Abstract
During herpes simplex virus type 1 (HSV-1) latent infection in human peripheral sensory ganglia, the major viral gene transcribed is the latency-associated transcript (LAT) gene. In order to facilitate the study of this gene, we generated a transgenic mouse that contains the DNA fragment that transcribes the LAT RNAs (2.0 kb and its 1.5-kb spliced transcript) under control of the cytomegalovirus promoter. The tissue distribution of these transcripts and their effects upon HSV-1 replication, latency, and reactivation in the transgenic-mouse model were examined. Different steady-state amounts of both transcripts were found in various tissues. While the highest levels of the 2.0-kb RNA were detected in heart and skeletal muscle, the 1.5-kb transcript was found at elevated levels in the brain and at much higher levels in the trigeminal ganglia (TG). Replication of both the wild-type and a LAT-negative mutant virus was suppressed in primary embryonic fibroblasts obtained from LAT-expressing transgenic mice compared to that in cells obtained from normal mice. HSV-1 DNA amounts in latently infected TG of transgenic mice were similar to those in normal mice. Reactivation of latent HSV-1 LAT-negative mutants by explant cocultivation of TG from transgenic mice was more efficient than reactivation from normal-mouse TG. Considering our present and previous results, we propose that the significantly higher steady-state level of the 1.5-kb RNA in the TG may link this transcript to latency functions and that by inhibition of virus replication, the LAT gene may protect ganglion cells and thereby increase the probability of reactivation.
Herpes simplex virus type 1 (HSV-1) establishes lifelong latent infection in human peripheral sensory ganglia (for reviews, see references 26 and 40), usually in the absence of viral replication (38). From a quiescent, nonreplicating state, it can reactivate to produce recurrent mucocutaneous disease. HSV-1 latency is present in the majority of the adult population (50) and is not accompanied by damage to peripheral sensory ganglion cells. During latency, a limited part of the viral genome, the LAT (latency-associated transcript) gene, is transcriptionally active, and two colinear LATs, 2.0 and 1.5 kb in size, accumulate in latently infected nervous tissues (34, 36, 41). The 2.0-kb LAT is a stable intron derived from a less abundant 8.3-kb RNA (28, 53) and is further spliced to produce the 1.5-kb LAT. While the 2.0-kb transcript is observed during productive and latent infections, the 1.5-kb RNA appears to be specific for neuronal tissue harboring the latent viral genome (13, 42). Both transcripts contain several large open reading frames, have been detected in the nucleus and cytoplasm (1, 12, 13, 29), and bind to polyribosomes (12). Although no protein products have yet been identified, there is an indication of biological function for the largest LAT open reading frame (41).
Viruses incapable of expressing the LATs have a defective reactivation phenotype (15, 37, 45, 47). Whether this gene exerts its function during reactivation or the defective reactivation phenotype is due to impaired ability of LAT-negative viruses to establish latent infection is still unsettled. However, improved reactivation efficiency was correlated with HSV-1 ability to establish latent infection in a higher percentage of neurons (45, 46). Neuronal cell lines that express the LAT gene were previously generated, and it was demonstrated that HSV-1 replication in these cells is partially blocked and that the viral immediate-early (IE) genes are repressed in the presence of the LATs. It was therefore suggested that the LAT gene might have a role in facilitating the establishment of HSV-1 latency (23).
Several questions associated with HSV-1 latent infection in vivo await elucidation. Is splicing of the 1.5-kb RNA specific to the nervous system? What cellular or tissue functions, mandatory for the latent state, are mediated by the LAT gene? Can LAT suppress HSV-1 replication and improve establishment of latency, and by what mechanism? Does this gene encode proteins?
To address these questions, we have generated a transgenic mouse in which the HSV-1 DNA fragment that transcribes the 2.0- and 1.5-kb LATs is controlled by the cytomegalovirus (CMV) IE promoter. A recent study examined the effect of the HSV-2 LAT transgene upon HSV-2 infection and latency in transgenic mice and did not identify any phenotypic difference from the parental mouse strain (49).
The present work reports differential processing of both the 2.0- and 1.5-kb LATs in various transgenic-mouse tissues. It indicates that splicing of the 2.0-kb LAT to generate the 1.5-kb transcript takes place in every tissue but its steady-state level is significantly higher in neuronal tissues. We also studied the effects of the LAT transgene upon the replication of the HSV-1 wild type, as well as an HSV-1 LAT-negative mutant, in primary fibroblasts obtained from the transgenic mice and on the efficiency of explant reactivation from the trigeminal ganglia (TG). Replication of both viruses was suppressed in the LAT-expressing cells; the latent-HSV-1 copy numbers were similar in LAT-expressing and normal TG, while reactivation from transgenic-mouse TG was more efficient than from normal-mouse TG.
MATERIALS AND METHODS
Viruses.
HSV-1 strain F was obtained from B. Roizman, University of Chicago, Chicago, Ill. HSV-1 FS1001K, a KOS strain-derived LAT-negative mutant (9), was kindly provided by N. Fraser, The Wistar Institute. The viruses were propagated and titrated on CV-1 cells as described before (38), using 0.5 mg of human gamma globulins/ml.
Generation of LAT transgenic mice.
The pLAT-expressing vector (23), which contains the LAT coding sequences (from the PvuI site at position 118802 to the MluI site at position 121651) under the control of the CMV IE promoter, was cleaved with AflIII and BamHI (Fig. 1A, II) to separate the LAT transgene from vector sequences. The appropriate transgene DNA fragment was electrophoresed on a 1% agarose gel, purified on an ion-exchange column (Elutip-d column; Schleicher and Schuell), ethanol precipitated, and dissolved in the injection buffer (7.5 mM Tris, 0.2 mM EDTA, pH 7.5) to a final concentration of 15 ng/ml. The LAT transgene was injected into pronuclei of fertilized ova derived from (C57Black × BALB/c)F1 mice according to standard procedures (16). After injection, ova were transferred at the two-cell stage to the oviducts of surrogate pseudopregnant F1 females. Transgenic mice generated by this procedure were identified by slot blot hybridization of high-molecular-weight DNA extracted from tail biopsy specimens.
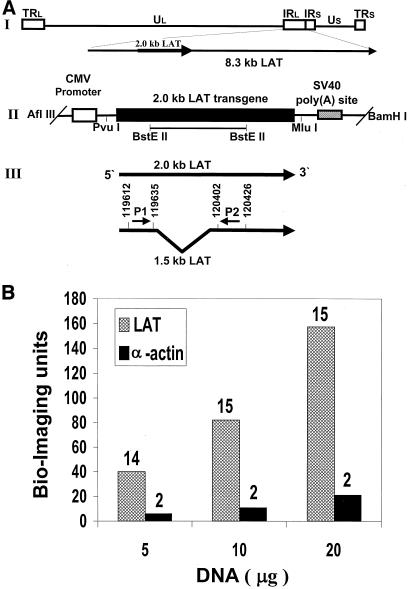
FIG. 1.
