Abstract
Control of viremia in natural human immunodeficiency virus type 1 (HIV-1) infection in humans is associated with a virus-specific T-cell response. However, still much is unknown with regard to the extent of CD8+ cytotoxic T-lymphocyte (CTL) responses required to successfully control HIV-1 infection and to what extent CTL epitope escape can account for rises in viral load and ultimate progression to disease. In this study, we chose to monitor through full-length genome sequence of replication-competent biological clones the modifications that occurred within predicted CTL epitopes and to identify whether the alterations resulted in epitope escape from CTL recognition. From an extensive analysis of 59 biological HIV-1 clones generated over a period of 4 years from a single individual in whom the viral load was observed to rise, we identified the locations in the genome of five CD8+ CTL epitopes. Fixed mutations were identified within the p17, gp120, gp41, Nef, and reverse transcriptase genes. Using a gamma interferon ELIspot assay, we identified for four of the five epitopes with fixed mutations a complete loss of T-cell reactivity against the wild-type epitope and a partial loss of reactivity against the mutant epitope. These results demonstrate the sequential accumulation of CTL escape in a patient during disease progression, indicating that multiple combinations of T-cell epitopes are required to control viremia.
Viral disease prevention typically is accomplished by global campaigns with vaccines that prevent systemic infection by inducing virus-specific humoral and cellular immunity. The only human immunodeficiency virus (HIV) vaccine that accomplishes this level of protection in experimental animals is based on a live, attenuated virus (11), but this approach already has encountered major setbacks (2, 3, 5, 15, 30). Alternatives such as DNA vaccines and vaccines based on viral vectors, such as poxviruses, prevent AIDS but not viral infection in monkey models (4, 31, 46). Their protection against AIDS is based on the prevention of CD4+-T-cell decline through a reduction of the viral load below the detection level in the first year after seroconversion, mimicking what occurs naturally in humans with long-term nonprogressing HIV type 1 (HIV-1) infections (13, 32, 33).
Control of viremia in monkeys infected experimentally with simian-human immunodeficiency virus or simian immunodeficiency virus (45) and in human HIV-1 infection (25, 35, 38, 39) is related directly to virus-specific cytotoxic T-lymphocyte (CTL) response levels. Evidence from HIV-exposed but seronegative African commercial sex workers suggests that CTLs may protect against infection (23). HIV-specific CTL responses have been associated with the viremic phase shortly after virus transmission and prior to antibody development and are believed to control the number of cells producing HIV (7, 28), thereby determining the viral load set point that predicts clinical outcome. Given that viruses replicate and genomic mutations accumulate even when the viral load is undetectable (52), T-cell epitopes may change, allowing for virus escape from a controlling immune response.
Immunodominant responses generally are effective, but HIV generates variants at an extremely rapid pace, exposing CTLs to a large pool of mutants that can all impair immune responsiveness. Escape from CTL control is indicated by a mutation that occurs in the T-cell epitope and becomes fixed in the virus population, resulting in an in vivo competitive advantage for viruses with this mutation and the reduction of the T-cell response to the wild-type epitope. Escape mutants can arise early or late in HIV infection (8, 24, 40, 41, 43) and even can be transmitted (21). Identifying HIV-1 escape mutants may reveal the T-cell response(s) that plays a role in controlling viral load (37).
To identify T-cell responses associated with the control of viremia, we studied an HIV-1 seroconverter whose viral load was low at the set point but rose steadily during study follow-up. This patient steadily lost CD4+-T-cell counts and eventually lost control of viremia (17). We obtained 59 biological virus clones from peripheral blood mononuclear cells (PBMCs) collected over a 4-year period. The clones were representative of the virus population in vivo, and the number of productively infected cells in the original PBMC samples paralleled the HIV RNA load in serum. Complete RNA genome sequencing was performed on seven HIV-1 clones selected from three time points (early, intermediate, and late) during infection. Based on these sequences and the patient's HLA type (A3,32; B51,15; Cw3,6), we identified a large number of potential CTL epitopes and separated the stable epitopes from those with fixed mutations. For potential epitopes with fixed mutations over time, we evaluated the frequency distributions of wild-type versus mutant epitopes in all 59 clones. Sequential PBMC samples were analyzed by using a gamma interferon (IFN-γ) ELIspot assay for general T-cell responses with HIV-1 region-specific peptide pools and for epitope-specific T-cell responses with peptides resembling wild-type or mutant epitopes. Using this approach, we identified fixed mutations in five epitopes and the loss of T-cell reactivity against four of these epitopes. These results demonstrate the loss of multiple T-cell responses in an HIV-1-infected individual who progressed to disease, suggesting that combinations of T-cell responses initially control virus replication.
MATERIALS AND METHODS
Patient.
Patient H671 entered the Amsterdam Cohort Studies on 5 June 1986 and tested seropositive for HIV-1-specific antibodies on 22 March 1995. Serum samples and PBMCs were collected every 3 months. The viral load was determined by using the NucliSens assay or HIV-1 RNA QT NASBA (Organon Teknika B.V., Boxtel, The Netherlands), and CD4+-cell counts were determined by flow cytometry with FACscan and monoclonal antibodies (both from Becton Dickinson, San Jose, Calif.). The CD4+-cell count at the set point was 1,120 cells/ml. By linear regression analysis, the intercept of CD4+-cell decline was found to be 1,154 cells/ml (standard deviation, 100), and the negative slope was found to be 58 cells/ml (standard deviation, 65). Testing by standard PCR and sequencing techniques of patient H671 for CCR5, CCR2, and SDF-1 revealed wild-type homozygosity for CCR2 and SDF-1 but heterozygosity for CCR5Δ32. The patient's complete HLA type was A3,32; B51,15; CW3,6; DR4,8; DQ7. He still undergoes monitoring and has not received antiretroviral therapy despite a rise in viral load. Patient H671 was infected with a zidovudine (AZT)-resistant virus, as indicated by mutations at codon positions 41 and 215 (12) of reverse transcriptase (RT). Since the 215Y mutation reverts in the absence of drug pressure, it served as a marker for characterizing the sequential virus clones in PBMCs versus serum (18, 19).
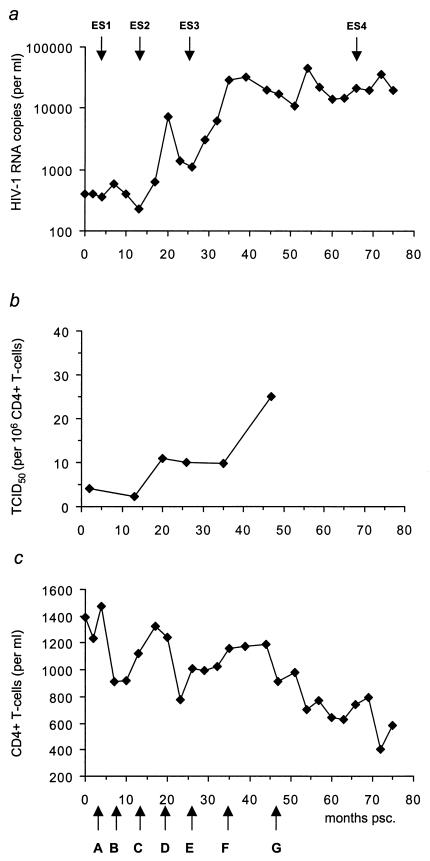
Figure 1 shows the HIV-1 RNA copy numbers, the 50% tissue culture infective doses per 106 CD4+ cells, reflecting the number of productively infected cells, and the CD4+-cell counts during follow-up.
FIG. 1.
Clinical parameters for patient H671. Longitudinal analyses of serum HIV RNA load (a), frequency of productively infected cells (50% tissue culture infective doses [TCID50]) (b), and CD4+ T cells (c) are shown. CD4+ T cells and viral load were measured every 3 months during follow-up. Arrows on the x axis indicate time points (A through G) at which biological clones were obtained. A total of 59 replication-competent HIV clones were obtained: 7 at time point A, 2 at B, 2 at C, 13 at D, 11 at E, 11 at F, and 13 at G (see also Fig. 4 and 5). Clones obtained at time points A, D, and G were selected for whole-genome sequencing. T-cell responses (see Fig. 4) were measured at four time points (ES1 to ES4).
