Abstract
The wild mouse species most closely related to the common laboratory strains contain proviral env genes of the xenotropic/polytropic subgroup of mouse leukemia viruses (MLVs). To determine if the polytropic proviruses of Mus spretus contain functional genes, we inoculated neonates with Moloney MLV (MoMLV) or amphotropic MLV (A-MLV) and screened for viral recombinants with altered host ranges. Thymus and spleen cells from MoMLV-inoculated mice were plated on Mus dunni cells and mink cells, since these cells do not support the replication of MoMLV, and cells from A-MLV-inoculated mice were plated on ferret cells. All MoMLV-inoculated mice produced ecotropic viruses that resembled their MoMLV progenitor, although some isolates, unlike MoMLV, grew to high titers in M. dunni cells. All of the MoMLV-inoculated mice also produced nonecotropic virus that was infectious for mink cells. Sequencing of three MoMLV- and two A-MLV-derived nonecotropic recombinants confirmed that these viruses contained substantial substitutions that included the regions of env encoding the surface (SU) protein and the 5′ end of the transmembrane (TM) protein. The 5′ recombination breakpoint for one of the A-MLV recombinants was identified in RNase H. The M. spretus-derived env substitutions were nearly identical to the corresponding regions in prototypical laboratory mouse polytropic proviruses, but the wild mouse infectious viruses had a more restricted host range. The M. spretus proviruses contributing to these recombinants were also sequenced. The seven sequenced proviruses were 99% identical to one another and to the recombinants; only two of the seven had obvious fatal defects. We conclude that the M. spretus proviruses are likely to be recent germ line acquisitions and that they contain functional genes that can contribute to the production of replication-competent virus.
Laboratory mice contain numerous proviruses related to the mouse leukemia viruses (MLVs), now classed as gammaretroviruses. These proviruses include the two major host range subgroups, ecotropic and xenotropic/polytropic, defined by env sequence variation and receptor usage. The use of type-specific hybridization probes shows that the common inbred strains carry, on average, zero to five copies of ecotropic env genes, most of which are associated with full-length proviruses that can be expressed as infectious virus (14, 21). Proviral copies of the nonecotropic xenotropic and polytropic MLV env genes are present in larger numbers, with more than 30 copies of each type being distributed over the laboratory mouse genome (8). Very few of the xenotropic proviruses can produce infectious virus (19), while both xenotropic and polytropic proviruses contribute to the generation of pathogenic mink cell focus-forming (MCF) viruses following recombination with replication-competent ecotropic virus (11, 27, 31).
The wild mouse species present a different picture. Few wild mouse species harbor infectious virus that are related to the MLVs found in the common strains of laboratory mice (20). The AKV-type ecotropic MLV has been found in Mus musculus molossinus and M. m. musculus of China (13), and viruses of xenotropic host range have also been isolated from M. m. molossinus (4). Viruses with ecotropic host range and divergent env genes have also been found in Asian and Californian mice (the Fv4-related viruses) (9, 13) and in the European species M. spicilegus (formerly M. hortulanus) (36). The AKV- and Fv4-type env genes, but not the M. spicilegus virus, are endogenous in their host species.
Many of the wild mouse species closely related to laboratory strains contain proviral xenotropic/polytropic env genes, but in contrast to the laboratory strains that have multiple copies of both env types, the xenotropic and polytropic proviruses are largely segregated into different Mus species (3, 20). Xenotropic MLV env genes are the predominant type in M. m. musculus of Eastern Europe and Asiatic Russia and in M. castaneus and its natural hybrid, M. m. molossinus. The polytropic MLV env genes are found in M. m. domesticus of western Europe and throughout the New World and Australia. A more recent analysis of the nonecotropic proviruses of several wild mouse species determined that the wild mouse nonecotropic proviruses show greater genetic variation than their laboratory mouse counterparts (33).
Further studies of these wild mouse nonecotropic viruses can contribute to our understanding of the generation of oncogenic viruses in mice as well as provide insight into the coevolution of virus and host. Are the proviral elements in these species relics of ancient infections, or are they more recently acquired proviruses that still contain functional genes? In an effort to address such issues, we decided to focus on M. spretus because of its extensive use in genetics, its ability to interbreed with laboratory mouse strains (1), its simple provirus content (20), and its susceptibility to MLV infection (17, 18). In this study, we introduced exogenous virus into neonates and screened for recombinant viruses. We isolated recombinant viruses from all inoculated mice, and we show that these viruses contain substantial env substitutions that result in altered host range properties. We also show that the endogenous env genes in this mouse are highly homologous, suggesting a recent origin.
MATERIALS AND METHODS
Viruses, cells, and mice.
All virus stocks were obtained from J. W. Hartley (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Md.) and included the NB-tropic Moloney ecotropic MLV (MoMLV), amphotropic MLV (A-MLV; strain 4070A), the xenotropic isolate NFS-Th-1 (X-MLV), and Moloney HIX MCF MLV, which was originally obtained from P. J. Fischinger (National Cancer Institute, National Institutes of Health) (7).
An inbred line of M. spretus mice, SPRET/Ei, was obtained from The Jackson Laboratory (Bar Harbor, Maine). Neonatal SPRET/Ei mice were inoculated intraperitoneally with MoMLV or A-MLV. Inoculated mice were sacrificed 7 to 22 weeks after inoculation, and cell suspensions of spleen and thymus cells were assayed for infectious virus. Cells from MoMLV-inoculated mice were used to infect cultured SC-1 (10), M. dunni (22), and mink lung cells (ATCC CCL 64), and cells from A-MLV-inoculated mice were used to infect ferret cells (5). Infections were done in the presence of Polybrene (4 μg/ml; Aldrich, Milwaukee, Wis.). Cells were passaged and tested for infectious virus by testing for reverse transcriptase (RT). RT-positive cultures were tested for ecotropic virus by examining plaque formation on rat XC cells (28) and for nonecotropic virus by focus formation on mink S+L− cells (26). For the XC test, mouse cells were irradiated 4 days after virus infection and overlaid with 106 XC cells/plate. Plates were stained 3 days later and examined for plaques of syncytia. For the S+L− assay, infected mouse, mink, or ferret cells were irradiated and overlaid with 6 × 105 mink S+L− cells, and foci were counted 5 to 7 days later. In a few cases, the titer of nonecotropic virus was determined after we infected various mouse cell lines and mink, ferret, and canine kidney cells (MDCK; ATCC CCL 34). These cells were obtained from J. W. Hartley.
