Abstract
The human cytomegalovirus (HCMV) UL36-38 immediate early (IE) locus encodes proteins required for its growth. The UL37 promoter drives production of an unspliced and several alternatively spliced RNAs. The UL37 exon 1 (UL37x1) unspliced RNA is abundant from IE to late times of HCMV infection, whereas the UL37 spliced RNAs are markedly less abundant. Production of the UL37x1 unspliced RNA requires polyadenylation (PA) at nucleotide 50998, which lies within intron 1, upstream of the UL37 exon 2 (UL37x2) acceptor. The physical proximity of its cis elements suggests steric hindrance between PA and splicing machineries for UL37 pre-mRNA. To test this possibility, we generated site-specific mutants in Target 1 PA and RNA splicing cis elements and compared the PA and splicing efficiencies of mutant RNAs with those of wild-type RNA. The mutually exclusive processing events of UL37x1 PA and UL37x1-UL37x2 splicing have been accurately recapitulated in transfected permissive human fibroblasts (HFFs) expressing a Target 1 minigene RNA, which contains the required splicing and PA cis elements. Two mutants in the invariant PA signal dramatically decreased UL37x1 PA as expected and, concomitantly, increased the efficiency of UL37x1-UL37x2 RNA splicing. Consistent with these results, changes to consensus UL37x1 donor and UL37x2 acceptor sites increased the efficiency of UL37x1-UL37x2 RNA splicing but decreased the efficiency of UL37x1 PA. Moreover, HCMV infection of HFFs increased the abundance of the PA cleavage stimulatory factor CstF-64, the potent splicing suppressor PTB, and the hypophosphorylated form of the splicing factor SF2 at 4 h postinfection. Induction of these factors further favors production of the UL37x1 unspliced RNA over that of the spliced RNAs. Taken together, these results suggest that there is a convergence in UL37 RNA regulation by cis elements and cellular proteins which favors production of the UL37x1 unspliced RNA during HCMV infection at the posttranscriptional level.
The human cytomegalovirus (HCMV) UL36-38 immediate early (IE) locus encodes proteins that play important roles for antiapoptosis, viral DNA replication, and growth (12, 14, 15, 21, 23, 24, 38, 40). All known UL37 proteins have antiapoptotic activities and dually traffic to the endoplasmic reticulum and mitochondria (2, 21, 23, 24). The UL37 exon 1 (UL37x1) open reading frame (ORF) encodes the amino termini of at least three UL37 IE isoforms, including two integral membrane N-glycoproteins, gpUL37 and gpUL37M (2, 11, 12, 21, 28). The UL37x1 ORF is required for HCMV growth in humans as more than 90% of its residues are invariant in numerous primary strains (23). Mutants made by Y. Dong, R. Sharon-Friling, and T. Shenk indicate that UL37x1 is essential for HCMV replication in fibroblasts (T. Shenk, personal communication). Although the UL37 exon 3 (UL37x3) ORF is nonessential for HCMV growth in cultured human diploid fibroblasts (HFFs) (7, 21), its carboxyl-terminal transmembrane and cytosolic tail are predicted to be important for HCMV pathogenesis in vivo, as they have been shown to be for mouse CMV M37 (24, 29).
The UL36-38 locus encodes at least five transcripts from three different transcriptional promoters (Fig. 1). An unspliced (UL37x1) and several spliced (UL37 and UL37M) RNAs, initiated at the UL37 IE promoter, are generated by alternative polyadenylation (PA) or splicing. As they are 5′ coterminal, UL37 RNAs appear to be differentially regulated after the initiation of transcription during HCMV infection (21, 28, 47, 48). The UL37x1 unspliced RNA is abundant at IE times and remains abundant until late times of infection, whereas the abundance of the UL37 spliced RNA is low.
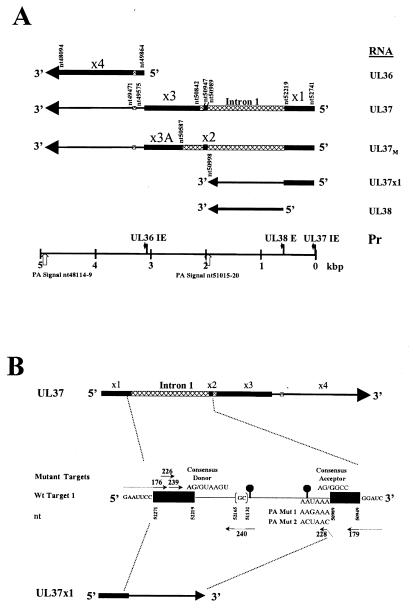
FIG. 1.
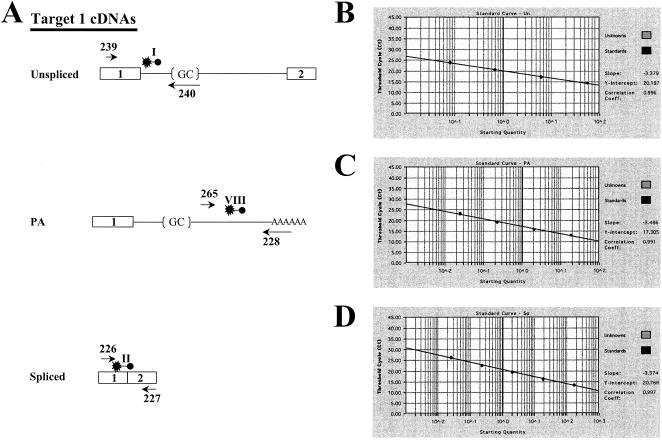
(A) HCMV UL36-38 transcripts. The RNA map indicates the direction of transcription from the HCMV genome, exons (solid boxes), introns (cross-hatched boxes), 3′ untranslated region (thin lines), and poly(A) tails (arrowheads). The bent black and straight white arrows on the scale bar represent UL37 IE, UL38 E, and UL36 IE promoters and UL37x1-UL38 and UL37-UL36 PA signals, respectively. A splice variant, UL37M, differing in a 3′ ss (exon 3A [×3A]) has been identified in transfected cells and HCMV-infected HFFs (21, 41). (B) Target 1 consensus donor, consensus acceptor, and PA mutants. The sequences in Target 1, UL37, and UL37x1 RNAs are represented in a 5′-to-3′ orientation. Target 1 spans part of UL37x1 (nt 52219 to 52271), intron 1 (nt 52218 to 52165 and nt 51132 to 50990) and UL37x2 (nt 50989 to nt 50949). The mutations in the consensus donor, consensus acceptor, PA Mut 1, and PA Mut 2 constructions as well as the physical location and polarity of the primers used for RT-PCR are shown. The two potential BPs in intron 1 (nt −30 and nt −115) are represented as lollipop structures.
UL37, UL37M, and UL37x1 RNAs share exon 1 but differ downstream of the first 5′ splice site (ss) (Fig. 1). The unspliced UL37x1 RNA, which encodes both the UL37x1 and UL38 ORFs, is polyadenylated within intron 1 at nucleotide (nt) 50998, downstream of the UL38 gene and just upstream of UL37 exon 2 (UL37x2). The UL37x1 donor is spliced to the UL37x2 acceptor, thereby removing the UL38 early gene promoter, the UL38 ORF, and the UL37x1 PA signal, all located within UL37 intron 1. The UL37x2 donor is alternatively spliced to an upstream (UL37) or a downstream (UL37M) acceptor in UL37x3 (21, 28, 41, 47). Alternative splicing from two new UL37 donors (di and dii) to the UL37x3A acceptor has recently been detected in HCMV-infected HFFs (41). Finally, the UL37x3 donor is spliced to the exon 4 acceptor in UL37, UL37M, and UL36 spliced IE RNAs.
The mechanisms regulating the differential production of the HCMV UL37 unspliced and alternatively spliced IE RNAs are not presently known. PA at the UL37x1 site is dominant throughout HCMV infection as the site is used for processing of UL37x1 IE and of UL38 early RNAs (28, 47). The initiating step for RNA PA is the initial recognition of the PA signal by the cleavage and PA specificity factor (CPSF) after its release from the transcription preinitiation complex (17, 34, 50). The complex is stabilized by binding of the cleavage stimulatory factor (CstF) trimer complex via CstF-64 to downstream U or G/U elements and of cleavage factor CFI (31, 43, 55) (see Fig. 7A). An additional component of the initiation complex is RNA polymerase II, which delivers the cleavage factors CPSF and CstF to the elongating pre-mRNA (32). Spacing of the CPSF, CstF, and CFI complexes on the pre-mRNA define its cleavage site (31). Addition of CFII and poly(A) polymerase (PAP) to the PA complex allows cleavage to occur, at which point CFI, CFII, and the CstF complex are released. A short tail of 10 to 15 A residues is added to the cleaved site and is followed by binding of poly(A) binding protein II (PABII) and then by addition of the full-length poly(A) tail by PAP (43, 50, 55). The signals used for UL37x1 and UL38 early RNA PA at nt 50998 are consensus and include the invariant sequence AAUAAA at nt 50115 to 50120, the cleavage site 17 nt downstream of the PA signal at nt 50998, and a downstream U-rich element 17 to 24 nt downstream of the cleavage or PA site within UL37x2 (11, 28). Moreover, this 3′ processing site is used throughout HCMV infection (28, 47, 48).
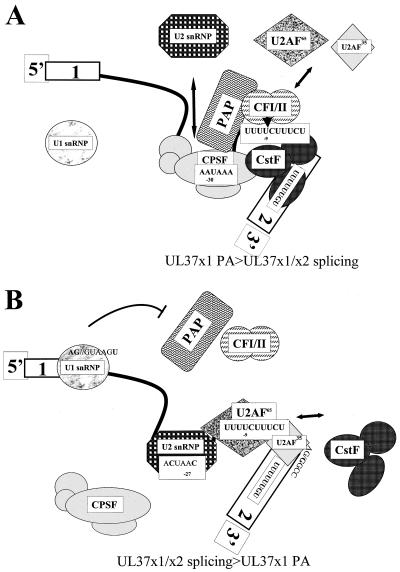
FIG. 7.
