Abstract
Prevention and treatment of infection by West Nile virus (WNV) and other flaviviruses are public health priorities. We describe a reporting cell line that can be used for high-throughput screening of inhibitors against all targets involved in WNV replication. Dual reporter genes, encoding Renilla luciferase (Rluc) and neomycin phosphotransferase (Neo), were engineered into a WNV subgenomic replicon, resulting in Rluc/NeoRep. Geneticin selection of BHK-21 cells transfected with Rluc/NeoRep yielded a stable cell line that contains persistently replicating replicons. Incubation of the reporting cells with known WNV inhibitors decreased Rluc activity, as well as the replicon RNA level. The efficacies of the inhibitors, as measured by the depression of Rluc activity in the reporting cells, are comparable to those derived from authentic viral infection assays. Therefore, the WNV reporting cell line can be used as a high-throughput assay for anti-WNV drug discovery. A similar approach should be applicable to development of genetics-based antiviral assays for other flaviviruses.
Many flaviviruses are significant human pathogens, including the four serotypes of dengue virus, yellow fever virus, Japanese encephalitis virus, tick-borne encephalitis virus, St. Louis encephalitis virus, and West Nile virus (WNV) (7). Flavivirus virions are spherical in shape with a diameter of 40 to 60 nm. The nucleocapsid of about 30 nm in diameter consists of capsid and genomic RNA and is surrounded by a lipid bilayer in which the viral envelope and membrane proteins are embedded (5). The flavivirus genome is a single-stranded RNA of positive polarity, approximately 11 kb in length. The genomic RNA contains a 5′ untranslated region (5′ UTR), a single open reading frame (ORF), and a 3′ UTR (Fig. 1A). The ORF encodes 10 viral proteins: three structural (capsid [C], premembrane [prM] or membrane [M], and envelope [E]) proteins and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins (6). The nonstructural proteins are primarily involved in viral replication. NS1 and its potential interaction with NS4A are required for RNA replication (26, 27). The hydrophobic NS2A was recently shown to function during virion assembly and release of infectious viral particles (24). NS2B forms a complex with NS3 and is a required cofactor for the serine protease function of NS3 (2, 9, 11, 17). NS3 is a multifunctional protein which exhibits enzymatic activities of a serine protease (in the presence of NS2B), 5′-RNA triphosphatase, NTPase, and helicase (3, 5, 25, 39-41). The functions of the membrane-associated NS4A and NS4B are not known. NS5 contains activities of an RNA-dependent RNA polymerase (RdRp) (1, 18, 38) and a methyltransferase (16, 23). Upon flavivirus infection, the plus-sense genomic RNA is transcribed into a complementary minus-sense RNA, which in turn serves as the template for the synthesis of more plus-sense genomic RNA (10, 15, 31, 35). The synthesis of plus- and minus-sense RNAs is asymmetric; plus-sense RNA is produced in 10- to 100-fold excess over minus-sense RNA (15, 31, 35).
FIG. 1.