For the RT assay, samples of culture fluid were centrifuged for 5 min at 2,000 to 3,000 rpm in a Sorall Biofuge (Kendro Laboratory Products) to remove cells. RT activity in the supernatants was then measured as described by Wilson and Eiden (37).
Southern blotting.
High-molecular-weight DNA was isolated from virus-infected cultures by standard protocols. DNAs were digested with restriction enzymes, electrophoresed on 0.4% agarose gels, transferred to nylon membranes (Hybond N+; Amersham, Piscataway, N.J.), and hybridized with radiolabeled probes. The hybridization probes represented env-specific segments for MoMLV (MOenv probe), nonecotropic xenotropic/polytropic MLVs (XMenv), MCF (polytropic) MLV (MCFenv), and A-MLV (Aenv) (Fig. 1). The DNA segments used as hybridization probes were amplified from high-molecular-weight DNAs extracted from virus-infected cells by using the following PCR primer pairs: for the MOenv probe (216 bp), 5′-GGACAAGATCCAGGGCTTACA-3′ (forward primer) and 5′-TACTAAGTTTAGCAGCCTATT-3′ (reverse primer); for the XMenv probe (419-bp env of the polytropic virus Moloney HIX MCF MLV), 5′-TTCAATGTTACTTGGAGAGTT-3′ (forward primer) and 5′-GCATCGACCCCCCGGTGTGGC-3′ (reverse primer); for the MCFenv-specific probe (96 bp), 5′-TGGGGACAATGACCGATG-3′ (forward primer) and 5′-GGAGTGCGACACCCGAGT-3′ (reverse primer); and for the Aenv-specific probe (140 bp), 5′-ACCGCCAATGCCACCTCCCT-3′ (forward primer) and 5′-GCGGGGTACTTGCAGCCATAC-3′ (reverse primer).
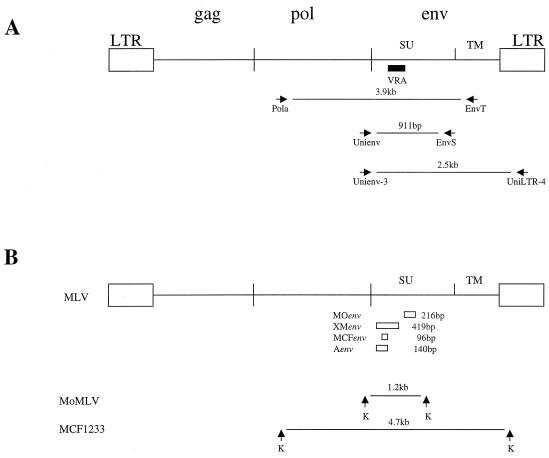
FIG. 1.
(A) General structure of MoMLV. The black box indicates the position of VRA in SU in env. The arrows identify the PCR primers and their products. (B) The open boxes represent the four env-specific probes used for hybridization. The arrows represent the locations of restriction sites for KpnI (K) that flank the sequences corresponding to the env probes in MoMLV and the polytropic virus MCF1233.
The PCRs were carried out in a GeneAmp PCR system 9700 machine (PE Applied Biosystems, Foster City, Calif.). The PCR (20-μl final volume) contained 100 ng of the template, 5 U of TaqPlus Precision polymerase mixture (Stratagene, La Jolla, Calif.), and 20 pmol of the primers. The PCRs were performed for 30 cycles with a 30-s DNA denaturation step at 95°C, a 30-s annealing step at 55°C, and a 1-min extension step at 72°C.
Cloning and sequencing.
High-molecular-weight DNA was prepared from M. dunni, mink, or ferret cells that were infected with virus isolates and was used as the PCR substrate. Four primers were used to amplify the viral pol and env genes and the long terminal repeat (LTR) U3 region (Fig. 1). Primers Unienv-3 (5′-GGATACACGCCGCTCACGTA-3′) and UniLTR-4 (5′-CGGGCGACTCAGTCTATCGG-3′) have been described previously (34). The forward primer Pola (5′-TGCTACAGTACGTGGATGAC-3′) and the reverse primer EnvT (5′-TGGAGTTGCTCGAATTGTTTG-3′) were designed from conserved regions within the viral regions encoding RT and TM, respectively.
The PCRs were performed for 30 cycles with a 1-min DNA denaturation step at 95°C, a 1-min annealing step at 55°C, and a 3-min extension step at 68°C. The PCR products were cloned into the pCR2.1-TOPO vector (Invitrogen Co., Carlsbad, Calif.) and sequenced.
Isolation and sequencing of the M. spretus env proviral sequences.