Structure of the HSV-1 LAT transgene and analysis of its copy number in transgenic mice. (A) Structure of the HSV-1 LAT transgene. (I) Schematic representation of HSV-1 genome and LAT transcripts. The unique long (UL) and short (US) regions are represented as lines, and the internal and terminal long and short repeats (IRL, TRL, IRS, and TRS) are represented as open boxes. The locations of the LAT gene and its 8.3- and 2.0-kb LATs are indicated. (II) DNA fragment inserted into mice. The 2.0-kb LAT gene, the locations of the CMV promoter and simian virus 40 (SV40) poly(A) site, the relevant restriction sites, and the location of the BstEII-cleaved LAT gene probe are indicated. (III) Locations of the primers used for RT-PCR and for quantitative RT-PCR. The arrows indicate the two colinear 2.0- and 1.5-kb LATs and their orientations. (B) LAT copy number analysis. DNAs (5, 10, and 20 μg), extracted from tail biopsy specimens from transgenic mice, were slot blotted to two different filters: one was hybridized with the LAT DNA probe (BstEII cleaved; 977 bp [panel A, II]), while the other filter was hybridized with a mouse α-actin cDNA fragment (PstI cleaved; 1,100 bp) (27) as a diploid gene control (LAT and α-actin, respectively). As a negative control, the same amounts of DNA derived from nontransgenic mice were blotted to the filters. The specific activities of the two probes were similar (3 × 109 ± 0.3 × 109 cpm/μg of DNA). Computerized image analysis and quantification of radioactive signals were performed with a Bio-Imaging analyzer (45). The LAT transgene copy numbers were estimated relative to the α-actin radioactive signal and are indicated above the columns.
Replication of HSV-1 in primary fibroblast cultures.
Mouse embryonic fibroblast (MEF) cultures were prepared from embryos collected on day E16 of gestation. The embryos were surgically removed, separated from maternal tissues and yolk sacs in phosphate-buffered saline, and then decapitated and degutted, and the bodies were incubated in 0.08% trypsin-0.007% EDTA solution with shaking at 37°C for 30 min. The suspension was allowed to settle, the supernatant was collected, and 1% fetal calf serum (FCS) was added. The pellet was then incubated with trypsin for an additional 30 min, the supernatant was collected and centrifuged for 7 min at 1,000 × g, and the cells were resuspended in Dulbecco's modified Eagle's medium with 10% FCS. After 18 h, the medium was replaced (3).
Primary fibroblast cells from the second passage were seeded (0.5 × 105 per plate). A day later, the cultures were infected with HSV-1 strain F and with the KOS strain-derived LAT-negative FS1001K mutant. Cells and media were harvested 24 h postinfection and subjected to three cycles of freeze and thaw, and virus titers were determined on CV-1 cells as described before (23). All experiments were done with triplicate plates. As control cultures, MEFs were derived from LAT-negative siblings obtained from the same litter.
Cell viability assay (MTT).
The cell viability assay is based on the ability of cellular mitochondria to convert MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide [Sigma]) into a blue formazan product and was performed according to the procedure of Miller and McDevitt (25). Absorbance (492 nm) was measured in an enzyme-linked immunosorbent assay plate reader (Organon Teknika) with a reference wavelength of 630 nm.
Inoculation of mice.
Eight-week-old transgenic and normal mice were anesthetized by intraperitoneal injection of ketamine (85 mg/kg) and xylazine (3 mg/kg). Both corneas were scarified, and a total inoculum of 2 × 105 PFU of the FS1001K LAT-negative mutant was applied to the corneas as described previously (37). The entire study with mice was approved by the Institutional Committee for Ethical Control of Experimentation with Animals.
HSV-1 reactivation from TG by explant cocultivation.
Following primary infection, the mice were maintained for at least 30 days. TG were explanted into 24-well plates, each well containing 4 × 104 CV-1 cells in 0.5 ml of Dulbecco's modified Eagle's medium including 10% FCS, and transferred to a new well every 3 days. To monitor reactivation, 0.1 ml of medium was transferred every day to another well containing cells as described above in 0.4 ml of fresh medium. The wells were examined daily for evidence of infectious viral particles by observing the cytopathic effect in the cell monolayer. For quantification of reactivated virus, another 0.1 ml of medium was removed and used for titration as described before (38).
DNA slot blot.
High-molecular-weight DNA was extracted from mouse organ samples, using the GenElute Mammalian Genomic DNA Miniprep kit (Sigma). Genomic DNA (10 μg) was then manipulated as described before (23).
Preparation of radioactively labeled DNA probes.
The DNA probe for the LAT gene was cleaved from the pLAT plasmid using the BstEII restriction enzyme (positions 120091 to 121068) (Fig. 1A, II). The probe was labeled as described before (23). The specific activity of the probes was ∼108 cpm/mg of DNA.
RNA purification and Northern blot analysis.
Individual tissues were dissected from transgenic mice and homogenized, and total RNA was obtained using the Tri reagent (Molecular Research Center, Inc.) according to the manufacturer's instructions. Northern blot analysis was performed according to the method of Spivack and Fraser (34).
RT-PCR.
Total RNA (0.15 μg), extracted from the various tissues, was subjected to reverse transcription (RT)-PCR using the Access RT-PCR system (Promega) according to the manufacturer's instructions. The following oligonucleotide primers that flank the intron within the 2.0-kb LAT (Fig. 1A, III) were prepared according to the published sequence of HSV-1 (31): P1, 5′-GACTCTGTTACTTACCCGTCCGAC-3′ (HSV-1 bases 119612 to 119635), and P2, 5′-GAAAGCATCCTGCCACTGGCATGGA-3′ (bases 120402 to 120426). RT was performed with primer P2.
Quantitative RT-PCR.
Quantitative RT-PCR was performed in two steps. First, 1 μg of total RNA from different tissues was subjected to RT for 45 min at 48°C using the P2 primer (Fig. 1A, III) and avian myeloblastosis virus reverse transcriptase (Promega). When the reaction was complete, the enzyme was inactivated for 2 min at 94°C. This step was followed by quantitative PCR in a LightCycler (Roche Molecular Biochemicals) in a total volume of 20 μl consisting of 3.5 μl of the RT product, 2 μl of LightCycler FastStart DNA Master SYBR Green I (Roche Molecular Biochemicals), P1 and P2 primers (1 μM each), and 4 mM MgCl2. The amplification conditions consisted of a hot-start preincubation step of 10 min at 95°C, followed by 45 cycles of denaturation for 15 min at 95°C, annealing (a touchdown from 65 to 60°C with a step size of 0.5°C per cycle), and elongation for 12 s at 72°C. Melting peaks were used to determine the specificity of the PCR.
DNA purification.
Following primary infection, mice were maintained for at least 30 days. High-molecular-weight DNA was extracted from pairs of ganglia, using the GenElute Mammalian Genomic DNA Miniprep kit; 80 ng of DNA was subjected to quantitative real-time PCR.
Quantitative PCR.
The viral genome copy number in latently infected TG was determined by quantitative DNA PCR, using an Applied Biosystems (Foster City, Calif.) PCR system. Reactions were performed in 50-μl volumes containing 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 80 ng of ganglion DNA, 500 nM (each) forward and reverse primers, and 100 nM probe. The primers and probe were based on the HSV-1 gB sequence: forward primer, 5′-CCGTCAGCACCTTCATCGA-3′; reverse primer, 5′-CGCTGGACCTCCGTGTAGTC-3′; and probe, 5′-FAM-CCACGAGATCAAGGACAGCGGCC-TAMRA-3′ (Applied Biosystems) (19).
