Abstract
The internal ribosome entry site (IRES) elements of cardioviruses (e.g., encephalomyocarditis virus [EMCV] and foot-and-mouth disease virus) are predicted to have very similar secondary structures. Among these complex RNA structures there is only rather limited complete sequence conservation. Within the J domain of the EMCV IRES there are four highly conserved nucleotides (A704, C705, G723, and A724)., which are predicted to be unpaired and have been targeted for mutagenesis. Using an IRES-dependent cell selection system, we have isolated functional IRES elements from a pool of up to 256 mutants. All changes to these conserved nucleotides resulted in IRES elements that were less efficient at directing internal initiation of translation than the wild-type element, and even some of the single point mutants were highly defective. Each of the mutations adversely affected the ability of the RNAs to interact with the translation initiation factor eIF4G.
Picornaviruses are a family of positive-sense RNA viruses that are responsible for important diseases of humans (e.g., poliomyelitis) and animals (e.g., foot-and-mouth disease). Picornavirus RNAs function like mRNAs and encode a single large polyprotein. The RNAs lack the cap structure (m7GpppN) found at the 5′ terminus of all cytoplasmic eukaryotic mRNAs; indeed the genomic RNAs have a short virus-encoded peptide (3B/VPg) covalently attached to the 5′ terminus, but this modification is rapidly lost within cells. The initiation of protein synthesis on picornavirus RNA occurs by a cap-independent mechanism and is directed by an internal ribosome entry site (IRES). Picornavirus IRES elements are mostly about 450 nucleotides (nt) in length and are predicted to contain extensive secondary structure (see review [4]). There are two major classes of picornavirus IRES elements, with apparently unrelated sequences and predicted secondary structures. However, they do share the presence of a polypyrimidine tract located about 20 nt upstream of the AUG codon at which ribosomes are believed to associate with the RNA (4, 25). The enterovirus and rhinovirus IRES elements (including the poliovirus IRES) constitute a class that functions rather inefficiently and inaccurately within the rabbit reticulocyte lysate (RRL) in vitro translation system. The activity of these IRES elements in this system is stimulated and corrected by the addition of HeLa cell proteins (6, 9). The second major class of picornavirus IRES elements includes the cardiovirus and aphthovirus IRES elements (e.g., the encephalomyocarditis virus [EMCV] and foot-and-mouth disease virus [FMDV] sequences). These IRES elements function very efficiently in the RRL system. The EMCV IRES comprises domains H to L within the 5′ untranslated region of the viral RNA (11).
Two other types of picornavirus IRES have been identified; they are from hepatitis A virus (7) and from porcine teschovirus 1 Talfan (17). In contrast to the other picornavirus IRES elements, the hepatitis A virus IRES requires the intact eIF4F complex for activity (1, 5). The teschovirus IRES displays properties similar to the cardiovirus and aphthovirus IRES elements. However, it is significantly shorter than these IRES elements, has a different predicted secondary structure (40), and lacks a polypyrimidine tract near the 3′ end (17).
Using reconstitution assays, it has been possible to determine which proteins are required, along with the small ribosomal unit, for the assembly of 48S initiation complexes on the cardiovirus and aphthovirus IRES elements (28, 31). The translation initiation factors eIF2, eIF3, eIF4A, and eIF4F are all needed by these IRES elements for 48S complex assembly, and this process is also stimulated by eIF4B (28). The cap-binding complex, eIF4F, comprises eIF4E (the cap-binding protein), eIF4A (an RNA helicase), and eIF4G (13). It has been shown that the central region of eIF4G (residues 653 to 1128, renumbered according to the full-length eIF4GI sequence of 1,600 residues) (8), together with eIF4A, is sufficient to replace eIF4F in assays for the formation of the 48S complex on the EMCV IRES (29). This result is consistent with the fact that many picornavirus IRES elements continue to function when cap-dependent protein synthesis is inhibited following the cleavage of eIF4G induced by the expression of the FMDV leader (L) protease or the enterovirus and rhinovirus 2A proteases (see, e.g., references 3 and 33). Each of these proteases induces cleavage of eIF4G on the C-terminal side of the binding site on eIF4G for eIF4E (the cap-binding protein) and, hence, separates the IRES-binding region from the eIF4E interaction site.
Toe print analyses identified a 3′ boundary for the eIF4G binding site on the EMCV IRES at nucleotide C786 (28). Consistent with this observation, binding sites for eIF4G on the EMCV and FMDV IRES elements have been mapped to the J and K domains of these IRES elements by a variety of methods (19, 23, 36). Both eIF4GI and eIF4GII (see reference 14) have been shown to interact with the FMDV IRES (24) and the EMCV IRES (20), and so they will be collectively termed eIF4G. Specific nucleotides within the EMCV J and K domains that are protected from chemical attack by eIF4G binding have been identified (19, 20). These bases include a “bulge” of A nucleotides (A770 to A774) together with various other nucleotides (including A687, A688, A724, and U725) within the J-domain. Other studies have also demonstrated the importance of the EMCV IRES J and K domains (15, 18, 32). Introduction of point mutations or small deletions within this region of the viral RNA decreased translation initiation (15, 32). Such modifications within the full-length viral cDNA resulted in the production of minute- or small-plaque-phenotype viruses (15). It was possible to isolate large-plaque revertants, which generally had additional mutations within this region of the IRES that compensated for the loss of translation activity.