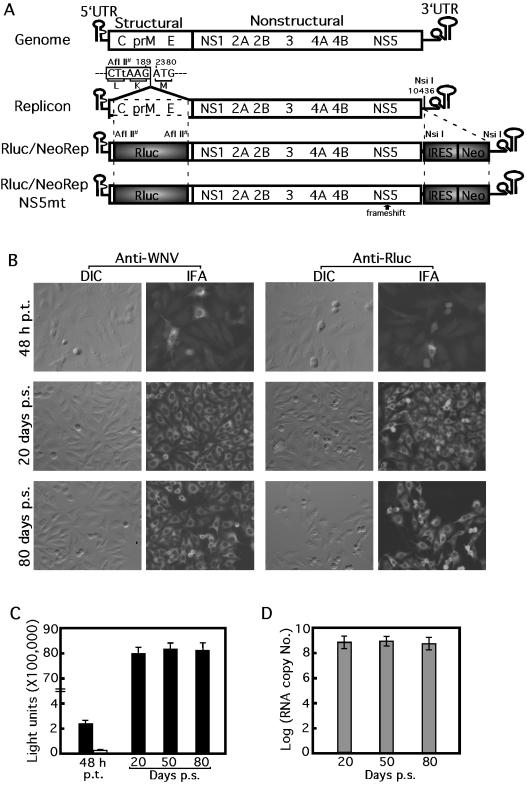
Construction and characterization of a stable cell line containing a dual reporting replicon of WNV. (A) WNV genome, subgenomic replicons, and dual reporting replicons. Compared with the full-length WNV genome, the wild-type replicon contained an in-frame deletion of the structural region (dotted open box) from nt 190 to 2379. An AflII site was generated (indicated by #) at the junction of the deletion by a silent coding mutation G186T (in lowercase). In Rluc/NeoRep, the Rluc was fused in frame with the ORF in the position where the structural genes were deleted; the IRES-Neo fragment was inserted into the NsiI site (nt 10436) in the upstream region of the 3′ UTR of the replicon. Rluc/NeoRepNS5mt contains a frameshift insertion of a nucleotide (U) between nt 8027 and 8028 to knock out the active site of the NS5 RdRp gene. The numbering of the nucleotide position is according to the sequence with GenBank accession no. AF404756. The drawing is not to scale. (B) IFAof cells at 48 h p.t. with Rluc/NeoRep (top panel) and IFA of Rluc/NeoRep-transfected cells at 20 and 80 days p.s. under G418 (lower two panels). The same field as imaged by differential interference contrast (DIC) and IFA staining with Texas red is presented to show percentages of cells containing the replicating Rluc/NeoRep. The expression of viral and Rluc proteins is as indicated. For the IFA, approximately 105 cells were seeded into four-chamber slides (Nalge, Naperville, Ill.), reacted with WNV immune mouse ascites fluid (1:100 dilution; American Type Culture Collection, Manassas, Va.) or with a mouse anti-Rluc monoclonal antibody (1:200 dilution; Chemicon) as a primary antibody, and further reacted with goat anti-mouse immunoglobulin G conjugated with Texas red (1:400 dilution; Kirkegaard & Perry Laboratories, Gaithersburg, Md.) as a secondary antibody. (C) Rluc activity in cells at various time points p.t. or p.s. as indicated. Rluc activities derived from the Rluc/NeoRep-containing cells are represented by filled bars. Background level of Rluc activity from cells at 48 h after transfection with the replication-defective replicon, Rluc/NeoRepNS5mt, is indicated by a hollow bar. Signals from 4 × 104 equivalent cells were presented for the Rluc quantification. (D) Replicon RNA copy numbers in cells at various time points p.s. as indicated. Signals from 105 equivalent cells were estimated for the Rluc/NeoRep RNA quantification.
WNV was originally isolated in 1937 from the blood of a febrile patient in the West Nile district of northern Uganda (37) and was subsequently found in many other regions, including additional areas of Africa, the Middle East, Europe, Russia, India, Indonesia, and, most recently, North America (7). Since its appearance in the United States in 1999, WNV has caused significant human, equine, and avian disease and has resulted in over 4,156 known human cases, including at least 284 human deaths (for updates, see http://www.cdc.gov/ncidod/dvbid/westnile/surv&controlCaseCount03.htm). The WNV epidemic in the United States in 2002 represents the largest meningoencephalitis outbreak in the Western Hemisphere and the largest WNV outbreak ever reported (8). Neither a vaccine nor an effective therapy for WNV infection in humans is available, although inactivated-WNV- and DNA-based vaccines have been developed for use in equines (13, 29). It is therefore a public health priority to develop effective means of prevention and treatment of WNV infections.
Development of high-throughput assays is essential for antiviral drug discovery. The present antiviral assay for WNV is based on viral infection of cultured cells, followed by monitoring of compound inhibition of viral replication through observation or quantification of cytopathic effects, or quantification of viral RNA by reverse transcription-PCR (RT-PCR) (20, 30). The low-throughput nature of the viral infection assay limits its use for efficiently screening compound libraries. The recent establishment of genetic systems of WNV (35, 36, 43) has provided foundations for the development of novel antiviral assays. Here we report a stable cell line that can be used for high-throughput screening of inhibitors of WNV replication.