SPRET/Ei genomic DNA was digested to completion with EcoRV, separated on 0.4% or 0.6% agarose gels, and transferred to nylon membranes. Hybridization with the MCF-specific env probe MCFenv (24) identified seven proviruses (Fig. 2A). DNA enriched for each fragment was gel purified with a gel extraction kit (QIAGEN, Valencia, Calif.) and used as a PCR substrate. The PCR (20-μl final volume) contained 0.5 μg of gel-purified DNA, 20 pmol each of the sense and antisense primers, and 2.5 U of AmpliTaq Gold DNA polymerase (PE Applied Biosystems). The reactions were performed for 35 cycles with a 30-s DNA denaturation step at 95°C, a 30-s annealing step, and a 1-min extension step at 72°C. The annealing temperature in the first cycle was 63°C and was subsequently reduced by 1°C each cycle for the next 8 cycles and was then maintained at 55°C for the remaining 27 cycles. The PCR primers used to amplify the M. spretus proviruses included Unienv-3 and UniLTR-4 (described above) and a reverse primer, EnvS (5′-CTCCCGTCCCAGGTTGT-3′) (Fig. 1).
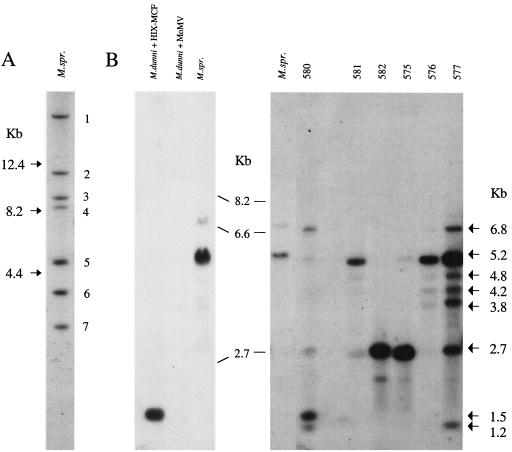
FIG. 2.
Southern blot analysis of DNAs from M. spretus (M.spr.) and from M. dunni cells infected with spleen cells from MoMLV-infected M. spretus. (A) M. spretus liver DNA digested with EcoRV and hybridized with MCFenv. The seven fragments are numbered by size. (B) KpnI-digested DNAs hybridized with XMenv (left) or MCFenv (right). Marker sizes are indicated between the two. The left blot includes M. spretus liver DNA and M. dunni cells infected with ecotropic MoMLV or Moloney HIX MCF MLV. The right blot includes M. spretus liver DNA and M. dunni cells infected with spleen cells from individual M. spretus mice inoculated with MoMLV as neonates. Lanes are identified by the mouse numbers given in Table 1.
The PCR products were cloned into the pCR2.1-TOPO vector and sequenced.
Nucleotide sequence accession numbers.
Sequences for the five recombinants produced in this study have been deposited in GenBank under accession numbers AY277733 to AY277737. Sequences for the seven provirus clones have been deposited under accession numbers AY277726 to AY277732.
RESULTS
Virus isolation from MoMLV-inoculated mice.
M. spretus mice were selected for inoculation because they carry few proviral env genes compared to inbred laboratory strains (20). These mice lack endogenous ecotropic and xenotropic MLV env sequences. The inbred M. spretus line used in this study, SPRET/Ei, carries only seven proviral env genes that are recognized by an MCF env-specific hybridization probe (Fig. 2A). Hybridization with the more broadly reactive XMenv probe identified the same seven fragments (data not shown). Previous studies had shown that cells from these mice are susceptible to infection by ecotropic, xenotropic, and polytropic MCF MLVs (17, 18). These mice also lack Fv1 restriction and carry a novel resistance allele at the Fv2 locus (17).
A total of 11 neonates in two SPRET/Ei litters was inoculated intraperitoneally with MoMLV, and one litter of seven mice was inoculated with A-MLV (Table 1). All but two of the MoMLV-inoculated mice survived. The surviving mice were sacrificed and examined for replicating virus 7 to 22 weeks after inoculation. Single-cell suspensions of spleen and thymus cells from the inoculated mice were plated on indicator cells: SC-1, M. dunni, and mink cells for the MoMLV-inoculated mice and ferret cells for the A-MLV-inoculated mice. These cells were selected to screen for host range variants of the inoculated virus, since MoMLV does not infect mink cells and infects M. dunni very inefficiently (22), and A-MLV infects ferret cells inefficiently (5).
TABLE 1.
SPRET/Ei mice inoculated with ecotropic MoMLV or A-MLV
| Inoculated virus | Mouse | No. of wks to sacrifice | Log10 ecotropic virus titera
|
|
|---|---|---|---|---|
| M. dunni | SC-1 | |||
| MoMLV | 575 | 22 | 0.8 | 3.3 |
| 576 | 20 | 2.7 | 4.4 | |
| 577 | 22 | 0.8 | 3.0 | |
| 578 | 22 | NT | NT | |
| 579 | 20 | 2.4 | 2.3 | |
| 580 | 17 | 2.8 | 3.5 | |
| 581 | 19 | 2.5 | 2.0 | |
| 582 | 19 | 0.1 | 3.9 | |
| 583 | 17 | 3.4 | 3.8 | |
| A-MLV | 496-1 | 9 | ||
| 496-2 | 9 | |||
| 496-4 | 10 | |||
| 496-5 | 10 | |||
| 497-1 | 7 | |||
| 497-2 | 7 | |||
| 497-3 | 9 | |||
Virus was collected from SC-1 or M. dunni cultures infected with spleen or thymus cells of virus-infected mice and titers were determined by XC plaque assay after infection of fresh SC-1 and M. dunni cells. NT, not tested.
Cultures were initially tested for evidence of replicating virus by the RT assay. All nine of the MoMLV-inoculated mice yielded virus that replicated in mouse and mink cells. High titers of ecotropic virus were detected by the XC plaque assay in infected SC-1 cells as well as in M. dunni cells. Virus replicating in the infected M. dunni cultures was collected, and titers were determined on SC-1 and M. dunni cells (Table 1). The ecotropic viruses isolated from four of these mice differed from MoMLV in host range in that they replicated as efficiently on M. dunni as on SC-1 cells. These M. spretus ecotropic isolates were not characterized further, but similar variants of MoMLV had been isolated from inoculated M. spicilegus in a related study, and the altered host range of these isolates was attributed to amino acid insertions or substitutions in the variable region A (VRA) within the surface (SU) region of env (16).
