Abstract
Human immunodeficiency virus type 1 (HIV-1) has evolved a number of strategies to resist current antiretroviral drugs and the selection pressures of humoral and cellular adaptive immunity. For example, R5 strains, which use the CCR5 coreceptor for entry and are the dominant viral phenotype for HIV-1 transmission and AIDS pathogenesis, are relatively resistant to neutralization by antibodies, as are other clinical isolates. In order to overcome these adaptations, we raised nucleic acid aptamers to the SU glycoprotein (gp120) of the R5 strain, HIV-1Ba-L. These not only bound gp120 with high affinity but also neutralized HIV-1 infectivity in human peripheral blood mononuclear cells (PBMCs) by more than 1,000-fold. Furthermore, these aptamers were able to neutralize the infectivity of R5 clinical isolates of HIV-1 derived from group M (subtypes A, C, D, E, and F) and group O. One aptamer defined a site on gp120 that overlaps partially with the conserved, chemokine receptor-binding, CD4-induced epitope recognized by monoclonal antibody 17b. In contrast to the antibody, the site is accessible to aptamer in the absence of CD4 binding. Neutralizing aptamers such as this could be exploited to provide leads in developing alternative, efficacious anti-HIV-1 drugs and lead to a deeper understanding of the molecular interactions between the virus and its host cell.
While current antiretroviral drugs have prolonged the quality of life for many human immunodeficiency virus type 1 (HIV-1)-positive individuals, they do not eliminate the virus (15, 37). The rapid emergence of drug-resistant HIV-1 strains has encouraged continued efforts to find novel antiretroviral agents with modalities different from those currently in use. One approach is to target the stage at which virus infects host cells. The entry of HIV-1 into target cells, its cellular tropism, and elements of the pathogenesis of AIDS are largely determined by the virion surface glycoprotein, gp120 (8, 9, 30). Variation in the hypervariable loops of gp120, particularly the V3 loop, determines the cellular tropism of the virus by governing the interaction with chemokine receptors such as CCR5 and CXCR4 (E. A. Berger, R. W. Doms et al., Letter, Nature 391:240, 1998). Strains that infect via CCR5, known as R5 strains, are preferentially transmitted from host to host (41), dominate the asymptomatic stage of infection (18, 36), and are sufficient to cause AIDS (33).
Elucidation of the three-dimensional crystal structure of gp120 (26, 27) coupled with site-directed mutagenesis (12, 24, 40) has revealed the remarkable way in which HIV-1 has evolved to protect its functionally conserved regions, thereby evading host antibody responses (26, 27). For example, regions of gp120 that interact with coreceptors are masked by extensive glycosylation and surface loops, limiting the ability of the immune system to mount a broad-spectrum neutralizing antibody response. These features partly explain the failure of candidate vaccine antigens based on recombinant HIV-1 surface envelope glycoprotein, gp120, to elicit antibodies that neutralize HIV-1 primary isolates (PIs). Consequently, one rational anti-HIV-1 strategy would be to generate ligands directed to these core regions of gp120. Having previously developed artificial nucleic acid ligands called “aptamers” against rat CD4 and streptavidin with useful functional properties (25, 45), we reasoned that aptamers, by virtue of their small size compared to antibodies, could access core regions of gp120, thereby blocking infection. We have recently described the isolation and structural characterization of 2′F nucleic acid aptamers that bind gp120 of the X4 molecular clone, HXB2 (42). These proved not to neutralize the infectivity of the virus, nor to bind to the gp120 of clinically relevant R5 strains. Accordingly, we now describe the isolation of aptamers selected explicitly for their ability to bind gp120 of the HIV-1 R5 strain Ba-L (HIV-1Ba-L) and neutralize infectious virus.
MATERIALS AND METHODS
Virus stocks.
All HIV-1 strains used in this study were obtained through the AIDS Research and Reference Reagent Program, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md. HIV-1Ba-L was contributed by S. Gartner, M. Popovic, and R. Gallo (17); HIV-1ADA was contributed by H. Gendelman (19); and HIV-1IIIB was contributed by R. Gallo (2)
MAbs.
Anti-gp120 monoclonal antibodies (MAbs) 17b (44), 48d (46), 2G12 (5) immunoglobulin G1 (IgG1) b12 (6), C108G (48), and 2/11C and A32 (51) and polyclonal human HIV Ig (38) were obtained from the National Institutes of Health AIDS Reagent Program (www.aidsreagent.org). MAb 19b was kindly provided by James Robinson, Department of Pediatrics, University of Connecticut, Farmington. Anti-gp120-CD4 complex MAb CG10 (20) and recombinant CD4-Ig were obtained from the NIBSC Centralized Facility for AIDS Reagents. Antibodies 17b, 48d, and CG10 map to the CD4-i region of gp120, overlapping the chemokine receptor binding site. CD4-Ig and IgG1 b12 bind to the CD4 binding site on gp120, 2G12 binds to carbohydrates on the “silent face,” C108G binds to the V2 loop, 2/11C and A32 bind to discontinuous epitopes in C1 to C3, and 19b binds to a conformational epitope on the V3 loop. Anti-FLAG M2 and antimouse IgG horseradish peroxidase (HRP)-conjugated MAbs were obtained from Sigma.
