Abstract
Late in infection, parvovirus minute virus of mice (MVMp) induces the lysis of mouse A9 fibroblasts. This effect depends on the large nonstructural phosphoprotein NS1, which plays in addition a major role in viral DNA replication and progeny particle production. Since the NS1 C-terminal region is subjected to late phosphorylation events and protein kinase C (PKC) family members regulate NS1 replicative activities, the present study was conducted to determine the impact of PKCs on NS1 cytotoxic functions. To this end, we performed site-directed mutagenesis, substituting alanine residues for two consensus PKC-phosphorylation sites located within the NS1 C-terminal region, T585 and S588. Although these substitutions had no detectable effect on virus multiplication in a single-round infection, the NS1-585A mutant virus was significantly less toxic to A9 cells than wild-type MVMp, whereas the NS1-588A mutant virus was endowed with a higher killing potential. These alterations correlated with specific changes in the late phosphorylation pattern of the mutant NS1 proteins compared to the wild-type polypeptide. Since the mutations introduced in this region of the viral genome also made changes in the minor nonstructural protein NS2, a contribution of this polypeptide to the above-mentioned phenotypes of mutant viruses cannot be excluded at present. However, the involvement of NS1 in these phenotypes was directly supported by the respective reduced and enhanced capacity of NS1-585A and NS1-588A recombinant proteins for inducing morphological alterations and cell detachment in transfected A9 cultures. Altogether, these data suggest that late-occurring phosphorylation of NS1 specifically regulates the cytotoxic functions of the viral product and that residues T585 and S588 contribute to this control in an antagonistic way.
The autonomous parvovirus minute virus of mice (MVMp) is a small icosahedral particle with a linear single-stranded DNA (ssDNA) of 5.1 kb as a genome. MVMp replication depends on host cell factors specifically present in S phase, resulting in a restriction of its propagation in proliferating tissues. This feature is likely to contribute to the oncotropism displayed by this virus, although additional factors appear to be required (51). The viral DNA comprises two overlapping transcription units. The right-hand unit encodes two capsid proteins, VP1 and VP2, which are expressed under the control of the P38 promoter, while the left-hand unit, driven by the P4 promoter, produces the two nonstructural proteins NS1 and NS2. The large nonstructural protein NS1 is endowed with multiple functions necessary for progeny virus production, while the small nonstructural proteins NS2, expressed in three distinct isoforms according to their unique C termini, are dispensable for MVMp propagation in nonmurine cell types (15, 35).
The functions and regulation of the NS1 protein have been investigated in great detail. This protein combines multiple enzymatic and nonenzymatic functions, such as ATP binding and hydrolysis (6, 56), homo-oligomerization (43, 46), site-specific binding to a cognate DNA recognition motif (10), DNA unwinding (40, 56), site-specific endonuclease activity (40), promoter trans regulation (27), and specific interactions with a number of cellular proteins (7, 17, 23, 25, 57). As a result of these properties, NS1 is involved in a variety of processes during virus propagation, ranging from viral DNA replication (12, 13, 30, 39, 52) and regulation of the expression of homo- and heterologous genes (28, 49, 53) to the induction of cytopathic effects (1, 5, 8, 34, 44, 54). The multifunctional character of NS1 led to the suggestion that posttranslational modifications such as phosphorylation might coordinate the different activities of this protein (9). In keeping with this view, it was shown that phosphorylation plays an essential role in the regulation of distinct properties of NS1 (18, 37, 38, 42), enabling the polypeptide to initiate viral DNA amplification (26, 41, 42). Using NS1 proteins modified by site-directed mutagenesis in various in vitro and in vivo assays, it was possible to assign at least part of this regulation to two members of the protein kinase C (PKC) family, namely, PKCλ and PKCη (8, 18, 26, 37, 42).
The functions and properties of the small NS2 proteins are more elusive to date. Roles have been ascribed to NS2 in viral DNA amplification, viral RNA translation, capsid formation, and packaging of virion ssDNA (11, 36). Although no intrinsic enzymatic function has been attributed to these polypeptides thus far, NS2 products were shown to physically interact with specific cellular partner proteins. Like NS1, NS2 is subjected to phosphorylation-mediated regulation. In particular, the distribution of NS2 proteins within the cell is regulated according to their phosphorylation state (14), and the interaction of these polypeptides with members of the 14-3-3 family of cellular proteins is dependent on distinct phosphorylation events. Most importantly, NS2 was linked to the control of nucleo/cytoplasmic trafficking through its interaction with the nuclear export factor CRM1 (2), which appeared to regulate the egress of progeny virus particles from the nucleus into the cytoplasm (20, 33). Although tolerated on their own by the host cell, NS2 proteins have been shown to modulate cytotoxic functions of NS1 in human cells (3). It is still unclear whether the increase in NS1 toxicity observed in the presence of NS2 results from a synergistic action of both viral proteins or from a cross-modulation of their functioning.
The NS1 phosphorylation pattern is complex (38) and shows striking differences between the productive phase (characterized by viral DNA replication and progeny particle formation) and the late phase (when NS1-dependent cytopathic effects become apparent) of a synchronized infection (9). These changes argue for a role played by phosphorylation in the regulation of NS1 late cytotoxic functions. This possibility was investigated in the present study by localizing major late phosphorylation events toward the C terminus of NS1 and substituting an alanine residue for either of two consensus PKC phosphorylation sites (T585 and S588) within this region. These substitutions proved to result in the sought-after alteration of the NS1 late phosphorylation pattern compared to the wild-type polypeptide. These mutations were then tested in the context of infectious MVMp clones for their effects on progeny virus production and virus-induced toxicity in mouse A9 fibroblasts. Since the mutations introduced to generate NS1-585A and -588A also affected NS2 (D93G and K96S, respectively), the mutant forms of NS1 were further tested in the absence of any other viral product in the form of recombinant proteins expressed in transfected A9 cells. Altogether, our results indicate that the above-mentioned mutations had no detectable effect on the replicative functions of NS1, while either reducing (NS1-585A) or increasing (NS1-588A) the cytotoxic activity of the viral product. These data demonstrate that NS1 cytotoxic and replicative functions can be dissociated through the targeting of residues regulating the late phosphorylation of the viral polypeptide.
MATERIALS AND METHODS
Viruses and cells.
Mouse A9 fibroblasts were grown in minimal essential medium (MEM) supplemented with 5% heat-inactivated fetal calf serum (FCS) and appropriate antibiotics. 293T cells were maintained in Dulbecco modified Eagle medium containing 10% FCS. Titers of virus stocks were determined by DNA hybridization assays on monolayer cultures of A9 cells (24). Multiplicities of infection (MOIs) are expressed as replicative center-forming units (CFU) per cell. To achieve similar MOIs with wild-type and mutant virions, virus stocks were normalized for their content in ssDNA as determined by Southern blotting. To avoid selection of revertants, only primary virus stocks were used in our experiments without further amplification.
Synchronization was achieved by serum deprivation as previously described (9). Briefly, cells grown in monolayer cultures were incubated for 3 days in MEM containing 0.3% serum. Starved cells comprising more than 90% of G0/G1 arrested cells, as measured by fluorescence-activated cell sorting analysis, were infected with MVMp wild type or mutant (10 CFU/cell) and incubated for 15 h before release into S phase by the addition of 20% serum. Primary stocks of wild-type and mutant MVMp were produced in 293T by calcium phosphate transfection with pdBMVp-derived molecular clones (20). Cells were harvested 3 days posttransfection, and the viruses were collected by repeated cycles of freezing and thawing in vTE (50 mM Tris-HCl [pH 8.3], 0.5 mM EDTA).
Construction of mutant MVMp variants.
Site-directed mutagenesis of MVMp DNA at PKC consensus phosphorylation sites was performed by chimeric PCR as described previously (39), with pdBMVp as a template and the N- and C-terminal primers 5′-CGGCAGAATTCAAACTAAAAAAGAAGTTTCTATTAAAACTACACTTAAAGAGCT-3′ and 5′-CGGTTCCGCACCGAAGCACGC-3′, together with two overlapping internal primers harboring the mutation. These mutated primers were, for pMVMp585A/93G (replacing threonine 585 with alanine in NS1 and aspartic acid 93 with glycine in NS2), 5′-AGGCGTACTTTTCGGTGCCGTGAATGGTGAGCG-3′ and 5′-CGCTCACCATTCACGGCACCGAAAAGTACGCCT-3′, and, for pMVMp588A/96S (replacing serine 588 with alanine in NS1 and lysine 96 with serine in NS2), 5′-TGGCTGAGAGGCGTAGCTTTCGGTGTCGTGAATGGT-3′ and 5′-ACCATTCACGACACCGAAAGCTACGCCTCTCAGCCA-3′. The BstEII- and XhoI-cleaved mutant PCR fragments replaced the corresponding restriction fragment (nucleotides [nt] 1885 to 2070) in the infectious MVMp molecular clone pdBMVp. The same fragment substitution was used to introduce either mutation into the NS sequence of the expression plasmid pP4-NS1X-P4-EGFP (8). The nature of all PCR products was confirmed by sequencing (Microsynth GmbH).
