Abstract
Noroviruses (NORs) are an important cause of acute gastroenteritis. Recent studies of NOR receptors showed that different NORs bind to different histo-blood group antigens (HBGAs), and at least four distinct binding patterns were observed. To determine the structure-function relationship for NORs and their receptors, two strains representing two of the four binding patterns were studied. Strain VA387 binds to HBGAs of A, B, and O secretors, whereas strain MOH binds to HBGAs of A and B secretors only. Using multiple sequence alignments, homology modeling, and structural analysis of NOR capsids, we identified a plausible “pocket” in the P2 domain that may be responsible for binding to HBGA receptors. This pocket consists of a conserved RGD/K motif surrounded by three strain-specific hot spots (N302, T337, and Q375 for VA387 and N302, N338, and E378 for MOH). Subsequent mutagenesis experiments demonstrated that all four sites played important roles in binding. A single amino acid mutation at T337 (to A) in VA387 or a double amino acid mutation at RN338 (to TT) in MOH abolished binding completely. Change of the entire RGD motif to SAS abolished binding in case of VA387, whereas single amino acid mutations in that motif did not have an apparent effect on binding to A and B antigens but decreased binding to H antigen. Multiple mutations at the RGK motif of MOH (SIRGK to TFRGD) completely knocked out the binding. Mutation of N302 or Q375 in VA387 affected binding to type O HBGA only, while switch mutants with three amino acid changes at either site from MOH to VA387 resulted in a weak binding to type O HBGAs. A further switch mutant with three amino acid changes at E378 from MOH to VA387 diminished the binding to type A HBGA only. Taken together, our data indicate that the binding pocket likely exists on NOR capsids. Direct evidence of this hypothesis requires crystallography studies.
Noroviruses (NORs) are the most important cause of nonbacterial epidemics of acute gastroenteritis, affecting individuals of all ages, in both developing and developed countries (7, 10). NORs are icosahedral, single-stranded, positive-sense RNA viruses whose capsids are composed of 180 copies of a single major structural protein (19, 22, 35). The viral capsid proteins expressed by baculovirus in insect cells self-assemble into virus-like particles (VLPs) (20, 21). These VLPs are morphologically and antigenically indistinguishable from authentic virions (20, 21), providing a useful tool for development of immunological assays and for study of receptor-virus interaction. Data from cryoelectron microscopy and X-ray crystallography showed that the viral capsid protein folds into two major domains, the S and P domains (35, 36). The S domain forms the interior shell, while the P domain builds up arch-like structures that protrude from the shell. Morphogenesis studies showed that the S domain contains elements required for assembly of the capsid, whereas intermolecular contacts between dimeric subunits of the P domain increase the stability of the capsid (1). The P domain is further divided into P1 and P2 domains, with the latter located at the most exterior surface of the capsid. In contrast to the S and P1 domains, the P2 domain has a high sequence variation and therefore is believed to be critical in immune recognition and receptor binding.
NORs have been recently found to recognize human histo-blood group antigens (HBGAs) as receptors (14, 16-18, 26, 30). The recognition of HBGAs by NORs was demonstrated to be strain specific. So far, four distinct binding patterns of NORs, which were defined by the ABO, Lewis, and secretor types of human hosts (16), have been described. Human HBGAs are complex carbohydrates linked to glycoproteins or glycolipids that are present on the red blood cells and mucosal epithelial cells or as free antigens in biological fluids, such as blood, saliva, intestinal contents, and milk (29). These antigens are synthesized by sequential additions of monosaccharides to the antigen precursors by several glycosyltransferases that are genetically controlled and known as the ABO, Lewis, and secretor gene families (29).
The prototype Norwalk virus (NV) represents one of the four binding patterns, and it binds to HBGAs of type A and O secretors but not of nonsecretors (16, 26). Human volunteer studies showed that saliva from volunteers with nonsecretor status did not bind to NV and that nonsecretors were naturally resistant to NV infection following the challenge (26). In our studies, NV did not bind to saliva of type B secretors (16). A retrospective study of volunteers challenged with NV showed that type B individuals had a lower rate of infection by NV than individuals of other blood types following the challenge (17). The other three binding patterns recognize A, B, and O secretors (strain VA387), A and B secretors (MOH), and Lewis-positive secretors and nonsecretors (strain VA207) (16). By analogy, we predict that each of the three binding patterns may have its own host ranges defined by human blood types, although direct evidence linking HBGAs with infection of these strains remains lacking.
In this study, we addressed the question of structural determinants of NOR capsid binding to HBGA receptors. Using computational approaches, we identified a putative receptor binding site on the surface of the P2 domain. Mutagenesis data revealed that this putative binding pocket is indeed involved in binding to HBGAs. More importantly, single amino acid changes within this pocket knocked out the binding completely, whereas shifting mutations resulted in changes of binding patterns, highlighting the importance of this newly identified site for virus-host interaction.
MATERIALS AND METHODS
Protein sequence analysis and computer modeling.
Multiple alignments of known NOR capsid sequences were carried out with OMIGA 2.0 software (Oxford Molecular Ltd.). The crystal structure of the prototype NV capsid protein (PDB code 1I HM) (35) was used to built homology models for other NOR strains. The initial sequence-to-structure alignments and the refined three-dimensional models of the NOR capsids with minimized side-chain conformations were obtained using the 3D-PSSM (6, 24) and MODELLER (32, 40) servers and programs as well as our own LOOPP (33) program.
Construction of mutant NOR capsids by site-directed mutagenesis.