The cardio- and aphthovirus IRES elements are about 50% identical in sequence, but some regions of the two are highly conserved (see references 4 and 16). One region exhibiting a high level of sequence conservation is within the apical region of the I domain, which contains a GNRA tetraloop. Other highly conserved sequences that interact with eIF4G are located within the J and K domains (as noted above). We have developed previously an IRES-dependent cell selection system, which permits the isolation of functional IRES elements from a pool of molecules containing many different mutants (34). Using this system, we showed that the 3′ A residue in the GNRA tetraloop of the EMCV IRES is absolutely required for EMCV IRES function. Furthermore, it was found that elements containing an RNRA motif (R is a purine, A or G) at this position display very high activity (34). However, in contrast, elements containing YNYA sequences (Y is a pyrimidine, C or U) at this position are severely defective. We have now used the same IRES-dependent cell selection system to explore the functional requirement for some of the highly conserved nucleotides (A704, C705, G723 and A724) within the J domain of the IRES. As indicated above, eIF4G protects A724 from chemical modification, but the importance of these nucleotides for the activity of the IRES has not been studied previously. The role of each of these conserved nucleotides in determining the activity of the IRES and its ability to interact with translation initiation factors has now been determined.
MATERIALS AND METHODS
Construction of cell selection plasmids.
All plasmids were constructed using standard methods as described previously (35) and by manufacturers. The plasmids pGUS/RXB/HOOK (lacking an IRES) and pGUS/EMC/HOOK (containing the EMCV IRES) express, from the T7 promoter, dicistronic mRNAs encoding β-glucuronidase (GUS) and a cell surface-targeted protein, HOOK, which contains a c-myc epitope tag (Invitrogen). The translation of the HOOK coding sequence is dependent on the presence and activity of the EMCV IRES (26, 34).
Generation of J-domain mutants by overlap PCR.
Two complementary degenerate oligonucleotides (MWG Biotech) with the structures dCCAGANNAGATCCCATACAATGGGNNACCTTCTG GG (reverse) and dAAGGTNNCCCATTGTATGGGATCTNNTCTGGGGCCT (forward) (where N indicates that a mixture of all four bases was present) were employed as primers, together with the T7 primer and the M13 reverse sequencing primer (Stratagene), respectively, in two separate PCRs using pSKEMCRB (10) as the template. The products (500 and 240 bp, respectively) were purified by agarose gel electrophoresis, and then a mixture of these products was used as the template in another PCR employing just the T7 and M13 primers to produce a pool of mutant EMCV IRES cDNA fragments. The product (725 bp) was digested with EcoRI and BamHI to create a 550-bp fragment that was gel purified and ligated into the EcoRI- and BamHI-digested pGUS/RXB/HOOK vector. Over 800 colonies were generated, 28 of which were analyzed individually, and all had the correct structure. The residual colonies were grown as two pools, and plasmid DNA was isolated using a Maxiprep kit (Qiagen). These pools of plasmids were called pGUS/Jpool1/HOOK and pGUS/Jpool2/HOOK.
Specific mutants were prepared in a similar way, using forward and reverse primers of the same structure as shown above except that the required unique sequences were employed rather than the highly degenerate primers. In each case, the construction of the PCR fragment back into the pGUS/RXB/HOOK vector was performed as described above. Sequencing throughout the complete IRES elements was performed on an ALFexpress DNA sequencer, using an AutoRead sequencing kit (Amersham Biosciences) with a Cy5-labeled primer (dGTCACCAGTGGAACCTGGAA; MWG Biotech) complementary to part of the HOOK sequence.
DNA transfection and transient expression assays.
COS-7 or BHK-38 cells (35-mm-diameter dishes) were infected with the recombinant vaccinia virus vTF7-3 (12), which expresses the bacteriophage T7 RNA polymerase. The cells were then transfected with plasmid DNA (up to 2.5 μg) by using Lipofectin and Optimem (Invitrogen) as described previously (2, 33). After 20 h, cells were harvested in Ca- and Mg-free phosphate-buffered saline containing 3 mM EDTA and processed to select cells that expressed the myc-tagged sFv (HOOK) protein as described previously (34). Briefly, the suspended cells were washed in Dulbecco modified Eagle medium containing 1% fetal bovine serum, incubated with anti-myc-tag antibody (9E10; Santa Cruz Biotechnology), washed, and then incubated, on a rotating wheel, with sheep anti-mouse immunoglobulin G (IgG)-coated Dynabeads M-450 (Dynal). Cells displaying HOOK on their surfaces were captured on a magnetic particle concentrator (Dynal) and washed. When required, cell extracts for protein analyses were prepared in buffer C (50 mM Tris [pH 8.0], 0.12 M NaCl, 0.5% NP-40).
RNA extraction and reverse transcription (RT)-PCR.
The RNA from selected cells was extracted using Trizol (0.5 ml; Invitrogen) according to the manufacturer's protocol. Residual plasmid DNA in the extracted RNA was removed by treatment with RQ1 RNase-free DNase (Promega). Random hexamers (Roche) were employed to synthesize cDNA from this RNA, using Moloney murine leukemia virus reverse transcriptase (Invitrogen). A PCR was performed, using this cDNA as the template, with the primers 5′-dCTGCACTCAATGTACACCGA-3′ (forward) and 5′-dGACGAACCCAGTGCATTCC-3′ (reverse), which anneal within the GUS and HOOK sequences, respectively. The expected fragment of approximately 1,000 bp was generated, and it was then digested with BamHI and EcoRI to produce a 550-bp product that corresponds to the EMCV IRES cDNA. This product was gel purified and ligated into similarly digested pGUS/RXB/HOOK vector, as describe above, to create a pool of plasmids enriched for the presence of a functional IRES element.
SDS-PAGE and immunoblot analysis.
Cell extracts were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 10% gel) (21), and specific proteins were detected by immunoblotting with Immobilon P membranes. Mouse anti-myc monoclonal antibody 9E10 (1:2;000 dilution; Santa Cruz Biotechnology) or rabbit anti-β-glucuronidase (1:5,000 dilution; 5′-3′, Inc.) or antiactin (1:1,000 dilution; Sigma) primary antibodies were used, followed by the appropriate peroxidase-labeled anti-species IgG secondary antibodies (1:3,000 dilution; Amersham Biosciences) and chemiluminescence reagents (Amersham Biosciences).
TNT reactions.
