Abstract
The glycoprotein (G) of vesicular stomatitis virus (VSV) is responsible for binding of virus to cells and for mediating virus entry following endocytosis by inducing fusion of the viral envelope with the endosomal membrane. The fusion peptide of G is internal (residues 116 to 137) and exhibits characteristics similar to those of other internal fusion peptides, but recent studies have implicated the region adjacent to the transmembrane domain as also being important for G-mediated membrane fusion. Sequence alignment of the membrane-proximal region of G from several different vesiculoviruses revealed that this domain is highly conserved, suggesting that it is important for G function. Mutational analysis was used to show that this region is not essential for G protein oligomerization, transport to the cell surface, or incorporation into virus particles but that it is essential for acid-induced membrane fusion activity and for virus infectivity. Deletion of the 13 membrane-proximal amino acids (N449 to W461) dramatically reduced cell-cell fusion activity and reduced virus infectivity approximately 100-fold, but mutation of conserved aromatic residues (W457, F458, and W461) either singly or together had only modest effects on cell-cell fusion activity; recombinant virus encoding these mutants replicated as efficiently as wild-type (WT) VSV. Insertion of heterologous sequences in the juxtamembrane region completely abolished membrane fusion activity and virus infectivity, as did deletion of residues F440 to N449. The insertion mutants showed some changes in pH-dependent conformational changes and in virus binding, which could partially explain the defects in membrane fusion activity, but all the other mutants were similar to WT G with respect to conformational changes and virus binding. These data support the hypothesis that the membrane-proximal domain contributes to G-mediated membrane fusion activity, yet the conserved aromatic residues are not essential for membrane fusion or virus infectivity.
Vesicular stomatitis virus (VSV) is the prototypic member of the Vesiculovirus genera of the Rhabdovirus family. The genome of the virus is a single molecule of negative-sense RNA that encodes five major proteins, glycoprotein (G), matrix protein (M), nucleoprotein (N), large protein (L), and phosphoprotein (P). The G protein mediates both virus attachment to the host cell and fusion of the viral envelope with the endosomal membrane following endocytosis (12, 29).
Results of mutational analyses of residues 118 to 136 of the G protein ectodomain as well as results from hydrophobic photolabeling experiments with VSV provided evidence that this region is the internal fusion peptide and that it inserts into target membranes at acidic pH (11, 16, 27, 33, 49, 51). It has also been shown that insertions or substitutions in the region between residues 395 to 418 affect membrane fusion activity of G protein (27, 40). Double mutants with substitutions in both the fusion peptide and residues 395 to 418 had an additive effect upon fusion inhibition (39), indicating that the C-terminal region of the ectodomain also plays an important role in the fusion activity of G.
Jeetendra et al. recently showed that the membrane-proximal 42 amino acids (aa) (residues 421 to 461) of the G protein ectodomain together with the transmembrane (TM) and cytoplasmic tail (called the G-stem or GS) can potentiate the membrane fusion activity of heterologous viral fusion proteins when the two proteins are coexpressed (22). Further, Jeetendra et al. also showed that this domain binds to membranes independent of the ectodomain and mediates hemifusion (22). These data suggested that the membrane-proximal domain of VSV G protein is also important for membrane fusion activity. Studies with a number of viral fusion proteins, such as human immunodeficiency virus type 1 (HIV-1) envelope protein gp41 (38), paramyxovirus SV5 fusion protein F (52), and human parainfluenza virus type 2 fusion protein F (44), showed that the region immediately adjacent to the membrane-spanning domain plays a critical role in the fusogenic activities of these proteins. For example, in HIV-1 gp41, the membrane-proximal domain contains several tryptophan residues which are invariant between lentivirus envelope proteins. Deletion of this membrane-proximal region or substitution of the conserved Trp residues blocked the cell-cell fusion activity of gp41 (38). Likewise, the juxtamembrane region immediately preceding the membrane-anchoring domain is partially conserved in the G proteins of vesiculoviruses (2, 9), suggesting a possible functional role. Therefore, it was important to investigate whether this region contributes to the membrane fusion activity of G protein. The studies described in this report utilized mutational analyses of the membrane-proximal region of VSV G glycoprotein and examined the effect of these mutations on the structure and function of G protein. By using a combination of deletion, insertion, and substitution mutations we showed that the juxtamembrane region comprising the GS domain is not essential for oligomerization or transport of the glycoprotein but is important for the membrane fusion activity of VSV G protein. We have also recovered recombinant viruses encoding the mutant G proteins and determined the effects of the mutations on G incorporation into virions, virus binding to cells, and virus infectivity.
MATERIALS AND METHODS
Plasmids and oligonucleotide-directed mutagenesis.
The gene encoding the G protein of VSV, serotype Indiana, strain San Juan, was cloned into the eukaryotic expression vector pXM to produce the plasmid pXM-G as described earlier (51). Mutants E452A, G456D, W457A, F458A, and W461A were constructed by oligonucleotide-directed mutagenesis (51), and the mutated regions were cloned into pXM-G(AXB) (32). Double and triple mutants G456DW457A (DA), W457AW461A (WW-AA), W457AF458AW461A (AAA), and G456DW457DW461A (DAA) were generated using a QuikChange site-directed mutagenesis kit according to the manufacturer's instructions (Stratagene). The pXM-G(AXB) plasmid was used as the template. Two complementary synthetic oligonucleotide primers (from McMaster University Central Facilities) containing the desired mutation were used for mutagenesis with Pfu Turbo DNA polymerase. The deletion and insertion mutants were constructed as described earlier (32). Constructs GΔ9 and Δ F440-N449 were made by deleting aa 453 to 461 and aa 440 to 449 of the VSV G ectodomain. Constructs G10DAF and GΔ9-10DAF were made by inserting nine amino acids (residues 311 to 319) from the juxtamembrane region of decay acceleration factor (DAF) (6) between aa 464 and 465 of VSV G(AXB) and GΔ9, respectively. We call these constructs 10DAF because, due to insertion of a restriction site, the vector G(AXB) contains additional serines at the TM junction. The DAF sequence in G10DAF and GΔ9-10DAF does not contain the signal for a glycophosphatidylinositol addition; therefore, the proteins do not have a glycophosphatidylinositol anchor (32). The chimera G(+9)gBG was constructed by inserting aa 721 to 726 and aa 773 to 795 of herpes simplex virus type 1 (HSV-1) glycoprotein gB between aa 464 of the ectodomain and aa 483 of the cytoplasmic tail of VSV G such that the membrane-anchoring (TM) domain of VSV G (residues 465 to 482) was replaced by the third TM domain of HSV-1 gB protein (21). This chimera contains an extra serine residue at the ecto-TM domain junction to maintain the reading frame of VSV G protein.
The constructs W457-A (W1A), W461-A (W2A), W457W461-AA (WW-AA), GΔ13, GSrev11, and GSrev11-AA were made using an overlap PCR method with the plasmid pVSV 9.1(+) (24) as the template. The sequences of the primers used are available upon request. The overlap PCR products were then purified on a 6% polyacrylamide gel, electroeluted, digested with unique restriction enzymes KpnI and NheI, and used for subcloning into pVSV-FL(+)2 (24) (previously digested with the same enzymes). The sequences were then confirmed by dideoxynucleotide sequencing. The G genes having the desired mutations were also subcloned into a modified form of the eukaryotic expression vector pCAGGS-MCS as MluI and NheI fragments. The constructs pVSV-DAA, -AAA, -G10DAF, -G(+9)gBG, -GΔ9, -GΔ9-10DAF, and -ΔF440-N449 were generated [using the corresponding pXM plasmid containing the mutant G genes as templates and subcloning this region into pVSV-FL(+)2] by amplifying the region of the G gene between the KpnI site and the 3′ end.
Surface expression and syncytium formation assays.
Indirect immunofluorescence assays were used to examine surface expression of the various G proteins. Cells were transfected as described in the figure legends and then fixed with 3% paraformaldehyde. The cells were then probed with a G-specific monoclonal antibody (MAb), MAb I1 (25), followed by a rhodamine-conjugated goat anti-mouse (or anti-rabbit) secondary antibody (Jackson ImmunoResearch Laboratories, Inc.). To quantify surface expression of G protein, we either used flow cytometric analysis on virus-infected cells or performed lactoperoxidase-catalyzed iodination of transfected COS-1 cells as described earlier (27). For flow cytometry, BHK-21 cells (5 × 105) in 35-mm-diameter plates were infected with either wild-type (WT) VSV or the appropriate G-complemented mutant virus at a multiplicity of infection of 10. At 6 h postinfection, the cells were removed from the plates with phosphate-buffered saline (PBS) containing 50 mM EDTA and pelleted by centrifugation at 1,250 × g for 5 min. The cells were then fixed in suspension using 3% paraformaldehyde for 20 min at room temperature. The cells were washed two times with PBS-glycine to remove the fixative. The cells were then incubated in PBS-glycine-0.5% bovine serum albumin (Sigma-Aldrich) for 30 min at room temperature. Following blocking with bovine serum albumin, the cells were probed with I1 MAb as a primary antibody and a rhodamine-conjugated goat anti-mouse antibody. The cells were then analyzed by flow cytometry to quantitate the levels of surface expression of the various mutant G proteins.