A series of mutant NOR capsids of strains VA387 (23) and MOH (11) were constructed by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, Calif.). The capsid genes of VA387 and MOH were cloned into pGEM-T vector (Promega, Madison, Wis.). Primers for site-directed mutagenesis were designed according to manufacturer's instruction with at least 15 nucleotides at both ends to the nearest mutated nucleotides. For strain VA387 the following primers were used: GTTGTCCAACCACAAAGTGCCAGTTGCACGACTGATGGC (NGR/SAA), CCAAGGTGTTTTACGGTCCACGTGCAGAC (NGR/NAR), GTCAATATCTGCACCTTCAGTGCGGCTGTCACCCACATTGCAG (RGD/SAS), CTGCACCTTCGCAGGGGATGTCACCCAC (RGD/AGD), CTGCACCTTCAGAGCGGATGTCACCCAC (RGD/RAG), CTGCACCTTCAGAGGGAAAGTCACCCACATTGCAG (RGD/RGK), GTCATGACTATATAATGGCTTTGGCATCTCAAAATTGG (N/A), GCTCACCCAAACCGCAAGAGAGGATGGC (T/A), and GACACAAACAATGATTTTGCAACTGGCCAAAACACG (Q/A). For strain MOH, the following primers were used: GTTGTTCAGCCACAGAGTGCTAGCGTCACATTAGATGGG (NGR/SAS), TGTAACATTTGCACCTTCGAGGGGGACGTGACAGGGCAG (SIRGK/TFRGD), CACATGTGGAACATGAACCTCACAAACCTAAATGGG (LEI/MNL), GGTGTGCTCAGCCAGACAACCAGAGGCGAAAGCAAC (RN/TT), and AACACCAATGATTTTCAAACCCAACCAACAAAATTC (VEN/FQT). Three chimera capsids of MOH with an 11-amino acid (aa) replacement from VA387 at site II (chimera 1) and/or site IV (chimera 2) were made by overlapping PCRs. These were achieved by designing a pair of primers that can anneal to the positions adjacent to the regions of replacement and have the overlapping sequences (mutated sequences) at their 5′ ends (ATTTTGAGATGCCAAATTCATTATATAGTCATGATTAGGGACCTGCCCTGTCAC and CATGACTATATAATGAATTTGGCATCTCAAAATAACCTAAATGGGACGCAATTTG for chimera 1; CGTGTTTTGGCCAGTTTGAAAATCATTGTTTGTCCAAGTTCCAATTTGCACTAAG and ACAAACAATGATTTTCAAACTGGCCAAAACACGTTCACCCCAATTGGTTTGAAT for chimera 2). PCR products from these two primers, together with a primer at the beginning or end of the coding region, were gel purified and used together as templates for a second PCR using primers at both ends of the coding region of the capsid gene. Chimera 3, which contains mutations of chimera 1 and 2, was prepared by the same method, with chimera 1 as the starting construct. Mutated sequences were validated by sequencing.
Expression and purification of mutated capsid proteins.
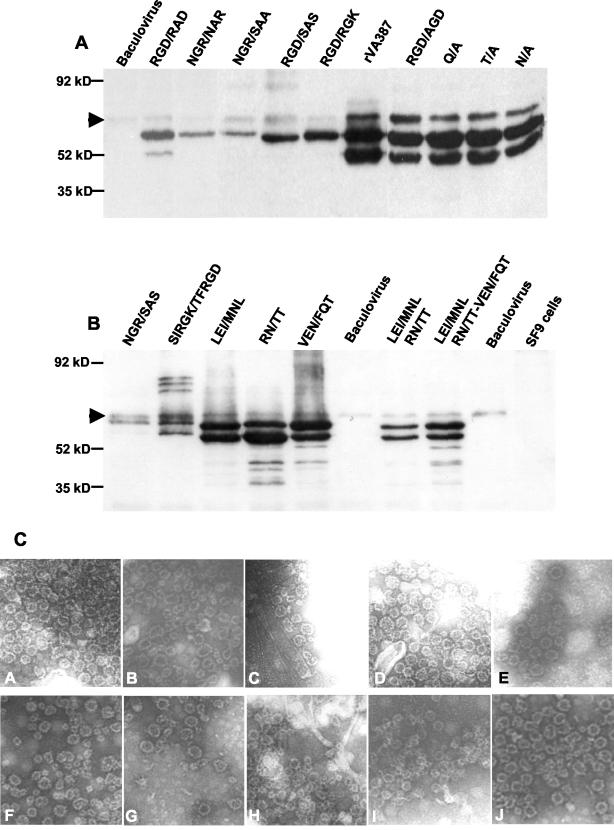
Mutated capsids were expressed in Spodoptera frugiperda (Sf9) cells using the Bac-to-Bac baculovirus expression system (Invitrogen, Carlsbad, Calif.) according to the manufacturer's manual. Briefly, the mutated capsid genes were subcloned into pFastBac1 donor plasmid and transposed into bacmid. Sf9 cells were then infected with wild-type baculovirus or the baculovirus recombinant bacmids containing the mutated capsid genes. Infected cells were harvested between the third and the sixth generation. After a few freeze-thaw cycles, the cell lysates were centrifuged at 5,000 × g for 15 min to separate the cell debris. The clear supernatants were centrifuged again at 10,000 × g for 30 min to spin down large protein complexes or baculovirus particles. VLPs in the supernatant were then purified by centrifugation at 100,000 × g for 150 min. For further purification of the VLPs, the resuspended pellets were separated using sucrose step gradient (5 to 45%) centrifugation, as described previously (20, 21). The recombinant proteins were stored in 1× phosphate-buffered saline, pH 7.4, at −70°C. To quantify the expressed proteins, a small aliquot of purified samples was separated on a sodium dodecyl sulfate-10% polyacrylamide gel. A series of dilutions of the quantitated recombinant wild-type VLPs of the two strains (VA387 and MOH) were loaded on the same gel as standards. Proteins were then transferred to a nitrocellulose membrane for Western blot analysis with hyperimmune antibodies raised in guinea pigs against the wild-type capsids of VA387 or MOH, respectively. The protein concentrations in the samples were determined by comparing the signal intensities using the image analysis software Scion Image (4.0.2; Scion Corporation) after immunodetection. Because we found background bands from wild-type baculovirus in some Western blot experiments (see Fig. 4), we did not use enzyme immune assays to determine the concentrations of mutated capsids.
FIG. 4.
Western blot analysis of mutant capsid proteins expressed in Sf9 culture. (A) Mutants from VA387; (B) mutants from MOH. Each sample contained partially purified VLPs corresponding to an equal amount of original insect culture. The proteins were detected by hyperimmune guinea pig antibodies against recombinant wild-type VA387 (rVA387) (A) and MOH (rMOH) (B) capsids. In most cases, two major bands at ∼58 and ∼50 kDa were observed for each recombinant capsid. Arrows show the background bands from baculovirus. (C) VLPs from mutated capsids. A, VA387 wild type; B, VA387 RGD/AGD mutant; C, VA387 RGD/RAD mutant; D, VA387 N/A mutant; E, VA387 T/A mutant; F, VA387 Q/A mutant; G, MOH LEI/MNL mutant; H, MOH TN/TT mutant; I, MOH VEN/FQT mutant, J, MOH wild type. Magnification, ×31,500.
