Abstract
Human EED, a member of the superfamily of WD-40 repeat proteins and of the Polycomb group proteins, has been identified as a cellular partner of the human immunodeficiency virus type 1 (HIV-1) matrix (MA) protein (R. Peytavi et al., J. Biol. Chem. 274:1635-1645, 1999). In the present study, EED was found to interact with HIV-1 integrase (IN) both in vitro and in vivo in yeast. In vitro, data from mutagenesis studies, pull-down assays, and phage biopanning suggested that EED-binding site(s) are located in the C-terminal domain of IN, between residues 212 and 264. In EED, two putative discrete IN-binding sites were mapped to its N-terminal moiety, at a distance from the MA-binding site, but EED-IN interaction also required the integrity of the EED last two WD repeats. EED showed an apparent positive effect on IN-mediated DNA integration reaction in vitro, in a dose-dependent manner. In situ analysis by immunoelectron microscopy (IEM) of cellular distribution of IN and EED in HIV-1-infected cells (HeLa CD4+ cells or MT4 lymphoid cells) showed that IN and EED colocalized in the nucleus and near nuclear pores, with maximum colocalization events occurring at 6 h postinfection (p.i.). Triple colocalizations of IN, EED, and MA were also observed in the nucleoplasm of infected cells at 6 h p.i., suggesting the ocurrence of multiprotein complexes involving these three proteins at early steps of the HIV-1 virus life cycle. Such IEM patterns were not observed with a noninfectious, envelope deletion mutant of HIV-1.
Integration of the proviral DNA into the host cell genome has been considered an obligatory step of the human immunodeficiency virus type 1 (HIV-1) life cycle, as for all known retroviruses (20, 35). The host DNA sites for retroviral integration have been long debated. Although it has been reported that integration events are statistically more frequent in inactive than active chromatin (11, 81, 82), other data suggest that retrovirus integration preferentially occurs in transcriptionally active chromatin (65) and in regions of DNA distortion (57-59). Recent studies have confirmed that transcriptional units are preferred targets for both murine leukemia virus (MLV) and HIV-1 (66, 83), but refined mapping of integration sites has revealed significant differences between HIV-1 and MLV, the latter preferentially integrating near the start of transcription (83). Whatever the scenario followed by the different types of retroviruses in their host cells, it is reasonable to assume that proteins which control the state of cellular chromatin would play a key role in the integration of HIV proviral DNA and, possibly, in further steps of the virus life cycle. The viral RNA genome is retrotranscribed by the reverse transcriptase (RT) into a double-stranded DNA molecule, which is the substrate for the viral integrase (IN) in the integration reaction. The RT reaction starts within the central core of extracellular virions, before they bind to the surface of the host cell, and is completed after the virus enters the cell (reviewed in references 5, 30, and 35). The final reaction takes place after partial uncoating of the HIV-1 particle, within a protein-nucleic acid complex termed preintegration complex (PIC). PIC competent for integration reaction in vitro has been isolated from HIV-1-infected cells (20, 43, 48, 49).
It is generally recognized that PIC is composed of linear double-stranded DNA, RT, IN, matrix protein (MA), nucleocapsid protein (NC), and auxiliary protein Vpr (reviewed in references 13, 16, 30, and 71). One of these components, the MA protein, has been identified as an effector of various biological functions in the virus life cycle (reviewed in reference 23). Since the MA carries a potential nuclear localization signal, it has been hypothesized to mediate the transport of PIC to the cell nucleus and its traverse of the nuclear pore complex (6, 7, 26, 27). However, the observation that a mutant HIV-1 virus with most of the MA domain deleted was still capable of infecting nondividing cells (60) argued against a major role of the basic MA sequences in the nuclear import of the PIC, and other data seem to assign this function to the Vpr protein (13, 16, 22, 71), IN (2, 52), and the central DNA flap (18, 85). Several cellular proteins such as high-mobility-group proteins (19, 28), the barrier-to-autointegration factor (41), IN interactor 1 (Ini1) (40, 48, 84), and human lens epithelium-derived growth factor (LEDGF/p75) (45), have been identified as partners of IN and/or cofactors of integration reaction, and at least one of them, the barrier-to-autointegration factor, has been found to be associated with PIC of MLV and HIV-1 (43, 77). These proteins could participate, directly or indirectly, in the cellular trafficking of PIC and/or in provirus integration.
One of the cellular partners of the HIV-1 MA protein has been identified as EED (54), the human homolog of the mouse embryonic ectoderm development (eed) gene product (51, 67, 68, 74), and a member of the WD-40 repeat superfamily of proteins (50). The eed gene is highly conserved in humans and mice and is also homologous to the Drosophila esc gene. Both eed and esc belong to the family of widely conserved Polycomb group (Pc-G) of genes (17, 51). EED and many other proteins of the Pc-G family act as transcriptional repressors and gene silencers (1, 3, 17, 64, 67, 68, 74). The product of the Pc-G genes in eukaryotes are involved in chromatin remodeling and, in particular, in the maintenance of the silent state of chromatin (reviewed in reference 55) and the reduction of DNA accessibility (21). EED has also been found to colocalize with histone H1 in heterochromatin subdomains of neuronal cell nuclei containing inactive DNA (1).
Previous studies have indicated that MA, RT, and IN proteins are each a component of HIV-1 PIC (26, 27, 49). Since MA has been found to bind to cellular EED (54), we explored the possibility of a protein-protein interaction between EED and IN and of the occurrence of a ternary complex of EED, MA, and IN. We found that IN and EED proteins were indeed able to interact in vitro, as well as in vivo, in yeast. In human cells infected with HIV-1 (HeLa CD4+ cells or MT4 lymphoid cells), in situ analysis by immunoelectron microscopy (IEM) showed that EED, IN, and MA proteins colocalized within the nucleus, with a maximum number of colocalization events occurring at 6 h after infection. Such patterns of nuclear colocalization were not observed in cells incubated with a noninfectious, envelope deletion mutant of HIV-1. Our data suggest that EED, a cellular partner of both MA and IN, would play a functional role in the HIV-1 life cycle, acting at early steps of virus infection.
MATERIALS AND METHODS
Cells, HIV-1 viruses and virus infection.
Human embryonic kidney cells (HEK-293) and epithelial cells (P4P56) (HeLa CD4+, lck+, LTR-LacZ) (52) were grown in Dulbecco modified Eagle medium (Gibco), supplemented with glutamine, antibiotics, and 10% fetal calf serum. MT4 lymphoid cells were grown in RPMI medium with the same supplements. The construction of HIV-1 BRU infectious molecular clone expressing the viral IN fused to the Flag epitope has been described in a previous study (53). It is abbreviated as BRU-FlagWT in the present study. BRU-FlagΔEnv mutant was derived from the BRU-Flag virus, by a deletion in the env gene (KpnI-BglII, nucleotides 6343 to 7600 of the sequence of the NL43/2 clone). Viruses were produced by transfection of the corresponding plasmids as previously described (69). Supernatants were analyzed for HIV-1 p24 antigen content, and infectivity titers are expressed here in nanograms of p24 per milliliter (53). Aliquots of MT4 and P4P56 cell samples (4 × 106 cells) were infected at a multiplicity of infection of 150 ng of p24 antigen per 106 cells, i.e., ca. 1,000 virus particles per cell, considering the latest estimate of 4,000 to 5,000 copies of CA per HIV-1 virion. In negative control experiments, cells were treated with 15 μM zidovudine (AZT), which was added 3 h before infection and maintained throughout the infection period. Cells were harvested at 0 (mock-infected cells), 1.5, 6, and 24 h postinfection (p.i.); fixed in 0.5% glutaraldehyde-4% paraformaldehyde; and processed for IEM.
Yeast two-hybrid assays.
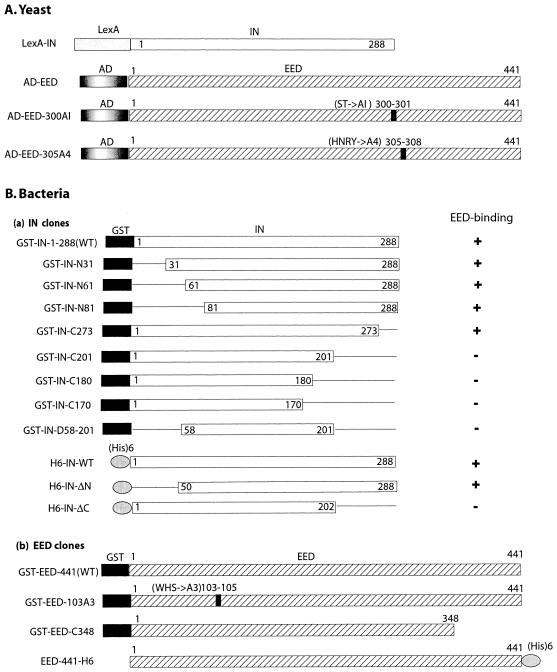
(i) Generation of the DNA-binding LexA-IN hybrid (Fig. 1A). The portion of the HIV-1BRU pol gene segment coding for the IN was cloned in frame with the DNA-binding LexA gene into the pBTM116 vector (47). The sequence of the construct was verified by DNA sequencing, and the expression of recombinant fusion protein in yeast was confirmed by gel electrophoresis and immunoblot analysis with anti-IN polyclonal antibody as described below. (ii) Generation of the pGAD-EED (Fig. 1A). The cDNA of wild-type (WT) EED (54) was inserted into the EcoRI and NotI sites of the Gal4 transcription activation domain vector pGAD3S2X, a modified version of the pGAD GH (Clontech) containing a NotI site in its polylinker. This resulted in a Gal4 activation domain AD-EED hybrid. In the hybrid mutant AD-EED-305A4 the tetrapeptide motif HNRY from positions 305 to 308 was replaced by the tetrapeptide AAAA, and in AD-EED-300AI the dipeptide motif ST at positions 300 and 301 was replaced by AI. (iii) The two-hybrid assays were performed as described in a previous study (54).
FIG. 1.
Schematic representation of genetic constructs of HIV-1 IN and EED, expressed in yeast (A) or in bacteria (B). The LexA (DNA-binding) domain and the Gal4 activating domain (AD) are shown by gray boxes, the GST domain by a black box, the His6 tag by a gray ellipse, the IN sequence by an open box, and EED by a hatched box. In panel Ba, deletions in the IN sequence are represented by a solid line. In panels A and Bb, the point mutations in the EED protein are shown as black bars, with the mutated amino acids indicated above. On the right of panel Ba, the binding of EED to IN protein (WT or deletants) is indicated by “+” or a “−.”
Bacterially expressed IN and EED recombinant proteins.