Cells.
Spodoptera frugiperda Sf9s cells were kindly provided by Ian Jones (Reading University, United Kingdom).
Human leukocytes were obtained from buffy coat fractions supplied by Bristol Hospital Services through the Oxford National Blood Services.
Oligonucleotides.
The following oligonucleotides were used (listed 5′→3′): Library, AATTAACCCTCACTAAAGGGAACTGTTGTGAGTCTCATGTCGAA(N)49TTGAGCGTCTAGTCTTGTCT; 5′ primer, AATTAACCCTCACTAAAGGGAA CTGTTGTGAGTCTCATGTCGAA; 3′ primer, TAATACGACTCACTATAGGGAGACAAGACTAGACGCTCAA; Env6309+ primer, AGCAGAAGACAGTGGC; and Env8023− primer, TAGTGCTTCCTGCTGCTCC.
Expression of HIV-1Ba-L gp120.
Sf9s cells were cultured at 28°C in SF 900 II serum-free insect medium (Gibco BRL) in suspension culture below 106 cells/ml. Sf9s cells were transfected with a mixture of 500 ng of p2BaC-gp120 (28) encoding HIV-1Ba-L SU glycoprotein (gp120) and linearized pAcBAK6 (Invitrogen) to generate recombinant virus following standard methods (23). Cells were infected at a multiplicity of infection (MOI) of 5 and incubated for 4 days at 28°C, at which time secretion of gp120 into the medium was optimal. gp120 was purified from clarified culture supernatants by using anti-FLAG M2 (Sigma) affinity chromatography, and fractions were evaluated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting. Protein was further purified by fast protein liquid chromatography (FPLC) gel filtration using Superdex 200 HR10/30 (Pharmacia) to exclude high-order aggregates and quantified with bicinchoninic acid (BCA) protein assay kit (Pierce, Chester, United Kingdom) according to the manufacturer's instructions.
In vitro transcription.
A total of 225 pmol of DNA template was added to a final 500-μl transcription reaction mixture comprising 1 mM 2′F UTP, 1 mM 2′F CTP (TriLink), 1 mM GTP, 1 mM ATP (Amersham-Pharmacia), 40 mM Tris-Cl (pH 7.5), 6 mM MgCl2, 5 mM dithiothreitol (DTT), 1 mM spermidine, and 1,500 U of T7 RNA polymerase (New England BioLabs) and incubated at 37°C for 16 h. Transcription was terminated by addition of 1 U of RNase-free DNase I (Sigma) per μg of DNA template used, and the reaction mixture was incubated for 15 to 30 min at 37°C, followed by phenol-chloroform extraction. The RNA was precipitated with ethanol, redissolved in water, separated from low-Mr contaminants with a Sephadex-G50 nick spin column (Pharmacia-Amersham), and quantified by determination of A260. RNA was refolded by heating in water to 95°C for 3 min and then cooling to room temperature for 10 min, at which temperature was added 1/5 volume of a 5× 10 mM final concentration of HBS buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, 2.7 mM KCl). Incubation continued at room temperature for 5 min.
In vitro selection of aptamers.
A BIAcore (Stevenage, United Kingdom) 2000 biosensor instrument was used. A total of 20,000 response units (RU) of HIV-1Ba-L gp120 was directly coupled to the carboxymethylated dextran surface of CM5 biosensor chips (research grade; BIAcore) using amine coupling through lysine residues following a standard protocol (21). RNA was prepared as described above in HBS. For the first round of selection, 20 μg of the RNA pool (a theoretical diversity of ≥1014 molecules) was injected at 1 μl/min at 37°C over the flow cell in which the gp120 was immobilized. Nonspecifically bound RNA was removed by injecting 100 μl of the HBS buffer at 5 μl/min. Bound RNA was eluted with 100 μl of 7 M urea at 5 μl/min and was deproteinized by extraction with phenol-chloroform and then precipitated with ethanol. Recovered RNA was reverse transcribed to cDNA and PCR amplified by using the 3′ and 5′ primers described for use under slightly mutagenic conditions. The RNA/protein ratio was increased by a factor of about 4 each round to increase the stringency of selection. At every selection round, the RNA was precleared against at least two uncoupled sensor chip flow cells to serve as controls and also to avoid the inadvertent selection of aptamers that might bind the chip matrix.
Analysis of aptamer-gp120 interaction by surface plasmon resonance (SPR) biosensor.
Affinity measurements were performed at 37°C. Five thousand to 7,500 RU of HIV-1 gp120 was covalently immobilized on the chip as described above. Aptamer or control nucleic acids were prepared at a range of concentrations (5 to 3,200 nM), injected at 5 μl/min (KININJECT procedure), and allowed to dissociate over 60 min. The ligand was regenerated by injecting 1 to 5 μl of freshly prepared 100 mM NaOH to dissociate any RNA that was still bound without affecting the ability of gp120 to bind soluble CD4 (sCD4). Data were analyzed by using the BIAevaluation 3.0 (BIAcore) and GraphPad Prism 3.00 (GraphPad Software, Inc.) and the Kd was calculated from the ratio of koff and kon.