NS1 metabolic 32P-labeling and phosphopeptide analyses.
Metabolic labeling, tryptic phosphopeptide analyses, and cyanogen bromide (CNBr) cleavage were performed as previously described (9) with minor modifications. Mouse A9 cells were synchronized in G0/G1, infected with wild-type or mutant MVMp (10 CFU/cell), and released into the S phase 15 h postinfection (p.i.). At the indicated times postrelease, cells were washed three times with phosphate-free MEM (ICN) and labeled with 32P-labeled orthophosphate (ICN; 10−10 Ci/cell) for 4 h at 37°C. Labeled cells were then washed in 20 mM HEPES (pH 7.5)-150 mM NaCl, harvested directly in radioimmunoprecipitation assay buffer (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1 mM EDTA, 0.1% sodium dodecyl sulfate [SDS], 1% sodium deoxycholate, 1% Triton X-100) containing a cocktail of protease and phosphatase inhibitors, and proteins were extracted for 30 min on ice. NS1 was specifically immunoprecipitated by using the αSP8 antiserum (4) and further purified by SDS-10% polyacrylamide gel electrophoresis (PAGE), followed by protein transfer onto a nitrocellulose membrane (Schleicher & Schuell, Dassel, Germany). For one-dimensional analyses, the membrane-bound 32P-labeled NS1 protein was excised and incubated with 75 mg of CNBr/ml in 70% formic acid for 2 h at room temperature, and the cleavage products were separated after extensive lyophilization by tripartite SDS-PAGE with 16.5% T-6% C as a separation gel. For two-dimensional analyses, membrane-bound 32P-labeled NS1 was digested with 50 U of trypsin for 18 h at 37°C, loaded on thin-layer cellulose plates, and fractionated first by electrophoresis (using pH 1.9 buffer) and then by chromatography (in phosphochromatography buffer).
Western blotting and enhanced-chemiluminescence detection.
Proteins were extracted in radioimmunoprecipitation assay buffer as described for metabolically labeled polypeptides, except that the washing step was performed in phosphate-buffered saline. Protein extracts (50 μg) were fractionated by bipartite SDS-PAGE (i.e., on 8 to 12% gels) and blotted on nitrocellulose membranes. Individual proteins were identified by using rabbit antiserum αSP8 for NS1 (1:2,000) and rabbit αSP6 (2) for NS2 (1:500). Protein-antibody complexes were detected with a 1:10,000 dilution of horseradish peroxidase-conjugated anti-rabbit immunoglobulin G (IgG; Promega) and revealed by enhanced chemiluminescence according to the manufacturer's instructions (Amersham Pharmacia Biotech).
DNA extraction and Southern blotting.
For analyses of the viral DNA intermediates produced after infection, cell extracts were prepared in a mixture (1:1) of vTE buffer and 2× Hirt buffer (20 mM Tris [pH 7.4], 20 mM EDTA, 1.2% SDS) and then digested with proteinase K (400 μg/ml) for 18 h at 46°C. After cellular genomic DNA was sheared by several passages through 0.5- and 0.4-μm needles, DNA samples (2 μg) were fractionated through 0.8% agarose gel electrophoresis. After DNA denaturation and transfer onto Hybond-N nylon membranes (Amersham Pharmacia Biotech), the viral intermediates were identified by hybridization with a 32P-labeled DNA probe corresponding to the EcoRV (nt 385)-EcoRI (nt 1084) fragment of MVMp NS gene. For analysis of the ssDNA content of viral stocks, samples corresponding to 107 CFU of infectious particles/ml were diluted in a 1:1 mixture of vTE and 2× Hirt buffer to a final volume of 50 μl, digested with proteinase K, subjected to phenol-chloroform extraction, and analyzed as mentioned above after DpnI digestion of any contaminating bacterially expressed infectious DNA clone plasmid.
Cytotoxicity assays. (i) LDH-release assay.
The lytic activity of MVM viruses was determined from the release of lactate dehydrogenase (LDH; a cytoplasmic cellular enzyme) from infected cells. LDH activity was measured by using a colorimetric assay (CytoTox 96; Promega Biotech, Madison, Wis.) according to the manufacturer's instructions. A9 cultures in 96-well plates (4 × 103 cells per well in 50 μl of culture medium) were infected by addition of 50 μl of medium containing wild-type or mutant MVMp (10 CFU/cell) for 1 h. After removal of the supernatant, cells were further kept in 100 μl of fresh MEM containing 5% serum. At 3 days p.i., LDH activity was measured in 50 μl of culture medium by using an enzyme-linked immunosorbent assay (ELISA) reader at 492 nm. After subtraction of the background value given by nonconditioned medium, the fraction of lyzed cells in individual infected or noninfected cultures was calculated from the ratio of the LDH activity in the conditioned medium to the total LDH activity of the corresponding culture. The total LDH activity was determined in triplicate cultures after cell lysis by the addition of 10× buffer containing 9% (vol/vol) Triton X-100.
(ii) MTT activity assay.
For the determination of cell viability, the metabolic activity of mitochondrial dehydrogenases was measured through the ability of these enzymes to produce a formazan dye through reduction of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT). The same cultures were used to determine LDH release and MTT activity. After removal of 50 μl of medium for LDH activity determination (see above), 10 μl of 5 mg of MTT (Sigma)/ml dissolved in PBS was added to the cultures, and incubation was continued for 4 h at 37°C in a CO2 incubator. After centrifugation of the plate, the supernatant (60 μl) was removed, and the cells were dried for 30 min at 37°C before they were lysed by the addition of 100 μl of isopropanol. The absorbance of the formazan dye was measured at 595 nm by using an ELISA plate reader.
The viability of infected cells was expressed as the ratio of the corresponding absorbance to that of noninfected cells taken arbitrarily as 100%.
(iii) Plaque formation assay.
A9 cells grown in monolayer cultures were infected with serial dilutions of wild-type or mutant MVMp stocks for 2 h, followed by replacement of the inoculum with a Bacto-Agar overlay (1.8% in MEM containing 5% FCS). At day 6 p.i., living cells were stained for 18 h by addition of neutral-red (0.2 mg/ml)-containing Bacto Agar, and the sizes and numbers of plaques were determined.
(iv) Determination of NS1-induced morphological alterations in transfected A9 cells.
A9 cultures grown on spot slides (4 × 103 cells/spot) were transfected with 50 ng (5 μl) of pP4-NS1x-P4-GFP (8) by using 1 μl of Lipofectamine in the presence of 50 μl of OPTI-MEM (Gibco-BRL). At 5 h posttransfection, the medium was changed to MEM containing 5% FCS, and living cells expressing green fluorescent protein (GFP) were monitored for their morphological changes at 24-h intervals.
Immunostaining of NS1-expressing cells.
For the detection of NS1-expressing cells, A9 cultures grown on spot slides were fixed in paraformaldehyde at the indicated times p.i. NS1 present in infected cells was detected by using a 1:50 dilution of the mouse monoclonal antibody 3D9 (2) and visualized with a 1:800 dilution of Cy3-conjugated anti-mouse immunoglobulin G. Stained cells were covered with Elvanol and examined by using a Leica microscope (original magnification of ×20).
RESULTS
Late phosphorylation of the carboxy-terminal region of NS1.
Previous investigations have shown that the phosphorylation pattern of NS1 changes in the course of a synchronized infection of A9 cells with MVMp (9). During the productive phase of infection, the majority of phosphorylation events, including the PKCλ-driven modification of residues T435 and S473 which is necessary for viral DNA amplification, mapped to a 18.5-kDa CNBr-fragment located within the internal helicase domain of the polypeptide (8, 9, 18). In order to investigate a possible role of NS1 phosphorylation in the regulation of the subsequent cytotoxic phase of infection, we first identified the regions of NS1 that are subjected to late phosphorlytion events. A9 cells were arrested in G0/G1 phase of the cell cycle by serum starvation, infected with MVMp (10 CFU/cell), and released into S phase by the addition of 20% FCS. Synchronization was assessed by fluorescence-activated cell sorting analysis, revealing that ca. 60% of the infected cell population entered S phase at 16 h postrelease (data not shown) and remained in S phase during the whole interval studied, as expected from the cytostatic effect of MVM infection (9, 44). To determine the late phosphorylation pattern of NS1, metabolic 32P labeling of an MVM-infected cell population was performed at 48 h postrelease for an additional 4 h. For the sake of comparison, cells were similarly labeled at 20 h postrelease, which was taken as a representative time of the replicative phase of infection. Labeled cells were harvested directly into lysis buffer, and NS1 proteins were isolated by immunoprecipitation and subsequently purified by SDS-PAGE.
