Abstract
Among the many host cell-derived proteins found in human immunodeficiency virus type 1 (HIV-1), HLA class II (HLA-II) appears to be selectively incorporated onto virions and may contribute to mechanisms of indirect imunopathogenesis in HIV infection and AIDS. However, the amount of HLA-II on the surface of HIV-1 particles has not been reliably determined due to contamination of virus preparations by microvesicles containing host cell proteins, including HLA-II. Even rigorous sucrose density centrifugation is unable to completely separate HIV-1 from microvesicles. CD45, a leukocyte integral membrane protein, is found on microvesicles, yet appears to be excluded from HIV-1 particles. Exploiting this observation, we have developed a CD45-based immunoaffinity depletion method for removing CD45-containing microvesicles that yields highly purified preparations of virions. Examination of CD45-depleted HIV-1MN by high-pressure liquid chromatography, protein sequencing, and amino acid analyses determined a molar ratio of HLA-II to Gag of 0.04 to 0.05 in the purified virions, corresponding to an estimated average of 50 to 63 native HLA-II complexes (i.e., a dimer of α and β heterodimers) per virion. These values are approximately 5- to 10-fold lower than those previously determined for other virion preparations that contained microvesicles. Our observations demonstrate the utility of CD45 immunoaffinity-based approaches for producing highly purified retrovirus preparations for applications that would benefit from the use of virus that is essentially free of microvesicles.
Human immunodeficiency virus type 1 (HIV-1) and other retroviruses incorporate a variety of cellular proteins in the process of assembly and budding (reviewed in reference 26). Some of these proteins appear to play important roles in virus biology. HLA class II (HLA-II) is the most commonly observed and, perhaps, the most abundant protein found on the surface of HIV-1 produced from HLA-II+ cells (reviewed in references 25 and 41). While the mechanism for HLA-II incorporation remains unknown, experiments with site-directed HIV-1 mutants in lymphoid cells demonstrated that efficient HLA-II incorporation onto HIV-1 required the packaging of Env, suggesting an active mechanism for incorporation (32). Similarly, the finding that the DR isoform of HLA-II was preferentially incorporated over the DP and DQ forms, regardless of abundance, indicates specificity in this process (2, 8, 15, 37). The reason for this is not clear, though HIV-1 could specifically incorporate HLA-II to use for its advantage. For instance, the presence of HLA-II on HIV-1 can provide for modest increases in virion infectivity (7).
Among the more intriguing proposed roles for HLA-II on the surface of HIV-1 is its potential contribution to indirect immunopathogenesis by suppressing antiviral immune responses, thereby facilitating viral persistence and spread (2). If HLA-II on virions could functionally bind the T-cell receptor on CD4+ T cells in conjunction with HIV-1 surface glycoprotein (Env)-CD4 binding, then these interactions could mimic part of the normal stimulation signal sent between antigen-presenting cells and T cells. However, HIV-1 does not incorporate CD80 (12), a protein that, upon binding CD28 on the T cell, can provide a second costimulatory signal required for effective T-cell stimulation (18). Receptor engagement in the absence of costimulation can result in T-cell anergy (a loss of antigen responsiveness) or apoptosis (programmed cell death). Therefore, it is possible that HLA-II on the surface of virions could send just its single signal that downregulates T-cell function, causing immunodeficiency without T-cell infection (2). This mechanism is consistent with the observation that, in infected individuals, most apoptotic death occurs in cells that are not productively infected, i.e., bystanders (13). Supporting this hypothesis, virus-associated HLA-II can present superantigen to T cells, indicating that HLA-II on the virion surface retains its functional activity (35). In addition, we found that chemically inactivated, noninfectious HIV-1 containing conformationally and functionally intact Env and HLA-II protein complexes caused apoptosis in T cells, while similar virions without HLA-II did not (11). Interestingly, microvesicles, which contain HLA-II but not Env, did not produce this effect, indicating a role for the CD4-Env interaction in this process. Together, these results support a role for HLA-II on HIV-1 as a potential contributor to virus-mediated immunodeficiency.
Despite the potential importance of HLA-II on virions, the amount of this protein complex present on HIV-1 has not been determined. The primary impediment to quantitating the amount of a particular cellular surface protein on the virion is the presence of contaminating protein-laden particles, mostly microvesicles, in virus preparations. While these microvesicles are heterogeneous, a significant fraction have the same density as virus, and thus are present even in highly purified, sucrose density gradient-banded virus preparations from lymphoid cells (4, 15). Moreover, the array of cellular proteins in microvesicles closely matches that found in virus preparations, making it difficult to distinguish cellular proteins in the virions from those in copurifying microvesicles. For the study of proteins inside the retroviruses, these particles can be removed by using a subtilisin digestion procedure, which removes both exterior and microvesicle-associated proteins (27). However, since digestion removes surface proteins from virions, it is not a suitable method to study these types of proteins (28). While microvesicles and virions have similar cellular protein compositions, the incorporation of host plasma membrane proteins into virions appears to have some specificity as some proteins appear to be excluded from the virion (reviewed in reference 26).
CD45, an integral membrane protein also known as leukocyte common antigen, is present in microvesicles, yet does not appear to be incorporated into virions (12, 24), possibly being excluded by its large cytoplasmic tail. Based on this finding, we developed an immunoaffinity depletion procedure to remove CD45-containing particles and tested whether it could be an effective approach for removing microvesicles from virus preparations. Using anti-CD45 immunomagnetic microbeads to capture microvesicles, we were able to effectively remove microvesicle-associated proteins from samples, allowing for the biochemical quantitation of HLA-II on the remaining highly purified HIV-1 virions.
MATERIALS AND METHODS
Virus and microvesicle preparations.
HIV-1MN was produced from the cell lines CEM-SS (23) and CEMx174 T1 (36) and Clone 4, a clonal line derived from HIV-1MN-infected H9 cells that constitutively expresses virus (30). HIV-1IIIB (33) was produced from either H9 or primary peripheral blood mononuclear cells (PBMC) that were stimulated by a 48-h treatment with a 1:200 vol/vol dilution of phytohemagglutinin-M (Invitrogen, Carlsbad, Calif.) and subsequent maintenance in 20 U per ml of interleukin-2. Donor PBMC were obtained from a healthy HIV-1-seronegative donor under an NIH institutional review board-approved protocol. Simian immunodeficiency virus SIVMAC239/251 (14) and SIVMAC239 G4,5 (34) were propagated on CEMx174 T1 cells, and SIVCP-MAC (21) was propagated in Sup-T1 cells (38). Virus was purified by sucrose density ultracentrifugation as previously described (5). Microvesicles were similarly purified from the supernatants of uninfected H9, HuT78, CEM-SS, and CEMx174 T1 cells as previously described (4) (AIDS Vaccine Program, NCI at Frederick, Frederick, Md.).
CD45 affinity depletion.