EM.
Negative-stain electron microscopy (EM) with ammonium molybdate (1) was used to confirm the VLP formation of mutated capsids. The preparations of the partially purified NOR capsid proteins used in the receptor binding assays were used; they had been stored at −70°C and frozen and thawed several times before the EM studies. To ensure reproducible results, multiple meshes of each grid were examined and multiple pictures were taken for each mutant.
Characterization of NOR capsid binding to HBGAs by saliva binding enzyme immune assays.
Saliva binding enzyme immune assays were used to monitor recombinant VLP binding to HBGAs, as described previously (16). Briefly, saliva samples of known ABO, secretor and Lewis types were boiled at 100°C and centrifugation at 10,000 g for 5 min. The supernatant was then used to coat microtiter plates (Dynex Immulon; Dynatech, Franklin, Mass.) at a dilution of 1:1,000 in 1× phosphate-buffered saline (pH 7.4). After blocking with 5% dried milk (Blotto), known amounts of the wild-type recombinant capsid proteins or their mutated forms were added with a serial dilution. The bound capsid proteins were detected with hyperimmune guinea pig anti-NOR antisera, and by adding horseradish peroxidase-conjugated goat anti-guinea pig immunoglobulin G (ICN, Aurora, Ohio). The horseradish peroxidase activity was detected with a TMB kit (Kirkegaard & Perry Laboratories, Gaithersburg, Md.), and the signal intensities (optical density [OD]) were read with an enzyme immunoassay spectrum reader (Tecan, Durham, N.C.). In order to determine the binding affinity of the mutated capsids in relation with their wild-type one, all capsids were assayed for their binding to HBGAs of A-, B-, and O-type saliva within a comparable range of protein concentrations.
RESULTS
Identification of NOR capsid domains binding to HBGA receptors.
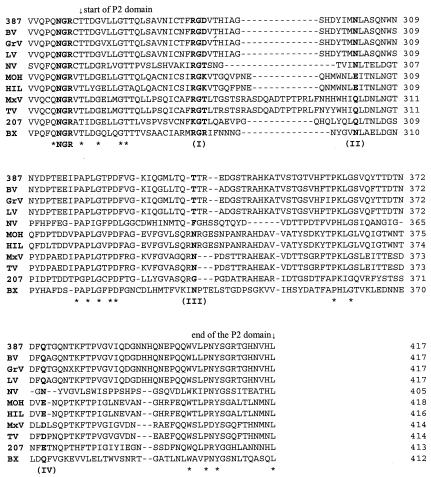
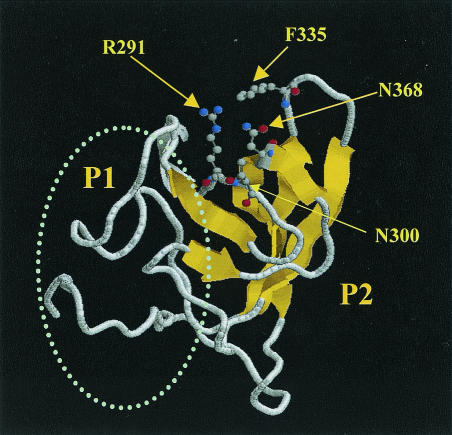
Because of the surface location and high genetic variation of the P domain of NOR capsid, our initial sequence analyses were focused on this region. Multiple alignments of the capsid sequences of NORs representing the four distinct binding patterns to HBGA receptors revealed a conserved RGD-like motif in the P2 domain (aa 288 to 290 in VA387) (Fig. 1). The RGD motif has been identified as a universal recognition site for cell-to-cell and cell-to-extracellular-environment interactions, such as receptor-ligand, signal transduction, enzyme-substrate, and hormone-target interactions (38). Typical receptor-ligand interaction includes foot-and-mouth disease virus (31), coxsackievirus, and adenovirus receptor recognition (46). To determine whether the RGD-like motif also plays a role in NOR receptor binding, we used a combination of multiple alignments and structural analysis to search for sites that are conserved between strains of similar binding patterns and at the same time are in close spatial proximity. The results showed that the conserved RGD-like motif is surrounded by a cluster of strain-specific residues, forming a pocket on the surface of the P2 domain (Fig. 2; structure is based on NV). The RGD-like motif is located at the bottom, and the hot spots N302, T337, and Q375 (VA387) surround the pocket. All four sites are in close spatial proximity. For convenience, the four sites are referred to as I, II, III, and IV, following their sequence order. The corresponding sites in MOH are RGK288-290, E302, N338, and E378, respectively (Fig. 1).
FIG. 1.
Sequence comparison of the P2 domain of NOR capsids. The strains representing the four binding patterns to human HBGAs are VA387, NV, MOH, and VA207. Four sites (I to IV) that are potentially responsible for building up a putative binding pocket are in bold. The NGR motif upstream of the P2 domain is also in bold. Strains VA387 (387) and Grimsby virus (GrV) bind to A-, B-, and O-type saliva. The binding patterns of the Bristol (BV) and Lordsdale viruses (LV) are unknown but they share over 95% amino acid identity with VA387 and Grimsby virus. The prototype NV is the only strain known to bind to A and O saliva. MOH and Mexico virus (MxV) bind to type A and B saliva. The binding patterns of their homologous strains Hillingdon virus (HIL) and Toronto virus (TV) remain to be determined. VA207 (207) binding to the Lewis epitope of secretors and nonsecretors. According to our preliminary results, a new cluster representative strain within genogroup I, Boxer (BX), also binds to the Lewis epitope (unpublished data), but additional characterization of this strain is necessary. The stars indicate conserved residues for all strains. Numbers on the right indicate the sequence position of capsid proteins.
FIG. 2.
Computational prediction of a plausible binding pocket on the surface of the NV capsid protein. The predicted pocket is located on top of the P2 domain and is composed of a conserved RGD-like motif (R291) and three strain-specific hot spots, N300, F335, and N368, that are located in close spatial proximity (see the text for details). Ball-and-stick models of the side chains indicate the critical residues surrounding the putative binding pocket. The oval represents the P1 domain of the capsid protein. The S domain is not shown. The Rasmol visualization program was used to prepare the figure.