Model for favored production of UL37 RNAs. (A) Preponderance of UL37x1 unspliced RNA in HCMV-infected cells. The figure represents processing of UL37 RNAs in HCMV-infected cells which greatly favors production of UL37x1 unspliced RNA, which is cleaved downstream of the PA signal (−26 to −31) at nt −9 with respect to the UL37x2 acceptor. Shown are binding of the CPSF complex at the PA signal via the action of CPSF-160 and binding of the CstF complex at the U-rich region in UL37x2. CFI-CFII binding to the UL37x1 cleavage site (nt −9) is also shown. Interactions between CPSF-160, CstF-77, PAP, and CFI are represented. The binding of splicing factor U1 snRNP is low because of the low-affinity 5′ ss. The entry of U2AF65 into the PPT upstream of UL37x2 is blocked by loading of the CFI-CFII complex, as is entry of U2 snRNP into the potential BP (nt −30) by CPSF binding to the UL37x1 PA signal. (B) Favoring of UL37x1-UL37x2 splicing over UL37x1 PA. The shift in balance observed with mutants in the consensus donor, PA mutants, and consensus acceptor is represented. In this model, the consensus donor binds U1 snRNP more efficiently than does the wt UL37x1 donor and inhibits PAP at the downstream UL37x1 PA site. The UL37x1 PA signal mutants block binding of the CPSF complex and thereby permit entry of U2 snRNP to the new consensus BP (at nt −27). Finally, the consensus acceptor is efficiently recognized by the native U2AF complex, permitting functional displacement of CFI-CFII and the CstF complex by splicing factors.
Splicing of pre-mRNAs requires spliceosome assembly, a complex structure containing 145 cellular proteins and snRNAs, and the recognition of three major splicing signals: the 5′ ss, the branch point (BP), and the 3′ ss (4, 52, 53, 56). Ser-Arg (SR) proteins, including SF2, are essential splicing factors and play roles in the regulation of alternative splicing (19). The UL37x1 donor (5′-CCA//GUAAGC-3′) and UL37x2 acceptor (5′-UUUUCUUUCUAG//UGC-3′) signals for splicing are weak as they are nonconsensus in 3 and 2 nt (underlined), respectively (10, 28, 47). In addition, the polypyrimidine tract (PPT) immediately upstream of UL37x2, predictably required for binding of U2AF65, subsequent recruitment of U2 snRNP to the BP, and binding of U2AF35 to the 3′ ss (53), contains the cleavage site at nt −9. This site is used for 3′ processing of UL37x1 IE and UL38 early RNAs and contains a potential core binding site (UCUU) for the polypyrimidine tract binding protein (PTB), a potent splicing suppressor (see Fig. 7B) (30, 37). Finally, the proximal consensus BP upstream, predictably used for splicing of the UL37x2 acceptor, is part of the UL37x1 PA signal.
This physical proximity of cis elements required for UL37x1 PA at nt 50998 and UL37x1-UL37x2 splicing suggests that cellular core PA (CPSF and CFI) and splicing factors (U2AF65 and U2 snRNP) may compete for binding of their cognate signals and determine the production of UL37x1 unspliced or UL37 spliced RNAs. We therefore undertook genetic analysis of UL37 pre-mRNA cis elements required for its PA and splicing to determine the effects of PA and splicing-complex competition on the balance of UL37 RNA processing. Moreover, as alteration of the levels and posttranslational modifications of PA and splicing factors can determine PA site usage and splicing efficiency (25, 27, 42, 45), we asked whether HCMV infection alters the levels of CstF-64, hypophosphorylated SF2, and PTB in the infected cell.
MATERIALS AND METHODS
Cell transfection.
HFFs were transfected using Lipofectamine or Lipofectamine 2000 (Invitrogen). Briefly, 7 × 106 to 9 × 106 HFFs, P-13 to P-20, were lipofected with 10 to 20 μg of Target 1 wild-type (wt) or mutant vectors and 2 to 5 μg of a mammalian expression vector for β-galactosidase (β-gal) (pCH110) as an internal transfection control as previously described (41). The lipofected cells were incubated at 37°C for 32 h and then treated with 100 μM anisomycin for 16 h prior to harvesting.
Target plasmids.
Target 1 wt minigene (p938 and p1021), under the control of the HCMV major IE promoter, includes the UL37x1 5′ ss (nt 52271 to 52219), UL37x2 3′ ss (nt 50989 to 50949), and immediately adjacent intronic sequences (nt 52218 to 52165 and nt 51132 to 50990) (41). Deletion of the intronic sequences removes the UL38 ORF (nt 52123 to 51128) but retains the UL37x1 PA signal (nt −31 to −26) and PA site (nt −9) of the UL37x2 acceptor. For in vitro transcription, the Target 1 unspliced minigene and UL37x1-UL37x2 spliced cDNA (p251 [46]) were PCR amplified and cloned into pBS II SK+ (Stratagene) generating p939 (unspliced), 1028 (unspliced), and p917 (spliced). UL37x1 cDNA with a poly(A) tail at nt 50998 was directly generated by reverse transcription (RT)-PCR of HCMV-infected-cell RNA by using primers 176 and 234 and was cloned into pBSII SK+ (p1009). pMC1871 DNA (Pharmacia) was used for in vitro transcription of β-gal sequences.
Site-specific mutagenesis.
The site-specific mutants in the UL37x1 5′ ss (p1034), 3′ ss (p1037), or UL37x1 PA signal (p1030, p1032, p1041, and p1043) in Target 1 expression vectors are listed in Table 1 and were generated using a QuikChange site-directed mutagenesis kit (Stratagene). The UL37x1 5′ ss (consensus donor) and UL37x2 3′ ss (consensus acceptor) mutants are more consensus than the wt sites and were therefore predicted to increase the efficiency of UL37x1-UL37x2 splicing. Two UL37x1 PA signal knockout mutants were generated: PA signal mutant 1 (PA Mut 1) (p1030 and p1041) carries a mutation (AAGAAA) known to abolish binding of CPSF to the invariant AAUAAA (6). PA Mut 2 (p1032 and p1043), carrying the mutant site ACUAAC, is also predicted to abolish CPSF binding as it alters the UL37x1 PA signal but, in addition, is more homologous to the consensus BP sequence (5′-YNYURAC-3′) (10). The identity of all site-specific mutants was confirmed by DNA sequencing of the complete minigenes by using a Beckman CEQ DTCS Quick Start kit and automated PCR sequencing as previously described (23).
TABLE 1.
Plasmids and templates used in this study
| Target 1 plasmid or template | Insert | Mutant sequencea | Reference | ||
|---|---|---|---|---|---|
| Plasmids | |||||
| p938, p1021 | Wt | 41 | |||
| p1030, p1041 | PA knockout Mut 1 | AAGAAA | 41 | ||
| p1032, p1043 | PA knockout Mut 2 | ACTAAC | This paper | ||
| p1034 | Consensus donor | AGGTAAGT | This paper | ||
| p1037 | Consensus acceptor | AGGGCC | This paper | ||
| In vitro transcription templates
|
|||||
| p939, p1028 | Unspliced wt | This paper | |||
| p917 | Spliced x1/x2 | This paper | |||
| p1009 | UL37x1 PA | This paper | |||
Mutated nucleotides are underlined.
RNA isolation.
Total cellular RNA was isolated by guanidinium isothiocyanate extraction and pelleting through 5.7 M cesium chloride cushions (13).
RT-PCR and quantification of alternatively processed Target 1 RNAs.
Total RNA (5 and 10 ng), previously treated with DNA-free (Ambion), was reverse transcribed as described previously (41, 49) using oligo(dT) primers (Invitrogen) and SuperScript II reverse transcriptase (Invitrogen). Control reactions without reverse transcriptase were assayed in parallel. PCR fragments were not observed in the absence of reverse transcriptase addition, confirming the use of RNA as templates (Y. Su and A. M. Colberg-Poley, unpublished results). cDNAs were serially diluted in threefold steps and 1 or 2 μl of each dilution was amplified by PCR using HotStar Taq DNA polymerase (Qiagen) and primers to detect unspliced and polyadenylated (primers 239-240), UL37x1-UL37x2 spliced (primers 176-179), and UL37x1 polyadenylated (primers 226-228) RNA species (Table 2). Control glyceraldehyde 3-phosphate dehydrogenase (GAPDH) RNA (Applied Biosystems) or β-gal RNA from cotransfected control pCH110 was amplified by RT-PCR using GAPDH primers (ABI) or primers 241-243, respectively. The products were detected by ethidium bromide staining following resolution by gel electrophoresis in nondenaturing 7% polyacrylamide gels. DNA (50- and 123-bp ladder) markers (Invitrogen) served as molecular size standards. Gel photographs were digitized using ScanWizard Pro version 1.21 and importation into Adobe Photoshop 5.0 LE and Microsoft PowerPoint 2000. The levels of each spliced, polyadenylated, and unspliced Target 1 RNA are reported as the reciprocal of the highest dilution of the cDNAs in which the product was last visually detected in the gel. The percentage of each (unspliced, polyadenylated, or spliced) Target 1 RNA species was determined by dividing the level of each Target 1 species by the sum total of the levels of all Target 1 RNA species and then multiplying by 100. The levels of unspliced RNA were deduced by subtracting the level of UL37x1 polyadenylated RNA from Target 1 unspliced or UL37x1 polyadenylated RNA.
TABLE 2.
Primers used in this study
| Primer | Assay(s) | HCMV sequence (nt)a | Reference |
|---|---|---|---|
| 176 | RT-PCR | 52251-52271 | 41 |
| 179 | RT-PCR | 50949-50968 | 41 |
| 226 | RT-PCR, TaqMan | 52240-52257 | This paper |
| 227 | RT-PCR, TaqMan | 50963-50979 | This paper |
| 228 | RT-PCR, TaqMan | 50998-51108 | This paper |
| 234 | Cloning | 50998-51010-T10 | This paper |
| 239 | RT-PCR, TaqMan | 52234-52249 | 41 |
| 240 | RT-PCR, TaqMan | 52165-52171/GC/51119-51132 | 41 |
| 241 | RT-PCR, TaqMan | 282-297b | This paper |
| 243 | RT-PCR, TaqMan | 392-413b | This paper |
| 265 | TaqMan | 51095-51116 | This paper |
Real-time PCR.