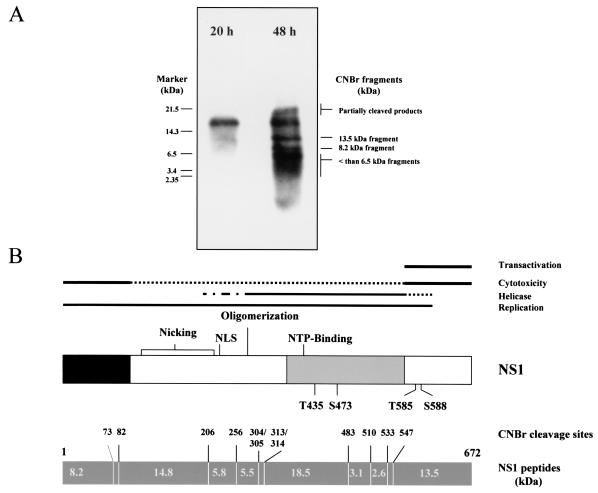
To identify the NS1 regions that are targets for phosphorylation during the respective time intervals, the purified metabolically 32P-labeled NS1 polypeptides were cleaved with CNBr at methionine residues, and the resulting peptides were fractionated by one-dimensional gel electrophoresis. Figure 1 illustrates the CNBr-cleavage pattern (Fig. 1B) and corresponding phosphorylation profile (Fig. 1A) of NS1 at the indicated time points in the course of infection. Although the 18.5-kDa NS1 fragment was the major target for phosphorylation during the replicative phase (20 h postrelease), as previously reported (9), a number of additional NS1 fragments became phospholabeled at later stages (Fig. 1A). Besides smaller polypeptides (<6.5 kDa), two fragments of 8.2 and 13.5 kDa were more particularly phosphorylated late in infection. These fragments had the sizes expected for the N and C termini of NS1, respectively (Fig. 1B). The C-terminal fragment was of special interest, given that a promoter-transregulating and cytopathic domain of NS1 had previously been mapped to this region (28), in keeping with a possible contribution of cellular gene expression disregulation to NS1 toxicity (1, 53, 54). Altogether, these data led us to focus the present study on the phosphorylation of the C-terminal domain of the viral polypeptide. It was further noticed that the C-terminal CNBr fragment of NS1 harbors two residues, T585 and S588, that are part of motifs showing >99% and >70% homology with consensus PKC phosphorylation sites, respectively, as revealed by computer analysis (Fig. 1B). The fact that NS1 functioning is tightly controlled by the PKC family of cellular protein kinases, at least regarding replicative activities (18, 26, 38, 41, 42), prompted us to use site-directed mutagenesis to alter these residues and determine the impact of these mutations on NS1 phosphorylation and cytotoxicity.
FIG. 1.
Determination of NS1 regions phosphorylated late in infection. (A) CNBr cleavage pattern of wild-type NS1 polypeptides that were metabolically 32P labeled 20 h (productive phase) or 48 h (cytopathic phase) after the release of serum-starved infected A9 cells into S phase. (B) Alignment of the functional map of NS1 with the predicted CNBr cleavage pattern of the polypeptide. Functional map: NLS, nuclear localization signal; NTP, nucleotide binding site; T435 and S473, previously determined PKCλ phosphorylation sites; T585 and S588, consensus PKC phosphorylation sites chosen as targets for mutagenesis in the present study. CNBr map: predicted cleavage sites (amino acid numbers) and sizes of generated peptides (in kilodaltons).
Generation of infectious MVMp viruses encoding mutant NS proteins.
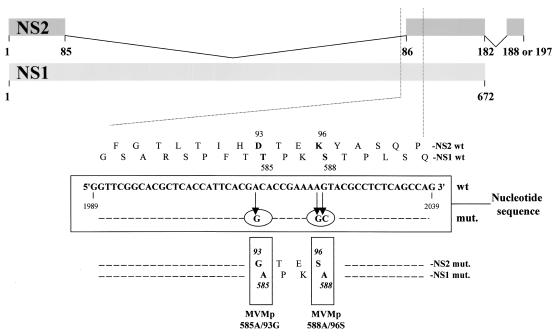
The infectious MVMp DNA clone pdBVMp (24) was altered by site-directed mutagenesis in order to substitute the nonpolar amino acid alanine for either of the above-mentioned NS1 residues, T585 or S588. As indicated in Fig. 2, the nucleotide sequence that codes for the NS1 region comprising these sites is also read in another frame to produce the NS2 proteins through alternative splicing. As a consequence, the mutations leading to the T585A and S588A substitutions in NS1 also resulted in the D93G and K96S changes in NS2, respectively.
FIG. 2.
Generation of MVMp585A/93G and MVMp588A/96S mutant viruses by site-directed mutagenesis. The upper diagram represents the NS1 and NS2 proteins, according to the splicing pattern of the respective transcripts and gives the amino acid sequence of both proteins in the region serving as a target for mutagenesis. The corresponding nucleic acid sequence is given in the central frame, together with the mutations (circled) introduced through chimeric PCRs to produce MVMp585A/93G and MVMp588A/96S, respectively. The resulting amino acid substitutions in NS1 and NS2 are shown at the bottom (boldface residues in the mutant versus wild-type sequences).
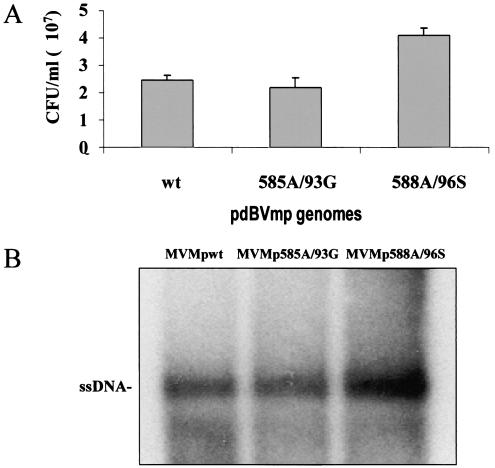
To determine whether the mutant MVMp DNA clones were still infectious, permissive human 293T cells were transfected with the same amount of wild-type (pdBVMp) or mutant (pMVMp585A/93G or pMVMp588A/96S) plasmids. Crude cell extracts were prepared 3 days posttransfection, and their virus titers were determined by replicative center assay, with monolayer cultures of A9 fibroblasts as indicators. Neither mutation was found to impair virus multiplication under these conditions, as was apparent from the similar (585A/93G) or even higher (588A/96S) yields of mutant compared to wild-type viruses (Fig. 3A). Besides these replicative titers, the amounts of single-stranded virion DNA in the various stocks were quantified by Southern blotting. The full virion contents of the stocks paralleled their CFU titers (Fig. 3B), indicating that the mutant and wild-type viruses had similar particle/infectivity ratios.
FIG. 3.
Production of wild-type and mutant MVMp virus stocks. Viruses were collected from 293T cells 3 days after transfection with either wild-type (wt; pdBMVp) or mutant (pMVMp585A/93G [585A/93G] and pMVMp588A/96S [588A/96S]) MVMp DNA clones. (A) Virus yields were determined by DNA hybridization assays after infection of A9 cell monolayers and are expressed in replicative CFU/ml of stock (average values and standard deviation bars from five independent transfection experiments). (B) Aliquots of the primary virus stocks (107 CFU) were compared for their contents in full virions as determined by Southern blotting detection of single-stranded genomic DNA (ssDNA).