Biological cloning.
To isolate biological virus clones and estimate the proportion of productively infected cells, limiting-dilution cultures were set up (26). Briefly, participant PBMCs (0.5 × 104 to 4 × 104 cells per well; 24 to 96 replicates per concentration) were cocultivated with phytohemagglutinin (PHA)-stimulated healthy donor PBMCs (105 cells per well) in 96-well microtiter plates. Every week for 5 weeks, 65 μl of each culture supernatant was collected for detection of p24 antigen by an in-house p24 antigen capture enzyme-linked immunosorbent assay. At the same time, half of the cells were transferred to new 96-well plates, and 105 fresh PHA-stimulated healthy donor PBMCs were added to propagate the cultures. The proportion of productively infected CD4+ T cells was estimated by the formula for a Poisson distribution as −1n(F0), where F0 is the fraction of negative cultures. PBMCs from wells with positive results were transferred to 25-ml culture flasks containing 5 × 106 fresh PHA-stimulated PBMCs in 5 ml of culture medium to grow virus stocks. Fifty-nine replication-competent biological clones were generated and grown on target PBMCs from a seronegative blood donor. Replication rates were determined as previously described (49).
Full-length HIV-1 sequencing.
Of the 59 biological clones, 7 (time points A, D, and G in Fig. 1c) were selected for complete genomic sequencing with PALM, a newly developed proportional amplification and labeling method (J. Zhang et al., European patent application WO 01/51661 A2, Jan. 2001). This protocol allows amplification of (long) nucleic acid sequences by use of primers containing nonspecific tails (NNNNNG), which enable random annealing on the target sequence. Two rounds of amplification with these primers yield a pool of random sequence amplimers. The protocol is completed by amplification with HIV-1-specific primers, sequencing, and analysis.
Briefly, viral RNA was isolated from seven selected virus supernatants by the method of Boom et al. (6). After denaturation for 10 min at 80°C, viral RNA was subjected to the first, nonspecific RT step. The reaction mixture (mixture A) had a total volume of 20 μl and contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 5 mM MgCl2, 4 μM deoxynucleoside triphosphates, 0.5 U of RNasin, 25 U of murine leukemia virus RT (all from Perkin-Elmer, Foster City, Calif.), and 50 ng of primer JZH2R (5′-GCTATCATCACAATGGACNNNNNG-3′). After incubation for 10 min at room temperature, 30 min at 42°C, and 5 min at 80°C, RT reactions were stopped by incubation for 30 min at 37°C with 0.5 U of RNase H (Roche Diagnostics, Almere, The Netherlands). cDNA strands were made (10 min on ice, 10 min at room temperature, and 10 min at 37°C) by combining 20 μl of mixture A with a total volume of 40 μl of mixture B, which contained 17.5 mM Tris-HCl (pH 7.5), 12.5 mM NaCl, 8.75 mM MgCl2, 4 μM deoxynucleoside triphosphates, 100 ng of primer JZH2R, and 2.6 U of Sequenase (Amersham Pharmacia, Piscataway, N.J.). Final amplifications (Perkin-Elmer 9700 PCR machine) were carried out (5 min at 95°C; 45 cycles of 20 s at 95°C, 30 s at 55°C, and 2 min at 72°C; 10 min at 72°C; and 10 min at 4°C) with a total volume of 50 μl containing mixture B, 100 ng of primer JZH1 (5′-GTATCATCACAATGGAC-3′), and 1.7 U of Amplitaq (Perkin-Elmer).
Randomly amplified viral DNA was analyzed on agarose gels before being subjected to multiple PCRs with multiple primer sets to allow amplification of the entire HIV-1 genome. PCR conditions were the same as those described above. Positive PCR products were sequenced by using a BigDye terminator cycle sequencing kit (ABI, Foster City, Calif.) and analyzed by using an ABI 377 automated sequencer (ABI). Sequences were manually edited and assembled with Autoassembler (PE Biosystems Software). Analysis of synonymous and nonsynonymous substitution rates across viral regions was conducted with MEGA software, version 2.1 (29), based on the method of Nei and Gojobori (36). A statisical comparison of synonymous and nonsynonymous substitution rates was performed by using the t test.
Peptides.
Synthetic peptides were custom ordered from SynPep Corporation (Dublin, Calif.). Peptide sequences were based on the HIV sequences most closely related to the clade B consensus sequences (Los Alamos National Laboratory, Los Alamos, N.Mex.; http://hiv-web.lanl.gov). Peptides were 15-mers overlapping by 11 amino acids (aa) unless specified otherwise. They were dissolved in dimethyl sulfoxide (DMSO) at 20 to 50 mg/ml and stored in small aliquots at −70°C. Peptide pools were composed of an equal volume of each peptide, with up to 120 peptides per pool. The final concentration for each peptide in a pool was 0.4 mg/ml.
IFN-γ ELIspot assay.
Wells of sterile 96-well microtiter plates containing polyvinylidene difluoride (MAIP S45; Millipore, Bedford, Mass.) were coated overnight at 4°C with 100 μl of mouse anti-human IFN-γ monoclonal antibody clone 1-D1K (MabTech, Stockholm, Sweden) diluted to 10 μg/ml in sterile Dulbecco phosphate-buffered saline (PBS; Gibco-BRL, Grand Island, N.Y.). Wells of coated plates were washed four times and blocked for 2 h in a humidified CO2 incubator at 37°C with 200 μl of complete RPMI 1640 medium complemented with 10% fetal bovine serum (R-10; Gibco-BRL). This blocking buffer was removed, and 2 × 100 μl of PBMCs diluted in R-10 was added per well to result in 2 × 105 and 1 × 105 cells/well. Peptides or peptide pools were diluted in R-10 and added at 25 μl/well, and the final concentration of each peptide in the pools was 2 to 3 μg/ml. Peptide-free DMSO diluent at the same concentration as DMSO in the peptide solutions was used as a negative control (mock antigen). Plates were incubated overnight in a humidified CO2 incubator at 37°C and then washed seven times with PBS containing 0.05% Tween 20 (Sigma, St. Louis, Mo.). Biotinylated anti-human IFN-γ monoclonal antibody clone 7-B6-1 (MabTech) was diluted to 1 μg/ml in assay diluent consisting of PBS and 0.5% bovine serum albumin (Sigma). The diluted antibody was added to the plates at 100 μl/well and incubated for 2 to 4 h at room temperature. Plates were washed four times with PBS-Tween, and 100 μl of alkaline phosphatase-conjugated antibiotin monoclonal antibody (Vector Laboratories, Burlingame, Calif.) diluted 1:750 in assay diluent was added to each well. Plates were incubated for 2 h at room temperature and washed four times with PBS-Tween.
To develop the spots, 100 μl of precipitating alkaline phosphatase substrate nitroblue tetrazolium-5-bromo-4-chloro-3-indolylphosphate (Pierce, Rockford, Ill.) was added to each well and incubated at room temperature until spots became visible (usually 5 to 10 min). Spots were counted by using a digital imager and automated spot-counting software (AutoImmun Diagnostika, Strassberg, Germany). The number of spots per well was normalized to 106 cells and averaged for each sample and antigen to give a final result of spot-forming cells per 106 cells.
Nucleotide sequence accession numbers.
The sequences described here have been submitted to GenBank and assigned accession numbers AY423381 through AY423387.
RESULTS
Generation of biological clones.
From a single HIV-1-infected individual, a total of 59 replication-competent (PBMC-derived) biological clones were generated from seven time points: 2, 7, 13, 20, 26, 35, and 47 months postseroconversion (psc) (Fig. 1c, A to G). The numbers of clones recovered from the different time points were as follows: A, 7; B, 2; C, 2; D, 13; E, 11; F, 11; and G, 13. The time points covered a 4-year period during which patient H671 showed an increasing viral load and declining CD4+-T-cell counts (Fig. 1a and c).
