Abstract
During a large serosurvey of wild-caught primates from Cameroon, we found 2 mona monkeys (Cercopithecus mona) out of 8 and 47 mustached monkeys (Cercopithecus cephus) out of 302 with human immunodeficiency virus (HIV)-simian immunodeficiency virus (SIV) cross-reactive antibodies. In this report, we describe the full-length genome sequences of two novel SIVs, designated SIVmon-99CMCML1 and SIVmus-01CM1085, isolated from one mona (CML1) and one mustached (1085) monkey, respectively. Interestingly, these viruses displayed the same genetic organization (i.e., presence of a vpu homologue) as members of the SIVcpz-HIV type 1 lineage and SIVgsn isolated from greater spot-nosed monkeys (Cercopithecus nictitans). Phylogenetic analyses of SIVmon and SIVmus revealed that these viruses were genetically distinct from other known primate lentiviruses but were more closely related to SIVgsn all across their genomes, thus forming a monophyletic lineage within the primate lentivirus family, which we designated the SIVgsn lineage. Interestingly, mona, mustached, and greater spot-nosed monkeys are phylogenetically related species belonging to three different groups of the genus Cercopithecus, the C. mona, C. cephus, and Cercopithecus mitis groups, respectively. The presence of new viruses closely related to SIVgsn in two other species reinforces the hypothesis that a recombination event between ancestral SIVs from the family Cercopithecinae is the origin of the present SIVcpz that is widespread among the chimpanzee population.
Throughout sub-Saharan Africa, many nonhuman primates are the natural hosts of simian immunodeficiency virus (SIV), and there is serological and/or molecular evidence for SIV infection in at least 33 African primate species (13, 22). Based on comparisons of their full-length genome sequences and the functional similarity of their genes, primate lentiviruses were classified into lineages which have mainly been named according to the chronological order of their discovery and their genetic characterization. Initially, six approximately equidistant lineages were described: (i) SIVcpz from chimpanzees, together with human immunodeficiency virus type 1 (HIV-1); (ii) SIVsm from sooty managabeys, together with HIV-2; (iii) SIVagm from different species of African green monkeys; (iv) SIVsyk from Sykes's monkeys; (v) SIVlhoest, SIVsun, and SIVmnd1 from l'Hoest's monkeys, sun-tailed monkeys, and mandrills, respectively; and (vi) SIVcol from guereza colobus (7). However, some fully characterized SIVs do not fall into the six-lineage classification because they appear to have mosaic genome structures. The first mosaic SIV was SIVagmSAB, isolated from a West African sabaeus monkey. Also, the recently described full-length genome sequences of SIVrcm from red-capped mangabeys in Nigeria, SIVmnd2 and SIVdrl from mandrills and drills, and SIVgsn from greater spot-nosed monkeys in Cameroon showed that these SIVs present discordant phylogenies, depending on the region of their genomes studied (4, 8, 14, 27).
The genomic organizations of these SIVs and HIVs are similar, showing a common genome structure comprising the gag, pol, vif, vpr, tat, rev, env, and nef genes, but viruses from the SIVcpz-HIV-1 lineage and SIVgsn carry a vpu gene, while viruses from the SIVsm-HIV-2 lineage, SIVrcm, SIVdrl, and SIVmnd2 have a vpx gene.
Phylogenetic studies of the primate lentiviruses provide evidence that both host-dependent evolution and cross-species transmissions were at the origin of viral transmission and evolution. On the one hand, phylogenetic relationships among these viruses indicate that some SIV lineages have coevolved with their hosts, as is the case for SIVagm in African green monkeys (1, 16, 20) and SIVlhoest and SIVsun within the Cercopithecus lhoesti superspecies (3). On the other hand, there are also multiple examples of cross-species transmissions from simians to humans and between different simian species. The most striking examples of cross-species transmissions are HIVs, the etiologic agents for AIDS in humans. Indeed, the fact that HIV-1 and HIV-2 belong to two different lineages of SIVs (SIVcpz from chimpanzees and SIVsm from sooty mangabeys, respectively) suggests that both are the result of several zoonotic transmissions. In the wild, cross-species transmissions of SIVs have also occurred between monkey species in Africa: for example, SIV infection of baboons and patas monkeys by viruses derived from the local sympatric species of African green monkeys have been reported (5, 17, 29). Also, mosaic SIV genomes are indirect evidence that both cross-species transmission and coinfection of viruses have occurred in the wild. Although based on sequence analysis only, we cannot define exactly which lineages are pure, the above-described observations show that recombination events have occurred between viruses in the wild and indicate that both cross-species transmission and coinfection with highly divergent viral strains are possible. None of these SIVs seems to induce an AIDS-like disease in its natural hosts, suggesting that they have been associated and evolved with their hosts over an extended period.
In order to elucidate the origins and the evolution of primate lentiviruses, additional molecular characterizations of SIVs are fundamental. Recently, a large serosurvey was performed in Cameroon to assess the extent of SIV infection in wild-caught primates (22). The results of the study revealed that SIV infection appears to be widespread among nonhuman primates in Africa and also that humans are still exposed to a plethora of primate lentiviruses through hunting and handling of primate bushmeat in Central Africa. Thus, the possibility of additional zoonotic transfers of primate lentiviruses from species other than chimpanzees and sooty mangabeys has to be considered. It is therefore important to obtain a complete and accurate assessment of all SIV-infected nonhuman primate species. The partial identification of new SIV lineages in primate bushmeat has revealed evidence of great SIV diversity, but analysis of the complete genomes of these new SIVs is necessary to determine their exact phylogenetic relationships to all other known SIVs.
Here, we describe the entire genomes of two of these novel SIVs: SIVmon, found in mona monkeys (Cercopithecus mona), and SIVmus, found in mustached monkeys (Cercopithecus cephus). The mona and mustached monkeys belong to the C. mona and C. cephus groups, respectively, of the genus Cercopithecus, which contains >26 species classified into seven groups (12). Mona monkeys are found in West Africa from eastern Ghana to western Cameroon, with the main population centered in Nigeria. Mustached monkeys are found in west central Africa, Cameroon, Congo, Equatorial Guinea, and Gabon. Both species inhabit a variety of habitats, including primary rainforests, secondary rainforests, gallery forests, and mangrove swamps.
Interestingly, SIVmon and SIVmus are closely related all across the genome to SIVgsn, isolated from greater spot-nosed monkeys, and display similar genomic organizations (i.e., the presence of a vpu homologue) (8), thus forming a new monophyletic lineage within the primate lentivirus family.
MATERIALS AND METHODS
Animals and serological tests.
Blood samples were obtained between 1998 and 2001 from 8 mona monkeys (C. mona) and 302 mustached guenons (C. cephus) originating from Cameroon; 2 and 273 were from bushmeat markets and 6 and 29 were kept as pets, respectively. For the bushmeat animals, blood was collected by intracardiac puncture as previously described. Information provided by the owners indicated that most of the animals had died 12 to 72 h prior to sampling. For the pet monkeys, blood was drawn by peripheral venipuncture after the animals were tranquilized with ketamine (10 mg/kg of body weight). All samples were stored at −20°C. Whole-blood samples were screened for the presence of HIV-SIV antibodies by the INNO-LIA HIV Confirmation test (Innogenetics, Ghent, Belgium). A subset of samples was also tested by an in-house peptide-based enzyme-linked immunosorbent assay that detects and differentiates antibodies against the V3 regions representative of the different SIV lineages, as previously described (8).
PCR amplification, cloning, and sequencing.
DNA was isolated from whole blood with the QIAamp blood kit (Qiagen) according to the manufacturer's instructions. PCR amplification was performed with an automated DNA thermal cycler (GeneAmp PCR system 2700).
