Abstract
The ability to elicit an immune response to a spectrum of human immunodeficiency virus type 1 (HIV-1) gene products from divergent strains is a desirable feature of an AIDS vaccine. In this study, we examined combinations of plasmids expressing multiple HIV-1 genes from different clades for their ability to elicit humoral and cellular immune responses in mice. Immunization with a modified Env, gp145ΔCFI, in combination with a Gag-Pol-Nef fusion protein plasmid elicited similar CD4+ and CD8+ cellular responses to immunization with either vector alone. Further, when mice were immunized with a mixture of Env from three clades, A, B, and C, together with Gag-Pol-Nef, the overall potency and balance of CD4+- and CD8+-T-cell responses to all viral antigens were similar, with only minor differences noted. In addition, plasmid mixtures elicited antibody responses comparable to those from individual inoculations. These findings suggest that a multigene and multiclade vaccine, including components from A, B, and C Env and Gag-Pol-Nef, can broaden antiviral immune responses without immune interference. Such combinations of immunogens may help to address concerns about viral genetic diversity for a prospective HIV-1 vaccine.
The genetic variation of human immunodeficiency virus type 1 (HIV-1) has created challenges for the development of a preventive AIDS vaccine (39). Not only would such a vaccine be expected to be safe and immunogenic, but it must also induce immune recognition of a broad spectrum of HIV isolates to prove highly effective (21). Though progress has been made with subtype-specific and Gag- or Env-based HIV vaccines (4, 8, 38), an alternative approach involves the utilization of multiple viral proteins from different clades that can maximize the breadth and potency of the antiviral immune response. An unresolved question for the development of such a multivalent HIV vaccine is whether this approach can elicit strong immune responses against individual gene products without cross-interference. In previous HIV vaccine studies, some multivalent DNA vaccine approaches induced suboptimal immune responses, likely due to interference among different viral antigens (15, 28). In this study, we have addressed this question by using gene-based vaccination techniques previously used in a variety of different vaccine studies (5, 25, 29, 32).
Env is a major target of both humoral and cellular immunity, while the viral genes for Gag, Pol, and Nef are potential targets of the CD8+ immune response. A modified form of HIV-1 envelope (Env), gp145ΔCFI, has been shown to improve antibody responses while maintaining its ability to induce cytotoxic-T-lymphocyte (CTL) responses (7). A fusion protein of Gag and Pol has also been developed that generates a protein from a single open reading frame that can be processed to present linear epitopes from at least four viral gene products: Gag, protease (PR), reverse transcriptase (RT), and integrase (IN) (11). To ensure that the pol region did not function in vivo, three point mutations were introduced, in PR, RT and IN, termed Pol(ΔPR ΔRT ΔIN). An additional viral protein, Nef, was included to expand its breadth, and representatives of clades A, B, and C were also generated.
The present study evaluated the immunogenicity of Env and Gag-Pol-Nef vaccine candidates alone or in combination. In addition, the ability to combine these immunogens from different clade isolates was also evaluated. The combination of Gag-Pol-Nef with Env elicited strong CD8 immunity to Env without compromising the CD4 or antibody response. In addition, combinations of Env from multiple clades help to expand the immune response to these alternative clades. The combination of multiple HIV genes from different clades may facilitate the generation of immune responses to diverse HIV strains.
MATERIALS AND METHODS
Gag-Pol-Nef immunogens.
Plasmids expressing HIV genes were synthesized by reverse translation (Genetics Computer Group, Inc., Madison, Wis.) of published sequences using codons expected for human cells. The methods used to make DNA plasmids expressing HIV-1 Gag-Pol-Nef polyproteins from different clades were similar to those previously described for Gag-Pol (11). To further inactivate viral proteins, additional inactivating mutations were inserted into PR, RT, and IN. The amino acid sequence of the Nef protein was not modified, but the NH2-terminal myristylation site required for its functional activity was not available, as it is synthesized as a fusion protein. The clade A, B, and C Gag-Pol-Nef plasmids were 9783, 9790, and 9786 nucleotides in length, respectively, and the clade A, B, and C Env plasmids are 6836, 6869, and 6829 nucleotides.
These genes were synthesized by preparation of oligonucleotides of 75 bp overlapping by 25 or of 60 bp overlapping by 20 and assembled by Pwo (Boehringer Mannheim) and Turbo Pfu (Stratagene) as described previously (7, 11). The cDNAs were cloned into the expression vector pVR1012 (7, 40). The protein sequence of each Gag polyprotein from the appropriate HIV-1 clade was used to create a synthetic version of the gag gene (gag/h) using codons preferred for expression in human cells. The synthetic gag/h gene contained sequences for all mature Gag proteins except for p1 and p6 (amino acids [aa] 433 to 500). The synthetic gag/h gene from clade A, B, or C was ligated in frame with codon-modified pol (pol/h), encoding aa 3 to 1003 from NL4-3 (GenBank accession number M19921). To inactivate the fusion proteins further, a PR mutation (Arg to Gly) was inserted at aa 553, an RT mutation (Asp to His) at aa 771, and an IN mutation (Asp to Ala) at aa 1209. A synthetic nef gene (nef/h) based on aa 1 to 206 from NL4-3 was fused to the 3′ end of pol/h by PCR to generate the appropriate Gag-Pol-Nef expression vector.
For the clade A Gag-Pol-Nef fusion protein, aa 1 to 432 from a CCR5-tropic clade A (GenBank accession number AF004885) were used and fused to the pol/h gene described above. In all three Gag-Pol-Nef plasmids, the same pol sequence was inserted, as this viral gene product is more than 90% conserved at the amino acid level among disparate clades. To add a matched Nef open reading frame, the stop codon in pol was removed, and synthetic clade A nef/h (GenBank accession number: AF069670) was fused to the 3′ end of pol/h by PCR to generate the clade A plasmid pVRC-4313. For the clade B Gag-Pol-Nef fusion protein, sequence encoding aa 1 to 432 from a CCR5-tropic clade B protein (GenBank accession number K03455) was used and fused to the pol/h described above. To add a clade B Nef protein, the stop codon from the Pol gene was removed and fused to a clade B synthetic Nef/h gene (aa 1 to 206) from HIV-1 PV22 (GenBank accession number K02083) to generate the clade B plasmid, pVRC-4306. For the clade C Gag-Pol-Nef fusion protein, aa 1 to 432 from a CCR5-tropic clade C (GenBank accession number U52953) were used and fused to the pol/h gene described above. The Pol stop codon was removed and fused to synthetic clade C Nef/h (aa 1 to 206) (GenBank accession number: U52953), designated pVRC-4311.
