Abstract
Recombination is thought to be an important source of genetic variation in herpesviruses. Several studies, performed in vitro or in vivo, detected recombinant viruses after the coinoculation of two distinguishable strains of the same herpesvirus species. However, none of these studies investigated the evolution of the relative proportions of parental versus recombinant progeny populations after coinoculation of the natural host, both during the excretion and the reexcretion period. In the present study, we address this by studying the infection of cattle with bovine herpesvirus 1 (BoHV-1). The recombination of two BoHV-1 mutants lacking either glycoprotein C (gC−/gE+) or E (gC+/gE−) was investigated after inoculation of cattle by the natural route of infection. The results demonstrated that (i) recombination is a frequent event in vivo since recombinants (gC+/gE+ and gC−/gE−) were detected in all coinoculated calves, (ii) relative proportions of progeny populations evolved during the excretion period toward a situation where two populations (gC+/gE+ and gC−/gE+) predominated without fully outcompeting the presence of the two other detected populations (gC+/gE− and gC−/gE−), and (iii) after reactivation from latency, no gC+/gE− and gC−/gE− progeny viruses were detected, although gC+/gE− mutants, when inoculated alone, were detected after reactivation treatment. In view of these data, the importance of gE in the biology of BoHV-1 infection and the role of recombination in herpesvirus evolution are discussed.
The genomes of large DNA viruses have generally evolved by three mechanisms: mutation during viral DNA replication, acquisition of cellular genes, and recombination between strains of the same viral species. The Herpesviridae form an extensive family of large DNA viruses that appear to have acquired a diverse collection of genes from their hosts at different times in the past (37). The rate of synonymous nucleotide substitution per site per year has been analyzed for different herpesviruses and was estimated to be 20 to 30 times that of the host genome (12, 19, 29). This low rate of nucleotide substitution most probably reflects the efficient proofreading activity of the herpesvirus DNA polymerase (7) and/or the importance of periods of latency in their biology. Recombination is thought to be an important source of genetic variation in herpesviruses, allowing combination of genetic features shared by different strains. Recombination between herpesviruses was first demonstrated in 1955 when wild-type herpes simplex virus type 1 (HSV-1) was recovered from mixed inoculations between pairs of temperature-sensitive mutants (54). Since then, herpesvirus recombination has been studied both in vitro and/or in vivo between distinguishable strains of HSV-1 and/or HSV-2 (5, 6, 20, 24, 28, 35, 43, 47, 48), pseudorabies virus (PrV) (8, 9, 15, 16, 17, 18, 23), feline herpesvirus 1 (14), and varicella-zoster virus (11). From in vitro studies it has been concluded that recombination is a frequent event, leading to the creation of viruses harboring new genetic combinations. Careful analyses of these recombinants has allowed a better understanding of the molecular mechanisms underlying recombination (49). However, in vitro approaches do not reflect what happens in vivo. In vivo reports, carried out mainly using heterologous animal models, led to the important finding that two avirulent strains can recombine to produce virulent viruses which were demonstrated, in most cases, to be lethal for coinoculated animals (15, 20, 23, 43).
However, the methods used to study in vivo recombination in the reports referenced above precluded the investigation of some important features linked to recombination. Indeed, these studies detected recombination events by characterization of isolated viruses taken from coinoculated animals that were either euthanized at different times postinoculation (p.i.) (16, 18, 24, 28) or died as a consequence of the restoration of virulence after recombination and/or complementation between two avirulent strains (15, 17, 20, 23, 35, 43). Such experimental approaches were incompatible with the analysis of the relative proportions of parental versus recombinant progeny populations during the course of the entire excretion period of a single animal. Moreover, herpesviruses can persist lifelong in a latent state in the infected host but can also periodically reactivate (21, 40). In regard to the latter property, no study has investigated the possibility of the survival of recombinants after reactivation from latency.
Bovine herpesvirus 1 (BoHV-1) is a major pathogen of cattle responsible for respiratory (infectious bovine rhinotracheitis) and genital (infectious pustular vulvovaginitis) diseases (21, 45, 46, 56). After primary infection by the intranasal route, BoHV-1 replicates in the nasal mucosa and can persist in a latent state in sensory neurons of the trigeminal ganglia (1) or in tonsils (55). BoHV-1 can be reactivated and reexcreted by means of several stimuli, including transport, parturition, or treatment with glucocorticoids (36, 56). Several features of the biology of BoHV-1 infection make this virus a good candidate as a model to study in vivo recombination between two strains. First, recombination can be studied in the natural host after coinoculation at the natural site of infection, the nasal mucosa. Second, in contrast to previous studies where sampling procedures to isolate progeny viruses implied the death of coinoculated animals, the nasal swabbing procedure allows monitoring the appearance and the evolution of recombinant viral populations during the entire excretion period of a single animal at the precise site of recombination, the nasal mucosa. Third, the experimental reactivation treatment with glucocorticoids performed on the same animal allows estimation of the survival of both parental and recombinant progeny populations after reactivation from latency.
In the present study, we used previously described PCR and immunofluorescence assays to characterize and quantify progeny viruses generated from coinoculation between two BoHV-1 mutants lacking either the glycoprotein C (gC) or gE open reading frames (ORFs) (41, 42). These assays allowed straightforward discrimination between parental and progeny recombinant viruses (43). Using this methodology, we investigated recombination after coinoculation of cattle by the intranasal route with these two parental BoHV-1 mutants. The present study is the first to investigate the rise of recombinant viruses in the natural host after coinoculation with two distinguishable strains and to monitor the evolution of these viruses during the excretion and reexcretion periods.
MATERIALS AND METHODS
Virus and cell culture.
