Abstract
The sequences of mink astroviruses collected from 11 farms in Denmark and Sweden were analyzed and found to be homologous with one another but different from those of other astroviruses. A species-specific reverse transcriptase-PCR for mink astrovirus was established and shown to be suitable for the analysis of clinical samples.
Preweaning diarrhea is a recurrent disease problem for mink farms in Europe. A case-controlled study revealed infection with astrovirus to be a significant risk factor (1). Astroviruses can cause diarrhea in a variety of mammalian and avian species (3). The virus genome consists of a positive-stranded, polyadenylated RNA of about 7 kb with three open reading frames (ORFs) designated ORF1a, ORF1b, and ORF2 (4). ORF1b encodes the RNA-dependent RNA polymerase (RdRp) (2) that is rather conserved among astroviruses infecting the same species.
Astroviruses seem to be extremely species specific, infecting only animals or cells of a single species. No adequate in vitro cultivation method is available for mink astrovirus, and our own experiences indicate that the virus presumably requires yet unrecognized proteases for cultivation. As few if any immunological reagents are commercially available for mink in general, diagnostic methods are currently not available for disease associated with this virus in mink.
Samples and RNA extraction.
Fecal samples were collected from mink kits from 11 farms in Denmark and Sweden as described elsewhere (1). Human astroviruses were detected by electron microscopy (5) in fecal samples submitted to the Swedish Institute of Infectious Disease Control. Turkey and porcine astrovirus samples were kindly provided by Linda J. Saif, Ohio State University, and seven recent mink samples were obtained from Tove Clausen, Danish Fur Breeders Research Center, Holstebro, Denmark. Seven archived intestinal samples from a single Swedish mink farm were also used in the diagnostic reverse transcriptase-PCR (RT-PCR). Suspensions of about 10% in phosphate-buffered saline were freshly prepared from all samples and stored at 4°C until use. Total RNA was extracted with TRIZOL (Gibco/BRL) according to the manufacturer's instructions.
Primers.
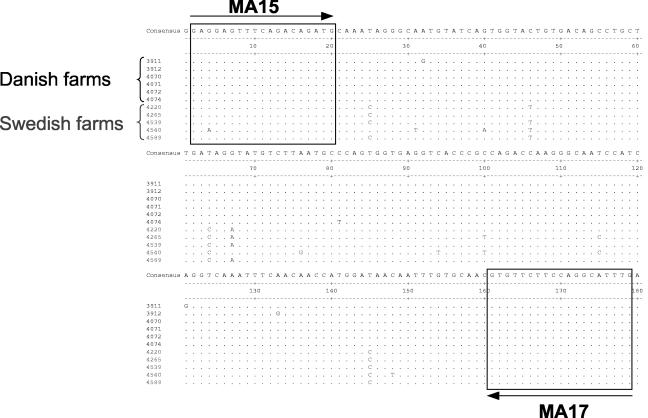
Based on the sequence obtained for a part of the RdRp region of a Swedish mink astrovirus (GenBank accession number AY179509), primers MA5 (reverse, 5′ GTC TTT TCG CTT CTC GTG TCT A 3′) and MA6 (forward, 5′ GCC ATA AAT TAG CGA ATC ACA C 3′) were designed. Sequences were obtained from 11 viruses (Fig. 1) and allowed for the design of completely conserved primers MA17 (reverse, 5′ GAG GAG TTT CAG ACA GAT G 3′) and MA15 (forward, 5′ CAA ATG CCT GGA AGA ACA C 3′) for use in a diagnostic RT-PCR for mink astrovirus infection.
FIG. 1.
Sequence alignment and locations of diagnostic RT-PCR primers. Sequences obtained with primers MA5 and MA6 for the viruses from six farms in Denmark and five farms in Sweden were aligned and slightly truncated to give a better overview. The target sequences for the primers MA15 and MA17, newly designed for use in the diagnostic RT-PCR, are boxed, and the arrows indicate the orientations of the primers.
RT-PCR.
Extracted RNA was reverse transcribed using Superscript II (Gibco/BRL) and either primer MA6 for initial sequence determination or primer MA17 for the diagnostic assay. Amplification of the initial fragments in the conserved RdRp regions was performed using Taq DNA polymerase (AmpliTaq; Perkin-Elmer) and primers MA5 and MA6, whereas the diagnostic PCR used the primer pair MA15 and MA17. The cycling profiles consisted of 2 min of denaturation at 94°C followed by 40 cycles of 94°C for 30 s, 53°C for 30 s, and 72°C for 30 s. For the diagnostic RT-PCR, the annealing temperature was changed to 50°C and the number of cycles was reduced to 30.
Cloning and sequence determination.
The remaining PCRs were run on preparative agarose gels, and products were purified with the QIAEX II kit (QIAGEN) and cloned into a plasmid vector by using the TOPO TA cloning kit (Invitrogen). Cycle sequencing was performed on plasmid DNA preparations with the BigDye Terminator cycle sequencing kit (Perkin-Elmer). For nucleotide sequence analysis, the DNASTAR package was used.
Phylogeny.