Alternative clade Env plasmid DNAs.
The sequences used to create the DNA plasmids encoding Env are derived from three HIV-1 CCR5-tropic strains of virus that have been modified to reduce potential cellular toxicity and increase immunogenicity by deletion of the fusion domain, the cleavage domains, and also by shortening of the interspace between heptad 1 (H1) and heptad 2 (H2), as described previously for clade B isolates (7). The synthetic protein sequence for the clade A Env polyprotein (gp160) was derived from 92rw020 (R5-tropic, GenBank accession number U51283) and designated clade A gp145ΔCFI/h. An XbaI site was inserted 18 nucleotides upstream from the ATG, together with a known Kozak sequence, and a BamHI site was created 1,912 nt downstream of the ATG for all Env expression vectors. This fragment was cloned into the XbaI-to-BamHI sites of pVR1012x/s sites. The fusion and cleavage domains from aa 486 to 519 and the interspace between H1 and H2 from aa 576 to 604 were deleted. The protein sequence of the clade B Env glycoprotein (gp160) from HXB2 (X4-tropic, GenBank accession number K03455) was used to create a synthetic version of the gene (X4gp160/h) by alteration of codons for better expression in human cells. The nucleotide sequence X4gp160/h shows little homology to the HXB2 gene, but the protein encoded is the same with the following aa substitutions: (aa 53, Phe→Leu; aa 94, Asn→Asp; aa 192, Lys→Ser; aa 215, Ile→Asn; aa 224, Ala→Thr; aa 346, Ala→Asp; and aa 470, Pro→Leu). To produce an R5-tropic version of the envelope glycoprotein (R5gp160/h), the region encoding HIV-1 envelope glycoprotein aa 205 to 361 from X4gp160/h was replaced with the corresponding region from the BaL strain of HIV-1 (GenBank accession number M68893, again using human-preferred codons). The full-length CCR5-tropic version of the envelope gene from pR5gp160/h was terminated after the codon for aa 704 to generate gp145/h. The fusion and cleavage domains from aa 503 to 536 and the interspace between H1 and H2 from aa 593 to 620 were then deleted. The protein sequence of the clade C Env polyprotein (gp145ΔCFI) from 97ZA012 (R5-tropic, GenBank accession number AF286227) was used to create a synthetic version of the gene (clade C gp145ΔCFI/h) with deletion of the fusion and cleavage domains from aa 487 to 520 and the interspace between H1 and H2 from aa 577 to 605.
Immunizations.
Mice received two 100-μl injections intramuscularly in each thigh at days 0, 14, and 42. Ten days after the final injection, mice were bled and sera were collected. Then the mice were sacrificed, spleens were removed, and the spleen cells were analyzed by intracellular cytokine flow cytometry (ICC) for CD4+ and CD8+ T-cell responses.
Flow-cytometric analysis of intracellular cytokines.
CD4+- and CD8+-T-cell responses were evaluated by using ICC for gamma interferon (IFN-γ) and tumor necrosis factor alpha (TNF-α). This sensitive assay was developed to study immune responses to HIV-1 (9, 10, 20, 23, 30). The assay was performed by removal of spleens, gentle homogenization to single-cell suspension, erythrocyte lysis with PharMLyse (BD-Pharmingen), washing with medium, and stimulation (107 cells/ml) at 37°C for 1 h with peptide pools (2.5 μg/ml for each peptide). All peptides used in this report were 15-mers overlapping by 11 amino acids that spanned the complete sequence of the HIV or negative control proteins tested. Anti-CD28 and anti-CD49d antibodies (BD-Pharmingen 553294 and 553153, respectively) were added (1 μg/ml) to the medium for costimulation. After an hour, brefeldin A (Sigma) was added to the medium (10 μg/ml) for an additional 5 h. After a total of 6 h, cells were washed and incubated with FC block (BD-Pharmingen) for 15 min on ice, fixed, and permeabilized with Cytofix/Cytoperm (BD-Pharmingen) according to the manufacturer's instructions. The cells were washed with phosphate-buffered saline (PBS) with 0.1% saponin (Sigma) followed by staining with the indicated fluorescence-labeled monoclonal antibodies against CD3, CD4, CD8, IFN-γ, and TNF-α (BD-Pharmingen) for 20 min on ice. After washing with PBS with 0.1% saponin, the cells were analyzed by fluorescence-activated cell sorting (FACS) to detect the IFN-γ- and TNF-α-positive cells in the CD4+ and CD8+ cell populations and analyzed with the program FlowJo (Tree Star, Inc.).
ELISAs.
To detect antibodies against Env proteins of different clades, enzyme-linked immunosorbent assay (ELISA) plates were coated with 100 μl of Galanthus nivalis lectin (10 μg/ml) overnight at 4°C. The lectin solution was removed from the wells and blocked with 200 μl of PBS containing 10% fetal bovine serum for 2 h at room temperature. The plates were washed twice with PBS containing 0.2% Tween 20 (PBS-T), and then 100 μl of supernatant from cells transfected with pVRC5304 (R5 gp140ΔCFI-Clade-A), pVRC2801 (R5 gp140ΔCFI-Clade-B), or pVRC5308 (R5 gp140ΔCFI-Clade-C) was added to each well, and wells were incubated for an hour at room temperature. The plates were washed with PBS-T five times, and then the sera from immunized mice from different groups were added with threefold dilutions for 1 h. The plates were washed with PBS-T five times, and then 100 μl of 1:5,000-diluted secondary antibody-conjugated horseradish peroxidase was added, and mixtures were incubated for 1 h and washed with PBS-T five times. Then 100 μl of substrate (Sigma Fast o-phenylenediamine dihydrochloride; catalog no. P-9187) was added to each well for 30 min. The reaction was then stopped by adding 100 μl of 1 N H2SO4, and the optical density (OD) reading was taken at 450 nm.