Two deletion mutants derived from the BoHV-1 strain Lam (33) were used in the present study. The Lam gC− mutant has a deletion in the gene encoding gC (22). From the gC gene the 1.4-kb NcoI-SplI fragment has been deleted, which starts at the start codon of the gC ORF and ends 172 bp upstream of the gC stop codon. Because the deletion causes a frameshift with respect to the last 57 amino acids, no gC peptides are encoded. In the Lam gE− mutant the entire gE ORF is deleted (50). Madin-Darby bovine kidney (MDBK; ATCC CCL-22) cells were grown in Earle minimal essential medium (Gibco-BRL) supplemented with 2% penicillin (P) (5,000 U/ml), streptomycin (S) (5,000 μg/ml), PS (Gibco-BRL), and 5% heat-inactivated fetal bovine serum (BioWhittaker) (MEM-FBS). Viral stocks were produced by separate infections of confluent MDBK cells with either the Lam gC− mutant or the Lam gE− mutant at a low multiplicity of infection (MOI) of 0.1 in Earle's minimal essential medium supplemented with 2% PS and 2% heat-inactivated horse serum (Serolab; International Medical) (MEM-HS). When a 90% cytopathic effect occurred, the culture medium was harvested and clarified twice by centrifugation at 1,000 × g. The resulting cell-free supernatant was divided into aliquots and frozen at −80°C (until used for inoculation) and titrated by plaque assay on MDBK cells as previously described (25). When indicated, embryonic bovine lung (DSMZ ACC 192) cells were used to produce BoHV-1 stocks according to the procedure described above for MDBK cells.
Experimental design.
Twenty calves (5 to 6 months old) from three BoHV-1-free dairy farms were used in the present study. The serological status of these farms had been confirmed through regular serological screening over the past 3 years by using two commercially available enzyme-linked immunosorbent assays: the Serelisa IBR/IPV Ab Monoblocking Assay (Synbiotics) and the BHV-1 gE Antibody Test (Herdcheck Idexx). Calves were transported to the experimental facilities 3 weeks before inoculation. As an additional precaution, serum samples were drawn weekly during this period and tested for the presence of BoHV-1 antibodies by using the enzyme-linked immunosorbent assays mentioned above. All calves were strictly isolated from each other during the course of the study, and precautions were taken to avoid spread of virus between calves. Ten calves (group A) were inoculated intranasally with 2 ml of a 1:1 mixture of Lam gC− and Lam gE− mutants containing 5 × 106 PFU of each mutant (1 ml of the mixture was slowly administered into each nostril). Five calves (group B) were inoculated intranasally with the Lam gC− mutant (107 PFU), and five calves (group C) were inoculated intranasally with the Lam gE− mutant (107 PFU). In order to reactivate putative BoHV-1 latent infection, animals were treated 12 weeks p.i. with dexamethasone (Dexavène; Schering-Plough) at the dose of 0.1 mg/kg of body weight per day administered intravenously for five consecutive days. To monitor virus excretion and reexcretion, nasal swabs were taken from each animal as described previously (25), divided into aliquots, and stored at −80°C until used. Swabs were performed daily during the first 14 days p.i. and during the post-reactivation treatment (PRT) period.
Animal care and experimental procedures were carried out in accordance with the Belgian law (A.R. 14/11/93) implementing the European Council directive number 86/609/ECC of 24 November 1986. The experimental design was accepted by the ethical committee of the Faculty of Veterinary Medicine of the University of Liège, Liège, Belgium.
Detection and titration of BoHV-1 in nasal secretions.
Detection of BoHV-1 in nasal swabs was performed by virus isolation on MDBK cells cultured in 6- and 24-well plates (Multiwell; Becton Dickinson). Each nasal fluid was incubated for 2 h, in duplicate, undiluted and 10-fold diluted. The inoculum was then removed, and MEM-HS was added. Monolayers were observed daily for 5 to 7 days to detect the cytopathic effect typical of BoHV-1. Nasal fluids found to be positive by this first method were further titrated by plaque assay in 24- and 96-well plates with 3 to 4 days of incubation at 37°C before they were stained with an aqueous-alcoholic solution of crystal violet (25). The virus titer was expressed as PFU/100 mg of nasal secretion.
Isolation and characterization of progeny viruses from nasal swabs.
Isolation and characterization of progeny viruses from nasal secretions was performed as described previously (42). Briefly, progeny viruses were isolated directly by plaque picking and further propagated individually. Isolates were then characterized by using a PCR, and an immunofluorescence-based approach as described previously (41, 42). The PCR method allows the simultaneous detection of both gC and gE sequences. To confirm PCR results suggesting gC+/gE+ or gC−/gE− recombinants, additional immunofluorescence assays were performed as described previously (42), with minor modifications. Plaques of the isolates that generated a gC+/gE+ PCR result were subjected to a double immunofluorescence staining with monoclonal antibodies (MAbs) raised against gC (MAb 1507 [30, 31]) and gE (MAb BH35 [2]). MAb 1507 is a mouse immunoglobulin G2a (IgG2a), used at a dilution of 1/10,000, and was detected by using fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse IgG2a antibodies (FITC-GAM-IgG2a; Imtech). MAb BH35 is a mouse IgG1, used at a dilution of 1/1,000, and was detected by using R-phycoerythrin (PE)-conjugated goat anti-mouse IgG1 antibodies (R-PE-GAM-IgG1; Imtech). The same assay was applied to plaques formed by isolates that generated a gC−/gE− PCR result. In addition, to control that these latter isolates were indeed BoHV-1, plaques were stained with MAb 3402 raised against BoHV-1 gD (30, 31). MAb 3402 is a mouse IgG2a, used at a dilution of 1/10,000, and was detected by using FITC-conjugated goat anti-mouse IgG2a antibodies (FITC-GAM-IgG2a; Imtech).
RESULTS
Parental strains replicated efficiently when inoculated alone.