For syncytium assays we used both virus-infected BHK-21 cells and plasmid-transfected COS-1 cells. For virus-infected cells, the medium was removed at 6 h postinfection, the cells were rinsed once with fusion medium [10 mM Na2HPO4, 10 mM HEPES, 10 mM 2-(N-morpholino)ethanesulfonic acid (MES) titrated to the indicated pH (5.9, 5.5 or 5.2) with HCl], and then bathed for 1 min in fresh fusion medium at room temperature. After 1 min, the fusion medium was replaced with fresh Dulbecco's modified Eagle's medium (DMEM) containing 5% fetal bovine serum (FBS) and cells were incubated at 37°C for 30 min. The cells were then fixed with 3% paraformaldehyde and processed for immunofluorescence as described above. Transfected COS-1 cells were processed as previously described (27).
Oligomerization transport kinetics and trypsin sensitivity assays.
The oligomeric state of the expressed G proteins was determined by sucrose density gradient centrifugation as described previously (27, 39). The endo H (endo-β-N-acetylglucosaminidase H) assays were performed on transfected cell lysates as described previously (27) and with virus-infected cell lysates as described previously (16) with a few modifications. BHK-21 cells in 35-mm-diameter plates were infected with the appropriate G-complemented mutant viruses at a multiplicity of infection of 10. At 6 h postinfection, the cells were rinsed once with methionine (Met)-free DMEM and then incubated in 2 ml of Met-free DMEM for 20 min. The medium was replaced with 2 ml of Met-free DMEM containing 55 μCi of [35S]Met (Translabel protein labeling mix; New England Nuclear) for the indicated amounts of time. Following the pulse period, the cells were either immediately lysed with 1 ml of detergent lysis buffer (10 mM Tris [pH 7.4], 66 mM EDTA, 1% Triton X-100, 0.4% deoxycholic acid, 0.02% sodium azide) or chased with DMEM-10% FBS medium containing 2 mM excess nonradioactive Met. Nuclei and cell debris were removed by centrifugation at 14,000 × g for 1 min on a tabletop microcentrifuge (IEC Centra). Immunoprecipitation was performed with anti-G tail rabbit polyclonal antibody (peptide antibody no. 3226) essentially as described previously (48) except that the postnuclear supernatants were made with 0.3% sodium dodecyl sulfate (SDS) and the antigen-antibody complexes were formed for 1 h at 37°C. One-half of the immunoprecipitates was digested with endo H (New England Biolabs) according to the manufacturer's instructions.
Trypsin sensitivities of the WT and mutant proteins were determined as described previously (15, 32). Briefly, transfected cells were labeled with [35S]Met and lysed with 1% Triton X-100 in 2× MNT (40 mM MES, 60 mM Tris, 200 mM NaCl, 2.5 mM EDTA) buffer at the indicated pH. The lysate was centrifuged at 14,000 × g for 5 min, and equivalent volumes of the supernatant were incubated in the absence or the presence of 10 μg of TPCK (l-1-tosylamide-2-phenylmethyl chloromethyl ketone)-trypsin for 30 min at 37°C. The digestion was stopped by the addition of aprotinin (100 U), and the mixture was centrifuged again at 14,000 × g for 2 to 5 min to remove any insoluble material. The supernatant was immunoprecipitated with anti-G (Indiana) antibody and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Recovery of viruses from cDNA.
Recovery of the viruses was done as described previously (22, 24, 43), with a few modifications. Briefly, confluent monolayers of BHK-21 cells in 35-mm-diameter plates were infected for 1 h at 31°C at a multiplicity of infection of 5 with a recombinant vaccinia virus encoding T7 RNA polymerase (vTF7-3) (17). The cells were then transfected with a DNA-liposome suspension consisting of 5 μg of pVSV(+)-G* (* indicates a G gene with mutations), 3, 5, 1, and 8 μg of plasmids containing the N, P, L, and G genes from VSVIND, respectively, and 90 μl of TransfectACE (36). After 3 h, the transfection mix was replaced with DMEM-10% FBS and cells were incubated at 37°C. The supernatants were collected after 48 h of incubation and filtered through a 0.2-μm-pore-size filter (Millex-GS; Millipore) to remove vaccinia virus. The filtrates were applied to BHK-21 cells that had been transfected with 2 μg of pCAGGS-GIND 24 h earlier. Recovery of the virus was assessed by examining the cells for the cytopathic effects that are typical of a VSV infection after 24 to 36 h. The recovered viruses were then plaque purified and passaged and their RNAs were isolated. The mutations in the G genes were confirmed by reverse transcription-PCR sequencing. The titers of the recovered viruses were determined by a plaque assay.
Budding assay.
The assay was done essentially as described previously (35). Confluent BHK-21 cells in 35-mm-diameter plates were infected with the respective mutant viruses at a multiplicity of infection of 10. Following adsorption, the residual inoculum was removed by rinsing the plate twice with serum-free DMEM (SF-DMEM) and washed two times in SF-DMEM with rocking at 37°C for 5 min each. The cells were then incubated at 37°C in 2 ml of SF-DMEM. After 16 h of incubation, the supernatants were harvested and clarified by centrifugation at 1,250 × g for 10 min. An aliquot of the supernatant was used in a plaque assay to determine the titers of the viruses. Virions from the remaining 1.5 ml of the supernatants were pelleted through a 20% sucrose cushion at 100,000 × g for 35 min. The viral pellet was resuspended in 50 μl of reducing sample buffer. One-fifth of each sample (10 μl) was resolved by electrophoresis on a 10% polyacrylamide gel. The gels were stained with Coomassie stain (GelCode Blue; Pierce Co.) per the manufacturer's instructions. The gels were destained and photographed using a Nikon digital camera and a 35- to 80-mm-focal-length Nikkor lens. Quantitation of viral protein was done using ImageQuant analytical software (Molecular Dynamics). Virus yield was determined by measuring the intensity of the N protein band.
Plaque assay.
BHK-21 cells (5 × 105) in a 6-well plate were infected with 10-fold serial dilutions of the virus. At 1 h postinfection, the inoculum was removed and the cells were overlaid with DMEM containing 0.9% agar and 5% FBS and incubated at 37°C for 36 h. After the incubation period, the number of plaques from at least two dilutions were counted and averaged. Virus titers were expressed as PFU per milliliter. The plaques were photographed with a Nikon digital camera and a 75- to 200-mm-focal-length Nikkor lens. The digital images were then magnified and printed on Kodak photo paper (10 by 20 cm). The sizes of at least 15 plaques per virus were determined and averaged.
RESULTS
Construction and expression of mutant G proteins.
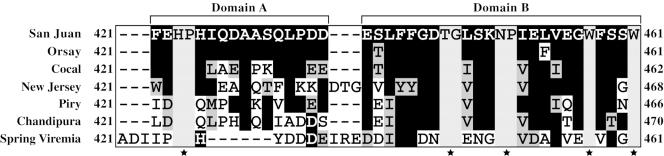
To determine which residues in the membrane-proximal region of VSV G are important for membrane fusion activity, we first performed a sequence alignment of the membrane-proximal domains from different vesiculoviruses (Fig. 1). The alignment showed that this region is well conserved across closely related vesiculoviruses, and we defined two distinct domains (labeled domains A and B) on the basis of the number of conserved residues in these subdomains (Fig. 1). The greatest conservation (>90%) was in the region between residues E437 and W461 (domain B). The amino acid sequences between F421 and D436 (domain A) for two strains of the VSV Indiana serotype (San Juan and Orsay) were identical, while there was >80% conservation between Cocal virus (which is a more distantly related virus classified as an Indiana-2 serotype) and the other two Indiana serotype viruses. However, domain A was not well conserved among the other viruses that were examined. Some of the interesting features in the alignment that attracted our attention were the tryptophan (W) residues at position 457 and 461, which are conserved across all vesiculoviruses examined. Apart from the tryptophan residues, several other amino acids (H423, P424, T444, G445, N449, and P450) were also conserved across the vesiculovirus sequences examined. The FFGDTG motif near the beginning of domain B and extending from aa 440 to 445 was conserved across most of the closely related members, with the TG residues being invariant across all vesiculoviruses examined.
FIG. 1.
