Abstract
The presence of various levels of anti-adenovirus serotype 5 (Ad5)-neutralizing antibodies in humans is thought to contribute to the inconsistent clinical results obtained so far in diverse gene transfer and vaccination studies and might preclude universal dosing with recombinant Ad5. Prescreening of individuals eligible for Ad5 or alternative serotype treatment and subsequently tailoring the vector dose might aid in ensuring the consistency of clinical parameters. For this purpose, a qualified Ad neutralization assay is required. Here we have tested the different protocols used to date to determine anti-Ad neutralizing activity. Based on simplicity, speed, high throughput, sensitivity, and robustness, we propose a qualified assay in which Ad neutralization is monitored by luciferase reporter gene expression.
Recombinant adenoviruses (rAd) based predominantly on serotype 5 (Ad5) and Ad2 are under investigation as vectors for gene delivery and vaccination in humans. The advantages of rAd5 over other recombinant vector systems currently available include high vector yields, excellent safety, and high transgene expression in a wide range of eukaryotic cells (9, 11). Recent investigations in rodents and nonhuman primates have confirmed that Ad vectors are powerful vaccine delivery vehicles (1, 19, 21). However, many humans have been preexposed to Ad5 (4, 6, 18) and, as a consequence, have high neutralizing activity against this virus. This fact is thought to hamper the clinical application of rAd5 vectors since it has been shown that neutralization results in less efficient gene transfer or induction of immune responses (2, 18, 23; E. A. Emini, Abstr. 9th Conf. Retroviruses Opportunistic Infect., abstr. L5, 2002). To overcome neutralization, a higher therapeutic dose of the rAd5 vector must be administered. However, anti-Ad5 activity varies significantly among individuals (4), and thus a single vector dose for all vaccinees is expected to lead to large differences in clinical outcomes. One strategy to circumvent the problem of inconsistent clinical results is to prescreen individual patients for their anti-Ad5 antibody titers and subsequently tailor the vector doses. To determine in vitro the anti-Ad5 antibody titers in human sera, a qualified Ad5 neutralization assay is required. Such a neutralization assay is also useful to monitor vaccination efficiency in experimental and clinical settings and allows worldwide standardization.
Currently, various assays are used to determine anti-Ad5 neutralizing activity, with the main differences among them being (i) input virus, (ii) cell type, and (iii) readout of neutralization. Either wild-type Ad (WT-Ad) or replication-deficient rAd5 is commonly used. With WT-Ad, cell lines that support replication are needed, such as Hep2, A549, and 293 cells. The readout is usually either performed microscopically by scoring the Ad-mediated cytopathic effect (CPE) (15), or it is quantifiable by staining for cell viability (3, 16). The results from such Ad replication inhibition assays are highly dependent on the timing of readout and usually take from 4 to 8 days. In another assay, replication-deficient Ad is used, and the inhibition of transgene expression is taken as a parameter for antiviral neutralization. For such Ad transgene expression inhibition assays, rAds carrying LacZ (14), GFP (green fluorescent protein) (20), or luciferase as reporter gene can be used. These differences in the assays used render published results of different studies difficult to interpret and compare, and thus demonstrate a need for standardization. Here we describe a head-to-head comparison of the different protocols that have been used to date to determine anti-Ad5 neutralization. For accuracy, robustness, simplicity, and sensitivity of the assay, we propose a neutralization assay based on rAd5 carrying luciferase with readout in terms of the inhibition of luciferase transgene expression.
MATERIALS AND METHODS
Control sera, human sera, and immunoglobulin G (IgG).
Ad5-neutralizing standard reference horse serum was prepared at the Centers for Disease Control and Prevention as described previously (10). The National Institute for Biological Standards and Controls (Potters Bar, Hertsmere, United Kingdom) second international standard antimeasles serum, human, and second international standard antipoliovirus serum, types 1, 2, and 3 (number 66/202), were used as positive controls. Another positive control, anti-Ad5 polyclonal antibody (ab6982), was obtained from Abcam, Ltd. (Cambridge, United Kingdom). Fetal bovine serum (FBS; Gibco BRL) was used as negative control serum.
Human serum samples were derived from healthy adult volunteers in Belgium. The samples were screened for antibodies present against WT-Ads (22). Several pools from Ad5-seropositive (at least 10 donors) and Ad5-seronegative (5 donors) samples were made and used for most of the assays described here.
IgG was purified from pools of human serum with the use of a monoclonal antibody (MAb) trap kit according to the manufacturer's protocol (Amersham Pharmacia Biotech, Uppsala, Sweden).
Cells and viruses.
A549 human lung carcinoma cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% heat inactivated FBS and 1% penicillin-streptomycin.