We previously reported that a subgenomic replicon of WNV, containing a large in-frame deletion (from nucleotides [nt] 190 to 2379 as indicated by dashed lines in Fig. 1A) of the C-prM-E structural region, replicates efficiently in BHK-21 cells (35). The replicon retained the N-terminal 31 amino acids (nt 97 to 189) of the C protein and the C-terminal 30 amino acids of the E protein (nt 2380 to 2469), to preserve a replication cis element (also known as the cyclization sequence) (19, 21, 44) and the correct processing and translocation of the remaining nonstructural polyprotein in the correct topology across the membrane of the endoplasmic reticulum (10), respectively. To develop a high-throughput antiviral assay, we engineered two reporter genes, Renilla luciferase (Rluc) and neomycin phosphotransferase (Neo), into the original WNV replicon, resulting in Rluc/NeoRep (Fig. 1A). Translation of the Neo gene was driven by an encephalomyocarditis virus internal ribosomal entry site (IRES) in the upstream end of the 3′ UTR of the replicon. To prepare the IRES-Neo fragment, we amplified individual IRES and Neo through PCR from plasmid pIRES2-GFP (Clontech, Palo Alto, Calif.) and pcDNA3.1 (Invitrogen, Carlsbad, Calif.), using primer F-IRES and R-IRES and primers F-Neo and R-Neo, respectively (Table 1). Fragments of IRES and Neo were then fused to yield IRES-Neo through overlapping PCR (32). The fused IRES-Neo was inserted into the NsiI site (nt 10436) in the 3′ UTR of the original replicon. Next, the Rluc gene was PCR amplified from plasmid pRL-SV40 (Promega, Madison, Wis.) using primer F-Rluc and R-Rluc (Table 1) and then fused in frame into the AflII site of the replicon (35). The Rluc gene was selected as a reporter for assay development because of its relatively small size (936 bp) and its robust enzymatic activity.
TABLE 1.
Primers used for construction of WNV reporter replicons
| Primera | Primer sequenceb | Amplified fragment |
|---|---|---|
| F-IRES | ataattATGCATCCGCCCCTCTCCCTC (NsiI) | IRES |
| R-IRESc | GCAATCCATCTTGTTCAATCATGGTATTATCATCGTGTTTTTCAAAGG | |
| F-Neoc | CCTTTGAAAAACACGATGATAATACCATGATTGAACAAGATGGATTGC | Neo |
| R-Neoc | acaaccATGCATCAGAAGAACTCGTCAAGAAG (NsiI) | |
| F-Rluc | tacactCTTAAGATGGCTTCCAAGGTGTACGA (AflII) | Rluc |
| R-Rluc | cacaagCTTAAGCTGCTCGTTCTTCAGCACG (AflII) |
The primers were named according to their polarity (F, forward sense primer; R, reverse antisense primer) followed by the name of the amplified fragment (IRES, Neo, or Rluc).
Sequences from IRES, Neo, Rluc, or WNV are in uppercase letters. Extra tail sequences to facilitate restriction enzyme digestion are in lowercase letters. Restriction endonuclease sites are underlined and indicated in parentheses.
Sequences from IRES and WNV are shown in regular uppercase. Sequence from the Neo gene is in italicized uppercase.
To test whether the dual reporting replicon is replication competent, Rluc/NeoRep RNA was in vitro transcribed and transfected into BHK-21 cells as previously described (35, 36). BHK-21 cells at 48 h posttransfection (p.t.) expressed both viral and reporter Rluc proteins, as evidenced by the positive cells from the immunofluorescence assay (IFA; top panel in Fig. 1B). Less than 10% of the cells were IFA positive, principally due to low transfection efficiency. Although the Rluc protein contained fusion tags at its N and C termini derived from the viral C protein and E protein, respectively (Rluc/NeoRep in Fig. 1A), a high level of Rluc activity was detected from cell lysates harvested at 48 h p.t. (a filled bar in Fig. 1C). By contrast, transfection of BHK-21 cells with an equal amount of a mutant replicon, containing a frameshift insertion of a nucleotide U between nt 8027 and 8028 to knock out the active site of the NS5 RdRp gene (Rluc/NeoRepNS5mt in Fig. 1A), yielded no signals in either IFA (data not shown) or Rluc assay at 48 h p.t. or longer (a hollow bar in Fig. 1C). These results suggested that the positive IFA and Rluc activity from cells at 48 h after transfection with the Rluc/NeoRep were due to translation of replicating viral RNA, not the translation of the input replicon RNA.