Sequence alignment of the membrane-proximal domains of vesiculovirus glycoproteins. The sequences shown are from San Juan (accession number J02428) (37) and Orsay strains; a VSV Indiana strain (accession number M11048) (19); a VSV New Jersey strain (accession number J02433) (18); a Cocal virus strain (accession number AF045556) (2); a Chandipura virus strain (accession number J04350) (28); a Piry virus strain (accession number D26175) (4); and a spring viremia of carp virus strain (accession number U18101) (3). Residues represented by black characters with a light-gray background were conserved among all the vesiculoviruses. Residues represented by white characters with a black background were identical in the virus sequences examined. Black characters with a dark-gray background indicate residues with similar properties. Stars at the bottom of the sequences represent residues that were invariant across the sequences examined.
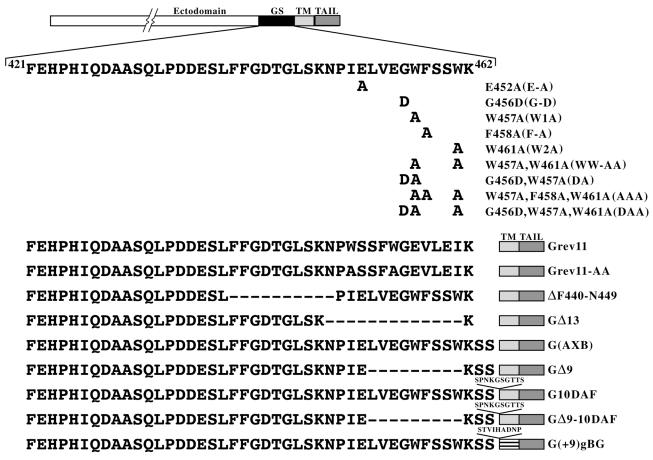
To determine the role of these conserved regions in the membrane fusion activity of G protein, we made substitutions, deletions, and insertions in this region (Fig. 2). The two invariant tryptophan (W) residues at position 457 and 461 were replaced with alanine. Similarly, the conserved glutamic acid (E452), glycine (G456), and phenylalanine (F458) were individually replaced with alanine or replaced with aspartic acid and alanine. Two double mutants were also constructed by replacing W457 and W461 with alanines as well as G456 and W457 with aspartic acid and alanine, respectively. In addition, two triple mutants were also generated by replacing W457, F458, and W461 with alanine and by replacing G456, W457, and W461 with aspartic acid and alanines, respectively. The mutants E452A, G456D, W457A, F458A, W461A, W457W-461-AA, G456D-W457A, G456D-W457A-W461A, and W457A-F458A-W461A are referred to as E-A, G-D, W1A, F-A, W2A, WW-AA, DA, DAA, and AAA, respectively, in the text.
FIG. 2.
Schematic representation of mutations in the membrane-proximal stem region of VSV G. A linear diagram of the full-length G protein (with the ectodomain, juxtamembrane GS region, TM, and cytoplasmic domains demarcated) is shown at the top of the panel. The sequence of the 42-aa stem region is also shown. The numbers at the beginning and end of the sequence indicate the positions of the amino acid residues from the N terminus of VSV GIND (San Juan strain). Amino acid K462 is believed to be the boundary between the TM domain and ectodomain (37). Mutations in the conserved tryptophan (W) (positions 457 and 461), the glutamic acid (E452), the glycine (G456), and the phenylalanine (F458) residues are shown. The sequences of the insertion, deletion, and inverted sequence mutants are also shown. The protein G(AXB) introduces two additional serines at the ectodomain TM junction.
In addition to the above-described point mutations, we also made deletion mutants (GΔ9 [lacking aa 453 to 461], GΔ13 [lacking residues 449 to 461], and ΔF440-N449 [lacking aa 440 to 449]) and several insertion mutants. The construct G(AXB) introduced two additional serines between K462 and S463 (described previously in reference 32), while G10DAF and GΔ9-10DAF contained 9 aa (residues 311 to 319) from DAF inserted between aa S464 and S465 of G(AXB) and GΔ9, respectively. The mutant G(+9)gBG had an insertion of nine residues between the ectodomain of G(AXB) and the TM domain of GgB3G (21). The remaining mutants that were examined have the sequence of the 11 aa adjacent to the TM domain in an inverted position (Gsrev11). The mutant GSrev11-AA has the same inverted 11 aa and also has the two W residues changed to alanine.
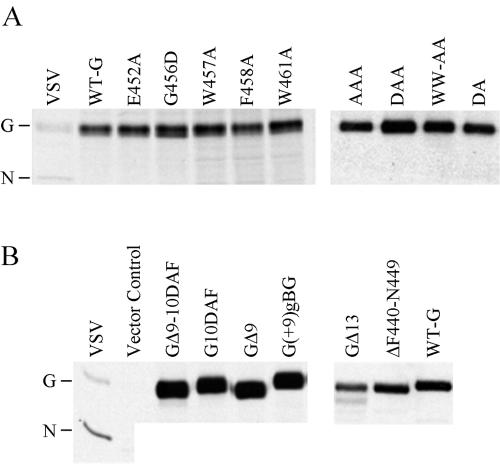
The mutant genes were expressed (using the pXM vector) in COS-1 cells (27), and the proteins were labeled with [35S]Met and then analyzed by immunoprecipitation with a polyclonal anti-G antibody followed by SDS-PAGE. All of the substitution mutants comigrated with the WT G protein; the intensities of bands corresponding to the WT and the mutants were similar, suggesting that the substitution of conserved residues and deletion or insertion of extra residues in the context of the G protein did not affect the expression or stability of the proteins (Fig. 3).
FIG. 3.
Expression and stability of the mutant proteins. COS-1 cells were transfected with plasmids encoding the indicated G proteins, and the proteins were labeled with [35S]Met and then analyzed by immunoprecipitation with a polyclonal anti-G antibody followed by SDS-PAGE. (A) Substitution mutants and WT G protein (WT-G). (B) Deletion and insertion mutants. The lanes labeled VSV represent immunoprecipitated proteins from cells that were infected with WT VSV and labeled with [35S]Met. The positions of the G and N proteins are indicated.
Transport and cell surface expression of mutant G proteins.
We next determined whether the proteins were transported to the cell surface, since both cell-cell fusion and virus budding requires localization to the plasma membrane. Using either flow cytometry or lactoperoxidase-catalyzed cell surface iodination, we found that the mutant G proteins were expressed on the cell surface at levels between 80 and >100% of WT G protein (Table 1).
TABLE 1.
Surface expression and membrane fusion activity of mutant G proteins
| Expressed protein | Relative surface expressiona
|
% Fusion activity
|
||
|---|---|---|---|---|
| Surface iodinationb | Flow cytometryc | Transfected cellsd | Infected cellse | |
| WT | 1.0 | 1.0 | 100 | 100 |
| E452A (E-A) | 1.17 | NDf | 50 | ND |
| G456D (G-D) | 0.82 | ND | 30 | ND |
| W457A (W1A) | 1.11 | 0.91 | 70 | 100 |
| F458A (F-A) | 1.25 | ND | 35 | ND |
| W461A (W2A) | 1.64 | 1.04 | 30 | 100 |
| WW-AA | 1.50 | 0.95 | 15 | 100 |
| G456DW457A (DA) | 1.35 | ND | 30 | ND |
| DAA | 1.85 | 1.03 | 5 | 100 |
| AAA | 1.80 | 1.15 | 5 | 100 |
| G(AXB) | 1.0 | 0.98 | 100 | 100 |
| GSrev11 | ND | 0.97 | ND | 100 |
| GSrev11-AA | ND | 1.01 | ND | 100 |
| GΔ9 | 2.08 | 1.01 | <1 | <1 |
| GΔ13 | 1.15 | 1.1 | <1 | <1 |
| ΔF440-N449 | 1.10 | 1.01 | <1 | 0 |
| G10DAF | 1.28 | 1.06 | <1 | 0 |
| GΔ9-10DAF | 2.70 | 1.16 | <1 | <1 |
| G(+9)gBG | 1.2 | 1.03 | <1 | 0 |
Relative surface expression was calculated using the following formula: [(% positive cells in mutant population) × (mean fluorescence intensity of mutant)]/[(% WT positive cells) × (mean fluorescence intensity of WT)].
COS cells were transfected with pXM vectors encoding the indicated G protein, the cells were surface iodinated, and the relative amount of surface expression was calculated as described previously (27).
BHK-21 cells were infected with G-complemented viruses and fixed at 6 h postinfection. The cells were then stained with the G-specific monoclonal antibody I1 and a rhodamine-labeled secondary antibody and then analyzed by flow cytometry.
Transfected COS cells were bathed in fusion medium buffered to pH 5.6, and cell-cell fusion was determined as described in Materials and Methods.
BHK-21 cells were infected with the respective virus mutants and incubated in fusion medium buffered to pH 5.9 as described in the legend for Fig. 5. Values are expressed as a percentage of WT fusion activity, which was set at 100%.