Mutant construction and confirmation of VLP formation.
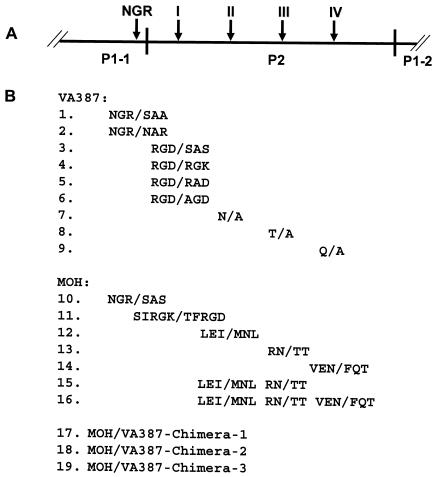
To assess the sequence specificity of NOR capsid protein in binding to HBGAs, a series of mutants with mutations at each of the four sites were constructed (Fig. 3) and the resulting recombinant capsid proteins were characterized for their binding to different HBGAs (see Fig. 5 to 8). To exclude the possible influence of a mutation on VLP formation, all recombinant capsid proteins used for the binding assays were derived from sucrose gradient fractions (30 to 40%) containing VLPs. Electron microscopy also confirmed the results by direct observation of VLPs of selected mutant capsids with mutations representing all four sites of the binding pocket (Fig. 4C). These mutant capsids were morphologically indistinguishable from the wild-type capsids, although degradation of VLPs was observed in some capsids, which could be due to storage or freeze-thawing of the proteins before the EM studies.
FIG. 3.
Schematic representation of mutation constructs of NOR capsids used in this study. (A) Graphic representation of the P domains with emphasis on P2 domain. Arrows indicate the positions of the four sites that are predicted to build up the binding pocket. The conserved NGR motif is also indicated. (B) Mutants with mutations in the NGR motif and the four sites of the VA387 and MOH capsid proteins.
FIG. 5.
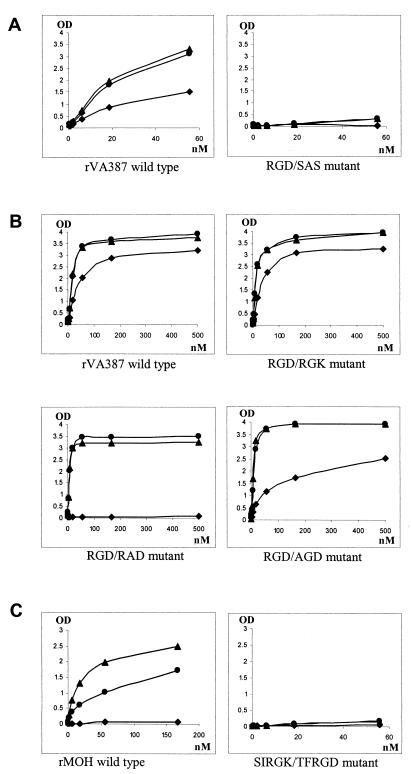
Binding curves of mutants with mutations related to the RGD-like motif. The x axes indicate the concentration of the capsid proteins, and the y axes indicate the ODs obtained from the saliva binding assay. (A) Mutants with amino acid changes of the entire RGD-like motif; (B) mutants with single amino acid change in the RGD-like motif; (C) mutant with longer sequences shifting from MOH to VA387. Data were averaged from at least two independent experiments. •, A antigen; ▴, B antigen; ⧫, H antigen (type O saliva).
FIG. 8.
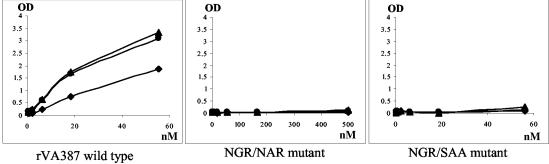
Binding curves of mutants from VA387 (A) and MOH (B) with mutations related to NGR motif. The axes are the same as in Fig. 5. Data were averaged from at least two independent experiments. •, A antigen; ▴, B antigen; ⧫, H antigen (type O saliva).
The RGD-like motif is important for NOR capsid binding to human HBGAs.
The RGD motif has been shown to be responsible for receptor-ligand interaction in other viruses. To determine whether this motif also plays a role in NOR receptor binding, recombinant mutants with mutations in the RGD-like motif were constructed for VA387 and MOH. One VA387 mutant with the entire RGD motif mutated to SAS abolished the binding completely (Fig. 5A). Mutants with only one amino acid change in the RGD motif (e.g., R288 to A or G289 to A) did not affect binding to A or B type saliva (Fig. 5B). However, these mutants had reduced affinities to O-type saliva comparing to the VA387 wild type. Modification of RGD to RAD led to a complete loss of binding to O-type saliva. Recent studies using synthetic oligosaccharides have shown that A, B, and H antigens (terminal carbohydrates) in corresponding saliva are responsible for NOR binding (16) (X. Jiang, P. W. Huang, W. M. Zhong, M. Tan, and T. Farkas, presented at the American Society for Virology Annual Meeting, University of California at Davis, 12 to 16 July 2003). MOH differs from VA387 only in the recognition of the H antigen. VA387 contains an RGD motif while MOH contains an RGK motif at site I. To test whether this difference plays a role in binding specificity, a switch mutant from RGD of VA387 to RGK of MOH was constructed. This mutant did not reveal significant change in binding to A, B, and H antigens (Fig. 5B). Another switch mutant involving a larger sequence alteration in the vicinity of the RGD/K motif, shifting from MOH (SIRGK) to VA387 (TFRGD), did not gain binding to the H antigen either. Instead, it lost binding to the A and B antigens (Fig. 5C). In conclusion, the RGD/K motifs are involved in NORs binding to HBGA receptors. Whether they are also responsible for strain-specific binding remains unclear.
Site III is critical for NOR capsid binding to human HBGAs.