MoMLV-inoculated SPRET/Ei mice also produced virus of nonecotropic host range that replicated in mink and M. dunni cells. All of the mink cultures infected with spleen or thymus cells from the nine inoculated mice showed evidence of virus replication by the RT assay. Fluids collected from these cultures induced foci on mink S+ L− cells. Mink cell-grown virus from mouse 581 (Sp581 MLV) was additionally tested for its ability to infect a broad range of cell lines (Table 2). Like typical polytropic MLVs, the Sp581 isolate replicated efficiently in mink, M. dunni, and ferret cells but did not replicate at all in canine cells. However, its low titer on mouse SC-1 cells is more typical of xenotropic than polytropic MLVs.
TABLE 2.
Replication of Sp581 MLV on cells of different species
| Cell type | Log10 virus titera
|
|||
|---|---|---|---|---|
| Sp581 | A-MLV | MCF MLV | X-MLV | |
| Mink | 3.7 | 5.1 | 4.8 | 5.0 |
| SC-1 | 1.9 | 4.8 | 3.5 | 1.7 |
| M. dunni | 3.7 | 4.0 | 4.8 | 3.9 |
| Canine | 0b | 3.2 | 0.3 | 4.8 |
| Ferret | 3.2 | 3.2 | 4.7 | 4.2 |
Virus-infected cultures were UV irradiated 4 days after infection and overlaid with mink S+L− cells. Foci were counted 5 to 7 days later. Viruses used for comparison were 4070A amphotropic, Moloney HIX MCF MLV, and X-MLV.
No virus-induced foci were detected in 0.2 ml of undiluted virus stock.
The seven A-MLV-inoculated mice were screened for recombinants with expanded host ranges by using ferret cells, since these cells had been reported to be infectible by polytropic virus but not amphotropic virus (5). However, we observed much higher than expected titers of A-MLV replication in ferret cells (Table 2), precluding the use of this approach to selectively amplify variant viruses.
Southern blot analysis.
Southern blotting was done to determine the nature and number of different viruses present in the cultures infected with M. spretus isolates. High-molecular-weight DNA extracted from the virus-infected cells was analyzed with probes specific for the env regions of MoMLV (probe MOenv), nonecotropic MLVs (XMenv), and polytropic MLV (MCFenv) (Fig. 1B). Digestion with BamHI, SstI, and KpnI recruits proviral copies of these different MLVs into common env-containing fragments (shown for KpnI in Fig. 1B) that differ in size for MoMLV and the germ line proviruses of M. spretus (Fig. 2B).
Southern blotting detected viral sequences in all infected mink and M. dunni cultures. All M. dunni cultures contained the expected MoMLV ecotropic env-reactive KpnI, PvuII, and SstI fragments (data not shown). All M. dunni and mink cultures also contained fragments identified by the MCFenv or XMenv probes (Fig. 2B); some DNAs contained more than one of these nonecotropic env-containing fragments. The sizes of the fragments containing these nonecotropic env sequences differed from those generated by the endogenous M. spretus proviruses and from MoMLV. For example, cleavage with KpnI produces major env-reactive fragments of 5.2 kb in M. spretus and 1.2 kb in MoMLV. DNAs from the six SPRET/Ei virus-infected cultures shown in Fig. 2B contain MCFenv-reactive fragments in addition to six novel fragments. Results for all three restriction enzymes identified novel env-reactive fragments in six of the nine samples. These results indicate that the replicating viruses in these cultures are distinct from the endogenous SPRET/Ei proviruses and are likely to be recombinants.
This same strategy identified novel proviruses in ferret cultures infected with spleen or thymus cells from the A-MLV-inoculated mice. DNAs were prepared from these cultures and were analyzed by Southern blotting after cleavage with SstI, BamHI, and HindIII. A-MLV was detected in all seven of these ferret cultures, and all seven DNAs also contained novel proviral segments detectable with the MCFenv probe (data not shown).
Cloning and sequencing.
Five proviral env genes were cloned and sequenced from four of the virus-infected mink, M. dunni, or ferret cultures. The cultures selected for cloning contained both MoMLV and A-MLV recombinants, as well as proviruses likely to have different env substitutions based on Southern blotting. Two of the three MoMLV-derived recombinants were cloned from infected mink cells (mice 581 and 582), and the third was cloned from M. dunni cells (mouse 583). Two different A-MLV recombinants were cloned from a single ferret cell culture (mouse 496-5).
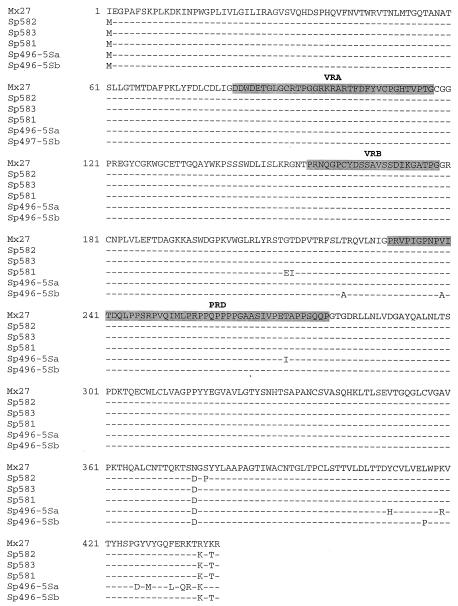
The entire proviral env gene with the U3 LTR was amplified from high-molecular-weight DNA by PCR with the primers Unienv-3 and UniLTR-4. The gene was cloned into the pCR2.1-TOPO vector and sequenced. Five of the seven sequenced PCR products were nonecotropic, and the remaining two samples were MoMLV. All five nonecotropic env genes were remarkably similar to one another and were most closely related to previously described polytropic MLVs (Fig. 3), such as the infectious MCF MLV MCF1233 (GenBank accession number U13766) and the endogenous provirus Mx27 (31). Except for the initiation codon, the 5′ ends of all five proviral SU genes were identical to that of Mx27. Amino acid substitutions were noted at only 12 positions in the 3′ ends of the SU genes. For one provirus, Sp496-5Sa, there were six amino acid substitutions relative to the sequence of Mx27 clustered at the end of SU. This region was identical to the A-MLV sequence, positioning the recombination breakpoint just proximal to this segment.