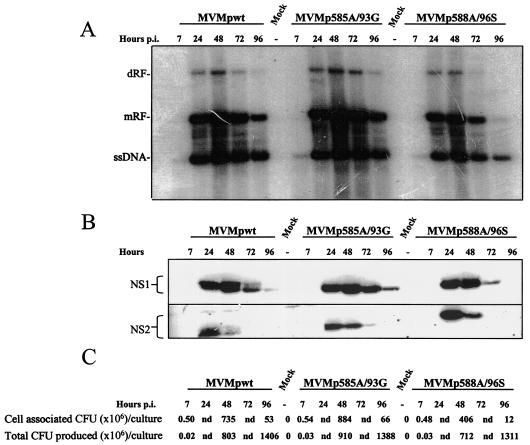
In order to compare the infectivities of the mutant and wild-type viruses in a more quantitative way, single-round infections were carried out. A9 mouse fibroblasts were used as host cells in order to allow the assessment of the effects of the amino acid substitution introduced not only in NS1 but also in NS2. Indeed, the NS2 proteins were previously shown to be required for MVMp replication in mouse cells while being dispensable in cells from other species (11, 35). Using the primary virus stocks described above, A9 fibroblasts were infected with 5 CFU of MVMp585A/93G, MVMp588A/96S, or wild-type MVMp per cell, respectively. Starting from 6 h p.i., treated cultures were incubated in the presence of 0.1 U of neuraminidase/ml in order to prevent new rounds of infection (16). At the indicated times p.i., adherent cells were harvested and processed for the analysis of viral DNA replication intermediates, proteins, and progeny particles by Southern blotting (Fig. 4A), Western blotting (Fig. 4B), and replicative center assays (Fig. 4C), respectively. All three viruses were growth competent, as is apparent from the time-dependent accumulation of NS proteins and replicative form DNAs (RF) eventually leading to the production of infectious progeny virions. Although the multiplication of the 585A/93G and wild-type viruses was similar, the 588A/96S mutant had a somewhat reduced replication proficiency and was distinguishable from the others by an accelerated loss of cell-associated viral DNA intermediates, progeny virions, and proteins late in infection. This was especially noticeable at 96 h p.i., at which time no NS1, low RF signals, and small amounts of cell-associated progeny virions could be detected in MVMp588A/96S-infected cultures, in contrast with the cell populations infected with the two other viruses. At this time period, cells infected with 588A/96S mutant viruses became either detached from the dish or lysed at an earlier time than did those infected with the other viruses. This feature would be in keeping with our working hypothesis, i.e., the mutations indeed targeted sites regulating the cytotoxic activity of NS proteins. The following experiments were conducted to further investigate this possibility.
FIG. 4.
Wild-type and mutant MVMp virus replication in a single-round infection. A9 cultures were infected with 5 CFU of primary stocks of either wild-type (wt) or mutant (585A/93G or 588A/96S) MVMp virus/cell and further incubated in culture medium supplemented with neuraminidase (from 6 h p.i. on) to prevent new rounds of infection. Cells were harvested at the indicated times p.i. and processed for the analysis of virus replication. (A) Southern blotting analysis of viral DNA intermediates. mRF, monomer RF; dRF, dimer RF; ssDNA, single-stranded genomic DNA. (B) Western blotting analysis of viral nonstructural protein production. The brackets encompass the phosphorylated and un(der)phosphorylated forms of the respective polypeptides. (C) Titration of progeny viruses by replication center assays on A9 indicator cell monolayers. Lane 1, amount of progeny viruses in adherent A9 monolayers; lane 2, total amount of viral particles present in adherent and detached cells, as well as in the culture medium; nd, not determined.
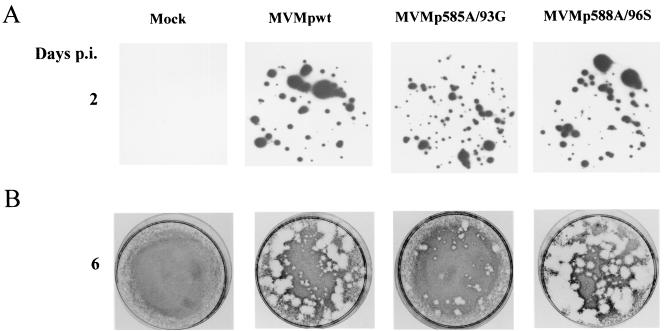
In a first step, the formation of lysis plaques in cell monolayers was analyzed since it reflects virus production and spreading. A9 indicator cultures were inoculated with equivalent amounts of full virions (corresponding to 10−3 CFU/cell) from wild-type and mutant stocks (Fig. 5A). Plaques developed in all cases, confirming the infectiousness of mutant viruses. However, there were significant differences in the size of the plaques generated by the individual viruses (Fig. 5B). In comparison with the wild-type virus, MVMp585A/93G gave rise to a majority of small or tiny plaques. In contrast, MVMp588A/96S generated mainly large plaques whose average diameter exceeded the size of the plaques formed by wild-type virus. Altogether, these data supported the view that the tested mutations resulted in the up (588A/96S)- or down (585A/93G)-modulation of virus spreading. Since the amounts of wild-type and mutant progeny viruses produced by infected A9 cultures (total yields from adherent and detached cells plus medium) were similar in a single-round infection (Fig. 4C), the question arose whether the enhanced (588A/96S) or reduced (585A/93G) dissemination of the mutants could be traced back to their hyper- or hypotoxicity, respectively.
FIG. 5.
Impact of 585A/93G and 588A/96S mutations on the ability of MVMp viruses to form lysis plaques in A9 cell monolyaers. A9 indicator cells were infected with primary stocks of either wild-type or mutant MVMp at a multiplicity of 10-3 CFU/cell and further processed for hybridization (replicative centers [A]) and plaque (B) assays. The figure is representative of five experiments performed with five different stocks of each virus.
Altered cytotoxicity of NS mutant viruses.
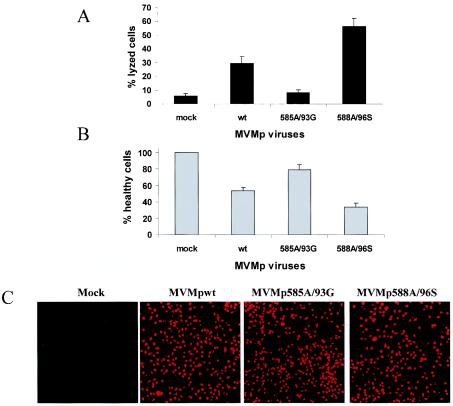
The question of whether the cytoxicity of NS mutant viruses is altered was investigated by using two complementary assays: (i) the virus-induced disruption of the plasma membrane integrity (cytolysis) was measured through the release of cytosolic LDH into the culture medium, and (ii) cells surviving virus infection were quantitated by determining their mitochondrial dehydrogenase activity in the so-called MTT test. The same A9 cultures were used for both assays, after infection with the respective viruses at a multiplicity of 10 CFU/cell. The results obtained in both tests were in good agreement and are summarized from 10 independent experiments in Fig. 6. Although the wild-type virus caused the release of ca. 29% of total LDH into the medium (representing an eightfold increase compared to the mock treatment), MVMp585A/93G and MVMp588A/96S were endowed with reduced (8%) and enhanced (56%) cytolytic activity, respectively (Fig. 6A). Conversely, the quantification of living cells (Fig. 6B) showed that the population's fraction surviving infection was higher for MVMp585A/93G (79%) and lower for MVMp588A/96S (33%) than for wild-type virus (53%). It was further ascertained that these differences were due to changes in the intrinsic cytotoxic activity of the mutant viruses and not in their competence for cell infection and viral protein expression. This was tested at the single-cell level by staining infected A9 cultures with a monoclonal antibody raised against the cytotoxic viral protein NS1. As illustrated in Fig. 6C, wild-type NS1 and either of the mutant forms were expressed in a similar fraction of infected cell populations. Together with the Western blotting detection of comparable amounts of wild-type and mutant proteins in the whole culture (Fig. 4B), these observations provided strong evidence to suggest that the mutant viruses studied were distinguishable from each other and from the parental virus by their intrinsic cytotoxic potential, with MVMp585A/93G being hypotoxic and MVMp588A/96S hypertoxic. It should be stated that the kinetics of LDH and virus release measured in nonsynchronized and synchronized A9 cells, respectively, showed no major difference between the wild-type and either mutant virus (data not shown), suggesting that the mutations affected the extent rather than timing of cell lysis.
FIG. 6.
Varying cytotoxicity of wild-type and mutant MVMp viruses. The indicated virus stocks were tested for their ability to jeopardize the survival of A9 fibroblasts after infection at a multiplicity of 10 CFU/cell and further incubation for 3 days. (A) The virus lytic activity was measured through quantification of the cytoplasmic LDH released into the medium, expressed as a percentage of total LDH (determined after lysis of the whole culture with detergent). (B) The cell-killing activity of the different viruses was assessed by determining the reduction in the number of living cells (still able to reduce MTT) in the infected population, a value expressed as the percentage of the value for mock-treated cultures. The same cultures were used for panels A and B. The data shown are means with standard deviation bars from 10 independent experiments carried out each in triplicate. (C) NS1 produced by wild-type and mutant MVMp viruses was detected by indirect immunofluorescence in parallel cultures. Images were obtained by using a ×16 magnification lens.
