Abstract
An earlier report showed that the expression of viral genes by a herpes simplex virus 1 mutant [HSV-1(vCPc0)] in which the wild-type, spliced gene encoding infected-cell protein no. 0 (ICP0) was replaced by a cDNA copy is dependent on both the cell type and multiplicity of infection. At low multiplicities of infection, viral gene expression in rabbit skin cells was delayed by many hours, although ultimately virus yield was comparable to that of the wild-type virus. This defect was rescued by replacement of the cDNA copy with the wild-type gene. To test the hypothesis that the delay reflected a dysfunction of ICP0 in altering the structure of host protein-viral DNA complexes, we examined the state of histone deacetylases (HDACs) (HDAC1, HDAC2, and HDAC3). We report the following. (i) HDAC1 and HDAC2, but not HDAC3, were modified in infected cells. The modification was mediated by the viral protein kinase US3 and occurred between 3 and 6 h after infection with wild-type virus but was delayed in rabbit skin cells infected with HSV-1(vCPc0) mutant, concordant with a delay in the expression of viral genes. (ii) Pretreatment of rabbit skin cells with inhibitors of HDAC activity (e.g., sodium butyrate, Helminthosporium carbonum toxin, or trichostatin A) accelerated the expression of HSV-1(vCPc0) but not that of wild-type virus. We conclude the following. (i) In the interval in which HSV-1(vCPc0) DNA is silent, its DNA is in chromatin-like structures amenable to modification by inhibitors of histone deacetylases. (ii) Expression of wild-type virus genes in these cells precluded the formation of DNA-protein structures that would be affected by either the HDACs or their inhibitors. (iii) Since the defect in HSV-1(vCPc0) maps to ICP0, the results suggest that this protein initiates the process of divestiture of viral DNA from tight chromatin structures but could be replaced by other viral proteins in cells infected with a large number of virions.
The infected-cell protein no. 0 (ICP0) of herpes simplex virus 1 (HSV-1) contains 775 amino acid residues. It is an α or immediate-early protein encoded in the three exons of the α0 gene. A ring finger is contained in the sequences encoded by exon 2 (reviewed in reference 47). The gene can be deleted, and at least in one cell line (U2-OS), the Δα0 mutant grows nearly as well as the wild-type virus (56). The phenotype of the α0 gene comprises two key features. In most cell lines, ICP0 null mutants grow sluggishly in a multiplicity-dependent fashion (49, 50). By itself, ICP0 transactivates genes introduced into cells by infection or transfection (9, 14, 37). Thus, at least one function of ICP0 is to enable efficient expression of viral genes. A second function of ICP0 emerged from observations that it mediates the proteasome-dependent degradation of sumoylated promyelocytic leukemia protein (PML), the degradation of several other (e.g., Sp100, CENP-C, CENP-A, DNA-dependent protein kinase, etc.) proteins and the dispersal of nuclear bodies known as PODs, ND10, etc. (5, 10-13, 27, 30, 31). The connection between the two apparent phenotypes of ICP0 emerged from two key observations. Thus, soon after initiation of viral gene expression, ICP0 localizes near the ND10 structures. Perhaps more important, all the viruses with mutations in the ring finger domain made thus far that disrupt the degradation of PML also failed to transactivate cotransfected genes. Thus, proximity of action and colocalization of the sites essential for these disparate functions support the hypothesis that degradation of ND10 and transactivation are at least covariant, if not genetically linked.
A more detailed analysis of the function expressed by ICP0 presents a much more complicated picture. Early studies revealed that ICP0 physically interacts with a large number of cellular proteins, including a transactivator known as BMAL1, cyclin D3, the elongation factor 1δ, and the ubiquitin-specific protease 7, and dynamically interacts, in a reversible manner, with proteasomes (13, 21-23, 32, 33, 54). Further analyses showed that ICP0 acts as a dual ubiquitin ligase. The HSV-1 ubiquitin-ligase site mapped in exon 3 (HUL-1) targets cdc34 for the proteasome-dependent degradation, whereas the ubiquitin-ligase site in exon 2 (HUL-2) mediates the proteasome-dependent degradation of PML and Sp100 (2, 17, 18, 52). The central question whether these two functions of ICP0 account for the phenotype of the gene does not have a satisfactory answer. For example, in cells infected with wild-type virus, ICP0 localizes initially in ND10 structures adjacent to or in proximity of PML. Subsequently, ICP0 spreads through the nucleus. After the onset of DNA synthesis or late gene expression, ICP0 is translocated into the cytoplasm. Overexpression of PML precludes the degradation of ND10 but has no effect either on the peregrinations of ICP0 during the infectious cycle or virus yield. Recent studies strongly suggest that ICP0 targets PML to preclude the inhibitory effect of exogenous interferon. Thus, wild-type or Δα0 viruses grow equally well in both PML−/− and PML+/+ cells (28, 29). However, PML+/+ cells pretreated with alpha interferon or gamma interferon become resistant to infection with either virus, whereas the susceptibility of PML−/− cells is practically unchanged (4). cdc34 is the ubiquitin conjugating enzyme involved in the turnover of cyclin D1. Both cyclin D1 and cyclin D3 are stabilized by wild-type virus but degraded by the ICP0 null mutant. HSV-1 interacts with cyclin D3 but not cyclin D1. Cyclin D3 plays a role in the translocation of ICP0 from nucleus to cytoplasm late in infection (53, 54). However, destruction of the binding site or deletion of key sequence responsible for the HUL-1 site decreases the yield or neurotoxicity of the virus by approximately 10- to 40-fold. Thus, the known functions encoded by the protein undoubtedly contribute to but do not fully account for the phenotype of the α0 gene.