Before use, anti-CD45-conjugated paramagnetic microbeads (Miltenyi Biotech, Auburn, Calif.) were bound to an LD column magnetized by a VarioMACS magnet (Miltenyi Biotech) and washed three times with cold TNE buffer (10 mM Tris-Cl [pH 7.5], 100 mM NaCl, and 1 mM EDTA). Microbeads were removed from the demagnetized column by plunging and resuspended in the same volume as initially supplied by the manufacturer. After optimizing the ratio of anti-CD45 microbeads to total protein, the following conditions were used. Virus and microvesicle preparations were incubated with 2 μl of prewashed microbeads per 1 μg of total protein for 1 h on ice. The sample-microbead mixture was then applied to a washed (two times with TNE) and magnetized MS or LD column (Miltenyi Biotech). The void volume and two washes with TNE (0.5 ml for MS columns and 2 ml for LD columns) were collected and combined to produce the flowthrough fraction and the next three additional wash volumes were also combined to produce the wash fraction. The column was then removed from the magnet and plunged with TNE buffer (1.5 ml for MS columns and 6 ml for LD columns) to recover the protein retained on the beads as the bead fraction. These fractions, as well as an equal aliquot of the untreated preparation (starting material) were pelleted at 50,000 × g for 1 h at 4°C in either a TLA100.1 rotor using a TL-100 centrifuge (Beckman Instruments, Palo Alto, Calif.) for the MS column or a SW41Ti rotor for the LD column and resuspended in 200 μl of TNE buffer.
Infectivity of CD45-depleted preparations.
One milliliter of clarified supernatant containing H9 T cells acutely infected with HIVNL4-3 (1) were mixed with 50 μl of anti-CD45 or anti-CD34 beads (Miltenyi Biotech), incubated at room temperature for 1 h, and then captured by a magnet held externally to the tube containing the sample. The supernatant was then removed and, along with an untreated control, titers were determined for infectivity in a single-round lacz-Tat transcomplementation reporter assay, using a HeLa-CD4 containing a long terminal repeat-promoted lacZ gene, and assayed for reverse transcriptase as previously described (17). This depletion procedure does not suffer the extensive loss of the column method and was as effective as the column procedure for removing CD45 from preparations (data not shown).
OptiPrep and sucrose gradients.
Step gradients were formed with either 12, 16, 20, or 24% OptiPrep (Accurate) or 20, 30, 40, 50, or 60% sucrose (Sigma) in phosphate-buffered saline (PBS) (wt/vol) by successive layering of density solutions in 1-ml fractions in an SW65.1Ti tube (Beckman Instruments). Samples were loaded onto the gradients in PBS and centrifuged at 220,000 × g for 1 h at 4°C. Fractions were removed by pipette and analyzed. The OptiPrep concentrations used here were somewhat higher than previously reported (10), as those conditions were insufficient to buoy virus or microvesicles in our experiments.
Protein analysis.
For viral protein immunoassays, samples were lysed in 1% Triton X-100 (Sigma, St. Louis, Mo.) for 1 h at 37°C, diluted, and assayed in duplicate with either HIV-1 p24CA or SIV p27CA antigen capture enzyme-linked immunoassay kits according to manufacturer's recommendations (AIDS Vaccine Program). Protein concentrations in samples were determined by the DC Protein Assay (Bio-Rad, Hercules, Calif.) after lysis with 0.4% (vol/vol) Sarkosyl in TNE or by amino acid analysis using a Hitachi L-8800 amino acid analyzer (Palo Alto, Calif.) as previously described (20). Reversed-phase high pressure liquid chromatography (HPLC) was performed as previously described using a 2.1 by 100 mm Poros R2/H narrow-bore column (Boehringer Mannheim, GmbH, Germany) (9), except for the analyses that separated HLA-II from actin. For the specialized HLA-II separation, a gradient (vol/vol) of acetonitrile in water with 0.1% (vol/vol) trifluoracetic acid was used as follows: 10 to 36% for 7 min, 36 to 42.5% for 5 min, 42.5% to 46.5% for 14 min, 46.5 to 80% for 9 min, and 80% for 5 min at 55°C. Proteins were detected by UV absorption at 206 and 280 nm. N-terminal protein sequence analysis was carried out with an Applied Biosystems Procise model 494 microsequencer (Foster City, Calif.) as previously described (19).
SDS-PAGE and immunoblot analyses.
Proteins analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were visualized with a silver-staining kit (Invitrogen) and used according to the manufacturer's recommendations. Immunoblotting onto Immobilon-P PVDF membranes (Millipore Corporation, Bedford, Mass.) was carried out as previously described (20), using enhanced chemiluminescence detection (Amersham Biosciences, Arlington Heights, Ill.) on Lumi-Film (Roche Applied Sciences, Indianapolis, Ind.). Semiquantitative immunoblotting was carried out by comparing samples of CD45-depleted material to a dilution series of untreated material. Only signals in the linear response range of the film were compared for quantitation purposes. Mouse monoclonal antibodies used were as follows: anti-CD45 (BD-Transduction Laboratories, San Diego, Calif.); anti-α-smooth muscle actin (Accurate, Westbury, N.Y.); anti-β-actin (Sigma); and anti-panactin (which detects all isoforms of actin), (Amersham Biosciences). Antisera, including rabbit HLA-IIα (DJ-39528) and HLA-IIβ (DJ-38857) and goat CA (p24CA/p28CA) (no. 81; 001090) and p17MA (no.83; 000794) were produced by the AIDS Vaccine Program, NCI-Frederick. Secondary sera used were horseradish peroxidase-conjugated antigoat and antirabbit antisera (Bio-Rad) and antimouse antiserum (Invitrogen). Reprobing of immunoblots was carried out by incubating the membrane in stripping buffer (100 mM β-mercaptoethanol, 2% wt/vol SDS, 62.5 mM Tris-HCl [pH 6.7]) at either 50 or 60°C for 30 min with agitation, followed by two 10-min washes in TBS (20 mM Tris-HCl [pH 7.6] and 137 mM NaCl) with 0.1% Tween-20 (Sigma). The filters were then blocked for 2 h and reacted with immune reagents as described above.
Electron microscopy.
Pelleted virions (containing 5 μg of CA) were stained with uranyl acetate and lead citrate before thin-section transmission electron microscopic analysis was done as previously described (16), using a Hitachi 7000 transmission electron microscope operated at 75 kV.
RESULTS
CD45 affinity depletion of microvesicles.
Since CD45 appears to be incorporated onto microvesicles and is apparently excluded from HIV-1 virions, microvesicles should be removed from virus preparations by a CD45 immunoaffinity approach. However, a significant fraction of microvesicles might not contain CD45 or might not have enough for anti-CD45-mediated capture and thereby could remain in the virion preparation. To determine the efficacy of using CD45 immunocapture to remove microvesicles, we incubated 50 μg of a microvesicle preparation produced from CEMx174 T1 cells with anti-CD45-conjugated paramagnetic microbeads. The mixture was then passed through a magnetic column to remove the CD45-antibody complexes, and the flowthrough fraction was collected. The beads retained on the column were washed (collected as the wash fraction) before being released from the column (collected as the bead fraction). To analyze the fate of the microvesicles, equivalent amounts of each fraction (by volume) and starting material were examined. CD45 immunoblot analysis of fraction samples showed that, although CD45 was present in the starting material (Fig. 1A), only a slight CD45 signal was detected in the column flowthrough fraction, indicating that the majority of the CD45 was removed. Based on semiquantitative immunoblotting, we estimate that less than 2.5% of the CD45 initially present remains after this treatment (data not shown). There was no detectable CD45 signal in the wash fractions. While there was a detectable CD45 signal in the bead sample, it was considerably weaker than that seen in the equivalent amount of the starting material. Therefore, some of the CD45 apparently was retained on the column, either attached to beads that remained trapped after demagnetization or simply nonspecifically bound to the inside surface of the column.