ND, not done.
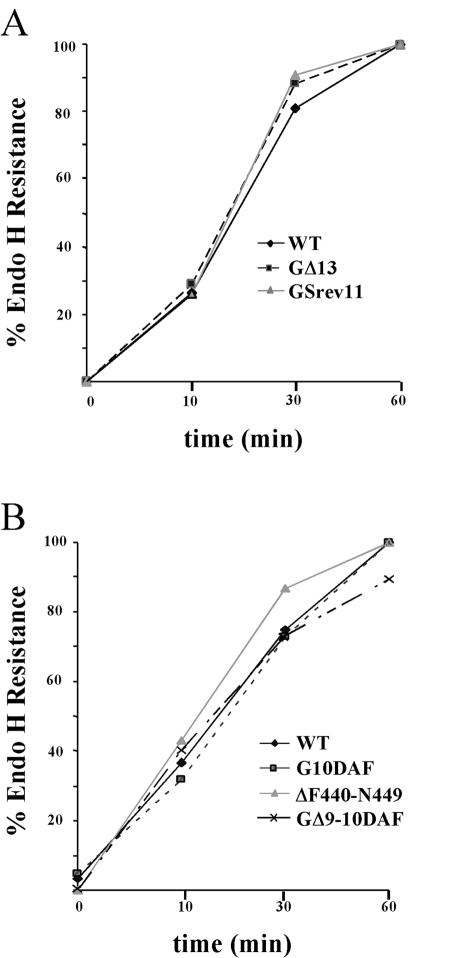
To determine whether mutations in the membrane-proximal region affected the intracellular transport or cell surface localization of G protein, we expressed WT and mutant G proteins in both COS-1 and BHK-21 cells. Transport from the endoplasmic reticulum (ER) to the Golgi complex was evaluated by examining the acquisition of endo H resistance. After a 15-min pulse, all the mutants were sensitive to endo H, showing that they were glycosylated with N-linked oligosaccharides. After a 1-h chase, the mutants were resistant to endo H digestion, indicating that all the mutant G proteins were transported from the ER to the Golgi with similar kinetics. Representative examples of data from some of the mutants expressed in BHK-21 cells are shown (Fig. 4).
FIG. 4.
Transport kinetics of WT and mutant G proteins. BHK-21 cells expressing WT G or the mutant proteins were labeled with [35S]Met for 15 min. The medium was removed, and medium containing excess unlabeled Met was added for 0, 10, 30, or 60 min. The G proteins were immunoprecipitated from cell lysates using an anti-G tail peptide antibody. One-half of the immunoprecipitates was digested with endo H. Proteins were resolved on an SDS-10% PAGE gel and visualized by fluorography. The amounts of endo H-resistant and -sensitive forms of the proteins were quantified using ImageQuant software (Molecular Dynamics). (A) Results of an experiment examining WT, GΔ13, and Gsrev11 proteins. (B) Results from a separate experiment comparing WT, G10DAF, ΔF440-N449, and GΔ9-10DAF proteins.
Recovery of recombinant viruses encoding the mutant G proteins.
Prior to characterization of the mutant G protein, we subcloned the cDNA for each of the mutants into a plasmid carrying the full-length VSV antigenome and recovered recombinant viruses from the plasmids according to published protocols (22) except that we coexpressed WT G protein during the initial recovery and subsequent amplification steps. This ensured that all viruses could be recovered, regardless of whether the G protein mutant was membrane fusion defective or not. The recombinant viruses were plaque purified on cells transiently expressing G protein, and G-complemented stocks were produced. After amplification, the virus phenotypes were examined by infecting BHK cells that did not express G protein.
Membrane fusion characteristics of mutant G proteins.
To examine the effect of the mutations on membrane fusion activity, we used a cell-cell fusion assay in which cells expressing either WT or the respective G mutants were treated with fusion medium buffered to pH 5.9 down to 4.8. When assayed in transiently transfected COS-1 cells, all of the substitution mutants showed reduced membrane fusion activity, ranging from 70 to 5% of WT activity (Table 1). However, when assayed in virus-infected BHK cells, many of these mutants (W1A, W2A, WW-AA, DAA, and AAA) produced extensive syncytia similar in size and extent to those seen in WT VSV-infected cells. The basis for this difference is not fully understood, but it may simply be a function of the level of G protein expression in virus-infected versus transiently transfected cells.
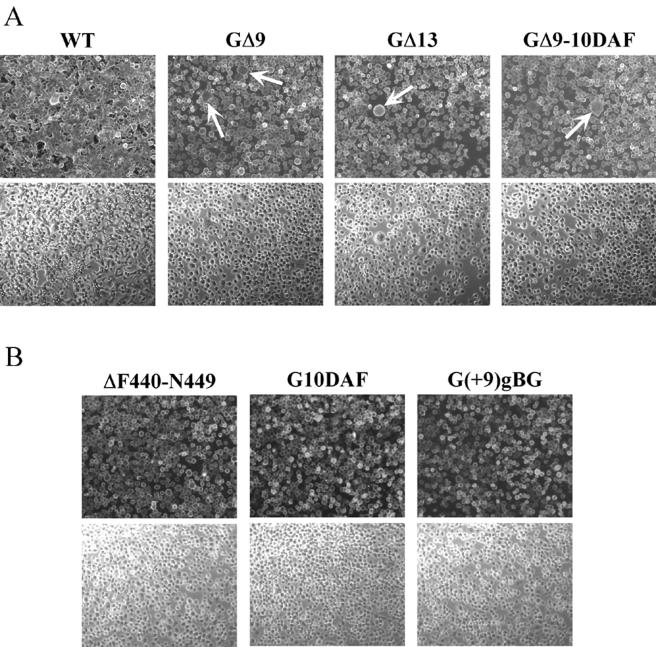
In contrast to the substitution mutants, the deletion and insertion mutants had very-low to undetectable membrane fusion activities in both COS and virus-infected BHK cells. When cells were exposed to pH 5.9, the mutants GΔ9, GΔ13, and GΔ9-10DAF produced very few syncytia, and those had only three to four nuclei (Fig. 5A, arrows). When cells expressing these proteins were bathed in medium buffered to pH 5.5, 5.2, or 4.8, neither the size nor the number of syncytia increased (data not shown), indicating that the defect in syncytium formation was not due to a shift in the pH threshold. In addition, prolonged incubation following exposure to the low pH trigger did not increase the number or size of syncytia seen (data not shown). When 9 or 10 aa were inserted between the boundary of the membrane-anchoring domain and the ectodomain [mutants G(+9)gB and G10DAF] and when residues F440 to N449 were deleted, there was a complete loss of membrane fusion activity (Fig. 5B). These data suggest that the sequence context of the juxtamembrane region is critical for fusion activity. In all of these cases, increasing the time of exposure to acidic pH or decreasing the pH of the fusion medium did not enhance membrane fusion activity.
FIG. 5.
Syncytium formation assays. Approximately 5 × 105 BHK-21 cells were infected at a multiplicity of infection of 10 for 1 h at 37°C. At 6 h postinfection, the cells were treated with fusion medium buffered to pH 5.9, 5.5, or 5.2 for 1 min at room temperature. The medium was replaced with DMEM-5% FBS, and the cultures were incubated at 37°C for 20 min to 1 h. Cells were then fixed and processed for indirect immunofluorescence using a G-specific MAb (I1). Rhodamine-conjugated goat anti-mouse antibody was used as the secondary antibody. Fluorescence and phase-contrast images were digitally captured using a Zeiss Axiocam fitted on a Zeiss Axiophot microscope with a 10× water-immersible ceramic objective. The images were then processed using Adobe Photoshop to adjust for brightness and contrast. (A) Syncytium formation induced in cells infected with rVSV-WT, -GΔ9, -GΔ13, and -GΔ9-10DAF strains after treatment with fusion medium buffered to pH 5.9. The arrows point to small syncytia in the mutant-infected cells. (B) Cells infected with rVSV-ΔF440-N449, -G10DAF, and -G(+9)gBG after treatment at pH 5.2. Upper and lower panels correspond to fluorescence and phase-contrast images, respectively.