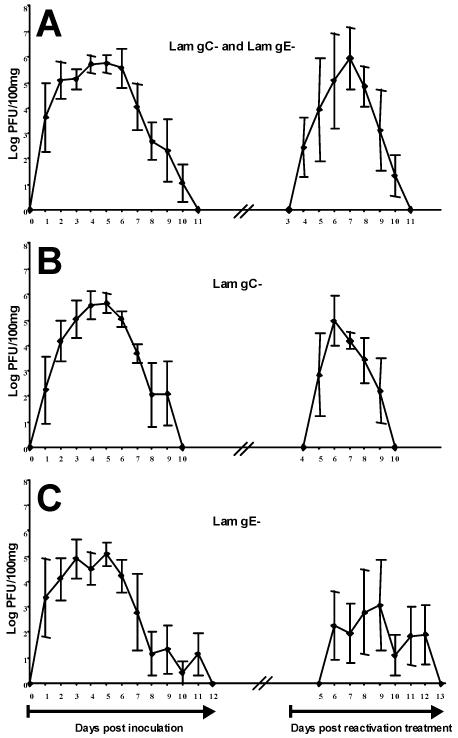
To study in vivo recombination between two strains of the same herpesvirus in the natural host, 10 calves were coinoculated intranasally with both BoHV-1 Lam gC− and Lam gE− mutants (group A) as described in Materials and Methods. However, because viral DNA replication is required for recombination (39), it was crucial to ensure that, in our experimental conditions, the two parental mutants used to study recombination replicate when inoculated alone. Two groups of five calves were therefore inoculated intranasally with either the Lam gC− parental mutant (group B) or the Lam gE− parental mutant (group C). Mean excretion virus titers are shown in Fig. 1. BoHV-1 replicated in the nasal mucosa of each calf, starting on day 1 p.i. for calves of group A and on day 1 or 2 for calves of groups B and C. BoHV-1 was recovered from nasal secretions between 8 and 10 days (group A), 7 and 9 days (group B), and 6 and 11 days (group C). The mean peak virus titers were detected on day 5 p.i. and were 105.7 ± 0.3 PFU/100 mg (group A), 105.6 ± 0.4 PFU/100 mg (group B), and 105.1 ± 0.4 PFU/100 mg (group C). These results indicate that our experimental conditions allow the replication of both parental mutants used to study recombination. Therefore, it can be deduced that the two parental mutants also replicated in the nasal mucosa of calves coinoculated with both mutants, thus meeting one of the conditions for recombination to occur.
FIG. 1.
Nasal BoHV-1 excretion and reexcretion. Mean (± the standard error of the mean) BoHV-1 excretion and reexcretion titers of 10 calves inoculated with both Lam gC− and Lam gE− mutants (A), five calves inoculated with Lam gC− mutant (B), and five calves inoculated with Lam gE− mutant (C). Nasal swabs were taken daily starting from day 0 (the day of inoculation) to day 14. BoHV-1 was reactivated experimentally 12 weeks p.i. by dexamethasone treatment as described in Materials and Methods. Nasal swabs were taken daily during the first 14 days PRT. The virus titers were determined by plaque assay and are expressed as the log PFU/100 mg of nasal secretions.
BoHV-1 reexcretion PRT analysis.
A reactivation treatment was performed 12 weeks p.i. The mean reexcretion viral titers are shown in Fig. 1. BoHV-1 was recovered PRT from nasal secretions of all calves in group A between 3 to 6 days. The mean peak virus titer was detected on day 7 PRT and was 105.9 ± 1.2 PFU/100 mg. In group B, calves shed the Lam gC− parental mutant for 4 days starting from day 4 or 5 up to day 8 or 9 PRT. The mean peak virus titer was detected on day 6 PRT and was 104.9 ± 1.0 PFU/100 mg. In group C, four of five calves reexcreted the Lam gE− parental mutant, whereas no virus was detected for the remaining calf. In calves experiencing reexcretion, the Lam gE− mutant was detected for a period of 1, 3, 4, or 5 days. The mean peak virus titer was detected on day 9 PRT and was 103.1 ± 1.8 PFU/100 mg.
Recombinant viruses were detected in nasal secretions of all coinoculated calves.
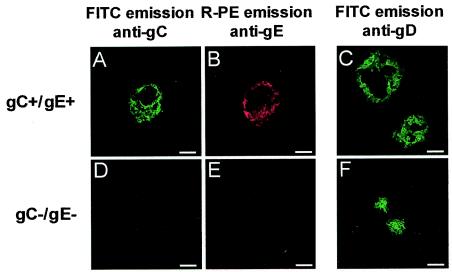
A first screening to detect recombinant viruses was performed on nasal swabs of each coinoculated calf taken on day 5 p.i. From each swab, 20 progeny viruses were plaque purified randomly and characterized for the presence or absence of gC and gE. Indeed, recombination between the Lam gC− (gC−/gE+) and the Lam gE− (gC+/gE−) parental mutants is theoretically expected to generate two distinguishable recombinant viruses: the first harboring both gC and gE (gC+/gE+) and the second with both genes deleted (gC−/gE−). Figure 2 shows the phenotypic characterization of both types of recombinant viruses generated during the course of this experiment. The results presented in Table 1 show that gC+/gE+ recombinant viruses were detected in all 10 coinoculated calves, whereas gC−/gE− recombinants were detected in 8 of 10 coinoculated calves. To investigate whether the lack of detection of gC−/gE− recombinants in nasal swabs of calves 9 and 10 was due to the limited number of progeny viruses characterized during this first screening, 20 additional progeny viruses were plaque purified and characterized. The results indicated that at least one progeny virus from each animal was a gC−/gE− recombinant (data not shown), demonstrating that gC−/gE− recombinants were detected in all coinoculated calves. These results demonstrate that recombination occurs frequently in vivo since both types of recombinants were detected in all coinoculated animals.
FIG. 2.
Phenotypic characterization of recombinant viruses. Progeny viruses identified by PCR as gC+/gE+ or gC−/gE− were further characterized by immunofluorescence. Plaques were stained for immunofluorescence detection of gC (A and D) and gE (B and E) as described in Materials and Methods. gC and gE were detected by determining the FITC and R-PE emission, respectively. (C and F) Independently, plaques generated from the same isolates were stained for immunofluorescence detection of gD and visualized by determining the FITC emission. Bars, 100 μm.