Statistical analysis.
For the simpler combination of plasmids listed in Table 1, Kruskal-Wallis tests were performed to test for overall differences in the three treatment groups' CD4+ and CD8+ response rates within each gene and clade combination at an α of 0.05. Within each of the two sets of tests (CD4+ and CD8+ responses), the Holm procedure was used to adjust the P values for multiple comparisons for each gene and clade combination. If the adjusted P value from the Kruskal-Wallis test for a given response-gene-clade combination was less than an α of 0.05, two-sided Wilcoxon tests were performed for all three possible pairs of different combinations (control vs. ABC(×4), control vs. ABC(×6), ABC(×4) vs. ABC(×6)). Again, the Holm procedure was used to adjust the P values for multiple comparisons. An adjusted P value less than an α of 0.05 was taken as evidence of a significant difference. An analogous approach was taken to test for differences among the groups immunized with Env and Gag-Pol-Nef plasmids (Fig. 1).
TABLE 1.
Experiment schema for analysis of plasmid combinations in micea
| Vaccine | Plasmid | Amt (μg) |
|---|---|---|
| VR1012 | 1012 | 50 μg |
| ABC(×4) | 1012-A-gp145ΔCFI | 8.3 μg |
| 1012-B-gp145ΔCFI | 8.3 μg | |
| 1012-C-gp145ΔCFI | 8.3 μg | |
| 1012-B-gag-pol-nef | 25 μg | |
| ABC(×6) | 1012-A-gp145ΔCFI | 8.3 μg |
| 1012-B-gp145ΔCFI | 8.3 μg | |
| 1012-C-gp145ΔCFI | 8.3 μg | |
| 1012-A-gag-pol-nef | 8.3 μg | |
| 1012-B-gag-pol-nef | 8.3 μg | |
| 1012-C-gag-pol-nef | 8.3 μg |
Different combinations of plasmids were tested in groups of 10 mice each. The first, second, and third immunizations were carried out at weeks 0, 2, and 6, and ICC was performed on spleen cells at week 7.
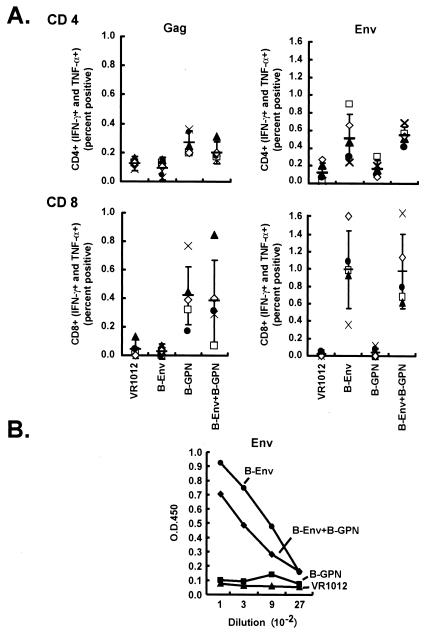
FIG. 1.
Comparison of immune response of multivalent plasmids with single gene approaches. Four groups of five mice each were immunized with the control vector alone (50 μg), Env (25 μg) with control vector (25 μg) as filler DNA, Gag-Pol-Nef (25 μg) with control vector (25 μg) as filler DNA, or Env (25 μg) with Gag-Pol-Nef (25 μg). Ten days after the final immunization, splenic cells were harvested and sensitized with B-Env peptide pool (158-peptide pool of clade B Env protein) and B-Gag peptide pool (122-peptide pool of clade B Gag protein). Six hours later, the cells were fixed, stained with monoclonal antibodies, and analyzed by FACS to detect the IFN-γ- and TNF-α-positive cells in the CD4-positive (A, top) and CD8-positive (A, bottom) population. (B) Mouse sera were collected to detect antibody against Env by ELISA. ELISA plates were prepared and coated as described in Materials and Methods with supernatant from cells transfected with pVRC2801 (R5 gp140delCFI-Clade-B) from clade B. Mouse sera from different groups were diluted starting from 1:100 to 1:2,700 before testing. The ELISA titers are shown for the group immunized with pVR1012 (▴), with pVR1012-B-Gag-Pol-Nef (GPN) and filler DNA (▪), with pVR1012-B-gp145ΔCFI and filler DNA(•), or with 1012-B-gp145ΔCFI + 1012-B-Gag-Pol-Nef (⧫). Each point represents the average OD reading from the five animals per group.
RESULTS
A combination of Env and Gag-Pol-Nef plasmids elicited CD4+ and CD8+ responses to Env and Gag similar to those obtained with single plasmids alone.