The phenotypes of PA Mut 1 and PA Mut 2 were verified using RNAs from independently transfected HFFs and real-time PCR. Standards for the detection of unspliced, spliced, and polyadenylated UL37 RNAs were generated by in vitro transcription of p939 (or 1028), p917, and p1009 cDNAs, respectively, using a MAXIscript kit (Ambion). UL37 RNAs, generated by in vitro transcription, were supplemented with 50 ng of uninfected HFF cell RNA and reverse transcribed as described above. Serially diluted UL37 cDNAs, corresponding to 166.67 to 0.025 pg of RNA standard, and transfected cell cDNA (25 to 100 ng) were analyzed in triplicate by real-time PCR using TaqMan Universal PCR Master Mix (Applied Biosystems) with final probe and primer concentrations of 250 and 900 nM, respectively. Matched PCR primers and MGB probes (Table 3) were designed to detect unspliced or polyadenylated (primers 239-240; FAM-probe I), UL37x1-UL37x2 spliced junction (primers 226-227; VIC-probe II), and polyadenylated (primers 265-228; FAM-probe VIII) UL37 RNAs by use of ABI Primer Express software. The reactions were amplified by incubation at 50°C (2 min) and 95°C (10 min) and then for 40 cycles at 95°C (15 s) and 60°C (1 min). Amplification was monitored by the ABI Prism 7700 sequence detection system (version 1.9).
TABLE 3.
Probes used in real-time PCR
| Probe | Target | Sequence | nt numbers |
|---|---|---|---|
| I | Unspliced | 5′-FAM-AAACCACAGACTCCGGGAT-MGBNFQ-3′ | 52180-52198 |
| II | Spliced | 5′-VIC-AGTCTCACCATGCCGCGG-MGBNFQ-3′ | 52219-52228 and 50982-50989 |
| VII | β-gal | 5′-TET-TCTACACCAACGTAACCTA-MGBNFQ-3′ | 299-317a |
| VIII | UL37x1 PA | 5′-FAM-CTATACCTCAGTTATCCC-MGBNFQ-3′ | 51045-51062 |
| GAPDH | GAPDH | 5′-JOE-CAAGCTTCCCGTTCTCAGCC-TAMRA-3′ | 3359-3378b |
Nuclear extracts.
Nuclear extracts were prepared from HeLa cell pellets (2 × 109 cells) (provided by the National Cell Culture Center) and from HFFs (total, ∼4 × 108 cells). Nearly confluent HFFs were infected with HCMV (strain AD169) at a multiplicity of 3 PFU/cell. The uninfected cells or infected cells at various times of infection were harvested by trypsinization, repeated washing in cold phosphate-buffered saline (PBS), and pelleting by centrifugation at 200 × g. The cell pellets were quickly frozen and stored in liquid N2. Nuclear extracts were isolated as described previously (18), except that the cells were homogenized for 20 strokes instead of 10. All solutions, tubes, pipettes, and equipment were prechilled. All work, except the centrifugation steps, was performed in a cold room. The nuclear extracts were aliquoted, frozen in liquid N2, and stored at −80°C until their use. Protein concentrations were determined by using a BCA kit (Pierce).
Western blot analysis.
Briefly, polypeptides were electrotransferred to a Hybond ECL membrane (Amersham) by using a semidry apparatus in Towbin's buffer at 200 mA for 1 h. Blots were treated with blocking buffer (PBS containing 0.00005% [vol/vol] Tween 20 [PBST] and 5% [wt/vol] dried milk powder) overnight at 4°C, washed five times with PBST (3 min per wash), and incubated with primary antibody (diluted in PBST containing 1% [wt/vol] bovine serum albumin) for 2 h at room temperature. Primary antibodies included mouse anti-SF2/ASF (1:500; Zymed), mouse anti-CstF-64 (monoclonal 3A7 [obtained from T. Shenk and C. MacDonald] diluted 1:50 in PBS with 1% milk powder [44]), goat anti-CPSF (sc-17289, 1:100; Santa Cruz), mouse anti-PTB (1:250; Zymed), rabbit anti-histone H2B (1:500; Chemicon), and goat anti-actin (I-19, sc1616, 1:250; Santa Cruz). The reacted membranes were washed again, incubated with secondary antibody, horseradish peroxidase-conjugated goat anti-mouse (Bio-Rad), goat anti-rabbit (Bio-Rad), or donkey anti-goat (Santa Cruz) as appropriate for 1 h at room temperature, washed for the final time, and then treated with ECL chemiluminescent detection reagents (Amersham) and exposed to film. The levels of induction were calculated using the ratios of density of scanned bands determined by the NIH Image program (version 1.62).
RESULTS
Mutation of the UL37x1 PA signal decreases UL37x1 PA and increases UL37x1-UL37x2 RNA splicing.
Target 1 RNA processing in transfected HFFs recapitulates HCMV UL37x1 RNA PA at nt 50998 and UL37x1-UL37x2 RNA splicing between nt 52219 and 50989 accurately (41). Based upon a minimal BP consensus (5′-YURAY-3′) sequence (35), Target 1 intron 1 has two potential BP sites (at nt −30 and −115) suitably spaced for UL37x1-UL37x2 splicing. However, only the proximal putative BP site (nt −30) is completely consensus to this minimal core and predicted to be used for the UL37x2 acceptor (41). This predicted BP site, nonetheless, is an integral residue of the UL37x1 PA signal (nt −26 to −31) used for processing of the UL37x1 and of UL38 unspliced RNA throughout HCMV infection (41, 47). To determine whether inefficient UL37x1-UL37x2 splicing results in part from competition of cellular splicing factors with the PA machinery for these overlapping sequences, the UL37x1 PA signal was altered at nt −29 to a mutant site (AAGAAA) in Target 1 PA Mut 1 RNA (Fig. 1B). This mutation is known to decrease CPSF-160 binding and reduce PA at downstream sites (6). PA of wt (Fig. 2A) and mutant (Fig. 2B) RNAs at the UL37x1 site (nt 50998) was monitored by RT-PCR of serially diluted cDNAs. PCR products of the predicted size were obtained (41). To determine the efficiency of PA, the relative abundance of each alternatively processed (unspliced, spliced, and polyadenylated) Target 1 RNA was determined by amplifying the same diluted samples with primers for each species. Based upon the RT-PCR amplification of diluted cDNAs, the percentage of each Target 1 and PA Mut 1 species was then determined and compared (Table 4). As predicted from the loss of the invariant UL37x1 PA signal and the CPSF-160 binding site, the efficiency of PA Mut 1 RNA PA at nt 50998 (1%) was decreased dramatically (10-fold) from the efficiency of the wt parent (10%) (Table 4). Concomitantly, the UL37x1-UL37x2 splicing of PA Mut 1 RNA (25%) increased 2.5-fold above the value obtained for wt Target 1 RNA (10%). The AAGAAA mutation is predicted to only block binding of CPSF to the AAUAAA signal and therefore block UL37x1 PA at nt 50998. As the reduction in UL37x1 PA at nt 50998 was accompanied by an increase in UL37x1-UL37x2 RNA splicing, we conclude that the cellular core PA factors and splicing factors compete for juxtaposed cis elements at the UL37x1 PA site on UL37 pre-mRNA and regulate the production of UL37x1 unspliced and UL37x1-UL37x2 spliced RNAs.
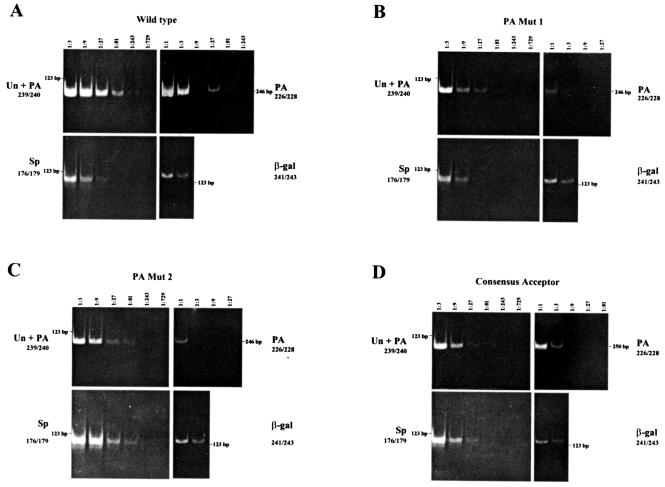
FIG. 2.
Mutation of the essentially invariant UL37x1 PA signal decreases UL37x1 PA and increases UL37x1-UL37x2 RNA splicing. RT-PCR was performed on serially diluted cDNAs from HFFs transfected with vectors expressing wt Target 1 (A), PA Mut 1 (AAGAAA) (B), PA Mut 2 (ACUAAC) (C), or consensus acceptor (AGGGCC) (D) and β-gal RNAs. Total RNAs (10 ng) were reverse transcribed using oligo(dT) primers, and the cDNAs were serially diluted in threefold steps. Target 1 unspliced or polyadenylated (Un + PA) (primers 239-240), UL37x1 polyadenylated (PA) (primers 226-228), and spliced (Sp) (primers 176-179) RNAs or cotransfected β-gal (primers 241-243) RNA in each indicated dilution were amplified by PCR using selected primers and resolved by electrophoresis in polyacrylamide gels. The relative abundance of each unspliced, polyadenylated, or spliced Target 1 RNA species was determined by the highest dilution which produced visibly detectable PCR product.
TABLE 4.