Ad vectors used include Ad5 WT, Ad5.Luciferase, Ad5.GFP, and Ad5.LacZ Ad35.dE3.Luciferase. The generation and purification of Ad5 rAd vectors have been described previously (5). Briefly, virus produced on PER.C6 cells was purified with a two-step cesium chloride purification protocol. After purification, the virus was aliquoted and stored at −80°C. Virus titers expressed in virus particles (VP) per milliliter were determined by high-pressure liquid chromatography.
Virus titration.
For each cell line and virus batch used, the infectious titer of the virus was determined. After a serial doubling dilution in medium, virus in concentrations ranging from 8,000 to 5 VP/cell was added to 104 cells/well in a flat-bottom 96-well plate in a total volume of 200 μl. After incubation for 24 h at 37°C and 10% CO2, the medium was discarded, and to each well 100 μl of phosphate-buffered saline (PBS) followed by 100 μl of Steady-Glo luciferase assay system reagent (Promega) was added. After incubation for 15 min at room temperature, 100 μl from each well was transferred to a black and white isoplate (Perkin-Elmer), and luminescence counts were measured on a 1450 Microbeta Trilux.
Ad neutralization assay.
Sera were heat inactivated at 56°C for 60 min before a serial doubling dilution was performed in a 96-well tissue culture plate. The dilutions covered a range from 12.5 to 0.006 μl of serum in a volume of 50 μl of Dulbecco's modified Eagle's medium (eventually resulting in dilutions from 1/16 to 1/32,768 in an end volume of 200 μl). No serum was added to the negative controls, which resulted in the maximum luciferase activity. This value was used to calculate the 90 and 50% neutralization values. To each well, 50 μl of virus solution was added with a number of VPs that was determined by the virus titration (i.e., 500 VP/cell for the batches described here). A cell suspension was made of 105 A549 cells/ml, and 100 μl was added to each well. Plates were incubated for 24 h at 37°C and 10% CO2 before readout.
Neutralization assay readouts.
Replication inhibition was determined by the method used to determine the neutralizing activity, as described previously (22). Briefly, sera were diluted, and 100 μl was dispensed in 96-well plates. Next, 50 μl of Ad stock diluted to 200 50% cell culture infective doses was added. Plates were incubated for 1 h before the addition of 50 μl (6 × 105/ml) of PER.C6 cell suspension, after which plates were further incubated overnight. The medium was replenished, and plates were incubated for another 4 days. On days 5 to 6, plates were analyzed with the dimethyl thiazoldiphenyl tetrazoleum assay (Promega) for the staining of viable cells.
Transgene expression was measured after incubation for 24 h. The method of readout for Ads carrying luciferase is described above.
For experiments with Ads carrying the GFP transgene, the medium was aspirated, 100 μl of PBS was added, and fluorescence was measured with a fluorescent plate reader (Fluoroskan Ascent FL; Labsystems) at wavelengths of 485 nm (excitation) and 527 nm (emission). Background fluorescence was equalized by wells containing cells only; maximum fluorescence was determined by wells with cells and rAd.GFP, without serum.
For experiments with Ads carrying the LacZ transgene, the medium was aspirated and cells were fixed with 1% formaldehyde-0.2% glutaraldehyde in 100 μl of PBS for 10 min at room temperature. After the cells were washed twice with 200 μl of PBS, they were incubated at 37°C in a 2.5 mM X-Gal (5-bromo-4-chloro-3-indolyl-β-galactosidase; Invitrogen, Grand Island, N.Y.) reaction mixture containing 5 mM K4Fe(CN)6, 5 mM K3Fe(CN)6, and 2 mM magnesium chloride in PBS. After 4 h of incubation, optical density was measured on an enzyme-linked immunosorbent assay plate reader (Power Wave 340; Bio-Tek Instruments Inc.) at 495 nm. Background absorbance was determined by wells containing cells only; maximum absorbance was determined by wells with cells and rAd.LacZ, without serum.
The 90% (or 50%) inhibition titer in serum corresponds to 10% (or 50%) of the maximum control value (no serum), interpolated in the serum dilution range.
Quantification of Ad genomes per cell by Q-PCR.
Total DNA was isolated from infected A549 cells with the DNeasy tissue kit (Qiagen, Germany). The quantitative (Q)-PCR protocol is derived from Klein et al. (12). The cytomegalovirus (CMV) promoter was used as the target sequence, which is present in all rAds used in this study. The primers used in this study were CMV-F353 (5′-CAT CTA CGT ATT AGT CAT CGC TAT TAC CA-3′) and CMV-R446 (5′-TGG AAA TCC CCG TGA GTC A-3′), and the probe used was CMV-2 (5′-VIC ACC GCT ATC CAC GCC CAT TGA TGT TAMRA-3′). A second pair of oligonucleotides and a probe recognizing 18S ribosomal DNA were added to the reaction to make possible the determination of the number of VP per cell (13). As standards for the determination of the Ad genomes and numbers of cells present, the CMV promoter containing plasmid pAdApt35IP1 and human genomic DNA were used, respectively. Amplification was performed in an ABI Prism 7700 sequence detection system (Perkin-Elmer).