TABLE 1.
Characterization of progeny viruses as parental or recombinant virusesa
| Calf no. | No. of progeny virus isolates
|
|||
|---|---|---|---|---|
| Parental
|
Recombinant
|
|||
| gC−/gE+ | gC+/gE− | gC+/gE+ | gC−/gE− | |
| 1 | 10 | 2 | 7 | 1 |
| 2 | 8 | 1 | 10 | 1 |
| 3 | 10 | 2 | 7 | 1 |
| 4 | 7 | 2 | 9 | 2 |
| 5 | 11 | 1 | 7 | 1 |
| 6 | 10 | 0 | 9 | 1 |
| 7 | 11 | 1 | 7 | 1 |
| 8 | 8 | 1 | 9 | 2 |
| 9 | 7 | 3 | 10 | 0 |
| 10 | 10 | 2 | 8 | 0 |
Twenty progeny viruses from nasal swabs taken on day 5 after coinoculation were isolated and characterized as described in Materials and Methods.
The cell line used for plaque purification and characterization does not influence the proportions of progeny virus populations.
The approach used in the present study to quantify the relative proportions of the four progeny populations postulates that the cell type used for the plaque purification procedure does not favor the infectivity of one (or more) of the four progeny populations. Because it was demonstrated that the requirements of gC in the attachment process of PrV vary between different cell types (32), it was crucial to address this issue. With that goal in mind, the four viruses were replicated and titrated by using embryonic bovine lung cells. Based on these titers, an appropriate dilution (i.e., allowing plaque purification) of a 1:1:1:1 mixture of the four progeny viruses was used to inoculate MDBK cells. After incubation, 113 plaques were purified and characterized as described above. The results indicated that 29 of 113 (25.7%) were gC−/gE+ viruses, 25 of 113 (22.1%) were gC+/gE− viruses, 23 of 113 (20.3%) were gC+/gE+ viruses, and 36 of 113 (31.9%) were gC−/gE− viruses. These results indicate that our methodology to isolate and characterize progeny viruses by using MDBK cells does not influence the relative proportions of progeny populations present in the nasal mucosa of coinoculated calves.
Evolution of viral populations during the course of the excretion period.
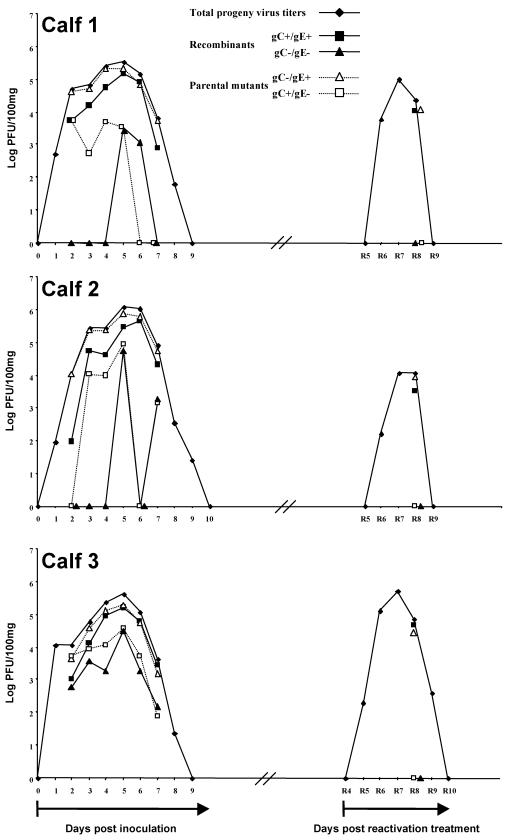
Three calves from group A (calves 1, 2, and 3) were selected randomly for this part of the study. Total progeny virus titers were determined, allowing the establishment of individual excretion curves (Fig. 3). Calves 1 and 3 excreted BoHV-1 from days 1 to 8 p.i., whereas calf 2 excreted BoHV-1 from days 1 to 9 p.i. Progeny viruses from BoHV-1-positive nasal swabs were isolated and characterized. The number of progeny viruses isolated from each nasal swab is listed in Table 2. Progeny virus isolation was not applied on days 1, 8, and 9 p.i. because either the amount of progeny viruses that were detected was too small or no progeny virus was detected. For a given day of the excretion period, each progeny population was first expressed as a relative percentage of the total number of isolates characterized. The percentages obtained were then expressed as log PFU/100 mg of nasal secretions after multiplication by the corresponding total progeny virus titer. After this procedure the total BoHV-1 excretion curves of calves 1, 2, and 3 were separated into four subcurves representing the evolution of the four progeny populations during the entire excretion period (Fig. 3).
FIG. 3.
Evolution of progeny virus populations in three calves coinoculated with both Lam gC− and Lam gE− mutants. Calves were coinoculated intranasally on day 0. An experimental reactivation treatment was performed at 12 weeks p.i. Nasal swabs were taken daily, and total virus titers were determined by plaque assay both p.i. and PRT. Progeny viruses from nasal secretions collected from day 2 to 7 p.i. and on day 8 PRT were further characterized to determine the relative titers of each progeny virus population and their evolution. Titers are expressed as the log PFU/100 mg of nasal secretions.
TABLE 2.
Number of progeny viruses isolated from nasal swabs collected during both the excretion and reexcretion periods
| Calf no. | No. of progeny virus isolates at day:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p.i.a
|
PRTb
|
|||||||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 5 | 6 | 7 | 8 | 9 | |
| 1 | -c | 123 | 125 | 112 | 107 | 130 | 105 | - | 0d | 0 | - | - | 117 | 0 |
| 2 | - | 119 | 132 | 126 | 108 | 109 | 177 | - | - | 0 | - | - | 89 | 0 |
| 3 | - | 135 | 121 | 121 | 130 | 131 | 56 | - | 0 | - | - | - | 116 | - |
Coinoculation was performed on day 0.
The first injection of dexamethasone was performed 12 weeks p.i.