Percentage of alternatively processed Target 1 PA mutant and consensus acceptor RNAs
| RNA | wt
|
PA Mut 1
|
PA Mut 2
|
Consensus acceptor
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Dila | % of RNAb | Dil | % of RNA | Change (fold)d | Dil | % of RNA | Change (fold) | Dil | % of RNA | Change (fold) | |
| Un + PA | 243 | 81 | 81 | 27 | |||||||
| Unc | 216 | 80 | 80 | 74 | 1.1 (↓) | 80 | 49.4 | 1.6 (↓) | 24 | 44 | 1.8 (↓) |
| PA | 27 | 10 | 1 | 1 | 10.0 (↓) | 1 | 0.6 | 16.7 (↓) | 3 | 6 | 1.7 (↓) |
| Sp | 27 | 10 | 27 | 25 | 2.5 (↑) | 81 | 50 | 5.0 (↑) | 27 | 50 | 5.0 (↑) |
| Total | 270 | 100 | 108 | 100 | 162 | 100 | 54 | 100 | |||
Dil, the endpoint dilution at which the processed species is visibly detectable following polyacrylamide gel electrophoresis and staining with ethidium bromide.
The percentage of each UL37 RNA species was calculated by dividing its endpoint dilution by the total and multiplying by 100.
The endpoint dilution for unspliced RNA was calculated by subtracting the endpoint dilution obtained with UL37x1 PA primers from the endpoint dilution obtained using primers which detect both UL37 unspliced and UL37x1 PA RNA.
↑, increase; ↓, decrease.
Mutation of the PA signal to a consensus BP sequence decreases PA and markedly increases UL37x1-UL37x2 splicing.
Although the potential UL37 BP at nt −30 is consensus for the minimal core 5′-YURAY-3′ sequence (35), this site is not optimal when considering adjacent residues for the larger BP consensus (5′-YNYURAC-3′) sequence (10). We therefore generated a second PA mutant (Target 1 PA Mut 2) in which the consensus UL37x1 PA signal (AAUAAA→ACUAAC) and, consequently, the CPSF binding site were abolished and also simultaneously created a fully consensus BP site suitably spaced (nt −27) from the UL37x2 acceptor (10). This mutation is therefore predicted to inhibit CPSF-160 binding and to block UL37x1 PA at site nt 50998. In addition, if the new BP is more suitable than the wt UL37 sequence, PA Mut 2 should increase the efficiency of UL37x1-UL37x2 splicing as well. PA at nt 50998 of the PA Mut 2 RNA substrate was monitored (Fig. 2C) and compared to that of the wt parent (Fig. 2A) as described above for PA Mut 1 RNA, and the relative abundance of each Target 1 alternatively processed RNA was determined (Table 4). As expected, PA of PA Mut 2 at nt 50998 (0.6%) was severely decreased (16.7-fold) below the levels obtained with the wt parent (10%). Moreover, and consistent with the results obtained with PA Mut 1, the splicing efficiency of PA Mut 2 RNA (50%) increased dramatically (fivefold) above that of the wt parent (10%). This change in the balance of spliced and unspliced UL37 RNAs is consistent with a possible competition between PA and splicing factors for the UL37x1 PA signal and splicing signals on UL37 pre-mRNA. Moreover, as the increase in PA Mut 2 RNA splicing (fivefold) was greater than that obtained with PA Mut 1 RNA (2.5-fold), these results suggest that incorporation of the highly consensus BP site further shifted the balance from UL37x1 RNA PA towards UL37x1-UL37x2 RNA splicing.
Mutation to an UL37x2 consensus acceptor increases UL37x1-UL37x2 RNA splicing and decreases UL37x1 PA.
If the cellular core PA and splicing machineries are competing for juxtaposed sites, PA may be favored because of the weak UL37x1-UL37x2 splicing signals. If this were the case, one would predict that an increase in the efficiency of UL37x1-UL37x2 splicing would result in decreased UL37x1 PA at nt 50998. The UL37x2 acceptor, just downstream of the UL37x1 PA signal and site, has two nonconsensus residues based on the consensus splice acceptor (PyPyPyPyPyPyPyPyNCAG//GU/G) (10). To increase the efficiency of UL37x1-UL37x2 RNA splicing, a consensus acceptor (AG//UG→AG//GG) with increased conformity to consensus acceptor sequences was generated and examined (Fig. 2D). This mutation is predicted to exclusively affect UL37x1-UL37x2 RNA splicing as it does not alter the UL37x1 PA signal, its cleavage site, or the U-rich sequence in UL37x2. Analogous to the experiments described above, serially diluted cDNAs were PCR amplified with primers to detect all alternatively processed Target 1 RNAs. Consistent with its increased conformity to consensus splice acceptors, splicing of the Target 1 consensus mutant acceptor RNA (50%) was more efficient (∼5-fold) than for the wt parent (10%) (Table 4). Conversely, PA of this mutant RNA (6%) was decreased 1.7-fold when compared to the wt parent (10%). These results demonstrate that the UL37x2 acceptor is partially responsible for inefficient UL37x1-UL37x2 RNA splicing. The shift from UL37x1 PA to UL37x1-UL37x2 RNA splicing by a mutation that is predicted to affect splicing exclusively suggests that the competition between splicing and PA factors for recognition signals occurs in UL37 pre-RNA processing.
Mutation to a UL37x1 consensus donor increases UL37x1-UL37x2 RNA splicing and decreases UL37x1 PA.
The UL37x1 donor, normally ∼1.2 kb upstream of the UL37x1 PA site, has three nonconsensus residues when compared to the consensus C/AAG//GTA/GAGT donor sequence (10). To determine whether a consensus splice donor would increase UL37x1-UL37x2 splicing efficiency and whether this increased splicing efficiency would analogously decrease UL37x1 PA, a UL37x1 consensus donor (CCA//GUAAGC→ CAG//GUAAGU) was generated and examined for UL37x1-UL37x2 splicing as well as for UL37x1 PA (Fig. 3). The percent of UL37x1-UL37x2 spliced RNAs in UL37x1 consensus donor-transfected cells (50%) increased fivefold above that obtained with the wt parent (10%) (Table 5). Conversely, PA of the Target 1 consensus donor (5.6%) decreased about 1.8-fold below the value obtained with the wt parent (10%). This result suggests that the wt UL37x1 nonconsensus donor is partially responsible for inefficient UL37x1-UL37x2 splicing. This distal upstream mutation of the UL37x1 donor is predicted to affect only UL37x1-UL37x2 RNA splicing and not any of the cis elements required for UL37x1 PA. Nonetheless, this mutation also decreased UL37x1 PA at nt 50998. These results are consistent with a model in which cellular PA and splicing factors compete for cis elements present on UL37 pre-mRNA.
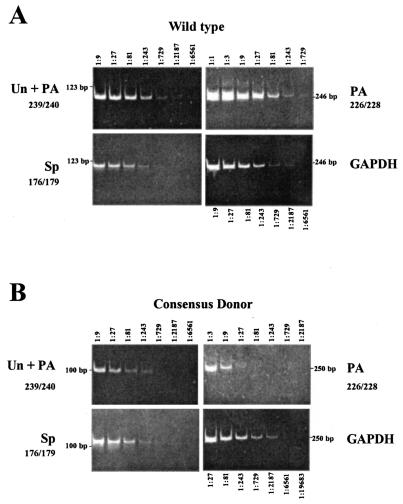
FIG. 3.
Mutation of the wt UL37 5′ ss to a consensus donor increases UL37x1-UL37x2 RNA splicing and decreases UL37x1 RNA PA at nt 50998. Transfected HFFs expressing wt Target 1 (A) or consensus donor (AGGUAAGU) (B) RNAs were assayed by RT-PCR. Total RNAs (5 ng) were reverse transcribed using oligo(dT), serially diluted, and analyzed for the presence of unspliced or polyadenylated RNA, UL37x1 polyadenylated, and UL37x1-UL37x2 spliced RNA as described in the legend to Fig. 2, except that GAPDH was used as a control. The relative abundance of each unspliced, polyadenylated, or spliced Target 1 RNA species was determined by the highest dilution which produced visibly detectable PCR product.
TABLE 5.
Percentages of alternatively processed Target 1 consensus donor RNAa
| RNA | wt
|
Consensus donor
|
|||
|---|---|---|---|---|---|
| Dil | % of RNA | Dil | % of RNA | Change (fold) | |
| Un + PA | 2,187 | 243 | |||
| Un | 1,944 | 80 | 216 | 44.4 | 1.8 (↓) |
| PA | 243 | 10 | 27 | 5.6 | 1.8 (↓) |
| Sp | 243 | 10 | 243 | 50.0 | 5.0 (↑) |
| Total | 2,430 | 100 | 486 | 100 | |
Dil, endpoint dilution; ↑, increase; ↓, decrease.
Real-time PCR quantification of alternatively processed Target 1 RNAs.
To accurately and specifically quantify the partially overlapping unspliced, spliced, and polyadenylated UL37 RNA species in transfected HFFs, we developed a real-time PCR. For these assays, we designed matched PCR primers and dual-labeled TaqMan probes to detect Target 1 unspliced or polyadenylated (primers 239-240; FAM-probe I), UL37x1-UL37x2 spliced junction (primers 226-227; VIC-probe II), and UL37x1 polyadenylated (primers 265-228; FAM-probe VIII) UL37 RNAs (Fig. 4A). To determine whether the TaqMan primer-probe combinations could detect the Target 1 alternatively processed RNAs, in vitro transcription vectors containing Target 1 unspliced (Fig. 4B), UL37x1 polyadenylated (Fig. 4C), and UL37x1-UL37x2 spliced (Fig. 4D) cDNAs were used to generate alternatively processed RNAs and known quantities were analyzed by using the designed primer-probe combinations. The real-time PCR quantification was sensitive, detecting each UL37 cDNA to levels corresponding to 0.025 pg of RNA.
FIG. 4.
(A) Real-time PCR primers and probes for detection of UL37 alternatively processed RNAs. The physical location of the primer-probe combinations used to detect UL37 unspliced, spliced, and polyadenylated RNAs in TaqMan assays are shown on each minigene substrate. (B to D) Detection of Target 1 unspliced (B), UL37x1 polyadenylated (C), and UL37x1-UL37x2 spliced (D) cDNAs. Cloned cDNAs corresponding to the alternatively processed RNAs were in vitro transcribed, reversed transcribed, and serially diluted. Triplicate samples were assayed by real-time PCR using primers 239-240 and probe I (B), primers 265-228 and probe VIII (C), and primers 226-227 and probe II (D). The amounts assayed ranged from 166.67 to 0.025 pg.