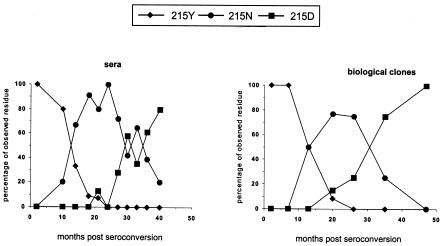
To verify whether the obtained biological clones were a fair representation of the circulating, biologically relevant virus (in serum), we analyzed the distribution of virus clones over time. As a marker we used the shift from AZT resistance to AZT sensitivity (RT position 215) in the absence of drugs. Figure 2 shows that the shift was evident, with the 215Y mutation in the RT gene being replaced by 215D and subsequently by 215N, and was seen in both the replication-competent biological clones and the virus population in serum. We conclude that the distribution of position 215 mutants over time among the biological clones is a direct reflection of the virus population found in serum.
FIG. 2.
Correlation of in vivo and in vitro virus quasispecies through evolution at position 215 of RT for patient H671. Serum-derived viruses (sera) were compared with replication-competent clones (biological clones) isolated at the same time points.
Overall T-cell responses to Gag, Pol, Nef, Rev, and Tat.
The overall T-cell responses were measured as IFN-γ responses over time with overlapping peptides as antigens. Initially, we determined the hierarchy of T-cell responses during the period when the viral load remained below 1,000 copies/ml (up to 17 months psc) and when it surpassed 10,000 copies/ml (39 or more months psc). In these experiments, we used peptide pools containing 20-mers overlapping by 10 aa. The strongest responses were found against the Gag pool, followed by Pol and Nef; negligible responses were observed for Tat and Rev (data not shown). While responses to Pol and Nef remained stable, responses to Gag increased over time (data not shown).
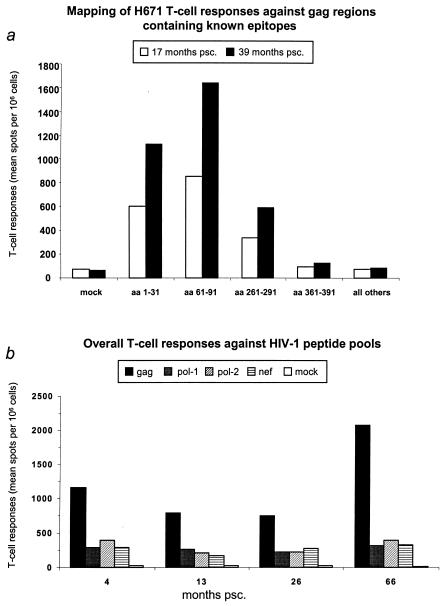
We subsequently mapped responsive regions in Gag to determine whether the observed increases in T-cell responses reflected a shift in epitope recognition. As shown in Fig. 3a, four major epitope-containing regions in Gag (aa 1 to 31, 61 to 91, 261 to 291, and 361 to 391) (27) were recognized, suggesting that the specificity of the T-cell responses had not changed. Based on these results, we next examined the T-cell responses against the most reactive gene products (Gag, Pol, and Nef) over the complete follow-up period by using 15-mer peptide pools overlapping by 11 aa. Again, we observed that the strong T-cell responses to Gag became even stronger despite declining CD4+-cell counts, while the responses to Pol and Nef remained relatively constant (Fig. 3b).
FIG. 3.
Patient H671 general T-cell responses. (a) Mapping of H671 T-cell responses against Gag regions containing known epitopes for the patient's haplotype (Los Alamos National Laboratory Immunology Database; http://hiv-web.lanl.gov) at 17 and 39 months psc. Amino acids (aa) 1 to 31; 61 to 91; 261 to 291, and 361 to 391 were spanned by four pools, and another pool spanned the rest of the Gag regions. Bars represent the mean IFN-γ spot-forming units/106 PBMCs in duplicate wells. (b) Profiles of overall H671 T-cell responses to HIV-1. Four time points (4, 13, 26, and 66 months psc) were screened by an IFN-γ ELIspot assay. The Pol region was divided, with Pol-1 representing aa 1 to 430 and Pol-2 representing aa 420 to 840. Bars represent the mean IFN-γ spot-forming units/106 PBMCs in duplicate wells.
Full-genome analysis of virus clones and CTL epitopes.
To obtain an indication as to whether immune pressure had affected regions important for immune recognition, we selected 7 of the 59 clones for full-genome sequencing. Clones were selected from three time points: two at 2 months psc (Fig. 1c, time point A), when the viral load was below 1,000 copies/ml; three at 20 months psc, when the viral load first rose above 1,000 copies/ml (time point D); and two at 47 months psc (time point G), when the viral load exceeded 10,000 copies/ml. Based on the HLA type of patient H671 (A3,32; B51,15; CW3,6; DR4,8; DQ7), we identified 29 potential CTL epitopes or reactive regions (27). We found 17 epitopes with no alterations over time in the seven complete genomes: 3 epitopes localized in Gag p17 and p24, 7 in Pol, 1 in Vif, 5 in gp120 and gp41, and 1 in Nef (Table 1). When we tested T-cell reactivity to a pool of nine peptides covering nonchanging epitopes, the reactivity was initially just above the background but rose over time (data not shown). In seven other epitopes, localized in Pol (three), Env (two), and Nef (two), mutations displayed a nonfixed character, either being unfixed over time or not being present in all clones at a certain time point (Table 2). Nonfixed mutations appeared throughout the infection. We found five previously defined epitopes or reactive regions with fixed mutations (Table 3): Gag p17 (A3-restricted epitope: RLRPRGGKKK), Pol-RT (B51: TAFTIPSI), Env gp120 (B15: SFNCGGEFF), Env gp41 (A32-restricted reactive 20-mer: VLSIVNQVRRQGYSPLSFQT), and Nef (B15: FFPDWKNYT). Mutations were considered fixed if they were identical and occurred in >80% of the clones taken from the same time point and if >80% of the clones recovered at later time points all contained the same nonsilent mutations. Fixed mutations which did not localize in the described epitopes were analyzed for HLA-binding motifs specific for the patient's haplotype to determine whether these mutations had arisen in undocumented epitopes. None of these mutations localized in possible binding regions specific for the patient's haplotype (data not shown).
TABLE 1.
Seventeen stable epitopes
| Region (positions) | Sequence | HLA type |
|---|---|---|
| p17 (18-26) | KIRLRPGGK | A3 |
| p24 (8-20) | GQMVHQAISPRTL | Cw3 |
| RT (42-50) | EKEGKISKI | B51 |
| RT (113-120) | DAYFSVPL | B51 |
| RT (158-166) | AIFQSSMTK | A3 |
| RT (269-277) | QIYPGIKVR | A3 |
| RT (309-317) | ILKEPVHGVY | B15 |
| RT (432-440) | EPIVGAETF | B51 |
| Vif (17-26) | RIRTWKSLVK | A3 |
| gp160 (37-46) | TVYYGVPVWK | A3 |
| gp160 (416-429) | LPCRIKQII | B51 |
| gp160 (557-565) | RAIEAQQHL | B15,51 |
| Nef (73-82) | QVPLRPMTYK | A3 |
| p24 (193-201) | NANPDCKTI | B51 |
| RT (293-301) | IPLTEEAEL | B51 |
| gp160 (78-86) | DPNPQEVVL | B51 |
| gp160 (308-322) | RIQRGPGRAFVTIGK | A3 |
TABLE 2.
Seven nonfixed epitopesa
| Region (positions of epitope boundaries) | Sequenceb | HLA type |
|---|---|---|
| Pol (192-201) | D L E I G Q H R T K | A3 |
| Mc | ||
| Pol (260-271) | L V G K L N W A S Q I Y | B15 |
| Xc | ||
| Pol (28-36) | L P P V V A K E I | B51 |
| Id | ||
| Env (770-780) | R L R D L L L I V T R | A3 |
| VcId | ||
| Env (835-843) | R A Y R A I L H I | B51 |
| Te | ||
| Fe Xd | ||
| Id | ||
| Nef (186-194) | D S R L A F H H V | B51 |
| Le Le | ||
| Re Me | ||
| Vd | ||
| Nef (190-198) | A F H H V A R E K | B51 |
| Le | ||
| Re Me | ||
| Vd | ||
| Xc Ic |
Consensus sequences and epitope boundaries were derived from HIV-1 HXB2.