Cultivation of human PBMCs.
These were isolated by Ficoll-Hypaque (Pharmacia-Amersham) density gradient centrifugation from heparinized buffy coats of normal, HIV-negative donors. The diluted, autologous plasma was saved, heat-inactivated, and clarified to provide autologous serum (AS) supplement for leukocyte culture. The PBMCs were washed six times in PBS (Sigma) at 4°C and were essentially free of platelets and granulocytes. In order to study HIV-1 neutralization in a different cell context, we used PBMC cultures cultivated either with or without mitogen activation and interleukin-2 (IL-2). In either culture system, the cells were maintained in X-VIVO-10 (BioWhittaker) containing 2% AS. The system without a mitogen and IL-2 produces a slowly proliferating mixed culture of lymphocytes and macrophages that in our hands supports a higher level of replication of primary isolates than mitogen-treated, cytokine-supplemented cultures. Mitogen-activated primary lymphoblasts were prepared by stimulating PBMCs with 1-μg/ml phytohemagglutinin (PHA; Wellcome Diagnostic) in X-VIVO-10 supplemented with 2% AS for 72 h. The cell cultures were then removed from PHA-containing medium and transferred to X-VIVO-10 supplemented with 2% AS and 20-U/ml IL-2 (Pharmacia) for infectivity and neutralization assays performed in 96-well plates.
Virus infectivity and neutralization assays. (i) Method 1.
Method 1 was applied for strains that grow to titers of 104 infectious units/ml in culture. Day 7 PBMCs seeded at 105 cells/well were infected with serially diluted virus that had been incubated with 100 nM anti-gp120 monoclonal aptamer or control aptamer SA19 (45). Eight replicates were used at each 10-fold dilution. At 16 h postinfection, the medium containing virus inoculum and aptamer was replaced with fresh, aptamer-free culture medium. Cultures were maintained for 14 days before preparing DNA for long terminal repeat (LTR) PCR as described previously (10).
(ii) Method 2.
Method 2 was applied for clinical isolates. Virus was quantified by the SPA Quan-T-RT assay for reverse transcriptase (RT) (reference TRK1022; Amersham), which makes use of the scintillation proximity assay (SPA) principle (4). Day 7 PBMCs seeded at105 cells/well of a 96-well plate were inoculated in duplicate with approximately 3,000 SPA counts of virus that had been preincubated with 100 nM anti-gp120 monoclonal aptamer or control aptamer, SA19, for 30 to 45 min. After 16 h, the inoculum was replaced with fresh, aptamer-free culture medium, the cells were cultured for a further 5 to 7 days, and then the supernatants were harvested and assayed for extracellular RT activity.
(iii) Method 3
Day 3, PHA-stimulated, IL-2-treated PBMCs seeded at 105 cells/well were infected with a 3-log10 50% tissue culture infectious dose (TCID50)/ml virus inoculum that had been preincubated with 50 μl of serially diluted neutralization agent (aptamer or antibody) for 30 to 45 min at room temperature. Sixteen hours postinfection, the inoculum was replaced with fresh, aptamer-free X-VIVO-10 medium containing 2% AS and 20 U of IL-2 per ml and cultured for a further 3 to 5 days. The extent of virus replication was determined by measuring extracellular p24 antigen content from the supernatants as previously described (32, 43).
Binding site mapping by antibody inhibition.
Binding sites were mapped by antibody inhibition by competitive enzyme-linked immunosorbent assay (ELISA) largely as previously described (32) with modifications as follows. Briefly, gp120 was captured in an Immulon II ELISA plate (Dynatech, Ltd.) using D7324 anti-gp120 COOH peptide antiserum (Aalto Bioreagents Plc.) After washing, bound gp120 was incubated with either 50 μl of HBS buffer or 10 μM soluble human CD4 in HBS buffer for 1 h at room temperature. The plate was washed, and 50 μl of aptamer B4 was added at a range of concentrations, in triplicate, in HBS binding buffer for 1 h. Anti-gp120 MAbs were added at a concentration previously determined to be within the linear range. After washing, bound antibody was detected using the ABC Elite amplification kit (Vector). One hundred percent binding was taken to be that seen in the absence of aptamer.
RESULTS
Selection of aptamers against HIV-1Ba-L gp120.
We selected 2′-fluoropyrimidine-containing RNA (2′-F-RNA) aptamers against the gp120 of the R5 strain, HIV-1Ba-L. The target protein was produced as previously described (28), and selection of aptamers was done by a modification of the SELEX protocol (13, 16) in which the target was immobilized on a BIAcore biosensor chip and enrichment was based on the slow dissociation rate of aptamers from target (Fig. 1A). We used 2′-F-RNA aptamers because they are not only resistant to nucleases, but also widen the spectrum of potential tertiary conformations (25) and give rise to more compact and rigid ligands with higher affinities than do unmodified RNA or NH2-substituted RNA aptamers (34).