To assess sequence diversity among mink astroviruses, we analyzed six Danish and five Swedish mink samples previously shown by electron microscopy to contain high numbers of virus particles (1). These samples were obtained from 11 different farms in Denmark and Sweden. When RT-PCR was performed using primers MA5 and MA6, small amounts of the expected 283-bp amplicon were obtained for 9 out of 11 samples, whereas the last two samples became positive only after reamplification. This indicated that the chosen primers were not yet optimal for amplification of all mink astroviruses, yet it allowed us to phylogenetically analyze the 11 virus strains. As expected, limited sequence diversity was observed when the 239-nucleotide-long sequences unique to the individual amplicons were compared. Sequence homologies were between 96.7 and 100%, with the exception of those for strain 4050, which were between 91.2 and 92.9%. Interestingly, we could clearly classify the viruses as Danish or Swedish (Fig. 1).
Diagnostic RT-PCR.
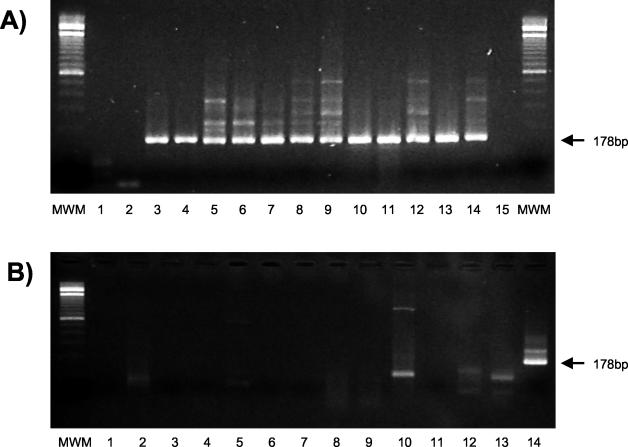
The aligned sequences from the 11 farms allowed us to design two new primers, termed MA15 and MA17 (Fig. 1). These primers were expected to recognize mink astrovirus with no or only few mismatches. After optimization of PCR parameters (MgCl2 concentration and annealing temperature) and sensitivity assessment by using a cloned sequence, we studied a range of clinical samples from Swedish and Danish mink (Fig. 2), not including the 11 samples originally used for phylogeny. A single round RT-PCR of only 30 cycles resulted in strong bands of the correct size of 178 bp (Fig. 2A), indicating the suitability of this assay for the analysis of clinical samples. Two of the recent samples from control farms that are free of the disease were negative. The species specificity of this PCR was tested against one turkey and two pig astroviruses as well as 10 human fecal samples previously found by electron microscopy to be strongly positive for astrovirus. None of these samples resulted in specific amplifications (Fig. 2B), indicating that the diagnostic RT-PCR is specific for infection with the mink astrovirus.
FIG. 2.
(A) Detection of mink astrovirus in Danish and Swedish mink samples. Lanes 1 to 7, recent Danish mink samples; lanes 8 to 14, archived Swedish mink samples from a single farm; lane 15, negative control. MWM, molecular weight marker (100-bp ladder with a stronger 600-bp fragment). Primers MA15 and MA17 produced a 178-bp fragment from the RdRp region of ORF1b of mink astroviruses. (B) Specificity of mink astrovirus RT-PCR. Lanes 1-10, human fecal samples previously found positive for human astroviruses by electron microscopy; lanes 11 and 12, pig astrovirus; lane 13, turkey astrovirus; lane 14, Swedish mink astrovirus.
Conclusions.
We have shown that mink astrovirus strains are genetically rather conserved and that the established diagnostic RT-PCR is species specific and suitable for analysis of clinical samples. Appearance of weak unspecific bands in addition to the strong expected product for some of the samples indicates possibilities for further improvement of the assay.
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in GenBank under accession numbers AY196095 to AY196105.
Acknowledgments
This work was supported by research grants received from the Danish Fur Breeders Research Centre.
REFERENCES
- 1.Englund, L., M. Chriel, H. H. Dietz, and K. O. Hedlund. 2002. Astrovirus epidemiologically linked to pre-weaning diarrhoea in mink. Vet. Microbiol. 85:1-11. [DOI] [PubMed] [Google Scholar]
- 2.Lewis, T. L., H. B. Greenberg, J. E. Herrmann, L. S. Smith, and S. M. Matsui. 1994. Analysis of astrovirus serotype 1 RNA, identification of the viral RNA-dependent RNA polymerase motif, and expression of a viral structural protein. J. Virol. 68:77-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Matsui, S. M., and H. B. Greenberg. 2001. Astroviruses, p. 875-893. In D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 4.Monroe, S. S., M. J. Carter, J. E. Herrmann, et al. 1995. Family Astroviridae, p. 364-367. In F. A. Murphy, C. M. Fauquet, D. L. Bishop, et al. (ed.), Virus taxonomy: classification and nomenclature of viruses. Springer-Verlag, New York, N.Y.
- 5.Qiao, H., M. Nilsson, E. R. Abreu, K. O. Hedlund, K. Johansen, G. Zaori, and L. Svensson. 1999. Viral diarrhea in children in Beijing, China. J. Med. Virol. 57:390-396. [PubMed] [Google Scholar]