To determine whether the real-time PCR could detect the processed UL37 species in mixtures of mRNAs, known quantities of Target 1 unspliced, UL37x1 polyadenylated, and UL37x1-UL37x2 in vitro-transcribed RNAs were mixed and quantified (Table 6). Primers 265-228 and probe VIII, specific for UL37 RNA polyadenylated at nt 50998, accurately quantified 0.06 to 42.36 pg. In addition, primers 226-227 and probe II, specific for the detection of UL37x1-UL37x2 RNA, accurately quantified 44.75 to 0.09 pg of spliced RNA within the mixture. These results show the accuracy and exquisite sensitivity of real-time PCR detection of the processed UL37 spliced and polyadenylated RNAs. In spite of these properties of TaqMan analysis, quantification of the phenotypes of the Target 1 consensus donor and consensus acceptor by using real-time PCR was not possible. These assays require probe II specific for the UL37x1-UL37x2 spliced junction, and the engineered 5′ and 3′ ss mutations span the probe II binding site, precluding detection of their UL37x1-UL37x2 spliced products by real-time PCR.
TABLE 6.
Sensitivity and specificity of real-time PCR quantification for UL37x1 polyadenylated and UL37x1-UL37x2 spliced RNAs
| Added amt (pg)a
|
Detected amt (pg)b
|
|||
|---|---|---|---|---|
| Un | PAc | Spd | PA | Sp |
| 55.56 | 0.08 | 0.08 | 0.06 | 0.09 |
| 6.17 | 0.69 | 0.69 | 0.91 | 0.65 |
| 0.69 | 6.17 | 6.17 | 6.52 | 4.87 |
| 0.08 | 55.56 | 55.56 | 42.36 | 44.75 |
Standards were generated by in vitro transcription of cloned UL37 unspliced (Un), UL37x1 PA (PA), and UL37x1-UL37x2 cDNAs. Known amounts of the RNAs were reverse transcribed, and the corresponding cDNAs were serially diluted and added to standard reactions.
Amount detected by real-time PCR analysis using the corresponding primer-probe combinations as shown in Fig. 4.
PA, Target 1 polyadenylated at nt 50998.
Sp, spliced Target 1 (UL37x1-UL37x2).
Real-time PCR quantification of Target 1 alternatively processed RNAs.
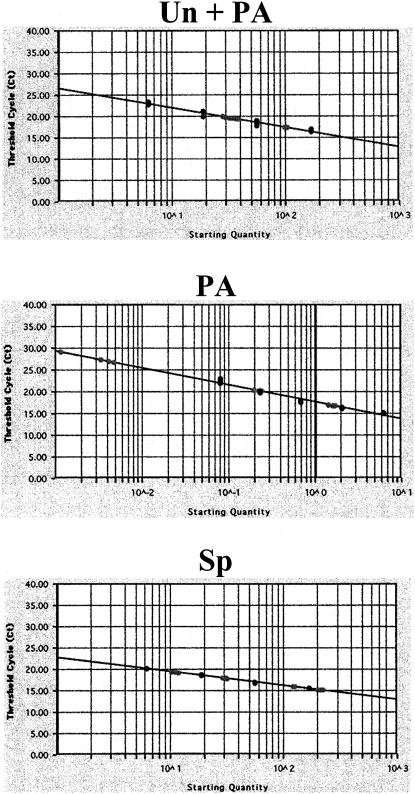
Because the PA mutations in PA Mut 1 and PA Mut 2 are not contained in any of the UL37 primer-probe sequences, real-time PCR could be used to verify the semiquantitative RT-PCR results. The abundances of alternatively processed UL37 RNAs in HFFs transfected with independently derived wt (p1021) and mutant (p1041 and p1043) clones were determined using real-time PCR (Fig. 5). Consistent with the results obtained with semiquantitative RT-PCR, PA of PA Mut 1 RNA at nt 50998 was decreased compared with that for the wt parent (0.2 pg; 0.14%) (Table 7). The abundance of PA Mut 1 RNA polyadenylated at nt 50998 was below the detection level of this real-time PCR assay. Conservatively, we estimated this detection value at the lowest control PA RNA detected in parallel (<0.08 pg; <0.04%). This represents a >3.5-fold reduction in PA at the UL37x1 site. As previously found by RT-PCR, the abundance of the UL37x1-UL37x2 spliced RNA, determined by real-time PCR, was increased 2.2-fold in PA Mut 1-transfected HFFs (51.2%) compared with the wt (22.9%).
FIG. 5.
Real-time PCR quantification of UL37 alternatively processed RNAs in PA Mut-transfected HFFs. HFFs were transfected with vectors expressing wt Target 1 (p1021), PA Mut 1 (p1041), or PA Mut 2 (p1043) and β-gal RNAs. Transfected cell RNAs (25 to 100 ng) were reverse transcribed and assayed in triplicate for Target 1 unspliced or polyadenylated, UL37x1 polyadenylated, and UL37x1-UL37x2 spliced RNAs using the primers and probes described in the legend to Fig. 4. Standard RNAs (black circles) were generated by in vitro transcription of cloned unspliced, UL37x1 polyadenylated, and spliced cDNAs. Unknown quantities (gray circles) were determined by interpolation. The slopes of the Un + PA, PA, and spliced curves are −4.535, −3.899, and −3.241, respectively. The y intercepts were 26.604 (Un + PA), 17.789 (PA), and 22.830 (Sp). The correlation coefficients were 0.957 (Un + PA), 0.968 (PA), and 0.998 (Sp).
TABLE 7.
Real-time PCR quantification of Target 1 alternatively processed RNAs
| RNAa | Expt 1
|
Expt 2
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| wt
|
PA Mut 1
|
wt
|
PA Mut 2
|
|||||||
| Amt (pg) | % of RNA | Amt (pg) | % of RNA | Change (fold)b | Amt (pg) | % of RNA | Amt (pg) | % of RNA | Change (fold) | |
| Un + PA | 110.4 | 89.0 | 304.3 | 105.5 | ||||||
| Un | 110.2 | 77.0 | 89.0 | 48.8 | 1.6 (↓) | 302.7 | 44.4 | 105.5 | 14.2 | 3.1 (↓) |
| PA | 0.2 | 0.14 | <0.08 | <0.04 | >3.5 (↓) | 1.6 | 0.2 | <0.08 | <0.01 | >20.0 (↓) |
| Sp | 32.7 | 22.9 | 93.5 | 51.2 | 2.2 (↑) | 377.6 | 55.4 | 638.3 | 85.8 | 1.5 (↑) |
| Total | 143.1 | 100 | 182.5 | 100 | 681.9 | 100 | 743.8 | 100 | ||
Un, unspliced; Sp, spliced.
↑, increase; ↓, decrease.
Analogously, the abundance of the PA Mut 2 polyadenylated at nt 50998 was also undetectable (<0.08 pg; <0.01%) in transfected HFFs. This reduction from the wt parent (1.6 pg; 0.2%) represents a >20-fold reduction in PA at the UL37x1 PA site. Consistent with the RT-PCR assay of PA Mut 2, there was a concomitant increase of 1.5-fold in UL37x1-UL37x2 splicing in PA Mut 2-transfected HFFs (85.8%) above that observed in wt-transfected cells (55.5%). The reproducibility of mutant phenotypes has been verified by repeatedly measuring, using real-time PCR, the efficiency of splicing and PA of the PA mutant RNA substrates. The decreased efficiency of PA Mut 1 and PA Mut 2 RNA processing at the UL37x1 PA site is reproducibly accompanied by increased UL37x1-UL37x2 RNA splicing (2.2- to 2.6-fold and 1.5- to 7.8-fold, respectively). Taken together, these results suggest that binding of the PA machinery at the UL37x1 PA site sterically hinders binding of the splicing machinery required for UL37x1-UL37x2 RNA splicing.
Induction of PA factors in HCMV-infected cells.
The abundance of UL37x1 unspliced RNA in HCMV-infected HFFs is far greater than that of UL37 spliced RNAs (28, 47, 48; C. N. Davis and A. M. Colberg-Poley, unpublished results). As the abundance of Target 1 spliced (UL37x1-UL37x2) RNA was equivalent to or greater than that of Target 1 RNA polyadenylated at the UL37x1 site in transfected HFFs treated with anisomycin, we tested whether the processing of UL37 pre-mRNA is also regulated during HCMV infection by the abundance of cellular proteins that regulate PA site usage (CstF-64), suppress splicing (PTB), or regulate splicing (SF2). For these analyses, we relied on several observations from the literature and from our laboratory. First, altered CstF-64 levels regulate PA site usage in the immunoglobulin M (IgM) heavy gene in differentiating B lymphocytes (42, 45). Second, we identified a potential PTB core-binding site just upstream of the UL37x2 acceptor, and PTB is a potent suppressor of splicing (30, 37). Finally, adenovirus infection and vaccinia virus infection increase the levels of hypophosphorylated SF2, thereby reducing the splicing of cellular pre-mRNAs and favoring virus gene expression over that of the cell (25-27, 54). Regulation of RNA splicing by SF2 is dependent on the expression level of SF2 and its phosphorylation status as the splicing activity of hypophosphorylated SF2 is greatly reduced (27, 54).
We therefore examined whether HCMV infection altered the balance between cellular PA and splicing machineries, which, in turn, might affect the production of UL37 unspliced and spliced RNAs. The abundance of key PA factors (CstF-64 and CPSF), PTB, and SF2 in nuclear extracts from HCMV-infected HFFs was examined (Fig. 6). HCMV infection initially decreased the abundance of the essential PA factor, CstF-64, at 2 h postinfection (∼35.7-fold) and then increased its levels ∼6.6-fold above uninfected HFF levels at 4 h postinfection (Fig. 6A). Analogously but less dramatically, the levels of CPSF-73 and CPSF-30 factors decreased at 2 h postinfection and increased very slightly at 4 and 6 h postinfection but not to levels of uninfected HFFs. PTB abundance increased at 4 h (3.7-fold) and 6 h (1.9-fold) after HCMV infection, similar to the results obtained with CstF-64 and CPSF. In contrast, the abundance of actin was slightly decreased at 2 h (1.8-fold), 4 h (1.6-fold), 6 h (1.7-fold), and 8 h (4.5-fold) of HCMV infection.