Consensus sequences are shown above variant sequences. Only variant residues are shown for the latter; when no variant residue is noted, the wild-type residue was found.
Variant residue which arose at late time points (47 months psc).
Variant residue which arose at intermediate time points (20 months psc).
Variant residue which arose at early time points (2 months psc).
TABLE 3.
Five epitopes with fixed mutations over timea
| Region (positions of epitope boundaries) | mo psc (no. of clones)b | Sequencec | HLA type |
|---|---|---|---|
| Gag p17 (20-28) | R L R P G G K K K | A3 | |
| 2 (7/7) | - - - - - - - - - | ||
| 120 (12/13) | - - - - - - - - R | ||
| 47 (13/13) | - - - - - - - - R | ||
| Pol-RT (128-135) | T A F T I P S I | B51 | |
| 2 (7/7) | - - - - - - - - | ||
| 20 (13/13) | - - - - - - - - | ||
| 47 (13/13) | - - - - - - -T | ||
| Env gp120 (375-383) | S F N C G G E F F | B15 | |
| 2 (7/7) | - - - - - - - - - | ||
| 20 (12/13) | - - - - R - - - - | ||
| 47 (12/13) | - - - - R - - - - | ||
| Nef (120-128) | F F P D W K N Y T | B51 | |
| 2 (6/7) | Y - - - - Q - - - | ||
| 2 (1/7) | Y - - - - H - - - | ||
| 20 (3/13) | Y - - - - H - - - | ||
| 20 (3/13) | Y - - - - Q S - - | ||
| 20 (6/13) | Y L - - - Q S - - | ||
| 20 (1/13) | Y - - - - D - - - | ||
| 47 (12/13) | Y - - - - D - - - | ||
| 47 (1/13) | Y - - - - Q S - - | ||
| Env gp41 (691-710) | V L S I V N Q V R R Q G Y S P L S F Q T | B15 | |
| 2 (7/7) | - - - - - - - - - - - - - - - - - - - - | ||
| 20 (7/13) | - - - - - - K - - - - - - - - - - - - - | ||
| 47 (13/13) | - - - - - - K - - - - - - - - - - - - - |
Escape from T-cell recognition would be visualized by a higher ratio of nonsynonymous substitutions to synonymous substitutions (dN/dS ratio) in regions with CTL recognition sites than in regions without CTL recognition sites. We analyzed the seven full genomes from the three time points to determine whether mutational pressure was exerted on CTL epitopes. dN/dS ratios were calculated for different HIV-1 regions between CTL epitopes and non-CTL epitopes (background). For the Nef gene, the dN/dS ratio in the CTL recognition sites and the dN/dS ratio in the background sequences were approximately equal, both within the group of seven sequences (Nefintra, 0.79 vs. 0.84) and between early (early and middle time points) and later sequences (Nefinter, 0.64 vs. 0.94). These results indicate that selective pressure to substitute at nonsynonymous nucleotide positions in the CTL recognition sites in Nef was absent. For the Gag, Pol, and Env genes, the dN/dS ratio in the CTL recognition sites was higher than that in the background sequences, both within the group of seven sequences and between sequences (Gagintra, 0.39 vs. 0.30; Gaginter, 0.85 vs. 0.38; Polintra, 0.55 vs. 0.13; Polinter, 1.0 vs. 0.23; Envintra, 2.0 vs. 0.82; and Envinter, 1.56 vs. 0.81). The high dN/dS ratio in the CTL recognition sites in Pol was remarkably high (0.55). The dN/dS ratio in the entire Pol region was determined before to range from 0.09 to 0.19 within a subtype as well as between subtypes (10).
The numbers of nonsynonymous mutations over the entire period were significantly higher in epitopes located in Pol and Nef sequences than in background sequences, as determined by the t test (P < 0.001). Nonsynonymous mutations in Env epitopes were borderline significant, as determined by the t test (P = 0.056), whereas no significance for Gag epitopes was found. The higher dN/dS ratio in the CTL recognition sites indicates that positive selection for amino acid substitutions is operational at these domains and that the mutations are indeed CTL escape mutations. Finally, we found no fixed mutations in the flanking regions (15 aa) of the five CTL recognition sites (data not shown).
Epitope frequency distribution in 59 biological clones and T-cell responses to wild-type versus mutant epitopes.
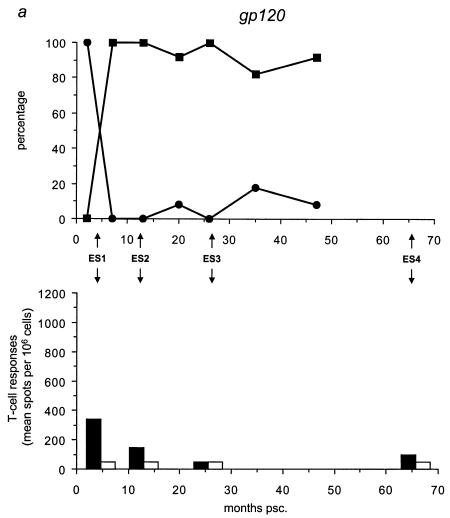
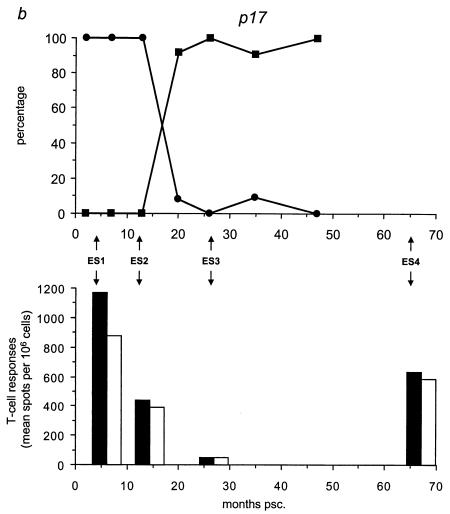
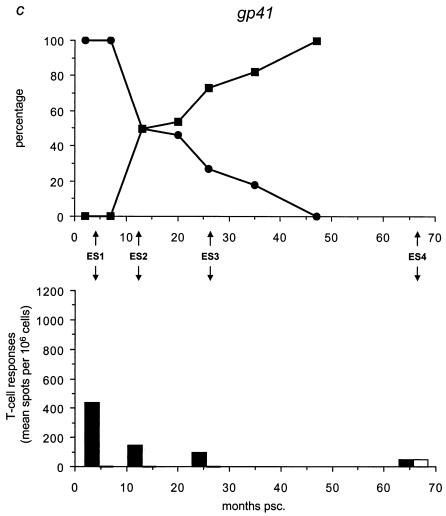
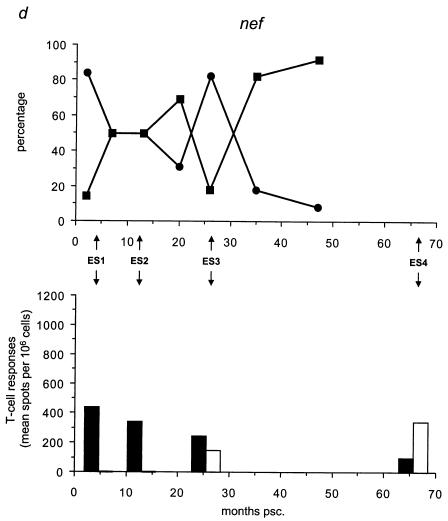
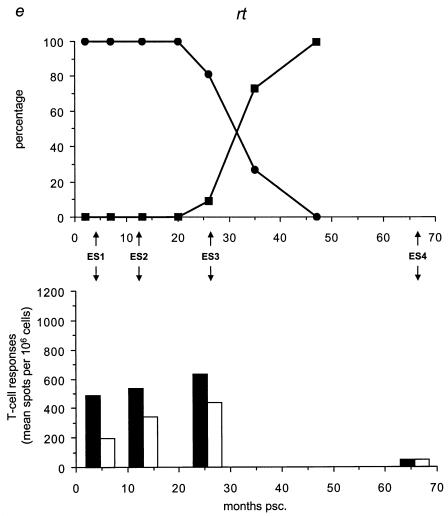
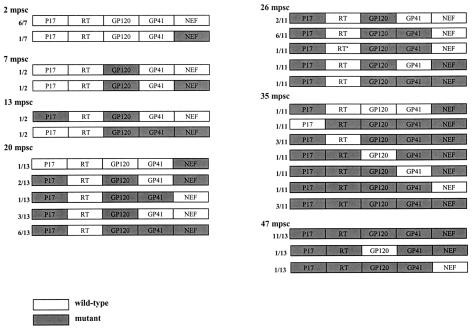
We analyzed the frequency distribution of the wild-type versus mutant epitopes by sequencing for the five different epitopes all 59 biological clones (Table 3; Fig. 4, upper panels) collected at time points A through G (Fig. 1c). We found that the mutant gp120 epitope was the first to completely replace the wild-type epitope at 7 months psc, resulting in a genotype that oscillated around a completely mutated virus pool from that date onward (Fig. 4a). At 20 months psc, the wild-type p17 epitope was replaced by the mutant epitope, resulting in the same oscillating pattern (Fig. 4b). The mutant gp41 genotype appeared at 10 months psc, resulting in a completely mutated pool at 47 months psc (Fig. 4c). The Nef epitope showed a more fluctuating course over time, with a nearly totally (92%) mutated virus pool at 47 months psc (Fig. 4d), coincident with the total replacement of the wild-type RT epitope (Fig. 4e).