To establish a stable cell line containing a persistently replicating dual reporting replicon, we transfected BHK-21 cells with the Rluc/NeoRep and selected the transfected cells under Geneticin (G418). Briefly, approximately 8 × 106 BHK-21 cells were electroporated with 10 μg of Rluc/NeoRep RNA at settings previously described (36), recovered in cuvettes for 10 min, resuspended in 50 ml of Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), and transferred to a T-150 flask. After an overnight recovery, the cells were subjected to G418 selection (1 mg/ml) in DMEM with 10% FBS. Medium was replaced every 2 to 3 days with fresh G418. After 10 days of selection, the majority of the cells died, presumably because these cells were untransfected or because they expressed an insufficient amount of Neo. However, many surviving cells were observed. Individual foci were cloned, expanded, and stored in 10% dimethyl sulfoxide (DMSO) and 40% FBS in liquid nitrogen. Cells were continually passaged under G418 selection for over 80 days (over 25 passages).
To test the stability of the replicon-containing cell lines, cells at various time points postselection (p.s.) were examined for viral and Rluc protein expression and for replicon RNA copy numbers. (i) IFA showed that all cells expressed viral and Rluc proteins on days 20 and 80 p.s. (lower two panels in Fig. 1B). Western blotting of the cell lysates harvested on day 80 p.s. also showed expression of viral and Rluc proteins of the expected molecular masses (data not shown). (ii) Stable and high levels of Rluc activity were detected from cells collected on days 20, 50, and 80 p.s. (Fig. 1C). For the Rluc assay, 106 cells were lysed in 500 μl of lysis buffer; 20 μl of lysate was measured for Rluc activity according to the instructions of the manufacturer (Promega) using a Lumat luminometer (EG & G Berthold). Approximately 8 × 106 light units were consistently detected from 4 × 104 cells, about 200 light units per cell. (iii) A consistent level of replicon RNA was maintained in cells at 20, 50, and 80 days p.s. (Fig. 1D). Real-time RT-PCR, targeting the viral NS1 region, was used to quantify replicon RNA as previously reported (34, 35). Total RNA extracted from 106 cells was eluted in 50 μl of RNase-free water using RNeasy kits (Qiagen); 5 μl of RNA was quantified by real-time RT-PCR, using in vitro-transcribed RNA as standards. We estimated 109 replicon molecules in 105 cells, or about 104 replicon RNAs per cell. These results demonstrated that the reporting cell line is stable and contains persistently replicating Rluc/NeoRep.
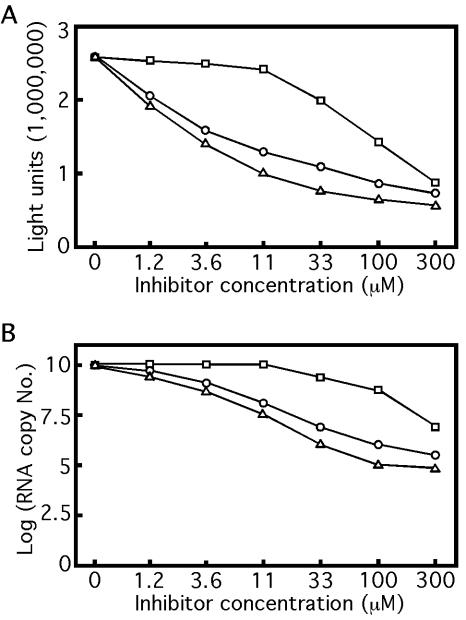
To examine whether the reporting cell line can be used as an antiviral assay, we tested three known WNV inhibitors in the cells: 6-azauridine, an inhibitor of orotidine monophosphate decarboxylase, and mycophenolic acid and ribavirin, inhibitors of IMP dehydrogenase (14, 20, 30). Ribavirin has also been reported to function as a mutagen, causing lethal levels of mutagenesis within the virus population (12). All three compounds were purchased from Sigma (St. Louis, Mo.). Initially, we performed the assay in 12-well plates. Approximately 2.5 × 105 cells were seeded into each well, containing DMEM with 2.5% FBS (without G418). Compounds were dissolved in DMSO and added to cells at various concentrations in medium with a final DMSO concentration of 1%. Cells not treated with compounds were still treated with 1% DMSO as a negative control. After 24 h of compound treatment at 37°C, cells were assayed for Rluc activity. As shown in Fig. 2A, the Rluc activity decreased with increasing concentration of each compound. Based on the Rluc curves, the EC50s (the compound concentration required to inhibit 50% of the Rluc activity) were estimated to be 5.4, 11, and 140 μM for mycophenolic acid, 6-azauridine, and ribavirin, respectively. These EC50s correlate well with the EC50s derived from the standard viral infection assay (8.4, 6.1, and 178 μM, respectively) (30). Longer (48-h) or shorter (12-h) treatment of the cells with the same compounds