Since NS1 and NS2 proteins were both affected by the site-directed mutagenesis performed, either viral product may conceivably contribute to the altered cytotoxicity of the mutant viruses. Some previous observations argued for NS1 mediating the effects of the present mutations. First, the NS1 protein alone was demonstrated to be toxic for both murine and human transformed cells, while NS2 alone was ineffective in this respect (3, 8, 29, 34). Second, the differential toxicity of the mutant viruses was shown not only for A9 fibroblasts (see above) but also for transformed human cells (data not shown). Given that NS2 is dispensable for virus production in the latter cells, the altered NS1 protein constituted a prime candidate for the effector of the varying cytopathogenicity of the mutant viruses. It should be stated, however, that NS2 was found to work synergistically with NS1 to reinforce the cytotoxic activity of the latter protein in human cells (3, 29). Therefore, it cannot be fully ruled out that the introduced mutations acted, at least in part, by affecting the capacity of NS2 for modulating NS1-dependent cytopathic effects. This possibility is not supported, however, by the previously reported failure to detect this cooperative effect of NS2 in rodent cells (34).
Altered phosphorylation of mutant versus wild-type NS1.
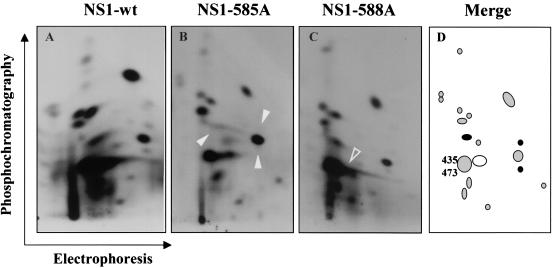
As stated above, NS1 was mutated at putative PKC target sites within the C-terminal region which gets phosphorylated late in infection. This led us to search for distinct changes in the late phosphorylation profile of mutant versus wild-type NS1. At 48 h after MVMp infection and release into S phase, synchronized A9 cells were metabolically labeled with orthophosphate and processed for NS1 isolation, trypsin digestion, and two-dimensional peptide fractionation by electrophoresis and phosphochromatography. Figure 7 shows the tryptic phosphopeptide maps of the NS1 proteins generated by wild type (Fig. 7A), 585A/93G (Fig. 7B), and 588A/96S (Fig. 7C) MVMp, together with their schematic representation (Fig. 7D). Compared to the wild-type protein, NS1-585A lacked three distinct phosphopeptides (arrowheads in panel B; black spots in panel D), while a single one was missing in NS1-588A (open arrowhead in panel C; circled white spot in panel D). These results confirmed that the late phosphorylation of NS1 was altered as a result of an alanine substitution for either of the consensus PKC target residues tested. The disappearance of a single phosphopeptide from the NS1-588A map argues for the possibility that the S588 residue is located within this peptide and is subjected to phosphorylation in vivo. It cannot be ruled out, however, that the S588A substitution may affect the phosphorylation of another amino acid by simply altering the conformation of the protein. The same restriction applies to the T585A substitution, especially since three distinct phosphopeptides were sensitive to this single amino acid replacement. However, this discrepancy is still compatible with the in vivo phosphorylation of T585, assuming that the corresponding peptide undergoes further modifications or this residue controls the phosphorylation of other amino acids by serving as a docking site for a kinase or for structural reasons. However that may be, our data indicate that T585 and S588 are directly or indirectly involved in the late phosphorylation of NS1, making this posttranslational modification a likely candidate for the regulation of parvovirus cytotoxicity.
FIG. 7.
Alteration of the late phosphorylation pattern of T585A and S588A mutant forms of NS1. A9 cultures arrested in G0/G1 were infected with indicated viruses, released into the cell cycle for 48 h and metabolically 32P labeled. NS1 proteins were isolated, purified, and processed for the analysis of tryptic phosphopeptides by two-dimensional electrophoresis and/or chromatography. Compared to wild-type NS1 (A), the NS1-585A (B) and NS1-588A (C) mutants lacked specific phosphorylation event(s), as evidenced by the disappearance of distinct phosphopeptide(s) (arrowheads). (D) Schematic representation of the “late” NS1 tryptic phosphopeptide pattern, in which the spots absent from the NS1-585A and NS1-588A maps are indicated with filled black and open circles, respectively, and the previously assigned PKCλ phosphorylation targets T435 and S473 are positioned.
Altered capacity of the mutant forms of NS1 for disrupting host cell morphology.
Although the mutations introduced in the MVMp genome affected both NS1 and NS2 proteins, the ensuring modulation of the cytotoxic activity of mutant viruses was likely to be ascribable to the changes brought to the NS1 protein (see above). However, the possible contribution of an as-yet-unknown function of NS2 could not be ruled out by above experiments. To further dismiss this possibility, an assay was devised to measure the cellular effects of NS1 in the absence of both viral DNA amplification and production of other viral proteins including NS2. This assay makes use of the plasmid pP4-NS1x-P4-GFP which combines the NS1 effector and GFP reporter proteins, allowing living NS1-expressing cells to be monitored after transfection (8). A9 cultures growing on spot slides were transfected with pP4-NS1x-P4-GFP constructs expressing either wild-type or mutant (585A or 588A) NS1 proteins, or with pP4-GFP as control. Successfully transfected cells (detected through GFP fluorescence) were examined over a period of 7 days.
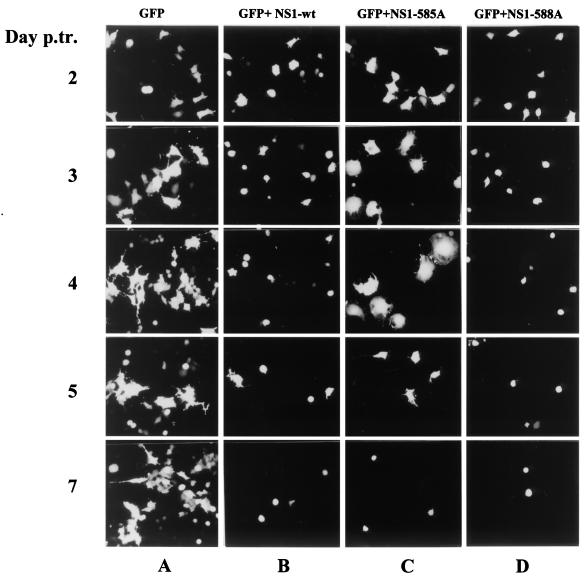
In the absence of NS1, no significant morphological alterations were detected in the majority of GFP-expressing A9 cells during the whole time interval (Fig. 8, column A). In contrast, when wild-type NS1 was coexpressed with GFP, transfected cells rounded up already 2 days after transfection, and detached from their substrate starting from day 3 (column B). A similar cytopathogenicity was achieved by NS1-588A, although the detachment of GFP-positive cells was somewhat faster and more pronounced compared to wild type (column D). The phenotype of the NS1-585A mutant was strikingly different in that transfected cells started by enlarging to a significant extent for 3 to 4 days, and their eventual rounding up was delayed until day 5 (column C). It should be stated that the production of NS1 in GFP-positive cells was verified by indirect immunofluorescence staining (data not shown) and that the wild-type and mutant proteins gave indistinguishable signals, indicating that the delayed cytopathogenicity of NS1-585A was a genuine property of this mutant and not the result of its poor expression. Altogether, these observations revealed a striking parallel between, on the one hand, the respectively accelerated and retarded induction of cell collapse by NS1-588A and NS1-585A proteins in the present transfection assay, and on the other hand, the hyper- and hypotoxicity of corresponding MVMp588A/96S and MVMp585A/93G viruses in the infection experiments described above (Fig. 6). Therefore, it seems likely that the changes observed in the cytotoxic/cytolytic potential of the mutant viruses can be assigned, at least in part, to the modulation of the intrinsic NS1 toxic function(s).
FIG. 8.
Induction of morphological alterations by wild-type and mutant NS1 proteins. A9 cells competent for transfection with plasmid pP4-NS1x-P4-GFP were identified on the basis of GFP fluorescence and examined over a period of 7 days to detect phenotypic alterations induced by wild-type (B), 585A (C), or 588A (D) NS1. Parallel cultures transfected with plasmid pP4-GFP served as negative control (A). Images of GFP-expressing cells were obtained by using a ×16 magnification lens.