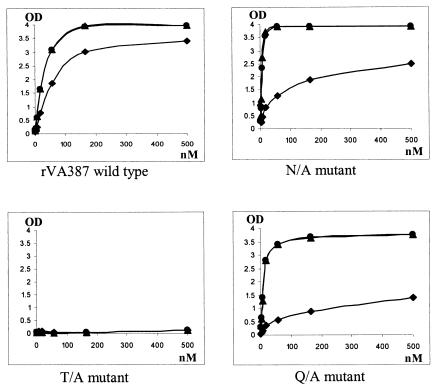
Sites II, III, and IV are located at the opening of the predicted binding pocket, and they are not conserved among strains representing different receptor binding patterns. Thus, they may be responsible for the strain specificity to HBGAs. However, site-directed mutagenesis analysis did not support the idea that site III is responsible for binding specificity. One VA387 mutant with a single amino acid change from T337 to A (Fig. 3) completely lost its binding to the HBGAs of A, B, and O types (Fig. 6). A double amino acid switch mutant at this position from RN of MOH to TT of VA387 not only did not gain binding to the H antigen but also lost binding to A and B antigens (Fig. 7). Two more mutants with mutations at site III plus mutations at sites II and/or IV (Fig. 3) also led to the same results. Taken together, our data indicated that site III plays a critical role in NOR capsid binding to human HBGAs, since even a single amino acid change can result in complete loss of binding.
FIG. 6.
Binding curves of mutants with single amino acid modification at sites II, III, and IV. The axes are the same as in Fig. 5. Data were averaged from at least two independent experiments. •, A antigen; ▴, B antigen; ⧫, H antigen (type O saliva).
FIG. 7.
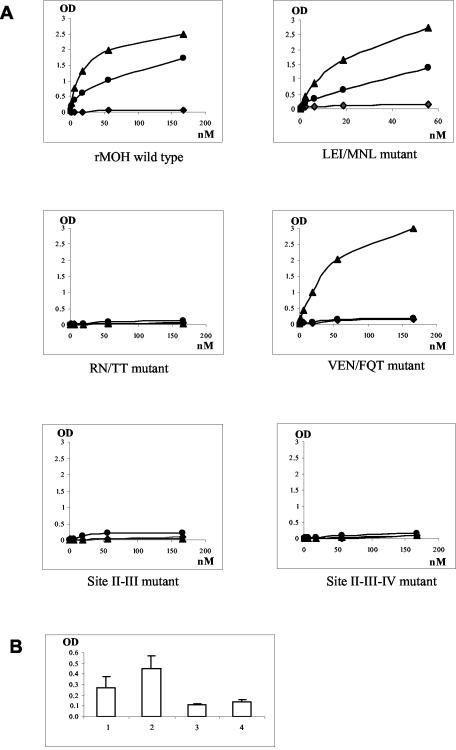
Binding curves of shift mutants with sequence modifications from MOH to VA387 at sites II, III, and IV (A) and comparison of binding to H antigen with that of the wild type (B). (A) The axes are the same as in Fig. 5. Data were averaged from at least two independent experiments. •, A antigen; ▴, B antigen; ⧫, H antigen (type O saliva). (B) 1, LEI/MNL mutant; 2, VEN/FQT mutant; 3, RN/TT mutant; 4, MOH wild-type capsid. The y axis shows the OD of the saliva-binding assay.
Sites II and IV may play a role in binding specificity to HBGAs.
Two mutant capsids with mutations at sites II (N302 to A) and IV (Q375 to A) (Fig. 3) were constructed for VA387. Both mutant capsids did not reveal a significant change in their binding to the A and B antigens but did exhibit decreased binding to the H antigen in O-type saliva, suggesting that sequences at these two sites might play a role in the binding specificity to human HBGAs. In addition, we constructed shift mutants from MOH to VA387 at these sites and found that shift mutants with three amino acid changes at either site (LEI to MNL at site II or VEN to FQT at site IV) resulted in weak binding to type O saliva (Fig. 7A and B). Moreover, the second mutant (VEN/FQT) lost its binding to type A, but not to type B, antigens. In view of the fact that both mutants gained only weak binding to O saliva, we constructed switch mutants with larger sequence changes to see if stronger binding to O saliva could be obtained. Among three chimeras with an 11-aa shift in the vicinity of sites II and/or IV from MOH to VA387 (Fig. 3), none resulted in a gain of binding to type O saliva; instead, all lost binding completely to the types A and B antigens (data not shown). In conclusion, both sites II and IV are important for binding specificity, although additional sites may also be involved. The loss of binding to HBGAs in the case of the three chimera mutants is most likely due to interruption of the required structure of the binding pocket.
The NGR motif is required for receptor binding.
Sequence comparison of NOR capsid proteins revealed a highly conserved NGR motif that was found in all known human and animal enteric NORs. Moreover, the asparagine residue is conserved in all caliciviruses. This motif is located at 20 aa upstream of the RGD-like motif and at the interface between the P1-1 and P2 domains. Therefore, this motif is likely to play an important role in structure and function of NORs. The NGR motif has been found to be involved in the interaction with integrin for rotavirus (9). To test if this motif is also related to NOR receptor binding, the same knockout mutants (NGR/SAA) were constructed for both VA387 and MOH. Both mutants resulted in low yields of the recombinant capsid proteins and VLPs in the insect cells, less than one-quarter of that of other recombinant capsid proteins made in this study (Fig. 4). Saliva binding assay of these mutants showed no detectable binding activity (Fig. 8; data not shown). To further dissect the motif for its role in receptor binding, another mutant (NGR/NAR) with a single amino acid modification was made for strain VA387. Again, a low yield of the protein and no detectable binding to HBGAs were observed. Based on the above observations, we speculated that the NGR motif is important for the maintenance of capsid structure. The loss of the binding to HBGAs in NGR-related mutants is probably due to a local or global conformation change(s) of the capsids, which directly or indirectly affects the conformation of the binding pocket.
DISCUSSION
This study characterized the structure of the NOR capsids and their binding to HBGAs. Using computational analyses, we identified a putative binding pocket in the P2 domain that could mediate binding to different HBGA receptors. Site-directed mutagenesis analyses provided further evidence supporting the binding pocket hypothesis. In particular, the following observations supported our conclusion. First, all four sites in the P2 domain that were predicted to be involved in the formation of the binding pocket by computational analysis were found to influence the ability of NORs to bind to different HBGAs based on site-directed mutagenesis analysis. Second, the two strains of NORs characterized in this study have distinct binding patterns, but the outcomes of binding of their mutant capsids were very similar, suggesting that a common structure is formed for the two capsids despite differences in their primary sequences. Third, two of the four sites, the RGD-like motif and site III, were found to be critical for binding. Changes at either site, particularly the single amino acid change at site III, resulted in complete loss of binding. Finally, switch mutants with sequence shifting between the two strains at sites II and IV, as well as the RGD-like motif, revealed changes of binding patterns at certain levels, suggesting that these sites contribute to a certain extent to binding specificity. Thus, we believe that the computer-predicted binding pocket likely exists. At least four sites are involved in the pocket formation and receptor binding, although an additional site(s) may also be important. The facts that the switch mutants with a 3-aa shift from MOH to VA387 at sites II and IV did not result in significant binding to H antigen and that the switch mutants with mutations involving larger regions (11 and 22 aa) at these two sites resulted in a complete loss of binding suggested that the binding determinants are associated with a very strictly defined structure.