First, degenerate consensus primers (DR1-DR2 or DR1-PolOR for the first round and DR4-DR5 or Polis4-UNIPOL2 for the second round) were used to amplify a small fragment of pol (194 and 650 bp, respectively). The PCR conditions were as previously reported (6, 7). The PCR products were cloned into the pGEM-TEasy vector (Promega) and sequenced. Then, based on the known pol fragment sequence (DR4-DR5 for SIVmon-99CMCML1 or DR4-UNIPOL2 for SIVmus-01CM1085), two specific primers were generated to amplify unintegrated circular forms in a first PCR round: CM1 (5′-GAACCGTTCAGAAAATACACGG-3′) and CM2 (5′-CAAGGGAATGCTGTAGAAGGCG-3′) or CEP1 (5′-AAGAAGCCATACACAAAATCAGGG-3′) and CEP2 (5′-TCTGGGTTTGCTTTCCTAATCTCTTC-3′) for SIVmon-99CMCML1 and SIVmus-01CM1085, respectively. Several nested PCRs were then performed to generate overlapping fragments spanning the entire genome, using combinations of SIVmon-CML1-specific and/or SIV consensus primers: DR4 and UNIPOL2, SPBS (5′-GGCGCCCGAACAGGGACTTG-3′) and RevMon (5′-AAACACAGTTGGGGACCCTTTCCAGCCCTG-3′), and CMLgagas1 (5′-AGCACCTCTGGAACCTTCCCTCTTGG-3′) and MonPF2 (5′-GGGGTACCCTACAACCCACAAAGCCAAGG-3′). For SIVmus, the full-length genome sequence was obtained directly by a second round of amplification using the primers CEP3 (5′-AGGTCACTTATTTGGAAACAGCAG-3′) and CEP4 (5′-TTGGCTGCTGTGAACTGGAAGATGG-3′). PCRs were performed using a Long Expand or Expand High Fidelity PCR kit (Roche Molecular Biochemicals), including a hot start (92°C for 3 min) with the following cycling conditions: 10 cycles of denaturation at 92°C for 10 s, annealing at 57°C for 30 s, and extension at 68°C for 7 min, followed by 20 cycles with extension at 68°C for 7 min with an increment of 20 s per cycle. Amplification was completed by a final extension at 68°C for 10 min. The cycling conditions for inner PCRs were 10 cycles of denaturation at 94°C for 15 s, annealing at 55°C (with primers SPBS and RevMon or at 58°C with primers CMLgagas1 and MonPF2) for 30 s, extension at 72°C for 2 or 3 min 30 s, followed by 20 cycles with an increment of 5 s per cycle of the extension time at 72°C, followed by 1 cycle of 7 min of extension at 72°C. PCR products were purified and cloned into the pGEM-TEasy vector (Promega) or the pCR-XL-TOPO vector (TOPO XL PCR cloning kit [Invitrogen]). Long PCR amplification products for SIVmus-01CM1085 were cleaved with EcoRI, and the resulting fragments were subcloned into pBluescript KS(+) cleaved by EcoRI or into the pGEM-TEasy vector after being blunt ended. DNAs were sequenced using cycle-sequencing and dye terminator methodologies (ABI PRISM Big Dye Terminator Cycle Sequencing Ready Reaction kit with AmpliTaq FS DNA polymerase [PE Biosystems, Warrington, England] on an automated sequencer [ABI 373, model Stretch; Applied Biosystems]) using the Genome Priming System GPS-1 (New England BioLabs, Beverly, Mass.). To reconstitute the full-length genome sequence, overlapping sequences were assembled into contiguous sequences by using Sequencher software (Gene Codes Corp.).
RNA folding.
The RNA secondary structures of the Tat-transactivation response element (TAR) were predicted using RNA mfold software version 3.1 by M. Zuker and D. H. Turner (http://bioinfo.math.rpi.edu/∼mfold/rna/form1.cgi) with the folding temperature fixed at 37°C (19, 31).
Sequence alignments.
Nucleotide alignments were constructed for the gag, pol, vif, env, and nef genes of SIVmon-99CMCML1 and SIVmus-01CM1085 plus 26 SIV and HIV strains representative of the major lentivirus lineages available from the Los Alamos HIV sequence database (http://hiv-web.lanl.gov/): HIV1-ANT70, HIV1-MVP5180, HIV1-YBF30, HIV1-U455, SIVcpzUS, SIVcpzGAB, SIVcpzCAM3, SIVcpzANT, SIVcpzTAN1, HIV2-ALI, HIV2-D205, SIVsmH4, SIVsyk173, SIVsun, SIVlhoest, SIVmnd1, SIVcol, SIVagmVER155, SIVagmGRI677, SIVagmVER906, SIVagmTAN1, SIVagmSAB1C, SIVmnd2, SIVrcmNG411, SIVgsn-99CM71, and SIVgsn-99CM166. The nucleotide sequences from the different genes were first translated and then aligned using Clustal W (28) implemented in DAMBE (30) and translated back to nucleotides in order to get a codon-based alignment. The alignments were then edited manually where necessary. A codon-aligned nucleotide sequence alignment was constructed excluding the overlapping sequences between gag and pol, pol and vif, and env and nef. Gaps were removed from the final alignment. A similar alignment was also obtained for translated amino acid sequences.
Homology searches were conducted using the Blast, FASTA, and HMMER programs with the following databases: PROSITE, Pfam, Prodom, and GenBank.
Diversity plot analysis.
Diversity plot analysis was performed on the concatenated nucleotide or amino acid sequence alignment, and the following strains were used in the analysis: SIVsyk173, SIVsun, SIVlhoest, SIVmnd1, SIVsmH4, SIVcpzUS, SIVcpzGAB, SIVcpzANT, SIVcpzTAN1, SIVagmVER155, SIVagmTAN1, SIVagmSAB1C, SIVcol, SIVmnd2, and SIVrcmNG411. Simplot and bootscan analyses were performed with the SIMPLOT program version 2.5 on first and second codon position alignments, grouping or not grouping the sequences per lineage, with a sliding window of 500 nucleotides (nt) moved in steps of 20 nt or 200 amino acids (aa) moved in steps of 10 aa.
Phylogenetic analysis.
The nucleotide substitution model best fitting our data set was evaluated through hierarchical likelihood ratio tests with the program MODELTEST version 3.06 (23). Phylogenetic analyses were performed with PAUP* version 4.0b8 written by David L. Swofford. Neighbor-joining and maximum-likelihood trees were calculated using the nucleotide substitution model selected, and the parameters were estimated through maximum likelihood (11). Bootstrap analysis (1,000 replicates) was applied to the neighbor-joining tree, whereas P values were obtained for the maximum-likelihood tree with the zero branch length test.
Nucleotide sequence accession numbers.
The complete sequences of SIVmus-01CM1085 and SIVmon-99CMCML1 can be obtained from GenBank under accession numbers AY340700 and AY340701, respectively.
RESULTS
Serology.
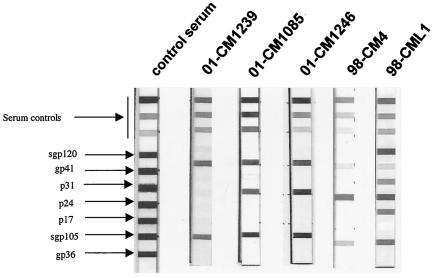
Sera from 8 mona and 302 mustached monkeys were screened for SIV infection using an INNO-LIA HIV-1/HIV-2 test, in which a nylon strip is coated with HIV-1 and HIV-2 recombinant proteins (p31, p24, p17, gp41, and gp36) and synthetic peptides (gp120 and gp105) as discrete bands (22). We identified two mona monkeys with HIV cross-reacting antibodies. In addition the sera from both animals also presented strong reactivity to the SIVcpzANT V3 loop peptide. Among the sera from mustached monkeys, 46 of 302 samples tested had antibodies that cross-reacted strongly with at least one HIV protein, and 4 of 45 HIV cross-reactive samples reacted exclusively with the SIVcpzANT peptide and not with HIV-1 N, SIVsm, SIVagm, SIVsyk, SIVlhoest, and SIVmnd V3 peptides.