These data show that the conserved membrane-proximal aromatic residues (tryptophan and phenylalanine) and the glycine residue are not critical for VSV G-induced membrane fusion in the context of virus-infected cells, but when these mutants were expressed transiently in COS-1 cells, some reduction in activity was seen. Also, inverting the sequence of the 11 membrane-proximal residues had no significant influence on the fusion activity. While the linear order of the nine membrane-proximal amino acids did not seem to affect membrane fusion activity, these residues are important, since deleting them reduced the fusion activity by >99%. Also, deletion of 13 aa had a similar effect on cell-cell fusion mediated by G protein. However, deletion of the region between F440 and N449 which includes the conserved FFGDTG motif completely abolished fusion activity, showing that this subdomain is important for the fusion activity of G. Results with the insertion mutants G10DAF and G(+9)gBG showed that although these insertions do not affect surface expression, they completely abolished fusion activity, indicating that the spacing of the membrane-proximal domain from the TM domain is very important for membrane fusion activity. Indeed, Odell et al. had shown previously that insertion of 9 aa in the mutant G(+9)gBG eliminated membrane fusion activity while the construct GgB3G had WT G protein fusion activity (32). Taken together, these results suggest that the region immediately adjacent to the membrane-anchoring domain is essential for the membrane fusion activity of VSV G protein.
Trimer stability of the mutant G proteins.
Although all of the mutant proteins were expressed on the cell surface, indicating they could fold and oligomerize sufficiently in the ER to be transported to the plasma membrane, Fredericksen and Whitt have shown previously that some mutations can affect trimer stability in sucrose density gradients without affecting transport (14, 15). To determine whether the mutations in the membrane-proximal region that reduced membrane fusion activity affected trimer stability, we utilized sucrose density gradient centrifugation at acidic and neutral pH. For the mutants that were examined, all showed sedimentation patterns that were similar to those of WT G, with the exception of G(+9)gBG, which was slightly less stable to centrifugation in gradients buffered to pH 5.6 (Table 2).
TABLE 2.
Oligomerization and trypsin sensitivity of G protein mutantsa
| Protein | Trimer stability at pH 5.6b | Trypsin resistance atc:
|
||
|---|---|---|---|---|
| pH 7.4 | pH 6.5 | pH 5.6 | ||
| WT | + | − | ++ | +++ |
| W1A | + | − | ++ | +++ |
| W2A | + | − | ++ | +++ |
| WW-AA | + | − | ++ | +++ |
| DAA | + | − | ++ | +++ |
| G(AXB) | + | − | ++ | +++ |
| GSrev11 | + | − | ++ | +++ |
| GSrev11-AA | + | − | ++ | +++ |
| GΔ9 | + | − | + | +++ |
| GΔ13 | + | − | ++ | +++ |
| ΔF440-N449 | + | − | ++ | +++ |
| G10DAF | + | − | + | +++ |
| GΔ9-10DAF | + | − | ++ | +++ |
| G(+9)gBG | ± | − | − | ++ |
Confluent monolayers of BHK-21 cells were infected with either WT or mutant viruses at a multiplicity of 10. At 6 h postinfection, the cells were radioactively labeled and chased as described in Materials and Methods.
For the trimer assays, the cells were lysed in 2× MNT buffered to pH 5.6. Lysates were then centrifuged through a 5 to 20% sucrose density gradient buffered to the same pH. Fractions were collected from the bottom, and G proteins were immunoprecipitated with a polyclonal anti-VSV antiserum.
For the trypsin sensitivity assays, cells were lysed in 1× MNT buffered to the indicated pH. Lysates were clarified by centrifugation to remove cell debris and nuclei. The supernatants were then treated with or without TPCK-trypsin, and the G proteins were immunoprecipitated with a polyclonal anti-VSV antibody. Immunoprecipitated proteins were resolved by SDS-PAGE, visualized by fluorography, and quantified by scanning densitometry. +++, 95 to 100% resistant; ++, 50 to 95% resistant; +, 10 to 50% resistant; −, <10% resistant.
Low-pH-induced conformational changes in the mutant G protein.
Previously, Fredericksen and Whitt had shown that G protein becomes resistant to trypsin digestion at low pH, presumably due to conformational changes induced by acidic pH (15). To investigate whether the changes in the fusogenic activities of the membrane-proximal mutants were due to alterations in low-pH-induced conformational changes, we examined the pH-dependent resistance to trypsin digestion of mutant and WT G proteins (Table 2). Most of the mutants showed trypsin resistance profiles similar to that observed for WT G protein. For example, at pH 6.5 approximately 75 to 80% of the mutant and WT G proteins were resistant to trypsin digestion. The mutants GΔ9 and G10DAF were somewhat less resistant (48 and 40%, respectively) to trypsin digestion at pH 6.5, whereas the mutant G(+9)gBG protein showed a drastic change in the resistance pattern and was completely sensitive to digestion at pH 6.5 and only partially resistant at pH 5.6.
Characterization of recombinant virus growth.
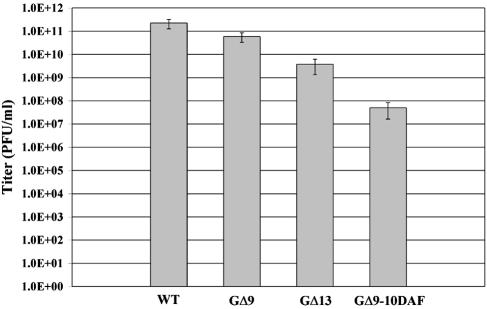
On the basis of the cell-cell fusion assay results, we predicted that recombinant virus encoding the mutants which exhibited WT G fusion activity would show growth patterns similar to those of WT VSV and that those that had undetectable membrane fusion activity would not be able to grow without complementation using WT G. As predicted, when grown on BHK-21 cells all of the point mutants, the insertion mutant G(AXB), and the sequence reversal mutants GSrev11 and GSrev11-AA produced titers similar to those of WT VSV. However, to our surprise some of the viruses that showed a >99% reduction in cell-cell fusion activity (e.g., GΔ9, GΔ13, and GΔ9-10DAF) were able to grow and spread on BHK-21 cells (albeit to lower titers) without the need for expression of G protein for complementation (Fig. 6). The deletion mutant GΔ9 gave titers that were consistently 10-fold lower than those of WT virus, while the deletion mutant GΔ13 had titers that were approximately 100-fold lower (Table 3). The titer of the GΔ9-10DAF was ∼10,000-fold lower than that of WT VSV. In accordance with the reduced viral titers, plaque formation by these mutants required 48 to 60 h whereas WT VSV and the other mutants produced plaques by 24 to 30 h postinfection and the plaques produced by the mutants were approximately two- to ninefold smaller than the WT plaques (Table 3). To determine whether any compensating mutations occurred in the G genes of GΔ9, GΔ13, or GΔ9-10DAF during recovery or subsequent amplification that might explain the ability to grow in the apparent absence of detectable membrane fusion activity, we performed reverse transcription-PCR sequencing on the entire G gene of these viruses. No mutations other than those specifically introduced were detected, which indicated that the phenotypes of the viruses were due to the designed mutations.
FIG. 6.
Virus infectivity assay. BHK-21 cells were infected with either WT or G-complemented mutant viruses at a multiplicity of infection of 10 for 1 h; the cells were then washed three times with growth medium. At 16 h postinfection, an aliquot of the supernatant was taken and plaque assays on BHK-21 cells were used to determine virus titers. At 36 h postinfection, the number of plaques were counted and averaged between at least two dilutions to determine the titers. Virus titers shown are averages from three independent experiments. Standard deviations are also shown.
TABLE 3.
Growth characteristics of mutant G virusesa
| Virus | Average plaque size (cm) | Relative plaque size (%) | Titer (PFU/ml) | Budding efficiency (%) | Specific infectivityb |
|---|---|---|---|---|---|
| WT | 0.86 | 100 | 2.8 × 1011 | 100 | 2.8 × 1011 |
| GΔ9 | 0.46 | 54 | 4.4 × 1010 | 78 | 5.6 × 1010 |
| GΔ13 | 0.31 | 36.5 | 3.5 × 109 | 79 | 4.4 × 109 |
| GΔ9-10DAF | 0.11 | 13.6 | 4.75 × 107 | 62 | 7.7 × 107 |
Confluent monolayers of BHK-21 cells were infected with either WT or mutant virus at a multiplicity of infection of 10. At 16 postinfection, the viral supernatants were collected and clarified by centrifugation at 1,250 × g for 5 min. Virions were collected by ultracentrifugation from 1.5 ml of the supernatant, and the proteins in the viral pellet were quantified following SDS-PAGE as described in Materials and Methods. The remaining 0.5 ml of the supernatant was used with plaque assays to obtain titers. The number of plaques were counted and averaged between at least two dilutions to determine the titers. Images of the plaques were obtained, and the sizes of 15 plaques per virus were measured. Budding efficiency of the virus was determined by quantifying the intensity of the N protein band from a Coomassie-stained gel.
Specific infectivity = (titer of the virus)/(budding efficiency of the virus).
The remaining mutants, which included G10DAF, G(+9)gBG, and ΔF440-N449, were noninfectious and unable to grow in BHK-21 cells without coexpression of WT G protein, which is consistent with their lack of cell-cell fusion activity. The lack of infectivity was not due to differences in the amount of G protein incorporated into virions, since all the mutant G proteins were incorporated at levels similar to that of the WT protein (Fig. 7).