The full-length, WT HIV-1 BRU IN, and various deletion mutants of IN were expressed in bacteria as glutathione S-transferase (GST) fusion proteins (GST-IN) (56). Their schematic structure and domains are presented in Fig. 1B, along with their acronyms. Likewise, full-length WT human EED was expressed as a GST fusion protein (GST-EED-441) by using the pGEX-KG plasmid (32). EED C-terminal deletion mutant was generated by insertion of a TGA stop codon at position 349 of GST-EED-441, yielding mutant protein GST-EED-C348. In the substitution mutant GST-EED-103A3, the tripeptide motif WHS from positions 103 to 105 was replaced by the tripeptide AAA. Various forms of EED and IN were also also expressed as His6-tagged proteins (Fig. 1B) by using the pT7-7 IPTG-inducible promoter (14). This was the case for the full-length versions of EED (tagged at its C terminus and abbreviated EED-441-H6) and of IN (tagged at its N terminus and abbreviated H6-IN-WT) and for the N and C terminally deleted versions of IN (abbreviated H6-IN-ΔN and H6-IN-ΔC, respectively). Mutagenesis was performed by using PCR and splicing by overlap extension (34, 38). Mutant sequences were verified by DNA sequencing.
Purification of recombinant tagged proteins and affinity pull-down assays.
Bacterial cells expressing the desired protein were centrifuged and resuspended at 1 to 2 g (wet weight) per 2 to 5 ml of lysis buffer (10 mM Tris-HCl [pH 8.0], 5 mM MgCl2, 1 mM EDTA, 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride) and then disrupted by sonication. After incubation for 30 min in the presence of a broad-spectrum endonuclease (Serratia marcescens Benzonase; Sigma) at 100 U/ml of sample, the lysates were clarified by centrifugation at 100,000 × g for 45 min. GST fusion proteins were purified by adsorption on a glutathione-Sepharose gel and recovered by elution with a glutathione-containing buffer or by thrombin cleavage of the GST linker by using a commercial kit (Bulk GST Purification Module; Pharmacia Biotech). For purification of His-tagged proteins, the bacterial cell lysate supernatant was applied to a Ni2+-nitrilotriacetic acid (NTA)-agarose column (Qiagen, Inc.) equilibrated in sodium phosphate buffer (SPB; 16 mM Na2HPO4, 4 mM NaH2PO4 [pH 6.8]) containing 0.5 M NaCl (SPB-500) and 1% Triton X-100. Unbound and weakly adsorbed proteins were washed from the affinity gel with 10 column volumes of SPB-500 containing 5 and 30 mM imidazole, successively, and then His-tagged proteins were eluted at 250 mM imidazole in SPB-500. For in vitro binding reactions, 100-ml cultures of Escherichia coli expressing GST fusion protein were centrifuged, resuspended in 400-μl aliquots of binding buffer (BB; phosphate-buffered saline containing 0.2% Nonidet P-40 and a cocktail of protease inhibitors [Boehringer Mannheim]), sonicated, and clarified by centrifugation. Aliquots (20 μl) of a glutathione-Sepharose bead suspension were added to the bacterial cell lysates, followed by incubation for 30 min at 4°C. After an extensive rinsing in BB, the affinity beads were mixed with 100 to 200 μl (100 μg of total protein) of cell lysates (from bacteria or insect cells) containing nontagged or differentially tagged partner protein and 900 μl of BB, followed by incubation for 2 h at room temperature. The beads were then washed three times with 500 μl of BB, resuspended, and boiled in 50 μl of sodium dodecyl sulfate (SDS) sample buffer. Alternatively, His-tagged proteins were retained on Ni2+-NTA-agarose beads, and affinity beads were processed as described above by using a magnetic pull-down device (Qiagen). Protein pull-down experiments were also performed with affinity-purified proteins, e.g., GST-IN plus EED-441-H6 or H6-IN-WT plus GST-EED. Pull-down proteins were analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting.
Gel electrophoresis and immunoblotting analysis.
PAGE of SDS-denatured protein samples and immunoblotting analysis have been described in detail in previous studies (8, 12, 54). Briefly, proteins were electrophoresed in SDS-denaturing, 10% polyacrylamide gel and electrically transferred to nitrocellulose membrane. Blots were blocked in 5% skimmed milk in Tris-buffered saline (TBS) containing 0.05% Tween 20 (TBS-T), rinsed in TBS-T, and then successively incubated with primary rabbit or mouse antibody (working dilutions of 1:1,000 to 1:2,000) and the complementary peroxidase- or phosphatase-labeled anti-immunoglobulin G (IgG) conjugate (1:1,000).
Antibodies.
For detection of EED, IN, and MA proteins by blotting, immunofluorescence, or IEM, the following antibodies were used. Antiserum against EED was prepared in rabbits by injection of affinity-purified EED protein (54). Anti-IN rabbit polyclonal antibody was also laboratory-made (42), as well as our anti-GST rabbit antiserum. For Flag-tagged IN (53), mouse monoclonal antibody (MAb) anti-Flag M2 (Kodak) or rabbit polyclonal anti-Flag (Zymed Laboratories, South San Francisco, Calif.) were used. MA protein was detected with MAb anti-MAp17 (Epiclone 5003 [Cylex, Inc., Columbia, Md.] mapped to epitope 121-DTGHSSQVSQNY-132) (12) or rabbit polyclonal anti-MA antibody (laboratory-made) (39). His-tagged proteins were detected by using monoclonal anti-His.Tag antibody (Novagen) specific for N-terminal, C-terminal, and internal histidine clusters.
Phage-displayed peptide libraries and biopanning.
Two filamentous phage-displayed peptide libraries were used, one consisting of random hexapeptides (kindly provided by G. Smith [75]), the other of dodecapeptides (BioLabs). Biopanning of immobilized protein ligate and specific ligand elution of phages have been described in previous studies (36, 37, 39). Purified protein (e.g., IN) was immobilized on enzyme-linked immunosorbent assay plates used as the solid support. Elution of phages was carried out by using the purified partner protein as a soluble competing ligand (in this case, EED). In the reverse biopanning experiment, purified EED was coated onto the plate, and IN was used as the soluble competing ligand. The hexapeptide or dodecapeptide phagotopes were identified by manual DNA sequencing of the recombinant fUSE5 pIII protein (36, 37) by using the dideoxynucleotide chain termination method, the required oligonucleotide primers, and the Sequenase kit version 2.0 (Amersham Biosciences).
In vitro HIV-1 IN assays.
Recombinant, bacterially expressed, N-terminally His-tagged HIV-1 IN protein was purified by affinity chromatography (44). IN-mediated strand transfer was assayed in vitro by using a restriction fragment from a modified version of plasmid pU3U5 ((9) as the IN substrate (donor DNA) and plasmid pBSK-zeo (a 3-kbp pBSK-derived construct in which the amp gene has been replaced by the zeocine-resistance gene) as target DNA. The donor DNA (300-bp fragment) contained the 20 terminal base pairs of the HIV-1 5′ and 3′ long terminal repeat (LTR) at each ends, with two-base 5′ overhangs generated by NdeI cleavage. Integration reactions (20 μl, final volume) were performed by using 10 ng of 32P-labeled donor DNA, 100 ng of target DNA, and 2 to 5 pmol of IN (200 to 500 nM, final concentration) in 20 mM HEPES (pH 7.5) buffer containing 50 mM NaCl, 30 mM MgCl2, 1 mM dithiothreitol, 15% dimethyl sulfoxide, and 8% PEG-8000. EED and IN proteins (diluted in reaction buffer) were mixed and preincubated at 0°C for 30 min in molar ratios varying from 1:0.5 to 1:16 in terms of IN:EED stoichiometry. Control reactions were performed with mock samples consisting of the same chromatographic fraction as the one containing IN but from bacterial lysates harboring a void plasmid. Donor and target DNAs were incubated with protein mix for an additional 30 min at 0°C. Reaction buffer was then added to the mixture to a final volume of 20 μl, and the reaction was allowed to proceed for 90 min at 37°C. Reaction products were phenol extracted, ethanol precipitated, and electrophoresed in 1.2% agarose gels. Agarose gels were dried, autoradiographed, and quantitatively analyzed by using a PhosphorImager SI-475 (Molecular Dynamics).
EM and IEM.
Cell specimens were included in metacrylate resin, sectioned and processed for conventional electron microscopy (EM) or IEM according to previously described methods (8, 29, 54). Cellular localization and possible colocalization of EED, IN, and MA proteins were analyzed in HIV-infected cells at different times p.i. by using single, double, or triple labeling with the relevant primary antibody, followed by the corresponding (anti-mouse or anti-rabbit IgG) colloidal gold-conjugated complementary antibodies. For IEM, rabbit IgG from laboratory-made antisera was purified by affinity chromatography by using a protein A-Sepharose column. MA protein was detected by using mouse monoclonal or rabbit polyclonal anti-MA antibodies, EED with rabbit antibody, and IN with anti-Flag MAb or rabbit anti-IN. In double-labeling experiments, when a 5-nm-gold-conjugated anti-mouse IgG antibody was used, a 10-, a 15-, or a 20-nm-gold-conjugated anti-rabbit IgG antibody was simultaneously used on the same cell section. In the triple-labeling experiments, since two primary antibodies belonged to the same species (e.g., the two rabbit anti-MA and anti-EED antibodies), both sides of the specimens on the grids were successively reacted. One side was simultaneously incubated with anti-MA rabbit antibody and anti-Flag(IN) MAb, followed by 10-nm-gold-conjugated anti-rabbit IgG antibody and 5-nm-gold-conjugated anti-mouse IgG antibody. The other side was then incubated with anti-EED rabbit antibody, followed by 20-nm-gold-conjugated anti-rabbit IgG antibody. To verify that there was no undesired cross-reaction of the 20-nm-gold-conjugated anti-rabbit IgG antibody with primary rabbit IgG bound to antigens on the first side of the section, stereoscopic views were taken upon tilting of the microscope goniometer. The 20-nm-gold grains could be seen, by using stereoscopic glasses, in a plane different from that of 5- and 10-nm gold grains (as exemplified in Fig. 9). Specimens were examined under a Hitachi HU7001 or a JEOL 1200-EX electron microscope, the latter equipped with a MegaView II High-Resolution TEM camera and a Soft Imaging System of analysis (Eloïse, Roissy, France).
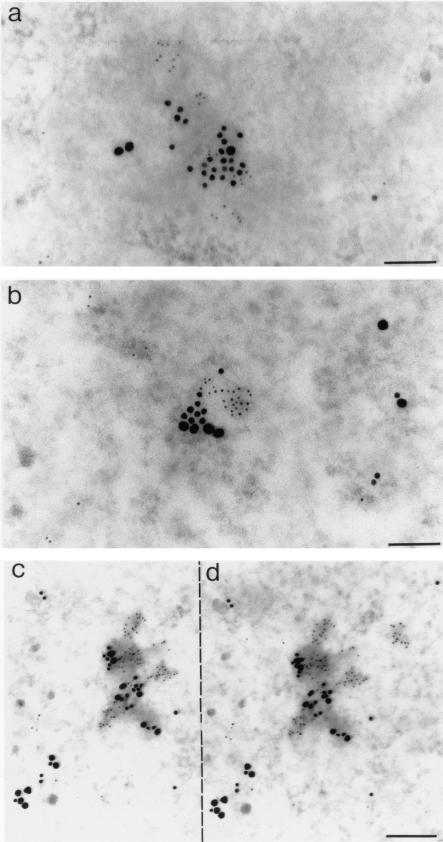
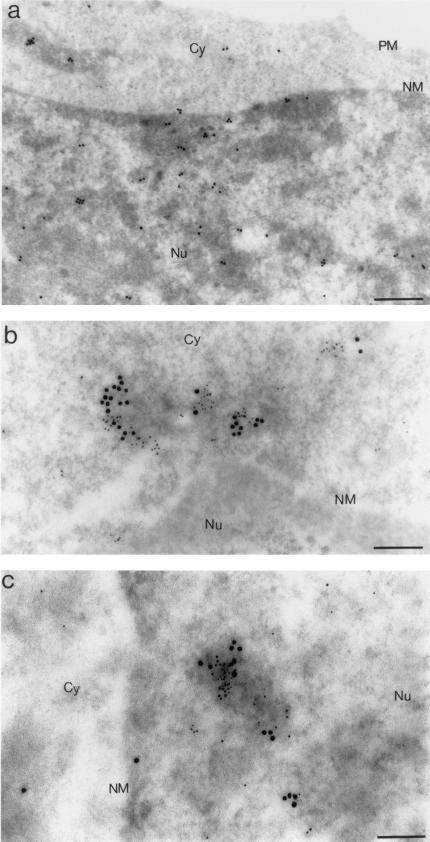
FIG.9.