To examine whether combined immunization with Env and Gag-Pol-Nef plasmids would enhance or inhibit antigen-specific responses, the CD4, CD8, and antibody responses to Env were analyzed. Four groups of mice with five mice per group were immunized with the control vector alone, Env with control vector as filler DNA, Gag-Pol-Nef with control vector as filler DNA, or Env with Gag-Pol-Nef. Ten days after the final DNA immunization, animals were sacrificed, and splenocytes were incubated with overlapping Gag peptide pools. Intracellular IFN-γ and TNF-α expression in stimulated CD4+ or CD8+ lymphocytes were analyzed by flow cytometry, and positive cells were enumerated. Cells from mice immunized with Gag-Pol-Nef alone and those immunized with the combination of Env and Gag-Pol-Nef responded similarly to Gag stimulation (Fig. 1A, left). Likewise, lymphocytes from mice vaccinated with Env alone and those with a combination of Env and Gag-Pol-Nef responded similarly to incubation with Env peptide pools (Fig. 1A, right). Based on statistical analysis, there was no difference in CD4 response to Gag between the Gag-Pol-Nef group and the combined Env and Gag-Pol-Nef group (P = 0.1746). Also, there was no difference in the CD4 response to Env between the Env group and the combined Env and Gag-Pol-Nef group (P value = 0.6905). In the case of CD8 responses to Gag, there was also no statistical difference between Gag and the combined Env and Gag-Pol-Nef group (P value = 1.0), and in the case of the CD8 responses to Env, there was also no statistical difference between Env and the combined Env and Gag-Pol-Nef group (P value = 1.0). Similarly, antibody to Env showed similar titers in both groups (Fig. 1B). There was no statistical difference between Env and the combined Env and Gag-Pol-Nef group (P > 0.05) in antibody response to Env at all four dilutions. This result suggested that combination plasmid vaccination did not cause immune interference but instead led to expanded breadth of the immune response. To determine whether the addition of alternative clades would prove similarly immunogenic, more complex plasmid combinations were evaluated.
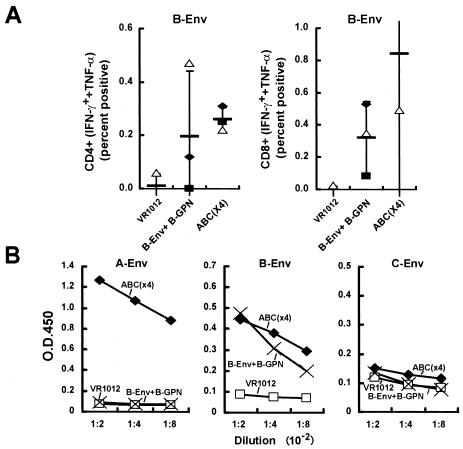
Combination of Env clades and Gag-Pol-Nef vaccination elicits immune responses similar to those obtained with single-clade immunogens.
We next determined whether the inclusion of multiple Env immunogens would affect the breadth and potency of the immune response. Mice were immunized with a negative control plasmid, combined Env and Gag-Pol-Nef (both from clade B), or Env from clades A, B, and C with Gag-Pol-Nef from clade B, termed ABC(×4). In the ABC(×4) group, the three Env proteins were retained in equal proportions, and the ratio of all Env proteins to all Gag-Pol-Nef proteins was kept constant (1:1, wt/wt). Both the combined Env and Gag-Pol-Nef group and the ABC(×4) group induced CD4+ and CD8+ responses similar to those obtained with clade B Env (Fig. 2A). Some minor variations in immune responses were seen between groups; however, both the clade B and the ABC(×4) groups showed comparable CD4+ and CD8+ responses to clade B Env peptide stimulation by intracellular flow analysis. For antibody responses, ABC(×4) showed a measurable response to clade A Env stimulation but, as expected, not in the clade B-immunized group, which did not contain clade A Env. More importantly, immunization with the clade B immunogens gave rise to titer responses to clade B Env similar to those obtained with ABC(×4), again showing that the mixture of clades did not inhibit the responses to a single-clade (clade B) Env component, despite the relative dilution of the immunogen. Neither the ABC(×4) nor the clade C Env alone (data not shown) induced a high-titer antibody response, possibly because of the lack of highly reactive epitopes in mice (Fig. 2B). These results indicated that the addition of multiple Env proteins from alternative clades to Gag-Pol-Nef did not interfere with T-cell or humoral immunity and instead added breadth to the immune response.
FIG. 2.
T-cell and antibody responses in mice immunized with Gag-Pol-Nef and clade B Env compared to Gag-Pol-Nef and clade A, B, C Env proteins. Mice (n = 3) were immunized with a total of 50 μg of control vector, Gag-Pol-Nef and clade B Env (1:1 ratio), or Gag-Pol-Nef and Env from clades A, B, and C (1:0.33:0.33:0.33 ratio). (A) Ten days after the final immunization, splenic cells were harvested and sensitized with a B-Env peptide pool (158 peptide pool of clade B Env protein). For controls, an Ebola glycoprotein peptide pool (22 peptides) or unstimulated cells served as a negative control, and phorbol myristate acetate was used as the positive control (data not shown). Six hours later, the cells were fixed, stained with monoclonal antibodies, and analyzed by FACS to detect the IFN-γ- and TNF-α-positive cells in the CD4 and CD8 positive populations (A). The symbols depict the individual results for the three mice in each group. The thin horizontal bar represents the average of the three data points, with a standard deviation error bar. (B) Sera from the three groups of animals were collected 10 days after the third immunization, and ELISA was performed to detect the antibody against the respective clade Env proteins as described in Materials and Methods. Mouse sera from different groups were diluted from 1:200 to 1:800 for testing. Each bar represents the average OD reading from the three mice per group.
Comparison of different multiple-clade immunogens.
We next compared different combinations of plasmids that could elicit immune responses to multiple immunogens. Mice were immunized with the control plasmid and two combinations of plasmids (Table 1), including a combination of six plasmids, designated ABC(×6), because it covered Gag, Nef, and Env from clades A, B, and C with Pol from clade B, or the ABC group with four components, ABC(×4), in which the Gag-Pol-Nef fusion protein from clade B was used alone, rather than with the Gag-Pol-Nef proteins from clades A and C. As above, the three Env clades were retained in similar ratios and amounts in both formulations, and the ratio of all Env proteins to all Gag-Pol-Nef proteins was kept constant (1:1, wt/wt).