FIG. 3.
Comparison of the deduced amino acid sequences of five recombinant viruses with Mx27 (31). Variable regions corresponding to VRA, VRB, and the PRD are shaded. The recombination breakpoint for Sp496-5Sb lies in the carboxy terminus of SU in env; the sequence is identical to A-MLV in this region.
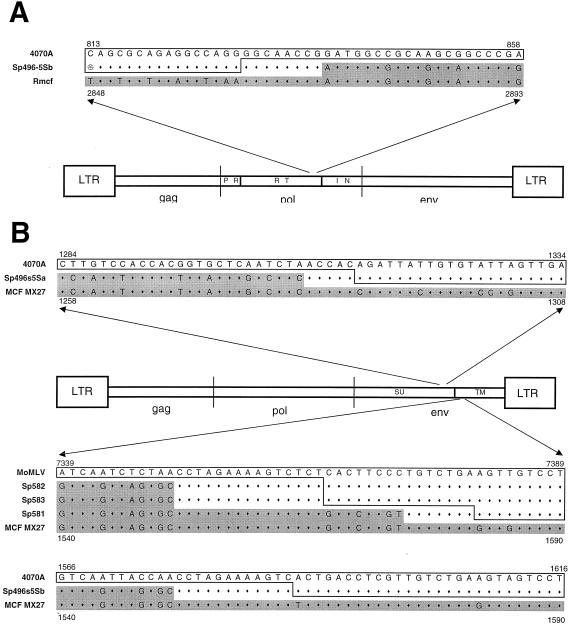
For the remaining four env genes, the entire SU gene was clearly M. spretus derived. The 3′ recombination breakpoints could be identified for all five clones (Fig. 4B). All three of the MoMLV recombinants and one of the A-MLV recombinants had breakpoints within a 30-bp segment of TM just downstream of the fusion domain. The breakpoint of the second A-MLV recombinant was in the 3′ end of SU, as indicated above.
FIG. 4.
Location of the recombination breakpoints in recombinant proviruses. (A) The 5′ recombination breakpoint for A-MLV derivative Sp496-5Sb lies within pol; (B) positions of the 3′ recombination breakpoint for all five env recombinants. The arrows position the sequences within the viral genome. The sequence for each recombinant is sandwiched between that of its progenitor (MoMLV or A-MLV; outlined) and the endogenous MCF sequence (grey shading) of Mx27 (31) or the Rmcf provirus (15). Numbers provide the locations for the indicated segments. The sequences of the M. spretus-derived recombinants are outlined or shaded to indicate parentage; the regions containing the recombination sites are neither outlined nor shaded.
The 5′ recombination site was determined for only one of these viruses, Sp496-5Sb. For this provirus, the Pola primer was used to amplify and clone 2.6 kb of pol along with the SU gene. Few pol genes associated with endogenous polytropic env genes have been sequenced, but the 3′ end of this pol gene was identical to that of the polytropic provirus associated with the Rmcf resistance gene (15). The region containing the recombination breakpoint could be identified in the 5′ end of RNase H (Fig. 4A).
Thus, four of the five recombinants had acquired endogenous sequences that included the complete wild mouse SU gene along with the segment of TM encoding the fusion domain. At least one of these recombinants also contained a wild mouse integrase (IN) gene.
Characterization of endogenous M. spretus sequences.
The five M. spretus recombinants acquired their novel env sequences by recombining with one or more of the seven SPRET/Ei polytropic proviruses (Fig. 1). To determine the extent of variability among these proviruses and to attempt to identify the progenitor(s) of the recombinant viruses, we sequenced the env genes of seven cloned proviruses from SPRET/Ei mice. One of these, SPR, represented an endogenous provirus cloned directly from SPRET/Ei DNA. The remaining six proviruses were cloned from gel-purified individual proviruses. EcoRV-digested SPRET/Ei DNA was electrophoresed on a 0.4% agarose gel, and DNA was extracted from gel slices containing the widely separated proviruses (Fig. 2A). Individual clones were sequenced from proviral fragments 3, 4, 5, and 6, along with two clones from fragment 7. The provirus in fragment 1 failed to amplify with the Unienv-3 and UniLTR-4 primers and was not analyzed. The fragment 2 provirus did not contain the internal EcoRI env site found in all of the recombinants and was not characterized further. Independent clones isolated from fragment 7 differed in the presence of the EcoRI site; two clones with and two without the site were selected for sequencing.