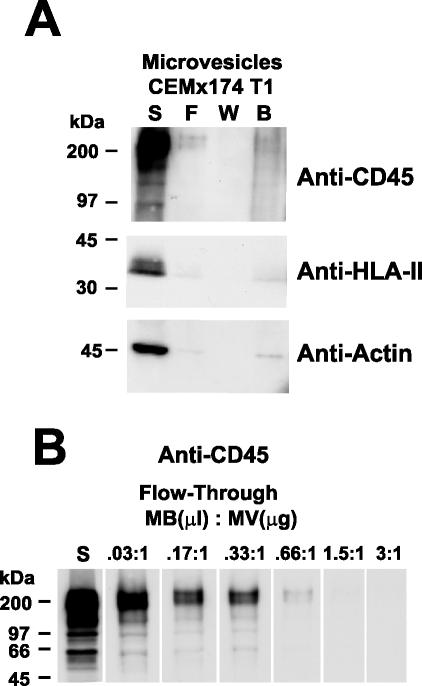
FIG. 1.
Immunoblot analysis of CD45-depleted microvesicles produced from CEMx174 T1. (A) Analysis of fractions from CD45 depletion of 50 μg (total protein) of microvesicles is shown. Samples are identified above their respective lanes as follows: starting material, S; flowthrough, F; wash, W; and bead, B. The antibody or antiserum used for each blot is indicated at the right. (B) Analysis of the CD45 content of starting (S) and flowthrough samples from a series of depletions using various ratios of CD45 microbeads (microliters) to microvesicle protein (micrograms) is presented. The ratio used for each depletion is indicated above its respective lane. Molecular mass standards are indicated at the left.
The removal of microvesicles should also be reflected in the removal of cellular proteins other than CD45. An immunoblot using antibodies to both the α and β chains of HLA-II revealed that this protein was effectively absent from the CD45-depleted microvesicle preparation, likely due to the removal of CD45+/HLA-II+ microvesicles (Fig. 1A). Based on semiquantitative immunoblotting, we estimate that at least 95% of the HLA-II associated with microvesicles was removed from the flowthrough fraction (data not shown). These data indicate that microvesicles that contain HLA-II also contain CD45. Thus, CD45 depletion should remove microvesicle-associated HLA-II from virion preparations. Immunoblotting for actin, another major component of microvesicles, demonstrated that this protein was also efficiently removed by CD45 depletion (Fig. 1A). Colorimetric quantitation of the total protein in the starting versus the flowthrough samples revealed that CD45 depletion removed 89% of the protein from microvesicle preparations. Similar results were obtained with other microvesicle preparations (data not shown). Therefore it appears that the majority of microvesicles are removed by this procedure. Removal of CD45 was dependent on the ratio of anti-CD45 beads to microvesicles (Fig. 1B), demonstrating that the anti-CD45 beads were proportionally removing the microvesicles.
OptiPrep fails to efficiently resolve microvesicles from virions.
The results above suggest that CD45 immunoaffinity depletion is a promising approach for removing microvesicles from virus preparations. In conjunction with our evaluations of CD45 depletion methods, we also examined other methods for separating microvesicles from virus. While sucrose density gradient centrifugation is unable to efficiently separate these two classes of particles (4, 15), the use of OptiPrep medium (iodixanol) for density separation has been reported to be able to effectively remove microvesicles from virions (10). To evaluate the ability of this approach to resolve microvesicles, preparations of HuT78 microvesicles or HIV-1IIIB from H9 cells were centrifuged in sucrose or OptiPrep step density gradients. Fractions from the gradients containing microvesicles were analyzed by SDS-PAGE. The results revealed that the microvesicle proteins, especially actin, were present in the 30 to 50% sucrose fractions (Fig. 2A), as previously observed (references 4 and 15 and unpublished data). For the OptiPrep gradient, these same proteins were also found over a wide range, being present in the 12 to 24% fractions. Thus, neither sucrose nor OptiPrep gradients was able to resolve microvesicles into a small region of the gradient. Fractions from the gradients containing virus were analyzed by immunoblotting with p17MA antiserum to detect the fractions containing HIV-1. The results showed that the virus was present in the 30% sucrose and the 16% OptiPrep fractions (Fig. 2B), densities where a significant portion of the microvesicles equilibrate (Fig. 2A). Since CD45 is found on microvesicles and not on virus, a CD45 blot revealed that this microvesicle marker was present in the same fractions as virus in both sucrose and OptiPrep gradients, revealing little separation between these two types of particles (Fig. 2B). Analysis of other virus preparations produced similar results, detecting no additional separation between viral and microvesicle proteins (data not shown). The presence of CD45 in the same fractions as virions (Fig. 2B) as well as the SDS-PAGE results for microvesicles (Fig. 2A) show that neither of these methods can satisfactorily remove the majority of microvesicles from virion preparations.
FIG. 2.
OptiPrep analysis of preparations. (A) Silver-stained SDS-polyacrylamide gels of gradient fractions from OptiPrep and sucrose centrifugation experiments with HuT78 microvesicles. MV, lane containing a sample of the microvesicle preparation before centrifugation. (B) Immunoblots of gradient fractions from OptiPrep or sucrose centrifugation experiments with HIV-1IIIB preparations produced from H9. The antibody or antiserum used for each blot is indicated above the lanes. For both panels, the percentage of density medium in the gradient fraction is indicated above each lane. Molecular mass standards are indicated at the left.
Removal of CD45+ microvesicles from HIV-1 and SIV preparations.
Having established the conditions for essentially complete immunoaffinity depletion of CD45+ particles from microvesicle preparations, we determined if this approach could be used to deplete CD45+ microvesicles from virus preparations. Fifty micrograms (total protein) of sucrose-purified HIV-1MN (2.3 μg of CA) or SIVMAC239/251 (0.1 μg of CA) produced from CEMx174 T1 cells was subjected to the CD45 depletion procedure and portions of fractions equivalent to the starting material (by volume) were analyzed by CD45 immunoblotting. Though CD45 was present in the starting material (Fig. 3), no CD45 signal was detected in the column flowthrough fraction, indicating that, similar to our microvesicle experiments, essentially all of the CD45 in the starting material was removed. Reacting this blot to CA antiserum (reactive to HIV-1 p24CA and SIV p27CA) showed that the intensity of the signal in the flowthrough samples was somewhat diminished from that of the starting material (Fig. 3). Immunoassays showed that approximately half of the CA (either HIV-1 or SIV) applied to the columns was recovered. However, only approximately 12% of the CA was found in the bead fraction. Since the depletion procedure was designed to optimize removal of CD45+ particles and not to maximize recovery of the CD45− fraction that contains virus, it is likely that the balance of the virions were nonspecifically trapped in the column, similar to our observations with microvesicles. Immunoblotting for HLA-II revealed that the flowthrough samples produced a reduced signal compared to the starting material. Since virtually all CD45 had been removed from the flowthrough material, this remaining HLA-II signal is likely due to the HLA-II that is present on the virions. The more intense HLA-II signal detected in the HIV-1 compared to the SIV flowthrough samples probably reflects the difference in the amounts of virus present in the starting samples (∼20-fold more HIV-1 CA than SIV CA).