Among the mona monkeys, the two samples considered positive (reacting strongly with at least one HIV antigen) did not present the same pattern in the INNO-LIA test; one of them (CML1) showed an HIV-1-HIV-2-like profile with strong reactivities against Gag, Pol, and Env proteins from HIV-1 and HIV-2 (gp120 plus gp36), while the second reacted strongly only with HIV-1 Gag and Env HIV-2 (gp36) proteins and weakly with Env HIV-1 (gp41). Among the mustached monkeys, the LIA patterns were very varied, showing an HIV-1, HIV-2, HIV-indeterminate, or HIV-1-HIV-2-like profile (Fig. 1).
FIG. 1.
Antibody profiles observed in C. mona and C. cephus using a line immunoassay (INNO-LIA HIV Confirmation). The five HIV-1 antigens are synthetic peptides for the exterior envelope glycoprotein (sgp120), as well as recombinant proteins for the transmembrane envelope glycoprotein (gp41) and integrase (p31), core (p24), and matrix (p17) proteins. The HIV-2 antigens are synthetic peptides for the exterior envelope glycoprotein (sgp105), as well as recombinant gp36 protein. Plasma samples which recognized at least one HIV antigen with an intensity equal to or greater than the assay ± cutoff line were scored as positive, samples which exhibited weaker but still visible reactivities with at least two HIV antigens were scored as indeterminant, and samples which yielded no reactivity or only a single band of less than plus-minus intensity were scored as negative. The 3+, 1+, and ± bands evident in the top portions of all test strips control for sample addition (the presence of plasma immunoglobulin) and test performance (binding of secondary antibody).
Similar genomic organizations of SIVmon, SIVmus, SIVgsn, and the SIVcpz-HIV-1 lineage.
As previously described, in order to confirm the presence of HIV-like viruses in these animals, PCR amplification of viral sequences was performed using pol lentivirus consensus primers, but the quality of the DNA extracted and a combination of other factors (viral load and primer mismatches) allowed us to amplify only one (99CM-CML1) of the mona and three (01CM-S1239, 01CM-S1085, and 01CM-S1246) of the mustached monkey samples cross-reacting with HIV antigens (22). Based on the sequences of the pol fragments of the new SIVs designated SIVmon-99CMCML1 and SIVmus-01CM-S1085, we designed specific primers to amplify unintegrated circular DNA in a first round. The complete genome sequence of SIVmon-99CMCML1 was then obtained by successive nested PCRs and cloning and sequencing of the different overlapping fragments amplified. The complete genome sequence of SIVmus-01CM-S1085 was successfully amplified by nested PCR, the long PCR products were cleaved, and the different fragments were cloned. The SIVmon (9,448-bp) and SIVmus (9,420-bp) genomes include the three structural genes (gag, pol, and env) and the five accessory genes (vif, vpr, tat, rev, and nef) common to all primate lentiviruses. In addition, these two genomes contain an open reading frame (ORF) (261 and 228 nt, respectively) in the central region of the genome at the location of vpu in the HIV-1-SIVcpz lineage.
The results of BLAST and Pfam searches against different databases revealed that the putative proteins encoded by these ORFs have significant, although limited, similarity to several Vpu proteins. Notably, for SIVmus, the best matches were with the Vpu proteins of SIVgsn-99CM71 and SIVgsn-99CM166, with 50.6 and 43.2% identity in a 73- and 74-aa overlap, respectively, and to a lesser degree with the Vpu proteins of HIV-199FR-MP129 and SIVcpzCAM3, with 36 and 31.7% identity in a 61- and 75-aa overlap, respectively. For the ORF of SIVmon, the best matches with the lowest Expect value (significance threshold) were with the Vpu proteins of SIVcpzGAB1 and SIVcpzANT, with 33.7 and 28.4% identity in an 86- and 81-aa overlap, respectively. The identity between the ORFs of SIVmon and SIVmus was 28.9%. These data, as well as the hydropathy profile displayed by the deduced protein of these ORFs, unambiguously identified these ORFs as a vpu gene (not shown). Thus, the genomic organizations of SIVmon and SIVmus are similar to those of members of the SIVcpz-HIV-1 lineage and the recently described SIVgsn from a greater spot-nosed monkey (Cercopithecus nictitans), all harboring the additional accessory gene vpu in the center of the genome (8).
Long terminal repeat (LTR) sequences from SIVmon and SIVmus were 728 and 754 bp in length, respectively, and possessed all the characteristic features of those of other primate lentiviruses, including one NF-κB and one potential SP-1 binding site. The LTR sequences of SIVmon and SIVmus were most closely related to the SIVgsn LTR, with 74 and 75.5% nucleotide identity, respectively, and despite differences in their primary sequences, the predicted TAR secondary structures were very similar to each other. The SIVmon and SIVmus TARs are capable of folding into two stable stem-loop structures (ΔG = −45.5 and −43.5 kcal/mol, respectively) and contain the highly conserved 6-nt loop sequence 5′-CUGGGA-3′, which is essential for in vivo HIV-1 transactivation responsiveness (26). Therefore, as previously reported for SIVgsn, these TAR structures closely resemble those described for SIVsyk (8).
SIVmon and SIVmus are closely related to SIVgsn.
A comparison of the predicted proteins of SIVmon and SIVmus with those of representatives of the other primate lentivirus lineages is shown in Tables 1 and 2 and revealed that SIVmon and SIVmus were distinct from all other known SIVs. The primate lentivirus most closely related to SIVmon and SIVmus was SIVgsn, isolated from greater spot-nosed monkeys, showing at least 60 and 64% amino acid identity, respectively, for all proteins except Vif. This was not really surprising, since mona, mustached, and greater spot-nosed monkeys are all members of the genus Cercopithecus, and furthermore, it reflects the results of the phylogenetic analyses of a small fragment in pol (22).
TABLE 1.
Percent amino acid identities between SIVmon-99CMCML1 and other SIVs
| SIV | % Identity to SIVmonCML1 and other SIVs in:
|
|||||
|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpr | Env | Nef | |
| SIVgsn-99CM166 | 68 | 67 | 50 | 63 | 61 | 70 |
| SIVsyk | 55 | 55 | 25 | 23 | 42 | 35 |
| SIVsmPBj | 55 | 54 | 25 | 22 | 38 | 35 |
| SIVmnd1 | 48 | 51 | 21 | 23 | 29 | 33 |
| SIVlhoest | 46 | 50 | 23 | 22 | 28 | 32 |
| SIVsun | 45 | 50 | 19 | 21 | 27 | 34 |
| SIVagmGRI | 53 | 53 | 25 | 23 | 38 | 36 |
| SIVcpzUS | 50 | 53 | 23 | 28 | 45 | 33 |
| SIVcpzANT | 50 | 51 | 23 | 22 | 47 | 31 |
| SIVcol | 38 | 47 | 20 | 21 | 30 | 26 |
TABLE 2.