In situ analysis by IEM of IN, EED, and MA proteins by using triple immunogold labeling of EM sections of BRU-FlagWT-infected human cells taken at 6 h p.i. (a) MT4 cell nucleoplasm; (b to d) HeLa CD4+ cell (P4P56) nucleoplasm. In a first step, one side of the grid carrying the cell pellet section was simultaneously reacted with mouse anti-Flag(IN) MAb and rabbit anti-MA antibody and then with a mix of 5-nm-gold-conjugated anti-mouse IgG antibody and 10-nm-gold-conjugated anti-rabbit IgG antibody. In a second step, the other side of the grid was incubated with rabbit anti-EED antibody, detected by using a 20-nm-gold-conjugated anti-rabbit IgG antibody. (c and d) Stereoscopic view of a portion of P4P56 cell nucleus, taken upon tilting the microscope goniometer, showing clusters of grains 5 nm (IN), 10 nm (MA), and 20 nm (EED) in diameter, in association with electron-dense nucleoplasmic material. By using stereoscopic glasses and aiming at the dotted line separating panels c and d, one can see that the 20-nm gold grains belong to a plane different from that of the 5- and 10-nm grains and that there was no cross-labeling between anti-MA (top side of the specimen) and anti-EED primary IgG (bottom side of the specimen) by the secondary anti-IgG antibody. Bars: 100 nm (a and b), 166 nm (c and d).
RESULTS
The larger isoform of human EED protein is now considered to be comprised of 441 residues. An ortholog of human and murine EED has been identified in Xenopus and is called Xeed (73). As in human EED, the initiator methionine in Xeed corresponds to the Met95 codon in the murine ortholog (mEED) sequence and terminates at residue Arg441, corresponding to Arg535 in mEED (17, 54). From sequence analogy with insect ESC proteins and mammalian Gβ protein (51), the full-length sequence of EED larger isoform contains seven potential WD repeats (54). Compared to WT EED, mutant EED-Ct348 had lost its last two WD repeats. (see Fig. 1 and the legend to Table 2).
TABLE 2.
Mimotopes of EED in phages eluted from INa
| Group | Phagotope sequenceb | No. of independent phages isolated | Homologous motif in EED |
|---|---|---|---|
| I | VAEWHG | 5 | 99-VQFNWH-104 |
| FFGLTK | 3 | 96-LFGV-99 | |
| FGQVWS | 2 | 96-LFGVQFNWHS-105 | |
| GANWPS | 2 | 102-NWHS-105 | |
| QGWFWL | 1 | 100-QFNW-103 | |
| MFEVEF | 1 | 96-LFGVQF-101 | |
| II | ATVLYG | 1 | 224-TLVAIFG-230 |
| VRYGP- | 1 | 226-VAIFG-230 | |
| AIAYIG | 1 | 227-AIFGG-231 | |
| ALFLVV | 1 | 227-AIFGGV-232 |
Peptide motifs in IN-bound phagotopes showing homology with the EED sequence are underlined.
The amino acid sequence of the human EED WT protein is shown below. The seven putative WD repeats are underlined. The N-terminal domain within residues 31 to 63 shows some sequence homology with consensus PEST motifs.
MSEREVSTAP AGTDMPAAKK QKLSSDENSN PDLSGDENDD AVSIESGTNT ERPDTPTNTP NAPGRKSWGK GKWKSKKCKY 80SFKCVNSLKE DHNOPLFGVO FNWHSKEGDP LVFATVGSNR VTLYECHSQG EIRLLQSYVD ADADENFYTC AWTYDSNTSH 160PLLAVAGSRG IIRIINPITM QCIKHYVGHG NAINELKFHP RDPNLLLSVS KDHALRLWNI QTDTLVAIFG GVEGHRDEVL 240SADYDLLGEK IMSCGMDHSL KLWRIYSKRM MNAIKESYDY NPNKTNRPFI SQKIHFPDFS TRDIHRNYVD CVRWLGDLIL 320SKSCENAIVC WKPGKMQDDI DKIKPSESNV TILGRFDYSO CDIWYMRFSM DFWOKMLALG NOVGKLYVW DLEVEDPHKAK 400 CTTLTHHKCG AAIROTSFSR DSSILIAVCD DASIWRWDRL R-(441).
Interaction of human EED protein with HIV-1 IN in yeast two-hybrid assays.
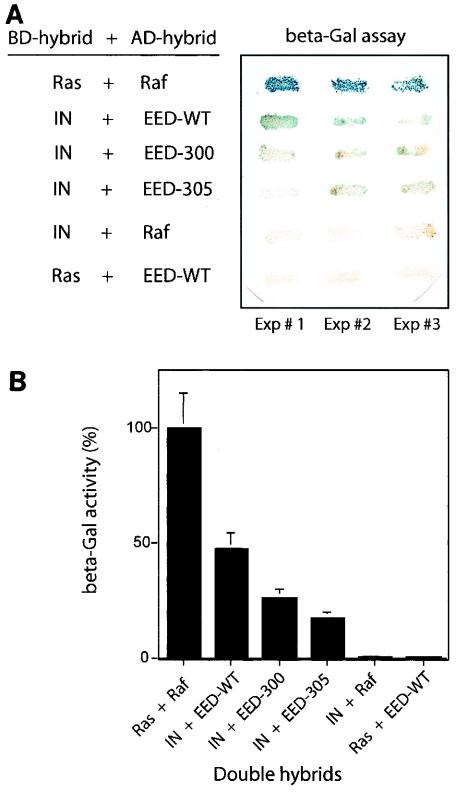
The HIV-1BRU IN, fused to the DNA-binding LexA protein (BD hybrid), was used in yeast two-hybrid assays to test the possible interaction with EED protein, coexpressed as the Gal4 transcription activation domain-fusion protein AD-EED. WT EED and IN proteins interacted in yeast with a significant affinity (Fig. 2A), which was ∼2-fold lower than that shown by the strong interactors Raf and Ras used in positive control samples (Fig. 2B). A weaker, but still positive β-galactosidase signal (one-third to one-fourth of the Ras-Raf control level; see Fig. 2B) was also observed with AD-EED-394AI and AD-EED-399A4 hybrids, two AD-fused EED mutants defective in MA protein interaction (54). This finding suggested that the IN-binding region in EED differed from the MA-interacting site, mapped to residues 294 to 309 (54) (see also the Table 2 footnotes) but that mutations in the MA-binding domain had some negative effect on the binding of EED to IN in yeast cells. The possibility that EED could have activated transcription of the reporter gene independently of a bona fide interaction with IN-BD hybrid could be excluded since no β-galactosidase signal over the background was detected with the pair of hybrids AD-EED and BD-Ras (Fig. 2) (54).
FIG. 2.
Interaction of EED and HIV-1 IN in vivo. (A) The affinity of EED (WT or mutants EED-300 and EED-305) for IN was analyzed in yeast by using a two-hybrid assay. IN (or Ras) was cloned in fusion with LexA protein (BD hybrid), and EED (or Raf) was cloned in fusion with the Gal4 activation domain (AD hybrid). The β-galactosidase activity of yeast transformants plated on medium without histidine and replica plated on Whatman filters is shown. The results of three independent experiments are shown. Growth in the absence of His and blue color in the β-Gal assay are indicative of interaction between hybrid proteins in yeast cells. Positive controls involved BD-Ras and AD-Raf hybrids as strong interactors, and negative controls consisted of BD-IN and AD-Raf and of BD-Ras and AD-EED hybrids, respectively. (B) Quantification of β-galactosidase activity in yeast transformants harboring EED and IN hybrids is expressed as the percentage of the BD-Ras plus AD-Raf activity. beta-Gal, β-galactosidase.
Mapping of EED-IN interacting sites by phage biopanning.
When phages are panned on immobilized EED protein and specific ligand-elution is performed with an excess of affinity-purified IN, the phagotopes isolated would theoretically be mimotopes of peptide motifs of IN, since IN acted as a competitor of phages bound to IN-binding sites on EED (36, 37, 39, 54). Two phage libraries were used—one displaying random hexapeptides and the other displaying random dodecapeptides—on the basis that longer peptides could adopt a conformational structure that would better mimic that of protein interacting sites. About one-third of the phagotopes recovered from both libraries (15 of 50) could be grouped according to recurrent peptide motifs that showed some homology with the C-terminal sequence of IN overlapping tryptophan-235, within residues 212 to 264 (Table 1). For example, motif VLPPK, homologous to 259-VVPRRK in IN, was found several times. However, we observed a high level of degeneration and scatter of all of the phagotopes isolated, suggesting that the EED-binding region in IN had a high degree of three-dimensional organization and/or was constituted of discontinuous epitopes.
TABLE 1.
Mimotopes of IN in phages eluted from EEDa
| Library | Phagotope sequenceb | No. of independent phages isolated | Homologous motif in IN |
|---|---|---|---|
| 16-mer | WSNIVV | 1 | 243-WKGEGAVV-250 |
| EVGPGW | 1 | 229-DSRDPLW-235 | |
| AQNFGQ | 1 | 220-IQNFR-224 | |
| LQTFRQ | 1 | 220-IQNFR-224 | |
| 12-mer | VLPPKPMRQPVA | 3 | 259-VVPRRK-264 |
| GIQVANPPRLYG | 2 | 257-IKVVPR-262 | |
| TTGLPLWSFNPS | 1 | 233-PLW-235 | |
| SPWRLPTPLT | 1 | 232-DPLWKGP-238 | |
| QLPFKLGPARID | 1 | 232-DPLWKGPAKL-241 | |
| SHPWNAQRELSV | 1 | 230-SRDPLWK-236 | |
| FSHELSWKPRKA | 1 | 234-LWKGPAKL-241 | |
| VPTNVQLQTPRS | 1 | 212-ELQKQIT-218 |
Peptide motifs in EED-bound phagotopes showing some homology with the IN sequence are underlined. Note that in a few phages, e.g., SPWRLPTPLT, the phagotope is sometimes shorter than the expected 12-mer.
The amino acid sequence of the HIV-1-BRU IN protein is shown below. The N-terminal domain is made up of the first 50 residues; the C-terminal domain is made up of last 88 residues. The residues involved in the N-terminal zinc finger-like domain are in boldface, the catalytic triad D, D, and E in the core domain is in boldface italics, and the putative region of EED binding (residues 212 to 264) is underlined.