FIG. 6.
(A) Induction of the essential PA factor, CstF-64, at IE times of HCMV infection. HFFs were infected with HCMV (3 PFU/cell), and nuclear extracts were prepared at 2, 4, 6, and 8 h postinfection. Nuclear extracts were also prepared from uninfected (un) HFFs. Ten micrograms of nuclear proteins was resolved by electrophoresis in sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis, transferred to membranes, and reacted with antibodies against CstF-64 (1:50), CPSF (1:100), PTB (1:250), or actin (1:250). (B) Increased abundance of hypophosphorylated SF2 at IE times of HCMV infection. Western blots of nuclear proteins (10 μg/lane) from HCMV-infected and uninfected controls were prepared and resolved as described for panel A and reacted with anti-SF2 (1:500) and anti-histone (1:500) antibodies.
The pattern of induction of the essential splicing SR protein, SF2, following HCMV infection (Fig. 6B) closely paralleled the pattern observed for CstF-64 and PTB (Fig. 6A). The levels of hypophosphorylated SF2 increased dramatically at 4 h (∼6.7-fold) after HCMV infection. The levels of hypophosphorylated SF2 in control HeLa nuclear extracts, which are active in splicing, were low. Conversely, control histone levels were comparable at 4 h (1.2-fold increase) and slightly increased at 8 h (2.9-fold) after HCMV infection. Taken together, these results suggest that at IE times of infection, there is a shift in the balance of PA and splicing factors which favors production of unspliced UL37x1 IE RNA and not splicing of the UL37 pre-mRNA.
DISCUSSION
Alternative processing of UL37 RNAs results in unspliced and differentially spliced RNAs, which encode multiple protein isoforms with predictably distinct functions. Although UL37 transcripts are 5′ coterminal, production of UL37x1 unspliced RNA is greatly favored during all temporal phases of HCMV infection of HFFs (28, 47, 48). Moreover, using semiquantitative RT-PCR and real-time PCR, we have determined that the UL37x1 unspliced RNA is ∼40-fold more abundant than the UL37x1-UL37x2 spliced RNA in HCMV-infected HFFs at 8 h postinfection (C. N. Davis and A. M. Colberg-Poley, unpublished results). This situation is highly unusual because PA signals, even strong signals, within introns are not usually recognized (55). The RNA cis elements that direct accurate UL37x1-UL37x2 splicing and UL37x1 PA are included in Target 1. Nonetheless, the dramatic preponderance of the UL37x1 unspliced RNA is not reproduced in Target 1-transfected HFFs, indicating that, in addition to the UL37 pre-mRNA cis elements regulating its processing, additional factors present during HCMV infection affect the balance between PA and splicing of the UL37 pre-mRNA.
In this study, we examined two posttranscriptional mechanisms potentially underlying the favored production of UL37x1 unspliced over UL37 spliced RNAs during HCMV infection. We used the Target 1 minigene to examine regulation by competition for cis elements, which determine the production of either UL37x1 unspliced RNA or UL37x1-UL37x2 spliced RNA. We also examined the abundance of key PA factors and factors which might negatively regulate splicing in HCMV-infected HFFs.
The 3′-end processing of the UL37x1 RNA requires initial recognition of the AAUAAA element (at nt 51015 to 51020) by CPSF-160 (41). The CPSF complex is stabilized by interactions with CstF and CFI (31, 34, 43) and with the phosphorylated serines of the C-terminal heptad repeat domain of RNA polymerase II (32). The stability of this commitment complex correlates with the efficiency of poly(A) site usage (5, 51). CstF binds at variable G/U or U-rich sequences downstream of CPSF. A U-rich element, sufficient to direct specific binding of CstF-64 (31) and downstream of the UL37x1 PA signal, 17 to 25 nt downstream of the cleavage site, lies within UL37x2 (11, 28). Because of its sequence and position, we predict that this U-rich element in UL37x2 binds CstF-64, facilitating UL37x1 and UL38 RNA PA at nt 50998. We also note that the abundance of CstF-64 increases markedly at 4 h postinfection with HCMV, consistent with the high production of UL37x1 unspliced RNA at IE times of infection (28, 47, 48). Cst-F64 levels have been implicated in the switch from membrane-bound to secreted IgM heavy chain that occurs during B-cell differentiation (45). Exon size also affects competition between splicing and PA factors in Ig selection (36). CFII and PAP then join the PA complex to form an active 3′-end processing complex (5).
UL37x1-UL37x2 RNA splicing joins nt 52219 to nt 50989 and has been accurately recapitulated using Target 1 minigene RNA in transfected cells (41). RNA splicing requires the recognition of the 5′ ss, BP, and 3′ ss. The 5′ ss, defined by the consensus sequence 5′-AGGURAGU-3′, is recognized by U1 snRNP (10). UL37x1 5′ ss is weak because it has three nonconsensus residues. The second element, the BP site (YURAY), is recognized by U2 snRNP (4, 10, 35). Based on this minimal core BP sequence, there are two possible BP sequences (nt −30 and nt −115) in Target 1 (41). However, the proximal site is favored because it is consensus and located immediately upstream of the authentic UL37x2 AG cleavage site. Conversely, the nt −115 BP site conforms less well to the consensus and is located upstream of four additional AG/PPT sites, which are not the authentic UL37x2 cleavage site. Cleavage of the downstream AG dinucleotide, essential for splicing, generally occurs at the first AG downstream of the BP (39, 57). We therefore predict that the BP site at nt −30 is used for Target 1 UL37x1-UL37x2 splicing. U2 snRNP entry into the intronic BP site is facilitated by recognition of the PPT by U2AF65. The PPT is closely spaced (usually nt −5 to −15) to the 3′ ss by U2AF65 (52, 53). Native U2AF is a heterodimer of U2AF65 and U2AF35, which recognizes the 3′ ss consensus (YAG/GU) (52, 53).
Competition for UL37 pre-mRNA cis elements.
Competition between splicing and PA factors has been documented previously in cellular and viral systems (3, 20, 22). However, splicing factors most often inhibit PA site utilization. In one exception, the adenovirus E3 transcription unit has closely spaced PA and splicing elements, which favor PA downstream of the 3′ ss (8). The distinct organization of the cis elements in UL37 pre-mRNA and the phenotypes of the Target 1 mutants suggest that competition of PA and splicing factors occurs at the site of unspliced UL37x1 RNA PA within intron 1. Because of the physical proximity of cis elements, it is unlikely that the PA and splicing machineries can simultaneously bind the UL37 pre-mRNA. Therefore, we propose a model of posttranscriptional processing in which PA and splicing complexes compete for their respective cis elements on UL37 pre-mRNA and favor the production of UL37x1 unspliced RNA (Fig. 7). In this model and consistent with the known sequence of events (31), CPSF-160 initially binds to the invariant AAUAAA motif and the complex is stabilized by interactions with the CstF complex formed on the downstream U-rich element in UL37x2. Furthermore, CFI binds at its cleavage site between the two PA complexes. Binding of the large CPSF complex to the PA signal (at nt −26 to −31) is predicted to block entry of U2 snRNP into the consensus BP site at nt −30. Moreover, binding of CFI to the cleavage site (nt −9) is predicted to block U2AF65 recognition of the PPT and, consequently, the recruitment of U2 snRNP to the BP as well as U2AF35 binding at the 3′ ss.
In the second part of the model, shown in Fig. 7B, the shift favoring splicing following the mutation of the 5′ ss, PA signal, and 3′ ss is represented. To determine whether the PA signal blocks U2 snRNP entry into the BP, we generated PA mutants and assayed the efficiency of the splicing and PA of the mutant substrate. The first mutant (PA Mut 1) contains a PA mutant signal (AAGAAA) known to block binding of the CPSF-160 factor (6). This PA mutant is known not to alter the UL37x1-UL37x2 splice site (41). As expected from its sequence, PA of the Target 1 PA Mut 1 RNA at nt 50998 is severely reduced. In addition, UL37x1-UL37x2 splicing of the mutant substrate is increased. The increase in splicing efficiency can be accounted for if the UL37 BP overlaps with the UL37x1 PA signal. As the CPSF complex cannot load efficiently on the site, it no longer blocks U2 snRNP entry into the consensus BP site at nt −30. We note that the increase of the PA Mut 1 substrate is observed in spite of the fact that the AAGAAA mutation makes the potential BP at nt −30 less consensus than the wt UL37 sequence.
We therefore generated a second PA mutant (PA Mut 2), which is predicted to abolish CPSF binding at the UL37x1 PA site and increase the conformity to consensus BP in its place. This mutation was used to also test the suitability of the BP spacing at nt −30 for UL37x1-UL37x2 splicing. As the BP sequence in PA Mut 2 is more consensus than those of the wt and PA Mut 1, PA Mut 2 is predicted to increase the affinity of U2 snRNP for the new BP if the spacing is correct. Indeed, we observed that UL37x1-UL37x2 RNA splicing of PA Mut 2 was increased about twofold above that of PA Mut 1 and fivefold above that of the wt parent. The results with Target 1 PA Mut 2 indicate that the key splicing cis elements, predictably its BP, in Target 1 are juxtaposed with the UL37x1 PA signal and site.