DISCUSSION
The NS1 protein of MVMp is involved in multiple processes necessary for progeny particle production, such as viral DNA replication (30, 39), trans regulation of the viral promoters (49), and cell killing (1, 5, 8, 34, 45). To timely coordinate all of these different functions, NS1 was proposed to be regulated through phosphorylation, an assumption strongly supported by the changes occurring in the NS1 phosphorylation pattern during the progression of an infectious cycle (9). It was shown that NS1 is indeed regulated for its replicative activities by distinct phosphorylation events driven by members of the PKC family, in particular PKCλ and PKCη (18, 26, 42). The present study gives further evidence to support that NS1 is independently regulated by phosphorylation with regard to its cytotoxic function. Indeed, the targeting of consensus PKC phosphorylation sites located in the C-terminal part of NS1 through site-directed mutagenesis allowed us to modulate the lytic and toxic activities of MVMp without affecting the production of infectious virions in a single round infection. These results demonstrate that at least some of the cytotoxic activites of MVMp are intrinsic properties of the viral nonstructural proteins and not merely side effects arising from virus replication. Furthermore, the mutations changing the balance between MVMp replication and cytotoxicity were found to result in the alteration of the NS1 phosphorylation pattern. Although the direct involvement of NS1 phosphorylation in this phenotypic change remains to be proven, a temporal correlation was observed between the phosphorylation events concerned and the appearance of cytotoxic effects, which both took place at a late stage of infection. Altogether, these data strongly argue for the fact that the cytotoxicity of NS1 represents a regulated process and that the residues targeted in the present study take part in this regulation by controlling directly and/or indirectly the late phosphorylation of the viral polypeptide.
The substitution of alanine for the NS1 residue S588 correlated with the disappearance of a single phosphopeptide from the late phosphorylation map of NS1, suggesting that S588 may constitute an in vivo phosphorylation site, in agreement with its inclusion in a consensus PKC target motif. In contrast, the T585A substitution resulted in the loss of three phosphopeptides from the NS1 map. Several explanations could be put forward to reconcile this observation with the possibility that T585 is also a direct target for phosphorylation. The T585-containing phosphopeptide may undergo additional posttranslational modifications leading to its resolution into several spots after two-dimensional fractionation. Alternatively, T585 may serve as a phosphorylation-dependent docking site, allowing NS1 to interact with a cellular kinase, which is responsible for the phosphorylation of the viral polypeptide at other positions. Such phosphorylation-dependent docking sites were reported for the superfamily of AGC-kinases, leading them to interact with and become activated by the kinase PDK-1 (21). The present data do not rule out, however, that the mutations tested only affected NS1 phosphorylation in an indirect way, by altering the structure of the viral protein irrespective of the modification of concerned residues.
Due to the overlap of NS1- and NS2-coding sequences, the present disruption of specific phosphorylation motifs in NS1 could not be achieved without changing the primary structure of the NS2 proteins. Therefore, a possible contribution of these NS2 alterations to the phenotype of mutant viruses needs to be considered. However, this putative role of NS2, if any, is likely to be of minor importance regarding the cytotoxicity of the mutant viruses in murine cells. (i) MVMp DNA mutagenesis was directed so as to result in the loss of distinct phosphorylation sites in NS1. In contrast, the consequent substitutions brought about to NS2 did not hit any known or suspected regulatory elements of these proteins. (ii) The only functions assigned thus far to NS2 concern distinct events of the virus productive cycle (11, 35, 36), which were not impaired as a result of the mutations tested. The mutant forms of NS2 therefore appear to be functional. In particular, the mutant viruses did not show any deficiency in the nuclear egress of NS2 proteins (data not shown), an effect recently reported to occur when the nuclear export signal of these protein was mutated (20, 33). A role for NS2 polypeptides in the phenotype of the present virus mutants would thus imply a new NS2 function that has not been described as yet in murine cells. (iii) NS2 polypeptides have little toxicity by themselves and could only contribute to the altered cytopathogenicity of mutant viruses in an indirect way, e.g., by modulating NS1 cytotoxicity. Although detected in human cells (3, 29), such a cooperative effect of NS2 could not be demonstrated in rodent cells (34). (iv) The up- or downmodulation of the lytic and toxic activities of MVMp mutants was detected in cells of both mouse and human origins. Whereas NS1 is absolutely required, NS2 proteins are dispensable for MVMp multiplication and propagation in human cells (35). This makes NS1 the prime candidate for mediating the effects of viral mutations in the latter cells. (v) The hypo- and hypertoxicity of MVMp585A/93G and MVMp588A/96S viruses in infection experiments, respectively, paralleled the reduced and enhanced efficiency of corresponding NS1-585A and NS1-588A proteins in inducing the rounding up and detachment of transfected cells. Altogether, these considerations strongly suggest that the up- or downmodulation of the toxic and lytic activities of the MVMp mutants generated in the present study can be traced back, at least in part, to the modified NS1 proteins encoded by the respective viruses. This does not rule out, however, that other experimental conditions may reveal a role of NS2 in MVMp-induced murine cell death.
It is presumably a matter of speculation how the late phosphorylation of NS1 may regulate the capacity of the viral product for inducing cell killing. NS1 expression in permissive cells was found to result in various molecular disturbances, including the shutoff of host cell macromolecular syntheses (15), the trans regulation of cellular promoters (53, 54), DNA damage (45), and changes in the synthesis and phosphorylation of cellular proteins (1). Affected cells undergo morphological alterations (8), mitotic cycle arrest (44), and apoptotic death (47, 48), which must be properly timed to avoid interference with virus replication and allow efficient progeny virus production. Therefore, there are several levels at which the substitutions introduced into the NS1 protein may conceivably act to modulate the cytotoxicity of the viral product. The fact that MVMp585A/93G and MVMp588A/96S were both indistinguishable from the wild-type virus regarding their replication and the production of progeny virions in a single-round infection argues for the full proficiency of the mutant forms of NS1 in the known enzymatic activities of the protein. This leads us to propose that the varying cytotoxicity of the NS1 mutants may be assigned to their differential interaction with partner cellular proteins. This would be in keeping with our assumption that the effects of tested substitutions results from their interference with NS1 phosphorylation. Indeed, the formation of multiprotein complexes is known to depend on the phosphorylation state of some of the partners involved (21). NS1 was reported to interact in specific ways with a number of cellular polypeptides, such as sp1 (25), TBP and TFIIA (alpha and beta) (31), RPA (7), SMN (57), or SGT (17). Given their inclusion in the previously identified transactivation domain of NS1 (Fig. 1) (27), the T585 and/or S588 residues may control the ability of NS1 to disregulate distinct cellular promoters, a property that was found to correlate with virus-induced cell killing (28, 53). The association of NS1 with other, nonexclusive cellular targets besides transcription factors may also be sensitive to the substitutions introduced in the viral protein. In particular, the herein described morphological alterations and loss of adherence displayed by NS1-expressing cell cultures are suggestive of direct or indirect interactions of the viral product with components of the cytoskeleton. Besides effector proteins, cellular polypeptides involved in NS1 regulation need to be considered among the interacting partners of the viral product. These regulators may in particular control the nucleo/cytoplasmic distribution of NS1 (43) and the resulting contacts of the viral protein with its potential nuclear (e.g., transcription factors) and cytoplasmic (e.g., cytoskeleton components) targets. It is worth noting in this respect that C-terminal deletions of NS1 were reported to concomitantly impair the cytoplasmic transport (43) and cytotoxicity (28) of this protein. It would therefore be most interesting to determine whether the mutant forms of NS1 generated in the present study differ from the original protein with regard to their binding to known and/or new cellular partner proteins. This investigation is in progress in our laboratory in an effort to unravel the molecular mechanisms underlying NS1 cytotoxicity.
These ongoing studies are also expected to clarify the reason for the intriguing antagonistic effects (hyper- versus hypotoxicity) of the alanine substitutions for two potential late phosphorylation sites of NS1. The opposite phenotypes of the T585A and S588A mutants lead us to hypothesize that a sequential phosphorylation of NS1 may take place late during an MVMp infection. According to this scenario, residue S588 would be phosphorylated in the first place so as to decrease the general cytotoxicity of the viral NS1 protein and maintain a cellular environment permissive for virus replication. Phosphorylation of residue T585 would occur at a later stage to activate the toxic function of NS1 and bring about a degenerative cell condition favorable to the release of progeny virus particles. Alternatively, residue S588 may be targeted by a cellular antiviral process tending to limit virus cytopathogenicity and spreading through the downmodulation of the NS1 products. Another possible clue to the differential effect of the mutations studied would lie in the above-mentioned multiple NS1 functions that may cooperate in the eventual collapse of infected cells and be independently regulated, in particular through phosphorylation. Accordingly, a combination of distinct (de)phosphorylation events may be involved in the control of the overall cytotoxic activity of the NS1 protein. The underlying mechanism of the antagonistic phenotypes of the herein described NS1 mutants is currently a matter of speculation and deserves to be investigated in greater detail.