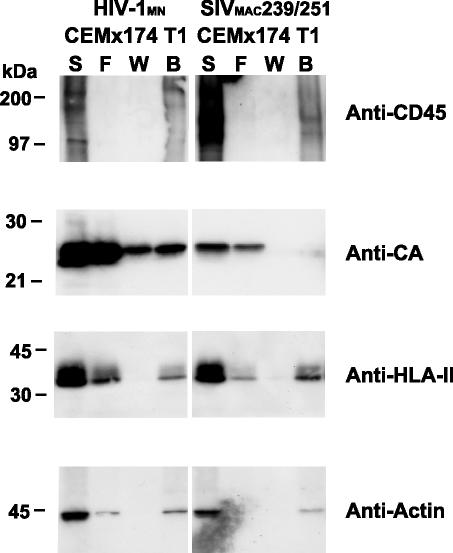
FIG. 3.
Immunoblots of CD45-depleted HIV and SIV. Immunoblots of fractions from CD45 depletion of 50 μg HIV-1MN and SIVMAC239/251 produced from CEMx174 T1 cells are presented. Samples are identified above their respective lanes as follows: starting material, S; flowthrough, F; wash, W; and bead, B. The antibody or antiserum used for each blot is indicated at the right. Molecular mass standards are indicated at the left.
Actin is another cellular protein that is found in both virions and microvesicles (4, 15, 29). Similar to the HLA-II results, the panactin immunoblot signal was decreased in but not eliminated from the HIV-1 flowthrough fraction and was undetectable in the corresponding SIV sample (Fig. 3). Our inability to detect actin in the SIV flowthrough fraction is likely due to the relatively low amount of virus in the SIV preparation and the sensitivity of the actin immunoblot. Taken together, these results demonstrate that CD45 affinity depletion is effective in removing microvesicles, including those containing HLA-II, from purified virion preparations.
Env remains intact after CD45 immunoaffinity depletion.
Previously, we have shown that the association of gp120SU with gp41 in the Env complex on HIV-1 and SIV virions is relatively stable: Env remains intact during many experimental manipulations, including the sucrose gradient centrifugation procedure used to purify and concentrate virions (9). To determine if Env remains intact after CD45 immunoaffinity depletion, samples of starting and flowthrough fractions from HIV-1MN produced from CEMx174 T1 cells were examined by immunoblot with both CA antiserum and gp120SU antibody. The relative intensities of the CA and gp120SU bands within each sample were similar (Fig. 4). Therefore, although there was somewhat less material in the flowthrough than the starting samples (in part due to loss of virus), there was little, if any, difference in the amount of gp120SU in the virions themselves, demonstrating that gp120SU is not lost from virions during the CD45 depletion process. An analysis of CD45-depleted HIVIIIB produced from stimulated PBMC produced a similar result: Env remained intact throughout this process.
FIG. 4.

Env stability of CD45-depleted HIV. Immunoblotting with both CA antiserum and gp120SU antibody of CD45-depleted HIV-1MN samples produced from CEMx174 T1 cells are presented. Starting material (S) and flowthrough (F) samples are identified above their respective lanes. Molecular mass standards are indicated at the left.
Infectivity of CD45-depleted virions.
To determine the infectivity of CD45-depleted HIV-1 virions with high-titer virus, clarified HIV-1NL4-3-infected H9 cell supernatants (from fresh harvests to maintain virus titer) were mixed with anti-CD45 beads that were later captured by magnet (without a column). The titers of the supernatants were determined along with those of untreated supernatants or those treated in the same manner with irrelevant antibody-conjugated beads against CD34, a marker which is not expressed on H9 cells. This depletion procedure was used rather than the column method as it can rapidly and effectively remove CD45 from small amounts of unconcentrated supernatants (data not shown). Compared to the untreated samples, there was no significant loss of virions in the bead-depleted samples as measured by reverse transcriptase assay (data not shown). The single-round Tat transcomplementation lacZ assay titers (n = 3) for all of the samples were similar: titers (blue cell-forming units per milliliter) for the untreated samples were 4.8 × 104 ± 0.6 compared to those of the CD45-depleted supernatants, 5.0 × 104 ± 1.2, and the CD34-depleted supernatants, 3.6 × 104 ± 0.4. These results show that the virions remain infectious after CD45 depletion, implying that Env remains functional. Additionally, these data are consistent with the absence of CD45 on HIV-1 particles.
Separation of microvesicles from virions.
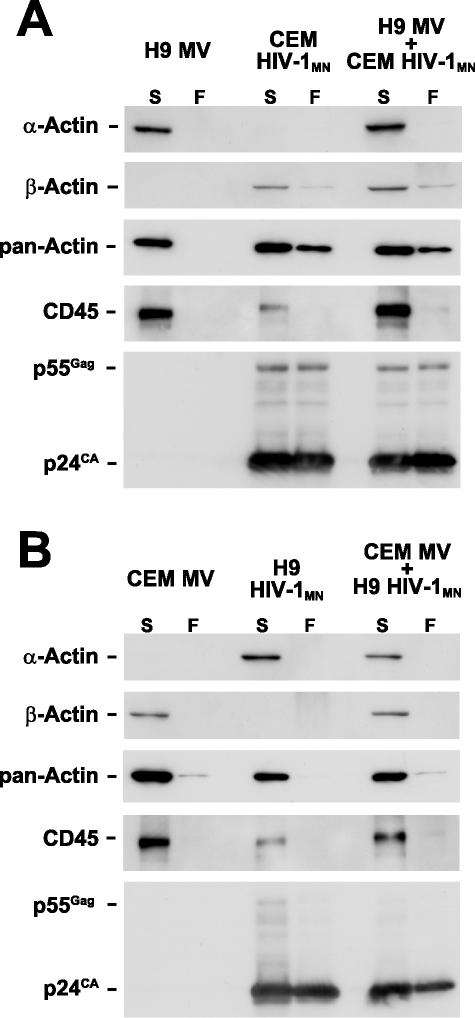
To formally demonstrate that this approach can remove microvesicles from virions, we took advantage of our previous finding that particles (i.e., microvesicles and virions) produced from different lymphocyte cell lines can contain different isoforms of actin (29). H9 cells produce particles with smooth muscle α-actin, while CEM-SS cells produce those with β-actin. This difference in actin isoform expression can be used as a marker in virus-microvesicle mixing experiments to distinguish the fates of the different particles after CD45 depletion. HIV-1MN virus produced from CEM-SS cells (containing β-actin), microvesicles from H9 cells (α-actin), and a mixture of the two preparations was subjected to the CD45 depletion procedure. Immunoblot analysis revealed that the α-actin signal from the microvesicles in the flowthrough fractions of the microvesicles and virus-microvesicle mixture was undetectable in the depleted samples while present in the untreated samples (Fig. 5A), demonstrating that microvesicles were removed from the mixture. Blotting the same membrane with β-actin antibody showed that a residual amount of this isoform was still present in the virus-containing samples after treatment, as expected since the virions produced from CEM-SS contain β-actin (29) (Fig. 5A). Reacting the blot with the panactin antibody that detects all actin forms confirmed the isoform-specific blot results. Depletion and analysis of the reciprocal mixture, HIV-1MN produced from H9 cells (α-actin) and mixed with microvesicles from CEM-SS (β-actin), produced the expected results, demonstrating that microvesicles were removed as detected by the loss of β-actin in the mixture (Fig. 5B). The α-actin signal was not present in the flowthrough lane that contained HIV-1 produced from H9, though the panactin blot showed that some actin still remained in the virus samples as expected. This loss of detectable α-actin signal is likely due to an insufficient amount of virus and the level of detection with this antibody. For both experiments, CD45 and CA immunoblots demonstrated that CD45 was effectively removed by immunoaffinity depletion and that most of the virions were recovered in the flowthrough fraction (Fig. 5). Together, these data confirm the results above, showing that microvesicles can be effectively removed from these virion preparations by CD45 immunoaffinity depletion.