In the course of a search for a model to explore the phenotype of α0 more fully, it was observed that a HSV-1 mutant [HSV-1(vCPc0)], in which the genomic version of the α0 gene was replaced with a cDNA copy, is in most respects similar to the wild-type virus. However, in rabbit skin cells (RSC), and to a lesser extent in HEp-2 cells, initiation of viral gene expression was delayed in a multiplicity-dependent fashion. Ultimately, even at a low multiplicity of infection, accumulation of viral proteins and virus yield caught up with that of the wild-type virus. The defect mapped to the α0 gene, inasmuch as restoration of the genomic version cured the defect. A biochemical event mapped to the silent interval involved a proteasome-dependent degradation inasmuch as MG132, an inhibitor of proteasomal degradation administered during this interval, further blocked viral gene expression. No such silent interval could be mapped to the wild-type virus (43).
One hypothesis concordant with this observation that we wanted to explore further is that upon entry into the nucleus, viral DNA is subjected to two competing events: silencing of viral genome as a consequence of association with histones and transcription of key viral genes required for efficient expression of viral genes. According to this scenario, a key, but not the sole determinant of this competition is ICP0. This hypothesis envisions that ICP0 made by the cDNA mutant [HSV-1(vCPc0)] is ineffective, either because a key spliced version of ICP0 is precluded from being synthesized or because a putative promoter element necessary for efficient expression of ICP0 in RSC is located in the first intron of the wild-type α0 gene. A key prediction of this hypothesis is that in RSC, inhibitors of deacetylation of histones would accelerate HSV-1(vCPc0) gene expression. We report here that this is in fact the case.
MATERIALS AND METHODS
Cells and viruses.
Vero, HeLa, HEp-2, and SK-N-SH cell lines were obtained from American Type Culture Collection, the human 143TK− cell line was obtained from Carlo Croce, RSC were originally obtained from J. McClaren, and the telomerase-transformed human foreskin fibroblast (HFF) cells were obtained from Thomas Shenk. The cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% (SK-N-SH and HFF) or 5% fetal bovine serum (HEp-2, 143TK−, and RSC) or 5% newborn calf serum (Vero and HeLa). HSV-1(F) and HSV-2(G) are the prototype HSV-1 and HSV-2 strains, respectively, used in this laboratory (8). Mutant viruses HSV-1(vCPc0), R7910 (Δα0), R2621 (ΔUL41), R7041(ΔUS3), R7356 (ΔUL13), R7353 (ΔUL13/ΔUS3), R7802 (Δα22), and repaired virus HSV-1(vCPc0)R were described elsewhere (23, 36, 38, 41-46). HSV-1(KOS) and HSV-1 (17) were obtained from P. A. Schaffer (Harvard, Boston, Mass.) and J. Subak Sharpe (Institute of Virology, Glasgow, United Kingdom), respectively.
Preparation of cell lysates, electrophoretic separation of proteins, and immunoblotting.
Replicate cell cultures in 25-cm2 flasks were either mock infected or infected with 5 PFU of virus per cell and maintained at 37°C in medium 199V consisting of a mixture of medium 199 supplemented with 1% calf serum. In some experiments, cells were exposed to HDAC inhibitors (6 mM sodium butyrate, 150 ng of trichostatin A [Sigma] per ml, or 70 ng of Helminthosporium carbonum toxin [Biomol] per ml) 11 h before infection, or 10 μM proteasome inhibitor MG132 (Biomol) at 3 h after infection. Cells were harvested at 18 to 24 h after infection, rinsed three times with phosphate-buffered saline containing protease inhibitor cocktail (Roche), and then solubilized in 200 μl of disruption buffer (50 mM Tris-HCl [pH 7], 2% sodium dodecyl sulfate, 710 mM β-mercaptoethanol, 3% sucrose). Fifty-microliter aliquots of lysates were boiled for 5 min, and the solubilized proteins were subjected to electrophoresis in an 11% denaturing polyacrylamide gel, transferred to a nitrocellulose sheet, blocked with 5% nonfat milk, allowed to react first with primary antibody and then with an appropriate secondary antibody conjugated to alkaline phosphatase (Bio-Rad), and visualized according to the manufacturer's instructions.
Antibodies.
Monoclonal antibodies to ICP0 and ICP4 were purchased from the Goodwin Cancer Research Institute (Plantation, Fla.). The mouse monoclonal antibody against US11 and rabbit polyclonal antibodies W2 (against the carboxyl-terminal region of ICP22) were described previously (1, 26, 48). Rabbit polyclonal antibodies against histone deacetylases (HDACs) (HDAC1, HDAC2, and HDAC3) were purchased from Sigma.
RESULTS
HDAC1 and HDAC2 are posttranslationally processed in HSV-1(F)-infected cells.
In this series of experiments, we examined the electrophoretic mobility of HDAC1, HDAC2, and HDAC3 in cells infected with HSV-1(F). Replicate cultures of SK-N-SH, Vero, RSC, 143TK−, HFF, HEp-2, or HeLa cells were harvested at 3 and either 18 or 24 h after infection with HSV-1(F), processed as described in Materials and Methods, and reacted with antibodies to the specific HDAC proteins. The results shown in Fig. 1 were as follows.
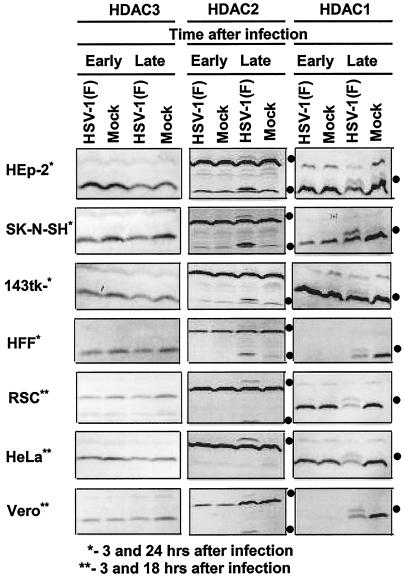
FIG. 1.