yielded EC50s severalfold higher than those obtained with the 24-h incubation (data not shown). One of the major differences between the replicon-containing cell assay (described here) and the standard viral infection assay is that, in the replicon-containing cell assay, the compound is added to cells inside which replicon RNAs are already under full replication, whereas in the standard viral infection assay, compound can be added either simultaneously or prior to viral infection of cells. In the replicon-containing cell assay, the Rluc signal is determined by multiple factors, including the doubling time of the replicon-containing cells (approximately 12 to 16 h), the potency and stability of the tested compound, the stability of replicon RNA, and the half-life of Rluc enzyme (about 3 to 6 h; Promega). As a consequence of these combinatory factors, at 12 h post-compound treatment, although the compound may start to inhibit replicon replication, the preexisting Rluc enzyme has not decayed to a level low enough to show a dramatic inhibitory effect of the compound. At 48 h post-compound treatment, the replicon-containing cells have doubled three to four times; if the inhibitory effect of the compound could not outcompete the increase of replicon RNA resulting from the cell number increase, the sensitivity of the assay would decrease, thereby increasing the EC50s of the tested compounds. The experimental results clearly showed that incubation of the cells with compound for 24 h yields the highest assay sensitivity. To increase the throughput of the assay, we performed the experiments in a 96-well format with approximately 5 × 104 cells seeded into each well; similar results as described above were obtained (data not shown). These results indicated that the Rluc activity from the cell line could be used for screening inhibitors of WNV in a high-throughput fashion.
FIG. 2.
The reporting cell line containing Rluc/NeoRep can be used as a high-throughput assay for antiviral drug discovery. Replicon-containing cells were treated with mycophenolic acid (▵), 6-azauridine (○), and ribavirin (□) at indicated concentrations for 24 h. The inhibition of viral replication by each compound was measured by the Rluc activity (A) and the replicon RNA copy number (B). One representative experiment of three is shown.
Next, we performed real-time RT-PCR to verify that the reduction in Rluc activity reflected the compound inhibition of viral RNA replication. After 24 h of compound treatment, cells from the 12-well plates were assayed for RNA amounts as described above, except that 12 μl from the 50-μl extracted RNA was used in the real-time RT-PCR assay. Similar to the Rluc results, decreasing amounts of replicon RNA were observed with increasing concentrations of inhibitor (Fig. 2B). The relative potencies of the three tested compounds are also similar to those observed with the Rluc results, in the decreasing order of mycophenolic acid, 6-azauridine, and ribavirin (compare Fig. 2A to B). However, the EC50s derived from the RNA copy numbers were much larger than those derived from the Rluc activity results: approximately 100 μM for mycophenolic acid and more than 300 μM for 6-azauridine and ribavirin (Fig. 2B). The higher sensitivity of the Rluc-based assay than that of the RNA copy-based assay is likely due to enzymatic signal amplification by the Rluc. Finally, to exclude the possibility that the reduction of Rluc activity and viral RNA copy number was due to cytotoxicity of the compounds, we performed an XTT [2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide, a substrate for mitochondrial dehydrogenase used for determination of cell viability] assay on parental BHK-21 cells (20). In accordance with previous reports (20, 30), no toxicity was observed at 300 μM for any of the three compounds (data not shown). Overall, the above results demonstrated that the Rluc activity reflects the replication level of viral RNA inside the cells and that the reporting cell line can serve as a high-throughput assay for screening inhibitors of WNV replication.