FLDGIDKAQD EHEKYHSNWR AMASDFNLPP VVAKEIVASC DKCQLKGEAM 50
HGQVDCSPGI WQLDCTHLEG KVILVAVHVA SGYIEAEVIP AETGQETAYF 100
LLKLAGRWPV KTIHTDNGSN FTSTTVKAAC WWAGIKQEFG IPYNPQSQGV 150
VESMNKELKK IIGQVRDQAE HLKTAVQMAV FIHNFKRKGG IGGYSAGERI 200
VDIIATDIQT KELOKOITKI ONFRVYYRDS RDPLWKGPAK LLWKGEGAVV 250
IODNSDIKVV PRRKAKIIRD YGKQMAGDDC VASRQDED-(288)
Reverse biopanning experiments were then performed to obtain mimotopes of the IN-binding site(s) in EED. The 6-mer library was panned on immobilized IN, and specific ligand elution was performed with an excess of affinity-purified, His-tagged EED-441-H6 protein as the specific competitor for IN-bound phages. The most represented group of phages (14 phages out of 24 independent clones isolated) showed homology with hydrophobic and aromatic motifs within residues 96 to 104, and the other group (4 phages) showed homology with hydrophobic and aromatic motifs within residues 224 to 232 (Table 2). These two putative IN-binding regions were located in the N-terminal moiety of EED and apparently did not overlap with the MA-binding site at positions 294 to 309, which confirmed the results of our two-hybrid assays in yeast (Fig. 2).
EED-IN interaction in vitro analyzed by mutagenesis and pull-down assays.
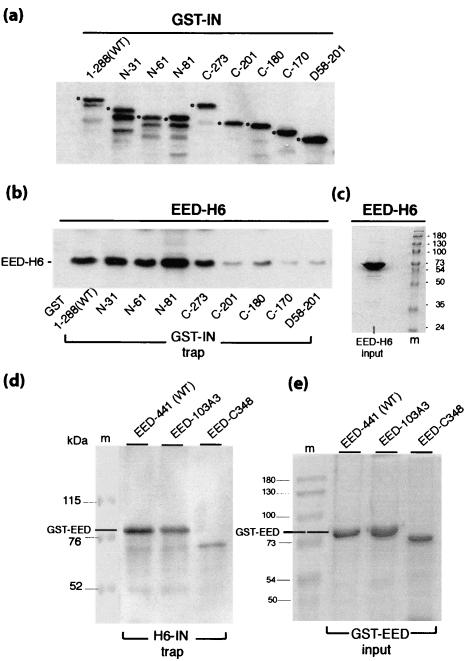
Samples of bacterial cell lysates containing various forms of GST-IN fusion protein (Fig. 3a) were incubated with EED-441-H6 protein, and the resulting complexes were isolated on a glutathione-agarose affinity gel. As shown in Fig. 3b, the binding of full-length WT IN to EED in vitro confirmed the data from our yeast two-hybrid assays and implied that IN and EED proteins could directly interact with each other, without any requirement for a third partner provided by the yeast cell. The possibility that some nucleic acid or protein bridge could link IN and EED in this assay was ruled out on the basis of the following experimental arguments. (i) IN interacted with EED in vitro not only in crude bacterial lysates but also as two affinity-purified recombinant proteins. (ii) In all cases, the bacterial cell lysates from which each protein was recovered were first treated with a broad-spectrum endonuclease from S. marcescens (Benzonase; Sigma) that is specific for both single- and double-stranded RNA and DNA substrates. (iii) UV spectrum analyses performed on EED and IN recombinant proteins revealed no detectable nucleic acid contamination, as shown by A280/A260 ratios consistently found to be greater than 1.9.
FIG. 3.
Interaction of EED and HIV-1 IN in vitro. The affinity between EED and IN and their respective mutants was analyzed by GST and histidine pull-down assays with glutathione-agarose (a to c) and Ni2+-NTA-agarose (d and e) as affinity gels. In panels a to c, aliquots of GST-fused full-length IN (i.e., positions 1 to 288), N-terminal (N), C-terminal (C), and double-deletion (D) mutants, as displayed in panel a, were immobilized on a glutathione-agarose gel, and the affinity gels were incubated with aliquots of bacterial cell lysates containing 10 μg each of full-length, His-tagged EED (EED-H6). The amount of EED-H6 protein retained on GST-INglutathione-agarose gel was then evaluated by SDS-PAGE and immunodetection on a blot with anti-His tag MAb as shown in panel b. In panel c is shown EED-H6 input (10-μg protein load; Coomassie blue staining). Note that in panel a, the pattern of anti-GST reacting bands is highly suggestive of a major proteolytic cleavage site located in the C-terminal domain of IN, within region 288-273: a prominent lower band migrating with a constant apparent molecular mass ∼2 kDa lower than that of the original GST-IN gene products (marked by solid dots) is visible in samples of full-length and N-deleted forms of GST-IN, whereas this band is absent from the C-truncated or double-truncated GST-IN clones. In panels d and e, the GST-fused, full-length EED, point mutant EED-103A3 and C-terminal deletion mutant EED-C348 were incubated with aliquots of His-tagged IN (H6-IN) adsorbed onto an Ni2+-agarose gel, and the amount of GST-EED protein retained on the H6-IN-Ni2+-agarose gel was determined by SDS-PAGE and immunoblotting with anti-GST polyclonal antibody (panel d). (e) GST-EED protein input (2-μg protein load per well; Coomassie blue staining).
In order to map the EED-binding region(s) in IN, eight individual deletions scanning the N- and C-terminal structural domains of IN and a double mutant mimicking the IN central core (56) were generated in GST-tagged IN protein (Fig. 1Ba), and the EED-binding capacity of these mutants was evaluated in pull-down assays. Deletions in the N-terminal domain of IN had no negative effect on its binding to EED, and even a slight enhancing effect was observed (Fig. 3b, compare clones 1-288 and N-81), implying that the presence of the N-terminal domain negatively influenced the apparent affinity of IN for EED in vitro. The C-terminal deletion mutant GST-IN-C-273 bound to EED at WT levels, whereas further deletion of residues 273 to 202, as in mutant C-201, reduced the binding to background levels. Likewise, the double-mutant D58-201, which represented the IN core domain (80), bound to EED to insignificant levels (Fig. 3b). The C-terminal region from residues 202 to 273 included the two overlapping sequences from positions 220 to 250 and positions 212 to 264 defined by our biopanning data (Table 1), and this finding confirmed that the C-terminal domain of IN was essential for the IN-EED interaction in vitro.
Since the most represented phagotope retained on immobilized IN corresponded to the sequence 99-VQFNWH-104 in EED (Table 2), a nonconserved mutation in three prominent amino acid residues of this motif was constructed: the bulky tripeptide motif WHS at position 103 to 105 was replaced by a stretch of three alanine residues to generate the EED-103A3 mutant (see Fig. 1Bb). Since this mutation could alter the immunoreactivity of the resulting EED mutant protein, the EED-103A3 mutant was assayed for IN binding as the fusion protein GST-EED-103A3, in comparison with its GST-EED WT counterpart. H6-IN-WT was used as the bait, Ni2+-agarose gel as the trap, and EED proteins were detected by their GST tag by using anti-GST antibodies. EED-103A3 mutant was still capable of binding to IN, although with a lower efficiency (two- to threefold; Fig. 3d and e). This suggested that at least one of the IN-binding sites mapped to residues 103 to 105 in EED but that additional IN contact sites could also be present in other domains of EED, as suggested by our phage biopanning (Table 2).
One of the striking features of Pc-G gene eed products is their high degree of conservation at the protein level in mammals, implying that all amino acid residues of EED are important for their biological functions (51, 68). Thus, it has been shown that the integrity of the last WD repeat of Pc-G proteins is highly critical and that even short deletions at the C terminus are detrimental to their activity (51, 76). Consistent with this, the C-terminal deletion mutant EED-C348 was found to bind to the IN-affinity gel at very low levels (Fig. 3d), indicating that the C-terminal fourth of EED was crucial for its interaction with IN, directly or indirectly, via EED conformation and structure stabilization.
Influence of EED on HIV-1 IN activity in vitro.
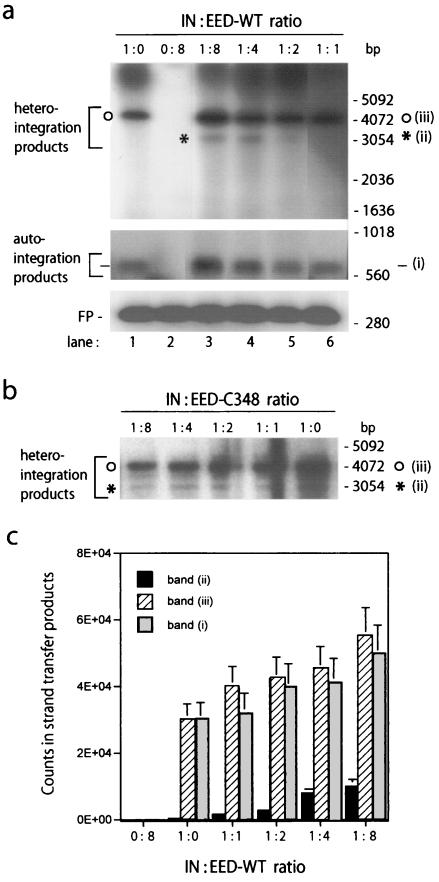
The enzymatic activity of IN was assayed in reactions of DNA integration in vitro in the presence or absence of affinity-purified recombinant EED protein. In IN assays, homologous integration (or autointegration) refers to the integration of one 5′+3′-LTR-containing DNA donor fragment into another (Fig. 4a, band i), whereas heterologous integration (or heterointegration) corresponds to the insertion of one (or more) 5′+3′-LTR-containing fragment(s) into one plasmid target (9). In the absence of EED, several discrete bands of heterointegration products were usually observed: a major band (Fig. 4a, band iii) corresponded to single-tagged circles, resulting from one single integration event per plasmid target, mediated by one LTR (one-ended integration) (10); faster-migrating minor band(s) represented concerted integration events involving two LTRs (Fig. 4a, band ii) and resulting from the insertion of two donors into one target DNA molecule with subsequent linearization (two one-ended concerted integration) and, more rarely, from the two-ended concerted integration of a single donor fragment into target DNA (10). When IN reactions were performed under conditions in which the major band of single-tagged circles (band iii) was the only detectable signal (Fig. 4a, control lane 1), the addition of WT EED provoked an increase in the intensity of autointegration prod-ucts (band i) and heterointegration products corresponding to single-tagged circles (band iii) (Fig. 4a). The effect became detectable for EED/IN ratios over 2:1 and seemed to occur in a dose-dependent manner, with a twofold augmentation obtained at a ratio of EED to IN of 8:1 (Fig. 4c). However, a 20- to 30-fold augmentation of the putative two one-ended concerted integration products (band ii) was observed within EED/IN ratios ranging from 4:1 to 8:1 (Fig. 4a and c). The same patterns were observed with affinity-purified, WT EED-441-H6 and GST-EED-441, the latter being cleaved off the GST domain. No detectable enhancing effect was observed with corresponding chromatographic fractions from mock-expressing bacterial cells (not shown) or with the deletion mutant protein EED-C348, which provoked a slight and probably nonspecific negative effect at high concentrations (Fig. 4b). This pattern confirmed the interaction between EED and IN proteins in vitro, with a secondary influence on the enzymatic activity of IN.