Electrophoretic profiles of HDAC1, HDAC2, and HDAC3 in wild-type HSV-1(F) virus-infected cells at early and late times after infection. Replicate cultures of HEp2, SK-N-SH, 143TK−, HFF, RSC, HeLa, or Vero cells in 25-cm2 flasks were either mock infected or infected with 5 PFU of HSV-1(F) per cell. The cells were harvested at early (3 h) or late (18 h [**] or 24 h [*]) times after infection. Proteins were solubilized in disruption buffer and electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibodies to HDAC1, HDAC2, and HDAC3 as described in Materials and Methods. The positions of posttranslationally modified protein bands are indicated by the solid circles to the right of the blots.
(i) There was no change in the electrophoretic mobility of HDAC3 in any of the infected cell lines tested. Additional bands of proteins reactive with specific HDAC1 and HDAC2 antisera were apparent in the electrophoretically separated lysates of HSV-1(F)-infected cells harvested late in infection.
(ii) The anti-HDAC1 antibody reacted with two proteins (apparent Mrs of 68,000 and 66,000). Whereas the 66,000-Mr protein was present in the lysates of both mock- and HSV-1(F)-infected cells, the 68,000-Mr protein could be detected only in the lysates of HSV-1(F)-infected cells harvested late in infection. A similar and in some instances slightly faster-migrating band was also present in lysates of cells harvested late after mock infection.
(iii) The anti-HDAC2 antibody reacted with three protein bands in HSV-1(F)-infected cells. The most rapidly migrating band (apparent Mr of 46,000) appears to be a cross-reacting viral protein inasmuch as a similar, but faster-migrating band (apparent Mr of 45,000) was detected in lysates of cells infected with HSV-2(G) (Fig. 2, band V). The slowest-migrating band was detected in lysates of virtually all HSV-1(F)-infected cells harvested at late but not at early times after infection.
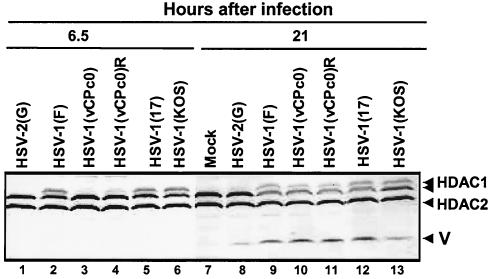
FIG. 2.
Electrophoretic profiles of HDAC1 and HDAC2 in RSC infected with wild-type and mutant viruses at early and late times after infection. Replicate cultures of RSC in 25-cm2 flasks were either mock infected (lane 7) or infected with wild-type HSV-1(F), HSV-1 (17), HSV-1(KOS), or HSV-2(G), mutant HSV-1(vCPc0), or repaired HSV-1(vCPc0)R. Cell cultures were infected with 10 PFU of virus per cell. Cells were harvested at 6.5 h (lanes 1 to 6) or 21 h (lanes 7 to 13) after infection. Proteins were solubilized in disruption buffer and electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibodies to HDAC1 and HDAC2 as described in Materials and Methods. The positions of HDAC1 and HDAC2 and of bands reacting with HDAC2 antibody (V) are indicated to the right of the gel.
In these experiments, a reduction in the accumulation of HDAC1, but not HDAC2 or HDAC3, was apparent in all HSV-1(F)-infected cells harvested late in infection.
Effects of various HSV strains on the modification of HDAC1 and HDAC2.
The primary objective of these studies was to determine whether the deacetylase enzymes were responsible for the delay in the expression of HSV-1(vCPc0) mutant in RSC. Specifically, the experiments described in this section were performed to determine whether HDAC1 and HDAC2 were also posttranslationally modified in cells infected with HSV-1(vCPc0). In light of the mixed parentage of HSV-1(vCPc0) mutant, we included both HSV-1(KOS) and HSV-1 (17) and the repaired virus HSV-1(vCPc0)R. In these experiments, replicate RSC cultures were harvested at 6.5 or 21 h after infection with various viruses (10 PFU of virus per cell). The harvested cells were processed as described above. The results were as follows.
(i) Lysates of cells infected with HSV-2(G) and probed with anti-HDAC1 antibody could not be differentiated from those of mock-infected cells (Fig. 2, lanes 1, 7, and 8). The anti-HDAC2 antibody reacted with a band (V) migrating slightly faster (Mr of 45,000) than the corresponding band formed in lysates of HSV-1(F)-infected cells (Fig. 2, lanes 8 and 9). This result was reproducible in other experiments and suggests that the fast-migrating band reacting with anti-HDAC2 antibody may be of viral origin.
(ii) The slow-migrating HDAC1 band (Mr of 68,000) was present in all wild-type HSV-1-infected cell lysates other than those infected with HSV-2(G). Since this band was absent in lysates of RSC harvested at 3 h after infection (Fig. 1), the results suggest that the modification took place between 3 and 6.5 h after infection. This band was absent from lysates of RSC harvested 6.5 h after infection with the HSV-1(vCPc0) mutant but was present in lysates of cells harvested at late times (Fig. 2, lanes 3 and 10). This finding is consistent with the delay in the initiation of expression of viral genes in RSC infected with this mutant.
Effects of selected HSV-1 mutants on the processing of HDAC1.