In addition to their use in tackling this fundamental question, mutant viruses such as those generated in the present study also have an applied potential regarding cancer therapy. Parvoviruses are indeed endowed with an oncosuppressive capacity that was demonstrated in various animal models, including recipient mice implanted with human neoplastic cells (for a review, see reference 51). This oncosuppression is thought to result from both the oncolytic and immunomodulating properties of concerned parvoviruses (for review, see reference 50). However, in many instances, the protection provided by natural parvoviruses is only partial or temporary and eventually gets overtaken by tumor growth (19, 22). Efforts are therefore made to enhance the anticancer potency of these viruses. One possible strategy consists in improving the efficiency or selectivity of parvoviruses through mutations that do not impair virus multiplication and propagation. A modulation of H-1 virus selectivity was recently achieved by modifying the viral P4 promoter so as to make its activation dependent on a signaling pathway that is specifically activated in colon cancer cells (32). The present work exemplifies another approach in which MVMp mutagenesis was aimed at increasing the capacity of this virus for killing target cells while preserving its competence for replication. Mutations giving rise to hypertoxic virus mutants can indeed be expected to stimulate the lysis of infected tumor cells and the release of immunogenic tumor-associated antigens, thereby tipping the balance between tumor growth and virus propagation in favor of the latter. An alternative strategy for enhancing the antineoplastic activity of parvoviruses involves supplementing them with therapeutic transgenes. Indeed, parvoviral vectors transducing specific cyto/chemokines proved to have reinforced capacity for suppressing certain tumors, compared to their parental viruses (22, 55). Transgene expression achieved by these vectors was found to be transient, most likely due to the viral toxicity for transduced cells (55). Therefore, virus mutants with a reduced or delayed cytopathogenicity, as identified in the present study, may also be of benefit to this type of approach by sustaining the expression of added transgenes for an extended period of time. It follows that the MVMp mutants described here may serve as a paradigm of how autonomous parvoviruses could be improved for their various applications to the treatment of cancer. This prospect is currently evaluated by comparing mutant and wild-type viruses or vector derivatives for their respective protective effects in animal tumor models.
Acknowledgments
We are indebted to S. Cotmore and P. Tattersall (Yale University, New Haven, Conn.) for providing us with the infectious MVMp DNA clone and to D. J. Pintel (University of Missouri, Columbia) for the generous gift of monoclonal mouse anti-NS1 antibody. We are also thankful to N. Salomé and M. Klein (Deutsches Krebsforschungszentrum, Heidelberg, Germany) for providing us with polyclonal anti-NS1 and anti-NS2 antibodies.
L. Daeffler was supported by the Alexander von Humboldt foundation and the European Commission (Fifth Framework Program, Marie Curie Action).
REFERENCES
- 1.Anouja, F., R. Wattiez, S. Mousset, and P. Caillet-Fauquet. 1997. The cytotoxicity of the parvovirus minute virus of mice nonstructural protein NS1 is related to changes in the synthesis and phosphorylation of cell proteins. J. Virol. 71:4671-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodendorf, U., C. Cziepluch, J. C. Jauniaux, J. Rommelaere, and N. Salomé. 1999. Nuclear export factor CRM1 interacts with nonstructural proteins NS2 from parvovirus minute virus of mice. J. Virol. 73:7769-7779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandenburger, A., D. Legendre, B. Avalosse, and J. Rommelaere. 1990. NS-1 and NS-2 proteins may act synergistically in the cytopathogenicity of parvovirus MVMp. Virology 174:576-584. [DOI] [PubMed] [Google Scholar]
- 4.Brockhaus, K., S. Plaza, D. J. Pintel, J. Rommelaere, and N. Salomé. 1996. Nonstructural proteins NS2 of minute virus of mice associate in vivo with 14-3-3 protein family members. J. Virol. 70:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caillet-Fauquet, P., M. Perros, A. Brandenburger, P. Spegelaere, and J. Rommelaere. 1990. Programmed killing of human cells by means of an inducible clone of parvoviral genes encoding nonstructural proteins. EMBO J. 9:2989-2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christensen, J., S. F. Cotmore, and P. Tattersall. 1995. Minute virus of mice transcriptional activator protein NS1 binds directly to the transactivation region of the viral P38 promoter in a strictly ATP-dependent manner. J. Virol. 69:5422-5430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, J., and P. Tattersall. 2002. Parvovirus initiator protein NS1 and RPA coordinate replication fork progression in a reconstituted DNA replication system. J. Virol. 76:6518-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corbau, R., V. Duverger, J. Rommelaere, and J. P. Nüesch. 2000. Regulation of MVM NS1 by protein kinase C: impact of mutagenesis at consensus phosphorylation sites on replicative functions and cytopathic effects. Virology 278:151-167. [DOI] [PubMed] [Google Scholar]
- 9.Corbau, R., N. Salomé, J. Rommelaere, and J. P. Nüesch. 1999. Phosphorylation of the viral nonstructural protein NS1 during MVMp infection of A9 cells. Virology 259:402-415. [DOI] [PubMed] [Google Scholar]
- 10.Cotmore, S. F., J. Christensen, J. P. Nüesch, and P. Tattersall. 1995. The NS1 polypeptide of the murine parvovirus minute virus of mice binds to DNA sequences containing the motif [ACCA]2-3. J. Virol. 69:1652-1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cotmore, S. F., A. M. D'Abramo, Jr., L. F. Carbonell, J. Bratton, and P. Tattersall. 1997. The NS2 polypeptide of parvovirus MVM is required for capsid assembly in murine cells. Virology 231:267-280. [DOI] [PubMed] [Google Scholar]
- 12.Cotmore, S. F., J. P. Nüesch, and P. Tattersall. 1993. Asymmetric resolution of a parvovirus palindrome in vitro. J. Virol. 67:1579-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cotmore, S. F., J. P. Nüesch, and P. Tattersall. 1992. In vitro excision and replication of 5′ telomeres of minute virus of mice DNA from cloned palindromic concatemer junctions. Virology 190:365-377. [DOI] [PubMed] [Google Scholar]
- 14.Cotmore, S. F., and P. Tattersall. 1990. Alternate splicing in a parvoviral nonstructural gene links a common amino-terminal sequence to downstream domains which confer radically different localization and turnover characteristics. Virology 177:477-487. [DOI] [PubMed] [Google Scholar]
- 15.Cotmore, S. F., and P. Tattersall. 1987. The autonomously replicating parvoviruses of vertebrates. Adv. Virus Res. 33:91-174. [DOI] [PubMed] [Google Scholar]
- 16.Cotmore, S. F., and P. Tattersall. 1989. A genome-linked copy of the NS-1 polypeptide is located on the outside of infectious parvovirus particles. J. Virol. 63:3902-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cziepluch, C., E. Kordes, R. Poirey, A. Grewenig, J. Rommelaere, and J. C. Jauniaux. 1998. Identification of a novel cellular TPR-containing protein, SGT, that interacts with the nonstructural protein NS1 of parvovirus H-1. J. Virol. 72:4149-4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dettwiler, S., J. Rommelaere, and J. P. Nüesch. 1999. DNA unwinding functions of minute virus of mice NS1 protein are modulated specifically by the lambda isoform of protein kinase C. J. Virol. 73:7410-7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dupressoir, T., J. M. Vanacker, J. J. Cornelis, N. Duponchel, and J. Rommelaere. 1989. Inhibition by parvovirus H-1 of the formation of tumors in nude mice and colonies in vitro by transformed human mammary epithelial cells. Cancer Res. 49:3203-3208. [PubMed] [Google Scholar]
- 20.Eichwald, V., L. Daeffler, M. Klein, J. Rommelaere, and N. Salomé. 2002. The NS2 proteins of parvovirus minute virus of mice are required for efficient nuclear egress of progeny virions in mouse cells. J. Virol. 76:10307-10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frödin, M., T. L. Antal, B. A. Dummler, C. J. Jensen, M. Deak, S. Gammeltoft, and R. M. Biondi. 2002. A phosphoserine/threonine-binding pocket in AGC kinases and PDK1 mediates activation by hydrophobic motif phosphorylation. EMBO J. 21:5396-5407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giese, N. A., Z. Raykov, L. DeMartino, A. Vecchi, S. Sozzani, C. Dinsart, J. J. Cornelis, and J. Rommelaere. 2002. Suppression of metastatic hemangiosarcoma by a parvovirus MVMp vector transducing the IP-10 chemokine into immunocompetent mice. Cancer Gene Ther. 9:432-442. [DOI] [PubMed] [Google Scholar]