Real-time PCR amplification and detection were performed using a PRISM 7900HT sequence detector (Applied Biosystems) with optimized cycle conditions: 50°C for 2 min, 95°C for 10 min, 50 cycles at 95°C for 15 s, and 60°C for 1 min. The relative copy number was calculated using a standard curve generated from plasmid pgB that was serially 10-fold diluted to contain from 104 to 1011 copies of the desired gene in 5 μl and subjected to TaqMan PCR with the same set of primers. By comparing the threshold cycle of each sample to the threshold cycle of the standards, the copy number for each reaction was estimated. The results were normalized for genomic DNA using the commercially available TaqMan 18S rRNA Control Reagents kit (Applied Biosystems) and a serial fivefold dilution of mouse genomic DNA. All samples were run in triplicate. TG DNA samples from 11 ganglion pairs per group were assessed.
RESULTS
Generation and characterization of a LAT gene transgenic mouse line.
All viable progeny arising from (C57Black × BALB/c)F1 mouse oocytes microinjected with the LAT transgene (Fig. 1A, II) were screened by DNA hybridization. Three founders containing the LAT transgene were identified and designated MLAT1, MLAT2, and MLAT3. The founders gave rise to offspring in crosses with parental nontransgenic mice and transmitted the transgene in a Mendelian fashion (∼50% hemizygotes). The male and female transgenic mice of all founders did not differ from the nontransgenic parental mice in development and growth and had similar weights at 1, 2, and 3 months of age (data not shown). By extracting DNA and RNA from tails, the three founder mouse lines were screened for the LAT gene copy number (by DNA hybridization) and LAT RNA expression (by Northern blot analysis), using the BstEII-cleaved LAT DNA probe (Fig. 1A, II). The transgene copy number per cell was analyzed by quantitative hybridization of tail-extracted DNA (Fig. 1B). Filters blotted with increasing amounts of DNA were hybridized with the LAT DNA probe or with a mouse α-actin cDNA fragment (PstI cleaved; 1,100 bp) (27) as a diploid gene control. The LAT transgene copy number was estimated relative to the α-actin radioactive signal (diploid) using a Bio-Imaging analyzer (48). We found that MLAT1 had a relatively high LAT gene copy number (13 or 14) (Fig. 1B) and a high LAT expression level (Fig. 2A), while MLAT2 and MLAT3 had low LAT gene copy numbers (∼2 or 3 copies) and a low LAT expression level (MLAT2) or no LAT expression (MLAT3) (data not shown). In all further experiments, we characterized the MLAT1 founder line and used its heterozygous offspring.
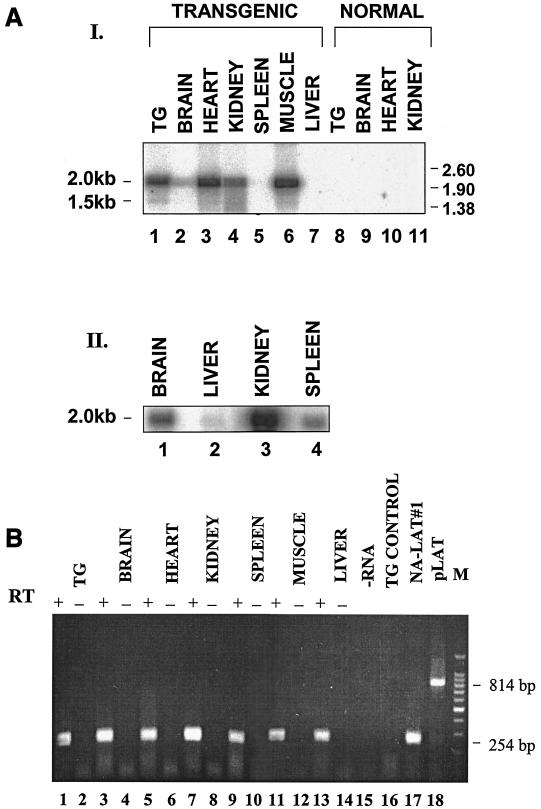
FIG. 2.
Analysis of HSV-1 2.0- and 1.5-kb LATs in transgenic-mouse tissues. (A) Northern blot analysis. Total RNAs were obtained from mouse tissues. The positions of RNA markers (in kilobases) are indicated on the right. The locations of the 1.5- and 2.0-kb LATs are indicated on the left. The DNA probe used was the BstEII-BstEII DNA fragment (Fig. 1A, II). (I) RNA (0.5 μg) was loaded in each lane. Lanes 1 to 7, transgenic-mouse tissues; lanes 8 to 11, normal-mouse tissues. To verify that equal amounts of RNA were loaded, the same blotted membrane was rehybridized with a 5′-end-labeled single-stranded DNA probe specific for the 18S rRNA, as described before (20) (data not shown). (II) RNA(10 μg) was loaded in each lane. (B) RT-PCR analysis. Shown are RT-PCR products from total RNAs extracted from various tissues (lanes 1, 3, 5, 7, 9, 11, and 13). A 254-bp RT-PCR DNA product represents the 1.5-kb spliced RNA, while a larger, 814-bp product represents the unspliced 2.0-kb transcript (18). For each sample, a control PCR was performed with no RT enzyme added (−) (lanes 2, 4, 6, 8, 10, 12, and 14). Lane 15, control RT-PCR with sample without RNA (−RNA). Lane 16, RT-PCR of RNA extracted from TG of normal mouse. Lane 17, RT-PCR of RNA extracted from LAT-expressing neuronal cells (20). Lane 18, PCR of the pLAT plasmid containing the LAT gene. Lane M, 100-bp DNA ladder (Promega). The PCR products were analyzed on a 1.8% agarose gel stained with ethidium bromide.
A complete autopsy was performed on two MLAT1 transgenic mice and one normal parental mouse. Samples of the lungs, heart, liver, spleen, kidneys, pancreas, adrenal glands, gastrointestinal tract, internal sex organs, skeletal muscle, TG, and cerebral cortex were histologically examined. The tissues were fixed in 4% formalin and embedded in paraffin, and 5-μm-thick sections were obtained. The slides were stained with hematoxylin and eosin for histopathological examination. No abnormality was detected, except for a single focus of liver cell necrosis observed in one of the transgenic mice.
Analysis of the 2.0- and 1.5-kb LATs in transgenic-mouse tissues.
The presence of the LAT RNA in various tissues was determined using Northern blot analysis (Fig. 2A) and RT-PCR (Fig. 2B). Total RNA was extracted from TG, brain, heart, kidney, spleen, skeletal muscle, and liver; resolved by gel electrophoresis; Northern blotted; and hybridized with the LAT probe (Fig. 1A, II). The 2.0-kb LAT was readily identified in all tissues examined, but in different amounts (Fig. 2A, I and II). Using Northern blot analysis, previous studies detected the 1.5-kb LAT only in the TG of HSV-1-infected mice and human individuals (31, 33, 48). By RT-PCR, it was detected in other neuronal (brainstem) (13) and quasineuronal (adrenal gland) tissues (42). Thus, the transgenic-mouse model provides a unique opportunity to study the steady-state level of the 1.5-kb LAT in nonneuronal tissues as well. Our Northern blot analysis demonstrated that the 1.5-kb RNA can be clearly detected only in TG tissue of the transgenic mouse (Fig. 2A, I, lane 1).