FIG. 4.
Effect of EED WT (a and c) and deletion mutant EED-C348 (b) on IN-mediated integration reaction in vitro. The respective IN/EED stoichiometric ratios are indicated at the tops of panels a and b. Control reactions were performed in the absence of EED (lane 1) or in the absence of IN (lane 2). The positions of autointegration products (band i) and heterologous integration products (bands ii and iii) are indicated by brackets on the left. As inferred from previous studies (10, 31), the minor band (band ii [✽]) is likely to represent the product of two one-ended concerted integration followed by linearization, whereas the major band (band iii [○]) represents single-tagged target circles. (b) Integration reaction performed in the presence of EED-C348 mutant. Only heterointegration products are shown. (c) Quantification of counts recovered from auto- and heterointegration products of IN reactions performed in the presence of EED-WT (mean of three experiments ± the standard deviation).
In situ analysis of IN, EED, and MA proteins in HIV-1-infected cells.
Human epithelial HeLa CD4+ cells (P4P56 cells) (52) and lymphoid MT4 cells were infected with BRU-FlagWT, a subclone of HIV-1BRU expressing a Flag-tagged IN (53). Negative control samples consisted of mock-infected cells or cells incubated with BRU-FlagΔEnv, a noninfectious env-deleted mutant used as control for nonspecific HIV-cell interaction. Cells were harvested at 1.5, 6, or 24 h p.i. and processed for EM and IEM. For IEM analysis, cell specimens were reacted with monoclonal anti-Flag(IN), polyclonal anti-EED, and monoclonal anti-MA MAbs and, occasionally, with anti-CA monoclonal or anti-MA polyclonal antibodies, as indicated. Antibodies were used in single-, double-, or triple-immunolabeling reactions with their corresponding complementary gold-tagged anti-IgG antibody.
At 1.5 h p.i. in mock-infected or BRU-FlagΔEnv-reacted cells, the cytoplasm and nucleus were poorly labeled with anti-MA, anti-CA, and anti-Flag(IN) antibodies, and the immunolabeling corresponded to background reactivity (not shown). In BRU-FlagWT-infected cells, however, virus particles were visible in numbers at the cell surface as roughly spherical structures of 120 to 140 nm in diameter that were double labeled with anti-MA and anti-CA antibodies (Fig. 5a). These plasma membrane-associated particles were no longer seen at later times, i.e., at 6 and 24 h p.i.. Within the cells, anti-Flag(IN) antibody-labeled globular structures that were 60 to 80 nm in diameter were visible in significant numbers in the nucleoplasm and frequently seen in the vicinity of nuclear pores (Fig. 5b). Their immunoreactivities and sizes were compatible with those of PICs, which have been reported to be ca. 56 nm in average diameter (49). At 1.5 h p.i., EED labeling was found to be evenly distributed between the cytoplasm and the nucleus and randomly dispersed in both compartments, with no apparent difference between control cells, mock-infected cells, and BRU-FlagΔEnv-treated cells (IEM images not shown; refer to data in Fig. 10a and b).
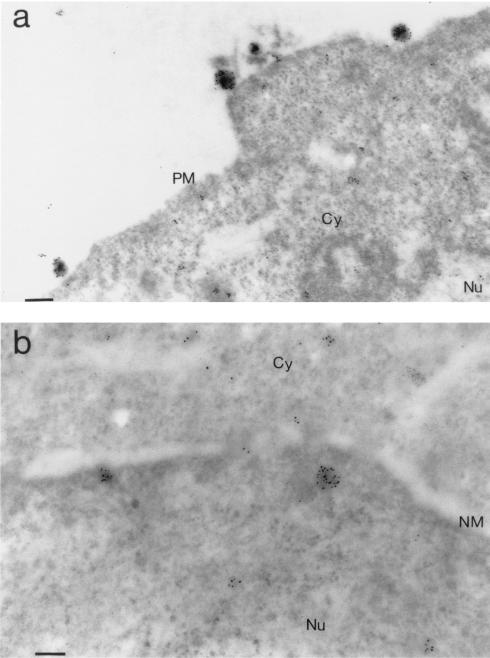
FIG. 5.
IEM analysis of human MT4 (a) and HeLa-CD4+ (P4P56) (b) cells infected with Flag-tagged IN-containing HIV-1 (BRU-FlagWT; multiplicity of infection of 1,000 virions/cell) obtained at 1.5 h p.i. In panel a, MA and CA proteins were detected by a mix of anti-MA and anti-CA MAbs, followed by 10-nm-colloidal-gold-tagged anti-mouse IgG antibody. The immunogold-labeled globular structures bound to the cell surface had diameters compatible to those of HIV-1 virions (110 to 130 nm). In P4P56 cells (panel b), the Flag-tagged IN protein was detected by using anti-FLAG MAb (M2), followed by 10-nm-colloidal-gold-tagged anti-mouse IgG antibody. Note that the two gold-labeled globular structures that were 60 to 80 nm in diameter visible in the nucleus in the vicinity of the nuclear membrane had dimensions compatible with those of the PIC. PM, plasma membrane; C, cytoplasm; N, nucleus; NM, nuclear membrane; PM, plasma membrane. Bars: 166 nm (a), 100 nm (b).
FIG. 10.
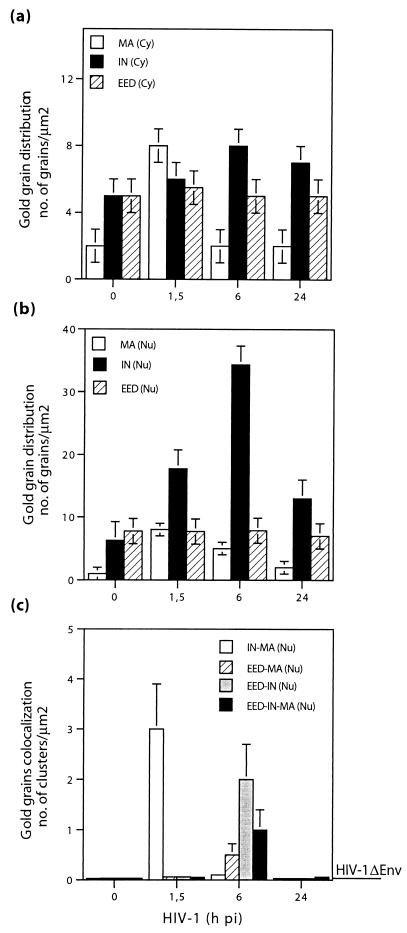
Quantitative image analysis of intracellular distribution (a and b) and nuclear colocalization (c) of EED, MA, and IN proteins in HIV-1-infected MT4 cells. Cells infected with BRU-FlagWT, or its noninfectious, envelope-deleted version BRU-FLAGΔEnv, were harvested at different times p.i. as indicated on the x axis. (a) Cytoplasm; (b and c) nucleus. In panels a and b, gold grains were counted on cell sections after single labeling with affinity-purified anti-EED, anti-MA, or anti-IN rabbit antibodies, respectively, followed by 10-nm-gold-tagged anti-rabbit IgG antibody. Background labeling for IN and MA proteins was given by the number of grains per square micrometer on mock-infected cell sections (zero point). In panel c are shown theresults of triple-labeling experiments (see, for example, Fig. 9). The baseline on the right corresponds to the data obtained with BRU-FLAGΔEnv (HIV-1ΔEnv).
Double labelings of MT4 and P4P56 cells were then performed to detect a possible colocalization of IN, EED, and MA, pairwise, in the cytoplasm or nucleus at later times p.i. In control cells (mock-infected or BRU-FlagΔEnv-treated cells), double immunolabeling with anti-EED (revealed by 10-nm-gold-tagged conjugate) and anti-IN (5-nm gold grains) or with anti-EED (10-nm gold grains) and anti-MA (5-nm gold grains), respectively, showed that grains of both diameters were randomly distributed in the cytoplasm and nucleoplasm, with no indication of a preferred localization or colocalization at 6 and 24 h p.i. (Fig. 6a and 7a).
FIG. 6.
In situ analysis by IEM of EED and MA proteins in HIV-1-infected MT4 cells taken at 6 h p.i. (a) Mock-infected cells; (b) BRU-FlagWT-infected cells. Cell sections were simultaneously labeled with rabbit anti-EED, detected by using a 10-nm-colloidal-gold-conjugated anti-rabbit IgG antibody, and with anti-MA MAb, detected by a 5-nm-colloidal-gold-conjugated anti-mouse IgG antibody. In panel a, a general view of the cell is shown. In panel b, an area of the nucleoplasm is presented, showing colocalization of 5-nm (MA) and 10-nm (EED) gold grains, associated with electron-dense material. Cy, cytoplasm; Nu, nucleus; PM, plasma membrane; NM, nuclear membrane. Bars: 200 nm (a), 100 nm (b).
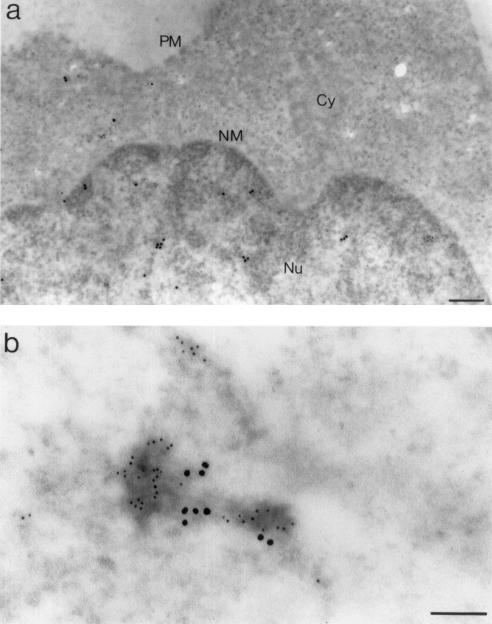
FIG. 7.
In situ analysis by IEM of EED and IN proteins in HIV-1-infected human cells at 6 h p.i. Mock-infected MT4 cells (a) and BRU-FlagWT-infected MT4 cells (b) and HeLa-CD4+ cells (P4P56) (c) are shown. Cell sections were simultaneously labeled with anti-EED rabbit antibody, detected by using a 10-nm-colloidal-gold-conjugated anti-rabbit IgG antibody, and with anti-Flag(IN) MAb (M2), detected by using a 5-nm-colloidal-gold-conjugated anti-mouse IgG antibody. Panel a presents a general view of the cell section. Panels b and c show enlargements of sections of nuclear membrane with flanking areas of nucleoplasm and cytoplasm. Cy, cytoplasm; Nu, nucleus; PM, plasma membrane; NM, nuclear membrane. Bars: 333 nm (a), 142 nm (b), 125 nm (c).