The two strains characterized in this study have multiple determinants for individual HBGAs. Our recent studies indicated that MOH recognizes the A (glucose residue) and B (N-acetyl-glucosamine residue) antigens, while VA387 recognizes the H antigen (1,2-fucosyl residue) in addition to the A and B antigens (16; Jiang et al., Am. Soc. Virol. Annu. Meet.). One question is how variations in the binding pocket contribute to recognition of these substrates. As demonstrated by our results, site III is responsible for binding but not for strain specificity, and single or double amino acid changes at this site resulted in loss of binding to all epitopes (A, B, and/or H) for both strains. The other three sites corresponded to certain levels of strain-specific binding but did not account fully for the binding specificity. Therefore, we hypothesize that the binding specificities of NOR capsids may not be determined by a linear epitope or a single residue; instead, they may involve conformational epitopes and combination of several residues surrounding the pocket.
In our recent NOR receptor studies, we have observed that saliva samples from type A individuals could block VA387 and MOH binding to type B saliva and vice versa, suggesting that both A and B antigens share the same binding site. Once this site is occupied, it is no longer accessible to other molecules. Thus, we assume that each capsid protein has a single pocket. The overall shape and chemical characteristics (e.g., charge distribution) of the pocket is likely to be different for different strains due to different side chains of the residues surrounding the pocket, providing fitness to a specific group of HBGAs.
The identification of the RGD-like motif in the NOR capsids is an important finding, as it led to the prediction of the binding pocket in the P2 domain. The location of the motif at the bottom of the pocket suggested that it interacts directly with HBGAs. NORs are genetically diverse, but the RGD-like motif is conserved among all NORs and the lagoviruses. The first amino acid of the motif of NORs is a basic and positively charged arginine (R) or lysine (K) residue, except for the bovine Newbury strain, which contains a valine (V). In lagoviruses it is either arginine or serine. The second amino acid of the motif is highly conserved throughout the two genera, and the third amino acid is more variable. The recent discovery of NORs in animals raised the questions of zoonotic transmission or animal reservoirs for human NORs. However, direct evidence for interspecies transmission of NORs is still lacking. Recently, a large surveillance of bovine enteric caliciviruses in the United Kingdom between 1976 and 2000 showed that they represent a distinct genogroup III of NORs but did not pose a threat to human health (34). Human blood types are unique among most mammal species, and known NORs mainly infect humans. A recent study showed that the prototype NV does not recognize blood antigens of many nonhuman mammals except chimpanzees (18). Most interestingly, the rabbit calicivirus recognizes the human H type 2 antigen (39). Thus, the RGD-like motif could be a genetic marker for host specificity between animal and human caliciviruses. Two exceptions, however, have been found so far. One is the Jena strain of bovine NORs (27), which contains an off-location RGD motif relative to those in human and rabbit strains (data not shown). The other exception is the swine NOR, which belongs to genogroup II and reveals a RGT motif (44). Whether these exceptional animal strains are truly zoonotic or of human origin and whether they pose a threat to human health remains unclear.
In contrast to the RGD-like motif, the role of the NGR motif may be significantly different. This motif is conserved among all animal and human NORs. Therefore, a common pressure not related to the host receptors must have selected it. The complete knockout of binding by mutations in this region suggests that this motif is indispensable. The location of this motif, near but not in the binding pocket, suggests that this motif is not directly involved in interaction with HBGAs. Thus, the lack of binding to receptors by VLPs containing NGR mutations indicates that the NGR motif may involve local conformation changes that perturb the structure of the binding pocket. The NGR motif is also likely to be required for capsid assembly, since mutations in this motif lead to low yields of the capsid proteins. Alternatively, the low yields are due to a low expression of the proteins. Low expression of mutated capsid proteins not due to conformational stability has been reported for mutations in other regions of the NV capsid (1, 45).
This is the first study to dissect the structure of NOR capsid in relationship with receptor binding. The attachment and entry of a virus to host cells could be the first step of viral infection. A precise map of binding domains and an elucidation of the structure of the interface between the receptor and viral capsid would facilitate understanding the virus-host interaction. This may lead to a discovery or design of specific compounds as antiviral drugs to block the virus infection. A growing number of viral and bacterial pathogens have been linked with HBGAs that serve as receptors for infection (2-5, 8, 12, 15, 25, 28, 37, 41-43). Although different pathogens cause different illness, they may share common mechanisms of interaction with HBGA receptors. NOR capsids are formed by a single capsid protein, making this system simpler than those of many other viral and bacterial pathogens. Our recent studies have described four binding patterns of NORs. The target HBGAs within each pattern have been clearly defined (16) (unpublished data). Thus, NORs could provide a unique model of pathogen-host interaction for the human HBGA system. Elucidation of this model promises to lead to new strategies for therapeutic control of emerging pathogens.
In this study we characterized only four sites within the P2 domain of NOR capsid. Whether additional sites within or adjacent to the predicted pocket are also involved in binding remains unclear. In addition, this study characterized two strains with closely related binding patterns, and both strains belong to the same genogroup of NORs. So far, at least 20 genetic clusters within three genogroups of NORs have been identified (13). Strains within the same genogroup can target different HBGA receptors, and strains in different genogroups can also have the same targets. Therefore, characterization of additional strains representing more genotypes and binding patterns is necessary. Finally, according to the biosynthetic pathways of HBGAs, the target antigens for individual binding patterns have been predicted. Thus, experiments using synthetic oligosaccharides representing these antigenic epitopes should be performed. Due to the unavailability of some of the synthetic oligosaccharides, our study used saliva-binding assays only. Further studies using defined oligosaccharides to confirm our results are necessary.