To further examine the presence of the 1.5-kb LAT spliced product in the transgenic-mouse tissues, we applied the method of RT-PCR using primers flanking the 2.0-kb LAT intron sequence (Fig. 1A, III). The 254-bp RT-PCR product, representative of the 1.5-kb LAT, was identified in all tissues examined (Fig. 2B). Thus, splicing of the 2.0-kb transcript to produce the 1.5-kb LAT is not a neuronal-tissue-specific event but takes place in other tissues as well.
It should be noted that the RT-PCR did not amplify the larger, 814-bp DNA product, representative of the 2.0-kb LAT, since the Taq polymerase tends to selectively amplify the smaller fragments of overlapping DNA templates bearing different sizes (22, 35).
Quantitative analysis of the 2.0- and 1.5-kb LAT RNAs in various tissues.
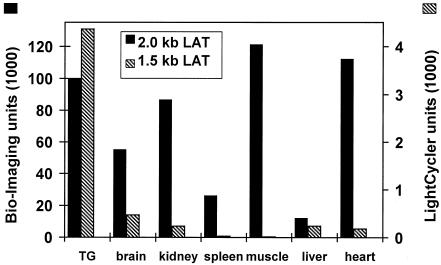
To determine the 2.0-kb LAT RNA levels in various tissues, a quantitative Northern blot analysis of radioactively labeled RNA was performed (Fig. 2A), using the Bio-Imaging analyzer. We revealed large amounts of the 2.0-kb LAT in TG, kidney, skeletal muscle, and heart compared to brain, spleen, and liver (Fig. 3).
FIG. 3.
Quantitative analysis of HSV-1 2.0- and 1.5-kb LATs in transgenic-mouse tissues. The relative amounts of the 2.0-kb LAT in the various tissues were determined by measurement of the radioactive signal from Northern-blotted RNA hybridized with a 32P-labeled probe, using the Bio-Imaging analyzer. The same blotted membrane was rehybridized with a 5′-end-labeled single-stranded DNA probe specific for the 18S rRNA, and the 2.0-kb LAT amounts were estimated relative to the 18S rRNA signals. The relative amounts of the 1.5-kb LAT were analyzed by real-time quantitative RT-PCR.
The 1.5-kb RNA levels were further analyzed by real-time quantitative PCR with the LightCycler system (Roche Molecular Biochemicals). We found 20-fold-larger amounts of the 1.5-kb LAT in the TG than in the kidney and heart and a 300-fold-higher level than in skeletal muscle of transgenic animals (Fig. 3). However, these four tissues expressed similar amounts of the 2.0-kb RNA (Fig. 3). Relatively larger amounts of the 1.5-kb RNA were also identified in the brain, another neuronal tissue that may harbor the latent viral genome (10, 39), than in the other, nonneuronal tissues, such as kidney, skeletal muscle, and heart, that also expressed higher levels of the 2.0-kb LAT (Fig. 3). Thus, (i) splicing of the 1.5-kb LAT takes place in every tissue, but its steady-state levels are higher in neuronal tissues, and (ii) no correlation was observed between 1.5- and 2.0-kb LAT levels. For example, levels of the 2.0-kb LAT in the liver were low compared to those in the kidney and heart, yet amounts of the 1.5-kb transcript were similar.
This may suggest that the limiting factor in the splicing process is tissue specific rather than the amount of the 2.0-kb precursor RNA.
HSV-1 replication in primary fibroblasts of transgenic mice.
It was previously shown that HSV-1 replication is suppressed in neuronal cell lines that express the LAT gene (23). Primary cell cultures have an advantage, since they do not express the antiapoptotic machinery characteristic of transformed cell lines (20). Therefore, we have cultured MEFs from transgenic and normal mice in order to compare viral replication.
MEFs from heterozygous transgenic embryos, as well as from the LAT-negative normal siblings obtained from the same litter, were prepared. Fibroblast cultures were infected with HSV-1 (strain F), and 24 h later, the plates were harvested and titrated on CV-1 cell monolayers. In parallel, cultures were subjected to cell quantification by the MTT viability assay. This analysis enabled us to accurately infect the different cell cultures at identical multiplicities of infection (MOI) by adjusting them to the number of viable cells.
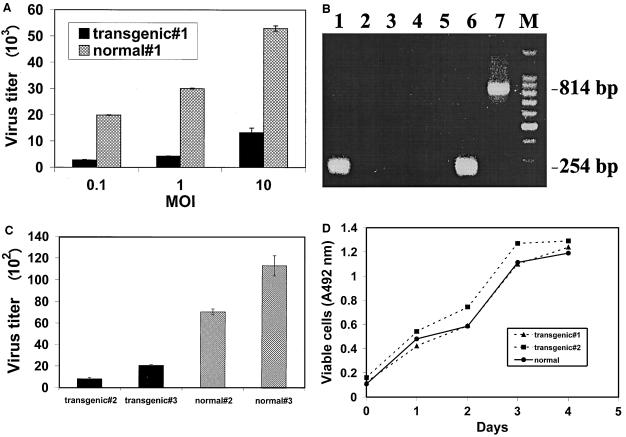
At MOI of 0.1 and 1, replication of the virus was suppressed sevenfold in the transgenic-mouse cells compared to normal cells, and at an MOI of 10, the LATs had a fourfold suppressive effect upon HSV-1 replication (Fig. 4A). The expression of LATs in these cells was confirmed by RT-PCR analysis (Fig. 4B).
FIG. 4.
LAT gene effects on HSV-1 replication in primary fibroblasts. (A) Wild-type HSV-1 replication in primary fibroblasts obtained from transgenic and normal embryos. The cells were infected for 24 h with HSV-1 (strain F) at MOI of 0.1, 1, and 10. The cultures were harvested and titrated on CV-1 cells. Presented are the mean values of experiments performed in triplicate ± standard errors of the mean (SEM). (B) LAT expression in primary fibroblasts obtained from transgenic and normal embryos. Shown are RT-PCR products from total RNAs extracted from transgenic (lane 1) and normal (lane 3) embryos. For each sample, a control PCR was performed with no RT enzyme added (lanes 2 and 4). Lane 5, control RT-PCR with sample without RNA. Lane 6, RT-PCR of RNA extracted from LAT-expressing neuronal cells (18). A 254-bp RT-PCR DNA product represents the 1.5-kb spliced RNA, while a larger, 814-bp product represents the unspliced 2.0-kb transcript (20). Lane 7, PCR of the pLAT plasmid containing the LAT gene. Lane M, 100-bp DNA ladder (Promega). The PCR products were analyzed on a 1.8% agarose gel stained with ethidium bromide. (C) HSV-1 LAT-negative mutant replication in primary fibroblasts obtained from transgenic and normal embryos. The cells were infected for 24 h with an HSV-1 LAT-negative mutant (FS1001K) at an MOI of 0.001. Triplicate cultures were harvested, pooled, and titrated in triplicate on CV-1 cells. Presented are the mean values of experiments ± SEM. (D) Proliferation rate of transgenic and normal primary fibroblasts. Cells were seeded (5 × 104 per plate) and subjected daily to the MTT cell viability assay.