Replicon-based cell lines have been previously reported for a number of flaviviruses, including Kunjin virus (22), hepatitis C virus (4, 28), and WNV (35). The novel antiviral assay described here serves as a proof-of-principle that a similar approach should be applicable to development of genetics-based antiviral assays for other flaviviruses (33). The replicon-based assay covers all targets during viral replication, including viral translation, and plus- and minus-sense RNA synthesis. However, because no structural genes and, therefore, no infectious viral particles are involved in the replicon system, the assay does not include targets involved in viral entry, genome encapsidation, and virion maturation. On the other hand, because no infectious virions are formed, the replicon-based assay can be performed in a biosafety level 2 laboratory, rather than in a biosafety level 3 containment chamber. One other advantage of the replicon-cell line assay is that inhibitors identified through such a cell-based assay should have a high success rate in subsequent animal experiments because the assay tests the cellular uptake of compounds and, potentially, the stability of the compounds inside the cells (33).
Since Rluc is the reporter for the system, the assay may select potential inhibitors of the Rluc enzyme rather than inhibitors of viral replication. Rluc inhibitors can be quickly eliminated by testing any hits derived from the replicon-based screening in a recombinant Rluc assay (Chemicon International, Temecula, Calif.). The mode of action of viral inhibitors can be identified through individual biochemical assays such as RdRp, protease, NTPase, or helicase activity. Alternatively, the mode of action of the compounds could be analyzed through selection of compound-resistant virus followed by mapping of the mutated gene(s) and back-engineering of specific mutations into an infectious clone for phenotypic verification. The full-length infectious clone of WNV (36, 43) and recombinant systems of WNV NS5 RdRp and NS3 NTPase-helicase (42) will facilitate these analyses.
Acknowledgments
We thank the Molecular Genetics Core and the Cell Culture Facility at the Wadsworth Center for sequencing and oligonucleotide synthesis and for maintenance of BHK-21 cells, respectively.
The work was funded in part by the National Institute of Allergy and Infectious Disease, National Institutes of Health under contract no. N01-AI-25490. M. K. Lo was supported by the Emerging Infectious Diseases Fellowship Program funded by the New York State Department of Health.
REFERENCES
- 1.Ackermann, M., and R. Padmanabhan. 2001. De novo synthesis of RNA by the dengue virus RNA-dependent RNA polymerase exhibits temperature dependence at the initiation but not elongation phase. J. Biol. Chem. 276:39926-39937. [DOI] [PubMed] [Google Scholar]
- 2.Arias, C. F., F. Preugschat, and J. H. Strauss. 1993. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology 193:888-899. [DOI] [PubMed] [Google Scholar]
- 3.Bartelma, G., and R. Padmanabhan. 2002. Expression, purification, and characterization of the RNA 5′-triphosphatase activity of dengue virus type 2 nonstructural protein 3. Virology 299:122-132. [DOI] [PubMed] [Google Scholar]
- 4.Blight, K. J., A. A. Kolykhalov, and C. M. Rice. 2000. Efficient initiation of HCV RNA replication in cell culture. Science 290:1972-1974. [DOI] [PubMed] [Google Scholar]
- 5.Borowski, P., A. Niebuhr, O. Mueller, M. Bretner, K. Felczak, T. Kulikowski, and H. Schmitz. 2001. Purification and characterization of West Nile virus nucleoside triphosphatase (NTPase)/helicase: evidence for dissociation of the NTPase and helicase activities of the enzyme. J. Virol. 75:3220-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinton, M. 2002. The molecular biology of West Nile virus: a new invader of the Western hemisphere. Annu. Rev. Microbiol. 56:371-402. [DOI] [PubMed] [Google Scholar]
- 7.Burke, D. S., and T. P. Monath. 2001. Flaviviruses, p. 1043-1126. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology, 4th ed., vol. 1. Lippincott William & Wilkins, Philadelphia, Pa.