In vitro transcription-translation (TNT) reactions were performed with plasmid DNA, using the T7 Quick Coupled Transcription/Translation system (Promega) and [35S]methionine as described by the manufacturer. Products were analyzed by SDS-PAGE, as described above, and autoradiography. Quantitation was performed using a phosphorimager (Bio-Rad) with QuantityOne software.
IRES binding assays.
A fragment (144 bp) of the EMCV cDNA, corresponding to the J to L domains of the IRES (see Fig. 1) flanked by HindIII and BamHI sites, was generated by PCR with the primers dGTATAAGCTTAGGGGCTGAAGGATGCC (forward; corresponding to the start of the J domain) and dGGAATGCACTGGGTTCGTC (reverse; complementary to HOOK). A selection of different mutant plasmids (see figures) was used as the template. In all cases, the PCR products were gel purified and ligated into the pGEM-T vector (Promega). From these plasmids, HindIII-BamHI fragments were isolated and inserted into similarly digested pSP64Poly(A) (Promega). The structures of the plasmids were verified by restriction enzyme digestion and sequence analysis using a Cy5-labeled primer (dTAGGCTTGTACATATTGTCG) that is complementary to the vector sequence.
FIG. 1.
Location of conserved nucleotides within the J domain of the EMCV IRES. (A) Schematic representation of the predicted secondary structure of the EMCV IRES. The domains are labeled as previously described (11), and the boundaries of each domain are indicated. Initiation of protein synthesis occurs at AUG-11. The locations of the conserved GNRA motif and the polypyrimidine tract (PPT) are indicated. (B) Predicted secondary structure of the distal stem-loop of the J domain within the EMCV IRES. The four conserved nucleotides that have been targeted for mutagenesis in this study are indicated, as are the two nucleotides (*) within this structure that are strongly protected from chemical modification by interaction with eIF4GI (19). Nucleotides that are highly conserved across all the cardiovirus and aphthovirus IRES sequences within this region are highlighted on a black background. A more detailed description of conserved nucleotides within the EMCV IRES is available elsewhere (4, 16).
Plasmids derived from the pSP64Poly(A) vector were linearized with EcoRI. The DNAs were purified by phenol extraction and ethanol precipitation and then used as templates for in vitro transcription reactions employing SP6 RNA polymerase (Ambion, SP6 Megascript kit). The RNA transcripts, containing a 3′ poly(A) tail of 30 nt, were purified according to the manufacturer's protocol.
Interaction of eIF4GI with mutant IRES transcripts.
The protein-RNA binding assay developed previously (36) was used. Briefly, oligo(dT) Dynabeads (0.5 ml; Dynal) were captured on a magnet, washed in 0.5× SSC (0.5 ml; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and binding buffer (10 mM Tris [pH 7.5], 100 mM KCl, 2 mM MgCl2; 0.2 ml), and resuspended in binding buffer (50 μl). An excess of in vitro-transcribed polyadenylated RNA transcripts (5 μg) was incubated with the Dynabeads at 4°C for 30 min on a rotating wheel. The bead-RNA complexes were washed twice with binding buffer (0.2 ml), and the immobilized RNA transcripts were then incubated with nuclease-treated RRL (25 or 50 μl; Promega) at 4°C for 60 min on a rotating wheel. The magnetic beads were captured, and the depleted RRL was removed. The bead-RNA-protein complexes were washed twice in binding buffer (0.2 ml), resuspended in SDS sample buffer, and incubated at 4°C for 10 min. These samples were analyzed by SDS-PAGE and immunoblotting for eIF4GI or eIF4A as described previously (36). For eIF4GI analysis, 7% gels were used, while 10% gels were employed for analysis of eIF4A.
UV melting analysis of RNA transcripts.
Transcription templates were produced from the derivatives of pSP64Poly(A) as described above except that the plasmids were linearized with BamHI. RNA transcripts [lacking a poly(A) tail] were synthesized by using an SP6 Megascript kit (Ambion). Each RNA (25 μg) was diluted with buffer (10 mM morpholinepropanesulfonic acid [pH 6.8], 120 mM KCl, 1 mM MgCl2; 1.0 ml). UV melting curves were determined on a Varian Cary 400 UV/visible light spectrophotometer equipped with a temperature control unit. Data points were collected at 0.5°C intervals between 20 and 98°C with a heating rate of 0.5°C/min (superimposable curves were obtained from multiple runs and at a rate of 1.0°C/min). The melting profiles obtained for RNAs are the first derivatives of the melting curves averaged over a 5°C window and normalized to the absorbance at 25°C.
RESULTS
Mutagenesis of conserved nucleotides with the EMCV IRES.
The predicted secondary structure (based on reference 30) of the EMCV IRES, illustrated in Fig. 1A, is labeled as described previously (11). Nucleotides that are highly conserved among all the cardio- and aphthovirus IRES elements are clustered within the apical region of the I domain and within the J and K domains (see references (4 and 16). The nucleotides within the distal region of the J-domain stem-loop that are conserved between all the cardiovirus and aphthovirus IRES sequences are illustrated in Fig. 1B. The binding site for eIF4GI on the EMCV IRES has been localized to the J and K domains (19). In the present study, we have analyzed the role of four highly conserved nucleotides that are predicted to be unpaired within the distal stem-loop region of the J domain. These nucleotides are A704, C705, G723, and A724 (Fig. 1B). Unpaired bases might be expected to be targets for interactions with other molecules. Indeed, nt A724 is protected from chemical modification in the presence of eIF4GI (19), but no analysis of the functional importance of this nucleotide has been described.