To further examine whether the LAT gene may function in trans to suppress HSV-1 replication, we used a mutant of another strain (HSV-1 FS1001K; KOS strain derived), in which 1.1 kb was deleted from the 5′ end of the 2.0-kb LAT coding sequence to abolish all LAT expression (9). MEFs from two other hetrozygote transgene-positive embryos, as well as from two other LAT− normal embryos derived from the same litter, were cultured, infected, and harvested after 24 h for titration. The results indicated that primary LAT-expressing cells suppressed virus replication at a range of 3.4 to 14 compared to viral replication in normal MEFs (Fig. 4C). A one-way analysis of variance revealed a statistically significant difference between the transgenic and the normal embryos in all experiments (P < 0.01). Thus, LAT may suppress viral replication in trans even out of the context of the HSV-1 genome.
The suppressive effect of the LAT gene upon HSV-1 replication was not due to its effect on cell viability and proliferation, since growth curves of MEFs from transgene-positive embryos were identical to that of a normal embryo (Fig. 4D).
HSV-1 reactivation from transgenic-mouse TG.
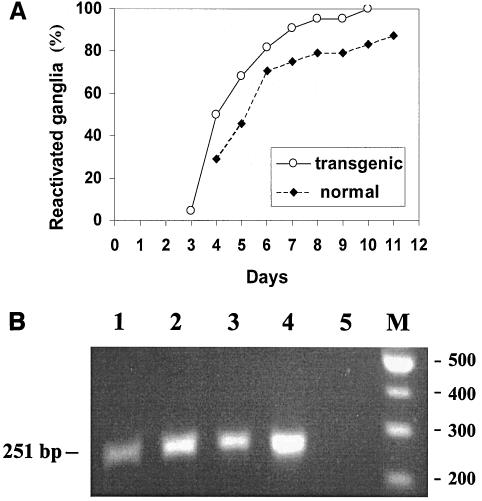
Since it was shown that HSV-1 reactivation ability in vivo correlated with expression of the LAT gene RNAs (45), we went on to examine the ability and efficiency of an HSV-1 LAT-negative mutant to reactivate from TG containing the LAT transgene. Transgenic and normal mice were infected as described above and maintained for at least 30 days postinfection. TG removed from the two groups of mice (22 transgenic and 24 normal TG) were explanted and cocultivated over a cell monolayer, and the cultures were examined daily for cytopathic effect. The results show that viruses reactivated earlier from TG explanted from the transgenic mice than from those from normal mice (Fig. 5A) and that the difference was statistically significant (P < 0.04 by hypothesis testing for incidence rate (one-side test)). At each time point postexplantation, more ganglia of the transgenic group than of the normal group yielded reactivated virus. After 10 days in culture, 100% of the ganglia from the transgenic group underwent reactivation compared to only 80% of normal ganglia. PCR analysis confirmed the presence of HSV-1 DNA in the three TG derived from normal animals that did not yield reactivated virus after 35 days (Fig. 5B).
FIG. 5.
Explant reactivation of latent HSV-1 from TG. (A) Reactivation of HSV-1 LAT-negative mutant from transgenic (n = 22) and normal (n = 24) TG. The monolayers were examined daily for cytopathic effects over a 35-day period. The results are presented as cumulative percentages of all ganglia that underwent reactivation. (B) Gel electrophoresis analysis of PCR products of DNA from nonreactivated TG. DNA was extracted from the three normal, LAT-negative nonreactivated ganglia 35 days postexplantation and subjected to PCR analysis using primers for the HSV-1 ICP27 IE gene. The primers were prepared according to the published sequence of HSV-1 (28): 5′-CCCTTTCTCCAGTGCTACCTGAA-3′ (bases 114919 to 114941) and 5′-GTGCGTGTCTAGGATTTCGATC-3′ (bases 115170 to 115149). Lanes 1 to 3, PCR products of DNAs extracted from the nonreactivated ganglia. Lane 4, PCR products of DNA extracted from a reactivated ganglion. Lane 5, PCR with no DNA template added to the reaction mixture. Lane M, 100-bp DNA ladder (New England Biolabs). The sizes of amplified sequences are presented.
In another experiment, we measured the viral titer produced in explanted TG on day 4 postexplantation, the day associated with the highest number of reactivated TG. Virus in the culture medium of each ganglion was measured separately by plaque assay on CV1 cells. TG from transgenic mice yielded 31 ± 18 viral plaques compared to 6 ± 3 plaques produced by normal reactivated ganglia (∼5-fold difference). We therefore conclude that HSV-1 is reactivated earlier by a higher percentage of ganglia and to a higher titer from transgenic than from normal ganglia.
Analysis of viral DNA copy numbers in transgenic-mouse ganglia.
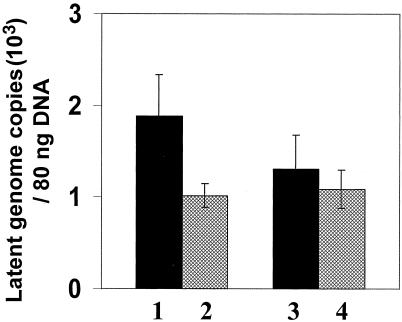
To determine whether the LAT gene effect upon reactivation is the outcome of improved HSV-1 ability to establish latency or is mediated by its impact upon the reactivation step per se, the latent viral copy number per TG was measured using quantitative real-time PCR. Total DNA was isolated from latently infected TG of transgenic as well as normal mice (11 TG pairs from each group) and subjected to real-time PCR for the HSV-1 gB gene. For the standard curve, we applied the plasmid pgB, including the corresponding HSV-1 gene. In parallel, the numbers of ribosomal genes were also measured in the same ganglion DNA preparations using quantitative real-time PCR. This measurement was used to calibrate the amounts of DNA applied in the test and to calculate the HSV-1 DNA copy number per ganglion. A comparison of the mean HSV-1 genome copy number per ganglion revealed no significant difference between the latently infected transgenic animals and the control animals (Fig. 6). The reported HSV-1 DNA copy number in the latently infected TG is variable, ranging from 103 to 106 copies per ganglion (7, 32, 33, 46). From the data obtained in this work, using quantitative real-time PCR, we estimated that there were ∼104 copies of HSV-1 DNA per ganglion.
FIG. 6.
Latent HSV-1 copy number in TG.Quantitative real-time PCR was performed on DNAs extracted from ganglia derived from transgenic and normal mice latently infected with an HSV-1 LAT-negative mutant. In parallel, the same DNA samples were analyzed for rRNA gene copy numbers for normalization of DNA amounts. Primers for the HSV-1 gB gene and for the rRNA gene were used (as described in Materials and Methods). The results represent HSV-1 copy numbers per 80 ng of DNA and are the means ± standard errors of the mean for 1 pair of ganglia in each group. Bars 1 and 2, HSV-1 copy numbers based on absorbance at 260 nm; bars 3 and 4, HSV-1 copy numbers following normalization for mouse rRNA gene. Solid bars, transgenic TG; cross-hatched bars, normal TG.