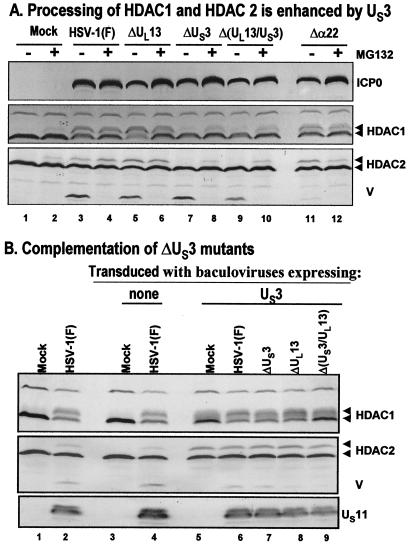
The objective of these studies was to determine whether the posttranslational modification of HDAC1 was mediated by the protein kinases encoded by US3 and UL13 open reading frames, respectively. In the first series of experiments, replicate cultures of RSC in 25-cm2 flasks were either mock infected or infected with wild-type HSV-1(F) or mutant virus R7356 (ΔUL13), R7041 (ΔUS3), R7353 (ΔUL13/ΔUS3), or R7802 (Δα22) (5 PFU of virus per cell). At 3 h after infection, one set of cultures were replenished with fresh untreated medium (Fig. 3A, − lanes), whereas a second set was replenished with medium containing 10 μM MG132 (+ lanes). The cells were harvested at 20 h after infection and solubilized in disruption buffer, and the lysates were subjected to electrophoresis in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to HDAC1 or HDAC2 and monoclonal antibody to ICP0. The Δα22 mutant was included inasmuch as earlier studies have shown that in some instances posttranslational modifications mediated by UL13 protein kinase required functional ICP22 protein (45, 46). The results shown in Fig. 3A were as follows. The 66,000-Mr HDAC1 band was present in lysates of all infected cells. The 68,000-Mr band was present in all lysates of cells treated with MG132 after mock infection or infection with either wild-type or mutant virus. This band was absent from untreated mock-infected cells and was present in much reduced amounts in untreated cells infected with mutants lacking the US3 gene (Fig. 3A, lanes 7 and 9). One hypothesis that could explain these data is that the 68,000-Mr band contained two distinct products. The processing of one product is mediated by US3 protein kinase. The other product resulting from a posttranslational modification through another pathway accumulated only in the presence of the proteasomal inhibitor. A corollary of this interpretation of the data is that the modification induced by US3 protein kinase is not subject to degradation by proteasomal pathway. Similar results were obtained with antibody directed against HDAC2.
FIG. 3.
Modification of HDAC1 and HDAC2. (A) Processing of HDAC1 and HDAC2 is enhanced by viral protein kinase US3. Replicate cultures of RSC in 25-cm2 flasks were either mock infected or infected with wild-type HSV-1(F) or mutant R7356 (ΔUL13), R7041 (ΔUS3), R7353 (ΔUL13/ΔUS3), or R7802 (Δα22). Cell cultures were infected with 5 PFU of virus per cell. At 3 h after infection, one set of cultures was replenished with fresh untreated medium (− lanes), whereas a second set was replenished with medium containing 10 μM MG132 (+ lanes). The cells were harvested 20 h after infection. Proteins were solubilized in disruption buffer, electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibodies to HDAC1 and HDAC2, and monoclonal antibody to ICP0. The positions of HDAC1 and HDAC2 and of bands reacting with HDAC2 antibody (V) and with monoclonal antibody to ICP0 are indicated to the right of the gel. (B) Complementation of ΔUS3 mutants by baculovirus expressing US3. Replicate RSC cultures in 25-cm2 flasks were transduced with baculoviruses that were either empty of inserts (lanes 3 and 4) or encoding the US3 protein kinase (lanes 5 to 9). At 12 h after transduction, cells were mock infected (lanes 3 and 5) or infected with wild-type HSV-1(F) (lanes 4 and 6) or mutant virus R7356 (ΔUL13), R7041 (ΔUS3), or R7353 (ΔUL13/ΔUS3) (lanes 7 to 9). Cell cultures were infected with 5 PFU of virus per cell. At 12 h after infection with wild-type HSV-1 or mutant viruses, the cultures were harvested and processed as described above except that cell lysates were reacted with anti-US11 antibody rather than ICP0. Lanes 1 and 2 show protein profiles of mock-infected and wild-type virus-infected cell cultures not previously exposed to baculoviruses.
In other studies, we tested mutants with deletions in γ134.5 and UL41. In RSC infected with these mutants, the processing of HDAC1 could not be differentiated from that of wild-type virus-infected cells (data not shown).
In the second series of experiments, replicate RSC cultures in 25-cm2 flasks were transduced as described previously (43) with baculoviruses that were either empty of inserts or encoding the US3 protein kinase. At 12 h after transduction, cells were mock infected or infected with wild-type HSV-1(F) or mutant virus R7356 (ΔUL13), R7041 (ΔUS3), or R7353 (ΔUL13/ΔUS3) (5 PFU of virus per cell). At 12 h after infection with wild-type HSV-1 or mutant viruses, the cultures were harvested and processed as described above except that cell lysates were reacted with anti-US11 antibody, rather than ICP0, as evidence of infection and accumulation of viral gene products. The results shown in Fig. 3B indicate that the US3 protein kinase expressed in baculovirus-transduced cells could mediate the modification of HDAC1 and HDAC2 (compare lanes 3 and 5) and complemented ΔUS3-infected cells (lanes 7 and 9) to enable the accumulation of the slow-migrating HDAC1 and HDAC2 protein bands. These results are consistent with the hypothesis that the US3 protein kinase mediates a posttranslational modification of HDAC1 and HDAC2.
Inhibitors of HDACs accelerate the expression of HSV-1(vCPc0) mutant but not that of wild-type virus in infected RSC.