A pool of mutants was generated by overlap PCR in which the four targeted nucleotides were each changed to all four bases by using degenerate oligonucleotides; thus, a collection of up to 256 possible mutants was created. Individual mutants are referred to by the nucleotides present at the four targeted positions; hence, a plasmid containing the wild-type (wt) IRES cDNA is referred to as AC/GA. The cDNA fragments were inserted into the vector pGUS/RXB/HOOK (Fig. 2A). Following transformation of E. coli over 800 colonies were produced. Twenty-eight of these colonies were picked, and plasmid DNA was isolated. As judged by digestion with restriction enzymes, each had the correct structure (data not shown), suggesting that a large proportion of the residual colonies that were used to generate two separate pools of plasmids (pGUS/Jpool/HOOK) contained the appropriate molecules. Direct sequencing of the plasmid pools demonstrated the expected unique sequence within the J domain except that each of the four bases was observed at the positions of the four targeted nucleotides as predicted (data not shown).
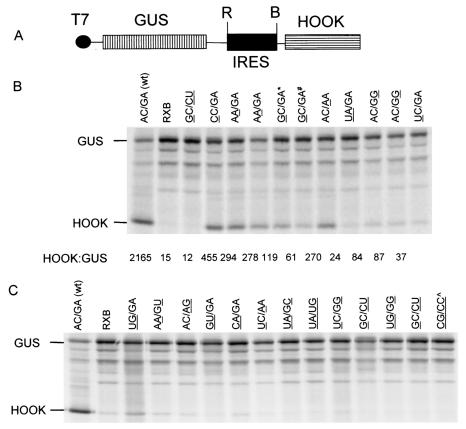
FIG. 2.
Analysis of J-domain mutant IRES elements in vitro. (A) Plasmids, as illustrated, were constructed from the pGUS/RXB/HOOK (RXB) vector (34) with the insertion of EMCV cDNA sequences as an EcoRI (R)-BamHI (B) fragment. (B) Plasmids containing cDNA rescued from COS-7 cells selected on the basis of HOOK expression were assayed in TNT reactions using [35S]methionine, as described in Materials and Methods, and analyzed by SDS-PAGE and autoradiography. Quantitation of HOOK and GUS expression was achieved with a phosphorimager, using QuantityOne software. The ratio of the levels of incorporation of methionine into HOOK and GUS was calculated and expressed as (HOOK/GUS) × 1,000. The mutations in the targeted region of the J domain are indicated by underlining. The plasmid pGUS/RXB/HOOK (lacking an IRES) and the mutant GC/CU (containing three mutations) were included as negative controls. Note that the GC/GA* mutant contains the second site mutation C537U and that the GC/GA# mutant contains the second site mutation C557U; both are in the I domain. Two isolates of the AA/GA and AC/GG mutants were obtained, and both are included to show the reproducibility of their properties and of the assay. (C) The results of an analysis of a collection of plasmids that expressed low levels of HOOK in the TNT assays (performed as for panel B) is shown. The sequences of these mutants at the four targeted nucleotides are shown with the mutated bases underlined. The mutant CG/CC^ contains a second site mutation, G564A, in the I domain.
Selection of functional IRES elements.
The pools of mutant plasmids were transfected into COS-7 cells that had been infected with the recombinant vaccinia virus vTF7-3 (12), which expresses the T7 RNA polymerase. Dicistronic RNA transcripts, produced from the plasmids in the cytoplasm of the cells, are translated to produce GUS and, when a functional IRES element is present, the c-myc-tagged HOOK protein. In an attempt to minimize cotransfection of plasmids, various quantities of DNA (20, 100, 500, and 2,500 ng) were used in the transfections. When the intact cells were harvested (after about 20 h), they were incubated with an anti-myc-tag monoclonal antibody (9E10) and then Dynabeads coated with anti-mouse IgG, which permitted the capture of cells that expressed HOOK and hence contained a functional EMCV IRES element. Significant cell selection was observed in each case (data not shown). From the selected cells, RNA was extracted, treated with RQ1 DNase (to remove residual plasmid DNA), and subjected to RT-PCR using primers specific for the GUS and HOOK sequences. The expected fragment of about 1,000 bp was generated in an RT-dependent manner, indicating that the plasmid DNA had been successfully removed (data not shown). This procedure ensured that the rescued cDNA sequences corresponded only to expressed sequences and could not have been derived from plasmid DNA adhering to the outside of cells (this could lead to apparent rescue of nonfunctional sequences [34]). The PCR fragment was digested with BamHI and EcoRI to release the IRES cDNA fragment,which was then ligated back into the pGUS/RXB/HOOK vector as before. From the colonies obtained, 40 were picked individually and analyzed, while the residual colonies (over 400) were pooled and used for a second round of this procedure.
Analysis of selected mutant IRES elements in vitro.
The plasmids obtained individually from the total pools and those generated following the cell selection and plasmid rescue procedures were analyzed initially using in vitro coupled TNT reactions within RRL. Since the wt EMCV IRES functions well in this system, the relative activity of the rescued sequences can be readily assessed. For each plasmid that appreciably expressed HOOK, the sequence of the EMCV cDNA was determined to identify the bases present at the targeted positions within the J domain and also to check for any second site changes introduced during the cloning procedures. Figure 2B shows the results obtained from a collection of plasmids containing IRES cDNA rescued from the selected cells. In all cases, the upstream open reading frame, encoding GUS, was efficiently translated. Significant expression of HOOK was detected from the wt EMCV IRES (designated AC/GA) and also from each of the selected mutants shown in Fig. 2B. Note that we detected little or no expression of HOOK from the plasmid pGUS/RXB/HOOK (as expected, since it lacks an IRES) or from an IRES with three of the four conserved bases modified (GC/CU), which was included here as a negative control. It may be that the pGUS/RXB/HOOK vector (RXB), lacking the IRES, is a less stringent negative control than the GC/CU mutant, since the presence of the defective IRES sequence may reduce reinitiation at the HOOK initiation codon after the termination of GUS translation. Quantitation of the GUS and HOOK bands was performed using a phosphorimager and the HOOK/GUS expression ratio (×103) was calculated in each case (Fig. 2B). It is interesting that the wt EMCV IRES depresses the translation of the upstream open reading frame in the TNT assay. None of the mutant IRES elements was as active as the wt element. However, it cannot be determined whether the most active mutant sequences (e.g., CC/GA and AA/GA) also competed significantly for translation of GUS in these assays; thus, the quantitation should be regarded only as a guide to the relative activities of the different mutants. It is apparent that the mutants showing the highest activity had only a single nucleotide different from the wt sequence, and this mutation could occur at each of the targeted nucleotides except A724. Some mutants with two mutations within the target site, or with a substitution at A724, displayed a lower but still detectable level of IRES activity. Among the functional IRES elements that were obtained from the selection and screen, the wt sequence was most frequently selected (33% of the plasmids found to have a functional IRES). Two of the mutants selected contained second site mutations on the same mutant background (GC/GA). The mutant with the higher activity had a mutation of C537 to U. This substitution has occurred within the EMCV-B strain (compare with the EMCV-R strain used here), and hence it may be that this mutation would not significantly affect the IRES activity. The results obtained from the selection and screening assay indicated that only a small proportion of the possible 256 mutants were functional in vitro and suggested that the conservation of this sequence is important to preserve the activity of the IRES.