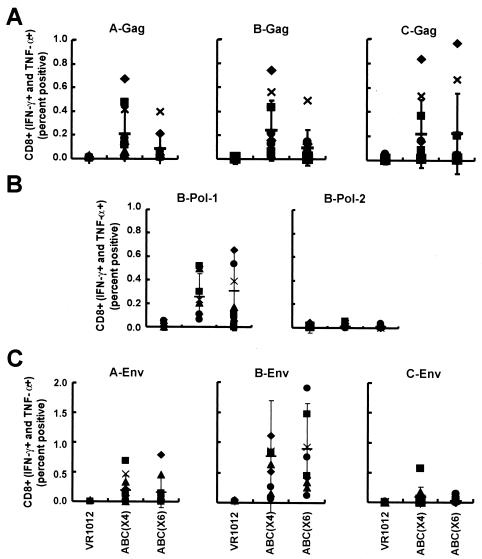
Both plasmid combination groups had similar CD8 responses to Gag, Pol, and Env from clade B but not from other clades (Fig. 3) and Table 2. Responses to Gag from clades A and B were significantly higher than the control (pVR1012) for both ABC(×6) and ABC(×4), but the differences between the response rates of the two treatment groups were not significantly different for either of these clades. CD8+ responses to Pol-1 and Env from clade B were significantly higher than the control (pVR1012) responses for both ABC(×6) and ABC(×4). However, CD8+ responses to Env from clade A were higher than the control for ABC(×6) only (P = 0.0316) (Fig. 3C and Table 2).
FIG. 3.
CD8+-T-cell responses to different clade and gene combination vaccine candidates by intracellular cytokine analysis. Three groups of mice were immunized with a control vector (VR1012), ABC(×4), or ABC(×6) as described in Table 1. Ten days after the final immunization, splenic cells were harvested and sensitized with the following peptide pools: A-Gag (125 peptides), B-Gag (122 peptides), C-Gag (105 peptides), A-Env (154 peptides), B-Env (158 peptides), C-Env (154 peptides), B-Pol-1 (120 peptides from the first half of clade B Pol), or B-Pol-2 (128 peptides from the second half of clade B Pol). Cells were stimulated and analyzed by FACS, with positive and negative controls as described in the legend to Fig. 2 to detect the IFN-γ- and TNF-α-positive cells in the CD8+ population. The symbols show the individual results for the 10 mice in each group. The thin horizontal bar is the average of the 10 data points, with standard deviation bars.
TABLE 2.
Summary of T-cell and antibody responses to vaccine candidates
| Analysis | Vaccine | Response to:
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A-Gag | B-Gag | C-Gag | A-Env | B-Env | C-Env | B-pd-1 | B-pd-2 | A-Nef | B-Nef | C-Nef | ||
| Intracellular cytokine staininga | ||||||||||||
| CD4 | ABC(×4) | + | + | − | + | + | + | + | + | − | + | − |
| ABC(×6) | + | + | + | + | + | ++ | − | + | − | − | − | |
| CD8 | ABC(×4) | + | + | − | − | + | − | + | − | − | − | − |
| ABC(×6) | + | + | + | + | + | − | + | − | − | − | − | |
| ELISAb | ABC(×4) | + | + | + | ||||||||
| ABC(×6) | + | + | + | |||||||||
CD4+- and CD8+-T-cell responses to different vaccine candidates. When Holm-adjusted Kruskal-Wallis tests indicated overall significant differences, the data from all possible pairs of groups were compared by Wilcoxon tests with a Holm adjustment for the multiple comparisons. −, no statistically significant difference from the control (P > 0.05); +, statistically significant difference from the control only (P < 0.05); ++, statistically significant difference from both the control and all other treatment groups (P = 0.05).
Antibody responses to different vaccine candidates. +, the average antibody titer of the group was more than 1:1,000.
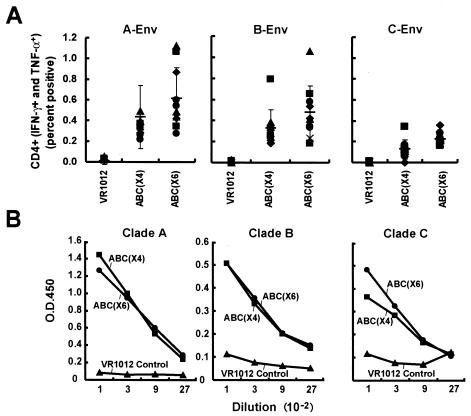
ABC(×6) and ABC(×4) induced similar CD4+ responses, in contrast to the control (pVR1012) plasmids in mice. Both stimulated higher CD4+ responses for Gag from clade A and B, Pol-2 from clade B, and Env from clades A and B (Table 2). ABC(×6) elicited significantly higher CD4+ responses to clade C Gag (P = 0.0138) than the control (pVR1012), while for Nef and Pol-1 from clade B, only ABC(×4) provoked significantly higher CD4+ responses than the control (P = 0.0097 for Nef and 0.0054 for Pol-1). CD4+ responses to Env from clade C were higher for ABC(×6) than ABC(×4) (P = 0.0418), although the responses for both groups were significantly higher than the control (Fig. 4 and Table 2). ABC(×4), but not ABC(×6), showed a response to Nef from clade B (Table 2). In summary, except for relatively minor differences in specificity, it appeared that ABC(×6) and ABC(×4) elicited comparable cell-mediated immune responses.
FIG. 4.
CD4+-T-cell and antibody responses to combination gene and clade vaccine candidates by intracellular flow cytometry and ELISA. Three groups of mice were immunized with the indicated control or combination vaccines as shown in Table 1. (A) Ten days after the final immunization, splenic cells were harvested and sensitized with the indicated peptide pools as described in the legend to Fig. 2. Individual responses are shown with the symbols, and the thin horizontal bar depicts the average of the 10 data points, with a standard deviation error bar. (B) Sera from the three groups of animals were collected 10 days after the third immunization, and ELISA was performed to detect the antibody against envelope as described in Materials and Methods. Mouse sera from different groups were diluted starting from 1:100 to 1:2,700 for testing. Each bar represents the average OD reading from the 10 mice per group.
Similar antibody responses in ABC(×4)-, ABC(×6)-, and single-clade-vaccinated mice to Env proteins from all clades.