FIG. 7.
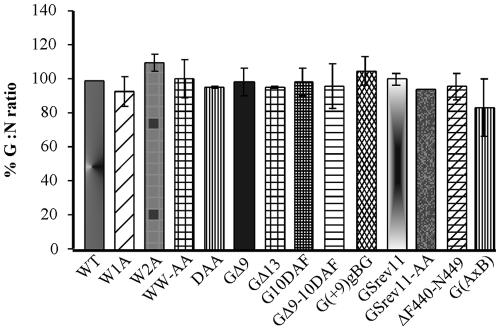
Incorporation of WT and mutant G proteins in virions. BHK-21 cells were infected with viruses encoding the WT or mutant proteins at a multiplicity of infection of 10 as described in the legend to Fig. 5. At 16 h postinfection, virus released into the supernatant was pelleted through a 20% sucrose cushion. The viral pellets were resuspended in sample buffer, and the proteins from one-fifth of the viral pellets were resolved by SDS-PAGE. The proteins were visualized by staining with Coomassie blue. Digital images of the gels were obtained using a Nikon camera with a 35- to 80-mm-focal-length Nikkor lens. Protein amounts were quantified by densitometry using Image Quant software (Molecular Dynamics). Relative amounts of G protein incorporated into virions were determined by calculating the ratio of G protein to N protein. The results are expressed as percentages relative to the G-to-N ratio found in the WT VSV control.
Virus binding to cells.
One explanation for the reduction or lack of membrane fusion activity, as well as for the loss of virus infectivity in some of the mutants, is that the mutations may have affected the ability of G protein to bind to cells. To determine whether any of the mutations affected virus binding, we incubated radiolabeled virions with BHK-21 cells in binding medium buffered to pH 7.0 or pH 5.9. Binding was done on ice to prevent endocytosis of the virions as well as to prevent fusion of the viral envelope with the cell membrane following exposure to low pH. Fredericksen and Whitt and others had shown previously that VSV binding is enhanced at acidic pH (13, 29). Except for GΔ13 and ΔF440-N449, all of the mutants examined showed binding to cells similar to that of WT VSV at both pH 7.0 and 5.9 (Fig. 8). Both GΔ13 and ΔF440-N449 consistently exhibited twofold-better binding at pH 7.0 compared to the WT; at pH 5.9, however, GΔ13 binding was approximately 10% less than that of WT VSV and ΔF440-N449 binding was reduced by 50% compared to that of the WT virus (Fig. 8). These data indicate that residues in the region between F440 and V454 contribute to virus binding and that the reduced amount of binding might be partially responsible for the defect in membrane fusion activity seen with these mutants. Insertion of 10 aa from DAF (G10DAF and GΔ9-10DAF) or 9 aa from HSV gB did not affect binding, indicating that the spacing of the residues between the TM and the ectodomain is not critical for binding. Evaluated on the basis of these results, it appears the defect in fusion activity and virus infectivity for G10DAF, G(+9)gBG, and GΔ9-10DAF most likely takes place at a postbinding step.
FIG. 8.
Virus binding assay. Radiolabeled virions (∼80,000 cpm) were resuspended in binding medium buffered to pH 7.0 or 5.9 and incubated at room temperature for 30 min. The suspensions were cooled on ice for 10 min and then added to prechilled confluent monolayers of BHK-21 cells. Virus binding was done for 3 h on ice. The medium was removed, and the amount of radioactivity was determined. This represented the unbound virus fraction. The cells were then washed three times with ice-cold binding buffer at the same pH used for binding, and the washes were collected for quantitation. Cells were lysed in PBS containing 1% Triton X-100, and the amount of radioactivity in the lysates (bound fraction) was determined. Virus binding was expressed as the percentage of the bound virus compared to the total input virus.
Specific infectivity of virus mutants.
In addition to affecting membrane fusion activity, the mutations may also reduce the amount of virus released from cells. To determine whether some of the reduction in virus titers observed with GΔ9, GΔ13, or GΔ9-10DAF was due to reduced virus budding, we determined the specific infectivity of each virus, which is calculated as the ratio of the virus titer to the relative amount of virus released compared to that of the WT virus. All three of the mutants that gave reduced virus titers produced between 60 and 90% of the amount of virus made from WT-infected cells (Table 3). Therefore, the defect in the ability of these mutants to spread in culture was primarily due to defects in membrane fusion activity rather than in virus budding.
DISCUSSION
Sequence analysis of the membrane-proximal ectodomains of viral fusion proteins from several virus families, including retroviruses, filoviruses, orthomyxoviruses, rhabdoviruses, alphaviruses, and flaviviruses, have shown that these regions have an unusually high concentration of aromatic residues and (especially) tryptophans (42). It was proposed that these membrane-proximal domains, and particularly the tryptophan residues, might play an important role in membrane fusion mediated by these glycoproteins. Mutational analysis of the conserved tryptophan motif in HIV-1 gp41 showed that these residues are important for incorporation of the HIV-1 envelope protein into the virions as well as for membrane fusion activity (38). It was later shown using a three-color assay that these W-A mutants could mediate transfer of both membrane and cytoplasmic dyes but were defective in fusion pore enlargement (31). Analysis using peptides derived from the membrane-proximal domain of the gp41 showed that this region could interact with membranes independent of the fusion peptide and cause membrane perturbation (42). The authors demonstrated that this C-terminal region of the gp41 ectodomain acts synergistically with the N-terminal fusion peptide in mediating membrane destabilization. Membrane-proximal domains of other viral glycoproteins have also been shown to be important for the mediating viral-envelope fusion. Deletion of the 12 aa domain between the second heptad repeat (HR2) and TM domain of human parainfluenza virus type 2 F protein completely abolished membrane fusion activity (44). Similarly, studies with the SV5 F protein also showed that the membrane-proximal domain is important for viral fusion (52).
Results seen with the three-dimensional structures of integral membrane proteins as well as sequence alignments of the regions immediately adjacent to the predicted membrane-anchoring regions of TM proteins (including a number of viral fusion proteins) show clustering of the polar-aromatic residue Trp at the lipid-water interface region of the protein (23, 44). Trp residues are believed to be able to bridge the gap between the aqueous and the hydrophobic environment present at the interface and to play an important role in correct folding, localization, and orientation of the protein at the interface (23). Results presented here show that in the case of VSV G protein, replacement of both of the Trp residues or replacement of the three aromatic residues in the juxtamembrane region with the hydrophobic residue Ala reduced the membrane fusion activity of G protein in transfected cells. However, viruses containing these mutant G proteins were as biologically competent as the WT VSV, indicating that in the context of the virus envelope the Trp residue may not be critical.
Mutational analysis using insertions (27) and substitutions (40) suggested that the C-terminal region of the VSV G ectodomain plays an important role in virus entry. Recently, Jeetendra et al. had shown that a truncation mutant of VSV G protein (called G-stem or GS), which consists of the membrane-proximal 42 aa of the ectodomain, the TM domain, and cytoplasmic tail, could mediate hemifusion and could potentiate the membrane fusion activity of several different heterologous viral glycoproteins (22). Furthermore, Jeetendra et al. showed that when GS was truncated from F421 down to N449, there was no loss of fusion potentiation activity. In this study we examined how mutations in the membrane-proximal domain of GS, when made in the context of the full-length G protein, affected protein folding, membrane fusion activity, incorporation into virions, and virus budding.
Our results show that none of the mutations in the membrane-proximal region affected stability, oligomerization, or transport of the full-length G proteins to the cell surface. Furthermore, we found that unlike HIV-1 gp41 (38), the conserved membrane-proximal W residues in VSV G are not critical for G protein incorporation into virus or for membrane fusion activity. Similarly, when the conserved glycine at position 456 was changed to aspartic acid or F458 was changed to alanine (either singly or in the context of the tryptophan mutants), no loss of G protein function was detected. In addition, when the specific order of the 11 aa closest to the membrane was reversed no change in cell-cell fusion or virus infectivity was observed.