FIG. 1.
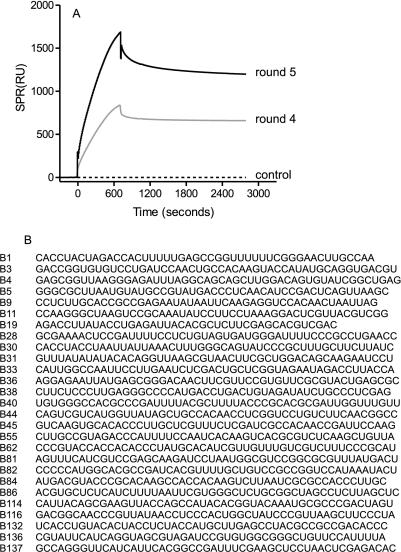
Isolation of aptamers that bind the gp120 of an R5 tropic strain, HIV-1Ba-L. (A) After four and five rounds of selection, the polyclonal pool of 2′F-RNA was analyzed for binding to gp120 by BIAcore. The round 5 pool was cloned, and monoclonal aptamers were sequenced. Panel B shows the sequences of the randomized portion of each aptamer.
Immobilized gp120 initially bound less than 0.1% of the applied RNA, but this rose to 1.5% at round 2, 16% at round 3, 60% at round 4, and 75% at round 5. The PCR-amplified DNA pool from the fifth round of selection was cloned with the TA cloning kit (Invitrogen). Each clone was screened by BIAcore analysis, and those that bound the gp120 with high affinity (Kd, 5 to 100 nM) as determined by BIAcore analysis were found to belong to at least 25 distinct sequence families (Fig. 1B). Taken together, these data imply that a large number of different sequences can fold to give gp120-binding aptamers.
Neutralization of subtype B, R5 strains of HIV-1 by aptamers.
Antibodies elicited during natural infection with HIV-1 are generally poorly neutralizing and are generated late in infection (35). Moreover, MAbs that recognize epitopes presented in the form of isolated gp120 are often unable to bind the same epitope in the context of the assembled Env trimer (31). This underlies the relative resistance of HIV-1 to neutralization by MAbs, which is particularly pronounced in PIs.
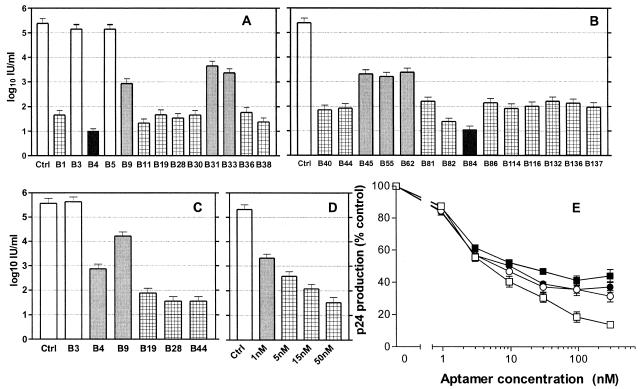
In this study, we therefore asked whether the aptamers isolated against HIV-1Ba-L gp120 could prevent or limit HIV-1 infectivity in target cells. Using an endpoint dilution and PCR-based TCID50 assay (10, 11), we showed that 25 of the 27 aptamer clones assayed neutralized homologous HIV-1Ba-L in PBMCs (Fig. 2A and B). Most of these aptamers neutralized HIV-1Ba-L by more than 1,000-fold, and one aptamer (B4) neutralized the virus by about 4 log10 in this cell system. Five HIV-1Ba-L-neutralizing aptamers tested, including aptamer B4, also cross-neutralized another R5 strain, HIV-1ADA (Fig. 2C). The gp120s of these two strains are only 84% identical, and they show differences in the degree of their macrophage tropism (11). Therefore, the ability of at least five aptamers to neutralize at least two HIV-1 strains suggests that they might recognize functionally important sites on gp120 that are conserved between at least the R5 members of this clade B subtype.
FIG. 2.
Neutralization of R5 HIV-1 strains by monoclonal aptamers. Virus was titrated by limiting dilution in PBMCs in the presence of a 100 nM concentration of the aptamers indicated. No reduction in infectivity is indicated by open columns, between 10- and 100-fold reduction of infectivity by shaded columns, 103- to 104-fold reduction of infectivity is indicated by hatched columns, and >104-fold reduction of infectivity is indicated by solid columns. Panels A and B show neutralization of HIV-1Ba-L, the strain from which the aptamers were raised. Seventeen aptamers neutralized the virus by 3 to 4 log10 IU/ml, and two aptamers (B4 and B84) inhibited HIV-1Ba-L entry by more than 4 log10 IU/ml. (C) Five of the aptamers tested so far also cross-neutralized another R5 strain, HIV-1ADA. (D) Aptamer B4 inhibited HIV-1Ba-L entry in a concentration-dependent manner and with an IC50 value of <1 nM. (E) Neutralization of HIV-1Ba-L by aptamers in PHA-stimulated, IL-2-treated PBMCs. The following aptamers were used: B4 (▪), B40 (□), B19 (○), and B28 (•). The extent of virus replication is represented as a percentage of p24 antigen produced in the absence of any inhibitor. All experiments were performed in triplicate, and error bars represent the standard error of the mean.