FIG. 5.
Separation of microvesicles and virions. Immunoblots of fractions from CD45 depletion of mixtures of microvesicles and virions are presented. Fractions analyzed are identified above the respective blots (MV, microvesicles) indicated by S for starting material or F for flowthrough. The antibody or antiserum used for each blot is indicated at the left.
Biochemical analysis of virions.
To better characterize treated virions, we individually CD45-depleted 500-μg preparations of HIV-1MN from H9 Clone 4 cells, HIV-1IIIB from phytohemagglutinin-stimulated PBMC, and SIVMAC239 G4,5 from Sup-T1 cells and analyzed the starting and flowthrough material by immunoblot analysis. As before, the data showed that CD45 was removed from the flowthrough fractions (Fig. 6A). Blotting for CA revealed that there was a decrease in CA signal that was confirmed by CA antigen capture immunoassay results, likely due to nonspecific trapping on the column. Nevertheless, SDS-PAGE analysis of starting and flowthrough samples normalized to equal amounts of CA clearly showed that a considerable amount of protein was removed from the treated material relative to that of the viral proteins (Fig. 6B). Quantitation of total protein content in the starting and depleted material by amino acid analysis showed that the majority of the total protein in the virus sample was removed by CD45 depletion (data not shown), mirroring the SDS-PAGE results.
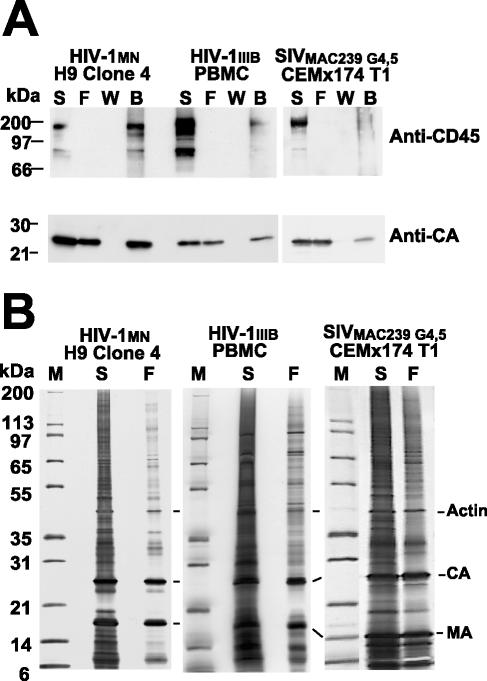
FIG. 6.
Analysis of CD45-depleted HIV and SIV. Analyses of fractions from CD45 depletion of 500 μg (total protein) of HIV-1 and SIV preparations are presented. (A) Immunoblots of fractions are displayed with the virus preparations and samples identified above their respective lanes as follows: starting material, S; flowthrough, F; wash, W; and bead, B. The antibody or antiserum used is indicated at the right. (B) Silver-stained SDS-polyacrylamide gels of the starting material (S) and flowthrough (F) samples from CD45 depletions are presented. Samples were normalized for CA to show the relative purity of the virions. Samples are identified above their respective lanes, and prominent proteins are identified at the right. Molecular mass standards, lane M in panel B, are indicated at the left.
To better characterize the proteins in these large-scale preparations, portions of the CD45-depleted and untreated HIV-1 and SIV samples examined above were analyzed by reversed-phase HPLC. To assess the purity of the virus preparations and to adjust for nonspecific loss of viral proteins during the CD45 depletion procedure, the chromatograms from the CD45-depleted and untreated samples are presented at different absorbance ranges to produce equal peak heights for the NC proteins (Fig. 7). This normalized comparison showed that, while the relative amounts of each of the Gag proteins were similar between the two profiles, there were substantial reductions in peak area for cellular proteins in the HPLC chromatograms of the CD45-depleted samples. While cellular proteins that contaminate virion preparations are found throughout the fractions, the levels for two especially prominent microvesicle-associated proteins, actin and HLA-II, were greatly reduced relative to Gag by this procedure (Fig. 7). This was especially apparent in the chromatograms from the HIV-1IIIB produced from PBMC, since this preparation contains a large amount of contaminating proteins, presumably from microvesicles.
FIG. 7.
HPLC Analysis of CD45-depleted HIV and SIV. A280 chromatograms of HPLC separations of untreated and treated HIV-1 and SIV samples are presented. Absorbance levels of the chromatograms were normalized to the NC peak height to allow for the assessment of the relative purity of the virus preparations in the starting (−) and CD45-depleted (+) material. Important peaks identified by immunoblot or protein sequence are labeled above their respective chromatograms.
Electron microscopy of virion preparations.
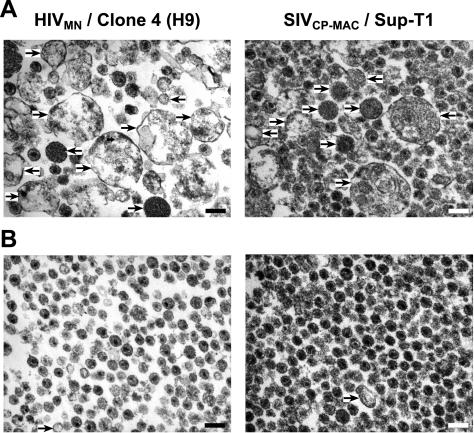
Samples of the HIV-1MN produced from H9 and SIVCP-MAC produced from Supt-T1, before and after CD45 depletion, were studied by transmission electron microscopy (Fig. 8). The untreated HIV-1 and SIV preparations contained a relatively small number of virions compared to the many nonviral particles. These results are consistent with other electron microscopic studies of virus preparations (4, 15). In contrast, the CD45-depleted virion preparations contained mostly virions and few, if any of these nonviral particles. These data confirm our biochemical data and directly show that CD45 depletion effectively removes microvesicles from virion preparations.