FIG. 4.
Temporal relationship between the frequency distribution of wild-type or mutant epitopes and T-cell reactivity against wild-type or mutant peptides. (Top panels) All 59 biological clones were analyzed for the five epitopes with fixed mutations in gp120, p17, gp41, Nef, and RT (a to e, respectively). Clones exhibiting wild-type (•) and mutant (▪) epitopes are indicated as a percentage of the total population for each time point and are plotted longitudinally. (Lower panels) PBMCs from patient H671 were tested for their reactivity against optimally sized wild-type and mutant synthetic peptides matching the wild-type (filled bars) and mutant (open bars) epitopes found in the virus population at four time points (4, 13, 26, and 66 months psc). The epitope frequency distribution and T-cell reactivity were aligned.
We evaluated T-cell responses to the five epitopes with fixed mutations by using optimally sized peptides that precisely matched the wild-type and mutant amino acid sequences (Fig. 4, lower panels) and PBMCs collected at 4, 13, 26, and 66 months psc (ES1 to ES4; Fig. 1a and 4). All of the shifts in epitope sequences were paralleled by a dramatic decrease in T-cell reactivity against all wild-type peptides (Fig. 4a and c to e), except for p17 (Fig. 4b), which showed an increase in T-cell reactivity after the mutations became fixed. The mutant peptides were recognized poorly or not at all by the T cells, except for Nef (Fig. 4d) and p17 (Fig. 4b); there was already a significant response to these mutant peptides before the switch. Due to the length of the gp41 epitope, it is possible that the sequence recognized and causing the response lies outside the region containing the mutation. However, the mutation becomes fixed in all the clones at the latest time point, coincident with a decrease in T-cell reactivity, suggesting a reactive region. Figure 5 shows an overview of the temporal accumulation of mutations in all 59 biological clones.
FIG. 5.
Schematic overviews of the appearance of escape mutations in the virus population. For each biological clone, the data indicate whether, at the time of isolation, escape mutations had occurred in one of the five epitopes with fixed mutations. The Nef epitope is represented as a mutant irrespective of the mutation that occurred (K→Q/H/D). For one clone at 26 months psc (mpsc), a unique I→L mutation was seen (asterisk).
Thus, in the patient tested, the virus population gradually shifted toward a population in which at 47 months psc, nearly the entire virus population was affected by mutations in five functional CTL epitopes or reactive regions, coincident with decreased T-cell reactivity; these findings are indicative of a loss of CTL control. These effects may have contributed to the increasing viremia.
DISCUSSION
Strong evidence has been obtained to indicate that cellular immune responses are key components of protective immunity in HIV-1 infection (16), and they have been the focus of many HIV-1 vaccine efforts. In the past, however, it has proven difficult to correlate ex vivo measured CTL responses with in vivo CTL-mediated antiviral effects, which are expected to control disease (53). The minimum set of epitopes that CTLs must recognize to limit the number of infected cells is presently still unknown. Whether such a set must be localized on a single viral protein or on multiple proteins is unclear; furthermore, variation and complexity within different HLA types complicate the identification of HIV-1 T-cell recognition patterns.
Through the mechanisms by which HIV-1 evades the suppressive activities of CTLs (epitope escape mutations) and the subsequent genomic imprint left by the virus, we have identified alterations in four predicted epitopes that inhibit CTL activity. HIV-1 mutational escape is a delicate interplay among CTL selective pressure, the viral fitness cost of mutations, and the tolerance of CTL for individual epitope mutations (53).
We present a set of HIV-1 epitope- or region-specific CTLs putatively involved in the control of viremia based on a longitudinal study of a single HIV-1-infected patient. The infecting virus had the characteristics of a low-fitness variant, including a 215Y mutation conferring AZT resistance (12, 18, 19). The absence of detectable peak viremia at seroconversion may be indicative of the transmission of a low-fitness virus population. This unusual course of infection, characterized by a steadily rising viral load over a 4-year period, made our study feasible, enabling an analysis of phenomena difficult to monitor during normal HIV-1 infection due to faster evolution. We hypothesized that virus escape from T-cell control through mutations in CTL epitopes paradoxically implies the successful clearance of cells harboring wild-type virus. We assumed that cells harboring viruses with a defined set of escape mutations are at least partly resistant to CTL-mediated lysis and thus produce more viruses. These notions correlate with the number of productively infected cells.
Based on the HLA type of the study individual and amino acid predictions from HIV-1 genomic DNA sequences, we identified 17 epitopes with no variations over time, yet the viral load finally rose and the number of productively infected cells increased. From the rise in viral load, despite apparent T-cell reactivity, we conclude that many epitopes are recognized by the immune system with little or no effect on virus clearance. The CTLs that respond to these epitopes may not confer effective lysis of infected cells or may do so but not at a level sufficient to effectively control viremia. Our mapping of the strength of CTL recognition of known epitopes in Gag suggests that these CTLs in some cases may be immunodominant but probably not sufficiently effective. Seven predicted epitopes that showed nonfixed variations were identified. These mutations probably were unrelated to the selective pressure exerted by epitope-specific T cells because viruses with these mutations had no advantage over viruses without them. However, we cannot rule out the possibility that these nonfixed mutations affected to some degree immune recognition of the epitope, yet the pressure was not sufficient to lead to fixed mutations. As with the 17 epitopes described above, we found epitopes relatively unaffected by T-cell pressure to be localized in many viral gene products, including Gag, Pol, Env, Vif, and Nef.