We went on to investigate the potency of neutralization by aptamers. In the same PBMC ID50 system used before, aptamer B4 inhibited HIV-1Ba-L entry with a 50% inhibitory concentration (IC50) value of less than 1 nM (Fig. 2D). In order to make a valid comparison with previously reported neutralization studies of MAbs, we studied four aptamers in a PHA-stimulated, IL-2-supplemented culture of primary lymphoblasts and used output of viral p24 antigen to measure the effectiveness of aptamers (Fig. 2E). Under these conditions, aptamers produced less profound neutralization, with IC50 values ranging from 5 nM to approximately 20 nM.
Neutralization of diverse clinical isolates.
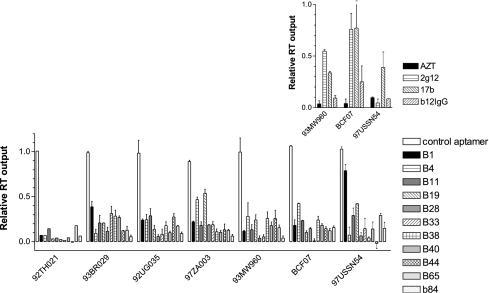
Outside the Americas, Europe and Australasia, subtype B of HIV is not predominant, with subtype C dominating the larger, heterosexually transmitted epidemic in sub-Saharan Africa and Asia. In addition, subtype A is common in South Asia, and subtypes A, D, F, G, H, I, J, and K circulate in southern Africa. All of these subtypes belong to group M, but isolates belonging to groups N and O are also found in Africa. Variation between env sequences of strains of different group M subtypes is typically 24% at the nucleotide level and greater still between groups. Accordingly, we screened a panel of six group M, non-B subtype clinical isolates and one group O isolate for sensitivity to neutralization by 11 representative aptamers. The results (Fig. 3) show that most of the aptamers tested produced an 80% or greater degree of inhibition of infection under these conditions, favorably comparable with the best available neutralizing antibodies tested under the same conditions (Fig. 3, inset).
FIG. 3.
Neutralization of diverse clinical isolates of HIV-1 by aptamers. Clinical isolates of HIV-1 representing five subtypes of group M and one group O strain were preincubated in duplicate with an aptamer before inoculation of cultures of mature PBMCs. Unbound virus and aptamer were washed off, and the cultures were incubated for a further 5 to 7 days, after which culture supernatants were assayed for HIV particles by using the SPA-RT assay (Amersham). Background-corrected values were expressed as a proportion of those obtained with parallel cultures in which aptamer was omitted. Error bars represent the standard error. The inset shows corresponding values for cells treated with 10 μM zidovudine (AZT) during infection or virus pretreated with anti-gp120 MAbs.
Binding of aptamer B4 to gp120 affects the conserved CCR5-binding surface.
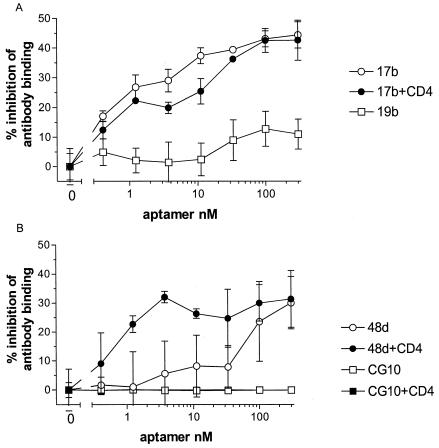
A likely mechanism of neutralization by aptamer was by inhibition of virus-receptor interactions; however, using SPR biosensor analysis, we found that aptamer B4 did not interfere with the binding of sCD4 to monomeric, immobilized gp120 (data not shown). We next tested whether aptamer B4 would interfere with the binding of MAbs whose epitopes had been previously mapped on gp120. The MAbs we chose were all known to neutralize R5 strains of clade B, because we hypothesized that aptamer B4 was likely to bind close to a conserved neutralization epitope rather than to a nonneutralizing epitope. Of the 10 MAbs tested, only 1, 17b, was convincingly inhibited by aptamer in the absence of CD4 (Fig. 4A). The level of inhibition was significant, but only 50% at the highest concentration of aptamer, suggesting that either the two molecules bound to spatially related but distinct surfaces or the binding of aptamer B4 produced a subtle allosteric change in the epitope of 17b, reducing the level of antibody binding without abolishing it. The 17b epitope is partially obscured from antibody in the absence of CD4 binding by the V1 and V2 hypervariable loops of gp120 (46, 51) and overlaps with the binding site for the major coreceptors of the virus, CCR5 and CXCR4 (50). The binding of a second antibody, 48d, which like 17b maps to the CD4i epitope, was inhibited by high concentrations of aptamer in the absence of CD4 but by nanomolar concentrations in the presence of CD4 (Fig. 4B). A third CD4i-binding antibody, CG10, was not inhibited by aptamer B4.