- 8.Centers for Disease Control and Prevention. 2002. Provisional surveillance summary of the West Nile virus epidemic—United States, January-November 2002. Morb. Mortal. Wkly. Rep. 51:1129-1133. [PubMed] [Google Scholar]
- 9.Chambers, T. J., A. Grakoui, and C. M. Rice. 1991. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J. Virol. 65:6042-6050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambers, T. J., C. S. Hahn, R. Galler, and C. M. Rice. 1990. Flavivirus genome organization, expression, and replication. Annu. Rev. Microbiol. 44:649-688. [DOI] [PubMed] [Google Scholar]
- 11.Chambers, T. J., A. Nestorowicz, S. M. Amberg, and C. M. Rice. 1993. Mutagenesis of the yellow fever virus NS2B protein: effects on proteolytic processing, NS2B-NS3 complex formation, and viral replication. J. Virol. 67:6797-6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crotty, S., D. Maag, J. J. Arnold, W. Zhong, J. Y. Lau, Z. Hong, R. Andino, and C. E. Cameron. 2000. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat. Med. 6:1375-1379. [DOI] [PubMed] [Google Scholar]
- 13.Davis, B., G. Chang, B. Cropp, J. Roehrig, D. Martin, C. Mitchell, R. Bowen, and M. Bunning. 2001. West Nile virus recombinant DNA vaccine protects mouse and horse from virus challenge and expresses in vitro a noninfectious recombinant antigen that can be used in enzyme-linked immunosorbent assays. J. Virol. 75:4040-4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Clercq, E. 1993. Antiviral agents: characteristic activity spectrum depending on the molecular target with which they interact. Adv. in Virus Res. 42:1-55. [DOI] [PubMed] [Google Scholar]
- 15.Diamond, M. S., D. Edgil, T. G. Roberts, B. Lu, and E. Harris. 2000. Infection of human cells by dengue virus is modulated by different cell types and viral strains. J. Virol. 74:7814-7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Egloff, M. P., D. Benarroch, B. Selisko, J. L. Romette, and B. Canard. 2002. An RNA cap (nucleoside-2′-O-)-methyltransferase in the flavivirus RNA polymerase NS5: crystal structure and functional characterization. EMBO J. 21:2757-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falgout, B., R. H. Miller, and C. J. Lai. 1993. Deletion analysis of dengue virus type 4 nonstructural protein NS2B: identification of a domain required for NS2B-NS3 protease activity. J. Virol. 67:2034-2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guyatt, K. J., E. G. Westaway, and A. A. Khromykh. 2001. Expression and purification of enzymatically active recombinant RNA-dependent RNA polymerase (NS5) of the flavivirus Kunjin. J. Virol. Methods 92:37-44. [DOI] [PubMed] [Google Scholar]
- 19.Hahn, C. S., Y. S. Hahn, C. M. Rice, E. Lee, L. Dalgarno, E. G. Strauss, and J. H. Strauss. 1987. Conserved elements in the 3′ untranslated region of flavivirus RNAs and potential cyclization sequences. J. Mol. Biol. 198:33-41. [DOI] [PubMed] [Google Scholar]
- 20.Jordan, I., T. Briese, N. Fischer, J. Y. Lau, and W. I. Lipkin. 2000. Ribavirin inhibits West Nile virus replication and cytopathic effect in neural cells. J. Infect. Dis. 182:1214-1217. [DOI] [PubMed] [Google Scholar]
- 21.Khromykh, A. A., H. Meka, K. J. Guyatt, and E. G. Westaway. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 75:6719-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khromykh, A. A., and E. G. Westaway. 1997. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J. Virol. 71:1497-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koonin, E. V. 1993. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J. Gen. Virol. 74:733-740. [DOI] [PubMed] [Google Scholar]