To determine whether low conformity of the UL37x1 5′ ss to the consensus sequence results in weak recognition by U1 snRNP and permits UL37x1 PA at nt 50998, we generated a UL37x1 consensus donor. By virtue of its highly consensus sequence, this mutant should be more effective at recruiting U1 snRNP to the 5′ ss and thereby increase the efficiency of UL37x1-UL37x2 RNA splicing. Indeed, the consensus donor increased UL37x1-UL37x2 splicing even though the strong UL37x1 PA signal was intact. Moreover, this mutation might have a secondary effect, as a strong U1 snRNP interaction upstream of a poly(A) site can inhibit PA at that site (1). Splicing factor U1 70K, a component of U1 snRNP, when bound adjacent to and upstream of a poly(A) signal, is able to inhibit pre-mRNA 3′-end formation in the bovine papilloma late gene (20, 22). Binding of a U1 snRNP-5′ ss complex blocks PA at the site of 3′-end formation by binding to and inhibiting the activity of PAP (22). Therefore, the improved recruitment of U1 snRNP to the UL37x1 consensus donor may decrease PAP activity at the downstream UL37x1 PA site. The invariance of the wt UL37x1 donor in multiple HCMV primary strains suggests that a weak U1 snRNP binding site plays an important role in regulating the balance of UL37x1 unspliced and UL37 spliced RNAs for HCMV growth in vivo (W. A. Hayajneh and A. M. Colberg-Poley, unpublished data).
The 3′ ss contains two elements: the PPT and the 5′-YAGGU-3′ sequence. Recognition of the consensus 5′-YAGGU-3′ sequence, which contains the acceptor site, requires native U2AF, a heterodimer of U2AF65 and U2AF35 (52, 53). The PPT tract located with the proper spacing to the UL37x2 acceptor contains the site of UL37x1 RNA cleavage, the site predicted to bind CFI and CFII (34). Thus, the UL37 intron 1 PPT required for U2AF65 binding and UL37x1-UL37x2 splicing may be blocked by the cleavage complex required for the production of UL37x1 unspliced RNA and UL38 early RNA. As the intron 1 PPT is also required for efficient UL37x1-UL37x2 splicing, we instead generated a consensus acceptor which is predicted to increase affinity of the native U2AF heterodimer for the site. The increased splicing of the Target 1 consensus acceptor is consistent with improved recognition of the UL37x2 acceptor by native U2AF. Decreased PA at the UL37x1 PA site suggests that the enhanced binding of U2AF at the PPT/3′ ss was able to functionally displace the CFI-CFII complex from the UL37x1 cleavage site.
Interestingly, there is a documented case in which binding of cellular proteins to PPT upstream of a splice acceptor can inhibit splicing (33). The Drosophila melanogaster sex-lethal (SXL) protein, present in female embryos, binds to PPT but lacks the RS domain in U2AF65 required to recruit U2 snRNP to the branch site. Thus, SXL can block splicing of an intron simply by occupying the PPT and preventing binding of U2AF65. In addition, SR proteins regulate adenovirus pre-mRNA splicing by binding to an intronic suppressor close to its BP and blocking entry of U2 snRNP into the BP and spliceosome (26). In the model, we propose a similar mechanism, however, involving steric hindrance by binding of CPSF and CFI-CFII complexes that blocks the binding of U2AF65 to the PPT, U2AF35 to the 3′ ss, and U2 snRNP entry into the UL37 intron 1 BP.
Alteration of the balance between cellular PA and splicing factors by HCMV infection.
Expression of IgM heavy chain is regulated during B-cell differentiation by use of two alternative PA sites (42, 45). mRNA coding for membrane-bound IgM is produced early by processing at a downstream site. The upstream PA site is located within an intron and is removed by splicing. When differentiation occurs, mRNA for the secretory IgM is produced by use of the upstream PA site. It has been proposed that increased use of the secretory PA site correlates with the levels of CstF-64 (42, 45). In addition, splicing is regulated by SR proteins such as SF2 whose level and phosphorylation state alter its activity (27, 54). Moreover, splicing suppressors such as PTB can regulate splicing (30, 37).
As the ratio of UL37x1-UL37x2 to UL37x1 unspliced RNA is higher in transfected cells than in HCMV-infected cells (C. N. Davis and A. M. Colberg-Poley, unpublished data), the regulation of UL37 pre-mRNA processing by juxtaposition of its cis elements appears to be only partially responsible for the favoring of UL37x1 unspliced RNA. The use of the minigenes enabled us to dissect the effects of the juxtaposed cis elements on UL37 pre-mRNA processing by cellular machineries in uninfected cells. With the information gleaned from these experiments, we can now investigate the effects of HCMV infection at different times on the alternative processing of the juxtaposed UL37 cis elements.
The abundance, phosphorylation, and nuclear localization of splicing factors and PA factors are known to regulate alternative RNA processing. We therefore examined the abundance of key PA and splicing factors in HCMV-infected cells and tested whether UL37x1 RNA PA at nt 50998 was favored by induction of PA factors. The induction of CstF-64, PTB, and hypophosphorylated SF2 occurred at IE times (4 h postinfection) of HCMV infection and lasted briefly. UL37x1 unspliced RNA is first detected at 4 h postinfection (47, 48), consistent with the observed induction of these factors favoring UL37x1 PA. This finding is analogous to the functional inactivation of SF2 by adenovirus and by vaccinia virus at late times of infection (25-27). Cytoplasmic expression of vaccinia genes, which lack introns, is favored over cellular gene expression by this shutoff of host cell RNA splicing. Analogously, inactivation of SF2 by hypophosphorylation in adenovirus-infected cells results in favored splicing of adenovirus IIIa pre-mRNA (27). However, in contrast to adenovirus- and vaccinia virus-infected cells (25), both hyperphosphorylated and hypophosphorylated SF2 are induced at IE times of infection in HCMV-infected cells. This finding may result from a continued requirement for functional splicing machinery until late times of HCMV infection. To determine whether induction of PA factors occurs at other times of HCMV infection, we are presently examining the abundances of CstF-64, SF2, and PTB in nuclear extracts from HCMV-infected cells at early and late times of infection. Moreover, we found that PTB, an inhibitor of RNA splicing (30, 37), was induced with similar kinetics to that observed for CstF-64 and hypophosphorylated SF2. The UL37 intron 1 PPT has a potential PTB core-binding site overlapping with the site required for U2AF65 binding. Our results are consistent with those of a microarray analysis of HCMV-infected cell mRNA during a 48-h time course (9). Interestingly, there were increases in the abundances of Cst-F-64 (2.5-fold) and PTB (1.9-fold) mRNAs at 4 h after HCMV infection of HFFs. Thus, in addition to the spacing of cis elements that regulate UL37 pre-mRNA processing at IE times of infection, HCMV infection induces a second mechanism at the level of PA and splicing factors which further favors the production of the UL37x1 unspliced RNA. Because the induction of CstF-64 (2.5-fold) and the induction of PTB (1.9-fold) RNA abundances previously detected by microarray assays (9) are considerably lower than the observed induction of the encoded proteins, we speculate that the induction of Cst-F-64 and PTB proteins occurs in part at the posttranslational level.
Taken together, these results suggest that the production of UL37x1 unspliced RNA results from a combination of competition by splicing and PA factors for UL37 pre-mRNA cis elements and the induction of factors favoring PA and inhibiting splicing. The balance between UL37x1-UL37x2 splicing and PA determines the mode of RNA processing and ultimately determines the UL37 proteins which are produced in the HCMV-infected cell. Thus, these results indicate that there is a convergence of cis elements and cellular PA and splicing factors to regulate production of UL37x1 unspliced RNA during HCMV infection.
Acknowledgments
We thank Dan Tenney for his critical comments on the manuscript, Tom Shenk for providing the personal communication about the HCMV UL37x1 mutant, and Clint MacDonald and Tom Shenk for generously providing us with the anti-CstF-64 antibody.
This work was supported by Public Health Service grant AI-46459 from the National Institute of Allergy and Infectious Diseases and by Children's Research Institutional Funds to A.M.C.-P.