Acknowledgments
The first three authors contributed equally to this paper.
The research described in this article was supported by the National Institute of Allergy and Infectious Diseases (RO1 AI37093-6).
We thank Irene Hofmann for technical assistance.
REFERENCES
- 1.Bertolotti-Ciarlet, A., L. J. White, R. Chen, B. V. Prasad, and M. K. Estes. 2002. Structural requirements for the assembly of Norwalk virus-like particles. J. Virol. 76:4044-4055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell, C. C., F. Z. Aly, V. S. James, D. M. Weir, A. Collier, A. W. Patrick, C. G. Cumming, D. Wray, and B. F. Clarke. 1989. Blood group, secretor status and oral carriage of yeasts among patients with diabetes mellitus. Diabetes Res. 12:101-104. [PubMed] [Google Scholar]
- 3.Blackwell, C. C., S. J. May, R. P. Brettle, C. J. MacCallum, and D. M. Weir. 1987. Secretor state and immunoglobulin levels among women with recurrent urinary tract infections. J. Clin. Lab. Immunol. 22:133-137. [PubMed] [Google Scholar]
- 4.Boren, T., P. Falk, K. A. Roth, G. Larson, and S. Normark. 1993. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science 262:1892-1895. [DOI] [PubMed] [Google Scholar]
- 5.Boren, T., S. Normark, and P. Falk. 1994. Helicobacter pylori: molecular basis for host recognition and bacterial adherence. Trends Microbiol. 2:221-228. [DOI] [PubMed] [Google Scholar]
- 6.Bower, M. J., F. E. Cohen, and R. L. Dunbrack, Jr. 1997. Prediction of protein side-chain rotamers from a backbone-dependent rotamer library: a new homology modeling tool. J. Mol. Biol. 267:1268-1282. [DOI] [PubMed] [Google Scholar]
- 7.Bresee, J., M. Widdowson, S. Monroe, and R. Glass. 2002. Foodborne viral gastroenteritis: Challenges and opportunities. Clin. Infect. Dis. 35:748-753. [DOI] [PubMed] [Google Scholar]
- 8.Correa, P., and B. A. Schmidt. 1995. The relationship between gastric cancer frequency and the ratio of gastric to duodenal ulcer. Aliment. Pharmacol. Ther. 9:13-19. [PubMed] [Google Scholar]
- 9.Coulson, B. S., S. L. Londrigan, and D. J. Lee. 1997. Rotavirus contains integrin ligand sequences and a disintegrin-like domain that are implicated in virus entry into cells. Proc. Natl. Acad. Sci. USA 94:5389-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fankhauser, R. L., S. S. Monroe, J. S. Noel, C. D. Humphrey, J. S. Bresee, U. D. Parashar, T. Ando, and R. I. Glass. 2002. Epidemiologic and molecular trends of “Norwalk-like viruses” associated with outbreaks of gastroenteritis in the United States. J. Infect. Dis. 186:1-7. [DOI] [PubMed] [Google Scholar]
- 11.Farkas, T., T. Berke, G. Reuter, G. Szucs, D. O. Matson, and X. Jiang. 2002. Molecular detection and sequence analysis of human caliciviruses from acute gastroenteritis outbreaks in Hungary. J. Med. Virol. 67:567-573. [DOI] [PubMed] [Google Scholar]
- 12.Glass, R. I., J. Holmgren, C. E. Haley, M. R. Khan, A. M. Svennerholm, B. J. Stoll, K. M. Belayet Hossain, R. E. Black, M. Yunus, and D. Barua. 1985. Predisposition for cholera of individuals with O blood group, possible evolutionary significance. Am. J. Epidemiol. 121:791-796. [DOI] [PubMed] [Google Scholar]
- 13.Green, J., J. Vinje, C. I. Gallimore, M. Koopmans, A. Hale, and D. W. Brown. 2000. Capsid protein diversity among Norwalk-like viruses. Virus Genes 20:227-236. [DOI] [PubMed] [Google Scholar]
- 14.Harrington, P. R., L. Lindesmith, B. Yount, C. L. Moe, and R. S. Baric. 2002. Binding of Norwalk virus-like particles to ABH histo-blood group antigens is blocked by antisera from infected human volunteers or experimentally vaccinated mice. J. Virol. 76:12335-12343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hooton, T. M. 2000. Pathogenesis of urinary tract infections: an update. J. Antimicrob Chemother. 46(Suppl. 1):1-7, 63-65. [PubMed]
- 16.Huang, P. W., T. Farkas, S. Marionneau, W. M. Zhong, N. Ruvoën-Clouet, L. K. Pickering, A. Morrow, M. Altaye, D. Newburg, J. LePendu, and X. Jiang. 2003. Noroviruses bind to human ABO, Lewis and secretor histo-blood group antigens: Identification of four distinct strain-specific patterns. J. Infect. Dis. 188:19-31. [DOI] [PubMed] [Google Scholar]
- 17.Hutson, A. M., R. L. Atmar, D. Y. Graham, and M. Estes. 2002. Norwalk virus infection and disease is associated with ABO histo-blood group type. J. Infect. Dis. 185:1335-1337. [DOI] [PubMed] [Google Scholar]
- 18.Hutson, A. M., R. L. Atmar, D. M. Marcus, and M. K. Estes. 2003. Norwalk virus-like particle hemagglutination by binding to h histo-blood group antigens. J. Virol. 77:405-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang, X., D. Graham, K. Wang, and M. Estes. 1990. Norwalk virus genome cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 20.Jiang, X., D. O. Matson, G. M. Ruiz-Palacios, J. Hu, J. Treanor, and L. K. Pickering. 1995. Expression, self-assembly, and antigenicity of a Snow Mountain agent-like calicivirus capsid protein. J. Clin. Microbiol. 33:1452-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang, X., M. Wang, D. Y. Graham, and M. K. Estes. 1992. Expression, self-assembly, and antigenicity of the Norwalk virus capsid protein. J. Virol. 66:6527-6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genomic organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 23.Jiang, X., W. M. Zhong, N. Wilton, T. Farkas, E. Barrett, D. Fulton, R. Morrow, and D. M. Matson. 2002. Baculovirus expression and antigenic characterization of the capsid proteins of three Norwalk-like viruses. Arch Virol. 147:119-130. [DOI] [PubMed] [Google Scholar]
- 24.Kelley, L. A., R. M. MacCallum, and M. J. Sternberg. 2000. Enhanced genome annotation using structural profiles in the program 3D-PSSM. J. Mol. Biol. 299:499-520. [DOI] [PubMed] [Google Scholar]