FIG. 8.
Electron microscopic analysis of CD45-depleted HIV-1 and SIV. Representative fields (×40,000) from 5 μg of untreated (A) and treated (B) preparations of HIV-1MN and SIVCP-MAC produced from H9 and Sup-T1 cell lines, respectively, analyzed by thin-section transmission electron microscopy are presented. Arrows highlight some of the potential microvesicles in the field and the scale bar (lower right in field) corresponds to 200 nm.
Quantitation of HLA-II on HIV-1MN.
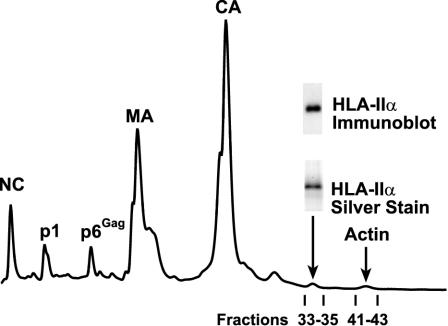
Our ability to produce relatively pure HIV-1 from lymphoid lines allows us to directly quantitate the amount of HLA-II on the virion. CD45-depleted HIV-1MN produced from H9 cells was separated by HPLC with a column elution gradient that was especially developed to isolate relatively pure fractions containing either the α-chain of HLA-II or actin. The fraction containing the HLA-II α-chain was identified by immunoblotting and the purity of this fraction was demonstrated by the presence a single band in SDS-PAGE analysis (Fig. 9). Protein sequencing of this band identified this protein specifically as the α-chain of the HLA-DR isoform, consistent with several previous studies suggesting specific HLA-DR incorporation (reviewed in references 2, 5, and 41). Amino acid analysis of this fraction produced the expected composition of amino acids for the α-chain of HLA-DR. Since this analysis directly measures the molar amount of each amino acid in the sample, we calculated the total pmoles of protein in this fraction (Table 1). A similar analysis was carried out on the fractions containing several of the Gag proteins (MA, CA, NC, and p6Gag). These results were averaged to determine the molar amount of Gag present in this preparation (Table 1). Based on amino acid analyses of two independent CD45 depletions of HIV-1MN produced from H9 cells, we found that there are between 100 to 125 α-chain HLA-II proteins per virion, assuming an estimate of 2,500 Gag molecules per virion (5, 22, 31, 40). Since the HLA-II complex is made up of a dimer of heterodimeric α- and β-chains (i.e., 2 α-chains per native HLA-II), this corresponds to 50 to 63 complexes on the surface of HIV-1MN produced from H9 cells. It should be noted that present estimates of the Gag content range from 1,200 to 5,000, thus, this estimate would vary given other Gag-to-virion ratios. Using the same methodology and Gag molecules per virion assumption, amino acid quantitation of the actin peak determined that there were 50 to 68 molecules of actin per virion (Table 1).
FIG. 9.
Quantitation of HLA-II. An HPLC chromatogram (A280) of treated HIV-1MN is presented. SDS-PAGE and immunoblotting results from the HLA-II-containing fractions are presented above the fraction. The fractions containing the Gag proteins and actin that were subjected to amino acid analysis are indicated above the respective peaks.
TABLE 1.
Quantitation of HLA-II on CD45-depleted HIV-1MN
| Protein | Amt (pmol) | Protein/Gag | Proteins/viriona (HLA-II complexes)b |
|---|---|---|---|
| Expt 1 | |||
| Gagc | 225 | 1 | 2,500 |
| HLA-II α | 12 | 0.05 | 125 (63) |
| Actin | 5 | 0.02 | 50 |
| Expt 2 | |||
| Gagc | 165 | 1 | 2,500 |
| HLA-II α | 6.8 | 0.04 | 100 (50) |
| Actin | 4.4 | 0.027 | 68 |
Assumes 2,500 Gag molecules/virion (other estimates suggest a range of ∼1,200 to 5,000).
Native HLA-II molecules, two HLA-II α chains per complex.
Based on average value for MA, CA, NC, and p6Gag fractions.
DISCUSSION
The study of cellular proteins incorporated into retroviruses is significantly hindered by the presence of microvesicles and their associated proteins in virus preparations purified by conventional methods. Using a variety of analyses, we have demonstrated that our novel approach based on CD45 immunoaffinity depletion is able to substantially remove microvesicles from density-purified virus preparations. The depletion of CD45-containing particles from microvesicle preparations also removes essentially all of the HLA-II, demonstrating effective removal of microvesicle-associated HLA-II. By analyzing CD45-depleted virus preparations, we estimate that an average of 50 to 63 HLA-II complexes per virion, specifically HLA-DR, are incorporated onto HIV-1MN produced from H9 cells. While this level of HLA-II is considerably lower than previously estimated (2), the prior estimate was offered with the caveat that the total amount of HLA-II was measured in virus preparations that were known to contain microvesicles. In fact, the virus preparation used in the previous study contains considerably more nonviral protein than the HIV-1MN preparations presented here (4; unpublished data). Therefore, these differences are likely due to the large amount of contaminating microvesicles in the earlier samples. Although our estimate of the average number of HLA-II on the virion surface is low, the total amount of HLA-II may not be uniformly distributed among the virions. Therefore, a small percentage of virions in the population might contain a relatively high number of HLA-II molecules.
From our biochemical analyses, we have estimated that the amount of Env on HIV-1MN produced from H9 cells is ∼14 trimers per virion (9). Based on these results and those presented here, the average amount of HLA-II present on this virus is approximately four times greater than the amount of Env on the surface of HIV-1MN. This relative difference is consistent with a previous proposal that there are more HLA-II complexes on HIV-1 than Env (2). While we have found that the efficient incorporation of HLA-II into virions required Env (32), the data presented here do not support a mechanism based on a simple binding of one HLA-II to one Env. Further experiments utilizing CD45 depletion and HIV-1 Env mutants should assist in defining the mechanism for HLA-II incorporation.
Clearly, our data confirm the observation that HLA-II and other proteins in microvesicles are considerable contaminants in virus preparations produced from lymphoid cells. In addition to HLA-II, this method should be useful for examining other cellular proteins that are on the surface of the virus. We have previously developed a method for purifying the cellular proteins inside virions by subtilisin digestion of the proteins on the exterior (27). With these two methods, we have developed approaches that allow for biochemical analyses of the proteins on the surface or inside retroviruses by producing preparations that are more highly purified than those produced by conventional methods. Furthermore, since the CD45 depletion process retains virion surface proteins, this procedure is potentially useful for the production of relatively pure virion preparations for immunological and vaccine studies. The presence of HLA molecules, especially those on microvesicles, in SIV-based vaccines produced from human cells has obscured the interpretation of their efficacy (3, 39). Removal of contaminating microvesicles should assist in the clearer interpretation of these types of studies.