Sera from ABC(×4), ABC(×6), or single-clade groups were tested for antibody responses by using a lectin-capture HIV-1 Env protein ELISA system. The sera from the two test groups showed similar responses to Env protein to all three clades (Fig. 4B). Antibody titers against clade A Env protein from both ABC(×4) and ABC(×6) groups were higher than the titers of antibody against clade B and clade C Env protein; however, there was no significant difference between the two groups in terms of their antibody response magnitude. This result suggested that addition of Gag and Nef immunogens from clade A and clade C to ABC(×4) groups did not interfere with the antibody responses against Env from clade B.
DISCUSSION
One requirement of a highly effective AIDS vaccine is the need to induce both neutralizing antibodies and cellular immunity to the many strains of HIV-1 that circulate throughout the world. In this report, we have evaluated the ability of plasmid DNA vaccines to elicit immune responses to multiple gene products of HIV-1 from alternative clades of virus. The goal was to elicit both antibody and T-cell responses against various HIV genes from these different clades. Env, Gag, Pol, and Nef were chosen as targets because they represent the major expressed proteins during viral infection. A mutant Env with deletions in the cleavage site, fusion domain, and a region between the heptad repeats was used for its ability to elicit a more potent humoral immune response while retaining its ability to stimulate Env-specific CTLs (7).
A variety of previous studies have shown that CTLs contribute to the control of viremia and protect against the progression of HIV disease (6, 12, 16, 17, 26, 27, 31, 33-36). Processed forms of Gag, Pol, Nef, and Env presented on class I major histocompatibility complex antigens can serve as the targets of CTLs that recognize and lyse HIV-1 infected cells, in this way contributing to the efficacy of a preventive vaccine. It is hoped that if the T-cell response is sufficiently robust, these cells will kill HIV-infected cells before the virus can replicate and establish a reservoir of infection in vivo. For a globally effective vaccine, it will be necessary to elicit CTL that react with strains from multiple clades. Though there may be some cross-clade reactivity after immunization with a single clade (e.g., see reference 13), there is also evidence of disparities in such immune responses (e.g., see reference 9). It therefore appeared desirable to include representatives of the major classes of virus in a DNA vaccine to induce cross-clade immunity. However, the main concern of such a cocktail is whether it will cause interference between gene-specific immune responses. Interference among immune responses to various viral genes has been seen previously in murine HIV immunization studies (15, 28). Recently, studies of modifications to HIV DNA vaccines, including different combinations of viral genes, altered RNA structure or codon usage, and/or stimulatory cytokine genes, have shown more encouraging results in mice (41). More importantly, some approaches have shown promise in challenge studies using nonhuman primates (1, 3, 14, 19, 22, 37), though complete protection against infection has been difficult to achieve. Additional modifications were therefore incorporated in this study in an attempt to improve efficacy.
When the immune responses to different combinations of Env and Gag-Pol-Nef were compared, there was no decrease in the humoral and cellular response to clade B mutant Env plasmid and Gag-Pol-Nef plasmids when mixed compared with the responses to the two plasmids individually (Fig. 1). When the complexity was increased to four components, including gp145ΔCFI from three clades and Gag-Pol-Nef from clade B, there was no interference with the humoral response to B-Env while the immune response to other clades was enhanced.
When the complexity of the vaccine was increased to six components, ABC(×6), containing the same Env-gp145ΔCFI from different clades as in group ABC(×4) plus the Gag-Pol-Nef fusion protein from clades A and C, minor differences in immunogenicity were seen. Analysis of the Gag response showed that ABC(×4) elicited CD4+ and CD8+ responses to clades A and B, while ABC(×6) improved the response to clade C Gag peptides. The lack of CD4+ and CD8+ responses to clade C Gag in ABC(×4) probably is due to the absence of clades A and C Gag; however, ABC(×4) containing only clade B Gag could induce both CD4+ and CD8+ responses to clade A Gag even though it shares only 85% homology in amino acid sequence. This result suggested that clades A and B Gag share some common CD4+ and CD8+ epitopes but differ more substantially from clade C Gag in mice. In contrast, the CD4+ and CD8+ responses against Env between ABC(×4) and ABC(×6) were similar: both groups elicited comparable CD4+ responses against all three clades and generated similar CD8+ responses against clade B Env. ABC(×6) also induced a significant CD8+ response to Env from clade A. Since both ABC(×4) and ABC(×6) contained the same combination of Env from clades A, B, and C, the similar CD4+ and CD8+ responses against Env from the two groups were not unexpected.
For Pol responses, both groups demonstrated CD4+ and CD8+ responses against Pol from clade B. The ABC(×4) group elicited a CD4+ response to both sets of Pol peptides, while ABC(×6) stimulated a CD4+ response only against one of the two Pol peptide pools (Table 2). Both groups induced CD8+ responses to the first half of the clade B Pol (Fig. 3B, left panel). For Nef, only the ABC(×4) group elicited a CD4+ response against Nef from clade B (Table 2). The poor anti-Nef response also may be due to the inability of BALB/c mice to recognize Nef epitopes, as other groups have reported that Nef is highly immunogenic in other strains of mice (15).
In addition, we attempted to determine whether CD4+ and CD8+ T-cell responses against multigenes would affect humoral responses. There was no significant difference among different groups in ELISA titers (summarized in Table 2). All the groups showed similar antibody titers to Env protein from clades A, B, and C (Fig. 4B). These data suggested that there was no interference among different clades of Env in antibody response. Equally importantly, there was no interference among various viral genes between humoral and cellular responses.