Identification of J-domain mutants which are severely defective in IRES activity.
From the screening of the pools of mutated plasmids, it was easy to isolate plasmids that expressed GUS efficiently in the TNT system but produced little or no HOOK (Fig. 2C). The results of the analysis of 13 different mutants are shown in Fig. 2C; the nucleotide sequence of the entire EMCV IRES cDNA was determined in each case. The examples presented indicate that each of the four bases was present at each of the four targeted positions within the pools of plasmids that had been generated; this is consistent with the nucleotide sequencing result described above. Each of these nonfunctional IRES mutants contained two or more substitutions compared to the wt (AC/GA) sequence. The mutant UG/GA, containing two nucleotide changes, showed a very low level of activity in the TNT system.
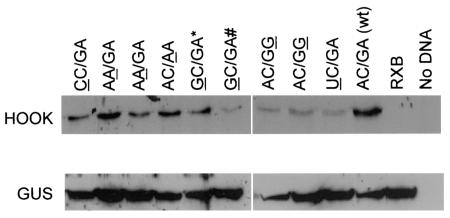
Analysis of selected IRES mutants within BHK cells.
The activity of the various selected IRES mutants was also determined within BHK cells. The plasmids were transfected into vTF7-3-infected BHK cells; after 20 h the cells, were harvested and total-cell extracts were prepared, which were analyzed by SDS-PAGE and immunoblotting for the expression of GUS and HOOK proteins. Expression of HOOK was detected from each of the mutant IRES elements that had scored as functional in the TNT screen (Fig. 3). The level of HOOK expression differed among the constructs and was strongest from the wt IRES, consistent with the results from the TNT assay (Fig. 2B). No HOOK expression was observed in cells from the plasmid pGUS/RXB/HOOK (Fig. 3). However, it was apparent that the cells had been successfully transfected with each plasmid, since GUS was efficiently expressed in each case (Fig. 3).
FIG. 3.
Activity of selected J-domain mutant IRES elements within BHK cells. Plasmids encoding the dicistronic GUS-HOOK mRNAs with the indicated J-loop mutant IRES elements were transfected individually into BHK cells. After approximately 20 h, cells were harvested and total-cell extracts were prepared. Samples were analyzed by SDS-PAGE and immunoblotting to detect the GUS and HOOK proteins as indicated. The nucleotides that differ from the sequence of the wt IRES are underlined. The mutants GC/GA* and GC/GA# contain second site mutations as described in the legend to Fig. 2.
Some of the IRES elements scored as defective in the TNT assays were also assayed using the transient expression system within BHK cells (Fig. 4). The wt (AC/GA) IRES plus the mutants UA/GA, UG/GA, and AC/AG expressed HOOK at a level that permitted some cell selection to occur (as judged by the retention of actin on the beads [Fig. 4]). However, it is clear that the expression of HOOK (as determined in the total-cell extracts) from these mutants was much weaker than that from the wt IRES. The even lower level of HOOK expression from the AA/GU, GU/GA, and GC/CU mutants (defective in TNT assays) was insufficient to achieve cell selection, since little or no actin was retained on the beads. Overall, there is a good correlation between the activities of the mutant IRES elements in the TNT reactions and within BHK cells (compare Fig. 2 with Fig. 3 and 4). Low-level IRES activity from the UA/GA and UG/GA mutants had been detected in the TNT system (Fig. 2B and C), but the weak activity observed in cells from the AC/AG mutant was not detected above the background level in vitro.
FIG. 4.
Analysis of defective IRES elements within BHK cells. Plasmids encoding the dicistronic GUS-HOOK mRNAs containing the J-domain mutant IRES elements were transfected into BHK cells as described in the legend to Fig. 3. Total-cell extracts were analyzed for the expression of HOOK (A) and GUS (B). The retention of cells on Dynabeads following cell selection was determined by the presence of actin (C). In all cases, immunoblotting was used to detect the proteins as described for Fig. 3. The individual mutants analyzed are indicated, and the nucleotides that differ from the sequence of the wt IRES are underlined. The CG/CC^ mutant contains a second site mutation, G564A.
Single point mutant construction and assays.
It was apparent from the results shown above that even single nucleotide changes within the targeted sequence had significant effects on the activity of the IRES and that multiple modifications resulted in severely defective IRES elements. Hence, it was decided to generate each of the possible single-nucleotide mutants that had not already been isolated. These mutants were generated using PCR with specific mutagenic primers (see Materials and Methods). The products were assembled into the pGUS/RXB/HOOK vector as described above. The properties of the complete collection of single-site mutants in the TNT assay are shown in Fig. 5A. Some of the new mutants, e.g., GC/GA (which lacks the second site changes found in the two GC/GA mutant plasmids analyzed in Fig. 2) and AG/GA, displayed high IRES activity. However, other single point mutants (e.g., AC/GU) were almost inactive and expressed HOOK at a level similar to that of the pGUS/RXB/HOOK (no-IRES) control. It is interesting that when each nucleotide was changed individually to a U, the IRES displayed the lowest level of activity. Each of the three mutants with mutations at the A724 position was much less active than the wt.