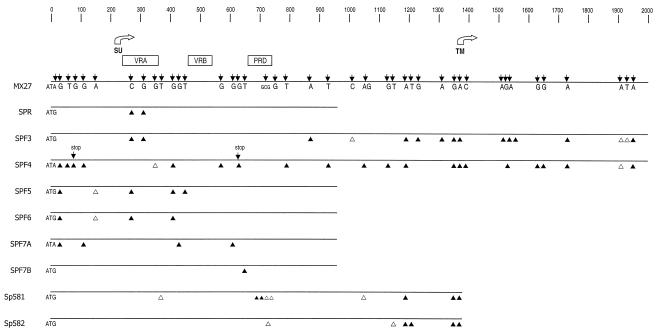
For the five selected proviruses, the 5′ env regions were amplified, cloned, and sequenced with primers Unienv-3 and EnvS, and for two of the proviruses, the entire SU gene was amplified with primers Unienv-3 and UniLTR-4. There was remarkably little sequence variation among these proviruses (Fig. 5). Base substitutions were identified at only 36 positions, of which all but five resulted in amino acid differences. More remarkably, only two of the proviruses contained sequence variations incompatible with function. Specifically, both of these env genes (SPF4 and SPF7A) did not begin with ATG, and one, SPF4, also contained two stop codons. Comparison of these seven proviral sequences with that of the recombinant viruses also showed minimal variation, although none was completely identical to any of the five recombinants. Pairwise comparisons between the endogenous proviruses and the recombinants identified a range of 3- to 15-bp differences. This finding suggests that the progenitor of these recombinants may have been one of the two unsequenced fragments, fragments 1 and 2. Alternatively, these differences might be due to rare sequencing errors, PCR-generated errors, or substitutions acquired by the recombinants during their multiple rounds of in vivo and in vitro replication.
FIG. 5.
Nucleotide sequence comparison of endogenous M. spretus nonecotropic proviruses. The entire env sequence was determined for two proviruses; only the 5′ half of env was sequenced for the remaining five proviruses. The M. spretus sequences were compared with that of Mx27. The SU and TM start sites are indicated by open arrows, and the VRA, VRB, and PRD regions are indicated by open boxes. Black arrows indicate the positions of base substitutions relative to the Mx27 sequence. Substitutions that result in amino acid changes are shown by filled triangles, and those that do not are shown by open triangles. Additional arrows indicate the two stop codons in SPF4.
Because none of the five recombinants included TM sequences past the heptad repeat region, we sequenced two of the SPRET/Ei proviruses through the end of TM to determine if these proviruses shared some fatal defect in this region that might account for its exclusion from recombinant virus. In both cases, there were no obvious defects such as deletions or stop codons, although there were nine base substitutions, of which seven produced amino acid substitutions.
The U3 LTR sequences in these two proviruses closely resemble the P-I-type LTRs described previously by Tomonaga and Coffin (33), and this resemblance is not surprising since this previous study determined this LTR type to be the most abundant in SPRET/Ei mice. Sequence comparisons show that the SPF3 provirus U3 region has only a 1-bp difference from the published SPR6 P-I sequence. SPF4 has a 4-bp deletion at the 5′ end of U3 and two substitutions within the 190-bp insertion that is commonly associated with endogenous polytropic LTRs (data not shown).
DISCUSSION
The wild mouse species M. spretus contains proviruses capable of contributing to the generation of infectious nonecotropic recombinant viruses following neonatal inoculation of either ecotropic or amphotropic MLVs. These recombinant viruses contain substantial substitutions that include the entire SU gene as well as the pol IN gene and the segment of the TM gene encoding the fusion peptide. The methods employed here were not designed to isolate all possible recombinants generated in these mice. We characterized only those recombinants that were capable of replication in M. dunni or mink cells and detectable with the hybridization probes and PCR primers that identified nonecotropic proviral sequences in M. spretus DNA.
Compared to the common strains of laboratory mice, M. spretus carries very few endogenous MLV env genes; only seven proviral MLV env genes are present, all of which are polytropic MLVs. In comparison, most laboratory strains carry several dozen nonecotropic polytropic and xenotropic env sequences (8). The seven sequenced proviral M. spretus env genes showed little sequence variation and few fatal mutations, suggesting that these germ line copies are a relatively recent acquisition rather than vestiges of ancient infections; this conclusion is consistent with the conclusions of a previous analysis of proviral LTR sequences in several wild mouse species, including M. spretus (34).
The original source of the polytropic proviruses of M. spretus is likely to be M. m. domesticus. M. m. domesticus commonly carries more than 30 copies of polytropic proviruses (20). This species is a major contributor to the fancy mouse colonies that gave rise to the laboratory mouse strains and is the presumed source of their polytropic env genes. M. spretus diverged from M. m. domesticus about 1 million years ago (29). These two species are sympatric over North Africa, the Iberian Peninsula, and southern France (2), but they have long been viewed as reproductively isolated species due to the sterility of the F1 males (1), the partial genetic incompatibilities between their genomes (30), and the different ecological niches they occupy; the aboriginal M. spretus resides in native vegetation, whereas the commensal M. m. domesticus resides in man-made structures. However, a recent study has now shown that some populations of these species show signs of interbreeding (25). The extent of the genetic exchange in these populations is limited, but the observed introgression could easily result in the transmission of invasive genes like transposable elements or genes likely to provide an immediate survival advantage (e.g., disease resistance genes). Thus, while there is evidence that at least a few of the shared proviruses in these two species may predate speciation (34), the acquisition of the proviruses described here may have been accomplished in the more recent past through either infection or inheritance.
We did not characterize the proviral structures associated with any of the M. spretus env genes. While some or all of these may represent full-length proviruses, none are capable of producing infectious virus. All attempts to isolate infectious virus from uninoculated mice were unsuccessful (C. A. Kozak, unpublished data). Also unsuccessful were attempts to isolate virus from M. spretus cells by treating cultured fibroblasts with halogenated pyrimidines or by mitogen stimulation of B cells (C. A. Kozak, unpublished data). The two sequenced SPRET/Ei proviral LTRs are likely to be transcriptionally inactive. Their U3 regions are virtually identical to those of the laboratory mouse polytropic env-associated LTRs. These LTRs lack some enhancer sequences and also contain a nearly complete copy of the 190-bp insertion that is characteristic of endogenous polytropic proviruses but is not found in the LTRs of any replication-competent MLVs (31). In the laboratory mouse, infectious polytropic virus generated during leukemogenesis either retains the LTR of its ecotropic progenitors or replaces this LTR with one of X-MLV origin (27, 31). M. spretus contains no X-MLV env sequences (20, 34), and the recombinant viruses isolated in this study retain the LTRs of their MoMLV or A-MLV progenitors.