In contrast to the point mutation results seen, we observed a dramatic reduction in cell-cell fusion activity with the deletion mutants GΔ9, GΔ13, and ΔF440-N449. For example, when the nine membrane-proximal residues (V454 to K462) were deleted (GΔ9), very few syncytia were observed (even when the pH of the fusion medium was reduced to 5.2 or the cultures were incubated for 1 h post-acid-pH triggering). Interestingly, there was only a 10-fold reduction in infectivity of GΔ9 virus and the deletion did not affect the incorporation of this protein into virions or virus binding to cells. We considered the 10-fold reduction in virus titer to be relatively modest, considering that the deletion had a dramatic effect upon the cell-cell fusion activity. It is possible that this mutant forms small fusion pores that cannot enlarge sufficiently to allow extensive polykaryon formation and yet are large enough to release the viral genome into the host cell and allow for infection. This has been observed previously with fusion-defective mutants of influenza virus hemagglutinin (HA), with which mutants that were defective in cell-cell fusion were infectious in the context of the virus, albeit at low levels (10). In studies with the SV5 F protein, deletion of the 7 aa proximal to the membrane altered the kinetics of fusion but did not affect the extent of overall fusion (1). It is possible that the GΔ9 mutant has a similar defect; thus, fusion may occur at a slower rate. Deletion of the membrane-proximal 13 aa (N449 to K462) had an effect on cell-cell fusion similar to that seen with the GΔ9 mutant. However, the decrease in virus infectivity was greater, since titers were reduced by ∼100-fold. This mutation also reduced virus binding to cells by ∼10% compared to WT virus binding when performed at acidic pH. The greatest effect on fusion activity was seen with the deletion construct ΔF440-N449, which resulted in no cell-cell fusion activity and the release of noninfectious virus. This mutation also resulted in a 50% reduction in virus binding activity, which could have been either a direct or an indirect effect.
Previously it was shown that the TM domain of G protein can be replaced with the third TM domain of the HSV-1 gB without affecting the fusion activity of VSV G protein (32). However, when an additional nine residues were inserted at the ectodomain-TM junction [as with, e.g., the construct G(+9)gBG], no membrane fusion occurred. Similarly, when 10 aa from DAF were inserted at the ectodomain-TM junction of VSV G, no cell-cell fusion or virus infectivity was observed. These data indicate that there is a sequence requirement for residues in the membrane-proximal domain and that the distance of this domain from the ectodomain-TM junction is important for membrane fusion activity. Consistent with this concept, when 9 aa were deleted from the fusion inactive G10DAF mutant (resulting in GΔ9-10DAF), membrane fusion activity was partially restored. These results are similar to those obtained with the SV5 F protein (52), in which the spacing between the TM region and the membrane-proximal domain is important for membrane fusion activity.
Other studies examining intracellular vesicle fusion involving SNARE proteins also showed that insertion of flexible linkers of various sizes between the membrane anchor domain and the membrane-proximal conserved coiled coil decreased fusion efficiency as the length of the spacer increased (30). Evaluated on the basis of the similarities between the results of these studies and our results with VSV G, it is possible that deletions or insertions in the membrane-proximal stem region of G affect fusion activity by preventing conformational changes required to drive membrane coalescence just prior to merger of the two apposed membranes. Alternatively, the deletions or insertions might alter the local environment between the apposing membranes in such a way that membrane fusion does not occur. Of note is that the insertions, and all but one of the deletions, did not affect G protein incorporation into virions or virus binding to cells, indicating that the defect is at a postbinding step. Additional studies will be required to decipher the basis of these defects. To summarize, our data indicate that the membrane-proximal GS domain is important for G protein-mediated fusion and for virus binding to host cells in the context of the full-length protein but that this region does not play a significant role in the transport, oligomerization- or acid-pH-induced conformational changes of VSV G protein.
Structural information obtained from studies with influenza virus HA (5, 8), Ebola virus glycoprotein GP2 (45, 46), SV5 F protein (1), and HIV-1 gp41 (47) have led to a model for membrane fusion wherein the viral glycoprotein, upon activation, undergoes conformational changes that result in the exposure of the fusion peptide and insertion of the fusion peptide into the target membrane. The conformational changes also result in a core coiled-coil structure that is formed by interactions between the heptad repeat adjacent to the fusion peptide and the heptad repeat close to the TM domain. Formation of this structure positions the fusion peptide and the TM domain close to each other and brings the target and viral membranes in close apposition. However, there is no structural evidence to indicate that VSV G forms trimeric coiled coils. Computational analyses using all the available algorithms failed to show any predicted coiled-coil motifs in VSV G protein. Also, destabilization of VSV G protein at neutral pH with heat or urea did not induce any conformational change leading to membrane fusion (50). In contrast, influenza virus HA protein, which requires a coiled-coil motif for fusion activity, can induce membrane fusion at neutral pH in the presence of heat or urea (7, 8). The results of structural as well as biochemical analyses of tick-borne encephalitis virus fusion protein E (34, 41) and Semliki Forest virus fusion protein E1 (20, 26) also demonstrated that the presence of a coiled-coil motif is not essential for membrane fusion. In similarity to VSV G, neither the tick-borne encephalitis E protein nor Semliki Forest virus E1 possesses any predicted coiled-coil motifs and, at neutral pH, membrane fusion activity cannot be induced by heat or urea (20, 41).
Earlier work (27) had shown that insertion of a 3-aa linker between residues 395 and 418 in VSV G protein, which is involved in low-pH-induced conformational changes, abolished G-mediated membrane fusion. Acid-pH-dependent conformational changes may bring the fusion peptide (aa 116 to 137) and the membrane-proximal region in close proximity to each other (39). Simultaneous insertion of the fusion peptide and the membrane-proximal region into the target membrane would result in increased binding, membrane destabilization, and lipid mixing. When mutations in a C-terminal region were combined with mutations in the fusion peptide, it was seen that the double mutations reduced membrane fusion activity additively, except for the mutant G131AG404A, which was 80% as fusogenic as the WT protein, indicating that the fusion peptide and the C-terminal domain might interact (39). Although mutational analysis has provided information on the regions of G protein involved in membrane fusion, it is clear that structural determination studies are now required to gain a better understanding of the VSV G-mediated membrane fusion mechanism. However, attempts to obtain crystals of G protein suitable for X-ray diffraction have not been successful to date. The difficulty in crystallizing VSV G protein may relate to the relative flexibility of the G protein structure and the reversible nature of pH-dependent conformational changes. Therefore, new innovative approaches will likely be required to gain insight into the molecular details of G-mediated membrane fusion.
Acknowledgments
The Department of Molecular Sciences, University of Tennessee Health Sciences Center, Memphis, Tenn., group and the Department of Biochemistry, McMaster University, Hamilton, Ontario, Canada, group made equal contributions.
We thank Carolyn Matthews and S. Culp-Stewart for expert technical assistance and Tim Higgins for help with figure preparation. Flow cytometry was performed by Felicia Waller in the Molecular Resource Center at UTHSC.
This work was supported by the Canadian Institute of Health Research and by NIH grant GM53726 to M.A.W.
REFERENCES
- 1.Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3:309-319. [DOI] [PubMed] [Google Scholar]
- 2.Bhella, R. S., S. T. Nichol, E. Wanas, and H. P. Ghosh. 1998. Structure, expression and phylogenetic analysis of the glycoprotein gene of Cocal virus. Virus Res. 54:197-205. [DOI] [PubMed] [Google Scholar]
- 3.Bjorklund, H. V., K. H. Higman, and G. Kurath. 1996. The glycoprotein genes and gene junctions of the fish rhabdoviruses spring viremia of carp virus and hirame rhabdovirus: analysis of relationships with other rhabdoviruses. Virus Res. 42:65-80. [DOI] [PubMed] [Google Scholar]
- 4.Brun, G., X. Bao, and L. Prevec. 1995. The relationship of Piry virus to other vesiculoviruses: a re-evaluation based on the glycoprotein gene sequence. Intervirology 38:274-282. [DOI] [PubMed] [Google Scholar]
- 5.Bullough, P. A., F. M. Hughson, J. J. Skehel, and D. C. Wiley. 1994. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Caras, I., G. Weddell, M. Davitz, V. Nussenzwlig, and D. W. Martin. 1987. Signal for attachment of a phospholipid membrane anchor in decay acceleration factor. Science 238:1280-1283. [DOI] [PubMed] [Google Scholar]
- 7.Carr, C. M., C. Chaudhry, and P. S. Kim. 1997. Influenza hemagglutinin is spring-loaded by a metastable native conformation. Proc. Natl. Acad. Sci. USA 94:14306-14313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carr, C. M., and P. S. Kim. 1993. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell 73:823-832. [DOI] [PubMed] [Google Scholar]
- 9.Coll, J. M. 1995. Heptad-repeat sequences in the glycoprotein of rhabdoviruses. Virus Genes 10:107-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cross, K. J., S. A. Wharton, J. J. Skehel, D. C. Wiley, and D. A. Steinhauer. 2001. Studies on influenza haemagglutinin fusion peptide mutants generated by reverse genetics. EMBO J. 20:4432-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durrer, P., Y. Gaudin, R. W. H. Ruigrok, R. Graf, and J. Brunner. 1995. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J. Biol. Chem. 270:17575-17581. [DOI] [PubMed] [Google Scholar]
- 12.Fan, D. P., and B. M. Sefton. 1978. The entry into host cells of Sindbis virus, vesicular stomatitis virus, and Sendai virus. Cell 15:985-992. [DOI] [PubMed] [Google Scholar]
- 13.Fredericksen, B. L., and M. A. Whitt. 1998. Attenuation of recombinant vesicular stomatitis viruses encoding mutant glycoproteins demonstrate a critical role for maintaining a high pH threshold for membrane fusion in viral fitness. Virology 240:349-358. [DOI] [PubMed] [Google Scholar]
- 14.Fredericksen, B. L., and M. A. Whitt. 1995. Mutations affecting the pH threshold for vesicular stomatitis virus G protein mediated membrane fusion, p. 122. In Scientific Programs and Abstracts, 14th Annual American Society for Virology Meeting. ASV, Austin, Tex.