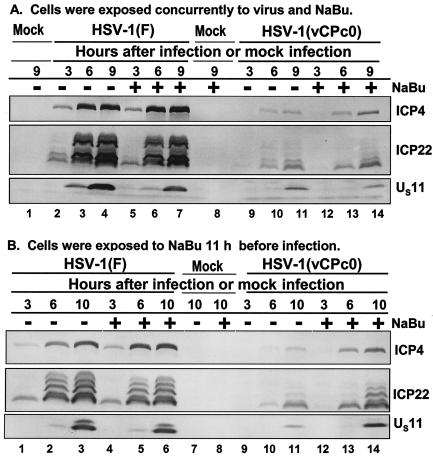
Several drugs and notably sodium butyrate, trichostatin A, and H. carbonum toxin inhibit the deacetylation of histones. If the delay in the expression of HSV-1(vCPc0) virus were due to temporary deacetylation of a chromatin-like structure, it could be expected that these drugs would accelerate viral gene expression in RSC infected with this virus. To test this hypothesis, two series of experiments were done. Replicate cultures of RSC in 25-cm2 flasks were either mock infected or infected with wild-type HSV-1(F) or mutant HSV-1(vCPc0) (5 PFU of virus per cell). In the experiment shown in Fig. 4A, a set of replicate cultures was exposed to 6 mM sodium butyrate at the time of infection. In Fig. 4B, the cells were exposed to the drug 11 h before infection and the exposure to drug was continued throughout the incubation period. The cells were harvested at the times indicated in Fig. 4. The results were as follows. Exposure of cells to sodium butyrate at the time of infection had no effect on the pattern of gene expression of either HSV-1(F) or HSV-1(vCPc0) virus in RSC (Fig. 4A). In contrast, exposure of RSC to sodium butyrate before infection accelerated the expression of ICP4 and ICP22 (shown) and ICP0 (not shown) by approximately 3 h (Fig. 4B). In this instance again, exposure of cells to sodium butyrate had no effect on the expression of these genes in cells infected with HSV-1(F).
FIG. 4.
Temporal pattern of accumulation of selected wild-type HSV-1(F) and mutant HSV-1(vCPc0) proteins in RSC treated with sodium butyrate (NaBu) (A) at the time of infection and (B) 11 h before infection. (A) Replicate cultures of RSC in 25-cm2 flasks were either mock infected (lanes 1 and 8) or infected with 5 PFU of wild-type HSV-1(F) per cell (lanes 2 to 7) or with 5 PFU of mutant HSV-1(vCPc0) per cell (lanes 9 to 14) in the absence (−) or presence (+) of 6 mM sodium butyrate. The cells were harvested at the indicated times after infection. Proteins were solubilized in disruption buffer, electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to ICP22 and monoclonal antibodies to ICP4 and US11. (B) Replicate cultures of RSC in 25-cm2 flasks were either mock treated (lanes 1 to 3, 7, and 9 to 11) or treated (lanes 4 to 6, 8, and 12 to 14) with 6 mM sodium butyrate. At 11 h after treatment, cells were either mock infected (lanes 7 and 8) or infected with 5 PFU of wild-type HSV-1(F) per cell (lanes 1 to 6) or with 5 PFU of mutant HSV-1(vCPc0) per cell (lanes 9 to 14) in the absence (−) or presence (+) of 6 mM sodium butyrate. Cells were harvested at the indicated times after infection. Proteins were solubilized in disruption buffer, electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to ICP22 and monoclonal antibodies to ICP4 and US11.
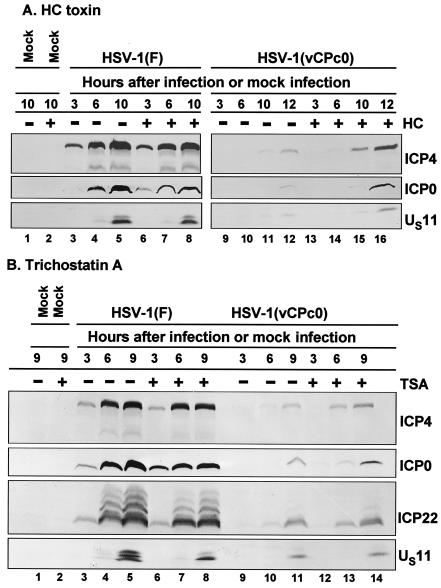
The experiments described above were repeated with H. carbonum toxin (70 ng/ml of medium) and trichostatin A (150 ng/ml of medium) except that the RSC were exposed to drug 11 h before infection. The results shown in Fig. 5 were consistent with those shown in Fig. 4 in that the expression of HSV-1(vCPc0) genes was accelerated in cells exposed to the drug (Fig. 5A, lanes 9 to 16, and Fig. 5B, lanes 9 to 14). H. carbonum toxin and trichostatin A had no effect on the gene expression of the wild-type virus (Fig. 5, lanes 3 to 8).
FIG. 5.
Temporal pattern of accumulation of selected wild-type HSV-1(F) and mutant HSV-1(vCPc0) proteins in RSC treated with H. carbonum (HC) toxin (A) and trichostatin A (B) 11 h before infection. (A) Replicate cultures of RSC in 25-cm2 flasks were either mock treated (−) or treated (+) with 70 ng of H. carbonum toxin per ml. At 11 h after treatment, cells were either mock infected (lanes 1 and 2) or infected with 5 PFU of wild-type HSV-1(F) per cell (lanes 3 to 8) or with 5 PFU of mutant HSV-1(vCPc0) per cell (lanes 9 to 16) in the absence (−) or presence (+) of 70 ng of H. carbonum toxin per ml. Cells were harvested at the indicated times after infection. Proteins were solubilized in disruption buffer, electrophoretically separated in 11% denaturing polyacrylamide gels, transferred to nitrocellulose sheets, blocked with 5% nonfat milk, and reacted with polyclonal antibody to ICP22 and monoclonal antibodies to ICP0, ICP4, and US11. (B) The experiment was repeated with 150 ng of trichostatin A (TSA) per ml, instead of H. carbonum toxin.