FIG. 5.
Functional analysis of all single point mutants at nt 704, 705, 723, and 724 within the J domain of the EMCV IRES. (A) Dicistronic GUS-HOOK vectors containing the indicated J-domain mutations within the EMCV IRES were used in RRL TNT reactions with [35S]methionine and analyzed by SDS-PAGE and autoradiography as described for Fig. 2. The mutated nucleotides are underlined. (B) The same collection of mutants was assayed in the BHK transient expression system as described for Fig. 3. Total cell extracts were prepared and analyzed by SDS-PAGE and immunoblotting with antibodies for GUS and HOOK as indicated.
Each of these single point mutants was also tested in the BHK transient expression system (Fig. 5B). As expected, all the plasmids expressed GUS with similar efficiencies, but there were significant differences in the levels of HOOK expressed. Consistent with the TNT results, the plasmids with mutations at nt A724 were most markedly defective. In particular, the mutants AC/GC and AC/GU were almost inactive in this assay. In contrast, nt A704 and G723 were most tolerant of change. However, as observed for the TNT assay, the most deleterious mutations involved introduction of a U at any position.
Role of the four conserved nucleotides in the interaction of the EMCV IRES with eIF4G.
Since the J and K domains of the EMCV IRES are known to interact with eIF4G, it was decided to determine whether the mutations within this region that clearly affected IRES activity also modified the interaction with eIF4G. It is clearly possible that the defect in IRES activity could be accounted for in a variety of different ways and, hence, the interaction with eIF4G would not necessarily be modified. An RNA-protein interaction assay developed previously (36) was used. The cDNA corresponding to the 3′ region (about 145 nt) of the wt and certain mutant IRES elements was amplified by PCR and inserted into the transcription vector pSP64Poly(A). RNA transcripts (in excess) that included a 3′ poly(A) tail of 30 nt were produced and immobilized onto oligo(dT) magnetic beads. The bead-RNA complexes were incubated with RRL, and the proteins that were retained on the washed beads were analyzed by SDS-PAGE followed by immunoblotting with antibodies against eIF4G and eIF4A. As expected, the wt EMCV IRES fragment captured both eIF4G and eIF4A (Fig. 6A and B), it is believed that the eIF4A is bound through its interaction with eIF4G (36). Each of the mutant sequences tested was less efficient in binding to eIF4G, but some binding was detected with the AA/GA mutant (Fig. 6A). The AA/GA mutant was the only mutant tested in this assay that also showed significant IRES activity in both the TNT and BHK transient expression systems. Consistent with this result, the interaction of the AA/GA mutant sequence with eIF4A was also apparent (Fig. 6B), but little or no interaction of eIF4A with the other mutant sequences was detected. Thus, even just a single point mutation (as in the AC/GU mutant) within the four targeted nucleotides can result in significant inhibition of IRES activity, and this was accompanied by a large reduction in the ability of the RNA to interact with eIF4G (and hence eIF4A).
FIG. 6.
RNA:protein binding assay to determine eIF4G and eIF4A interaction with mutant EMCV IRES transcripts. RNA transcripts corresponding to nt 679 to 823 of the wt and mutant EMCV IRES elements linked to a 30-nt poly(A) tail were immobilized onto oligo(dT) Dynabeads. The bead-IRES complexes were then incubated with RRL, and the depleted lysate was removed. The proteins bound to the beads were eluted in SDS sample buffer, and the presence of eIF4GI (A) and eIF4A (B) bound to the transcripts was analyzed using SDS-PAGE and immunoblotting with appropriate antibodies. The FMD-AS transcript is the full-length antisense FMDV IRES (36) and was used as a negative control. An aliquot of nondepleted RRL was included as a positive control. Underlining indicates the mutated nucleotides in the RNA transcripts. The mutant AA/GU# contains a second site mutation, A741U.
The mutations that affect the activity of IRES and the interaction with eIF4G may exert their effect either by directly modifying a site of RNA-protein interaction or, more indirectly, by modifying the local RNA structure in the vicinity of the protein binding site. In an attempt to determine whether the mutations analyzed here were significantly modifying the local RNA structure, the melting profiles of RNA transcripts were analyzed (as described previously [see, e.g., references 22 and 37]). RNA transcripts corresponding to the J-to-L region of the wt and mutant sequences were prepared as for the RNA-protein interaction studies except that the plasmids were linearized with BamHI prior to the production of runoff transcripts; hence, these RNAs lacked a poly(A) tail. The melting profiles were determined under salt conditions similar to those used for in vitro translation reactions. The results (data not shown) indicated that the wt (AC/GA) EMCV RNA unfolded with two major apparent transitions (i.e., peaks in the profile) with apparent Tms of 60° and 66°C. Melting profiles for the mutant transcripts AA/GA (high IRES activity) and AC/AG (low IRES activity) were very similar to that observed with the wt RNA. In contrast, the mutants AC/GU (low activity) and GC/CU (low activity) produced significantly altered melting profiles. Both showed a single apparent transition at 62°C, while the latter mutant also had a more pronounced broad transition at about 80°C. The AA/GU RNA (low IRES activity), which also had only a single apparent transition, was most greatly perturbed having a reduced Tm of 56°C. Thus, it is interesting that the AA/GA mutant, which still possessed significant IRES activity and continued to bind eIF4G, was little perturbed in structure. In contrast, a single point mutation at A724 (in AC/GU) that severely affected both activity and interaction with eIF4G could be shown by this technique to affect the local RNA stability. The multiple sequence changes in the AA/GU mutant clearly had the most drastic effect on RNA structure. The lack of apparent change in structure of the AC/AG mutant in this assay, despite the significant defect in activity of the mutant IRES, suggests a critical role for G723 or A724 in determining the activity of the IRES.