Our genome-wide screen for regions containing predicted epitopes revealed five mutational hot spots, four of which were previously optimally defined epitopes (gp120, p17, Nef, and RT) and one of which was described as a reactive region (gp41). Localized in five different gene products, these five regions were restricted by the complete set of class I alleles of our patient. The p17, gp41, and Nef epitopes were previously described (20, 44, 50), and the gp120 and RT epitopes were already associated with impaired epitope binding and/or viral escape (48, 51). The RT epitope was also previously associated with viral escape on a population level (34). The finding of viral escape mutations in multiple CTL epitopes that are temporally associated with a rise in viral load may imply that epitope-specific CTLs can clear cells harboring viruses lacking such mutations, thus keeping the viral load low. The observation that recognition of the wild-type epitope is lost as soon as the mutant peptide replaces it indicates that the set of five epitopes plays a role in the control of viremia. All five epitopes followed this pattern, with two variations. For the Gag p17 epitope, responses to both wild-type and mutant peptides were detectable but decreased and were shown to increase after the mutation became fixed. Since this is a region dense in targets for cellular immunity, this phenomenon may be due to cross-reactivity of CTL populations specific for an epitope located 2 aa upstream of the tested p17 epitope (50), but this notion remains to be investigated. Nevertheless, the loss of T-cell reactivity seen at up to 26 months psc with fixation of the mutation is indicative of viral escape. For the Nef epitope, the loss of reactive T cells against the wild-type peptide accompanied the emergence of a new T-cell response against the mutant peptide. Such emergence of a de novo T-cell population was unique in our set of escape mutants and might be specific for Nef. Indeed, new T-cell clones specific for mutant Nef epitopes have been reported (22, 47). Furthermore, the finding that not all of the gp120 and Nef populations (92%) are mutated at 47 months psc suggests that the effects of pressure on the wild-type epitope may not be within our window of analysis and/or that CTL pressure or avidity has not been constant (9).
Although the numbers of replication-competent clones recovered from time points B and C are not optimal, we still identified the initial appearance of mutations that become steadily fixed. The fact that the mutations do become fixed suggests to us that conclusions with regard to the timing of their appearance are indeed justified. The small number of replication-competent biological clones from these two time points (Fig. 1c, B and C) likely reflects the low viral load observed in the patient during this period. Escape from recognition of the Env gp41and Gag p17 epitopes correlated with the first rise in viral load and, interestingly, with the increase in the number of productively infected cells. As viral load rose above 10,000 copies/ml, escape in the Nef epitope clearly followed escape in the RT epitope. These results describe a sequential alteration of CTL escape that correlates with an increase in viremia.
The difficulty with the detailed analysis of HIV-1 pathogenesis in general is that pathogenesis is a multifactorial process where, besides immune recognition, host as well as viral factors can determine disease outcome. Solely holding the occurrence of escape from CTL recognition responsible for the rise in viral load is almost a certain violation of reality. Although the reversion from an AZT-resistant to an AZT-sensitive phenotype is known to result in increased viral fitness (12), the in vivo scenario is that wild-type residue 215 is controlled in many individuals, indicating that the virus can be controlled sufficiently by effective immune responses. If disease progression were solely due to the reversion at position 215, then no pressure at CTL domains would be evident. Antibody-specific neutralization, Th responses, and unknown changes (e.g., the detected mutations outside the described epitopes) in viral fitness likely also contribute to the increase in viral load and remain to be investigated. However, our identification of CTL epitopes escaping recognition suggests that strong pressure has been placed on these regions. Analysis of the ex vivo rate of replication of the biological clones demonstrated that the increase in viral load did not reflect increasing fitness of circulating viruses (data not shown). Finally, although the analyzed virus populations were derived from PBMCs and correlated with virus in sera, it remains to be investigated whether virus in other compartments, e.g., lymph nodes, is subject to the same phenomena. Analyzing only the described epitopes for the patient's haplotype poses a bias through underestimation of the quantity of CTL responses. It is more than likely that within an infected individual more epitopes than have been predicted are functional and that we have underestimated the number of epitopes in our patient. This possibility, however, does not affect our observation that within the patient studied, 17 epitopes remained constant and 5 epitopes belonging to different genes were shown to mutate from recognizable to nonrecognizable epitopes, correlating with a steady rise in the viral load.
T-cell responses in the patient's PBMC fraction were measured by using the IFN-γ ELIspot assay. This assay sheds no light on the responses of the different fractions constituting PBMCs, e.g., CD4+-T-helper cells (42), as opposed to CTL responses. This assay also does not give any indications as to the phenotype and/or maturational state of the CTLs. The use of the IFN-γ ELIspot assay with overlapping consensus peptide pools is known to underestimate true in vivo CD8 responses (14). However, autologous, epitope-specific peptides were synthesized for monitoring epitope-specific T-cell responses, thereby limiting erroneous measurements. Moreover, a recent study indicated that peptide pools ranging from 15 to 20 aa and with a 10- or 11-aa overlap yielded similar results in the IFN-γ ELIspot assay (14).
It is difficult to rank the contributions of the various CTL populations described to the control of viremia. Sequence analysis of biological clones from a progressing individual has revealed that CTL escape mutations do occur and that they occur in a cumulative manner. The virus population at the end of follow-up, when virus replication appears unconstrained, is composed almost entirely of viruses having all five mutant epitopes. Comparisons of the described phenomena with other situations where the immune system encounters new populations of virus, e.g., HIV-1 superinfection (1), and fails to react to provide sufficient clearance may provide more insights into immune escape mechanisms. Whether these findings can be extrapolated to HIV infection on a population level requires a thorough evaluation of key issues, e.g., HLA background.
In summary, through the unprecedented detection of the sequential accumulation of CTL escape mutations in multiple HIV-1 regions followed by the (nearly) total loss of T-cell reactivity, we can conclude that there seems to be selection pressure at these sites and that it can be associated with increased viremia. These data argue for the inclusion of multiple HIV-1 regions in any HIV vaccine aimed at CTL control of HIV replication.
Acknowledgments
We thank Lucy Philips and Debby van Baarle for critically reading the manuscript.
This work was supported by AIDSFONDS grant 6002.
REFERENCES
- 1.Altfeld, M., T. M. Allen, X. G. Yu, M. N. Johnston, D. Agrawal, B. T. Korber, D. C. Montefiori, D. H. O'Connor, B. T. Davis, P. K. Lee, E. L. Maier, J. Harlow, P. J. Goulder, C. Brander, E. S. Rosenberg, and B. D. Walker. 2002. HIV-1 superinfection despite broad CD8+ T-cell responses containing replication of the primary virus. Nature 420:434-439. [DOI] [PubMed] [Google Scholar]
