensu stricto invasive group, no hatching or shading; B. garinii invasive group, cross-hatched and shaded gray.
Fingerprinting studies (12, 25, 26) indicate that B. garinii is the most heterogeneous species, whereas B. afzelii is the most homogeneous one. We found 11 invasive groups for B. garinii. Within group G4, which was very heterogeneous, as indicated by the phylogenetic tree in Fig. 2, two different patterns were observed. These data confirmed that B. garinii is heterogeneous. However, surprisingly, we found eight invasive groups of B. afzelii and two different patterns within the A6 group. Thus, B. afzelii was as heterogeneous as B. garinii regarding the ospC gene.
Wang et al. (24) have shown that the genetic diversity of B. burgdorferi sensu stricto within a local population of ticks is almost equal to the worldwide genetic diversity. Our study revealed that ospC groups identified from human disseminated forms were found in different areas, suggesting that these groups are widely distributed. Moreover, the analysis of 61 Slovenian strains showed that most invasive groups were found in a local area (Table 2). These results were in agreement with those of Wang et al. (24). However, a heterogeneous distribution of the sequences of human origin was found. For instance, in data banks, 11 out of 14 sequences from humans in group G4 were from strains isolated in Germany. Regarding the Slovenian strains tested in this study, 30 (70%) out of the 43 invasive B. afzelii strains fell into group A5. These data suggested that all of the ospC groups had a wide range but some groups could be selected, in humans, in a restricted geographical area.
Each pathogenic Borrelia species is predominantly associated with a given late clinical manifestation: B. afzelii with cutaneous manifestations, B. garinii with neurologic manifestations, and B. burgdorferi sensu stricto with articular manifestations (1, 22). However, sequences from strains isolated from each clinical manifestation are scattered along the tree (data not shown). Fifteen strains included in our study were responsible for a clinical manifestation different from that expected (six B. afzelii strains isolated from CSF, five B. garinii strains from ACA, two B. burgdorferi sensu stricto strains from ACA, one B. garinii strain from LBC, and one B. burgdorferi sensu stricto strain from CSF; Table 2). Indeed, lateral transfer, which is common in the ospC gene (6, 9, 12, 13), could have been responsible for such interspecific organotropism. However, our results showed that ospC sequences from these particular isolates clustered together with strains isolated from expected clinical manifestations. For example, B. afzelii strains Srav and Svir, isolated from the CSF of a patient with NB, belonged to group A1, as did Spet 1793/01 and Sbri, which were isolated from a patient with ACA (Table 2). Moreover, sequence analysis revealed that B. burgdorferi sensu stricto strains P1F (arthritis) and IP1-2-3 (NB) in group B4 have exactly the same ospC sequence (Table 1 and Fig. 2). Furthermore, it was found that B. afzelii strain Shrv, which was involved in a genetic transfer from B. garinii, was responsible for a case of ACA in a Slovenian patient. Our data indicate that clinical presentation is not associated with a given ospC sequence.
Our results and those of others (17, 24) have demonstrated that SSCP can be used for epidemiological studies of tick isolates or first-stage EM isolates in order to evaluate the proportion of invasive strains and to predict evolution toward disseminated forms. Extensive geographical studies could determine the distribution and frequency of ospC groups according to the area.
OspC expression is induced during tick feeding (16), and it is the major outer surface protein expressed in early infection, but its role is still unknown. Our results show that invasive strains belong to restricted ospC groups, suggesting that OspC is one of the factors involved in Borrelia invasiveness. Further studies to determine the role of OspC in Borrelia invasiveness are in progress in our laboratory.
Acknowledgments
We thank D. Dykhuizen and L. Gern for reviewing the manuscript and I. Old for reading the manuscript.
REFERENCES
- 1.Assous, M., D. Postic, G. Paul, P. Nevot, and G. Baranton. 1993. Western blot analysis of sera from Lyme borreliosis patients according to the genomic species of the Borrelia strains used as antigens. Eur. J. Clin. Microbiol. Infect. Dis. 12:261-268. [DOI] [PubMed] [Google Scholar]
- 2.Baranton, G., D. Postic, I. Saint Girons, P. Boerlin, J.-C. Piffaretti, M. Assous, and P. A. D. Grimont. 1992. Delineation of Borrelia burgdorferi sensu stricto, Borrelia garinii sp. nov., and group VS461 associated with Lyme borreliosis. Int. J. Syst. Bacteriol. 42:378-383. [DOI] [PubMed] [Google Scholar]
- 3.Baranton, G., G. Seinost, G. Theodore, D. Postic, and D. Dykhuisen. 2001. Distinct levels of genetic diversity of Borrelia burgdorferi are associated with different aspects of pathogenicity. Res. Microbiol. 149-156. [DOI] [PubMed]
- 4.Caniça, M. M., F. Nato, L. Du Merle, J. C. Mazie, G. Baranton, and D. Postic. 1993. Monoclonal antibodies for identification of Borrelia afzelii sp. nov. associated with late cutaneous manifestations of Lyme borreliosis. Scand. J. Syst. Infect. Dis. 25:441-448. [DOI] [PubMed] [Google Scholar]
- 5.Dhôte, R., A. L. Basse-Guerineau, V. Beaumesnil, B. Christoforov, and M. Assous. 2000. Full spectrum of clinical, serological, and epidemiological features of complicated forms of Lyme borreliosis in the Paris, France, area. Eur. J. Clin. Microbiol. Infect. Dis. 809-815. [DOI] [PubMed]
- 6.Dykhuisen, D. E., and G. Baranton. 2001. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9:344-350. [DOI] [PubMed] [Google Scholar]
- 7.Foretz, M., D. Postic, and G. Baranton. 1997. Phylogenetic analysis of Borrelia burgdorferi sensu stricto by arbitrarily primed PCR and pulsed-field gel electrophoresis. Int. J. Syst. Bacteriol. 47:11-18. [DOI] [PubMed] [Google Scholar]
- 8.Hongyo, T., G. S. Buzard, R. J. Calvert, and C. Meghorst. 1993. ‘Cold SSCP’: a simple, rapid and non-radioactive method for optimized single-strand conformation polymorphism analyses. Nucleic Acids Res. 21:3637-3642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jauris-Heipke, S., G. Liegl, V. Preac-Mursic, D. Rossler, E. Schwab, E. Soutschek, G. Will, and B. Wilske. 1995. Molecular analysis of genes encoding outer surface protein C (OspC) of Borrelia burgdorferi sensu lato: relationship to ospA genotype and evidence of lateral gene exchange of ospC. J. Clin. Microbiol. 33:1860-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Johns, R. H., D. E. Sonenshine, and W. L. Hynes. 2000. Enhancement of OspC expression by Borrelia burgdorferi in the presence of tick hemolymph. FEMS Microbiol. Lett. 193:137-141. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and N. Masatoshi. 1993. MEGA: molecular evolutionary genetics analysis, version 1.01. The Pennsylvania State University, University Park.