In summary, the ABC(×4) vaccine regimen was able to induce substantial and balanced CD4+ and CD8+ T-cell responses to the viral antigens from different clades. Though ABC(×6) induced a comparable response, the lower complexity of ABC(×4) suggests that it is a better candidate for development for clinical production. The more complex combination, ABC(×6), could also be more variable because more epitopes are included, and the larger number of components could complicate production and quality control that might also affect immune responses. These factors suggest that more complexity does not always yield incremental immunity, as with ABC(×6). Though there was no significant loss of immunogenicity with the vaccine studied here, this complexity has been problematic in previous studies (15, 28). For this reason, immune analyses, such as those performed in this study, are needed to address these issues and facilitate the development of safe and effective vaccines. The results here suggest that a multigene HIV-1 DNA vaccine is feasible because the immune responses to individual genes do not cause interference when combined with one another. Because mouse serum is difficult to evaluate for neutralizing antibody activity due to its high background in neutralization assays, future studies in other animal species will address this issue. As the HIV-1 pandemic continues to grow, virus variability becomes increasingly problematic. Though a few subtypes of HIV-1 predominate in different regions of the world, a rising number of recombinant strains have been reported lately (18). Such viruses continually mutate and escape (2, 24) during different stages of infection. A multiepitope and multiclade immune response should help to reduce the likelihood of viral escape. The data presented in this report may therefore help to guide the development of improved vaccines against diverse strains of HIV.
Acknowledgments
The first four authors contributed equally to this study.
We thank Ann Gooch and Ati Tislerics for help with manuscript preparation, Karen Stroud and Toni Garrison for figure preparation, and members of the Nabel laboratory for helpful discussions.
REFERENCES
- 1.Amara, R. R., F. Villinger, J. D. Altman, S. L. Lydy, S. P. O'Neil, S. I. Staprans, D. C. Montefiori, Y. Xu, J. G. Herndon, L. S. Wyatt, M. A. Candido, N. L. Kozyr, P. L. Earl, J. M. Smith, H. L. Ma, B. D. Grimm, M. L. Hulsey, J. Miller, H. M. McClure, J. M. McNicholl, B. Moss, and H. L. Robinson. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69-74. [DOI] [PubMed] [Google Scholar]
- 2.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, and M. G. Lewis. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 4.Bojak, A., D. Hammer, H. Wolf, and R. Wagner. 2002. Muscle specific versus ubiquitous expression of Gag based HIV-1 DNA vaccines: a comparative analysis. Vaccine 20:1975-1979. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, M. C., J. Tartaglia, F. Verdier, P. Kourilsky, A. Lindberg, M. Klein, and P. Moingeon. 2000. Recombinant viruses as a tool for therapeutic vaccination against human cancers. Immunol. Lett. 74:11-25. [DOI] [PubMed] [Google Scholar]
- 6.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chakrabarti, B. K., W. P. Kong, B.-Y. Wu, Z.-Y. Yang, J. Friborg, Jr., X. Ling, S. R. King, D. C. Montefiori, and G. J. Nabel. 2002. Modifications of the human immunodeficiency virus envelope glycoprotein enhance immunogenicity for genetic immunization. J. Virol. 76:5357-5368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deml, L., A. Bojak, S. Steck, M. Graf, J. Wild, R. Schirmbeck, H. Wolf, and R. Wagner. 2001. Multiple effects of codon usage optimization on expression and immunogenicity of DNA candidate vaccines encoding the human immunodeficiency virus type 1 Gag protein. J. Virol. 75:10991-11001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorrell, L., B. E. Willcox, E. Y. Jones, G. Gillespie, H. Njai, S. Sabally, A. Jaye, K. DeGleria, T. Rostron, E. Lepin, A. McMichael, H. Whittle, and S. Rowland-Jones. 2001. Cytotoxic T lymphocytes recognize structurally diverse, clade-specific and cross-reactive peptides in human immunodeficiency virus type-1 gag through HLA-B53. Eur. J. Immunol. 31:1747-1756. [DOI] [PubMed] [Google Scholar]
- 10.Goepfert, P. A., A. Bansal, B. H. Edwards, G. D. Ritter, Jr., I. Tellez, S. A. McPherson, S. Sabbaj, and M. J. Mulligan. 2000. A significant number of human immunodeficiency virus epitope-specific cytotoxic T lymphocytes detected by tetramer binding do not produce gamma interferon. J. Virol. 74:10249-10255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, Y., W. Kong, and G. J. Nabel. 2001. Human immunodeficiency virus type 1-specific immunity after genetic immunization is enhanced by modification of Gag and Pol expression. J. Virol. 75:4947-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jin, X., D. E. Bauer, S. E. Tuttleton, S. Lewin, A. Gettie, J. Blanchard, C. E. Irwin, J. T. Safrit, J. Mittler, L. Weinberger, L. G. Kostrikis, L. Zhang, A. S. Perelson, and D. D. Ho. 1999. Dramatic rise in plasma viremia after CD8+ T cell depletion in simian immunodeficiency virus-infected macaques. J. Exp. Med. 189:991-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keating, S. M., R. C. Bollinger, T. C. Quinn, J. B. Jackson, and L. M. Carruth. 2002. Cross-clade T lymphocyte-mediated immunity to HIV type 1: implications for vaccine design and immunodetection assays. AIDS Res. Hum. Retroviruses 18:1067-1079. [DOI] [PubMed] [Google Scholar]
- 14.Kim, J. J., J. S. Yang, L. K. Nottingham, D. J. Lee, M. Lee, K. H. Manson, M. S. Wyand, J. D. Boyer, K. E. Ugen, and D. B. Weiner. 2001. Protection from immunodeficiency virus challenges in rhesus macaques by multicomponent DNA immunization. Virology 285:204-217. [DOI] [PubMed] [Google Scholar]