- 2.Baba, T. W., Y. S. Jeong, D. Pennick, R. Bronson, M. F. Greene, and R. M. Ruprecht. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820-1825. [DOI] [PubMed] [Google Scholar]
- 3.Barouch, D. H., J. Kunstman, M. J. Kuroda, J. E. Schmitz, S. Santra, F. W. Peyerl, G. R. Krivulka, K. Beaudry, M. A. Lifton, D. A. Gorgone, D. C. Montefiori, M. G. Lewis, S. M. Wolinsky, and N. L. Letvin. 2002. Eventual AIDS vaccine failure in a rhesus monkey by viral escape from cytotoxic T lymphocytes. Nature 415:335-339. [DOI] [PubMed] [Google Scholar]
- 4.Barouch, D. H., S. Santra, J. E. Schmitz, M. J. Kuroda, T. M. Fu, W. Wagner, M. Bilska, A. Craiu, X. X. Zheng, G. R. Krivulka, K. Beaudry, M. A. Lifton, C. E. Nickerson, W. L. Trigona, K. Punt, D. C. Freed, L. Guan, S. Dubey, D. Casimiro, A. Simon, M. E. Davies, M. Chastain, T. B. Strom, R. S. Gelman, D. C. Montefiori, M. G. Lewis, E. A. Emini, J. W. Shiver, and N. L. Letvin. 2000. Control of viremia and prevention of clinical AIDS in rhesus monkeys by cytokine-augmented DNA vaccination. Science 290:486-492. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout, B., K. Verhoef, J. L. van Wamel, and N. K. Back. 1999. Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains. J. Virol. 73:1138-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boom, R., C. J. Sol, M. M. Salimans, C. L. Jansen, P. M. Wertheim-van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borrow, P., H. Lewicki, B. H. Hahn, G. M. Shaw, and M. B. Oldstone. 1994. Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68:6103-6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borrow, P., H. Lewicki, X. Wei, M. S. Horwitz, N. Peffer, H. Meyers, J. A. Nelson, J. E. Gairin, B. H. Hahn, M. B. Oldstone, and G. M. Shaw. 1997. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3:205-211. [DOI] [PubMed] [Google Scholar]
- 9.Brander, C., K. E. Hartman, A. K. Trocha, N. G. Jones, R. P. Johnson, B. Korber, P. Wentworth, S. P. Buchbinder, S. Wolinsky, B. D. Walker, and S. A. Kalams. 1998. Lack of strong immune selection pressure by the immunodominant, HLA-A*0201-restricted cytotoxic T lymphocyte response in chronic human immunodeficiency virus-1 infection. J. Clin. Investig. 101:2559-2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornelissen, M., R. van den Burg, F. Zorgdrager, V. Lukashov, and J. Goudsmit. 1997. pol gene diversity of five human immunodeficiency virus type 1 subtypes: evidence for naturally occurring mutations that contribute to drug resistance, limited recombination patterns, and common ancestry for subtypes B and D. J. Virol. 71:6348-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daniel, M. D., F. Kirchhoff, S. C. Czajak, P. K. Sehgal, and R. C. Desrosiers. 1992. Protective effects of a live attenuated SIV vaccine with a deletion in the nef gene. Science 258:1938-1941. [DOI] [PubMed] [Google Scholar]
- 12.de Ronde, A., M. van Dooren, L. van Der Hoek, D. Bouwhuis, E. de Rooij, B. Van Gemen, R. de Boer, and J. Goudsmit. 2001. Establishment of new transmissible and drug-sensitive human immunodeficiency virus type 1 wild types due to transmission of nucleoside analogue-resistant virus. J. Virol. 75:595-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Wolf, F., I. Spijkerman, P. T. Schellekens, M. Langendam, C. Kuiken, M. Bakker, M. Roos, R. Coutinho, F. Miedema, and J. Goudsmit. 1997. AIDS prognosis based on HIV-1 RNA, CD4+ T-cell count and function: markers with reciprocal predictive value over time after seroconversion. AIDS 11:1799-1806. [DOI] [PubMed] [Google Scholar]
- 14.Draenert, R., M. Altfeld, C. Brander, N. Basgoz, C. Corcoran, A. G. Wurcel, D. R. Stone, S. A. Kalams, A. Trocha, M. M. Addo, P. J. Goulder, and B. D. Walker. 2003. Comparison of overlapping peptide sets for detection of antiviral CD8 and CD4 T cell responses. J. Immunol. Methods 275:19-29. [DOI] [PubMed] [Google Scholar]
- 15.Dyer, W. B., G. S. Ogg, M. A. Demoitie, X. Jin, A. F. Geczy, S. L. Rowland-Jones, A. J. McMichael, D. F. Nixon, and J. S. Sullivan. 1999. Strong human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte activity in Sydney Blood Bank Cohort patients infected with nef-defective HIV type 1. J. Virol. 73:436-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gandhi, R. T., and B. D. Walker. 2002. Immunologic control of HIV-1. Annu. Rev. Med. 53:149-172. [DOI] [PubMed] [Google Scholar]
- 17.Goudsmit, J., J. A. Bogaards, S. Jurriaans, H. Schuitemaker, J. M. A. Lange, R. A. Coutinho, and G. J. Weverling. 2002. Naturally HIV-1 seroconverters with lowest viral load have best prognosis, but in time lose control of viraemia. AIDS 16:791-796. [DOI] [PubMed] [Google Scholar]
- 18.Goudsmit, J., A. de Ronde, E. de Rooij, and R. de Boer. 1997. Broad spectrum of in vivo fitness of human immunodeficiency virus type 1 subpopulations differing at reverse transcriptase codons 41 and 215. J. Virol. 71:4479-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goudsmit, J., A. de Ronde, D. D. Ho, and A. S. Perelson. 1996. Human immunodeficiency virus fitness in vivo: calculations based on a single zidovudine resistance mutation at codon 215 of reverse transcriptase. J. Virol. 70:5662-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goulder, P. J., C. Brander, K. Annamalai, N. Mngqundaniso, U. Govender, Y. Tang, S. He, K. E. Hartman, C. A. O'Callaghan, G. S. Ogg, M. A. Altfeld, E. S. Rosenberg, H. Cao, S. A. Kalams, M. Hammond, M. Bunce, S. I. Pelton, S. A. Burchett, K. McIntosh, H. M. Coovadia, and B. D. Walker. 2000. Differential narrow focusing of immunodominant human immunodeficiency virus gag-specific cytotoxic T-lymphocyte responses in infected African and caucasoid adults and children. J. Virol. 74:5679-5690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goulder, P. J., C. Brander, Y. Tang, C. Tremblay, R. A. Colbert, M. M. Addo, E. S. Rosenberg, T. Nguyen, R. Allen, A. Trocha, M. Altfeld, S. He, M. Bunce, R. Funkhouser, S. I. Pelton, S. K. Burchett, K. McIntosh, B. T. Korber, and B. D. Walker. 2001. Evolution and transmission of stable CTL escape mutations in HIV infection. Nature 412:334-338. [DOI] [PubMed] [Google Scholar]
- 22.Haas, G., U. Plikat, P. Debre, M. Lucchiari, C. Katlama, Y. Dudoit, O. Bonduelle, M. Bauer, H. G. Ihlenfeldt, G. Jung, B. Maier, A. Meyerhans, and B. Autran. 1996. Dynamics of viral variants in HIV-1 Nef and specific cytotoxic T lymphocytes in vivo. J. Immunol. 157:4212-4221. [PubMed] [Google Scholar]
- 23.Kaul, R., F. A. Plummer, J. Kimani, T. Dong, P. Kiama, T. Rostron, E. Njagi, K. S. MacDonald, J. J. Bwayo, A. J. McMichael, and S. L. Rowland-Jones. 2000. HIV-1-specific mucosal CD8+ lymphocyte responses in the cervix of HIV-1-resistant prostitutes in Nairobi. J. Immunol. 164:1602-1611. [DOI] [PubMed] [Google Scholar]
- 24.Kelleher, A. D., C. Long, E. C. Holmes, R. L. Allen, J. Wilson, C. Conlon, C. Workman, S. Shaunak, K. Olson, P. Goulder, C. Brander, G. Ogg, J. S. Sullivan, W. Dyer, I. Jones, A. J. McMichael, S. Rowland-Jones, and R. E. Phillips. 2001. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J. Exp. Med. 193:375-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein, M. R., C. A. van Baalen, A. M. Holwerda, S. R. Kerkhof Garde, R. J. Bende, I. P. Keet, J. K. Eeftinck-Schattenkerk, A. D. Osterhaus, H. Schuitemaker, and F. Miedema. 1995. Kinetics of Gag-specific cytotoxic T lymphocyte responses during the clinical course of HIV-1 infection: a longitudinal analysis of rapid progressors and long-term asymptomatics. J. Exp. Med. 181:1365-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koot, M., A. B. van 't Wout, N. A. Kootstra, R. E. de Goede, M. Tersmette, and H. Schuitemaker. 1996. Relation between changes in cellular load, evolution of viral phenotype, and the clonal composition of virus populations in the course of human immunodeficiency virus type 1 infection. J. Infect. Dis. 173:349-354. [DOI] [PubMed] [Google Scholar]
- 27.Korber, B. T., C. Brander, R. Haynes, R. A. Koup, C. L. Kuiken, J. P. Moore, B. D. Walker, and D. Watkins (ed.). 2000. HIV molecular immunology 2000, part II. Los Alamos National Laboratory, Los Alamos, N.Mex.