- 12.Livey, I., C. P. Gibbs, R. Schuster, and F. Dorner. 1995. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol. Microbiol. 18:257-269. [DOI] [PubMed] [Google Scholar]
- 13.Marti Ras, N., D. Postic, and G. Baranton. 1997. Borrelia burgdorferi sensu stricto, a bacterial species “made in the USA”? Int. J. Syst. Bacteriol. 47:1112-1117. [DOI] [PubMed] [Google Scholar]
- 14.Masuzawa, T., T. Kurita, H. Kawabata, and Y. Yanagihara. 1994. Relationship between infectivity and OspC expression in Lyme disease Borrelia. FEMS Microbiol. Lett. 123:319-324. [DOI] [PubMed] [Google Scholar]
- 15.Orita, M., Y. Suzuki, T. Sekiya, and K. Hayashi. 1989. Rapid and sensitive detection of point mutations and DNA polymorphisms using the polymerase chain reaction. Genomics 5:874-879. [DOI] [PubMed] [Google Scholar]
- 16.Schwan, T. G., J. Piesman, W. T. Golde, M. C. Dolan, and P. A. Rosa. 1995. Induction of an outer surface protein on Borrelia burgdorferi during tick feeding. Proc. Natl. Acad. Sci. USA 92:2909-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Seinost, G., D. E. Dykhuisen, R. J. Dattwyler, W. T. Golde, J. J. Dunn, I. N. Wang, G. P. Wormser, M. E. Schriefer, and B. J. Luft. 1999. Four clones of Borrelia burgdorferi sensu stricto cause invasive infection in humans. Infect. Immun. 67:3518-3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sneath, P. H. A., and R. R. Sokal. 1973. Numeral taxonomy. W. H. Freeman & Co., San Francisco, Calif.
- 19.Stevenson, B., T. G. Schwan, and P. A. Rosa. 1995. Temperature-related differential expression of antigens in the Lyme disease spirochete, Borrelia burgdorferi. Infect. Immun. 63:4535-4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamaki, J., Y. Arimura, T. Koda, S. Fujimoto, T. Fujino, A. Wakisaka, and M. Kakinuma. 1993. Heterogeneity of HLA-G genes identified by polymerase chain reaction/single strand conformational polymorphism (PCR/SSCP). Microbiol. Immunol. 37:633-640. [DOI] [PubMed] [Google Scholar]
- 21.Theisen, M., B. Frederiksen, A.-M. Lebech, J. Vuust, and K. Hansen. 1993. Polymorphism in ospC gene of Borrelia burgdorferi and immunoreactivity of OspC protein: implications for taxonomy and for use of OspC protein as a diagnostic antigen. J. Clin. Microbiol. 31:2570-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Dam, A. P., H. Kuiper, K. Vos, A. Widjojokusumo, B. M. Dejonh, L. Spanjaard, A. C. P. Ramselaar, M. D. Kramer, and J. Dankert. 1993. Different genospecies of Borrelia burgdorferi are associated with distinct clinical manifestations of Lyme borreliosis. Clin. Infect. Dis. 17:708-717. [DOI] [PubMed] [Google Scholar]
- 23.Wang, G. Q., A. P. Van Dam, I. Schwartz, and J. Dankert. 1999. Molecular typing of Borrelia burgdorferi sensu lato: taxonomic, epidemiological, and clinical implications. Clin. Microbiol. Rev. 12:633-653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang, I. N., D. E. Dykhuisen, W. Qui, J. J. Dunn, E. M. Bosler, and B. J. Luft. 1999. Genetic diversity of ospC in a local population of Borrelia burgdorferi sensu stricto. Genetics 151:15-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilske, B., S. Jauris-Heipke, R. Lobentanzer, I. Pradel, V. Preac-Mursic, D. Rossler, E. Soutschek, and R. C. Johnson. 1995. Phenotypic analysis of outer surface protein C (OspC) of Borrelia burgdorferi sensu lato by monoclonal antibodies: relationship to genospecies and OspA serotype. J. Clin. Microbiol. 33:103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilske, B., V. Preac-Mursic, S. Jauris, A. Hofmann, I. Pradel, E. Soutschek, E. Schwab, G. Will, and G. Wanner. 1993. Immunological and molecular polymorphisms of OspC, an immunodominant major outer surface protein of Borrelia burgdorferi. Infect. Immun. 61:2182-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]