- 23.Harris, C. E., R. A. Boden, and C. R. Astell. 1999. A novel heterogeneous nuclear ribonucleoprotein-like protein interacts with NS1 of the minute virus of mice. J. Virol. 73:72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kestler, J., B. Neeb, S. Struyf, J. Van Damme, S. F. Cotmore, A. D'Abramo, P. Tattersall, J. Rommelaere, C. Dinsart, and J. J. Cornelis. 1999. cis requirements for the efficient production of recombinant DNA vectors based on autonomous parvoviruses. Hum. Gene Ther. 10:1619-1632. [DOI] [PubMed] [Google Scholar]
- 25.Krady, J. K., and D. C. Ward. 1995. Transcriptional activation by the parvoviral nonstructural protein NS-1 is mediated via a direct interaction with Sp1. Mol. Cell. Biol. 15:524-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lachmann, S., J. Rommelaere, and J. P. Nüesch. 2003. Novel PKCη is required to activate replicative functions of the major nonstructural protein NS1 of minute virus of mice. J. Virol. 77:8048-8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legendre, D., and J. Rommelaere. 1994. Targeting of promoters for trans activation by a carboxy-terminal domain of the NS-1 protein of the parvovirus minute virus of mice. J. Virol. 68:7974-7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Legendre, D., and J. Rommelaere. 1992. Terminal regions of the NS-1 protein of the parvovirus minute virus of mice are involved in cytotoxicity and promoter trans inhibition. J. Virol. 66:5705-5713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Legrand, C., J. Rommelaere, and P. Caillet-Fauquet. 1993. MVM(p) NS-2 protein expression is required with NS-1 for maximal cytotoxicity in human transformed cells. Virology 195:149-155. [DOI] [PubMed] [Google Scholar]
- 30.Li, X., and S. L. Rhode III. 1990. Mutation of lysine 405 to serine in the parvovirus H-1 NS1 abolishes its functions for viral DNA replication, late promoter trans activation, and cytotoxicity. J. Virol. 64:4654-4660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorson, C., J. Pearson, L. Burger, and D. J. Pintel. 1998. An Sp1-binding site and TATA element are sufficient to support full transactivation by proximally bound NS1 protein of minute virus of mice. Virology 240:326-337. [DOI] [PubMed] [Google Scholar]
- 32.Malerba, M., L. Daeffler, J. Rommelaere, and R. D. Iggo. 2003. Replicating parvoviruses which target colon cancer cells. J. Virol. 77:6683-6691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller, C. L., and D. J. Pintel. 2002. Interaction between parvovirus NS2 protein and nuclear export factor Crm1 is important for viral egress from the nucleus of murine cells. J. Virol. 76:3257-3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mousset, S., Y. Ouadrhiri, P. Caillet-Fauquet, and J. Rommelaere. 1994. The cytotoxicity of the autonomous parvovirus minute virus of mice nonstructural proteins in FR3T3 rat cells depends on oncogene expression. J. Virol. 68:6446-6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naeger, L. K., J. Cater, and D. J. Pintel. 1990. The small nonstructural protein (NS2) of the parvovirus minute virus of mice is required for efficient DNA replication and infectious virus production in a cell-type-specific manner. J. Virol. 64:6166-6175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Naeger, L. K., N. Salomé, and D. J. Pintel. 1993. NS2 is required for efficient translation of viral mRNA in minute virus of mice-infected murine cells. J. Virol. 67:1034-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nüesch, J. P., J. Christensen, and J. Rommelaere. 2001. Initiation of minute virus of mice DNA replication is regulated at the level of origin unwinding by atypical protein kinase C phosphorylation of NS1. J. Virol. 75:5730-5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nüesch, J. P., R. Corbau, P. Tattersall, and J. Rommelaere. 1998. Biochemical activities of minute virus of mice nonstructural protein NS1 are modulated In vitro by the phosphorylation state of the polypeptide. J. Virol. 72:8002-8012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nüesch, J. P., S. F. Cotmore, and P. Tattersall. 1992. Expression of functional parvoviral NS1 from recombinant vaccinia virus: effects of mutations in the nucleotide-binding motif. Virology 191:406-416. [DOI] [PubMed] [Google Scholar]
- 40.Nüesch, J. P., S. F. Cotmore, and P. Tattersall. 1995. Sequence motifs in the replicator protein of parvovirus MVM essential for nicking and covalent attachment to the viral origin: identification of the linking tyrosine. Virology 209:122-135. [DOI] [PubMed] [Google Scholar]
- 41.Nüesch, J. P., S. Dettwiler, R. Corbau, and J. Rommelaere. 1998. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J. Virol. 72:9966-9977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nüesch, J. P., S. Lachmann, R. Corbau, and J. Rommelaere. 2003. Regulation of minute virus of mice NS1 replicative functions by atypical PKCλ in vivo. J. Virol. 77:433-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nüesch, J. P., and P. Tattersall. 1993. Nuclear targeting of the parvoviral replicator molecule NS1: evidence for self-association prior to nuclear transport. Virology 196:637-651. [DOI] [PubMed] [Google Scholar]
- 44.Op De Beeck, A., F. Anouja, S. Mousset, J. Rommelaere, and P. Caillet-Fauquet. 1995. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 6:781-787. [PubMed] [Google Scholar]
- 45.Op De Beeck, A., and P. Caillet-Fauquet. 1997. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J. Virol. 71:5323-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pujol, A., L. Deleu, J. P. Nüesch, C. Cziepluch, J. C. Jauniaux, and J. Rommelaere. 1997. Inhibition of parvovirus minute virus of mice replication by a peptide involved in the oligomerization of nonstructural protein NS1. J. Virol. 71:7393-7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ran, Z., B. Rayet, J. Rommelaere, and S. Faisst. 1999. Parvovirus H-1-induced cell death: influence of intracellular NAD consumption on the regulation of necrosis and apoptosis. Virus Res. 65:161-174. [DOI] [PubMed] [Google Scholar]
- 48.Rayet, B., J.-A. Lopez-Guerrero, J. Rommelaere, and C. Dinsart. 1998. Induction of programmed cell death by parvovirus H-1 in U937 cells: connection with the tumor necrosis factor alpha signalling pathway. J. Virol. 72:8893-8903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhode, S. L., III, and S. M. Richard. 1987. Characterization of the transactivation-responsive element of the parvovirus H-1 P38 promoter. J. Virol. 61:2807-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rommelaere, J., and J. Cornelis. 2001. Autonomous parvoviruses, p. 100-129. In P. Hernáiz Driever and S. D. Rabkin (ed.), Replication-competent viruses for cancer therapy, vol. 22. Monographs in virology. Krager, Basel, Switzerland.
- 51.Rommelaere, J., and J. J. Cornelis. 1991. Antineoplastic activity of parvoviruses. J. Virol. Methods 33:233-251. [DOI] [PubMed] [Google Scholar]
- 52.Tullis, G. E., L. Labieniec-Pintel, K. E. Clemens, and D. Pintel. 1988. Generation and characterization of a temperature-sensitive mutation in the NS-1 gene of the autonomous parvovirus minute virus of mice. J. Virol. 62:2736-2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanacker, J. M., R. Corbau, G. Adelmant, M. Perros, V. Laudet, and J. Rommelaere. 1996. Transactivation of a cellular promoter by the NS1 protein of the parvovirus minute virus of mice through a putative hormone-responsive element. J. Virol. 70:2369-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vanacker, J. M., V. Laudet, G. Adelmant, D. Stehelin, and J. Rommelaere. 1993. Interconnection between thyroid hormone signalling pathways and parvovirus cytotoxic functions. J. Virol. 67:7668-7672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wetzel, K., P. Menten, G. Opdenakker, J. Van Damme, H. J. Grone, N. Giese, A. Vecchi, S. Sozzani, J. J. Cornelis, J. Rommelaere, and C. Dinsart. 2001. Transduction of human MCP-3 by a parvoviral vector induces leukocyte infiltration and reduces growth of human cervical carcinoma cell xenografts. J. Gene Med. 3:326-337. [DOI] [PubMed] [Google Scholar]
- 56.Wilson, G. M., H. K. Jindal, D. E. Yeung, W. Chen, and C. R. Astell. 1991. Expression of minute virus of mice major nonstructural protein in insect cells: purification and identification of ATPase and helicase activities. Virology 185:90-98. [DOI] [PubMed] [Google Scholar]
- 57.Young, P. J., K. T. Jensen, L. R. Burger, D. J. Pintel, and C. L. Lorson. 2002. Minute virus of mice NS1 interacts with the SMN protein, and they colocalize in novel nuclear bodies induced by parvovirus infection. J. Virol. 76:3892-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]