Percent amino acid identities between SIVmus1085 and other SIVs
| SIV | % Identity to SIVmus1085 and other SIVs in:
|
|||||
|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpr | Env | Nef | |
| SIVgsn-99CM166 | 70 | 70 | 54 | 74 | 64 | 68 |
| SIVsyk | 53 | 54 | 27 | 25 | 42 | 32 |
| SIVsmPBj | 52 | 56 | 22 | 24 | 37 | 32 |
| SIVmnd1 | 50 | 52 | 20 | 21 | 27 | 34 |
| SIVlhoest | 45 | 51 | 24 | 19 | 30 | 33 |
| SIVsun | 47 | 50 | 20 | 23 | 31 | 33 |
| SIVagmGRI | 53 | 53 | 28 | 26 | 36 | 34 |
| SIVcpzUS | 51 | 55 | 23 | 25 | 45 | 33 |
| SIVcpzANT | 48 | 52 | 22 | 22 | 46 | 31 |
| SIVcol | 40 | 49 | 19 | 19 | 30 | 26 |
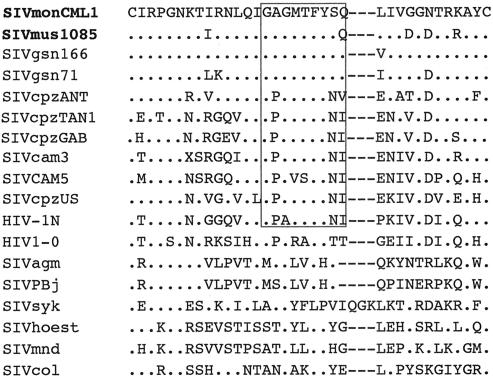
Although SIVmon, SIVmus, and SIVgsn have similar genome organizations (i.e., the presence of a vpu homologue) and are more closely related to SIVgsn than to any other known SIV across the genome, the lowest protein identities were observed with their predicted Vpu proteins (26.3 and 40.7% identity, respectively). High variability has been described for Vpu proteins among HIV-1 group M, O, and N and SIVcpz isolates. Hence, the comparison of Vpu sequences from several HIV-1 group M isolates identified only seven residues (W23, Q36, E51, D52, G54, N55, and G59; positions according to a consensus HIV-1 M sequence), plus the initiating methionine and the two conserved serines, as invariant. An attempt to align the Vpu amino acid sequences of SIVmon, SIVmus, SIVcpz, SIVgsn, and HIV-1 (M, N, and O) in order to look for the presence of known functional domains required for HIV-1 Vpu activity is shown in Fig. 2. Interestingly, three (W, D, and G) of the seven invariant residues were present in the Vpu protein of SIVmon. The two serines were also conserved, albeit not at the same positions (DSGXXXXS instead of the consensus DSGXXS for HIV-1), but as in HIV-1 M, N, and O and SIVcpz (except SIVcpzANT and SIVcpzTAN1 [only one]), they represent two potential casein kinase II phosphorylation sites. It has been reported that phosphorylation of serine residues in this conserved motif was involved in the downregulation of CD4 from the cell surface. In SIVmus Vpu, this motif was not found, and only one of the two conserved serines had no potential casein kinase II phosphorylation sites, as in the Vpu protein of SIVgsn. Finally, a GFXNP motif located at the C terminus of the protein and found in HIV-1 group O, SIVcpz, and SIVgsn was also present in SIVmon but was replaced by AFXNP in SIVmus.
FIG. 2.
Alignment of Vpu amino acid sequences of SIVmon-99CMCML1, SIVmus-01CM1085, SIVgsn-99CM71, SIVgsn-99CM166, SIVcpzANT, SIVcpzTAN1, SIVcpzGAB, SIVcpzCAM5, SIVcpzUS, HIV1-YBF30, HIV1-MVP5180, and HIV1-MD. Identical residues are in solid boxes. Similar amino acids are in shaded boxes. The dashes indicate gaps introduced to optimize alignment.
The similarity among SIVmon, SIVmus, and SIVgsn is also quite evident in an alignment of the surface unit part of the Env proteins of the three viruses: 18 of 18 cysteine residues and 14 of the 22 potential N-linked glycosylation sites were conserved. The V3 loop analog and the CD4 binding domain were also remarkably conserved (between one and five amino acid changes and five amino acid changes, respectively). In these two regions, the level of conservation was comparable to that observed between the two SIVgsn strains (8). The remarkable identity among the V3 loop sequences of SIVmon, SIVmus, and SIVgsn, as well as with those of SIVcpz and HIV1-N, is illustrated in an alignment shown in Fig. 3.
FIG. 3.
Amino acid alignment of the V3 loop region of gp120 from SIVmon-99CMCML1, SIVmus-01CM1085, and representatives of all SIV-HIV lineages. The crown region is boxed. The dashes indicate gaps introduced to optimize alignment, and dots indicate identical residues.
Phylogenetic relationships to other SIVs.
To examine in more detail the relationship of SIVmon and SIVmus to other primate lentiviruses, we performed diversity plot analyses. A gap-stripped concatenated alignment of the nonoverlapping regions of the gag, pol, vif, env, and nef genes of SIVmon, SIVmus, and representatives of other primate lentivirus lineages was generated, as both nucleotide and translated amino acid sequences. Since saturation always occurred at the third codon positions for the five genes, implying that only the first-plus-second codon position alignment was phylogenetically informative, we removed the third codon position (8). Therefore, we used either the amino acid alignment or the first-plus-second codon position nucleotide alignment for our analyses.
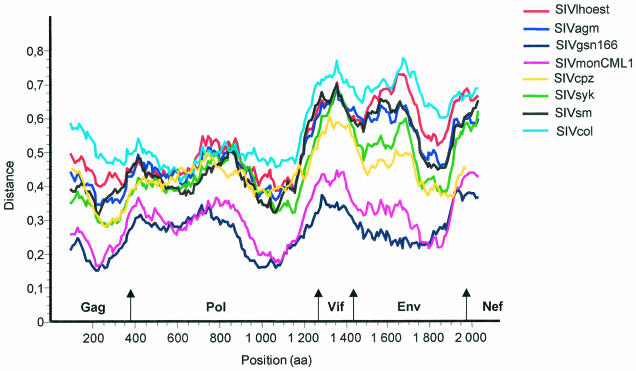
For the diversity plot analysis, the percent sequence diversity between sequence pairs was calculated for a window of 200 aa moved in steps of 10 aa along the alignment. We performed these analyses with group sequences or with individual sequences, since it has been reported that it would probably be more accurate to use a group of sequences rather than a single representative. All of the resulting diversity plots confirmed that SIVmon and SIVmus are most closely related to SIVgsn in all genome regions (Fig. 4). As for other viruses, SIVmon and SIVmus were highly divergent from SIVcol and equidistantly related to SIVlhoest, SIVagm, SIVsm, SIVcpz, and SIVsyk from Gag to Vif, while in the envelope, SIVmon and SIVmus were clearly more closely related to SIVcpz and then to SIVsyk than to the other SIVs. Similar patterns were obtained when individual sequences or a group of sequences per lineage with a threshold of 50% for the consensus were used. In order to determine if SIVmon and SIVmus display the same apparent mosaic structure as SIVgsn, we performed bootscanning analysis using 500-bp windows on the codon-aligned nucleotide sequence without the SIVgsn sequence. As illustrated in Fig. 5, the bootscan plots of SIVmon and SIVmus against representatives of the six major SIV lineages were slightly different but comparable, with the 5′ end region related to SIVsyk and SIVcol and the 3′ end related to SIVcpz.
FIG. 4.
Diversity plot analysis comparing SIVmus-01CM1085 with SIVmon-99CMCML1 and representatives of the six major lineages of the primate lentiviruses, i.e., SIVcpz, SIVmnd, SIVsyk, SIVsm, SIVagm, and SIVcol plus SIVgsn. The protein sequence differences are plotted for windows of 200 aa moved in steps of 10 aa.
FIG. 5.