RESULTS
Titration of rAd with the luciferase marker gene.
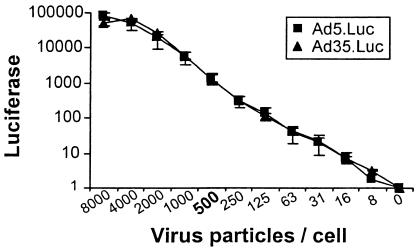
For an accurate readout of luciferase transgene expression, Ad was used at a VP/cell ratio that would yield sufficiently high luciferase activity but a suboptimal luciferase level so as not to reach the plateau of transgene expression. On the other hand, high VP/cell ratios require relatively more serum antibodies to demonstrate neutralization, which makes a neutralization assay less sensitive and not suited for small serum samples. Two rAds carrying the luciferase gene were titrated on A549 cells to determine the optimal VP/cell ratio. Figure 1 shows the titration curves of Ad5 and Ad35. The serotypes transduce A549 cells similarly, and for both viruses a concentration of 500 VP/cell, which is in the middle of the linear range, was selected for further experiments.
FIG. 1.
rAd Ad5.Luc and Ad35.Luc, each carrying the luciferase marker gene, were titrated on A549 cells. The x axis indicates VP per cell, ranging from 8,000 to 0, added to 104 A549 cells per well in a 96-well plate. Luciferase expression was measured after incubation for 1 day. For both serotypes, 500 VP/cell in the middle of the linear range was selected for further experimentation.
rAd5 neutralization measured by replication versus transgene inhibition.
Two essentially different detection methods for virus neutralization are the Ad replication inhibition assay and the transgene expression assay. Generally accepted replication inhibition assays as described in literature (2, 3, 8, 15, 17) are performed with a wide array of assay parameters. We have composed one protocol, which in our view is representative of the replication inhibition method and is suitable for comparison with a principally different assay like the luciferase-based assay. To meet the major criteria for a rapid, high-throughput assay, CPE scoring was automated by staining for viable cells and subsequent analysis of optical density (22).
To compare replication inhibition with transgene expression, a panel of human serum samples, negative FBS, and a serotype-specific horse serum pool as reference serum were tested for anti-Ad5 antibody titers by both protocols.
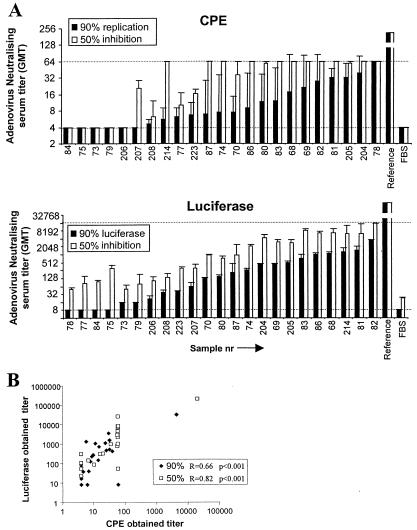
The results of the samples are shown in Fig. 2A in ascending order of titer in serum for each method. For the Ad5-specific reference serum, 90 and 50% neutralizing titers yielded 1/4,500 ± 4,300 and 1/20,000 ± 23,000, respectively, by replication inhibition (Fig. 2A, CPE); by luciferase inhibition assays, 90 and 50% neutralizing titers for this serum yielded 1/32,000 ± 4,200 and 1/209,000 ± 36,000, respectively. As shown in Fig. 2, the 50 and 90% inhibition titers correlate better within the transgene inhibition assay than in the replication inhibition assay (CPE). Both assays correlated well (based on Spearman correlation tests), as shown in Fig. 2B, although one sample scored opposite results, probably reflecting a technical error (Fig. 2A, sample 78). Several of the samples that were negative by measuring replication inhibition scored positive according to transgene expression (samples 73, 79, and 206), indicating that transgene expression is a more sensitive parameter than replication. Thus, based on sensitivity, amount of serum required, and time needed until readout, the transgene expression assay is preferred.
FIG. 2.