DISCUSSION
We have described previously the development and application of an IRES-dependent cell selection system coupled with a plasmid rescue system to identify functional mutants from within a large pool of mutant plasmids (34). We applied this system to one conserved motif (a GNRA tetraloop) within the I domain of the EMCV IRES. One drawback of the procedure was the presence of nonselected plasmids within the collection of those that were rescued. It is believed that they resulted from adherence of plasmid DNA to the outside of transfected cells or from cotransfection with a plasmid that encoded a functional IRES element. We have now modified the rescue procedure of nucleic acids from the selected cells in an attempt to reduce the background signal from plasmid DNA that adhered to the outside of cells. In the modified procedure described here, employing RT-PCR, only sequences that have been expressed as mRNA within the selected cells will be isolated. We still obtain plasmids that do not encode a functional IRES, but we also obtain a significant enrichment for functional IRES elements, which is particularly important when large pools of mutant plasmids are generated initially. Two other IRES-dependent cell selection strategies have been published recently (27, 39). Both of these strategies employed constructs that expressed, from within the nucleus, dicistronic mRNAs encoding two different fluorescent proteins. Cells containing plasmids in which both cistrons were expressed, were selected using fluorescent-activated cell sorting. These systems were used to analyze a large library of random synthetic sequences either of 9 or 18 nt (27) or of 50 nt (39) for IRES function. Owens et al. (27) used a retroviral vector to express the dicistronic constructs within mammalian cells, whereas protoplast fusion was utilized by Venkatesan and Dasgupta (39) for delivery of a library of plasmids into mammalian cells. Protoplast fusion has the advantage that it results in near-clonal delivery of plasmids into cells (38), and a relatively high level of enrichment was seen. For certain future applications of the HOOK selection system, it may be beneficial to explore other methods for delivering the GUS-HOOK vectors into cells to improve further the level of enrichment gained in the system.
Previous studies have demonstrated the general importance of the J and K domains for EMCV IRES function (15, 18, 32). Certain modifications could affect the interaction with the polypyrimidine tract binding protein (18), but the basis for other defects was undefined (15). Some of these mutations may have affected the interaction with eIF4G directly or indirectly through modification of the structure of this domain. In the present study, four nucleotides that are highly conserved within the J domain of the cardio- and aphthovirus IRES elements have been modified. The effect of these modifications on the activity of the IRES and on the ability of the RNA to interact with the translation initiation factor eIF4G has been determined. It is apparent that these unpaired AC/GA bases are conserved, because they are important for IRES activity; all the single point mutants were less active than the wt IRES. Replacement of any one of these four nucleotides by a U was highly deleterious, and nt A724 was particularly sensitive to any change. Thus, the AC/GU mutant was severely defective in IRES activity, and this was reflected in its inability to interact with eIF4G. It is also noteworthy that the RNA melting profile for this mutant was significantly perturbed (data not shown). Hence, apparently minor changes in sequence can have greater effects on the structure of the RNA than may be envisaged. In contrast, the RNA melting profile of the single point mutant AA/GA, which retained significant IRES function and still bound to eIF4G (albeit less strongly than the wt), showed little perturbation. Thus, presumably the C705-to-A change directly affected the ability of the RNA to bind to eIF4G and hence IRES activity. Similarly, the RNA melting profile of the AC/AG mutant exhibited little apparent change, but this mutant was severely defective in interacting with eIF4G and in IRES function; thus, G723 and/or A724 must be a critical site within the IRES.
For these J-domain mutants, the correlation between IRES activity and the ability to interact with eIF4G is consistent with the results of previous studies of the J domain of the FMDV IRES (23). These authors showed that modifications at the base of the J domain severely affected the ability of the IRES to interact with eIF4G and, hence, affected the activity of the IRES. Furthermore, it has been shown (19) by chemical probing studies of the EMCV IRES that the A bulge (nt 770 to 774) between the J and K domains plus various nucleotides at the base of the J domain (e.g., A687 and A688) together with A724 and U725 were protected from modification by eIF4G. It is apparent from our studies that nucleotide A724 is critical for the activity of the IRES. Thus, our studies have demonstrated the importance of this site of interaction with eIF4G for the biological function of the IRES. In a very recent report (20), it was indicated that eIF4G makes more extensive interactions with the J-K and L domains of the EMCV IRES than was previously recognized. Indeed it was shown that nt A704 is also weakly protected from chemical modification by eIF4G, a result entirely consistent with the data presented here that shows the importance of this nucleotide for IRES activity. It is, therefore, apparent that a number of nucleotides are critical for the interaction of the EMCV IRES with eIF4G. However, some of the mutants characterized here and elsewhere (see, e.g., references 20 and 23) may have their function modified as a result of significantly affecting the three-dimensional structure of the J and K domains.
Acknowledgments
A.T.C. and M.E.M.R. gratefully acknowledge receipt of their studentships from the BBSRC. G.L.C. has a Wellcome Trust Career Development Fellowship (reference no. 061444).
REFERENCES
- 1.Ali, I. K., L. McKendrick, S. J. Morley, and R. J. Jackson. 2001. Activity of the hepatitis A virus IRES requires association between the cap-binding translation initiation factor (eIF4E) and eIF4G. J. Virol. 75:7854-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belsham, G. J. 1997. Analysis of picornavirus internal ribosome entry site function in vivo, p. 323-340. In J. Richter (ed.), mRNA formation and function. Academic Press, New York, N.Y.