- 25.Klaamas, K., O. Kurtenkov, M. Ellamaa, and T. Wadstrom. 1997. The Helicobacter pylori seroprevalence in blood donors related to Lewis (a, b) histo-blood group phenotype. Eur. J. Gastroenterol Hepatol. 9:367-370. [DOI] [PubMed] [Google Scholar]
- 26.Lindesmith, L., S. Moe, S. Marionneau, N. Ruvoen, X. Jiang, L. Lindblad, P. Stewart, J. LePendu, and R. Baric. 2003. Human susceptibility and resistance to Norwalk virus infection. Nat. Med. 9:548-553. [DOI] [PubMed]
- 27.Liu, B. L., P. R. Lambden, H. Gunther, P. Otto, M. Elschner, and I. N. Clarke. 1999. Molecular characterization of a bovine enteric calicivirus: relationship to the Norwalk-like viruses. J. Virol. 73:819-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomberg, H., U. Jodal, H. Leffler, P. De Man, and C. Svanborg. 1992. Blood group non-secretors have an increased inflammatory response to urinary tract infection. Scand. J. Infect. Dis. 24:77-83. [DOI] [PubMed] [Google Scholar]
- 29.Marionneau, S., A. Cailleau-Thomas, J. Rocher, B. Le Moullac-Vaidye, N. Ruvoen, M. Clement, and J. Le Pendu. 2001. ABH and Lewis histo-blood group antigens, a model for the meaning of oligosaccharide diversity in the face of a changing world. Biochimie 83:565-573. [DOI] [PubMed] [Google Scholar]
- 30.Marionneau, S., N. Ruvoen, B. Le Moullac-Vaidye, M. Clement, A. Cailleau-Thomas, G. Ruiz-Palacios, P. W. Huang, X. Jiang, and J. Le Pendu. 2002. Norwalk virus binds to H type S 1/3 histo-blood group antigens present on gastro-duodenal epithelial cells of “secretor” individuals. Gastroenterology 122:1967-1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mason, P. W., E. Rieder, and B. Baxt. 1994. RGD sequence of foot-and-mouth disease virus is essential for infecting cells via the natural receptor but can be bypassed by an antibody-dependent enhancement pathway. Proc. Natl. Acad. Sci. USA 91:1932-1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meller, J., and R. Elber. 1998. Computer simulations of carbon monoxide photodissociation in myoglobin: structural interpretation of the B states. Biophys. J. 74:789-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meller, J., and R. Elber. 2001. Linear programming optimization and a double statistical filter for protein threading protocols. Proteins 45:241-261. [DOI] [PubMed] [Google Scholar]
- 34.Oliver, S. T., A. M. Dastjerdi, S. Wong, L. El-Attar, C. Gallimore, D. W. Brown, J. Green, and J. C. Bridger. 2003. Molecular characterization of bovine enteric caliciviruses: a distinct third genogroup of noroviruses (Norwalk-like viruses) unlikely to be of risk of humans. J. Virol. 77:2789-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prasad, B. V., M. E. Hardy, T. Dokland, J. Bella, M. G. Rossmann, and M. K. Estes. 1999. X-ray crystallographic structure of the Norwalk virus capsid. Science 286:287-290. [DOI] [PubMed] [Google Scholar]
- 36.Prasad, B. V., R. Rothnagel, X. Jiang, and M. K. Estes. 1994. Three-dimensional structure of baculovirus-expressed Norwalk virus capsids. J. Virol. 68:5117-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raza, M. W., C. C. Blackwell, P. Molyneaux, V. S. James, M. M. Ogilvie, J. M. Inglis, and D. M. Weir. 1991. Association between secretor status and respiratory viral illness. BMJ 303:815-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruoslahti, E., and M. D. Pierschbacher. 1986. Arg-Gly-Asp: a versatile cell recognition signal. Cell. 44:517-518. [DOI] [PubMed] [Google Scholar]
- 39.Ruvoen-Clouet, N., J. P. Ganiere, G. Andre-Fontaine, D. Blanchard, and J. Le Pendu. 2000. Binding of rabbit hemorrhagic disease virus to antigens of the ABH histo-blood group family. J. Virol. 74:11950-11954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sali, A., and J. P. Overington. 1994. Derivation of rules for comparative protein modeling from a database of protein structure alignments. Protein Sci. 3:1582-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sidebotham, R. L., J. H. Baron, J. Schrager, J. Spencer, J. R. Clamp, and L. Hough. 1995. Influence of blood group and secretor status on carbohydrate structures in human gastric mucins: implications for peptic ulcer. Clin Sci (Colch). 89:405-415. [DOI] [PubMed]
- 42.Stapleton, A., E. Nudelman, H. Clausen, S. Hakomori, and W. E. Stamm. 1992. Binding of uropathogenic Escherichia coli R45 to glycolipids extracted from vaginal epithelial cells is dependent on histo-blood group secretor status. J. Clin. Invest. 90:965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stroud, M. R., A. E. Stapleton, and S. B. Levery. 1998. The P histo-blood group-related glycosphingolipid sialosyl galactosyl globoside as a preferred binding receptor for uropathogenic Escherichia coli: isolation and structural characterization from human kidney. Biochemistry 37:17420-17428. [DOI] [PubMed] [Google Scholar]
- 44.Sugieda, M., and S. Nakajima. 2002. Viruses detected in the caecum contents of healthy pigs representing a new genetic cluster in genogroup II of the genus “Norwalk-like viruses”. Virus Res. 87:165-172. [DOI] [PubMed] [Google Scholar]
- 45.White, L. J., M. E. Hardy, and M. K. Estes. 1997. Biochemical characterization of a smaller form of recombinant Norwalk virus capsids assembled in insect cells. J. Virol. 71:8066-8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wickham, T. J., M. E. Carrion, and I. Kovesdi. 1995. Targeting of adenovirus penton base to new receptors through replacement of its RGD motif with other receptor-specific peptide motifs. Gene Ther. 2:750-756. [PubMed] [Google Scholar]