While we remove nearly all of the microvesicles by CD45 depletion, some still appear to be present (Fig. 8B). Our data suggest that the T-cell line-derived microvesicles that contain HLA-II also contain CD45, allowing us to remove microvesicle-associated HLA-II from these preparations by our immunoaffinity method. However, the potential presence of CD45− microvesicles necessitates that studies of host proteins other than HLA-II in virus preparations demonstrate that the protein of interest is similarly removed by CD45 depletion. Another limitation of this technique is that it depends on the presence of CD45 on the particle. While CD45 is predominantly present on hematopoietic cells, nonhematopoietic cells (e.g., HEK293T and HeLa) might not express CD45 and, therefore, likely produce microvesicles without CD45. Furthermore, hematopoietic cells can release microvesicle-like particles, exosomes, via the endocytic pathway that may not contain CD45 (6). Therefore, characterization of nonviral particles released by the virus-producing cells needs to be carried out to ensure that CD45 depletion is an appropriate procedure for removing the contaminating protein of interest from virus preparations. Other as-yet-undefined markers on CD45− particles could be used in a similar depletion method if they are excluded from virions.
While our method is effective at removing CD45-containing particles from virion preparations, our results do show a significant nonspecific loss of both nonviral and viral material. This could be due to the use of magnetic columns that were designed for cell separation and not created for small particle and protein work. Currently we are investigating procedural changes to eliminate this drawback.
While the use of OptiPrep gradients has been proposed as a method to separate microvesicles from virions, our experiments clearly found that density gradients, either sucrose- or OptiPrep-based, could not effectively resolve microvesicles from virions. This contrasts with the previous results of Dettenhofer et al. (10). A reexamination of their results shows that the primary protein contaminant removed from their virus preparations by OptiPrep gradient centrifugation was not a typical microvesicle protein, e.g., actin or HLA-II, rather it was a predominant ∼66-kDa protein that is not present in purified microvesicle preparations (Fig. 2A and reference 4). We speculate this protein was bovine serum albumin, a common contaminant from cell culture media. Thus, in the previous experiments, the OptiPrep gradients appear to remove free proteins and debris and not those associated with microvesicles. Based on our data, OptiPrep density centrifugation does not adequately resolve virions from microvesicles.
The ability to quantitate HLA-II on retroviruses promises to allow for better studies of the mechanism for HLA-II incorporation into virions. Results from these studies along with the potential to produce very pure virus preparations should allow for the improvement of and better understanding of viral vaccines and immunology studies.
Acknowledgments
We thank Julian Bess, Jr., Bill Bohn, and the late Mike Grimes for providing virus and microvesicle preparations from cell lines; Jeremy Miller, Terra Schaden-Ireland, and Rodman Smith for production and purification of virus from PBMC culture; James Hoxie for sharing the SIVMAC239/251 and SIVCP-MAC isolates; Ronald Desrosiers for sharing the SIVMAC239 G4,5 isolate; James Roser for SDS-PAGE analysis; Raymond Sowder II for protein sequencing; and Robert Gorelick for critical reading and helpful comments.
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract No. NO1-CO-12400.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
REFERENCES
- 1.Adachi, A., S. Koenig, H. E. Gendelman, D. Daugherty, S. Gattoni-Celli, A. S. Fauci, and M. A. Martin. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61:209-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur, L. O., J. W. Bess, Jr., R. C. Sowder II, R. E. Benveniste, L. D. Mann, J.-C. Chermann, and L. E. Henderson. 1992. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science 258:1935-1938. [DOI] [PubMed] [Google Scholar]
- 3.Arthur, L. O., J. W. Bess, Jr., R. G. Urban, J. L. Strominger, W. R. Morton, D. L. Mann, L. E. Henderson, and R. E. Benveniste. 1995. Macaques immunized with HLA-DR are protected from challenge with simian immunodeficiency virus. J. Virol. 69:3117-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bess, J. W., Jr., R. J. Gorelick, W. J. Bosche, L. E. Henderson, and L. O. Arthur. 1997. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology 230:134-144. [DOI] [PubMed] [Google Scholar]
- 5.Bess, J. W., Jr., P. J. Powell, H. J. Issaq, L. J. Schumack, M. K. Grimes, L. E. Henderson, and L. O. Arthur. 1992. Tightly bound zinc in human immunodeficiency virus type 1, human T-cell leukemia virus type 1, and other retroviruses. J. Virol. 66:840-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchard, N., D. Lankar, F. Faure, A. Regnault, C. Dumont, G. Raposo, and C. Hivroz. 2002. TCR activation of human T cells induces the production of exosomes bearing the TCR/CD3/zeta complex. J. Immunol. 168:3235-3241. [DOI] [PubMed] [Google Scholar]
- 7.Cantin, R., J.-F. Fortin, G. Lamontagne, and M. Tremblay. 1997. The presence of host-derived HLA-DR1 on human immunodeficiency virus type 1 increases viral infectivity. J. Virol. 71:1922-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantin, R., J.-F. Fortin, and M. Tremblay. 1996. The amount of host HLA-DR proteins acquired by HIV-1 is virus strain- and cell type-specific. Virology 218:372-381. [DOI] [PubMed] [Google Scholar]
- 9.Chertova, E., J. W. Bess, Jr., B. J. Crise, R. C. Sowder II, T. M. Schaden, J. M. Hilburn, J. A. Hoxie, R. E. Benveniste, J. D. Lifson, L. E. Henderson, and L. O. Arthur. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 76:5315-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dettenhofer, M., S. Cen, B. A. Carlson, L. Kleiman, and X. F. Yu. 2000. Association of human immunodeficiency virus type 1 Vif with RNA and its role in reverse transcription. J. Virol. 74:8938-8945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esser, M. T., J. W. Bess, Jr., K. Suryanarayana, E. Chertova, D. Marti, M. Carrington, L. O. Arthur, and J. D. Lifson. 2001. Partial activation and induction of apoptosis in CD4+ and CD8+ T lymphocytes by conformationally authentic noninfectious human immunodeficiency virus type 1. J. Virol. 75:1152-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esser, M. T., D. R. Graham, L. V. Coren, C. M. Trubey, J. W. Bess, Jr., L. O. Arthur, D. E. Ott, and J. D. Lifson. 2001. Differential incorporation of CD45, CD80 (B7-1), CD86 (B7-2), and major histocompatibility complex class I and II molecules into human immunodeficiency virus type 1 virions and microvesicles: implications for viral pathogenesis and immune regulation. J. Virol. 75:6173-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finkel, T. H., G. Tudor-Williams, N. K. Banda, M. F. Cotton, T. Curiel, C. Monks, T. W. Baba, R. M. Ruprecht, and A. Kupfer. 1995. Apoptosis occurs predominantly in bystander cells and not in productively infected cells of HIV- and SIV-infected lymph nodes. Nat. Med. 1:129-134. [DOI] [PubMed] [Google Scholar]