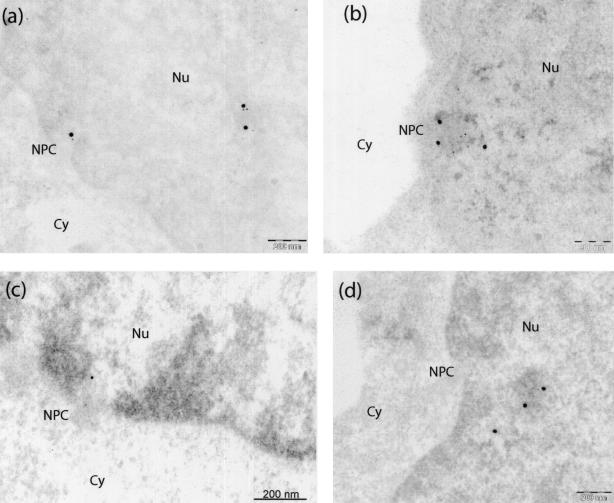
The results were different in BRU-FlagWT-infected cells (Fig. 6b). Many patches of electron-dense material were double labeled with anti-MA and anti-EED antibodies in the nucleus of infected cells taken at 6 h p.i. This finding confirmed the colocalization of EED and MA proteins already observed in recombinant protein coexpressing insect and human cells (54). Likewise, a pattern of colocalization was observed for IN and EED in BRU-FlagWT-infected MT4 (Fig. 7b) and P4P56 (Fig. 7c) cells taken at 6 h p.i. and double-immunolabeled with anti-IN and anti-EED antibodies. Colocalizations of IN and EED were mainly observed in the nucleus (Fig. 7c) but were also observed in the cytoplasm in close vicinity to the nuclear membrane (Fig. 7b). IN-EED colocalizations were frequently seen at or near the nuclear pores (Fig. 8). In most of these observations, the clusters of EED- and IN-bound grains were found to be associated with electron-dense material in the nucleoplasm or at the nuclear pores (Fig. 6 to 8). IN-EED colocalizations were no longer observed at 24 h p.i.
FIG. 8.
IEM analysis of colocalization of EED and IN proteins at or near the nuclear pores of BRU-FlagWT-infected MT4 cells obtained at 6 h p.i. The different panels present various areas of cell sections showing nucleoplasm and nuclear pores. Specimens were labeled with rabbit anti-EED, detected by using a 20-nm-colloidal-gold-conjugated anti-rabbit IgG antibody, and anti-Flag(IN) MAb, detected by using a 5-nm-colloidal-gold-conjugated anti-mouse IgG antibody. Cy, cytoplasm; Nu, nucleus; NPC, nuclear pore complex. Bars: 200 nm.
In order to detect a possible colocalization of the three proteins IN, EED, and MA, triple-immunolabeling experiments were also performed with anti-EED (revealed by 20-nm colloidal gold grain), anti-MA (10-nm colloidal gold grain), and anti-IN (5-nm colloidal gold grain) antibodies on sections of BRU-FlagWT-infected MT4 and P4P56 cells. Many clusters of gold grains of the three different diameters were found in nuclei at 6 h p.i. (Fig. 9). Such triple colocalizations were never observed in control samples, mock-infected cells, and BRU-FlagΔEnv-treated cells. Our IEM data therefore suggested that IN, MA, and EED proteins could form ternary complexes and/or participate together in higher-order complexes in vivo within the nucleus of HIV-1-infected cells at early times p.i.
Quantitative IEM analysis of the cellular distribution and colocalization of IN, EED, and MA proteins in HIV-1-infected cells.
To further analyze the distribution of EED, IN, and MA proteins in the cellular compartments in a semiquantitative and kinetic manner, sections of control or HIV-1-infected P4P56 or MT4 cells were separately reacted with rabbit anti-EED, anti-IN and anti-MA IgG, followed by 10-nm-gold conjugated anti-rabbit IgG antibody in single immunogold labeling experiments. Gold grains were counted by using EM in several sections of independent cells taken at time intervals during HIV-1 infection. At least 400 grains were counted on 20 different cell sections, and the variations of the density of grains (number of grains per unit surface area of cell section) in the cytoplasm (Fig. 10a) and nucleus (Fig. 10b) were compared during the course of infection. Backgound labeling was given by mock-infected cells reacted with the same primary and gold-conjugated secondary antibodies (zero time point).
In HIV-1-infected cells, MA labeling was detected as early as at 1.5 h p.i. in both cytoplasm (Fig. 10a) and nucleus (Fig. 10b). The MA signal decreased to background levels at 6 h p.i. in the cytoplasm but was still observed at significant levels in the nucleus until 6 h p.i. In the cytoplasm, the IN labeling was not significantly higher than the background level at any time p.i. (Fig. 10a). In the nucleus however, IN signal was detected at 1.5 h p.i. and reached a maximum level at 6 h p.i., suggesting a higher number, or better accessibility, of IN molecules at this time of the virus cycle (Fig. 10b; see also Fig. 5b). The level of EED labeling remained almost constant throughout the time period in both cytoplasmic and nuclear compartments (Fig. 10a and b). No significant variation and increase over the background labeling was observed in BRU-FlagΔEnv-infected P4P56 or MT4 cells for the three markers, MA, IN, and EED, during the same time period (background line; Fig. 10c).
Colocalization of EED, IN, and MA proteins was also quantitavely evaluated by EM by counting neighboring grains on sections from cells taken at different times p.i. and subjected to triple immunolabeling with specific monoclonal and polyclonal antibodies (see, for example, Fig. 9). This quantitative assay was based on the following principle. The size of an antibody molecule under EM is ca. 15 nm and, in our IEM analyses, primary and secondary gold-labeled antibody molecules were used. Thus, if one antibody is labeled with a 10-nm gold grain and the other one is labeled with a 20-nm grain and if both gold conjugations occurred at their respective Fc domains, the two gold grains that are 90 nm apart (15 + 15 + 10 + 15 + 15 + 20 nm) might theoretically mark two adjacent epitopes carried by the same protein molecule or, alternatively, two epitopes belonging to two neighboring molecules. Since epitopes on interacting proteins could lie at a certain distance from each other, one could reasonably consider that immunogold grains that are seen within a distance range of 100 to 120 nm would mark molecules that colocalize in the same cell compartment.
Using this quantitative method, we observed no significant colocalization of grains marking EED, MA, and IN in the cytoplasm and nucleus of mock-infected cells or in cells incubated with BRU-FlagΔEnv (Fig. 10c). Likewise, colocalization of gold grains marking IN, MA, and EED was rarely observed in the cytoplasm of BRU-FlagWT-infected cells at early times p.i. (e.g., 1.5 h p.i.), and no statistical analysis could be performed (results not shown). The results were different for the nuclei of BRU-FlagWT-infected cells, which showed clusters of double and even triple colocalizations with a significant frequency (Fig. 10c). An average value of three colocalization events per square micrometer of nucleoplasm area was found for the pair IN-MA at 1.5 h p.i., but no other pairwise colocalization (IN-EED or MA-EED) was found in significant numbers. At later times, no more colocalization of IN and MA was observed. At 6 h p.i., a mean value of two colocalization events per square micrometer was found for IN and EED, and triple colocalization of EED, IN and MA occurred at an average frequency of one event per square micrometer of nucleus section area (Fig. 10c). No detectable double or triple colocalizations were detectable at 24 h p.i. This finding suggested that the time point of 6 h p.i. represented the phase of the virus cycle when significant numbers of EED, IN, and MA molecules were in close vicinity within the nucleus of HIV-1-infected cells. Alternatively, it could mean that the intranuclear microenvironment that allowed for a maximal immunoreactivity and accessibility of the three proteins took place at 6 h p.i.
DISCUSSION
We found here that human EED, a Pc-G protein, can interact with HIV-1 IN both in vitro and in vivo in yeast. Using deletion mutagenesis and phage biopanning, we mapped the major EED-binding sites to the C-terminal domain of IN. In vitro, we observed an apparent positive effect of EED on IN-mediated integration reaction. We hypothesize that this effect was indirect: the interaction of EED with IN could promote the oligomerization of IN molecules, which would in turn favor the integration process. In situ analysis of EED and IN cellular localization was performed on HIV-1-infected human epithelial (HeLa CD4+) or lymphoid (MT4) cells by using IEM and differential immunogold labeling. We found that EED and IN colocalized within the nucleus of HIV-infected cells, a phenomenon that was mainly observed at early times p.i. (1.5 to 6 h). Triple-immunolabeling experiments showed that the MA protein, another viral protein partner of EED (54), was also detected in significantly frequent associations with both EED and IN proteins in the nucleus at early times p.i., suggesting the occurrence of ternary complexes involving EED, MA, and IN. Although the role of EED protein in the HIV-1 life cycle is not known, our data suggest that EED could be involved in some cellular function(s) necessary for and/or induced by early steps of the virus-host cell interaction. The fact that we never observed any cellular colocalization of EED, IN, and MA in cells infected with HIV-1 in the presence of virus inhibitor AZT (data not shown) suggests that the role played by EED in the virus cycle would require the step of proviral DNA synthesis. Since EED is a partner of two viral proteins—MA, which has both structural and functional roles at the early stages of the cycle (23), and IN, which is responsible for proviral integration—one might envisage EED as a possible participant in at least two major retroviral processes: the intracellular transport of incoming virions and/or the proviral integration.
In spite of a number of studies (2, 13, 16, 24-27, 35, 40, 48, 49, 52, 84, 85), the cellular and viral factors that control the reaction and the site(s) of integration of HIV-1 proviral DNA still remain incompletely elucidated. The integration process is better understood at the molecular level for retrotransposons, such as Ty5, which integrates into regions of silent chromatin via yeast proteins Sir (86). Like the Sir proteins in yeast, the products of many Pc-G genes are responsible for the maintenance of the silent state of chromatin in upper eukaryotes, likely by recruitment of histone deacetylases (reviewed in reference 55). Thus, the transcriptional repression by EED has been recently reported to involve histone deacetylation (79). Likewise, in murine neurons, EED has been shown to colocalize with histone H1 in transcriptionally inactive domains of perinucleolar heterochromatin (1). The function of Pc-G proteins as transcriptional repressors and gene silencers (17, 33, 51, 63, 67, 74) has been attributed in several cases to direct interaction with transcription factors rather than to direct DNA binding (3, 64). For example, EED has been found to bind to YY1, a vertebrate DNA-binding protein (4), and a stable, if only transient, interaction between EED, EZH2 (enhancer of Zeste 2) (70), YY1, and histone deacetylase has been suggested (62). An indirect physical link has therefore been established between EED and the DNA of target chromatin regions via the DNA-binding protein YY1 (62).
In light of the latest results on HIV-1 integration into transcriptionally active regions of the host genome (66, 83), an attractive hypothesis would be that intranuclear interaction occurring between the viral proteins MA and IN on one side and the cellular protein EED on the other side might deregulate silent cellular genes by releasing the binding of EED to YY1 (or other factors involved in the transcription machinery), by modifying the acetylation state of histones, or by both of these processes. This would then activate the integration process of the proviral DNA. However, it is important to note that YY1 is a multifunctional transcription factor which, under certain circumstances, can act as a transcriptional repressor (72).