The env substitutions in the M. spretus viruses are larger than those found in the prototypical oncogenic MCF viruses, such as Moloney HIX MCF MLV and MCF247. For these laboratory mouse viruses, the 3′ recombination site is within SU in env, just downstream of the proline-rich domain (PRD). In contrast, the position of the 3′ breakpoints for four of the M. spretus recombinants are within a 30-bp segment of TM, making this substitution about 300 bp larger than that of Moloney HIX MCF MLV. Sequence analysis of the TM genes of two endogenous copies did not identify an obvious defect that would preclude inclusion of these sequences in an infectious virus. This segment of TM may simply represent a hot spot of recombination or ensure production of a virus with compatible env domains.
The sizes of the substitutions found in the M. spretus recombinants more closely resemble those of the type (class) II MCF recombinants (12, 23, 32). These recombinants are nonthymic in origin and are not leukemogenic. Restriction mapping and T1 oligonucleotide fingerprinting has shown that these recombinants differ from the oncogenic type I recombinants in that they have larger env substitutions that extend through TM. These type II viruses, like the viruses isolated in the present study, also have reduced titers on SC-1 cells. Similarly, larger substitutions have also been reported in a class of recombinants termed ERV viruses (6). These viruses also carry substitutions that extend through TM, and these ERV viruses also have a very restricted host range. It is not clear what viral sequence is responsible for the altered host range in any of these virus isolates.
The fact that M. spretus can generate recombinant viruses raises the question of the pathogenic potential of these isolates, as well as the susceptibility of M. spretus to virus-induced disease. In the present study, no gross signs of disease were identified in the sacrificed animals. In an earlier study, however, several litters of M. spretus mice were inoculated with MoMLV and monitored for signs of disease. Two of the 11 mice showed evidence of lymphomas by gross pathology and histological examination (35), suggesting that these mice are not resistant to the induction of neoplastic disease by these viruses, which may provide a useful system for the further analysis of leukemogenesis.
Acknowledgments
We acknowledge Alicia Buckler-White for sequencing and Caroline Ball for editorial assistance in the preparation of the manuscript. We also thank J. W. Hartley for generously supplying cells and viruses.
REFERENCES
- 1.Bonhomme, F., S. Martin, and L. Thaler. 1978. Hybridation en laboratoire de Mus musculus et Mus spretus Lataste. Sep. Exper. 34:1140. [DOI] [PubMed] [Google Scholar]
- 2.Boursot, P., J.-C. Auffray, J. Britton-Davidian, and F. Bonhomme. 1993. The evolution of house mice. Annu. Rev. Ecol. Syst. 24:119-152. [Google Scholar]
- 3.Ch'ang, L.-Y., W. K. Yang, F. E. Myer, C. K. Koh, and L. R. Boone. 1989. Specific sequence deletions in two classes of murine leukemia virus-related proviruses in the mouse genome. Virology 168:245-255. [DOI] [PubMed] [Google Scholar]
- 4.Chattopadhyay, S. K., A. I. Oliff, D. L. Linemeyer, M. R. Lander, and D. R. Lowy. 1981. Genomes of murine leukemia viruses isolated from wild mice. J. Virol. 39:777-791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloyd, M. W., M. M. Thompson, and J. W. Hartley. 1985. Host range of mink cell focus-inducing viruses. Virology 140:239-248. [DOI] [PubMed] [Google Scholar]
- 6.Cloyd, M. W., and S. K. Chattopadhyay. 1986. A new class of retrovirus present in many murine leukemia systems. Virology 151:31-40. [DOI] [PubMed] [Google Scholar]
- 7.Fischinger, P. J., S. Nomura, and D. P. Bolognesi. 1975. A novel murine oncornavirus with dual eco- and xenotropic properties. Proc. Natl. Acad. Sci. USA 72:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel, W. N., J. P. Stoye, B. A. Taylor, and J. M. Coffin. 1990. A linkage map of endogenous murine leukemia proviruses. Genetics 124:221-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner, M. B. 1978. Type C viruses of wild mice: characterization and natural history of amphotropic, ecotropic and xenotropic MuLV. Curr. Top. Microbiol. Immunol. 79:215. [DOI] [PubMed] [Google Scholar]
- 10.Hartley, J. W., and W. P. Rowe. 1975. Clonal cell lines from a feral mouse embryo which lack host-range restrictions for murine leukemia viruses. Virology 65:128-134. [DOI] [PubMed] [Google Scholar]
- 11.Hoggan, M. D., R. R. O'Neill, and C. A. Kozak. 1986. Nonecotropic murine leukemia viruses in BALB/c and NFS/N mice: characterization of the BALB/c Bxv-1 provirus and the single NFS endogenous xenotrope. J. Virol. 60:980-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holland, C. A., J. Wozney, P. A. Chatis, N. Hopkins, and J. W. Hartley. 1985. Construction of recombinants between molecular clones of murine retrovirus MCF 247 and Akv: determinant of an in vitro host range property that maps in the long terminal repeat. J. Virol. 53:152-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ikeda, H., K. Kato, H. Kitani, T. Suzuki, T. Yoshida, Y. Inaguma, N. Yamamoto, J.-G. Suh, B.-H. Hyun, T. Yamagata, T. Namikawa, and T. Tomita. 2001. Virological properties and nucleotide sequences of Cas-E-type endogenous ecotropic murine leukemia viruses in South Asian wild mice, Mus musculus castaneus. J. Virol. 75:5049-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jenkins, N. A., N. G. Copeland, B. A. Taylor, and B. K. Lee. 1982. Organization, distribution, and stability of endogenous ecotropic murine leukemia virus DNA sequences in chromosomes of Mus musculus. J. Virol. 