Serum samples were tested by two different neutralization assays for the presence of neutralizing antibodies against rAd5. Virus infection was read out either by measurement of the luciferase transgene or by virus replication scoring with viable cell staining. Maximum virus infection was determined in control wells without serum. Serum titers were determined by the dilution at which 50 or 90% of cell viability or luciferase expression was observed. (A) The y axes indicate the serum dilution at which 50 or 90% infection inhibition was observed, relative to the maximum control value. Dotted lines indicate the lower limits of detection as defined by the maximum concentration of serum. Individual samples are indicated on the x axis in ascending order of 90% inhibition serum titer for each method. Ad5-positive (Reference) serum and FBS are included as positive and negative controls, respectively. (B) Serum titers obtained by transgene expression inhibition (y axis) and replication inhibition (x axis) are compared and analyzed for correlation coefficients.
Selection of the transgene.
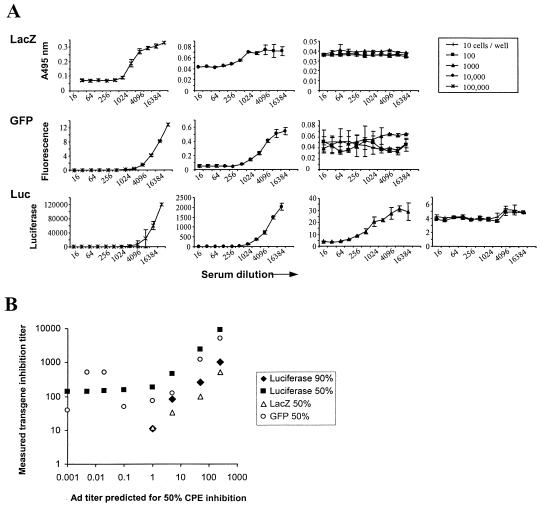
One important parameter dictating the usefulness of this assay is the possibility of using small volumes of serum for high-throughput analyses. This result can be achieved by a higher level of sensitivity and by reducing the scale of the assay (384-well plates, for example). Herewith, we determined the detection limits by testing three different transgenes and their corresponding readout systems in combination with low cell numbers (101 to 105 cells/well). Since the assays should meet criteria such as the ability for automated readout and suitability for high throughput, Ad5.LacZ-infected cells were automatically quantified by optical density measurements (7), which proved successful in that obtained results were representative for transduction inhibition as measured by counting infected cells with a microscope. Similarly, GFP expression could be detected by a fluorometer (20), with results that corresponded to analysis by microscope. The cell monolayer may interfere with the measurements, but this interference is controlled by wells in which cells only are seeded. The cells-only wells represent fluorescent background, whereas wells containing cells and GFP virus but not serum represent the maximum GFP fluorescence. Hence, the negative and positive controls are included in the assays to indicate the window of detection. From the results obtained (Fig. 3A), it could be concluded that the range between minimum and maximum expression of the LacZ protein and GFP was smaller than the range for luciferase, which makes luciferase more accurate. Furthermore, with LacZ and GFP, neutralizing antibodies could be detected when 104 cells, but not 103 cells, per well were seeded. In contrast, luciferase activity could still be detected when wells were seeded with 103 cells per well, which indicates that luciferase can be used in a smaller-scale (384-well plates) assay than GFP or LacZ. Nevertheless, we prefer to use the 96-well format with 104 cells per well, as relative standard deviations increase and sensitivity decreases (line shifts to the left) with lower cell numbers.
FIG. 3.
Neutralization determined by transgene expression inhibition with three different transgenes: LacZ, GFP, and Luc. (A) Virus and serum were incubated in fixed ratios and fixed VP/cell ratios but with various numbers of A549 cells per well. Neutralization curves calculated from triplicate measurements are shown for each virus and different numbers of cells per well. Different numbers of cells per well resulted in different transgene expression values, which cannot be depicted on the same y-axis scale. To visualize the effect of different cell numbers, the panels decrease in y-axis range from left to right. (B) A standard Ad5-neutralizing polyclonal antibody was diluted in negative human serum at different concentrations and analyzed for Ad5 neutralization with three different transgenes. Transgene inhibition-derived titers in serum were plotted against the expected neutralizing titer based on the polyclonal antibody specification of 1/25,000. For all transgenes, 50% titers are shown, but only the assay using luciferase provided 90% titers also.
We next determined what concentrations of Ad-neutralizing antibodies could be detected with the three different transgenes. Negative human serum was spiked with an anti-Ad5 antibody (Abcam) at different concentrations, which were subsequently tested for neutralization of Ad5.Luc, Ad5.GFP, and Ad5.LacZ recombinant viruses (Fig. 3B). According to specifications, the spiking agent had a titer of 1/25,000 for 50% replication inhibition of 1,000 VP. The antibody was diluted in negative serum, and the expected titer was calculated (x axis). Results from the three transgene inhibition assays were plotted and showed that similar ranges of neutralizing activity could be detected for all three. GFP results suffered from background interference in the lower range of the antibody dilution, possibly resulting in false positives. Only by using luciferase could both 50 and 90% inhibition titers be obtained, again suggesting this readout to be the most sensitive. In conclusion, all three tested transgenes can be used to determine neutralizing titers, but luciferase is preferred for its suitability for high throughput, a better minimum-maximum ratio, sensitivity, and specificity.