Alternatively, but not exclusively, there may be a role in the intracellular transport and nuclear translocation of the PIC for EED. As a component of a multifactor transport complex, EED might act as a shuttle protein to convoy the viral PIC to the nucleus via its binding to MA and IN. Only a few cellular proteins have been identified thus far as involved in the transport of HIV-1 PIC. A DNA targeting function has been assigned to the product of the INI1 gene (40), a cellular interactor of HIV-1 IN that seems to also act in late events in the viral life cycle (84). It has been shown that HIV-1 PIC recruits INI1 and PML proteins within the cytoplasm, and this complex could facilitate the integration via the recruitment of additional cell factors, such as the PML-binding CBP/p300 (78). Interestingly, a protein similar to EED, called WAIT-1 (WD protein associated with integrin cytoplasmic tails-1) (61), has been found to interact with the cytoplasmic domain of the integrin β7 subunits, and a shuttling of WAIT-1/EED between membrane-associated β7 integrins and the nucleus has been suggested (61). The β7 cytoplasmic domain is involved in major integrin functions such as receptor affinity and signaling (15, 46). It is noteworthy that, among the β7 subfamily members, αEβ7 integrin is restricted to a subset of gut-associated T lymphocytes and dendritic cells (61). The frequent occurrence of EED and IN double labeling at nuclear pore complexes, as observed in our IEM study, would support a shuttling function for EED. Further investigations by cell fractionation of HIV-1-infected cells, overexpression versus synthesis inhibition of EED in HIV-infected cells, and isolation of dominant-negative mutants of EED would help elucidate the role(s) of cellular EED protein in HIV-1 infection.
Acknowledgments
This work was supported by the Agence Nationale de Recherche sur le SIDA (ANRS; AC14-2 “HIV-1 Integrase and Preintegration Complex”). S.V. received an ANRS fellowship, and S.P. received a fellowship from the French ECS Association (Ensemble contre le SIDA).
We are deeply grateful to Simone Peyrol and Isabelle Leparc-Goffart (Centre d'Imagerie de la Faculté de Médecine Laennec) and to Paul Paulet (Centre Régional d'Imagerie Cellulaire de Montpellier) for significant contributions to the EM specimen processing, image digitalization, and photography. We are also grateful to Jean-Claude Cortay for valuable advice on pT7-7 cloning, fast-performance liquid chromatography, and protein purification. We thank Roger Monier, Catherine Dargemont, Robert Vigne, Etienne Decroly, Gilles Quérat, and Corinne Ronfort for fruitful discussions during this study. We are indebted to François Grateau, Hospices Civils de Lyon, for the financing of the MegaView II TEM camera and automatic MT-X ultramicrotome (RMC EM Products Group, Ventana Medical Systems, Inc., Tucson, Ariz.).
REFERENCES
- 1.Akhmanova, A., T. Verkerk, A. Langeveld, F. Grosveld, and N. Galijart. 2000. Characterisation of transcriptionally active and inactive chromatin domains in neurons. J. Cell Sci. 113:4463-4474. [DOI] [PubMed] [Google Scholar]
- 2.Bouyac-Bertoia, M., J. D. Dvorin, R. A. Fouchier, Y. Jenkins, B. E. Meyer, L. I. Wu, M. Emerman, and M. H. Malim. 2001. HIV-1 infection requires a functional integrase NLS. Mol. Cell 7:1025-1035. [DOI] [PubMed] [Google Scholar]
- 3.Breilling, A., B. M. Turner, M. E. Bianchi, and V. Orlando. 2001. General transcription factors bind promoters repressed by Polycomb group proteins. Nature 412:651-655. [DOI] [PubMed] [Google Scholar]
- 4.Brown, J. L., D. Mucci, M. Whiteley, M. L. Dirksen, and J. A. Kassis. 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a sequence-specific DNA binding protein with homology to the multifunctional mammalian transcription factor YY1. Mol. Cell 7:1057-1064. [DOI] [PubMed] [Google Scholar]
- 5.Brown, P. O. 1997. Integration. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y. [PubMed]
- 6.Bukrinsky, M. I., S. Haggerty, M. P. Dempsey, N. Sharova, A. Adzhubei, L. Spitz, P. Lewis, D. Goldfarb, M. Emerman, and M. Stevenson. 1993. A nuclear localization signal within HIV-1 matrix protein that governs infection of non-dividing cells. Nature 365:666-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bukrinsky, M. I., N. Sharova, T. L. McDonald, T. Pushkarskaya, W. G. Tarpley, and M. Stevenson. 1993. Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. USA 90:6125-6129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrière, C., B. Gay, N. Chazal, N. Morin, and P. Boulanger. 1995. Sequence requirement for encapsidation of deletion mutants and chimeras of human immunodeficiency virus type 1 Gag precursor into retrovirus-like particles. J. Virol. 69:2366-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carteau, S., S. C. Batson, L. Poljak, J.-F. Mouscadet, H. de Rocquigny, J.-L. Darlix, B. P. Roques, E. Käs, and C. Auclair. 1997. Human immunodeficiency virus type 1 nucleocapsid protein specifically stimulates Mg2+-dependent DNA integration in vitro. J. Virol. 71:6225-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carteau, S., R. J. Gorelick, and F. D. Bushman. 1999. Coupled integration of human immunodeficiency virus type 1 cDNA ends by purified integrase in vitro: stimulation by the viral nucleocapsid protein. J. Virol. 73:6670-6679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carteau, S., C. Hoffmann, and F. D. Bushman. 1998. Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target. J. Virol. 72:4005-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chazal, N., B. Gay, C. Carrière, J. Tournier, and P. Boulanger. 1995. Human immunodeficiency virus type 1 MAp17 deletion mutants expressed in baculovirus-infected cells: cis and trans effects on the Gag precursor assembly pathway. J. Virol. 69:365-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chicurel, M. 2000. Probing HIV's elusive activities within the host cell. Science 290:1876-1879. [DOI] [PubMed] [Google Scholar]
- 14.Cortay, J.-C., D. Nègre, M. Scarabel, T. M. Ramseler, N. B. Vartak, J. Reizer, and A. J. Cozzone. 1994. In vitro asymmetric binding of the pleiotropic regulatory protein, FruR, to the ace operator controlling glyoxylate shunt enzyme synthesis. J. Biol. Chem. 269:14885-14891. [PubMed] [Google Scholar]
- 15.Crowe, D. T., H. Chiu, S. Fong, and I. L. Weissmann. 1994. Regulation of the avidity of integrin α4β7 by the β7 cytoplasmic domain. J. Biol. Chem. 269:14411-14418. [PubMed] [Google Scholar]
- 16.Cullen, B. 2001. Journey to the center of the cell. Cell 105:697-700. [DOI] [PubMed] [Google Scholar]
- 17.Denisenko, O. N., and K. Bomsztyk. 1997. The product of the murine homolog of the Drosophila extra sex combs gene displays transcriptional repressor activity. Mol. Cell. Biol. 17:4707-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvorin, J. D., P. Bell, G. G. Maul, M. Yamashita, M. Emerman, and M. H. Malim. 2002. Reassessment of the roles of integrase and the central DNA flap in human immunodeficiency virus type 1 nuclear import. J. Virol. 76:12087-12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farnet, C. M., and F. D. Bushman. 1997. HIV-1 cDNA integration: requirement of HMG I(Y) protein for function of preintegration complexes in vitro. Cell 88:483-492. [DOI] [PubMed] [Google Scholar]
- 20.Farnet, C. M., and W. A. Haseltine. 1990. Integration of human immunodeficiency virus type 1 DNA in vitro. Proc. Natl. Acad. Sci. USA 87:4164-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.FitzGerald, D. P., and W. Bender. 2001. Polycomb group repression reduces DNA accessibility. Mol. Cell. Biol. 21:6585-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fouchier, R. A., B. E. Meyer, J. H. Simon, U. Fischer, A. V. Albright, F. Gonzales-Scarano, and M. H. Malim. 1998. Interaction of the human immunodeficiency virus type 1 Vpr protein with the nuclear pore complex. J. Virol. 72:6004-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freed, E. O. 1998. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology 251:1-15. [DOI] [PubMed] [Google Scholar]
- 24.Gallay, P., T. Hope, D. Chin, and D. Trono. 1997. HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. USA 94:9825-9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallay, P., V. Stitt, C. Mundy, M. Oettinger, and D. Trono. 1996. Role of the karyopherin pathway in human immunodeficiency virus type 1 nuclear import. J. Virol. 70:1027-1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallay, P., S. Swingler, C. Aiken, and D. Trono. 1995. HIV-1 infection of nondividing cells: C-terminal tyrosine phosphorylation of the viral matrix protein is a key regulator. Cell 80:379-388. [DOI] [PubMed] [Google Scholar]
- 27.Gallay, P., S. Swingler, J. Song, F. Bushman, and D. Trono. 1995. HIV nuclear import is governed by the phosphotyrosine-mediated binding of matrix to the core domain of integrase. Cell 83:569-576. [DOI] [PubMed] [Google Scholar]
- 28.Gao, K., R. J. Gorelick, D. G. Johnson, and F. Bushman. 2003. Cofactors for human immunodeficiency virus type 1 cDNA integration in vitro. J. Virol. 77:1598-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gay, B., J. Tournier, N. Chazal, C. Carrière, and P. Boulanger. 1998. Morphopoietic determinants of HIV-1 GAG particles assembled in baculovirus-infected cells. Virology 247:160-169. [DOI] [PubMed] [Google Scholar]
- 30.Goff, S. P. 2001. Intracellular trafficking of retroviral genomes during the early phase of infection: viral exploitation of cellular pathways. J. Gene Med. 3:517-528. [DOI] [PubMed] [Google Scholar]
- 31.Goodarzi, G., G. J. Im, K. Brackmann, and D. Grandgenett. 1995. Concerted integration of retrovirus-like DNA by human immunodeficiency virus type 1 integrase. J. Virol. 69:6090-6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 33.Gutjarhr, T., E. Frei, S. C., S. Baumgatner, A. H. White, and M. Noll. 1995. The polycomb-group gene, extra sex combs, encodes a nuclear member of the WD40 repeat family. EMBO J. 14:4296-4306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Higuchi, R., B. Krummel, and R. K. Saiki. 1988. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 16:7351-7367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hindmarsh, P., and J. Leis. 1999. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 63:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong, S. S., and P. Boulanger. 1995. Protein ligands of human adenovirus type 2 outer capsid identified by biopanning of a phage-displayed peptide library on separate domains of WT and mutant penton capsomers. EMBO J. 14:4714-4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hong, S. S., L. Karayan, J. Tournier, D. T. Curiel, and P. A. Boulanger. 1997. Adenovirus type 5 fiber knob binds to MHC class I α2 domain at the surface of human epithelial and B lymphoblastoid cells. EMBO J. 16:2294-2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horten, R. M., H. D. Hunt, S. N. Ho, J. K. Pullen, and P. L. R. 1989. Engineering hybrid genes without the use of restrictions enzymes: gene splicing by overlap extension. Gene 77:61-68. [DOI] [PubMed] [Google Scholar]