DISCUSSION
The structure and composition of HSV capsids preclude the presence of large amounts of viral proteins bound to viral DNA. Capsids do contain large amounts of spermine, sufficient to neutralize at least 40% of the viral DNA (15). Entering viral DNA could therefore be considered quasinaked and could be expected to become bound by cellular proteins. In most of the lytic infection models in cell culture or acute infection models in mice, nucleosome-like structures containing viral DNA sequences were barely detected (20, 25, 34). Nucleosome-like structures containing viral DNA sequences, including those of actively transcribed regions, were detected in brain stems with latent infections (7). These observations, however, do not exclude the possibility that viral DNA is bound by histones immediately after entry into the nucleus. Tests for nucleosome-like structures early after infection of permissive cells with wild-type viruses have not been successful, and one explanation for the failure is that rapid viral gene expression alters the conformation of the genome and precludes stable association with cellular proteins. The delay in viral gene expression observed in RSC infected with the mutant HSV-1(vCPc0) encoding the cDNA copy of the α0 gene appeared to be a more promising system for testing this hypothesis, particularly since the virus yield, infectivity, and overall behavior of the mutant could not be differentiated from that of wild-type virus in many different cell lines. The problem encountered in these assays was that the detection of nucleosome-like structures is not very efficient. At a low multiplicity of infection in which the delay in initiation of gene expression was sufficiently long to allow the formation of such structures, viral DNA was virtually undetectable in nucleosomes containing cellular DNA by Southern blotting of partial micrococcal nuclease digests. Although viral DNA was detected by PCR in nucleosomal structures in a chromatin immunoprecipitation assay using anti-acetylated histone 4 antibodies, we could not exclude contamination of nucleosomes with naked viral DNA (Y. Liang and B. Roizman, unpublished results). At high multiplicities of infection, the delay in initiation of gene expression was too short to be fully exploited. An alternative test of this hypothesis is described in this report. If entering viral DNA is bound by histones in chromatin-like structures, it could be expected that the transcription corepressor complexes which recruit HDACs would repress gene expression, whereas viral protein would alter the structure of viral DNA-protein complexes to enable efficient transcription of viral genes (39, 55). The results presented in this report are consistent with this hypothesis. The specific results are discussed below.
(i) Of the three HDAC enzymes examined in this report, HDAC1 and HDAC2 appear to undergo a posttranslational modification sometime between 3 and 6 h after infection of several different cell lines with wild-type virus. The time interval is based on the comparison of the results presented in Fig. 1 and 2 and suggests the possibility that the modification is mediated by a post-immediate-early gene expression. In RSC infected with HSV-1(vCPc0), the posttranslational modification of HDAC1 and HDAC2 was observed late in infection but not at early times, which is consistent with the observed delay in the initiation of HSV-1(vCPc0) gene expression. Of the HSV-1 mutants tested, only mutants lacking the US3 gene decreased but did not abolish the modification of HDAC1 and HDAC2. Although the modification correlates with viral gene expression, the data are not sufficient to conclude that the modification is the consequence of a physical interaction between HDAC1 or HDAC2 and a viral protein. We should note that an extensive literature indicates that the activity of HDAC1 and HDAC2 is regulated by Sumo-1 modification and by phosphorylation by cyclic AMP-dependent kinase and casein kinase 2 (6, 40, 51).
(ii) The inhibitors of histone deacetylases, sodium butyrate, trichostatin A, and the toxin derived from H. carbonum, each accelerated the expression of viral genes encoded by HSV-1(vCPc0) mutant. Neither of these drugs had an effect on the expression of wild-type virus in RSC. In the case of sodium butyrate, the accelerated expression was observed in cells pretreated with the drug but not in cells exposed to virus and drug at the same time. These results are consistent with the hypothesis that viral DNA entering the nucleus, like any nuclear DNA, would be expected to become associated with cellular proteins and assembled in nucleosome-like structures. Such structures could potentially become silenced or undergo only a basal level of transcription in the absence of potent activator capable of inducing the expression of other genes essential for a global transcription of the DNA. ICP0 appears to be this initiator. This conclusion is based on two lines of evidence. First, this property is consistent with the known phenotype of wild-type ICP0 described in the introduction. Second, the defect in HSV-1(vCPc0) maps to and is rescued by the genomic version of ICP0. The results suggest that in wild-type virus-infected RSC and other cell lines, functions encoded in ICP0 are expressed so rapidly after infection as to preclude an effective aggregation of DNA with cellular proteins. Thus, the inhibitors of histone deacetylation had no discernible effect on wild-type virus gene expression. In the absence of ICP0, the progression of viral gene expression is dependent on the accumulation of viral gene products resulting from low-level expression of a large number of viral genes introduced into the cell by high ratios of virions per cell.
The notion that ICP0 has an additional, unmapped function that affects the interaction of viral DNA with cellular proteins and in particular with histone-modifying enzymes resonates well with other observations on the role of these enzymes in the regulation of herpesviral gene expression. Thus, Murphy et al. (35) reported that inhibition of HDACs enabled expression of human cytomegalovirus immediate-early promoter in nonpermissive cells. Repression by HDACs have been implicated in the maintenance of Epstein-Barr virus and of human herpesvirus 8 expression (3, 16). More relevant here, Zhang and Jones (57) reported that the bovine homolog of ICP0 associates with HDAC1 to activate transcription. Our attempts to demonstrate direct interaction between ICP0 and HDAC1 or HDAC2 have not been successful (Liang and Roizman, unpublished). The failure to observe a direct interaction does not exclude the possibility that ICP0 interacts with HDAC1 or with other members of the protein complexes involved in the regulation of chromatin structure.
The RSC model for the study of HSV gene expression has implications for the mechanism by which latent HSV infections are established and maintained. Thus, delay in the expression of viral genes because of the absence of specific host factors in sensory neurons may lead to the establishment of chromatin-like structures described elsewhere (7, 34) and silencing of viral gene expression. Reactivation of latency in this model would require the transient availability of specific transcriptional factors (e.g., HCF-1 and Oct-1 [24]) and histone acetylation accelerated by newly made ICP0. This model is similar to that described by Hsia and Shi (19) for the disruption of chromatin structure and acetylation of histones bound to long terminal repeats of human immunodeficiency virus type 1 by thyroid hormone receptors.