FIG. 4.
Effect of aptamer B4 on the binding of MAbs to gp120. Recombinant Ba-L gp120 was captured on the surface of a microtiter plate by polyclonal antibody. If indicated, the bound gp120 was then allowed to interact with sCD4. The bound gp120 or gp120-CD4 complex was then incubated with various concentrations of aptamer B4 in triplicate. The gp120-aptamer or gp120-CD4-aptamer complexes were then incubated with the anti-gp120 MAbs indicated at a concentration previously determined to be in the linear detection range of each. Bound MAb was then detected with an anti-Ig-HRP conjugate system. The results are displayed as a percentage of reduction from the aptamer-free control value ± standard error.
DISCUSSION
We have presented a novel strategy for identifying conserved regions of the envelope of primary HIV-1 isolates that could be targeted by potentially therapeutic agents. The small size and biophysical properties of aptamers have enabled us to target conserved sites on HIV-1 gp120 whose ligation produced highly efficient neutralization of infectivity. Studies on the structure of gp120 derived from HIV strains of different coreceptor utilization indicate that, although the surface loops of gp120 are of variable sequence and variable topology, the core of the protein, containing the principal receptor-interacting surfaces, is relatively well conserved in tertiary structure (26). Quaternary interactions between variable surface loops of gp120 monomers seem to have evolved to enable the virus to escape from neutralization by antibody but, critically, expose the conserved core of gp120 to interference by smaller ligands, such as aptamers.
Two properties of aptamer neutralization deserve particular comment: potency and resistance to strain variation in envelope sequences. The ability of an aptamer like B4 to reduce the infectivity of R5 virus is remarkable when compared with the most potent antibodies, such as IgG1-b12 and 2G12. In our hands, these antibodies have IC50 values of >50 nM (data not shown) under conditions in which aptamers such as B4 have IC50 values of approximately 5 nM (Fig. 2E). Second, antibody-mediated neutralization is rapidly overcome by virus evolution both in vitro (29) and in vivo (1, 49), and this results in an extreme diversity of HIV envelope sequences in the wild, presenting a challenge to prophylaxis and therapy aimed at targeting the entry phases of HIV infection. Very encouragingly, 11 aptamers tested here were able not only to neutralize the infectivity of strains of HIV from the same subtype as the original target gp120, but also clinical isolates from five other common subtypes and even one group O strain. Strikingly, even the most cross-reactive, potent bivalent antibody, IgG-b12, was not as active as the most potent monomeric aptamers in this study.
Other therapeutic strategies designed to interfere with HIV entry include gp41-disrupting peptide analogs (3, 14, 22) and small-molecule ligands of CCR5 (47). The most promising, such as C34 (7) and T-20 (22), have IC50 values close to that of the aptamers described here (∼2 nM). However, virus variants resistant to inhibition by T-20 peptide have now been identified in clinical trials (52).
Our results give some clues to the mechanism of neutralization by aptamer. Aptamer B4 did not interfere with the interaction between CD4 and gp120 nor the binding of antibodies whose epitopes on gp120 comprise the V3 loop, the V1/V2 loops, and the carbohydrate on the “silent” face. The ability of potently neutralizing aptamer B4 to partially inhibit binding of MAbs 17b and 48d suggests that the mechanism of neutralization may be by interfering with the interaction of gp120 with its coreceptor, CCR5. In the absence of binding of CD4 to gp120, the 17b/48d epitopes are partially occluded on monomeric gp120 and fully occluded on functional, trimeric gp120, ensuring that it is only exposed to the antiviral effects of the humoral immune response very transiently during infection, if at all. In contrast, perhaps because of its smaller size or biophysical properties, aptamer B4 binds to its neutralization site in the absence of CD4 binding. The CD4-dependent binding of antibody 48d to gp120 was more potently inhibited by aptamer B4 in the presence of CD4, implying that the access of aptamer B4 to the 48d epitope may be partially blocked on uncomplexed gp120. Intriguingly, we found that CG10, a third antibody that maps to the CD4i region, was not inhibited by aptamer B4 at all. This is consistent with the location of its epitope closest to the base of the V1/V2 loops, and therefore the most occluded of these three epitopes (39, 40). Taken together with the evidence that the aptamer B4 binding site is highly conserved, this suggests that, as we had hoped, the aptamer approach has identified a more circumscribed and functionally important neutralization target on gp120 than has been possible previously when using antibodies.
Although the high cost of synthesis of modified nucleic acids, together with their poor bioavailability, makes it unlikely that aptamers in the form we describe here could be readily used as anti-HIV-1 therapeutic agents, they may well provide invaluable leads for structure-based drug design. The challenge now is to characterize the neutralization sites identified by these aptamers at a functional and structural level both in order to gain greater insights into the molecular biology of HIV-1 infection and to provide detailed structural leads for drug development.
Acknowledgments
This work was funded by the Wellcome Trust and South African MRC.