-, no progeny virus was isolated because BoHV-1 virus titers were too low.
0, BoHV-1 was not detected by plaque assay.
Although isolation and characterization of progeny viruses were not done on day 1 p.i., the results demonstrate that recombination occurred soon after coinoculation since recombinants were detected as early as 2 days p.i. Indeed, from nasal swabs collected on day 2 p.i., a gC+/gE+ recombinant population was detected in the three calves, whereas a gC−/gE− recombinant population was detected only in calf 3. For calves 1 and 2 the gC−/gE+ parental population predominates on day 2 p.i., whereas the gC+/gE− parental population was approximately 10-fold less abundant in calf 1 and was not detected in calf 2. In contrast, both parental populations were detected in approximately equal amounts in calf 3. Starting from these different proportions of progeny populations on day 2 p.i., the gC−/gE+ parental population evolved to predominate in each calf even in the case of calf 3, in which the two parental progeny viruses were detected in approximately equal amounts on day 2 p.i. In addition, the gC+/gE+ recombinant population evolved during the course of the excretion period to reach levels very similar to those of the gC−/gE+ parental population (see Fig. 3, day 6 p.i.). Finally, the gC−/gE− recombinant population was detected continuously during all of the excretion period for calf 3, whereas this recombinant population was detected only on days 5 and 6 p.i. for calf 1 or on days 5 and 7 p.i. for calf 2. Since the replication of the gC+/gE− mutant is required to give rise to gC−/gE− recombinants, it should be noted that this observation correlates with the fact that the parental gC+/gE− mutant was detected throughout the excretion period for calf 3 but not for calves 1 and 2.
Taken together, these results confirm that recombination is a frequent event in vivo that occurs shortly after coinoculation. Moreover, recombination between the two parental populations generated two new recombinant populations, and our results demonstrated that the relative proportions of the progeny populations evolved during the excretion period toward a situation where two populations predominated largely without fully outcompeting the other two populations.
Evolution of viral populations during the course of the reexcretion period.
Distinguishable strains of a herpesvirus can establish latency in the same tissue of their natural host (53). Moreover, the characterization of isolates taken from the latency sites of animals coinoculated with two different parental strains demonstrated in some cases that both recombinants and parental viruses reached the latency site (8, 43). However, the question of how the different progeny populations evolved after reactivation from latency remained. To address this issue, progeny viruses isolated from nasal swabs taken on day 8 PRT (Table 2) from calves 1, 2, and 3 were characterized. The results indicated that only two progeny virus populations were detected after reactivation (Fig. 3): the gC−/gE+ parental and the gC+/gE+ recombinant populations. In contrast, gC+/gE− and gC−/gE− progeny populations were not detected PRT, although the gC+/gE− mutant, when inoculated alone, was detected in the nasal secretions of four of five calves.
DISCUSSION
In this study we investigated the process of in vivo recombination between two distinguishable mutants of BoHV-1 after coinoculation of the natural host by the natural route of infection. To the best of our knowledge, the methodology used in the present study is the first to allow the investigation of both the rise and the survival of recombinant viruses after primary infection and reactivation from latency, the entire experiment being performed on the same animal. The results presented here demonstrate that (i) starting from two parental mutants, recombination generates a new progeny population composed of both parental (gC−/gE+ and gC+/gE−) and recombinant (gC+/gE+ and gC−/gE−) viruses; (ii) recombination is a frequent event in vivo since recombinants were detected in all coinoculated calves; (iii) the relative proportions of progeny populations evolve during the excretion period toward a situation in which two populations (gC+/gE+ and gC−/gE+) predominate largely without outcompeting completely the presence of the other two (gC+/gE− and gC−/gE−); and (iv), after reactivation from latency, no gC+/gE− and gC−/gE− progeny viruses could be detected, although gC+/gE− viruses, when inoculated alone, were observed.
BoHV-1 gC plays a role in viral attachment to target cells (27) and BoHV-1 gE in the cell-to-cell spread of the virus (38), as was demonstrated for homologue proteins of HSV-1 (10, 44) and PrV (32, 57). In addition, it was recently shown that PrV gE is involved in virus egress (3, 4, 13). In our experiments, both types of recombinants were detected in all coinoculated calves. The detection of high levels of gC+/gE+ recombinants was not surprising since they possess wild-type characteristics. In contrast, the detection of gC−/gE− recombinants, even at low levels, was surprising in light of two previous reports dealing with PrV gC−/gE− mutants. Although gC−/gE− and gC−/gE+ PrV mutants grew similarly when the cells were infected at a high MOI, the gC−/gE− virus had a considerable growth defect when the cells were infected at a low MOI (57). In addition, it has been shown recently that after inoculation by the intranasal route of naive pigs with high titers of a PrV gC−/gE− mutant, only half of the inoculated animals excreted the gC−/gE− virus during a short period of time (34). These results indicated that the simultaneous deletion of gC and gE has an important impact on the ability of these mutants to replicate both in vitro and in vivo. We hypothesize that because gC−/gE− recombinants rise obligatory in a cell coinoculated with both parental mutants (gC−/gE+ and gC+/gE−), they should be complemented phenotypically for both gC and gE, allowing them to interact with their environment as efficiently as wild-type virus. In particular, after egress from the cell, these recombinants possess a viral envelope containing gC, which is known to play a role in the primary attachment of the virus to the target cell (27, 46). Moreover, because gE is required for efficient cell-to-cell spread of BoHV-1 (38), the synthesis of gE in cells where gC−/gE− recombinants were generated could contribute to the survival of these recombinants by an easier propagation to uninfected cells. Additional investigations are required to demonstrate this hypothesis. For example, it will be of particular interest to compare the replication rate of BoHV-1 gC−/gE− mutants in the presence or absence of a wild-type virus both in vitro and in vivo by using the methodology described previously (42).