DISCUSSION
Steady-state levels of the 2.0- and 1.5-kb LATs differ in various tissues of transgenic mice. (i) The 2.0-kb LAT.
It was recently reported that, unlike typical introns, which are rapidly degraded following splicing, the 2.0-kb LAT intron has a half-life (∼24 h) similar to that of the more stable cellular mRNAs (43). Therefore, the different steady-state levels of the 2.0-kb LAT in the transgenic tissues observed in this work (Fig. 2A and 3) do not result from differences in stability but are probably due to the fact that transgenes controlled by the CMV IE promoter are differently transcribed in various tissues (4, 11, 52). This promoter was selected in order to enable constitutive expression of the LAT gene, as well as to prevent the impact of other HSV-1 genes upon the LAT promoter (5).
(ii) The 1.5-kb LAT.
The mechanism responsible for the accumulation of the 1.5-kb LAT in TG during HSV-1 latency might be either neuronal cell specific or determined by the virus, namely, the outcome of HSV-1 latency (51). The results of our experiments with the LAT transgenic model, demonstrating significantly greater accumulation of the 1.5-kb LAT in TG, argue for tissue specificity. Higher levels of the 1.5-kb LAT in the TG may reflect either more efficient splicing or higher stability of the transcript in this tissue.
Efficient splicing in neuronal tissue can be explained by the observation that the splicing donor site for the 1.5-kb LAT (GC rather than GT) constitutes a unique neuronal-tissue-specific signal (35). Other splicing control mechanisms and factors that operate exclusively in the nervous system (14) could also govern production of the 1.5-kb LAT in TG. Accumulation of the 1.5-kb LAT is not influenced by the amounts of 2.0-kb LAT, since large amounts of this transcript in nonneuronal tissues (e.g., kidney, muscle, and heart [Fig. 3]) were not associated with correspondingly large amounts of 1.5-kb LAT. Thus, although nonneuronal tissues are also capable of splicing the 2.0-kb LAT (Fig. 2B), though inefficient1y, the limiting factor in the splicing process is tissue specific rather than the amount of the 2.0-kb precursor RNA.
The finding of increased steady-state levels of the 1.5-kb LAT in TG is intriguing. It is reasonable to assume that these high levels have a functional significance, since latency is a neuronal-tissue-specific phenomenon.
LATs suppress HSV-1 replication in the transgenic-cell cultures.
HSV-1 is a lytic virus; therefore, prevention of its replication in neuronal tissue might be mandatory for the ability of the virus to establish latent infection. In a previous study, a neuronal cell line that stably expresses the HSV-1 LAT gene under the control of the CMV IE promoter was generated. Infection of these cells with an HSV-1 LAT-negative mutant, as well as with wild-type HSV-1, was suppressed by 2 to 3 orders of magnitude compared to control neuronal cells (23). These observations were tested in cultured primary fibroblasts (MEFs), in which the endogenous antiapoptotic machinery characteristic of cell lines is not operative (20). This is especially important for the analysis of the LAT gene, as it was reported to possess antiapoptotic activity (2, 17, 30) and the capacity to protect cells from death following HSV-1 infection of mice (44). We show here that replication of HSV-1 strain F, as well as of an HSV-1 KOS strain-derived LAT-negative mutant, is inhibited in primary fibroblasts derived from heterozygous LAT transgenic embryos compared to that in control cells obtained from normal mice (Fig. 4). Taking into consideration previous findings in LAT-expressing neuronal cell lines (24), this inhibition might take place at a step prior to IE gene expression.
The LAT gene facilitates HSV-1 reactivation.
Our results indicate that expression of the LAT gene in transgenic mice augments reactivation of the LAT-negative virus mutant (Fig. 5A). Enhancement was observed in the total number of reactivation-positive TG, kinetics of reactivation, and amount of infectious virus released from the ganglia. These findings are in accordance with those of other studies that demonstrated improved reactivation of wild-type virus compared to LAT-negative mutants (15, 18, 21, 37, 46, 47).
LAT transgene involvement in HSV-1 latency.
In order to directly study whether the LAT transgene can facilitate the entry of HSV-1 into latency, we infected groups of transgenic and normal animals with an HSV-1 LAT-negative mutant and compared the viral DNA copy numbers per ganglion. The data were analyzed by three statistical tests, Kolmogonov-Smirnov, Shapiro-Wilk, and Wilcoxon. Although there was appreciable variation among individual animals, no statistically significant differences between the amounts of latent viral DNA in the two groups could be detected (Fig. 6). The amounts of latent viral DNA after infection of normal mice with LAT-negative and wild-type HSV-1 have been investigated by several groups. While Thompson and Sawtell (46) reported ∼3-fold reduction in the amount of latent DNA after infection with LAT-negative compared to wild-type virus, no significant differences in the amounts of latent-virus DNA were reported in other studies (6, 7, 8, 30).
Differences between HSV-2 and HSV-1 LAT transgenic mice.
Our findings with HSV-1 differ from those reported in a recent study that examined the effects of the HSV-2 LAT transgene upon viral replication, latency, and reactivation from transgenic mice (49) and did not identify phenotypic differences compared to the parental mouse strain.
There are several notable differences between the LAT genes of the two viruses: HSV-2 has only one major LAT species, 2.2 kb in size, transcribed from a single latency-associated promoter. An equivalent of the HSV-1 spliced 1.5-kb LAT RNA is not found during HSV-2 latency. The HSV-1 LAT transgene described here also differs from the HSV-2 transgenic model in that LAT expression is not driven by either of the two native HSV-1 latency-associated promoters but by a constitutive CMV promoter.
While HSV-2 replication in fibroblasts obtained from the HSV-2 transgenic mouse was identical to replication in parental mice (49), we observed substantial inhibition of HSV-1 replication in primary fibroblasts of transgenic mouse embryos. These discrepancies might be inherent to the two different viruses, but they may also reflect variation between the sources of the primary cells. We used cells derived from degutted whole embryos, while Wang et al. (49) infected primary lung fibroblast cells.
The HSV-2 viral copy numbers in latently infected TG for both LAT transgenic and normal mice were similar (49). Our experiments with HSV-1 also indicated no significant differences between the amounts of latent viral DNA in infected transgenic and control groups. The reactivation of HSV-2 from latently infected TG of transgenic mice was identical to that from TG of wild-type mice, while reactivation of HSV-1 from its respective transgenic mouse was superior. This might be related either to the inherent differences between HSV-1 and HSV-2 or to the difference in reactivation techniques: we measured reactivation kinetics and viral titers in explant cultures of TG, while Wang et al. (49) used UV irradiation of the eyes, followed by extraction of TG and homogenization of the tissue 2 days later. In conclusion, the present work demonstrates that the steady-state levels of the spliced 1.5-kb LAT are increased in neuronal tissues of HSV-1 transgenic animals, a fact governed by cellular and not viral factors; that the LAT gene suppresses in trans HSV-1 replication; and that this is associated with enhanced reactivation ability. Based on these data, we propose that LAT is involved in the viral reactivation phase, perhaps through controlling viral replication and thus protecting the ganglion cells against death.