- 28.Koup, R. A., J. T. Safrit, Y. Cao, C. A. Andrews, G. McLeod, W. Borkowsky, C. Farthing, and D. D. Ho. 1994. Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68:4650-4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 30.Learmont, J. C., A. F. Geczy, J. Mills, L. J. Ashton, C. H. Raynes-Greenow, R. J. Garsia, W. B. Dyer, L. McIntyre, R. B. Oelrichs, D. I. Rhodes, N. J. Deacon, and J. S. Sullivan. 1999. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. A report from the Sydney Blood Bank Cohort. N. Engl. J. Med. 340:1715-1722. [DOI] [PubMed] [Google Scholar]
- 31.Letvin, N. L. 2002. Strategies for an HIV vaccine. J. Clin. Investig. 110:15-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lifson, A. R., N. A. Hessol, and G. W. Rutherford. 1992. Progression and clinical outcome of infection due to human immunodeficiency virus. Clin. Infect. Dis. 14:966-972. [DOI] [PubMed] [Google Scholar]
- 33.Mellors, J. W., C. R. Rinaldo, Jr., P. Gupta, R. M. White, J. A. Todd, and L. A. Kingsley. 1996. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science 272:1167-1170. [DOI] [PubMed] [Google Scholar]
- 34.Moore, C. B., M. John, I. R. James, F. T. Christiansen, C. S. Witt, and S. A. Mallal. 2002. Evidence of HIV-1 adaptation to HLA-restricted immune responses at a population level. Science 296:1439-1443. [DOI] [PubMed] [Google Scholar]
- 35.Musey, L., J. Hughes, T. Schacker, T. Shea, L. Corey, and M. J. McElrath. 1997. Cytotoxic-T-cell responses, viral load, and disease progression in early human immunodeficiency virus type 1 infection. N. Engl. J. Med. 337:1267-1274. [DOI] [PubMed] [Google Scholar]
- 36.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 37.O'Connor, D., T. Friedrich, A. Hughes, T. M. Allen, and D. Watkins. 2001. Understanding cytotoxic T-lymphocyte escape during simian immunodeficiency virus infection. Immunol. Rev. 183:115-126. [DOI] [PubMed] [Google Scholar]
- 38.Ogg, G. S., X. Jin, S. Bonhoeffer, P. R. Dunbar, M. A. Nowak, S. Monard, J. P. Segal, Y. Cao, S. L. Rowland-Jones, V. Cerundolo, A. Hurley, M. Markowitz, D. D. Ho, D. F. Nixon, and A. J. McMichael. 1998. Quantitation of HIV-1-specific cytotoxic T lymphocytes and plasma load of viral RNA. Science 279:2103-2106. [DOI] [PubMed] [Google Scholar]
- 39.Ogg, G. S., S. Kostense, M. R. Klein, S. Jurriaans, D. Hamann, A. J. McMichael, and F. Miedema. 1999. Longitudinal phenotypic analysis of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes: correlation with disease progression. J. Virol. 73:9153-9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phillips, R. E., S. Rowland-Jones, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1991. Hum. immunodeficiency virus genetic variation that can escape cytotoxic T cell recognition. Nature 354:453-459. [DOI] [PubMed] [Google Scholar]
- 41.Price, D. A., P. J. Goulder, P. Klenerman, A. K. Sewell, P. J. Easterbrook, M. Troop, C. R. Bangham, and R. E. Phillips. 1997. Positive selection of HIV-1 cytotoxic T lymphocyte escape variants during primary infection. Proc. Natl. Acad. Sci. USA 94:1890-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosenberg, E. S., J. M. Billingsley, A. M. Caliendo, S. L. Boswell, P. E. Sax, S. A. Kalams, and B. D. Walker. 1997. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 278:1447-1450. [DOI] [PubMed] [Google Scholar]
- 43.Rowland-Jones, S. L., R. E. Phillips, D. F. Nixon, F. M. Gotch, J. P. Edwards, A. O. Ogunlesi, J. G. Elvin, J. A. Rothbard, C. R. Bangham, C. R. Rizza, et al. 1992. Human immunodeficiency virus variants that escape cytotoxic T-cell recognition. AIDS Res. Hum. Retrovir. 8:1353-1354. [DOI] [PubMed] [Google Scholar]
- 44.Safrit, J. T., C. A. Andrews, T. Zhu, D. D. Ho, and R. A. Koup. 1994. Characterization of human immunodeficiency virus type 1-specific cytotoxic T lymphocyte clones isolated during acute seroconversion: recognition of autologous virus sequences within a conserved immunodominant epitope. J. Exp. Med. 179:463-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schmitz, J. E., M. J. Kuroda, S. Santra, V. G. Sasseville, M. A. Simon, M. A. Lifton, P. Racz, K. Tenner-Racz, M. Dalesandro, B. J. Scallon, J. Ghrayeb, M. A. Forman, D. C. Montefiori, E. P. Rieber, N. L. Letvin, and K. A. Reimann. 1999. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science 283:857-860. [DOI] [PubMed] [Google Scholar]
- 46.Shiver, J. W., T. M. Fu, L. Chen, D. R. Casimiro, M. E. Davies, R. K. Evans, Z. Q. Zhang, A. J. Simon, W. L. Trigona, S. A. Dubey, L. Huang, V. A. Harris, R. S. Long, X. Liang, L. Handt, W. A. Schleif, L. Zhu, D. C. Freed, N. V. Persaud, L. Guan, K. S. Punt, A. Tang, M. Chen, K. A. Wilson, K. B. Collins, G. J. Heidecker, V. R. Fernandez, H. C. Perry, J. G. Joyce, K. M. Grimm, J. C. Cook, P. M. Keller, D. S. Kresock, H. Mach, R. D. Troutman, L. A. Isopi, D. M. Williams, Z. Xu, K. E. Bohannon, D. B. Volkin, D. C. Montefiori, A. Miura, G. R. Krivulka, M. A. Lifton, M. J. Kuroda, J. E. Schmitz, N. L. Letvin, M. J. Caulfield, A. J. Bett, R. Youil, D. C. Kaslow, and E. A. Emini. 2002. Replication-incompetent adenoviral vaccine vector elicits effective anti-immunodeficiency-virus immunity. Nature 415:331-335. [DOI] [PubMed] [Google Scholar]
- 47.Singh, M. K., G. Janvier, V. Calvez, P. Coulaud, and Y. Riviere. 2001. A long-term follow-up of an HIV type 1-infected patient reveals a coincidence of Nef-directed cytotoxic T lymphocyte effectors and high incidence of epitope-deleted variants. AIDS Res. Hum. Retrovir. 17:1265-1271. [DOI] [PubMed] [Google Scholar]
- 48.Sipsas, N. V., S. A. Kalams, A. Trocha, S. He, W. A. Blattner, B. D. Walker, and R. P. Johnson. 1997. Identification of type-specific cytotoxic T lymphocyte responses to homologous viral proteins in laboratory workers accidentally infected with HIV-1. J. Clin. Investig. 99:752-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van 't Wout, A. B., H. Blaak, L. J. Ran, M. Brouwer, C. Kuiken, and H. Schuitemaker. 1998. Evolution of syncytium-inducing and non-syncytium-inducing biological virus clones in relation to replication kinetics during the course of human immunodeficiency virus type 1 infection. J. Virol. 72:5099-5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilson, C. C., R. C. Brown, B. T. Korber, B. M. Wilkes, D. J. Ruhl, D. Sakamoto, K. Kunstman, K. Luzuriaga, I. C. Hanson, S. M. Widmayer, A. Wiznia, S. Clapp, A. J. Ammann, R. A. Koup, S. M. Wolinsky, and B. D. Walker. 1999. Frequent detection of escape from cytotoxic T-lymphocyte recognition in perinatal human immunodeficiency virus (HIV) type 1 transmission: the Ariel project for the prevention of transmission of HIV from mother to infant. J. Virol. 73:3975-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, C. C., S. A. Kalams, B. M. Wilkes, D. J. Ruhl, F. Gao, B. H. Hahn, I. C. Hanson, K. Luzuriaga, S. Wolinsky, R. Koup, S. P. Buchbinder, R. P. Johnson, and B. D. Walker. 1997. Overlapping epitopes in human immunodeficiency virus type 1 gp120 presented by HLA A, B, and C molecules: effects of viral variation on cytotoxic T-lymphocyte recognition. J. Virol. 71:1256-1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong, J. K., M. Hezareh, H. F. Gunthard, D. V. Havlir, C. C. Ignacio, C. A. Spina, and D. D. Richman. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291-1295. [DOI] [PubMed] [Google Scholar]
- 53.Yang, O. O. 2003. Will we be able to 'spot' an effective HIV-1 vaccine? Trends Immunol. 24:67-72. [DOI] [PubMed] [Google Scholar]