Bootscanning of SIVmon-99CMCML1 (A) and SIVmus-01CM1085 (B) against representatives of the six major SIV lineages. Bootscan analysis was performed on the concatenated nucleotide alignment with the nonoverlapping regions of gag, pol, env, vif, and nef. Since saturation always occurred at the third codon position for all genes, the bootscan analysis was performed on first and second codon positions, with a sliding window of 500 nt with a 20-nt step increment and 1,000 bootstrap replicates. Below the graph, the bar shows the locations of the gene regions.
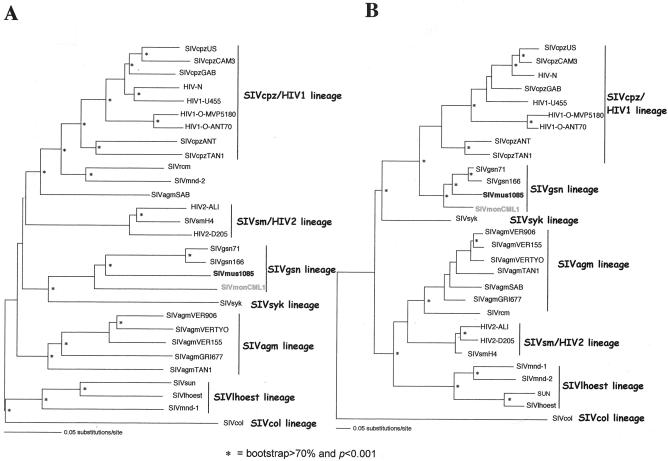
To confirm the close relationship among SIVmon, SIVmus, and SIVgsn, phylogenetic-tree analyses of the concatenated nucleotide alignment with the nonoverlapping regions of the gag, pol, and env genes were generated using the best-fitting nucleotide substitution model evaluated through the hierarchical likelihood ratio test implemented in the Modeltest program version 3.06 (see the figure legends for the parameters used). Tree topologies were derived by both neighbor-joining and maximum-likelihood analyses. As expected, these analyses revealed that SIVmon, SIVmus, and SIVgsn always clustered together, with SIVmon being the most external taxon of this group, but that their phylogenetic positions were significantly discordant in regard to the initial described primate lentivirus lineages in the different regions. In the gag and pol genes, SIVmon and SIVmus clustered with SIVgsn and the SIVsyk lineage, with 100% bootstrap support for neighbor joining and a P value of <0.001 for maximum likelihood (Fig. 6A), whereas in the env gene they clustered with the SIVcpz lineage, with 80% bootstrap support and a P value of <0.001 for neighbor joining and maximum likelihood, respectively (Fig. 6B).
FIG. 6.
Unrooted maximum-likelihood trees of the pol (A) and env (B) genes, including 26 SIV-HIV strains of the six major SIV lineages, as well as other SIVs for which full-length sequences are available (SIVrcm, SIVmnd2, and SIVgsn) and the new SIVmon-99CMCML1 and SIVmus-01CM1085 isolates. The pol and env trees were inferred using only first and second codon positions, with the TrN + Inv + Γ (α = 1.3854; I = 0.1862) and the HKY + Inv + Γ (α = 1.1796; I = 0.2508; Ti/Tv = 0.9724) model of nucleotide substitution, respectively. Horizontal branch lengths are drawn to scale, with the bar representing 0.1 nucleotide replacements per site. The neighbor-joining method gives a similar tree topology. An asterisk along a branch indicates that the branch has a P value of <0.001 in the maximum-likelihood analysis and a percentage of bootstrap replicates (out of 1,000) of >70 in the neighbor-joining tree. α, shape parameter of the Γ distribution; I, proportion of invariable sites; Ti/Tv, expected transition/transversion ratio.
SIVmon, SIVmus, and SIVgsn are members of the same primate lentivirus lineage.
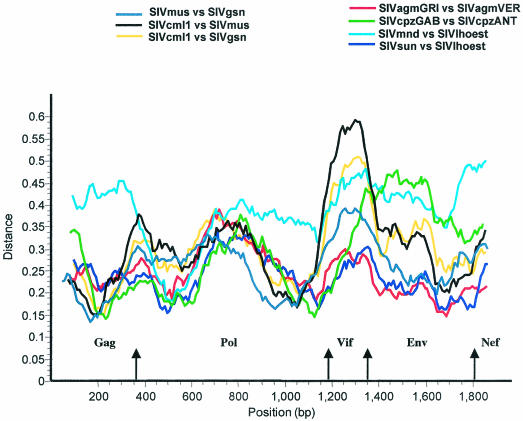
As shown in Table 3, the extent of amino acid identities among SIVmon, SIVmus, and SIVgsn for all genes were comparable overall to the values observed among other SIV strains from the same lineage, such as SIVlhoest-SIVsun-SIVmnd1 for the l'Hoest lineage, SIVagmVER-SIVagmGRI for the SIVagm lineage, or SIVcpzGAB-SIVcpzANT for the SIVcpz lineage. As can be seen in Fig. 7, the diversity plots show that in the 5′ part of the genome, the extent of sequence difference between SIVmon and SIVgsn was similar to that observed between SIVlhoest and SIVsun or between two SIVagm or two SIVcpz lineages. In Vif, except for SIVmon-SIVmus, the divergence is comparable to SIVlhoest-SIVmnd1, while in the envelope, the divergence is intermediate between SIVlhoest-SIVsun and two SIVagm strains and SIVmnd1-SIVlhoest and SIVcpzANT-SIVcpzGAB.
TABLE 3.
Percent amino acid identities within primate lentivirus lineages
| Comparison | % Identity in:
|
|||||
|---|---|---|---|---|---|---|
| Gag | Pol | Vif | Vpr | Env | Nef | |
| SIVmon-SIVgsn | 67.9 | 67.3 | 49.6 | 63 | 60.8 | 69.6 |
| SIVmon-SIVmus | 67.8 | 68.2 | 45.3 | 64.4 | 59.3 | 63 |
| SIVmus-SIVgsn | 70.4 | 70.2 | 54.4 | 74.4 | 64 | 68.3 |
| SIVmnd-SIVhoest | 54 | 67 | 38 | 65 | 51 | 44 |
| SIVlhoest-SIVsun | 71 | 73 | 52 | 63 | 67 | 56 |
| SIVagmTAN-SIVagmVER | 73 | 79 | 60 | 82 | 66 | 63 |
| SIVagmGRI-SIVagmVER | 71 | 71 | 53 | 70 | 68 | 70 |
| SIVcpzANT-SIVcpzGAB | 68 | 74 | 57 | 64 | 51 | 53 |
FIG. 7.
Diversity plot analysis comparing SIVmus-SIVmon, SIVmus-SIVgsn, and SIVmus-SIVgsn with SIVagmGRI-SIVagmVER, SIVmnd1-SIVlhoest, SIVsun-SIVlhoest, and SIVcpzGAB-SIVcpzANT. The protein sequence difference is plotted for windows of 200 aa moved in steps of 10 aa.
Therefore, taking all these analyses together, we can conclude that SIVmon, SIVmus, and SIVgsn most likely originated from a common SIV ancestor and that they represent a new primate lentivirus lineage, which we currently designate the SIVgsn lineage, consistent with the name of the first isolate of the lineage.
Relationship of new representatives of the SIVgsn lineage with SIVcpz isolates obtained from Pan troglodytes troglodytes and Pan troglodytes schweinfurthii.