Acknowledgments
This work was supported by grants from the Chief Scientist, Ministry of Health, Hadassah Medical Organization (Women's Health), Israeli Science Ministry, and by European Commission program no. 5, Quality of Life and Management of Living Resources. E.B. was supported by a fellowship grant from the ICRF.
We are grateful to Alik Honigman for reading the manuscript, to Blanka Shanitzki for technical assistance, and to Dalit Braun-Ginsburg for statistical analysis.
REFERENCES
- 1.Ahmed, M., and N. W. Fraser. 2001. Herpes simplex virus type 1 2-kilobase latency-associated transcript intron associates with ribosomal proteins and splicing factors. J. Virol. 75:12070-12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed, M., M. Lock, C. G. Miller, and N. W. Fraser. 2002. Regions of the herpes simplex virus type 1 latency-associated transcript that protect cells from apoptosis in vitro and protect neuronal cells in vivo. J. Virol. 76:717-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alexander, D. L., S. E. Eltom, and C. R. Jefcoate. 1997. A receptor regulation of CYP1B1 expression in primary mouse embryo-derived cells. Cancer Res. 57:4498-4506. [PubMed] [Google Scholar]
- 4.Baskar, J. F., P. P. Smith, G. Nilaver, R. A. Jupp, S. Hoffmann, N. J. Peffer, D. J. Tenney, A. M. Colberg-Poley, P. Ghazal, and J. A. Nelson. 1996. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell-type-specific statement in transgenic mice. J. Virol. 70:3207-3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Batchelor, A. H., and P. O'Hare. 1990. Regulation and cell-type-specific activity of a promoter located upstream of the latency-associated transcript of herpes simplex virus type 1. J. Virol. 64:3269-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bloom, D. C., J. M. Hill, G. Devi-Rao, E. K. Wagner, L. T. Feldman, and J. G. Stevens. 1996. A 348-base-pair region in the latency-associated transcript facilitates herpes simplex virus type 1 reactivation. J. Virol. 70:2449-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S.-H., M. F. Kramer, P. A. Schaffer, and D. M. Coen. 1997. A viral function represses accumulation of transcripts from productive-cycle genes in mouse ganglia latently infected with herpes simplex virus. J. Virol. 71:5878-5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devi-Rao, G. B., D. C. Bloom, J. G. Stevens, and E. K. Wagner. 1994. Herpes simplex virus type 1 DNA replication and gene expression during explant-induced reactivation of latently infected murine sensory ganglia. J. Virol. 68:1271-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fareed, M. U., and J. G. Spivack. 1994. Two open reading frames (ORF1 and ORF2) within the 2.0-kilobase latency-associated transcript of herpes simplex virus type 1 are not essential for reactivation from latency. J. Virol. 68:8071-8081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser, N. W., W. C. Lawrence, Z. Wroblewska, D. H. Gilden, and H. Koprowski. 1981. Herpes simplex type 1 DNA in human brain tissue. Proc. Natl. Acad. Sci. USA 78:6461-6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Furth, P. A., L. Hennighausen, C. Baker, B. Beatty, and R. Woychick. 1991. The variability in activity of the universally expressed human cytomegalovirus immediate early gene 1 enhancer/promoter in transgenic mice. Nucleic Acids Res. 19:6205-6208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldenberg, D., N. Mador, M. Ball, A. Panet, and I. Steiner. 1997. The abundant latency-associated transcripts of herpes simplex virus type 1 are bound to polyribosomes in cultured neuronal cells and during latent infection in mouse trigeminal ganglia. J. Virol. 71:2897-2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldenberg, D., N. Mador, A. Panet, and I. Steiner. 1998. Tissue specific distribution of the herpes simplex virus type 1 latency-associated transcripts on polyribosomes during latent infection. J. Neurovirol. 4:426-432. [DOI] [PubMed] [Google Scholar]
- 14.Grabowski, P. J., and D. L. Black. 2001. Alternative RNA splicing in the nervous system. Prog. Neurobiol. 65:289-308. [DOI] [PubMed] [Google Scholar]
- 15.Hill, J. M., F. Sedarat, R. T. Javier, E. K. Wagner, and J. G. Stevens. 1990. Herpes simplex virus latent phase transcription facilitates in vivo reactivation. Virology 174:117-125. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, B., F. Constantini, and E. Lacey. 1986. Manipulating the mouse embryo, p. 92-94. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Inman, M., G. C. Perng, G. Henderson, H. Ghiasi, A. B. Nesburn, S. L. Wechsler, and C. Jones. 2001. Region of herpes simplex virus type 1 latency-associated transcript sufficient for wild-type spontaneous reactivation promotes cell survival in tissue culture. J. Virol. 75:3636-3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Javier, R. T., J. G. Stevens, V. B. Dissette, and E. K. Wagner. 1988. A herpes simplex virus transcript abundant in latently infected neurons is dispensable for establishment of the latent state. Virology 166:254-257. [DOI] [PubMed] [Google Scholar]
- 19.Jerome, K. R., M.-L. Huang, A. Wald, S. Selke, and L. Corey. 2002. Quantitative stability of DNA after extended storage of clinical specimens as determined by real-time PCR. J. Clin. Microbiol. 40:2609-2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaCasse, E. C., S. Baird, R. G. Korneluk, and A. E. MacKenzie. 1998. The inhibitors of apoptosis (IAPs) and their emerging role in cancer. Oncogene 17:3247-3259. [DOI] [PubMed] [Google Scholar]
- 21.Leib, D. A., C. I. Bogard, M. Kosz-Vnenchak, K. A. Hicks, D. M. Coen, D. M. Knipe, and P. A. Scheffer. 1989. A deletion mutant of the latency-associated transcript of herpes simplex virus type 1 reactivates from the latent state with reduced frequency. J. Virol. 63:2893-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mador, N., A. Panet, D. Latchman, and I. Steiner. 1995. Expression and splicing of the latency-associated transcripts of herpes simplex virus type 1 in neuronal and non-neuronal cell lines. J. Biochem. 117:1288-1297. [DOI] [PubMed] [Google Scholar]
- 23.Mador, N., D. Goldenberg, O. Cohen, A. Panet, and I. Steiner. 1998. Herpes simplex virus type 1 latency-associated transcripts suppress viral replication and reduce immediate early gene mRNA levels in a neuronal cell line. J. Virol. 72:5067-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mador, N., A. Panet, and I. Steiner. 2002. The latency-associated gene of herpes simplex virus type 1 (HSV-1) interferes with superinfection by HSV-1. J. Neurovirol. 8:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Miller, R. R., and C. A. McDevitt. 1991. A quantitative microwell assay for chondrocyte cell adhesion. Anal. Biochem. 192:380-383. [DOI] [PubMed] [Google Scholar]
- 26.Millhouse, S., and B. Wigdahl. 2000. Molecular circuitry regulating herpes simplex virus type 1 latency in neurons. J. Neurovirol. 6:6-24. [DOI] [PubMed] [Google Scholar]