- 3.Belsham, G. J., and J. K. Brangwyn. 1990. A region of the 5′ noncoding region of foot-and-mouth disease virus RNA directs efficient internal initiation of protein synthesis within cells: interaction with the role of the L protease in translational control. J. Virol. 64:5389-5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 5.Borman, A. M., and K. M. Kean. 1997. Intact eukaryotic initiation factor 4G is required for hepatitis A virus internal initiation of translation. Virology 237:129-136. [DOI] [PubMed] [Google Scholar]
- 6.Brown, B., and E. Ehrenfeld. 1979. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97:396-405. [DOI] [PubMed] [Google Scholar]
- 7.Brown, E. A., A. J. Zajac, and S. M. Lemon. 1994. In vitro characterization of an internal ribosomal entry site (IRES) present within the 5′ nontranslated region of hepatitis A virus RNA: comparison with the IRES of encephalomyocarditis virus. J. Virol. 68:1066-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 22:4499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorner, A. J., B. L. Semler, R. J. Jackson, R. Hanecak, E. Duprey, and E. Wimmer. 1984. In vitro translation of poliovirus RNA: utilization of internal initiation sites in reticulocyte lysate. J. Virol. 50:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drew, J., and G. J. Belsham. 1994. trans complementation of defective foot-and-mouth disease virus internal ribosome entry site elements. J. Virol. 68:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duke, G. M., M. A. Hoffman, and A. C. Palmenberg. 1992. Sequence and structural elements that contribute to efficient encephalomyocarditis virus-RNA translation. J. Virol. 66:1602-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuerst, T. R., E. G. Niles, F. W. Studier, and B. Moss. 1986. Eukaryotic transient expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gingras, A. C., B. Raught, and N. Sonenberg. 1999. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68:913-963. [DOI] [PubMed] [Google Scholar]
- 14.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, M. A., and A. C. Palmenberg. 1996. Revertant analysis of J-K mutations in the encephalomyocarditis virus internal ribosome entry site detects an altered leader protein. J. Virol. 70:6425-6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson, R. J., and A. Kaminski. 1995. Internal initiation of translation in eukaryotes: the picornavirus paradigm and beyond. RNA 1:985-1000. [PMC free article] [PubMed] [Google Scholar]
- 17.Kaku, Y., L. S. Chard, T. Inoue, and G. J. Belsham. 2002. Unique characteristics of a picornavirus internal ribosome entry site from the porcine teschovirus-1 Talfan. J. Virol. 76:11721-11728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminski, A., and R. J. Jackson. 1998. The polypyrimidine tract binding protein (PTB) requirement for internal initiation of translation of cardiovirus RNAs is conditional rather than absolute. RNA 4:626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolupaeva, V. G., T. V. Pestova, C. U. T. Hellen, and I. N. Shatsky. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273:18599-18604. [DOI] [PubMed] [Google Scholar]
- 20.Kolupaeva, V. G., I. B. Lomakin, T. V. Pestova, and C. U. T. Hellen. 2003. Eukaryotic initiation factors 4G and 4A mediate conformational changes downstream of the initiation codon of the encephalomyocarditis virus internal ribosome entry site. Mol. Cell. Biol. 23:687-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Laing, L. G., and D. E. Draper. 1994. Thermodynamics of RNA folding in a conserved ribosomal RNA domain. J. Mol. Biol. 237:560-576. [DOI] [PubMed] [Google Scholar]
- 23.López de Quinto, S., and E. Martínez-Salas. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6:1380-1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.López de Quinto, S., E. Lafuente, and E. Martínez-Salas. 2001. IRES interaction with translation initiation factors: functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7:1213-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meerovitch, K., and N. Sonenberg. 1993. Internal initiation of picornavirus RNA translation. Semin. Virol. 4:217-227. [Google Scholar]
- 26.Mulder, J., M. E. M. Robertson, R. A. Seamons, and G. J. Belsham. 1998. Vaccinia virus protein synthesis has a low requirement for the intact translation initiation factor eIF4F, the cap-binding complex within infected cells. J. Virol. 72:8813-8819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owens, G. C., S. A. Chappell, V. P. Mauro, and G. M. Edelman. 2001. Identification of two short internal ribosome entry sites selected from libraries of random oligonucleotides. Proc. Natl. Acad. Sci. USA 98:1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pestova, T. V., C. U. T. Hellen, and I. N. Shatsky. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16:6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova, T. V., I. N. Shatsky, and C. U. T. Hellen. 1996. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell Biol. 16:6870-6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilipenko, E. V., V. M. Blinov, B. K. Chernov, T. M. Dmitrieva, and V. I. Agol. 1989. Conservation of the secondary structure elements of the 5′-untranslated region of cardiovirus and aphthovirus RNAs. Nucleic Acids Res. 17:5701-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. T. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 32.Roberts, L. O., and G. J. Belsham. 1997. Complementation of defective picornavirus internal ribosome entry site (IRES) elements by the co-expression of fragments of the IRES. Virology 227:53-62. [DOI] [PubMed] [Google Scholar]
- 33.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells: influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robertson, M. E., R. A. Seamons, and G. J. Belsham. 1999. A selection system for functional internal ribosome entry site (IRES) elements: analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5:1167-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 36.Stassinopoulos, I. A., and G. J. Belsham. 2001. A novel protein-RNA binding assay: functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA 7:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stein, A., and D. M. Crothers. 1976. Conformational changes of transfer RNA. The role of magnesium (II). Biochemistry 15:160-167. [DOI] [PubMed] [Google Scholar]
- 38.Tan, R., and A. D. Frankel. 1998. A novel glutamine-RNA interaction identified by screening libraries in mammalian cells. Proc. Natl. Acad. Sci. USA 95:4247-4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkatesan, A., and A. Dasgupta. 2001. Novel fluorescence-based screen to identify small synthetic internal ribosome entry site elements. Mol. Cell. Biol. 21:2826-2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Witwer, C., S. Rauscher, I. L. Hofacker, and P. F. Stadler. 2001. Conserved RNA secondary structures in Picornaviridae genomes. Nucleic Acids Res. 29:5079-5089. [DOI] [PMC free article] [PubMed] [Google Scholar]