Qualification of the transgene expression inhibition assay.
To validate the transgene expression inhibition assay, the intra-assay variation, or the standard deviation of eight measurements within one assay, was determined. The pool of serum tested showed a 90% luciferase inhibition titer of 769 ± 100 (data not shown). The intra-assay variation was thus 13%.
In addition, the assay was performed independently, five times in duplicate, to assess interassay reproducibility. A second Ad5-positive serum pool was aliquoted and stored at −20°C until use, and a neutralization assay was performed with the same virus batch and serum batch by the same operator on five separate days. Given the low standard deviations for the obtained luciferase values, we concluded that the assay is highly reproducible. In this experiment the serum dilution needed for 90% neutralization is 1,260 ± 220, resulting in an interassay variation of 17%. For serum samples measured by the replication inhibition assay, we calculated an interassay variation of 53%. These data show that the luciferase-based assay is highly reproducible with acceptable standard deviations.
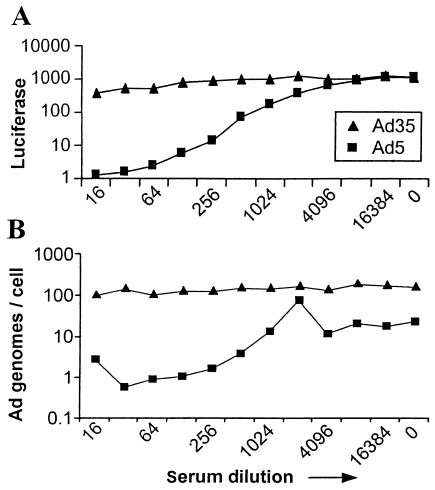
The assay is intended to determine the inhibition of virus infection by measuring luciferase activity. To determine whether serum antibodies decreased actual virus entry into target cells, and to exclude that other serum components killed target cells, thereby diminishing transgene expression, we combined transgene detection (measurement of luciferase activity) (Fig. 4A) with cellular Ad genome detection (by Q-PCR) (Fig. 4B). Simultaneous detection of the number of virus copies of Ad5 and Ad35 per cell and of luciferase activity showed that transgene expression was correlated with the number of Ad genomes per cell and that addition of other serum components decreased both the amount of luciferase and the number of cellular Ad copies. Serum does not interfere with Q-PCR results, as the positive controls with Ad35 are positive throughout the serum dilution. These results show that neutralization takes place mainly extracellularly, not after virus entry in the cellular vesicles, and that the assay specifically measures the inhibition of virus infection but not the secondary effects of serum.
FIG. 4.
Comparison between transgene expression and the number of Ad genomes per cell. A standard neutralization was performed with human Ad5-positive serum in combination with the vectors Ad5.Luc and Ad35.Luc at 500 VP/cell. (A) Cells were analyzed for luciferase activity. Ad5-positive serum shows a serotype-specific inhibition of Ad vector transduction. (B) Packaged DNA was isolated from A549 cells used in a neutralization assay as in panel A. Isolated DNA was used as a template for Ad-specific real-time PCR. The number of Ad5 genome copies per cell is decreased due to Ad5-specific serum. Ad35 genome copies are stable irrespective of concentrations in serum.
Standard control samples.
Naturally, qualification of an assay requires the presence of a standard positive control serum, one that is sufficiently characterized and readily available to the scientific and medical communities. One such standard could be the second international standard for antimeasles and antipoliovirus human serum, types 1, 2, and 3 (number 66/202), obtained from the National Institute for Biological Standards and Control, provided that this serum neutralizes Ad5. For this purpose we tested the standard serum and found that, indeed, it contains neutralizing antibodies against rAd5. This positive control serum was titrated by the transgene inhibition assay on neutralizing activity for 1/2,550 (50%) and 1/625 (90%), respectively. In addition, we obtained polyclonal Ad5-neutralizing antibodies (Abcam) with a reported 50% neutralizing activity of 1,000 VP at a dilution of 1/25,000. This serum was tested with the transgene inhibition assay and showed 90% luciferase inhibition at a dilution of 50,000 ± 9,000.
Robustness.