- 15.Fredericksen, B. L., and M. A. Whitt. 1996. Mutations at two conserved acidic amino acids in the glycoprotein of vesicular stomatitis virus affect pH-dependent conformational changes and reduce the pH threshold for membrane fusion. Virology 217:49-57. [DOI] [PubMed] [Google Scholar]
- 16.Fredericksen, B. L., and M. A. Whitt. 1995. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J. Virol. 69:1435-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuerst, T. R., E. G. Niles, R. W. Studier, and B. Moss. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. USA 83:8122-8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallione, C. J., and J. K. Rose. 1983. Nucleotide sequence of a cDNA clone encoding the entire glycoprotein from the New Jersey serotype of vesicular stomatitis virus. J. Virol. 46:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallione, C. J., and J. K. Rose. 1985. A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J. Virol. 54:374-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gibbons, D. L., A. Ahn, P. K. Chatterjee, and M. Kielian. 2000. Formation and characterization of the trimeric form of the fusion protein of Semliki Forest virus. J. Virol. 74:7772-7780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert, R., K. Ghosh, L. Rasile, and H. P. Ghosh. 1994. Membrane anchoring domain of herpes simplex virus glycoprotein gB is sufficient for nuclear envelope localization. J. Virol. 68:2272-2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeetendra, E., C. S. Robison, L. M. Albritton, and M. A. Whitt. 2002. The membrane-proximal domain of VSV G protein functions as a membrane fusion potentiator and can induce hemifusion. J. Virol. 76:12300-12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killian, J. A., and G. von Heijne. 2000. How proteins adapt to a membrane-water interface. Trends Biochem. Sci. 25:429-434. [DOI] [PubMed] [Google Scholar]
- 24.Lawson, N. D., E. A. Stillman, M. A. Whitt, and J. K. Rose. 1995. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA 92:4477-4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lefrancois, L., and D. S. Lyles. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157-167. [PubMed] [Google Scholar]
- 26.Lescar, J., A. Roussel, M. W. Wien, J. Navaza, S. D. Fuller, G. Wengler, and F. A. Rey. 2001. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell 105:137-148. [DOI] [PubMed] [Google Scholar]
- 27.Li, Y., C. Drone, E. Sat, and H. P. Ghosh. 1993. Mutational analysis of the vesicular stomatitis virus glycoprotein G for membrane fusion domains. J. Virol. 67:4070-4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Masters, P. S., R. S. Bhella, M. Butcher, B. Patel, H. P. Ghosh, and A. K. Banerjee. 1989. Structure and expression of the glycoprotein gene of Chandipura virus. Virology 171:285-290. (Erratum, 174:630, 1990.) [DOI] [PubMed] [Google Scholar]
- 29.Matlin, K., H. Reggio, A. Helenius, and K. Simons. 1982. The pathway of vesicular stomatitis entry leading to infection. J. Mol. Biol. 156:609-631. [DOI] [PubMed] [Google Scholar]
- 30.McNew, J. A., T. Weber, D. M. Engelman, T. H. Sollner, and J. E. Rothman. 1999. The length of the flexible SNAREpin juxtamembrane region is a critical determinant of SNARE-dependent fusion. Mol. Cell 4:415-421. [DOI] [PubMed] [Google Scholar]
- 31.Munoz-Barroso, I., K. Salzwedel, E. Hunter, and R. Blumenthal. 1999. Role of the membrane-proximal domain in the initial stages of human immunodeficiency virus type 1 envelope glycoprotein-mediated membrane fusion. J. Virol. 73:6089-6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Odell, D., E. Wanas, J. Yan, and H. P. Ghosh. 1997. Influence of membrane anchoring and cytoplasmic domain on the fusogenic activity of vesicular stomatitis virus glycoprotein G. J. Virol. 71:7996-8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pak, C. C., A. Puri, and R. Blumenthal. 1997. Conformational changes and fusion activity of vesicular stomatitis virus glycoprotein: [125I]iodonaphthyl azide photolabeling studies in biological membranes. Biochemistry 36:8890-8896. [DOI] [PubMed] [Google Scholar]
- 34.Rey, F. A., F. X. Heinz, C. Mandl, C. Kunz, and S. C. Harrison. 1995. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature 375:291-298. [DOI] [PubMed] [Google Scholar]
- 35.Robison, C. S., and M. A. Whitt. 2000. The membrane-proximal stem region of vesicular stomatitis virus G protein confers efficient virus assembly. J. Virol. 74:2239-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose, J. K., L. Buonocore, and M. A. Whitt. 1991. A new cationic liposome reagent mediating nearly quantitative transfection of animal cells. BioTechniques 10:520-525. [PubMed] [Google Scholar]
- 37.Rose, J. K., and C. J. Gallione. 1981. Nucleotide sequences of the mRNA's encoding the vesicular stomatitis virus G and M proteins determined from cDNA clones containing the complete coding regions. J. Virol. 39:519-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salzwedel, K., J. T. West, and E. Hunter. 1999. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J. Virol. 73:2469-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shokralla, S., R. Chernish, and H. P. Ghosh. 1999. Effects of double-site mutations of vesicular stomatitis virus glycoprotein G on membrane fusion activity. Virology 256:119-129. [DOI] [PubMed] [Google Scholar]
- 40.Shokralla, S., Y. He, E. Wanas, and H. P. Ghosh. 1998. Mutations in a carboxy-terminal region of vesicular stomatitis virus glycoprotein G that affect membrane fusion activity. Virology 242:39-50. [DOI] [PubMed] [Google Scholar]
- 41.Stiasny, K., S. L. Allison, C. W. Mandl, and F. X. Heinz. 2001. Role of metastability and acidic pH in membrane fusion by tick-borne encephalitis virus. J. Virol. 75:7392-7398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suárez, T., W. R. Gallaher, A. Agirre, F. M. Gõni, and J. L. Nieva. 2000. Membrane interface-interacting sequences within the ectodomain of the human immunodeficiency virus type 1 envelope glycoprotein: putative role during viral fusion. J. Virol. 74:8038-8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takada, A., C. Robison, H. Goto, A. Sanchez, K. G. Murti, M. A. Whitt, and Y. Kawaoka. 1997. A novel system for functional analysis of Ebola virus glycoprotein. Proc. Natl. Acad. Sci. USA 94:14764-14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong, S., F. Yi, A. Martin, Q. Yao, M. Li, and R. W. Compans. 2001. Three membrane-proximal amino acids in the human parainfluenza type 2 (HPIV 2) F protein are critical for fusogenic activity. Virology 280:52-61. [DOI] [PubMed] [Google Scholar]
- 45.Weissenhorn, W., L. J. Calder, S. A. Wharton, J. J. Skehel, and D. C. Wiley. 1998. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc. Natl. Acad. Sci. USA 95:6032-6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissenhorn, W., A. Carfi, K. H. Lee, J. J. Skehel, and D. C. Wiley. 1998. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2:605-616. [DOI] [PubMed] [Google Scholar]
- 47.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 48.Whitt, M. A., L. Buonocore, C. Prehaud, and J. K. Rose. 1991. Membrane fusion activity, oligomerization, and assembly of the rabies virus glycoprotein. Virology 185:681-688. [DOI] [PubMed] [Google Scholar]
- 49.Whitt, M. A., P. Zagouras, B. Crise, and J. K. Rose. 1990. A fusion-defective mutant of the vesicular stomatitis virus glycoprotein. J. Virol. 64:4907-4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao, Y., K. Ghosh, R. F. Epand, R. M. Epand, and H. P. Ghosh. 2003. Membrane fusion activity of vesicular stomatitis virus glycoprotein G is induced by low-pH but not by heat or denaturant. Virology 310:319-332. [DOI] [PubMed] [Google Scholar]
- 51.Zhang, L., and H. P. Ghosh. 1994. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J. Virol. 68:2186-2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou, J., R. E. Dutch, and R. A. Lamb. 1997. Proper spacing between heptad repeat B and the TM domain boundary of the paramyxovirus SV5 F protein is critical for biological activity. Virology 239:327-339. [DOI] [PubMed] [Google Scholar]