- 14.Frank, I., M. Piatak, Jr., H. Stoessel, N. Romani, D. Bonnyay, J. D. Lifson, and M. Pope. 2002. Infectious and whole inactivated simian immunodeficiency viruses interact similarly with primate dendritic cells (DCs): differential intracellular fate of virions in mature and immature DCs. J. Virol. 76:2936-2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gluschankof, P., I. Mondor, H. R. Gelderblom, and Q. J. Sattentau. 1997. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology 230:125-133. [DOI] [PubMed] [Google Scholar]
- 16.Gonda, M. A., S. A. Aaronson, N. Ellmore, V. H. Zeve, and K. Nagashima. 1976. Ultrastructural studies of surface features of human normal and tumor cells in tissue culture by scanning and transmission electron microscopy. J. Natl. Cancer Inst. 56:245-263. [DOI] [PubMed] [Google Scholar]
- 17.Gorelick, R. J., S. M. Nigida, J. W. Bess, Jr., L. E. Henderson, L. O. Arthur, and A. Rein. 1990. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J. Virol. 64:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenfield, E. A., K. A. Nguyen, and V. K. Kuchroo. 1998. CD28/B7 costimulation: a review. Crit. Rev. Immunol. 18:389-418. [DOI] [PubMed] [Google Scholar]
- 19.Henderson, L. E., M. A. Bowers, R. C. Sowder II, S. Serabyn, D. G. Johnson, J. W. Bess, Jr., L. O. Arthur, D. K. Bryant, and C. Fenselau. 1992. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processings, and complete amino acid sequences. J. Virol. 66:1856-1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson, L. E., R. C. Sowder II, G. Smythers, and S. Oroszlan. 1988. Chemical and immunological characterizations of equine infectious anemia virus gag-encoded proteins. J. Virol. 61:2587-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBranche, C. C., M. M. Sauter, B. S. Haggarty, P. J. Vance, J. Romano, T. K. Hart, P. J. Bugelski, and J. A. Hoxie. 1994. Biological, molecular, and structural analysis of a cytopathic variant from a molecularly cloned simian immunodeficiency virus. J. Virol. 68:5509-5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Layne, S. P., M. J. Merges, M. Dembo, J. L. Spouge, S. R. Conley, J. P. Moore, J. L. Raina, H. Renz, H. R. Gelderblom, and P. L. Nara. 1992. Factors underlying spontaneous inactivation and susceptibility to neutralization of human immunodeficiency virus. Virology 189:695-714. [DOI] [PubMed] [Google Scholar]
- 23.Nara, P. L., W. C. Hatch, N. M. Dunlop, W. G. Robey, L. O. Arthur, M. A. Gonda, and P. J. Fischinger. 1987. Simple, rapid, quantitative, syncytium-forming microassay for the detection of human immunodeficiency virus neutralizing antibody. AIDS Res. Hum. Retrovir. 3:283-302. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen, D. H., and J. E. Hildreth. 2000. Evidence for budding of human immunodeficiency virus type 1 selectively from glycolipid-enriched membrane lipid rafts. J. Virol. 74:3264-3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ott, D. E. 1997. Cellular proteins in HIV. Rev. Med. Virol. 7:167-180. [DOI] [PubMed] [Google Scholar]
- 26.Ott, D. E. 2002. Potential roles of cellular proteins in HIV-1. Rev. Med. Virol. 12:359-374. [DOI] [PubMed] [Google Scholar]
- 27.Ott, D. E., L. V. Coren, D. G. Johnson, B. P. Kane, R. C. Sowder II, Y. D. Kim, R. J. Fisher, X. Z. Zhou, K. P. Lu, and L. E. Henderson. 2000. Actin-binding cellular proteins inside human immunodeficiency virus type 1. Virology 266:42-51. [DOI] [PubMed] [Google Scholar]
- 28.Ott, D. E., L. V. Coren, D. G. Johnson, R. C. Sowder II, L. O. Arthur, and L. E. Henderson. 1995. Analysis and localization of cyclophilin A found in the virions of human immunodeficiency virus type 1 MN strain. AIDS Res. Hum. Retrovir. 11:1003-1006. [DOI] [PubMed] [Google Scholar]
- 29.Ott, D. E., L. V. Coren, B. P. Kane, L. K. Busch, D. J. Johnson, R. C. Sowder II, E. N. Chertova, L. O. Arthur, and L. E. Henderson. 1996. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 70:7734-7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ott, D. E., S. M. Nigida, Jr., L. E. Henderson, and L. O. Arthur. 1995. The majority of cells are superinfected in a cloned cell line that produces high levels of human immunodeficiency virus type 1 strain MN. J. Virol. 69:2443-2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Piatak, M., M. S. Saag, L. C. Yang, S. J. Clark, J. C. Kappes, K.-C. Luk, B. H. Hahn, G. M. Shaw, and J. D. Lifson. 1993. High levels of HIV-1 in plasma during all stages of infection determined by competitive PCR. Science 259:1749-1754. [DOI] [PubMed] [Google Scholar]
- 32.Poon, D. T., L. V. Coren, and D. E. Ott. 2000. Efficient incorporation of HLA class II onto human immunodeficiency virus type 1 requires envelope glycoprotein packaging. J. Virol. 74:3918-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Popovic, M., P. S. Sarin, M. Robert-Gurroff, V. S. Kalyanaraman, D. Mann, J. Minowada, and R. C. Gallo. 1983. Isolation and transmission of human retrovirus (human T-cell leukemia virus). Science 219:856-859. [DOI] [PubMed] [Google Scholar]
- 34.Reitter, J. N., R. E. Means, and R. C. Desrosiers. 1998. A role for carbohydrates in immune evasion in AIDS. Nat. Med. 4:679-684. [DOI] [PubMed] [Google Scholar]
- 35.Rossio, J. L., J. W. Bess, Jr., L. E. Henderson, P. Cresswell, and L. O. Arthur. 1995. HLA class II on HIV-1 particles is functional in superantigen presentation to human T cells: implications for HIV pathogenesis. AIDS Res. Hum. Retrovir. 11:1433-1439. [DOI] [PubMed] [Google Scholar]
- 36.Salter, R. D., and P. Cresswell. 1986. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 5:943-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schols, D., R. Pauwels, J. Desmyter, and E. De Clercq. 1992. Presence of class II histocompatibility DR proteins on the envelope of human immunodeficiency virus demonstrated by FACS analysis. Virology 189:374-376. [DOI] [PubMed] [Google Scholar]
- 38.Smith, S., M. Shatsky, P. Cohen, R. Warnke, M. Link, and B. Glader. 1984. Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines. Cancer Res. 44:5657-5660. [PubMed] [Google Scholar]
- 39.Stott, E. M. 1991. Anti-cell antibody in macaques. Nature(London) 353:393. [DOI] [PubMed] [Google Scholar]
- 40.Summers, M. F., L. E. Henderson, M. R. Chance, J. W. Bess, Jr., T. L. South, P. R. Blake, I. Sagi, G. Perez-Alvarado, R. C. Sowder II, D. R. Hare, and L. O. Arthur. 1993. Nucleocapsid zinc fingers detected in retroviruses: EXAFS studies of intact viruses and the solution-state structure of the nucleocapsid protein from HIV-1. Protein Sci. 1:563-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tremblay, M. J., J. F. Fortin, and R. Cantin. 1998. The acquisition of host-encoded proteins by nascent HIV-1. Immunol. Today 19:346-351. [DOI] [PubMed] [Google Scholar]