43:26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jung, Y. T., M. S. Lyu, A. Buckler-White, and C. A. Kozak. 2002. Characterization of a polytropic murine leukemia virus proviral sequence associated with the virus resistance gene Rmcf of DBA/2 mice. J. Virol. 76:8218-8224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jung, Y. T., and C. A. Kozak. 2003. Generation of novel syncytium-inducing and host range variants of ecotropic Moloney murine leukemia virus in Mus spicilegus. J. Virol. 77:5065-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozak, C. A. 1985. Analysis of wild-derived mice for Fv-1 and Fv-2 murine leukemia virus restriction loci: a novel wild mouse Fv-1 allele responsible for lack of host range restriction. J. Virol. 55:281-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozak, C. A. 1985. Susceptibility of wild mouse cells to exogenous infection with xenotropic leukemia viruses: control by a single dominant locus on chromosome 1. J. Virol. 55:690-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak, C. A., J. W. Hartley, and H. C. Morse III. 1984. Laboratory and wild-derived mice with multiple loci for production of xenotropic murine leukemia virus. J. Virol. 51:77-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak, C. A., and R. R. O'Neill. 1987. Diverse wild mouse origins of xenotropic, mink cell focus-forming, and two types of ecotropic proviral genes. J. Virol. 61:3082-3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozak, C. A., and W. P. Rowe. 1982. Genetic mapping of ecotropic murine leukemia virus-inducing loci in six inbred strains. J. Exp. Med. 155:524-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lander, M. R., and S. K. Chattopadhyay. 1984. A Mus dunni cell line that lacks sequences closely related to endogenous murine leukemia viruses and can be infected by ecotropic, amphotropic, xenotropic, and mink cell focus-forming viruses. J. Virol. 52:695-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lung, M. L., J. W. Hartley, W. P. Rowe, and N. H. Hopkins. 1983. Large RNase T1-resistant oligonucleotides encoding p15E and the U3 region of the long terminal repeat distinguish two biological classes of mink cell focus-forming type C viruses of inbred mice. J. Virol. 45:275-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Neill, R. R., A. S. Khan, M. D. Hoggan, J. W. Hartley, M. A. Martin, and R. Repaske. 1986. Specific hybridization probes demonstrate fewer xenotropic than mink cell focus-forming murine leukemia virus env-related sequences in DNAs from inbred laboratory mice. J. Virol. 58:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orth, A., K. Belkhir, J. Britton-Davidian, P. Boursot, T. Benazzou, and F. Bonhomme. 2002. Hybridation naturelle entre deux especes sympatriques de souris Mus musculus domesticus L. et Mus spretus Lataste. C. R. Biol. 325:89-97. [DOI] [PubMed] [Google Scholar]
- 26.Peebles, P. T. 1975. An in vitro focus-induction assay for xenotropic murine leukemia virus, feline leukemia virus C, and the feline-primate viruses RD-114/CCC/M-7. Virology 67:288-291. [DOI] [PubMed] [Google Scholar]
- 27.Quint, W., W. Boelens, P. van Wezenbeek, T. Cuypers, E. R. Maandag, G. Selten, and A. Berns. 1984. Generation of AKR mink cell focus-forming viruses: a conserved single-copy xenotrope-like provirus provides recombinant long terminal repeat sequences. J. Virol. 50:432-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rowe, W. P., W. E. Pugh, and J. W. Hartley. 1970. Plaque assay techniques for murine leukemia viruses. Virology 42:1136-1139. [DOI] [PubMed] [Google Scholar]
- 29.Sage, R. D., W. R. Atchley, and E. Capanna. 1993. House mice as models in systematic biology. Syst. Biol. 42:523-561. [Google Scholar]
- 30.Santos, J., X. Montagutelli, A. Acevedo, P. Lopez, C. Vaquero, M. Fernandez, M.-R. Arnau, M. Szatanik, E. Salido, J.-L. Guenet, and J. Fernandez-Piqueras. 2002. A new locus for resistance to γ-radiation-induced thymic lymphoma identified using inter-specific consomic and inter-specific recombinant congenic strains of mice. Oncogene 21:6680-6683. [DOI] [PubMed] [Google Scholar]
- 31.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 61:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, C. Y., J. D. Nuckols, C. Murphy, and D. Innes. 1993. Generation and pathogenicity of an NB-tropic SL3-3 murine leukemia virus. Virology 193:1013-1017. [DOI] [PubMed] [Google Scholar]
- 33.Tomonaga, K., and J. M. Coffin. 1998. Structure and distribution of endogenous nonecotropic murine leukemia viruses in wild mice. J. Virol. 72:8289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tomonaga, K., and J. M. Coffin. 1999. Structures of endogenous nonecotropic murine leukemia virus (MLV) long terminal repeats in wild mice: implication for evolution of MLVs. J. Virol. 73:4327-4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villar, C. J., T. N. Frederickson, and C. A. Kozak. 1988. Effect of the Gv-1 locus on Moloney ecotropic murine leukemia virus induced disease in inbred wild mice. Curr. Top. Microbiol. Immunol. 137:250-255. [DOI] [PubMed] [Google Scholar]
- 36.Voytek, P., and C. A. Kozak. 1989. Nucleotide sequence and mode of transmission of the wild mouse ecotropic virus, HoMuLV. Virology 173:58-67. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, C. A., and M. V. Eiden. 1991. Viral and cellular factors governing hamster cell infection by murine and gibbon ape leukemia viruses. J. Virol. 65:5975-5982. [DOI] [PMC free article] [PubMed] [Google Scholar]