- 39.Huvent, I., S. S. Hong, C. Fournier, B. Gay, J. Tournier, C. Carriere, M. Courcoul, R. Vigne, B. Spire, and P. Boulanger. 1998. Interaction and co-encapsidation of HIV-1 Vif and Gag recombinant proteins. J. Gen. Virol. 79:1069-1081. [DOI] [PubMed] [Google Scholar]
- 40.Kalpana, G. V., S. Marmon, W. Wang, G. R. Crabtree, and S. P. Goff. 1994. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science 266:2002-2006. [DOI] [PubMed] [Google Scholar]
- 41.Lee, M. S., and R. Craigie. 1998. A previously unidentified host protein protects retroviral DNA from autointegration. Proc. Natl. Acad. Sci. USA 95:1528-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leh, H., P. Brodin, J. Bischerour, E. Deprez, P. Tauc, J.-C. Brochon, E. LeCam, D. Coulaud, C. Auclair, and J.-F. Mouscadet. 2000. Determinants of Mg2+-dependent activities of recombinant human immunodeficiency virus type 1 integrase. Biochemistry 39:9285-9294. [DOI] [PubMed] [Google Scholar]
- 43.Lin, C.-W., and A. Engelman. 2003. The barrier-to-autointegration factor is a component of functional human immunodeficiency virus type 1 preintegration complexes. J. Virol. 77:5030-5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lutzke, R. A., and R. H. Plasterk. 1998. Structure-based mutational analysis of the C-terminal DNA-binding domain of the human immunodeficiency virus type 1 integrase: critical residues for protein oligomerization and DNA binding. J. Virol. 4841-4848. [DOI] [PMC free article] [PubMed]
- 45.Maertens, G., P. Cherepanov, W. Pluymers, K. Busschots, E. De Clercq, Z. Debyser, and Y. Engelborghs. 2003. LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278:372-381. [DOI] [PubMed] [Google Scholar]
- 46.Manié, S. N., A. Astier, D. Wang, J. S. Phifer, J. Chen, A. I. Lazarovits, C. Morimoto, and A. S. Freedman. 1996. Stimulation of tyrosine phosphorylation after ligation of β7 and β1 integrins on human B cells. Blood 87:1855-1861. [PubMed] [Google Scholar]
- 47.Margottin, F., S. P. Bour, H. Durand, L. Selig, S. Benichou, V. Richard, D. Thomas, K. Strebel, and R. Benarous. 1998. A novel human WD protein, h-beta TrCP, that interacts with HIV-1 Vpu connects CD4 to the ER degradation pathway through an F-box motif. Mol. Cell 1:565-574. [DOI] [PubMed] [Google Scholar]
- 48.Miller, M. D., and F. D. Bushman. 1995. Ini1 for integration? Curr. Biol. 5:368-370. [DOI] [PubMed] [Google Scholar]
- 49.Miller, M. D., C. M. Farnet, and F. D. Bushman. 1997. Human immunodeficiency virus type 1 preintegration complexes: studies of organization and composition. J. Virol. 71:5382-5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neer, E. J., C. J. Schmidt, R. Nambudripad, and T. F. Smith. 1994. The ancient regulatory-protein family of WD-repeat proteins. Nature 371:297-300. [DOI] [PubMed] [Google Scholar]
- 51.Ng, J., R. Li, K. Morgan, and J. Simon. 1997. Evolutionary conservation and predicted structure of the Drosophila extra sex combs repressor protein. Mol. Cell. Biol. 17:6663-6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petit, C., O. Schwartz, and F. Mammano. 2000. The karyophilic properties of human immunodeficiency virus type 1 integrase are not required for nuclear import of proviral DNA. J. Virol. 74:7119-7126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petit, C., O. Schwartz, and F. Mammano. 1999. Oligomerization within virions and subcellular localization of human immunodeficiency virus type 1 integrase. J. Virol. 73:5079-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peytavi, R., S. S. Hong, B. Gay, A. Dupuy d'Angeac, L. Selig, S. Bénichou, R. Benarous, and P. Boulanger. 1999. H-EED, the product of the human homolog of the murine eed gene, binds to the matrix protein of HIV-1. J. Biol. Chem. 274:1635-1645. [DOI] [PubMed] [Google Scholar]
- 55.Pirrotta, V. 1998. Polycombing the genome: Pcg, trxG, and chromatin silencing. Cell 93:333-336. [DOI] [PubMed] [Google Scholar]
- 56.Priet, S., J.-M. Navarro, N. Gros, G. Quérat, and J. Sire. 2003. Functional role of HIV-1 virion-associated uracil DNA glycosylase 2 in the correction of G:U mispairs to G:C pairs. J. Biol. Chem. 278:4566-4571. [DOI] [PubMed] [Google Scholar]
- 57.Pruss, D., F. D. Bushman, and A. P. Wolffe. 1994. Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. USA 91:5913-5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pruss, D., R. Reeves, F. D. Bushman, and A. P. Wolffe. 1994. The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269:25031-25041. [PubMed] [Google Scholar]
- 59.Pryciak, P. M., and H. E. Varmus. 1992. Nucleosomes, DNA-binding proteins, and DNA sequence modulate retroviral integration target site selection. Cell 69:769-780. [DOI] [PubMed] [Google Scholar]
- 60.Reil, H., A. A. Bukovsky, H. R. Gelderblom, and H. G. Goettlinger. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rietzler, M., M. Bittner, W. Kolanus, A. Schuster, and B. Holzmann. 1998. The human WD repeat protein WAIT-1 specifically interacts with the cytoplasmic tails of β7-integrins. J. Biol. Chem. 273:27459-27466. [DOI] [PubMed] [Google Scholar]
- 62.Satjin, D. P. E., K. M. Hamer, J. den Blaauwen, and A. P. Otte. 2001. The Polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol. Cell. Biol. 21:1360-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Satjin, D. P. E., and A. P. Otte. 1999. RING1 interacts with multiple Polycomb group proteins and displays tumorigenic activity. Mol. Cell. Biol. 19:57-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Saurin, A. J., Z. Shao, H. Erdjument-Bromage, P. Tempst, and R. E. Kingston. 2001. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412:655-660. [DOI] [PubMed] [Google Scholar]
- 65.Scherdin, U., K. Rhodes, and M. Breindl. 1990. Transcriptionally active genome regions are preferred targets for retrovirus integration. J. Virol. 64:907-912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schroder, A. R., P. Shinn, H. Chen, C. Berry, J. R. Ecker, and F. Bushman. 2002. HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110:521-529. [DOI] [PubMed] [Google Scholar]
- 67.Schumacher, A., C. Faust, and T. Magnuson. 1996. Positional cloning of a global regulator of anterior-posterior patterning in mice. Nature 383:250-253. [DOI] [PubMed] [Google Scholar]
- 68.Schumacher, A., O. Lichtarge, S. Schwartz, and T. Magnusson. 1998. The murine polycomb-group eed and its human orthologue: functional implications of evolutionary conservation. Genomics 54:79-88. [DOI] [PubMed] [Google Scholar]
- 69.Schwartz, O., A. Dautry-Varsat, B. Goud, V. Marechal, A. Subtil, J. M. Heard, and O. Danos. 1995. Human immunodeficiency virus type 1 Nef induces accumulation of CD4 in early endosomes. J. Virol. 69:528-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sewalt, R. G. A. B., J. van der Vlag, M. J. Gunster, K. M. Hamer, J. L. den Blaauwen, D. P. E. Satijn, T. Hendrix, R. van Driel, and A. P. Otte. 1998. Characterization of interactions between the mammalian Polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian Polycomb-group protein complexes. Mol. Cell. Biol. 18:3586-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sherman, M. P., and W. C. Greene. 2002. Slipping through the door: HIV entry into the nucleus. Microbes Infect. 4:67-73. [DOI] [PubMed] [Google Scholar]
- 72.Shi, Y., E. Seto, L.-S. Chang, and T. Shenk. 1991. Transcriptional repression by YY1, a human GLI-Kruppel-related protein, and relief of repression by adenovirus E1A. Cell 67:377-388. [DOI] [PubMed] [Google Scholar]
- 73.Showell, C., and V. T. Cunliffe. 2002. Identification of putative interaction partners for the Xenopus Polycomb-group protein Xeed. Gene 291:95-104. [DOI] [PubMed] [Google Scholar]
- 74.Simon, J. 1995. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr. Opin. Cell Biol. 7:376-385. [DOI] [PubMed] [Google Scholar]
- 75.Smith, G. P., and J. K. Scott. 1993. Libraries of peptides and proteins displayed on filamentous phage. Methods Enzymol. 217:228-257. [DOI] [PubMed] [Google Scholar]
- 76.Spillane, C., C. MacDouglas, C. Stock, C. Köhler, J.-P. Vielle-Calzada, S. M. Nunes, U. Grossniklaus, and J. Goodrich. 2000. Interaction of the Arabidopsis Polycomb group proteins FIE and MEA mediates their common phenotypes. Curr. Biol. 10:1535-1538. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki, Y., and R. Craigie. 2002. Regulatory mechanisms by which barrier-to-autointegration factor blocks autointegration and stimulates intermolecular integration of Moloney murine leukemia virus preintegration complexes. J. Virol. 76:12376-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turelli, P., V. Doucas, E. Craig, B. Mangeat, N. Klages, R. Evans, G. Kalpana, and D. Trono. 2001. Cytoplasmic recruitment of INI1 and PML on incoming HIV preintegration complexes: interference with early steps of viral replication. Mol. Cell 7:1245-1254. [DOI] [PubMed] [Google Scholar]
- 79.Van der Vlag, J., and A. P. Otte. 1999. Transcriptional repression mediated by the human polycomb-group protein EED involves histone acetylation. Nat. Genet. 23:474-478. [DOI] [PubMed] [Google Scholar]
- 80.Wang, J.-Y., H. Ling, W. Yang, and R. Craigie. 2001. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 20:7333-7343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Weidhaas, J. B., E. A. Angelichio, S. Fenner, and J. M. Coffin. 2000. Relationship between retroviral integration and gene expression. J. Virol. 74:8382-8389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Withers-Ward, E. S., Y. Kitamura, J. P. Barnes, and J. M. Coffin. 1994. Distribution of targets for avian retrovirus DNA integration in vivo. Genes Dev. 8:1473-1487. [DOI] [PubMed] [Google Scholar]
- 83.Wu, X., Y. Li, B. Crise, and S. M. Burgess. 2003. Transcription start regions in the human genome are favored targets for MLV integration Science 300:1749-1751. [DOI] [PubMed] [Google Scholar]
- 84.Yung, E., M. Sorin, A. Pal, E. Craig, A. Morozov, O. Delattre, J. Kappes, D. Ott, and G. V. Kalpana. 2001. Inhibition of HIV-1 virion production by a transdominant mutant of integrase interactor 1. Nat. Med. 8:920-926. [DOI] [PubMed] [Google Scholar]
- 85.Zennou, V., C. Petit, D. Guetard, U. Nerhbass, L. Montagnier, and P. Charneau. 2000. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell 101:173-185. [DOI] [PubMed] [Google Scholar]
- 86.Zou, S., and D. F. Voytas. 1997. Silent chromatin determines target preference of the Saccharomyces retrotransposon Ty5. Proc. Natl. Acad. Sci. USA 94:7412-7416. [DOI] [PMC free article] [PubMed] [Google Scholar]