- 15.Kjerrstrom, A., J. Hinkula, G. Engstrom, V. Ovod, K. Krohn, R. Benthin, and B. Wahren. 2001. Interactions of single and combined human immunodeficiency virus type 1 (HIV-1) DNA vaccines. Virology 284:46-61. [DOI] [PubMed] [Google Scholar]
- 16.Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuiken, C., B. Foley, B. Hahn, P. Marx, F. McCutchan, J. Mellors, J. Mullins, S. Wolinsky, and B. Korber. 2000. Human retroviruses and AIDS 1999. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 19.Letvin, N. L. 2002. Strategies for an HIV vaccine. J. Clin. Investig. 110:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maecker, H. T., H. S. Dunn, C. J. Pitcher, E. Khatamzas, C. J. Pitcher, T. Bunde, N. Persaud, W. Trigona, T. M. Fu, E. Sinclair, B. M. Bredt, J. M. McCune, V. C. Maino, F. Kern, and L. J. Picker. 2001. Use of overlapping peptide mixtures as antigens for cytokine flow cytometry. J. Immunol. Methods 255:27-40. [DOI] [PubMed] [Google Scholar]
- 21.Mascola, J. R., and G. J. Nabel. 2001. Vaccines for the prevention of HIV-1 disease. Curr. Opin. Immunol. 13:489-495. [DOI] [PubMed] [Google Scholar]
- 22.McKay, P. F., J. E. Schmitz, D. H. Barouch, M. J. Kuroda, M. A. Lifton, C. E. Nickerson, D. A. Gorgone, and N. L. Letvin. 2002. Vaccine protection against functional CTL abnormalities in simian human immunodeficiency virus-infected rhesus monkeys. J. Immunol. 168:332-337. [DOI] [PubMed] [Google Scholar]
- 23.Migueles, S. A., and M. Connors. 2001. Frequency and function of HIV-specific CD8+ T cells. Immunol. Lett. 79:141-150. [DOI] [PubMed] [Google Scholar]
- 24.Mortara, L., F. Letourneur, H. Gras-Masse, A. Venet, J. G. Guillet, and I. Bourgault-Villada. 1998. Selection of virus variants and emergence of virus escape mutants after immunization with an epitope vaccine. J. Virol. 72:1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moss, B. 1996. Genetically engineered poxviruses for recombinant gene expression, vaccination, and safety. Proc. Natl. Acad. Sci. USA 93:11341-11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moss, P. A. H., S. L. Rowland-Jones, P. M. Frodsham, S. McAdam, P. Giangrande, A. J. McMichael, and J. I. Bell. 1995. Persistent high frequency of human immunodeficiency virus-specific cytotoxic T cells in peripheral blood of infected donors. Proc. Natl. Acad. Sci. USA 92:5773-5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Musey, L., J. Hughes, T. Schacker, T. Shea, L. Corey, and M. J. McElrath. 1997. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337:1267-1274. [DOI] [PubMed] [Google Scholar]
- 28.Muthumani, K., S. Kudchodkar, D. Zhang, M. L. Bagarazzi, J. J. Kim, J. D. Boyer, V. Ayyavoo, G. N. Pavlakis, and D. B. Weiner. 2002. Issues for improving multiplasmid DNA vaccines for HIV-1. Vaccine 20:1999-2003. [DOI] [PubMed] [Google Scholar]
- 29.Nabel, G. J. 2001. Challenges and opportunities for development of an AIDS vaccine. Nature 410:1002-1007. [DOI] [PubMed] [Google Scholar]
- 30.Novitsky, V., N. Rybak, M. F. McLane, P. Gilbert, P. Chigwedere, I. Klein, S. Gaolekwe, S. Y. Chang, T. Peter, I. Thior, T. Ndung'u, F. Vannberg, B. T. Foley, R. Marlink, T. H. Lee, and M. Essex. 2001. Identification of human immunodeficiency virus type 1 subtype C Gag-, Tat-, Rev-, and Nef-specific ELISpot-based cytotoxic T-lymphocyte responses for AIDS vaccine design. J. Virol. 75:9210-9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 32.Ramsay, A. J., K. H. Leong, and I. A. Ramshaw. 1997. DNA vaccination against virus infection and enhancement of antiviral immunity following consecutive immunization with DNA and viral vectors. Immunol. Cell Biol. 75:382-388. [DOI] [PubMed] [Google Scholar]
- 33.Rowland-Jones, S. L., T. Dong, K. R. Fowke, J. Kimani, P. Krausa, H. Newell, T. Blanchard, K. Ariyoshi, J. Oyugi, E. Ngugi, J. Bwayo, K. S. MacDonald, A. M. McMichael, and F. A. Plummer. 1998. Cytotoxic T cell response to multiple conserved HIV epitopes in HIV-resistant prostitutes in Nairobi. J. Clin. Investig. 102:1758-1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rowland-Jones, S. L., D. F. Nixon, M. C. Aldhous, F. Gotch, K. Ariyoshi, N. Hallam, J. S. Kroll, K. Froebel, and A. McMichael. 1993. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet 341:860-861. [DOI] [PubMed] [Google Scholar]
- 35.Rowland-Jones, S. L., J. Sutton, K. Ariyoshi, T. Dong, F. Gotch, S. McAdam, D. Whitby, S. Sabally, A. Gallimore, T. Corrah, et. al. 1995. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat. Med. 1:59-64. [DOI] [PubMed] [Google Scholar]
- 36.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 37.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 38.Srivastava, I. K., K. VanDorsten, L. Vojtech, S. W. Barnett, and L. Stamatatos. 2003. Changes in the immunogenic properties of soluble gp140 human immunodeficiency virus envelope constructs upon partial deletion of the second hypervariable region. J. Virol. 77:2310-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Groen, G., P. N. Nyambi, E. Beirnaert, D. Davis, K. Fransen, L. Heyndrickx, P. Ondoa, G. van der Auwera, and W. Janssens. 1998. Genetic variation of HIV type 1: relevance of interclade variation to vaccine development. AIDS Res. Hum. Retrovir. 14(Suppl. 3):S211-S221. [PubMed] [Google Scholar]
- 40.Yang, Z., R. Delgado, L. Xu, R. F. Todd, E. G. Nabel, A. Sanchez, and G. J. Nabel. 1998. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science 279:1034-1037. [DOI] [PubMed] [Google Scholar]
- 41.zur Megede, J., G. R. Otten, B. Doe, H. Liu, L. Leung, J. B. Ulmer, J. J. Donnelly, and S. W. Barnett. 2003. Expression and immunogenicity of sequence-modified human immunodeficiency virus type 1 subtype B pol and gagpol DNA vaccines. J. Virol. 77:6197-6207. [DOI] [PMC free article] [PubMed] [Google Scholar]