BoHV-1 establishes latency in sensory neurons of trigeminal ganglia after primary infection of the nasal mucosa (1), and it has been demonstrated that a single host can support the latent infection of two distinguishable BoHV-1 strains (53). It was also demonstrated that the gE deletion does not affect the ability of BoHV-1 to replicate and to establish latency but leads to the reduction of the amount of sensory neurons infected in comparision to wild-type virus (51). In addition, it was reported that gE− strains can be reactivated and reexcreted from latency (26) even if, in this case, the process is also severely impaired compared to gE positive strains. Our results are in agreement with these previous observations since four of five calves reexcreted the Lam gE− mutant in significantly smaller amounts than the Lam gC− mutant when these mutants were inoculated alone. We then sought to investigate the survival of both gC+/gE− parental and gC−/gE− recombinant populations in the context of the simultaneous presence of parental gC−/gE+ and recombinant gC+/gE+ populations. From 312 isolates that were characterized PRT, no gE− viruses were detected in nasal swabs, although our results demonstrated that gC+/gE− viruses were detected when inoculated alone. Unfortunately, we were unable to investigate whether the lack of detection of gE− viruses PRT is attributable to their failure to reach the latency site or reexcrete from it.
Our study has also important implications in the context of the extensive use of gE deleted marker vaccines as a tool in BoHV-1 eradication programs (52), especially those evaluating the potential emergence of recombinants between BoHV-1 gE deleted marker vaccine and wild-type strains. The results presented here suggest that the epidemiological risk of the spread of recombinant viruses with the gE gene deleted from a latently infected animal to adjacent cattle is very slight.
The data obtained here indicate that in vivo BoHV-1 recombination is a frequent event. Indeed, all possible recombinant progeny viruses (even gC−/gE− viruses) were isolated from all coinoculated animals, indicating that recombination is an efficient mechanism for creating genetic diversity in BoHV-1. As indicated above, the rate of nucleotide substitution is relatively low in herpesviruses. The use of recombination is certainly a mechanism by which herpesviruses make the most of the genetic diversity created by mutation during viral DNA replication or by acquisition of cellular genes.
In conclusion, the present study is the first to investigate the rise of recombinant viruses after coinoculation, in the natural host, of two distinguishable strains and to monitor the evolution of these viruses during the excretion and reexcretion period. This study reveals the importance of gE in the biology of BoHV-1 infection in vivo and highlights the importance of recombination in the evolution of herpesviruses.
Acknowledgments
We thank J. Letchworth (University of Wisconsin-Madison) for providing monoclonal antibodies and J. T. van Oirschot and F. A. M. Rijsewijk (Institute for Animal Science and Health, Lelystad, The Netherlands) for providing viruses. We are grateful to M. A. McVoy and E. Baranowski for critical reading of the manuscript and to J.-P. Georgin and A. Brichaud for excellent technical assistance.
This study was financially supported by the Ministère des Classes Moyennes et de l'Agriculture, Administration Recherche et Développement. F.M. is a Research Fellow of the Fonds National Belge de la Recherche Scientifique (FNRS). A.V. is a Senior Research Associate of the FNRS. Purchase of the confocal microscope was supported by the following grants: FRFC 2.4532.98 from the FNRS, FNRS LOTTO 9.4592.97 from the Belgian National Lottery, and ARC 98/03-220 from the French Community of Belgium.
REFERENCES
- 1.Ackermann, M., E. Peterhans, and R. Wyler. 1982. DNA of bovine herpesvirus type 1 in the trigeminal ganglia of latently infected calves. Am. J. Vet. Res. 43:36-40. [PubMed] [Google Scholar]
- 2.Baranowski, E., J. Dubuisson, P.-P. Pastoret, and E. Thiry. 1993. Identification of 108K, 93K, and 42K glycoproteins of bovine herpesvirus-1 by monoclonal antibodies. Arch. Virol. 133:97-111. [DOI] [PubMed] [Google Scholar]
- 3.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, S. M., D. A. Ritchie, and J. H. Subak-Sharpe. 1973. Genetic studies with herpes simplex virus type 1: the isolation of temperature-sensitive mutants, their arrangement into complementation groups and recombination analysis leading to a linkage map. J. Gen. Virol. 18:329-346. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S. M., J. H. Subak-Sharpe, J. Harland, and A. R. MacLean. 1992. Analysis of intrastrain recombination in herpes simplex virus type 1 strain 17 and herpes simplex virus type 2 strain HG52 using restriction endonuclease sites as unselected markers and temperature-sensitive lesions as selected markers. J. Gen. Virol. 73:293-301. [DOI] [PubMed] [Google Scholar]
- 7.Crute, J. J., and I. R. Lehman. 1989. Herpes simplex-1 DNA polymerase: identification of an intrinsic 5′-3′ exonuclease with ribonuclease H activity. J. Biol. Chem. 264:19266-19270. [PubMed] [Google Scholar]
- 8.Dangler, C. A., L. M. Henderson, L. A. Bowman, and R. E. Deaver. 1993. Direct isolation and identification of recombinant pseudorabies virus strains from tissues of experimentally coinfected swine. Am. J. Vet. Res. 54:540-545. [PubMed] [Google Scholar]
- 9.Dangler, C. A., R. E. Deaver, and C. M. Kolodziej. 1994. Genetic recombination between two strains of Aujeszky's disease virus at reduced multiplicity of infection. J. Gen. Virol. 75:295-299. [DOI] [PubMed] [Google Scholar]
- 10.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dohner, D. E., S. G. Adams, and L. D. Gelb. 1988. Recombination in tissue culture between varicella-zoster virus strains. J. Med. Virol. 24:329-341. [DOI] [PubMed] [Google Scholar]
- 12.Dolan, A., F. E. Jamieson, C. Cunningham, B. C. Barnett, and D. J. McGeoch. 1998. The genome sequence of herpes simplex virus type 2. J. Virol. 72:2010-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuchs, W., B. G. Klupp, H. Granzow, C. Hengartner, A. Brack, A. Mundt, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita, K., K. Maeda, N. Yokoyama, T. Miyazawa, C. Kai, and T. Mikami. 1998. In vitro recombination of feline herpesvirus type 1. Arch. Virol. 143:25-34. [DOI] [PubMed] [Google Scholar]