- 24.Kummerer, B. M., and C. M. Rice. 2002. Mutations in the yellow fever virus nonstructural protein NS2A selectively block production of infectious particles. J. Virol. 76:4773-4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, H., S. Clum, S. You, K. E. Ebner, and R. Padmanabhan. 1999. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J. Virol. 73:3108-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindenbach, B., and C. Rice. 1997. trans-Complementation of yellow fever virus NS1 reveals a role in early RNA replication. J. Virol. 71:9608-9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lindenbach, B. D., and C. M. Rice. 1999. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J. Virol. 73:4611-4621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lohmann, V., F. Korner, J. Koch, U. Herian, L. Theilmann, and R. Bartenschlager. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110-113. [DOI] [PubMed] [Google Scholar]
- 29.Monath, T. 2001. Prospects for development of a vaccine against the West Nile virus. Ann. N. Y. Acad. Sci. 951:1-12. [DOI] [PubMed] [Google Scholar]
- 30.Morrey, J., D. Smee, R. Sidwell, and C. Tseng. 2002. Identification of active antiviral compounds against a New York isolate of West Nile virus. Antivir. Res. 55:107-116. [DOI] [PubMed] [Google Scholar]
- 31.Muylaert, I. R., T. J. Chambers, R. Galler, and C. M. Rice. 1996. Mutagenesis of the N-linked glycosylation sites of the yellow fever virus NS1 protein: effects on virus replication and mouse neurovirulence. Virology 222:159-168. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 33.Shi, P. Y. 2002. Strategies for the identification of inhibitors of West Nile virus and other flaviviruses. Curr. Opin. Investig. Drugs 3:1567-1573. [PubMed] [Google Scholar]
- 34.Shi, P. Y., E. B. Kauffman, P. Ren, A. Felton, J. H. Tai, A. P. Dupuis II, S. A. Jones, K. A. Ngo, D. C. Nicholas, J. Maffei, G. D. Ebel, K. A. Bernard, and L. D. Kramer. 2001. High-throughput detection of West Nile virus RNA. J. Clin. Microbiol. 39:1264-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi, P. Y., M. Tilgner, and M. K. Lo. 2002. Construction and characterization of subgenomic replicons of New York strain of West Nile virus. Virology 296:219-233. [DOI] [PubMed] [Google Scholar]
- 36.Shi, P. Y., M. Tilgner, M. K. Lo, K. A. Kent, and K. A. Bernard. 2002. Infectious cDNA clone of the epidemic West Nile virus from New York City. J. Virol. 76:5847-5856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smithburn, K. C., T. P. Hughes, A. W. Burke, and J. H. Paul. 1940. A neurotropic virus isolated from the blood of a native of Uganda. Am. J. Trop. Med. 20:471-492. [Google Scholar]
- 38.Tan, B. H., J. Fu, R. J. Sugrue, E. H. Yap, Y. C. Chan, and Y. H. Tan. 1996. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology 216:317-325. [DOI] [PubMed] [Google Scholar]
- 39.Warrener, P., J. K. Tamura, and M. S. Collett. 1993. RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J. Virol. 67:989-996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wengler, G., and G. Wengler. 1991. The carboxy-terminal part of the NS 3 protein of the West Nile flavivirus can be isolated as a soluble protein after proteolytic cleavage and represents an RNA-stimulated NTPase. Virology 184:707-715. [DOI] [PubMed] [Google Scholar]
- 41.Wengler, G., and G. Wengler. 1993. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology 197:265-273. [DOI] [PubMed] [Google Scholar]
- 42.Wong, S. J., R. H. Boyle, V. L. Demarest, A. N. Woodmansee, L. D. Kramer, H. Li, M. Drebot, R. A. Koski, E. Fikrig, D. A. Martin, and P.-Y. Shi. 2003. Immunoassay targeting nonstructural protein 5 to differentiate West Nile virus infection from dengue and St. Louis encephalitis virus infections and from flavivirus vaccination. J. Clin. Microbiol. 41:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yamshchikov, V. F., G. Wengler, A. A. Perelygin, M. A. Brinton, and R. W. Compans. 2001. An infectious clone of the West Nile flavivirus. Virology 281:294-304. [DOI] [PubMed] [Google Scholar]
- 44.You, S., and R. Padmanabhan. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 274:33714-33722. [DOI] [PubMed] [Google Scholar]