Acknowledgments
This study was supported in part by grants from the National Cancer Institute (CA87661, CA83939, CA71933, CA78766, and CA88860) of the U.S. Public Health Service.
REFERENCES
- 1.Ackermann, M., M. Sarmiento, and B. Roizman. 1985. Application of antibody to synthetic peptides for characterization of the intact and truncated α22 protein specified by herpes simplex virus 1 and the R325 α22− deletion mutant. J. Virol. 56:207-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutell, C., S. Sadis, and R. D. Everett. 2002. Herpes simplex virus type 1 immediate-early protein ICP0 and its isolated RING finger domain act as ubiquitin E3 ligases in vitro. J. Virol. 76:841-850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryant, H., and P. J. Farrell. 2002. Signal transduction and transcription factor modification during reactivation of Epstein-Barr virus from latency. J. Virol. 76:10290-10298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chee, A. V., P. Lopez, P. P. Pandolfi, and B. Roizman. 2003. PML mediates interferon-based anti-herpes simplex virus 1 effects. J. Virol. 77:7101-7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chelbi-Alix, M. K., and H. de The. 1999. Herpes virus induced proteasome-dependent degradation of the nuclear body-associated PML and Sp100 proteins. Oncogene 18:935-941. [DOI] [PubMed] [Google Scholar]
- 6.David, G., M. A. Neptune, and R. A. DePinho. 2002. SUMO-1 modification of histone deacetylase 1 (HDAC1) modulates its biological activities. J. Biol. Chem. 277:23658-23663. [DOI] [PubMed] [Google Scholar]
- 7.Deshmane, S. L., and N. W. Fraser. 1989. During latency, herpes simplex virus type 1 DNA is associated with nucleosomes in a chromatin structure. J. Virol. 63:943-947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ejercito, P., E. D. Kieff, and B. Roizman. 1968. Characterization of herpes simplex virus strains differing in their effects on social behavior of infected cells. J. Gen. Virol. 2:357-364. [DOI] [PubMed] [Google Scholar]
- 9.Everett, R. D. 1984. Transactivation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 3:3135-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Everett, R. D., W. C. Earnshaw, J. Findlay, and P. Lomonte. 1999. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 18:1526-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Everett, R. D., P. Freemont, P., H. Saitoh, M. Dasso, A. Orr, M. Kathoria, and J. Parkinson. 1998. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J. Virol. 72:6581-6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Everett, R. D., and G. G. Maul. 1994. HSV-1 IE protein Vmw 110 causes redistribution of PML. EMBO J. 13:5062-5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett, R. D., M. Meredith, A. Orr, A. Cross, M. Kathoria, and J. Parkinson. 1997. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 16:566-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gelman, I. H., and S. Silverstein. 1985. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc. Natl. Acad. Sci. USA 82:5265-5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gibson, W., and B. Roizman. 1971. Compartmentalization of spermine and spermidine in the herpes simplex virion. Proc. Natl. Acad. Sci. USA 68:2818-2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gwack, Y., H. Byan, S. Hwang, C. Lim, and J. Cloe. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 60. J. Virol. 75:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagglund, R., and B. Roizman. 2002. Characterization of the novel E3 ubiquitin ligase encoded in exon 3 of herpes simplex virus-1-infected cell protein 0. Proc. Natl. Acad. Sci. USA 99:7889-7894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hagglund, R., C. Van Sant, P. Lopez, and B. Roizman. 2002. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin conjugating enzymes. Proc. Natl. Acad. Sci. USA 99:631-666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsia, S.-C. V., and Y.-B. Shi. 2002. Chromatin disruption and histone acetylation in regulation of the human immunodeficiency virus type 1 long terminal repeat by thyroid hormone receptor. Mol. Cell. Biol. 22:4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jamieson, D. R., L. H. Robinson, J. I. Daksis, M. J. Nicholl, and C. M. Preston. 1985. Quiescent viral genomes in human fibroblasts after infection with herpes simplex virus type 1 Vmw65 mutants. J. Gen. Virol. 76:1417-1431. [DOI] [PubMed] [Google Scholar]
- 21.Kawaguchi, Y., R. Bruni, and B. Roizman. 1997. Interaction of herpes simplex virus 1 α regulatory protein ICP0 with elongation factor 1δ: ICP0 affects translational machinery. J. Virol. 71:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kawaguchi, Y., M. Tanaka, A. Yokoymama, G. Matsuda, K. Kato, H. Kagawa, K. Hirai, and B. Roizman. 2001. Herpes simplex virus 1 α regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc. Natl. Acad. Sci. USA 98:1877-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawaguchi, Y., C. Van Sant, and B. Roizman. 1997. Herpes simplex virus 1 α regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J. Virol. 71:7328-7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kristie, T. M., J. L. Vogel, and A. E. Sears. 1999. Nuclear localization of the C1 factor (HCF) in sensory neurons correlates with reactivation of HSV from latency. Proc. Natl. Acad. Sci. USA 96:1229-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leinbach, S. S., and W. C. Summers. 1980. The structure of herpes simplex virus type 1 DNA as probed by micrococcal nuclease digestion. J. Gen. Virol. 51:45-59. [DOI] [PubMed] [Google Scholar]
- 26.Leopardi, R., P. L. Ward, W. O. Ogle, and B. Roizman. 1997. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J. Virol. 71:1133-1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lomonte, P., K. F. Sullivan, and R. D. Everett. 2001. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J. Biol. Chem. 276:5829-5835. [DOI] [PubMed] [Google Scholar]