- 15.Glazenburg, K. L., R. J. Moormann, T. G. Kimman, A. L. Gielkens, and B. P. Peeters. 1994. In vivo recombination of pseudorabies virus strains in mice. Virus Res. 34:115-126. [DOI] [PubMed] [Google Scholar]
- 16.Glazenburg, K. L., R. J. Moormann, T. G. Kimman, A. L. Gielkens, and B. P. Peeters. 1995. Genetic recombination of pseudorabies virus: evidence that homologous recombination between insert sequences is less frequent than between autologous sequences. Arch. Virol. 140:671-685. [DOI] [PubMed] [Google Scholar]
- 17.Henderson, L. M., J. B. Katz, G. A. Erickson, and J. E. Mayfield. 1990. In vivo and in vitro genetic recombination between conventional and gene-deleted vaccine strains of pseudorabies virus. Am. J. Vet. Res. 51:1656-1662. [PubMed] [Google Scholar]
- 18.Henderson, L. M., R. L. Levings, A. J. Davis, and D. R. Sturtz. 1991. Recombination of pseudorabies virus vaccine strains in swine. Am. J. Vet. Res. 52:820-825. [PubMed] [Google Scholar]
- 19.Hughes, A. L. 2002. Origin and evolution of viral interleukin-10 and other DNA virus genes with vertebrate homologues. J. Mol. Evol. 54:90-101. [DOI] [PubMed] [Google Scholar]
- 20.Javier, R. T., F. Sedarati, and J. G. Stevens. 1986. Two avirulent herpes simplex viruses generate lethal recombinants in vivo. Science 234:746-748. [DOI] [PubMed] [Google Scholar]
- 21.Jones, C. 1998. Alphaherpesvirus latency: its role in disease and survival of the virus in nature. Adv. Virus Res. 51:81-133. [DOI] [PubMed] [Google Scholar]
- 22.Kaashoek, M. J., F. A. M. Rijsewijk, R. C. Ruuls, G. M. Keil, E. Thiry, P. P. Pastoret, and J. T. van Oirschot. 1998. Virulence, immunogenicity and reactivation of bovine herpesvirus 1 mutants with a deletion in the gC, gG, gI, gE, or in both the gI and gE gene. Vaccine 16:802-809. [DOI] [PubMed] [Google Scholar]
- 23.Katz, J. B., L. M. Henderson, and G. A. Erickson. 1990. Recombination in vivo of pseudorabies vaccine strains to produce new virus strains. Vaccine 8:286-288. [DOI] [PubMed] [Google Scholar]
- 24.Kintner, R. L., R. W. Allan, and C. R. Brandt. 1995. Recombinants are isolated at high frequency following in vivo mixed ocular infection with two avirulent herpes simplex virus type 1 strains. Arch. Virol. 140:231-244. [DOI] [PubMed] [Google Scholar]
- 25.Lemaire, M., F. Schynts, G. Meyer, and E. Thiry. 1999. Antibody response to glycoprotein E after bovine herpesvirus type 1 infection in passively immunized, glycoprotein E-negative calves. Vet. Rec. 144:172-176. [DOI] [PubMed] [Google Scholar]
- 26.Lemaire, M., F. Schynts, G. Meyer, J. P. Georgin, E. Baranowski, A. Gabriel, C. Ros, S. Belak, and E. Thiry. 2001. Latency and reactivation of a glycoprotein E negative bovine herpesvirus type 1 vaccine: influence of virus load and effect of specific maternal antibodies. Vaccine 19:4795-4804. [DOI] [PubMed] [Google Scholar]
- 27.Liang, X. P., L. A. Babiuk, S. van Drunen Littel-van den Hurk, D. R. Fitzpatrick, and T. J. Zamb. 1991. Bovine herpesvirus 1 attachment to permissive cells is mediated by its major glycoproteins gI, gIII, and gIV. J. Virol. 65:1124-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lingen, M., F. Hengerer, and D. Falke. 1997. Mixed vaginal infections of BALB/c mice with low virulent herpes simplex type 1 strains result in restoration of virulence properties: vaginitis/vulvitis and neuroinvasiveness. Med. Microbiol. Immunol. 185:217-222. [DOI] [PubMed] [Google Scholar]
- 29.Markine-Goriaynoff, N., J.-P. Georgin, M. Goltz, W. Zimmermann, H. Broll, H. M. Wamwayi, P.-P. Pastoret, P. M. Sharp, and A. Vanderplasschen. 2003. The core 2 β-1,6-N-acetylglucosaminyltransferase-mucin encoded by bovine herpesvirus 4 was acquired from an ancestor of the African buffalo. J. Virol. 77:1784-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall, R. L., L. L. Rodriguez, and G. J. Letchworth. 1986. Characterization of envelope proteins of infectious bovine rhinotracheitis virus (bovine herpesvirus 1) by biochemical and immunological methods. J. Virol. 57:745-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marshall, R. L., B. A. Israel, and G. J. Letchworth. 1988. Monoclonal antibody analysis of bovine herpesvirus-1 glycoprotein antigenic areas relevant to natural infection. Virology 165:338-347. [DOI] [PubMed] [Google Scholar]
- 32.Mettenleiter, T. C., L. Zsak, F. Zuckermann, N. Sugg, H. Kern, and T. Ben-Porat. 1990. Interaction of glycoprotein gIII with a cellular heparinlike substance mediates adsorption of pseudorabies virus. J. Virol. 64:278-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzler, A. E., H. Matile, U. Gassmann, M. Engels, and R. Wyler. 1985. European isolates of bovine herpesvirus 1: a comparison of restriction endonuclease sites, polypeptides, and reactivity with monoclonal antibodies. Arch. Virol. 85:57-69. [DOI] [PubMed] [Google Scholar]
- 34.Nauwynck, H. J., G. G. Labarque, and M. B. Pensaert. 1999. Efficacy of an intranasal immunization with gEgC and gEgI double-deletion mutants of Aujeszky's disease virus in maternally immune pigs and the effects of a successive intramuscular booster with commercial vaccines. J. Vet. Med. B 46:713-722. [DOI] [PubMed] [Google Scholar]
- 35.Nishiyama, Y., H. Kimura, and T. Daikoku. 1991. Complementary lethal invasion of the central nervous system by nonneuroinvasive herpes simplex virus types 1 and 2. J. Virol. 65:4520-4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pastoret, P. P., E. Thiry, and R. Thomas. 1986. Logical description of bovine herpesvirus type 1 latent infection. J. Gen. Virol. 67:885-897. [DOI] [PubMed] [Google Scholar]
- 37.Raftery, M., A. Muller, and G. Schonrich. 2000. Herpesvirus homologues of cellular genes. Virus Genes 21:65-75. [PubMed] [Google Scholar]
- 38.Rebordosa, X., J. Pinol, J. A. Perez-Pons, J. Lloberas, J. Naval, X. Serra-Hartmann, E. Espuna, and E. Querol. 1996. Glycoprotein E of bovine herpesvirus type 1 is involved in virus transmission by direct cell-to-cell spread. Virus Res. 45:59-68. [DOI] [PubMed] [Google Scholar]
- 39.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 40.Roizman, B., and P. E. Pellett. 2001. The family of Herpesviridae: a brief introduction, p. 2381-2497. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott/The Williams & Wilkins Co., Philadelphia, Pa.