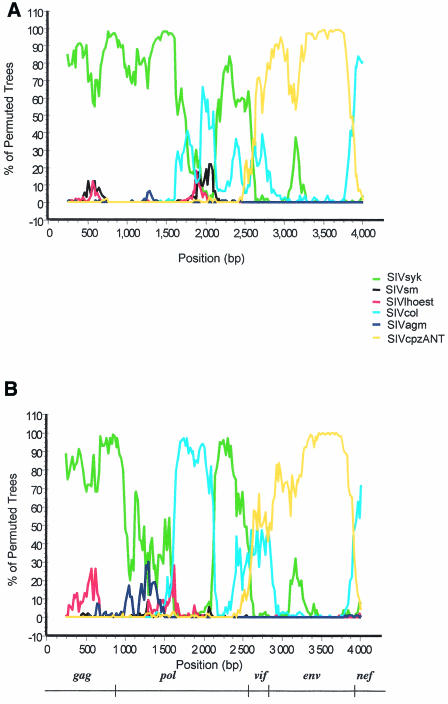
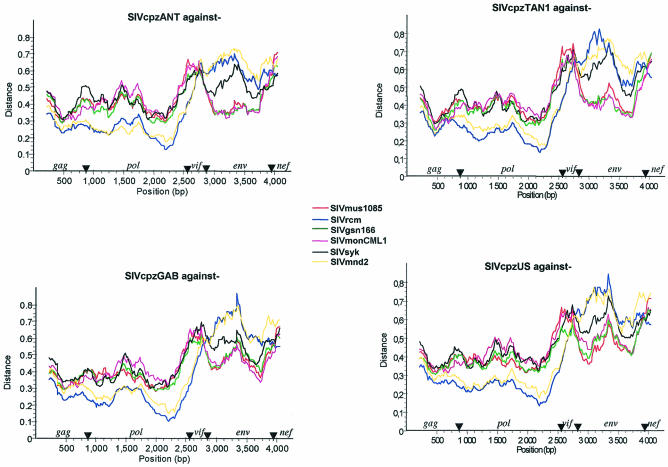
As already indicated, in the phylogenetic trees, the envelopes of SIVmon, SIVmus, and SIVgsn cluster with the SIVcpz lineage, branching off before SIVcpzANT-SIVcpzTAN1. However, since the position of SIVcpz depends on the gene examined (pol or env [Fig. 6]), we further investigated the recombinant structure of SIVcpz with these new SIV isolates. We performed diversity plot analyses of SIVcpz isolates obtained from the two chimpanzee subspecies known to be infected (SIVcpz, SIVcpz-GAB, and SIVcpzUS from P. t. troglodytes in west central Africa and SIVcpz-ANT and SIVcpz-TAN1 from P. t. schweinfurthii in east central Africa) against SIVmon, SIVmus, SIVgsn, SIVsyk, SIVmnd2, and SIVrcm. The recombinant origin of SIVcpz is clearly illustrated in Fig. 8, showing that across gag, pol, and vif, SIVcpz is more closely related to SIVrcm or SIVmnd2, while across env, SIVcpz is closest to SIVmon, SIVmus, and SIVgsn. Two divergent SIVcpz lineages are found in the two chimpanzee subspecies, but similar patterns of genetic relationships were obtained with SIVcpz from P. t. schweinfurthii and from P. t. troglodytes as the query sequences, suggesting that these viruses are descendents of the same recombinant.
FIG. 8.
Diversity plots of concatenated nucleotide alignments with the nonoverlapping regions of gag, pol, env, vif, and nef comparing SIVcpz from P. t. schweinfurthii (SIVcpzANT and SIVcpzTAN1) and SIVcpz from P. t. troglodytes (SIVcpzUS and SIVcpzGAB1) to SIVmus-01CM1085, SIVgsn-99CM166, SIVmon-99CMCML1, SIVsyk, SIVrcmNG411, and SIVmnd2. Simplot analysis was performed on first and second codon positions with a sliding window of 450 nt moved in steps of 20 nt.
DISCUSSION
Since the discovery of HIV-1 and HIV-2, surveys of nonhuman primate species have indicated that related viruses are widespread in a large number of African primate species (18, 21). Thirty species with serological evidence of SIV infection have already been identified, and in 27 this was confirmed by partial or full-length genome sequencing, highlighting the increasing complexity of phylogenetic relationships among primate lentiviruses. Additional molecular characterizations of SIVs are necessary to elucidate the evolution of this group of viruses and also to study SIVs to which humans are exposed. In this report, we describe the full-length sequences of two novel primate lentiviruses, SIVmon and SIVmus, isolated from mona and mustached monkeys, respectively, from Cameroon. One other SIVmon strain and two additional SIVmus strains have been partially characterized from mona and mustached monkeys in Nigeria and Cameroon, respectively, indicating that these monkeys are the natural hosts for the viruses (22; J. P. Clewley, K. L. Barlow, J. V. Parry, and J. Lewis, presented at the 8th Annual HIV Dynamics and Evolution Meeting, Paris, France, April 27 to 29, 2001). Full-length sequence analysis of SIVmon and SIVmus in our study revealed that these viruses were most closely related to SIVgsn, previously described in greater spot-nosed monkeys from Cameroon (8). In all three SIVs, we documented the presence of a vpu homologue, and they were all related to SIVsyk in gag-pol and to SIVcpz in env. These three monkeys are phylogenetically related species belonging to different groups within the genus Cercopithecus, the C. mona, C. cephus, and C. mitis groups. The genus Cercopithecus comprises the largest number of species in the nonhuman primate classification (12), and it is one of the most infected by SIV. According to different authors, the C. lhoesti superspecies forms a small clade distinct from other Cercopithecus species (9; F. Bibollet-Ruche, X. Pourrut, V. Courgnaud, E. Mpoudi, J. Mwenda, V. Hirsch, E. Delaporte, M. Peeters, F. Gao, G. M. Shaw, and B. H. Hahn, Abstr. 7th Conf. Retrovir. Opportunistic Infect., abstr. 223, 2000). Therefore, excluding the SIVlhoest-SIVsun lineage, the other SIVs from Cercopithecus species characterized so far, such as SIVsyk from Sykes' monkeys, SIVdeb from de Brazza monkeys, SIVgsn, and now SIVmon and SIVmus, are, at least in part of their genomes (gag and pol), clearly more closely related to each other than to other SIVs infecting monkeys not belonging to Cercopithecus species. Furthermore, as was previously discussed at the time of the analysis of SIVgsn (8), in different phylogenetic trees for Gag, Pol, or Env, the position of the SIVcpz-HIV-1 lineage depends on the gene examined, and thus, without the SIVcpz-HIV-1 lineage, SIVsyk, SIVdeb, SIVgsn, and now SIVmon and SIVmus cluster together (the last three always form a separate subcluster). This suggests that SIVgsn-SIVmon-SIVmus are more closely related to SIVsyk-SIVdeb across gag, pol, and consequently env than to the remaining lineages, implying a possible host-dependent evolution in Cercopithecus species. However, an interesting and intriguing finding is that these five viruses display two different genomic organizations with the presence of an additional accessory gene, vpu, for SIVgsn-SIVmus-SIVmon or without it for SIVsyk-SIVdeb. The origin of vpu has not yet been elucidated, and before the characterization of SIVgsn, this feature was characteristic of members of the SIVcpz-HIV-1 lineage. There are different modes of acquisition of a new gene during the evolution of a genome, such as recombination, duplication, and insertion. Thus, the identification of SIVs having a vpu homologue in certain monkeys belonging to the genus Cercopithecus could suggest that vpu is a new gene acquired during the evolution of SIV in some Cercopithecus species. Obviously, an alternative possibility could be that certain SIVs lost the vpu gene during evolution. vpu is not essential for replication, and extensive genetic diversity has been observed between sequences of vpu genes in HIV-1 and SIVcpz. Vpu has two known distinct biological functions, i.e., regulation of virus release from infected cells and induction of CD4 degradation (15). It has been demonstrated that the latter function requires phosphorylation of two conserved serine residues in a conserved DSGXXS motif in HIV-1 Vpu. Although we do not have direct evidence proving that the SIVmon vpu homologue is functional, and bearing in mind that proteins with divergent sequences can have similar functions, it is tempting to imagine that, unlike SIVmus-SIVgsn Vpu, SIVmon Vpu may have the ability to downregulate CD4. Indeed, SIVmon Vpu contains the motif DSGXXXXS, which represents two potential casein kinase II phosphorylation sites, which is not the case for SIVmus or SIVgsn Vpu. On the other hand, it has been reported that the HIV-2 Env protein is a functional complement to HIV-1 Vpu that enhances virion release (5a). Thus, it will be interesting to determine whether this function has evolved in HIV-2 Env in place of Vpu or whether it reflects a function that has been lost in HIV-1 because of Vpu. Moreover, it will be interesting to investigate if the envelopes of SIVmon, SIVmus, and SIVgsn contain duplicate Vpu-like activities (i.e., within Vpu itself and within the envelope) or only within Env or Vpu. In any case, studies of the functionality of these three new proteins are necessary in order to know whether these biological properties are a specific characteristic of HIV-1 acquired during adaptation and evolution in a new host or whether they are ancestral activities.