REFERENCES
- 1.Arendrup, M., C. Nielsen, et al. 1992. Autologous HIV-1 neutralizing antibodies: emergence of neutralization-resistant escape virus and subsequent development of escape virus neutralizing antibodies. J. Acquir. Immune Defic. Syndr. 5:303-307. [PubMed] [Google Scholar]
- 2.Barre-Sinoussi, F., J. C. Chermann, et al. 1983. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS). Science 220:868-871. [DOI] [PubMed] [Google Scholar]
- 3.Bewley, C. A., J. M. Louis, et al. 2002. Design of a novel peptide inhibitor of HIV fusion that disrupts the internal trimeric coiled-coil of gp41. J. Biol. Chem. 277:14238-14245. [DOI] [PubMed] [Google Scholar]
- 4.Bosworth, N., and P. Towers. 1989. Scintillation proximity assay. Nature 341:167-168. [DOI] [PubMed] [Google Scholar]
- 5.Buchacher, A., R. Predl, et al. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 6.Burton, D. R., C. F. Barbas III, et al. 1991. A large array of human monoclonal antibodies to type 1 human immunodeficiency virus from combinatorial libraries of asymptomatic seropositive individuals. Proc. Natl. Acad. Sci. USA 88:10134-10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, D. C., C. T. Chutkowski, et al. 1998. Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. USA 95:15613-15617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chavda, S. C., P. Griffin, et al. 1994. Molecular determinants of the V3 loop of human immunodeficiency virus type 1 glycoprotein gp120 responsible for controlling cell tropism. J. Gen. Virol. 75:3249-3253. [DOI] [PubMed] [Google Scholar]
- 9.Chesebro, B., J. Nishio, et al. 1991. Identification of human immunodeficiency virus envelope gene sequences influencing viral entry into CD4-positive HeLa cells, T-leukemia cells, and macrophages. J. Virol. 65:5782-5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collin, M., G. Herbein, et al. 1993. PCR analysis of HIV1 infection of macrophages: virus entry is CD4-dependent. Res. Virol. 144:13-19. [DOI] [PubMed] [Google Scholar]
- 11.Collin, M., P. Illei, et al. 1994. Definition of the range and distribution of HIV macrophage tropism using PCR-based infectivity measurements. J. Gen. Virol. 75:1597-1603. [DOI] [PubMed] [Google Scholar]
- 12.Cordonnier, A., L. Montagnier, et al. 1989. Single amino-acid changes in HIV envelope affect viral tropism and receptor binding. Nature 340:571-574. [DOI] [PubMed] [Google Scholar]
- 13.Ellington, A. D., and J. W. Szostak. 1990. In vitro selection of RNA molecules that bind specific ligands. Nature 346:818-822. [DOI] [PubMed] [Google Scholar]
- 14.Ferrer, M., T. M. Kapoor, et al. 1999. Selection of gp41-mediated HIV-1 cell entry inhibitors from biased combinatorial libraries of non-natural binding elements. Nat. Struct. Biol. 6:953-960. [DOI] [PubMed] [Google Scholar]
- 15.Finzi, D., J. Blankson, et al. 1999. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat. Med. 5:512-517. [DOI] [PubMed] [Google Scholar]
- 16.Fitzwater, T., and B. Polisky. 1996. A SELEX primer. Methods Enzymol. 267:275-301. [DOI] [PubMed] [Google Scholar]
- 17.Gartner, S., P. Markovits, et al. 1986. Virus isolation from and identification of HTLV-III/LAV-producing cells in brain tissue from a patient with AIDS. JAMA 256:2365-2371. [PubMed] [Google Scholar]
- 18.Gendelman, H. E., J. M. Orenstein, et al. 1989. The macrophage in the persistence and pathogenesis of HIV infection. AIDS 3:475-495. [DOI] [PubMed] [Google Scholar]
- 19.Gendelman, H. E., J. M. Orenstein, et al. 1988. Efficient isolation and propagation of human immunodeficiency virus on recombinant colony-stimulating factor 1-treated monocytes. J. Exp. Med. 167:1428-1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gershoni, J. M., G. Denisova, et al. 1993. HIV binding to its receptor creates specific epitopes for the CD4/gp120 complex. FASEB J. 7:1185-1187. [DOI] [PubMed] [Google Scholar]
- 21.Karlsson, R., A. Michaelsson, et al. 1991. Kinetic analysis of monoclonal antibody-antigen interactions with a new biosensor based analytical system. J. Immunol. Methods 145:229-240. [DOI] [PubMed] [Google Scholar]
- 22.Kilby, J. M., S. Hopkins, et al. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 23.King, L., and R. Possee. 1992. The baculovirus expression system: a laboratory guide. Chapman & Hall, London, United Kingdom.