- 27.Minty, A. J., M. Caravatti, B. Robert, A. Cohen, P. Daubas, A. Weydert, F. Gros, and M. E. Buckingham. 1981. Mouse actin messenger RNAs. Construction and characterization of a recombinant plasmid molecule containing a complementary DNA transcript of mouse alpha-actin mRNA. J. Biol. Chem. 256:1008-1014. [PubMed] [Google Scholar]
- 28.Mitchell, W., J. R. P. Lirette, and N. W. Fraser. 1990. Mapping of low abundance latency associated RNA in the trigeminal ganglia of mice latently infected with herpes simplex virus type 1. J. Gen. Virol. 71:125-132. [DOI] [PubMed] [Google Scholar]
- 29.Nicosia, M., J. M. Zabolotny, R. P. Lirette, and N. W. Fraser. 1994. The HSV-1 2-kb latency-associated transcript is found in the cytoplasm comigrating with ribosomal subunits during productive infection. Virology 204:717-728. [DOI] [PubMed] [Google Scholar]
- 30.Perng, G. C., C. Jones, J. Ciacci-Zanella, M. Stone, G. Henderson, A. Yukht, S. M. Slanina, F. M. Hofman, H. Ghiasi, A. B. Nesburn, and S. L. Wechsler. 2000. Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript. Science 287:1500-1503. [DOI] [PubMed] [Google Scholar]
- 31.Perry, L. J., and D. J. MaGeoch. 1998. The DNA sequences of the long repeat region and adjoining parts of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:2831-2846. [DOI] [PubMed] [Google Scholar]
- 32.Saldanha, C. E., J. Lubinski, C. Martin, T. Nagashunmugam, L. Wang, H. van Der Keyl, R. Tal-Singer, and H. M. Friedman. 2000. Herpes simplex virus type 1 glycoprotein E domains involved in virus spread and disease. J. Virol. 74:6712-6719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sawtell, N. M., D. K. Poon, C. S. Tansky, and R. L. Thompson. 1998. The latent herpes simplex virus type 1 genome copy number in individual neurons is virus strain specific and correlates with reactivation. J. Virol. 72:5343-5350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spivack, J. G., and N. W. Fraser. 1987. Detection of herpes simplex virus type 1 transcripts during latent infection in mice. J. Virol. 61:3841-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spivack, J. G., G. M. Woods, and N. W. Fraser. 1991. Identification of a novel latency-specific splice donor signal within the herpes simplex virus type 1 2.0-kilobase latency-associated transcript (LAT); translation inhibition of LAT open reading frames by the intron within the 2.0-kilobase LAT. J. Virol. 65:6800-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steiner, I., J. G. Spivack, D. R. O'Boyle II, E. Lavi, and N. W. Fraser. 1988. Latent herpes simplex virus type 1 transcription in human trigeminal ganglia. J. Virol. 62:3493-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steiner, I., J. G. Spivack, R. P. Lirette, S. M. Brown, A. R. MacLean, J. H. Subak-Sharpe, and N. W. Fraser. 1989. Herpes simplex virus type 1 latency-associated transcripts are evidently not essential for latent infection. EMBO J. 8:505-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steiner, I., J. G. Spivack, S. L. Deshmane, C. I. Ace, C. M. Preston, and N. W. Fraser. 1990. A herpes simplex virus type 1 mutant containing a nontransinducing Vmw65 protein establishes latent infection in vivo in the absence of viral replication and reactivates efficiently from explanted trigeminal ganglia. J. Virol. 64:1630-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steiner, I., N. Mador, I. Reibstein, J. G. Spivack, and N. W. Fraser. 1994. Herpes simplex virus type 1 latency in human and mouse central nervous systems. Neuropathol. Appl. Neurobiol. 20:253-260. [DOI] [PubMed] [Google Scholar]
- 40.Steiner, I., and P. G. E. Kennedy. 1995. Herpes simplex virus latent infection in the nervous system. J. Neurovirol. 1:19-29. [DOI] [PubMed] [Google Scholar]
- 41.Stevens, J. G., E. K. Wagner, G. B. Devi-Rao, M. L. Cook, and L. T. Feldman. 1987. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science 235:1056-1059. [DOI] [PubMed] [Google Scholar]
- 42.Tanaka, S., H. Minagawa, Y. Toh, Y. Liu, and R. Mori. 1994. Analysis by RNA-PCR of latency and reactivation of herpes simplex virus in multiple neuronal tissues. J. Gen. Virol. 75:2691-2698. [DOI] [PubMed] [Google Scholar]
- 43.Thomas, D. L., M. Lock, J. M. Zabolotny, B. R. Mohan, and N. W. Fraser. 2002. The 2-kilobase intron of the herpes simplex virus type 1 latency-associated transcript has a half-life of approximately 24 hours in SY5Y and COS-1 cells. J. Virol. 76:532-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thomas, S. K., C. E. Lilley, D. S. Latchman, and R. S. Coffin. 2002. A protein encoded by the herpes simplex virus (HSV) type 1 2-kilobase latency-associated transcript is phosphorylated, localized to the nucleus, and overcomes the repression of expression from exogenous promoters when inserted into the quiescent HSV genome. J. Virol. 76:4056-4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson, R. L., and N. M. Sawtell. 1997. The herpes simplex virus type 1 latency-associated transcript gene regulates the establishment of latency. J. Virol. 71:5432-5440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson, R. L., and N. M. Sawtell. 2001. Herpes simplex virus type 1 latency-associated transcript gene promotes neuronal survival. J. Virol. 75:6660-6675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trousdale, M. D., I. Steiner, J. G. Spivack, S. L. Deshmane, S. M. Brown, A. R. MacLean, J. H. Subak-Sharpe, and N. W. Fraser. 1991. In vivo and in vitro reactivation impairment of a herpes simplex virus type 1 latency-associated transcript variant in a rabbit eye model. J. Virol. 65:6989-6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsumoto, H. 1990. Autoradiography of new era replacing traditional X-ray film. Cell Technol. 9:456-462. [Google Scholar]
- 49.Wang, K., L. Pesnicak, E. Guancial, P. R. Krause, and S. E. Straus. 2001. The 2.2-kilobase latency-associated transcript of herpes simplex virus type 2 does not modulate viral replication, reactivation, or establishment of latency in transgenic mice. J. Virol. 75:8166-8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitley, R. J. 1990. Herpes simplex viruses, p. 1843-1886. In B. N. Fields, D. M. Knipe, et al. (ed.), Virology, 2nd ed. Raven Press, New York, N.Y.
- 51.Wu, T. T., Y. H. Su, T. M. Block, and J. M. Taylor. 1998. Atypical splicing of the latency associated transcripts of herpes simplex type 1. Virology 243:140-149. [DOI] [PubMed] [Google Scholar]
- 52.Zhan, Y., J. L. Brady, A. M. Johnston, and A. M. Lew. 2000. Predominant transgene statement in exocrine pancreas directed by the CMV promoter. DNA Cell Biol. 19:639-645. [DOI] [PubMed] [Google Scholar]
- 53.Zwaagstra, J. C., H. Ghiasi, S. M. Slanina, A. B. Nesburn, S. C. Wheatley, K. Lillycrop, J. Wood, D. S. Latchman, K. Patel, and S. L. Wechsler. 1990. Activity of herpes simplex virus type 1 latency-associated transcript (LAT) promoter in neuron-derived cells: evidence for neuron specificity and for a large LAT transcript. J. Virol. 64:5019-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]