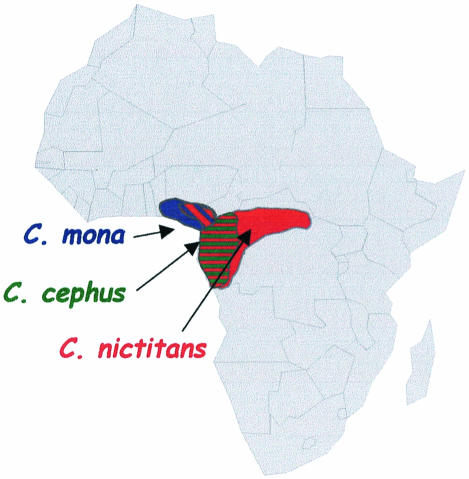
Overall, it seems that, with the exclusion of SIVs from the l'Hoest lineage, SIVs from the genus Cercopithecus are more closely related to each other than to other SIVs. Another important finding regarding the evolution of lentiviruses in these Cercopithecus species is the fact that SIVs which cluster together were not obtained from monkeys belonging to the same evolutionary group within the Cercopithecus species. More precisely, SIVsyk and SIVgsn, both isolated in representatives of the C. mitis group, are each more closely related to SIVs characterized from monkeys belonging to other evolutionary groups. In other words, SIVsyk is more closely related to SIVdeb from the de Brazza group, and SIVgsn is more closely related to SIVmon from the C. mona group and SIVmus from the C. cephus group. This is not in accordance with the hypothesis of host-dependent evolution of viruses infecting related species from the Cercopithecus genus. The close relationship among SIVmon, SIVmus, and SIVgsn could therefore be explained by possible ancient cross-species transmissions among mona, mustached, and greater spot-nosed monkeys, since these monkeys share the same habitats (Fig. 9). Furthermore, it has been reported that the mustached monkey species forms mixed-species associations with greater spot-nosed and mona monkeys (11), thus allowing an exchange of SIVs among them.
FIG. 9.
Map of Africa showing the natural ranges of mona monkeys (C. mona), mustached monkeys (C. cephus), and greater spot-nosed monkeys (C. nictitans).
So far, only two subspecies of chimpanzees (P. t. troglodytes and P. t. schweinfurthii) are known to be infected by SIVcpz (10), and the absence of SIV infection in other subspecies may suggest that chimpanzees acquired SIVcpz relatively recently (25). As SIVcpz infection among chimpanzees in the wild is not very well known but seems to be less frequent than in smaller monkey species and unevenly distributed (24), and as chimpanzees are known to prey on small monkeys, we previously proposed the hypothesis, which has been confirmed by extensive phylogenetic analyses, that a recombination event between ancestral SIVs from the family Cercopithecinae could be the origin of the current SIVcpz that is widespread in the chimpanzee population (2, 8). Although we cannot exclude the possibility that this recombination occurred in an as-yet-unidentified species, the characterization of the two new viruses SIVmon and SIVmus strengthens this hypothesis by substantially increasing the number of potential viruses closely related to SIVgsn spreading in different Cercopithecus species (Fig. 8).
Since the current habitats of mustached and mona monkeys do not overlap those of eastern chimpanzees, it will be interesting to characterize SIVs in Cercopithecus species in regions overlapping the habitat of eastern chimpanzees. It could also be possible that SIVcpz was introduced into the P. t. troglodytes population in west central Africa and further spread eastward into the P. t. schweinfurthii population.
It is difficult to determine if the predominant mode of viral transmission in lentivirus evolution is cospeciation or cross-species transmission, since cospeciation can be masked by subsequent successful interspecies transmissions or, conversely, successful interspecies transmissions in related species could be interpreted as cospeciation. Recently, it was shown in mandrills that two different SIVs can circulate in one primate species. In order to discover to what extent host-dependant evolution with a common SIV ancestor and/or cross-species transmissions between cohabiting primate species occurred, screening of geographically separated primate populations belonging to the same species will be necessary to determine to what extent cross-species infection, superinfection, and recombination occurred. Knowledge of primate behavior and past and recent geographic distributions of the different primate species could add important complementary information for the interpretation of SIV sequences.
All viruses are not equivalent for transmission and/or infectivity, and the factors contributing to successful simian-to-simian and simian-to-human transmission are not fully determined. Certainly, in most cases, these cross-species transmissions lead to a biological dead end. However, moving up the primate ladder in chimpanzees, recombination between coinfecting divergent SIVs from lower species may generate viruses with new biological properties, such as the ability to spread in humans.
REFERENCES
- 1.Allan, J. S., M. Short, M. E. Taylor, S. Su, V. M. Hirsch, P. R. Johnson, G. M. Shaw, and B. H. Hahn. 1991. Species-specific diversity among simian immunodeficiency viruses from African green monkeys. J. Virol. 65:2816-2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailes, E., F. Gao, F. Bibollet-Ruche, V. Courgnaud, M. Peeters, P. A. Marx, B. H. Hahn, and P. M. Sharp. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. [DOI] [PubMed] [Google Scholar]
- 3.Beer, B. E., E. Bailes, R. Goeken, G. Dapolito, C. Coulibaly, S. G. Norley, R. Kurth, J. P. Gautier, A. Gautier-Hion, D. Vallet, P. M. Sharp, and V. M. Hirsch. 1999. Simian immunodeficiency virus (SIV) from sun-tailed monkeys (Cercopithecus solatus): evidence for host-dependent evolution of SIV within the C. lhoesti superspecies. J. Virol. 73:7734-7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beer, B. E., B. T. Foley, C. L. Kuiken, Z. Tooze, R. M. Goeken, C. R. Brown, J. Hu, M. S. Claire, B. T. Korber, and V. M. Hirsch. 2001. Characterization of novel simian immunodeficiency viruses from red-capped mangabeys from Nigeria (SIVrcmNG409 and -NG411). J. Virol. 75:12014-12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bibollet-Ruche, F., A. Galat-Luong, G. Cuny, P. Sarni-Manchado, G. Galat, J. P. Durand, X. Pourrut, and F. Veas. 1996. Simian immunodeficiency virus infection in a patas monkey (Erythrocebus patas): evidence for cross-species transmission from African green monkeys (Cercopithecus aethiops sabaeus) in the wild. J. Gen. Virol. 77:773-781. [DOI] [PubMed] [Google Scholar]
- 5a.Bour, S., U. Schubert, K. Peden, and K. Strebel. 1996. The envelope glycoprotein of human immunodeficiency virus type 2 enhances viral particle release: a Vpu-like factor? J. Virol. 70:820-829. [DOI] [PMC free article] [PubMed]
- 6.Clewley, J. P., J. C. Lewis, D. W. Brown, and E. L. Gadsby. 1998. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J. Virol. 72:10305-10309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courgnaud, V., X. Pourrut, F. Bibollet-Ruche, E. Mpoudi-Ngole, A. Bourgeois, E. Delaporte, and M. Peeters. 2001. Characterization of a novel simian immunodeficiency virus from guereza colobus monkeys (Colobus guereza) in Cameroon: a new lineage in the nonhuman primate lentivirus family. J. Virol. 75:857-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Courgnaud, V., M. Salemi, X. Pourrut, E. Mpoudi-Ngole, B. Abela, P. Auzel, F. Bibollet-Ruche, B. Hahn, A. M. Vandamme, E. Delaporte, and M. Peeters. 2002. Characterization of a novel simian immunodeficiency virus with a vpu gene from greater spot-nosed monkeys (Cercopithecus nictitans) provides new insights into simian/human immunodeficiency virus phylogeny. J. Virol. 76:8298-8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dutrillaux, B., M. Muleris, and J. Couturier. 1988. Chromosomal evolution of Cercopithecinae. Cambridge University Press, Cambridge, United Kingdom.