- 24.Kowalski, M., J. Potz, et al. 1987. Functional regions of the envelope glycoprotein of human immunodeficiency virus type 1. Science 237:1351-1355. [DOI] [PubMed] [Google Scholar]
- 25.Kraus, E., W. James, et al. 1998. Novel RNA ligands able to bind CD4 antigen and inhibit CD4+ T lymphocyte function. J. Immunol. 160:5209-5212. [PubMed] [Google Scholar]
- 26.Kwong, P. D., R. Wyatt, et al. 2000. Structures of HIV-1 gp120 envelope glycoproteins from laboratory-adapted and primary isolates. Struct. Fold Des. 8:1329-1339. [DOI] [PubMed] [Google Scholar]
- 27.Kwong, P. D., R. Wyatt, et al. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin, C. L., A. K. Sewell, et al. 2000. Macrophage-tropic HIV induces and exploits dendritic cell chemotaxis. J. Exp. Med. 192:587-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKeating, J. A., J. Gow, et al. 1989. Characterization of HIV-1 neutralization escape mutants. AIDS 3:777-784. [DOI] [PubMed] [Google Scholar]
- 30.Mondor, I., M. Moulard, et al. 1998. Interactions among HIV gp120, CD4, and CXCR4: dependence on CD4 expression level, gp120 viral origin, conservation of the gp120 COOH- and NH2-termini and V1/V2 and V3 loops, and sensitivity to neutralizing antibodies. Virology 248:394-405. [DOI] [PubMed] [Google Scholar]
- 31.Moore, J. P. and D. D. Ho. 1995. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS 9(Suppl. A):S117-S136. [PubMed] [Google Scholar]
- 32.Moore, J. P., J. A. McKeating, et al. 1990. Characterization of recombinant gp120 and gp160 from HIV-1: binding to monoclonal antibodies and soluble CD4. AIDS 4:307-315. [DOI] [PubMed] [Google Scholar]
- 33.Mosier, D., and H. Sieburg. 1994. Macrophage-tropic HIV: critical for AIDS pathogenesis? Immunol. Today 15:332-339. [DOI] [PubMed] [Google Scholar]
- 34.Pagratis, N. C., C. Bell, et al. 1997. Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat. Biotechnol. 15:68-73. [DOI] [PubMed] [Google Scholar]
- 35.Pilgrim, A. K., G. Pantaleo, et al. 1997. Neutralizing antibody responses to human immunodeficiency virus type 1 in primary infection and long-term-nonprogressive infection. J. Infect. Dis. 176:924-932. [DOI] [PubMed] [Google Scholar]
- 36.Ping, L. H., J. A. Nelson, et al. 1999. Characterization of V3 sequence heterogeneity in subtype C human immunodeficiency virus type 1 isolates from Malawi: underrepresentation of X4 variants. J. Virol. 73:6271-6281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Piot, P., M. Bartos, et al. 2001. The global impact of HIV/AIDS. Nature 410:968-973. [DOI] [PubMed] [Google Scholar]
- 38.Prince, A. M., H. Reesink, et al. 1991. Prevention of HIV infection by passive immunization with HIV immunoglobulin. AIDS Res. Hum. Retrovir. 7:971-973. [DOI] [PubMed] [Google Scholar]
- 39.Rizzuto, C., and J. Sodroski. 2000. Fine definition of a conserved CCR5-binding region on the human immunodeficiency virus type 1 glycoprotein 120. AIDS Res. Hum. Retrovir. 16:741-749. [DOI] [PubMed] [Google Scholar]
- 40.Rizzuto, C. D., R. Wyatt, et al. 1998. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science 280:1949-1953. [DOI] [PubMed] [Google Scholar]
- 41.Samson, M., F. Libert, et al. 1996. Resistance to HIV-1 infection in caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 42.Sayer, N., J. Ibrahim, et al. 2002. Structural characterization of a 2′ F-RNA aptamer that binds HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun. 293:924-931. [DOI] [PubMed]
- 43.Simon, J. H., C. Somoza, et al. 1993. A rat CD4 mutant containing the gp120-binding site mediates human immunodeficiency virus type 1 infection. J. Exp. Med. 177:949-954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sullivan, N., Y. Sun, et al. 1998. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J. Virol. 72:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahiri-Alaoui, A., L. Frigotto, et al. 2002. High affinity nucleic acid aptamers for streptavidin incorporated into bi-specific capture ligands. Nucleic Acids Res. 30:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thali, M., J. P. Moore, et al. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trkola, A., S. E. Kuhmann, et al. 2002. HIV-1 escape from a small molecule, CCR5-specific entry inhibitor does not involve CXCR4 use. Proc. Natl. Acad. Sci. USA 99:395-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warrier, S. V., A. Pinter, et al. 1994. A novel, glycan-dependent epitope in the V2 domain of human immunodeficiency virus type 1 gp120 is recognized by a highly potent, neutralizing chimpanzee monoclonal antibody. J. Virol. 68:4636-4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wei, X., J. Decker, et al. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 50.Wu, L., N. P. Gerard, et al. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179-183. [DOI] [PubMed] [Google Scholar]
- 51.Wyatt, R., J. Moore, et al. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zollner, B., H. H. Feucht, et al. 2001. Primary genotypic resistance of HIV-1 to the fusion inhibitor T-20 in long-term infected patients. AIDS 15:935-936. [DOI] [PubMed] [Google Scholar]