- 41.Schynts, F., E. Baranowski, M. Lemaire, and E. Thiry. 1999. A specific PCR to differentiate between gE negative vaccine and wild-type bovine herpesvirus type 1 strains. Vet. Microbiol. 66:187-195. [DOI] [PubMed] [Google Scholar]
- 42.Schynts, F., A. Vanderplasschen, E. Hanon, F. A. M. Rijsewijk, J. T. van Oirschot, and E. Thiry. 2001. Use of PCR and immunofluorescence to detect bovine herpesvirus 1 recombinants. J. Virol. Methods 92:99-104. [DOI] [PubMed] [Google Scholar]
- 43.Sedarati, F., R. T. Javier, and J. G. Stevens. 1988. Pathogenesis of a lethal mixed infection in mice with two nonneuroinvasive herpes simplex virus strains. J. Virol. 62:3037-3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spear, P. G., M. T. Shieh, B. C. Herold, D. WuDunn, and T. I. Koshy. 1992. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv. Exp. Med. Biol. 313:341-353. [DOI] [PubMed] [Google Scholar]
- 45.Straub, O. C. 1990. Infectious bovine rhinotracheitis, p. 71-108. In Z. Dinter and B. Morein (ed.), Virus infections of ruminants. Elsevier Science Publishers, Amsterdam, The Netherlands.
- 46.Tikoo, S. K., M. Campos, and L. A. Babiuk. 1995. Bovine herpesvirus 1 (BHV-1): biology, pathogenesis, and control. Adv. Virus Res. 45:191-223. [DOI] [PubMed] [Google Scholar]
- 47.Timbury, M. C., and J. H. Subak-Sharpe. 1973. Genetic interactions between temperature-sensitive mutants of types 1 and 2 herpes simplex viruses. J. Gen. Virol. 18:347-357. [DOI] [PubMed] [Google Scholar]
- 48.Umene, K. 1985. Intermolecular recombination of the herpes simplex virus type 1 genome analysed using two strains differing in restriction enzyme cleavage sites. J. Gen. Virol. 66:2659-2670. [DOI] [PubMed] [Google Scholar]
- 49.Umene, K. 1999. Mechanism and application of genetic recombination in herpesviruses. Rev. Med. Virol. 9:171-182. [DOI] [PubMed] [Google Scholar]
- 50.van Engelenburg, F. A. C., M. J. Kaashoek, F. A. M. Rijsewijk, L. van den Burg, A. Moerman, A. L. Gielkens, and J. T. van Oirschot. 1994. A glycoprotein E deletion mutant of bovine herpesvirus 1 is avirulent in calves. J. Gen. Virol. 75:2311-2318. [DOI] [PubMed] [Google Scholar]
- 51.van Engelenburg, F. A., M. J. Kaashoek, J. T. van Oirschot, and F. A. M. Rijsewijk. 1995. A glycoprotein E deletion mutant of bovine herpesvirus 1 infects the same limited number of tissues in calves as wild-type virus, but for a shorter period. J. Gen. Virol. 76:2387-2392. [DOI] [PubMed] [Google Scholar]
- 52.van Oirshot, J. T., M. J. Kaashoek, F. A. M. Rijsewijk, and J. A. Stegeman. 1996. The use of marker vaccines in eradication of herpesviruses. J. Biotechnol. 44:75-81. [DOI] [PubMed] [Google Scholar]
- 53.Whetstone, C. A., and J. M. Miller. 1989. Two different strains of an alphaherpesvirus can establish latency in the same tissue of the host animal: evidence from bovine herpesvirus 1. Arch. Virol. 107:27-34. [DOI] [PubMed] [Google Scholar]
- 54.Wildy, P. 1955. Recombination with herpes simplex virus. J. Gen. Microbiol. 13:34-46. [DOI] [PubMed] [Google Scholar]
- 55.Winkler, M. T., A. Doster, and C. Jones. 2000. Persistence and reactivation of bovine herpesvirus 1 in the tonsils of latently infected calves. J. Virol. 74:5337-5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wyler, R., M. Engels, and M. Schwyzer. 1989. Infectious bovine rhinotracheitis/vulvovaginitis (BHV1), p. 1-72. In G. Wittmann (ed.), Herpesvirus disease of cattle, horses, and pigs. Kluwer Academic Publishers, Boston, Mass.
- 57.Zsak, L., F. Zuckermann, N. Sugg, and T. Ben-Porat. 1992. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J. Virol. 66:2316-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]