To determine the robustness of the luciferase-based assay, we investigated several factors that may influence the outcomes of the assay. One factor could be the cell line used. The luciferase neutralization assay was performed routinely on A549 cells as this cell line is highly permissive to Ad infection of both Ad5 and Ad35, serotypes that we frequently use. For several cell lines, including 3T3 (mouse fibroblasts), C2C12 (mouse myoblasts), and human and murine dendritic cells, we tested the Ad5- and Ad35-neutralizing activity of Ad5-positive serum (either human or mouse). As Ad vectors had different infectious titers for the different cell lines used, the maximum luciferase activity varied among different cells, as it is receptor dependent. Although each cell line showed that Ad5-positive serum neutralized Ad5 and did not neutralize Ad35, and vice versa (data not shown), 90 and 50% neutralization titers were shifted, depending on the maximum luciferase value.
Furthermore, we tested the effect of the sequence of events, i.e., whether A549 cells should be attached to the bottom of the wells before exposure to serum and virus or whether cells can be added after serum and virus are mixed, but no difference was observed (data not shown). Therefore, the cells can be added after diluting serum and adding VP, which is easier and faster. When large amounts of samples are to be tested, the time between the addition of cells to the virus-serum mix may vary. Therefore, we tested the effect of the incubation time (at room temperature) of serum and virus before cells are added. The incubation of serum and virus was varied from 0.5 to 60 min, but no differences in results were detected, allowing for the flexible timing of subsequent activities in the protocol.
Contribution of serum IgG.
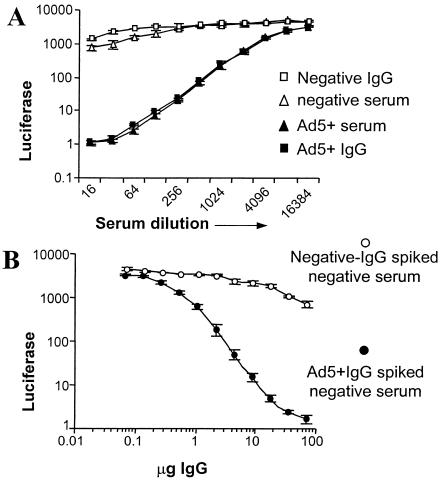
To demonstrate that the neutralizing effect of serum is mainly mediated through antibodies, the assay was performed with isolated IgG. The IgG isolation and purity were confirmed by gel electrophoresis and Coomassie blue staining (data not shown). Figure 5A shows the neutralization capacity of Ad5-positive and -negative serum from which the IgG was isolated and the neutralizing activity of the isolated IgG fractions. Ad5-positive serum and IgG isolated from the same serum batch show neutralization, whereas Ad5-negative serum and cognate IgG do not show neutralization. Figure 5B shows the results obtained when Ad5-negative serum was spiked with IgG isolated from Ad5-positive serum or with IgG isolated from Ad5-negative serum. The results demonstrate that neutralization activity can be transferred from positive to negative serum by using IgG antibodies.
FIG. 5.
The role of IgG in Ad neutralization. Results shown are the average of triplicate measurements performed with pooled human serum or isolated IgG and the vector Ad5.Luc. (A) Ad5-positive and -negative sera were compared with IgG isolated from positive or negative serum in an rAd5 neutralization assay. (B) IgG isolated from negative or positive serum pools was spiked in negative serum. Inhibition of luciferase activity was detected with increasing IgG concentrations.
DISCUSSION
In this report we compared Ad neutralization assays to define an optimal assay based on the criteria of simplicity, speed, and sensitivity. The principle of all the assays is the same: serum, virus, and cells are incubated, which allows antibodies present in the serum to neutralize the virus, thereby inhibiting infection. Subsequently, inhibition of virus infection can be detected either by assessing cell viability (dependent on virus replication) or by transgene expression. The outcomes of different forms of readout of virus infection vary widely in the literature, which makes comparisons among different studies difficult. To avoid setting up several different assays, we have composed one protocol, which in our view represents the replication inhibition method commonly used. A first comparison was made between replication and transgene inhibition assays. By using luciferase as a transgene, infection or inhibition of infection could be measured already after 24 h, and the assay appeared more sensitive and required smaller volumes of serum than the replication inhibition assay. Luciferase transgene expression was measured by an automated luminescence detector. In our experiments, replication inhibition was also automatically scored after MTT staining, but in the literature, many results are derived from subjective CPE scoring by microscopic examination, which is less quantitative and more error prone. Luciferase activity was also compared with automated quantitation of GFP and LacZ protein expression. Luciferase activity detection was more sensitive than other transgenes and required fewer target cells, which makes it suitable for use in 384-well plates for high throughput.