REFERENCES
- 1.Adami, G., and J. R. Nevins. 1988. Splice site selection dominates over poly(A) site choice in RNA production from complex adenovirus transcription units. EMBO J. 7:2107-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Barazi, H. O., and A. M. Colberg-Poley. 1996. The human cytomegalovirus UL37 immediate-early regulatory protein is an integral membrane N-glycoprotein which traffics through the endoplasmic reticulum and Golgi apparatus. J. Virol. 70:7198-7208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashe, M. P., L. H. Pearson, and N. J. Proudfoot. 1997. The HIV-1 5′ LTR poly(A) signal is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 16:5752-5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berglund, J. A., N. Abovich, and M. Rosbash. 1998. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes Dev. 12:858-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beyer, K., T. Dandekar, and W. Keller. 1997. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem. 272:26769-26779. [DOI] [PubMed] [Google Scholar]
- 6.Bienroth, S., E. Wahle, C. Suter-Crazzolara, and W. Keller. 1991. Purification of the cleavage and polyadenylation factor involved in the 3′-processing of messenger RNA precursors. J. Biol. Chem. 266:19768-19776. [PubMed] [Google Scholar]
- 7.Borst, E.-M., G. Hahn, U. H. Koszinowski, and M. Messerle. 1999. Cloning of the human cytomegalovirus (HCMV) genome as an infectious bacterial artificial chromosome in Escherichia coli: a new approach for construction of HCMV mutants. J. Virol. 73:8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brady, H. A., and W. S. M. Wold. 1988. Competition between splicing and polyadenylation reactions determines which adenovirus region E3 mRNAs are synthesized. Mol. Cell. Biol. 8:3291-3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Browne, E. P., B. Wing, D. Coleman, and T. Shenk. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319-12330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burge, C. B., T. Tuschl, and P. A. Sharp. 1998. Splicing of precursors to mRNAs by the spliceosomes, p. 525-560. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Chee, M. S., A. T. Bankier, S. Beck, R. Bohni, C. M. Brown, R. Cerny, T. Horsnell, C. A. Hutchison III, T. Kouzarides, J. A. Martignetti, E. Preddie, S. C. Satchwell, P. Tomlinson, K. M. Weston, and B. G. Barrell. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125-169. [DOI] [PubMed] [Google Scholar]
- 12.Colberg-Poley, A. M. 1996. Functional roles of immediate early proteins encoded by the human cytomegalovirus UL36-38, UL115-119, TRS1/IRS1 and US3 loci. Intervirology 39:350-360. [DOI] [PubMed] [Google Scholar]
- 13.Colberg-Poley, A. M., S. D. Voss, K. Chowdhury, and P. Gruss. 1985. Structural analysis of murine genes containing homoeo box sequences and their expression in embryonal carcinoma cells. Nature 314:713-718. [DOI] [PubMed] [Google Scholar]
- 14.Colberg-Poley, A. M., L. D. Santomenna, P. P. Harlow, P. A. Benfield, and D. J. Tenney. 1992. The HCMV US3 and UL36-38 immediate early proteins regulate gene expression. J. Virol. 66:95-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colberg-Poley, A. M., L. Huang, V. E. Soltero, A. Iskenderian, R.-F. Schumacher, and D. G. Anders. 1998. The acidic domain of pUL37x1 and gpUL37 plays a key role in transactivation of HCMV DNA replication gene promoter constructions. Virology 246:400-408. [DOI] [PubMed] [Google Scholar]
- 16.Colberg-Poley, A. M., M. B. Patel, D. P. P. Erezo, and J. E. Slater. 2000. Human cytomegalovirus immediate early regulatory proteins traffic through the secretory apparatus and to mitochondria. J. Gen. Virol. 81:1779-1789. [DOI] [PubMed] [Google Scholar]
- 17.Dantonel, J. C., K. G. Murthy, J. L. Manley, and L. Tora. 1997. Transcription factor TFIID recruits factor CPSF for formation of 3′ end of mRNA. Nature 389:399-402. [DOI] [PubMed] [Google Scholar]
- 18.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fu, X. D. 1995. The superfamily of arginine/serine-rich splicing factors. RNA 1:663-680. [PMC free article] [PubMed] [Google Scholar]
- 20.Furth, P. A., W. Choe, J. H. Rex, J. C. Byrne, and C. C. Baker. 1994. Sequences homologous to the 5′ splice sites are required for the inhibitory activity of papillomavirus late 3′ untranslated regions. Mol. Cell. Biol. 14:5278-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goldmacher, V. S., L. M. Bartle, A. Skaletskaya, C. A. Dionne, N. L. Kedersha, C. A. Vater, J.-W. Han, R. J. Lutz, S. Watanabe, E. D. Cahir McFarland, E. D. Kieff, E. S. Mocarski, and T. Chittenden. 1999. A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to bcl-2. Proc. Natl. Acad. Sci. USA 96:12536-12541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunderson, S. I., M. Polycarpou-Schwarz, and I. W. Mattaj. 1998. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell 1:255-264. [DOI] [PubMed] [Google Scholar]
- 23.Hayajneh, W. A., A. M. Colberg-Poley, A. Skaletskaya, L. M. Bartle, M. M. Lesperance, D. G. Contopoulos-Ioannidis, N. L. Kedersha, and V. S. Goldmacher. 2001. The sequence and antiapoptotic functional domains of the human cytomegalovirus UL37 exon 1 immediate early protein are conserved in multiple primary strains. Virology 279:233-240. [DOI] [PubMed] [Google Scholar]
- 24.Hayajneh, W. A., D. G. Contopoulos-Ioannidis, M. M. Lesperance, A. Venegas, and A. M. Colberg-Poley. 2001. The carboxyl terminus of the human cytomegalovirus UL37 immediate-early glycoprotein is conserved in primary strains and is important for transactivation. J. Gen. Virol. 82:1569-1579. [DOI] [PubMed] [Google Scholar]
- 25.Huang, T.-S., C. E. Nilsson, T. Punga, and G. Akusjarvi. 2002. Functional inactivation of the SR family of splicing factors during a vaccinia virus infection. EMBO Rep. 3:1088-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanopka, A., O. Muhlemann, and G. Akusjarvi. 1996. Inhibition by SR proteins of splicing of a regulated adenovirus pre-mRNA. Nature 381:535-538. [DOI] [PubMed] [Google Scholar]
- 27.Kanopka, A., O. Muhlemann, S. Petersen-Mahrt, C. Estmer, C. Ohrmalm, and G. Akusjarvi. 1998. Regulation of adenovirus alternative RNA splicing by dephosphorylation of SR proteins. Nature 393:185-187. [DOI] [PubMed] [Google Scholar]
- 28.Kouzarides, T., A. T. Bankier, S. C. Satchwell, E. Preddy, and B. G. Barrell. 1988. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology 165:151-164. [DOI] [PubMed] [Google Scholar]
- 29.Lee, M., J. Xiao, E. Haghjoo, X. Zhan, G. Abenes, T. Tuong, W. Dunn, and F. Liu. 2000. Murine cytomegalovirus containing a mutation at open reading frame M37 is severely attenuated in growth and virulence in vivo. J. Virol. 74:11099-11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, C.-H., and J. G. Patton. 1995. Regulation of alternative 3′ splice site selection by constitutive splicing factors. RNA 1:234-245. [PMC free article] [PubMed] [Google Scholar]
- 31.MacDonald, C., J. Wilusz, and T. Shenk. 1994. The 64-kilodalton subunit of CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14:6647-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maniatis, T., and R. Reed. 2002. An extensive network of coupling among gene expression machines. Nature 416:499-506. [DOI] [PubMed] [Google Scholar]
- 33.Merendino, L., S. Guth, D. Bilbao, C. Martinez, and J. Valcárcel. 1999. Inhibition of msl-2 splicing by Sex-lethal reveals interaction between U2AF35 and the 3′ splice site AG. Nature 402:838-841. [DOI] [PubMed] [Google Scholar]
- 34.Minvielle-Sebastia, L., and W. Keller. 1999. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 3:352-357. [DOI] [PubMed] [Google Scholar]
- 35.Moore, M. J. 2000. Intron recognition comes of AGe. Nat. Struct. Biol. 7:14-16. [DOI] [PubMed] [Google Scholar]
- 36.Peterson, M. L., M. B. Bryman, M. Peiter, and C. Cowan. 1994. Exon size affects competition between splicing and cleavage-polyadenylation in the immunoglobulin mu gene. Mol. Cell. Biol. 14:77-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh, R., J. Valcárcel, and M. R. Green. 1995. Distinct binding specificities and functions of higher eukaryotic polypyrimidine tract-binding proteins. Science 268:1173-1176. [DOI] [PubMed] [Google Scholar]
- 38.Skaletskaya, A., L. M. Bartle, T. Chittenden, A. L. McCormick, E. S. Mocarski, and V. S. Goldmacher. 2001. A cytomegalovirus-encoded inhibitor of apoptosis that suppresses caspase-8 activation. Proc. Natl. Acad. Sci. USA 98:7829-7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. W. J., E. B. Porro, J. G. Patton, and B. Nadal-Ginard. 1989. Scanning from an independently specified branch point defines the 3′ splice site of mammalian introns. Nature 342:243-247. [DOI] [PubMed] [Google Scholar]
- 40.Smith, J. A., and G. S. Pari. 1995. Expression of human cytomegalovirus UL36 and UL37 genes is required for viral DNA replication. J. Virol. 69:1925-1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Su, Y., J. R. Testaverde, C. N. Davis, W. A. Hayajneh, R. Adair, and A. M. Colberg-Poley. 2003. Human cytomegalovirus UL37 immediate early target minigene RNAs are accurately spliced and polyadenylated. J. Gen. Virol. 84:29-39. [DOI] [PubMed] [Google Scholar]
- 42.Takagaki, Y., and J. L. Manley. 1998. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol. Cell 2:761-771. [DOI] [PubMed] [Google Scholar]
- 43.Takagaki, Y., and J. L. Manley. 1997. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 17:3907-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takagaki, Y., J. L. Manley, C. C. MacDonald, J. Wilusz, and T. Shenk. 1990. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 4:2112-2120. [DOI] [PubMed] [Google Scholar]
- 45.Takagaki, Y., R. L. Seipelt, M. L. Peterson, and J. L. Manley. 1996. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 87:941-952. [DOI] [PubMed] [Google Scholar]
- 46.Tenney, D. J., and A. M. Colberg-Poley. 1990. RNA analysis and isolation of cDNAs derived from the human cytomegalovirus immediate early region at 0.24 map units. Intervirology 31:203-214. [DOI] [PubMed] [Google Scholar]
- 47.Tenney, D. J., and A. M. Colberg-Poley. 1991. Expression of the human cytomegalovirus UL36-38 immediate early region during permissive infection. Virology 182:199-210. [DOI] [PubMed] [Google Scholar]
- 48.Tenney, D. J., and A. M. Colberg-Poley. 1991. Human cytomegalovirus UL36-38 and US3 immediate early genes: temporally regulated expression of nuclear, cytoplasmic, and polysome associated transcripts during infection. J. Virol. 65:6724-6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tenney, D. J., L. D. Santomenna, K. B. Goudie, and A. M. Colberg-Poley. 1993. The human cytomegalovirus US3 immediate early protein lacking the putative transmembrane domain regulates gene expression. Nucleic Acids Res. 21:2931-2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wahle, E., and W. Keller. 1996. The biochemistry of polyadenylation. Trends Biochem. Sci. 21:247-250. [PubMed] [Google Scholar]
- 51.Weiss, E. A., G. M. Gilmartin, and J. R. Nevins. 1991. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 10:215-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu, S., and M. R. Green. 1997. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 16:4421-4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu, S., C. M. Romfo, T. W. Nilsen, and M. R. Green. 1999. Functional recognition of the 3′ splice site AG by the splicing factor U2AF35. Nature 402:832-835. [DOI] [PubMed] [Google Scholar]
- 54.Xiao, S.-H., and J. L. Manley. 1998. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J. 17:6359-6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, J., L. Hyman, and C. Moore. 1999. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63:405-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou, Z., L. J. Licklider, S. P. Gygi, and R. Reed. 2002. Comprehensive proteomic analysis of the human spliceosome. Nature 419:182-185. [DOI] [PubMed] [Google Scholar]
- 57.Zhuang, Y., and A. M. Weiner. 1990. The conserved dinucleotide AG of the 3′ splice site may be recognized twice during in vitro splicing of mammalian mRNA precursors. Gene 90:263-269. [DOI] [PubMed] [Google Scholar]