- 10.Gao, F., E. Bailes, D. L. Robertson, Y. Chen, C. M. Rodenburg, S. F. Michael, L. B. Cummins, L. O. Arthur, M. Peeters, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397:436-441. [DOI] [PubMed] [Google Scholar]
- 11.Gautier, J. P., and A. Gautier-Hion. 1969. Les associations polyspecifiques chez les Cercopithecidae du Gabon. Terre Vie 23:164-201. [Google Scholar]
- 12.Groves, C. 2001. Primate taxonomy. Smithsonian Institution Press, Washington, D.C.
- 13.Hahn, B. H., G. M. Shaw, K. M. De Cock, and P. M. Sharp. 2000. AIDS as a zoonosis: scientific and public health implications. Science 287:607-614. [DOI] [PubMed] [Google Scholar]
- 14.Hu, J., W. M. Switzer, B. T. Foley, D. L. Robertson, R. M. Goeken, B. T. Korber, V. M. Hirsch, and B. E. Beer. 2003. Characterization and comparison of recombinant simian immunodeficiency virus from drill (Mandrillus leucophaeus) and mandrill (Mandrillus sphinx) isolates. J. Virol. 77:4867-4880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jabbar, M. A. 1995. The human immunodeficiency virus type 1 Vpu protein: roles in virus release and CD4 downregulation. Curr. Top. Microbiol. Immunol. 193:107-120. [DOI] [PubMed] [Google Scholar]
- 16.Jin, M. J., H. Hui, D. L. Robertson, M. C. Muller, F. Barre-Sinoussi, V. M. Hirsch, J. S. Allan, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Mosaic genome structure of simian immunodeficiency virus from west African green monkeys. EMBO J. 13:2935-2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin, M. J., J. Rogers, J. E. Phillips-Conroy, J. S. Allan, R. C. Desrosiers, G. M. Shaw, P. M. Sharp, and B. H. Hahn. 1994. Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J. Virol. 68:8454-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lowenstine, L. J., N. C. Pedersen, J. Higgins, K. C. Pallis, A. Uyeda, P. Marx, N. W. Lerche, R. J. Munn, and M. B. Gardner. 1986. Seroepidemiologic survey of captive Old-World primates for antibodies to human and simian retroviruses, and isolation of a lentivirus from sooty mangabeys (Cercocebus atys). Int. J. Cancer 38:563-574. [DOI] [PubMed] [Google Scholar]
- 19.Mathews, D. H., J. Sabina, M. Zuker, and D. H. Turner. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288:911-940. [DOI] [PubMed] [Google Scholar]
- 20.Muller, M. C., N. K. Saksena, E. Nerrienet, C. Chappey, V. M. Herve, J. P. Durand, P. Legal-Campodonico, M. C. Lang, J. P. Digoutte, A. J. Georges, et al. 1993. Simian immunodeficiency viruses from central and western Africa: evidence for a new species-specific lentivirus in tantalus monkeys. J. Virol. 67:1227-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohta, Y., T. Masuda, H. Tsujimoto, K. Ishikawa, T. Kodama, S. Morikawa, M. Nakai, S. Honjo, and M. Hayami. 1988. Isolation of simian immunodeficiency virus from African green monkeys and seroepidemiologic survey of the virus in various non-human primates. Int. J. Cancer 41:115-122. [DOI] [PubMed] [Google Scholar]
- 22.Peeters, M., V. Courgnaud, B. Abela, P. Auzel, X. Pourrut, F. Bibollet-Ruche, S. Loul, F. Liegeois, C. Butel, D. Koulagna, E. Mpoudi-Ngole, G. M. Shaw, B. H. Hahn, and E. Delaporte. 2002. Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg. Infect. Dis. 8:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 24.Santiago, M. L., M. Lukasik, S. Kamenya, Y. Li, F. Bibollet-Ruche, E. Bailes, M. N. Muller, M. Emery, D. A. Goldenberg, J. S. Lwanga, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, D. P. Watts, A. E. Pusey, D. A. Collins, R. W. Wrangham, J. Goodall, J. F. Brookfield, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2003. Foci of endemic simian immunodeficiency virus infection in wild-living eastern chimpanzees (Pan troglodytes schweinfurthii). J. Virol. 77:7545-7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Santiago, M. L., C. M. Rodenburg, S. Kamenya, F. Bibollet-Ruche, F. Gao, E. Bailes, S. Meleth, S. J. Soong, J. M. Kilby, Z. Moldoveanu, B. Fahey, M. N. Muller, A. Ayouba, E. Nerrienet, H. M. McClure, J. L. Heeney, A. E. Pusey, D. A. Collins, C. Boesch, R. W. Wrangham, J. Goodall, P. M. Sharp, G. M. Shaw, and B. H. Hahn. 2002. SIVcpz in wild chimpanzees. Science 295:465. [DOI] [PubMed] [Google Scholar]
- 26.Sheline, C. T., L. H. Milocco, and K. A. Jones. 1991. Two distinct nuclear transcription factors recognize loop and bulge residues of the HIV-1 TAR RNA hairpin. Genes Dev. 5:2508-2520. [DOI] [PubMed] [Google Scholar]
- 27.Souquiere, S., F. Bibollet-Ruche, D. L. Robertson, M. Makuwa, C. Apetrei, R. Onanga, C. Kornfeld, J. C. Plantier, F. Gao, K. Abernethy, L. J. White, W. Karesh, P. Telfer, E. J. Wickings, P. Mauclere, P. A. Marx, F. Barre-Sinoussi, B. H. Hahn, M. C. Muller-Trutwin, and F. Simon. 2001. Wild Mandrillus sphinx are carriers of two types of lentivirus. J. Virol. 75:7086-7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J., D. Higgins, and T. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 11:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Rensburg, E. J., S. Engelbrecht, J. Mwenda, J. D. Laten, B. A. Robson, T. Stander, and G. K. Chege. 1998. Simian immunodeficiency viruses (SIVs) from eastern and southern Africa: detection of a SIVagm variant from a chacma baboon. J. Gen. Virol. 79:1809-1814. [DOI] [PubMed] [Google Scholar]
- 30.Xia, X., and Z. Xie. 2001. DAMBE: software package for data analysis in molecular biology and evolution. J. Hered. 92:371-373. [DOI] [PubMed] [Google Scholar]
- 31.Zuker, M., D. H. Mathews, and D. H. Turner. 1999. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide, p. 11-43. RNA Biochemistry and Bio/Technology, NATO ASI series. Kluwer Academic Publishers, Dordrecht, The Netherlands.