- 28.Lopez, P., R. J. Jacob, and B. Roizman. 2002. Overexpression of promyelocytic leukemia protein precludes the dispersal of ND10 structures and has no effect on accumulation of infectious herpes simplex virus 1 or its proteins. J. Virol. 76:9355-9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lopez, P., C. Van Sant, and B. Roizman. 2001. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J. Virol. 75:3832-3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maul, G. G., and R. D. Everett. 1994. The nuclear location of PML, a cellular member of the C3HC4 zinc-binding domain protein family, is rearranged during herpes simplex virus infection by the C3HC4 viral protein ICP0. J. Gen. Virol. 75:1223-1233. [DOI] [PubMed] [Google Scholar]
- 31.Maul, G. G., H. H. Guldner, and J. G. Spivack. 1993. Modification of discrete nuclear domains induced by herpes simplex virus type-1 immediate early gene-1 product (ICP0). J. Gen. Virol. 74:2679-2690. [DOI] [PubMed] [Google Scholar]
- 32.Meredith, M., A. Orr, M. Elliott, and R. D. Everett. 1995. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology 209:174-187. [DOI] [PubMed] [Google Scholar]
- 33.Meredith, M., A. Orr, and R. D. Everett. 1994. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology 200:457-469. [DOI] [PubMed] [Google Scholar]
- 34.Muggeridge, M. I., and N. W. Fraser. 1986. Chromosomal organization of the herpes simplex virus genome during acute infection of the mouse central nervous system. J. Virol. 59:764-767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, J. C., W. Fischle, E. Verdin, and J. H. Sinclair. 2002. Control of cytomegalovirus lytic gene expression by histone acetylation. EMBO J. 21:1112-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogle, W. O., and B. Roizman. 1999. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J. Virol. 73:4305-4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Hare, P., and G. S. Hayward. 1985. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J. Virol. 53:751-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Panagiotidis, C. A., E. K. Lium, and S. J. Silverstein. 1997. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J. Virol. 71:1547-1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pazin, M. J., and J. T. Kadonaga. 1997. What's up and down with histone deacetylation and transcription? Cell 89:325-328. [DOI] [PubMed] [Google Scholar]
- 40.Pflum, M. K. H., J. K. Tong, W. S. Lane, and S. L. Schreiber. 2001. Histone deacetylase 1 phosphorylation promotes enzymatic activity and complex formation. J. Biol. Chem. 276:47733-47741. [DOI] [PubMed] [Google Scholar]
- 41.Poon, A. P. W., W. O. Ogle, and B. Roizman. 2000. Posttranslational processing of infected-cell protein 22 mediated by viral protein kinases is sensitive to amino acid substitutions at distant sites and can be cell type specific. J. Virol. 74:11210-11214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Poon, A. P. W., and B. Roizman. 1997. Differentiation of the shutoff of protein synthesis by virion host shutoff and mutant γ134.5 genes of herpes simplex virus 1. Virology 229:98-105. [DOI] [PubMed] [Google Scholar]
- 43.Poon, A. P. W., S. J. Silverstein., and B. Roizman. 2002. An early regulatory function required in cell-type-dependent manner is expressed by the genomic but not by the cDNA copy of the herpes simplex virus 1 gene encoding the infected-cell protein no. 0. J. Virol. 76:9744-9755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Purves, F. C., R. M. Longnecker, D. P. Leader, and B. Roizman. 1987. Herpes simplex virus 1 protein kinase is encoded by open reading frame US3 which is not essential for virus growth in cell culture. J. Virol. 61:2896-2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Purves, F. C., W. O. Ogle, and B. Roizman. 1993. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc. Natl. Acad. Sci. USA 90:6701-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Purves, F. C., and B. Roizman. 1992. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc. Natl. Acad. Sci. USA 89:7310-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roizman, B., and D. M. Knipe. 2001. The replication of herpes simplex viruses, p. 2399-2459. In D. M. Knipe, P. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed. Lippincott-Williams and Wilkins, New York, N.Y.
- 48.Roller, R. J., and B. Roizman. 1992. The herpes simplex virus 1 RNA binding protein US11 is a virion component and associates with ribosomal 60S subunits. J. Virol. 66:3624-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sacks, W. R., and P. A. Schaffer. 1987. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J. Virol. 61:829-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stow, N. D., and E. C. Stow. 1986. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J. Gen. Virol. 67:2571-2585. [DOI] [PubMed] [Google Scholar]
- 51.Tsai, S.-C., and E. Seto. 2002. Regulation of histone deacetylase 2 by protein kinase CK2. J. Biol. Chem. 277:31826-31833. [DOI] [PubMed] [Google Scholar]
- 52.Van Sant, C., R. Hagglund, P. Lopez, and B. Roizman. 2001. The infected cell protein 0 of herpes simplex virus 1 dynamically interacts with proteasomes, binds and activates the cdc34 E2 ubiquitin conjugating enzyme and possesses in vitro E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. USA 98:8815-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van Sant, C., Y. Kawaguchi, and B. Roizman. 1999. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc. Natl. Acad. Sci. USA 96:8184-8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Sant, C., P. Lopez, S. J. Advani, and B. Roizman. 2001. Role of cyclin D3 in the biology of herpes simplex virus 1 ICP0. J. Virol. 75:1888-1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolffe, A. P. 1997. Transcriptional control. Sinful repression. Nature 387:16-17. [DOI] [PubMed] [Google Scholar]
- 56.Yao, F., and P. A. Schaffer. 1995. An activity specified by the osteosarcoma line U2OS can substitute functionally for ICP0, a major regulatory protein of herpes simplex virus type 1. J. Virol. 69:6249-6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y., and C. Jones. 2001. The bovine herpesvirus 1 immediate-early protein (bICP0) associates with histone deacetylase 1 to activate transcription. J. Virol. 75:9571-9578. [DOI] [PMC free article] [PubMed] [Google Scholar]