Having established the recombinant Ad carrying the luciferase gene as the most appropriate neutralization target, we next qualified the method for reproducibility, specificity, and robustness. Interassay and intra-assay variations of repeated measurements showed that the assay was highly reproducible, adding to the precision of the measurements. The robustness of the assay was determined by various factors in the protocol. The cell line used is of importance, since rAds require permissive cells, and, in the case of different viruses to be tested, all viruses should have a similar infectious titer for that cell line. A sufficiently high maximum level of luciferase activity is required for precise determination of the 90% reduction titer. Although in general similar results were obtained for serum tested on variable cell lines, we recommend a standard cell line for the assay, i.e., A549, as this cell line is highly permissive to infection by a large range of Ad serotypes (unpublished observations). Furthermore, timing of the protocol was not critical for the outcome, which adds to the interoperator reproducibility.
Two aspects of the specificity of the assay were investigated. First, the inhibiting effect of serum on the infectivity of the virus was confirmed by real-time PCR to detect the reduction of virus genome copies per cell. This shows that serum does not reduce cell viability or transgene expression but reduces the number of copies of the virus entering the cells. Second, inhibition of virus infection was mediated through at least IgG antibodies. The luciferase inhibition curves were similar for IgG and whole serum, except for minor differences at higher concentrations in serum. Also, negative serum shows marginal inhibition at higher concentrations in serum. By definition (with serotype-specific serum), Ad serotypes are not cross-neutralized by antibodies raised against other serotypes. But we cannot exclude the possibility that some of the donors which we designated Ad5 serotype-negative do have very low titers of Ad5 antibodies. Alternatively, serum may manifest nonspecific antiviral activity, as has been observed with high concentrations (1/4) in serum derived from Ad-naïve monkeys that also inhibit Ad gene transfer without previous Ad encounters (unpublished results).
Our results show that the luciferase-based transgene inhibition assay performs adequately for all criteria. As the neutralization assay is very sensitive, small amounts of virus, cells, and, most importantly, serum are enough to detect the presence of neutralizing activity. This sensitivity allows more determinations per serum sample and high-throughput screening of sera. The qualification performed in this study and the availability of a standard control demonstrate the suitability for the luciferase-based transgene inhibition assay as the standard assay. Since recombinant Ad is a highly potent vector for vaccination (1, 19, 21), a standard assay is imperative to support large-scale administration.
Acknowledgments
We thank Jolande Schoemaker and Richard Lonsdale for critically reading the manuscript and Marie-Pierre de Béthune and Laura Panitti from Tibotec-Virco NV for analyzing many samples.
REFERENCES
- 1.Bruna-Romero, O., G. Gonzalez-Aseguinolaza, J. C. Hafalla, M. Tsuji, and R. S. Nussenzweig. 2001. Complete, long-lasting protection against malaria of mice primed and boosted with two distinct viral vectors expressing the same plasmodial antigen. Proc. Natl. Acad. Sci. USA 98:11491-11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen, Y., D. C. Yu, D. Charlton, and D. R. Henderson. 2000. Pre-existent adenovirus antibody inhibits systemic toxicity and antitumor activity of CN706 in the nude mouse LNCaP xenograft model: implications and proposals for human therapy. Hum. Gene Ther. 11:1553-1567. [DOI] [PubMed] [Google Scholar]
- 3.Crawford-Miksza, L. K., and D. P. Schnurr. 1994. Quantitative colorimetric microneutralization assay for characterization of adenoviruses. J. Clin. Microbiol. 32:2331-2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.D'Ambrosio, E., N. Del Grosso, A. Chicca, and M. Midulla. 1982. Neutralizing antibodies against 33 human adenoviruses in normal children in Rome. J. Hyg. (London) 89:155-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fallaux, F. J., A. Bout, I. van der Velde, D. J. van den Wollenberg, K. M. Hehir, J. Keegan, C. Auger, S. J. Cramer, H. van Ormondt, A. J. van der Eb, D. Valerio, and R. C. Hoeben. 1998. New helper cells and matched early region 1-deleted adenovirus vectors prevent generation of replication-competent adenoviruses. Hum. Gene Ther. 9:1909-1917. [DOI] [PubMed] [Google Scholar]
- 6.Flomenberg, P., V. Piaskowski, R. L. Truitt, and J. T. Casper. 1995. Characterization of human proliferative T cell responses to adenovirus. J Infect. Dis. 171:1090-1096. [DOI] [PubMed] [Google Scholar]
- 7.Gahery-Segard, H., V. Molinier-Frenkel, C. Le Boulaire, P. Saulnier, P. Opolon, R. Lengagne, E. Gautier, A. Le Cesne, L. Zitvogel, A. Venet, C. Schatz, M. Courtney, T. Le Chevalier, T. Tursz, J. G. Guillet, and F. Farace. 1997. Phase I trial of recombinant adenovirus gene transfer in lung cancer: longitudinal study of the immune responses to transgene and viral products. J. Clin. Investig. 100:2218-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gall, J., A. Kass-Eisler, L. Leinwand, and E. Falck-Pedersen. 1996. Adenovirus type 5 and 7 capsid chimera: fiber replacement alters receptor tropism without affecting primary immune neutralization epitopes. J. Virol. 70:2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Havenga, M. J., R. Vogels, and A. Bout. 2001. Prospects for chimeric adenoviruses. IDrugs 4:179-188. [PubMed] [Google Scholar]
- 10.Hierholzer, J. C., Y. O. Stone, and J. R. Broderson. 1991. Antigenic relationships among the 47 human adenoviruses determined in reference horse antisera. Arch. Virol. 121:179-197. [DOI] [PubMed] [Google Scholar]
- 11.Jenne, L., G. Schuler, and A. Steinkasserer. 2001. Viral vectors for dendritic cell-based immunotherapy. Trends Immunol. 22:102-107. [DOI] [PubMed] [Google Scholar]
- 12.Klein, D., B. Bugl, W. H. Gunzburg, and B. Salmons. 2000. Accurate estimation of transduction efficiency necessitates a multiplex real-time PCR. Gene Ther. 7:458-463. [DOI] [PubMed] [Google Scholar]
- 13.Klein, D., P. Janda, R. Steinborn, M. Muller, B. Salmons, and W. H. Gunzburg. 1999. Proviral load determination of different feline immunodeficiency virus isolates using real-time polymerase chain reaction: influence of mismatches on quantification. Electrophoresis 20:291-299. [DOI] [PubMed] [Google Scholar]
- 14.Kuriyama, S., K. Tominaga, M. Kikukawa, T. Nakatani, H. Tsujinoue, M. Yamazaki, S. Nagao, Y. Toyokawa, A. Mitoro, and H. Fukui. 1998. Inhibitory effects of human sera on adenovirus-mediated gene transfer into rat liver. Anticancer Res. 18:2345-2351. [PubMed] [Google Scholar]
- 15.Malasig, M. D., P. R. Goswami, L. K. Crawford-Miksza, D. P. Schnurr, and G. C. Gray. 2001. Simplified microneutralization test for serotyping adenovirus isolates. J. Clin. Microbiol. 39:2984-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pauwels, R., J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter, and E. De Clercq. 1988. Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds. J. Virol. Methods 20:309-321. [DOI] [PubMed] [Google Scholar]
- 17.Piedra, P. A., G. A. Poveda, B. Ramsey, K. McCoy, and P. W. Hiatt. 1998. Incidence and prevalence of neutralizing antibodies to the common adenoviruses in children with cystic fibrosis: implication for gene therapy with adenovirus vectors. Pediatrics 101:1013-1019. [DOI] [PubMed] [Google Scholar]
- 18.Schulick, A. H., G. Vassalli, P. F. Dunn, G. Dong, J. J. Rade, C. Zamarron, and D. A. Dichek. 1997. Established immunity precludes adenovirus-mediated gene transfer in rat carotid arteries: potential for immunosuppression and vector engineering to overcome barriers of immunity. J. Clin. Investig. 99:209-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 20.Stallwood, Y., K. D. Fisher, P. H. Gallimore, and V. Mautner. 2000. Neutralization of adenovirus infectivity by ascitic fluid from ovarian cancer patients. Gene Ther. 7:637-643. [DOI] [PubMed] [Google Scholar]
- 21.Sullivan, N. J., A. Sanchez, P. E. Rollin, Z. Y. Yang, and G. J. Nabel. 2000. Development of a preventive vaccine for Ebola virus infection in primates. Nature 408:605-609. [DOI] [PubMed] [Google Scholar]
- 22.Vogels, R., D. Zuijdgeest, R. van Rijnsoever, E. Hartkoorn, I. Damen, M.-P. de Béthune, S. Kostense, G. Penders, N. Helmus, W. Koudstaal, M. Cechini, A. Wetterwald, M. Sprangers, A. Lemckert, O. Ophorst, B. Koel, M. van Meerendonk, P. Quax, L. Panitti, J. Grimbergen, A. Bout, J. Goudsmit, and M. Havenga. Replication-deficient human adenovirus type 35 vectors for gene transfer and vaccination: efficient human cell infection and bypass of preexisting adenovirus immunity. J. Virol. 77:8263-8271. [DOI] [PMC free article] [PubMed]
- 23.Xiang, Z., G. Gao, A. Reyes-Sandoval, C. J. Cohen, Y. Li, J. M. Bergelson, J. M. Wilson, and H. C. Ertl. 2002. Novel, chimpanzee serotype 68-based adenoviral vaccine carrier for induction of antibodies to a transgene product. J. Virol. 76:2667-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]