Abstract
The UL46, UL47, UL48, and UL49 genes, which encode major tegument proteins, are conserved in most alphaherpesvirus genomes. However, the relative importance of each of these proteins for replication of individual alphaherpesviruses appears to be different. Recently, we demonstrated that single deletions of UL47 or UL48 impair maturation and egress of pseudorabies virus (PrV) particles to different extents, whereas deletions of UL46 or UL49 have no significant effects on virus replication in cell culture (W. Fuchs, H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter, J. Virol. 76:6729-6742, 2002; M. Kopp, B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter, J. Virol. 76:8820-8833, 2002). To test for possible functional redundancy between the four tegument proteins, a quadruple gene deletion mutant (PrV-ΔUL46-49) was generated and characterized in vitro. Although plaque formation by this mutant was almost abolished and maximum titers were reduced more than 100-fold compared to those of parental wild-type virus, PrV-ΔUL46-49 could be propagated and serially passaged in noncomplementing porcine and rabbit kidney cells. Electron-microscopic studies revealed that nucleocapsid formation and egress of PrV-ΔUL46-49 from the host cell nucleus were not affected, but secondary envelopment of nucleocapsids in the cytoplasm was only rarely observed. The replication defect of PrV-ΔUL46-49 could be fully corrected by reinsertion of the UL46-to-UL49 gene cluster. Plaque sizes and virus titers were only slightly increased after restoration of only UL47 expression, whereas repair of only UL48 resulted in a significant increase in replication capacity to the level of a UL47 deletion mutant. In conclusion, we show that none of the UL46 to UL49 tegument proteins is absolutely required for productive replication of PrV. Moreover, our data indicate that the UL47 and UL48 proteins function independently during cell-to-cell spread and virus egress.
Pseudorabies virus (PrV, suid herpesvirus 1), the causative agent of Aujeszky's disease of pigs, is a member of the genus Varicellovirus of the Alphaherpesvirinae subfamily of the Herpesviridae (34, 43). It exhibits a type D herpesvirus genome (43), which consists of two unique regions (UL and US), with inverted repeat sequences (IR and TR) bracketing the US region (Fig. 1A). The PrV genome is ca. 143 kbp in size, and its complete nucleotide sequence was recently determined (B. G. Klupp et al., unpublished results). Most of the ca. 70 identified PrV genes exhibit homology to genes in other alphaherpesviruses such as varicella zoster virus (VZV) (8), herpes simplex virus type 1 (HSV-1) (31), and equine herpesvirus 1 (44). For the sake of clarity, most PrV gene designations conform with their HSV-1 counterparts. Gene arrangements in different alphaherpesvirus genomes were also shown to be widely similar (31, 44). However, PrV contains an internal inversion of the conserved UL27 to UL44 genes within the UL genome region (2, 5). Up to now, a related inversion ranging from UL22 to UL44 was identified only within the genome of a phylogenetically distant alphaherpesvirus, avian infectious laryngotracheitis virus (ILTV) (51). Next to the inverted region, the PrV genome contains a conserved cluster of four open readings frames (ORFs) named UL46, UL47, UL48, UL49 (Fig. 1A), which were shown to encode major components of the virion tegument (4, 5, 9, 16, 17, 27). Homologues of these four genes have also been found in all hitherto-characterized alphaherpesviruses, including the avian pathogens Marek's disease virus (MDV) (48) and ILTV (51), but not in beta- or gammaherpesvirus genomes. In ILTV, however, gene clustering is disrupted since the UL47 homologous gene is present in the US genome region (47, 51).
FIG. 1.
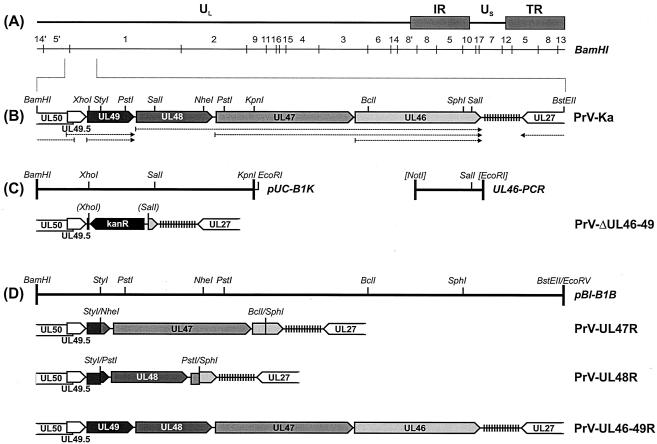
Generation of virus mutants. (A) Schematic map of the PrV genome, which consists of a UL region and a US region which is bracketed by inverted repeat sequences (IR and TR). The positions of BamHI restriction fragments are indicated. (B) Enlarged map of the investigated part of the genome of wild-type PrV-Ka with relevant restriction sites. The positions of repetitive sequences (vertical lines) and the transcriptional organization (dotted arrows) of the UL50 to UL27 genes (pointed rectangles) are also shown. (C) A cloned BamHI-KpnI fragment of the PrV-Ka genome (pUC-B1K) and a PCR product containing artificial restriction sites (UL46-PCR) were utilized for construction of the deletion mutant PrV-ΔUL46-49 in which the UL46 to UL49 genes were replaced by a kanamycin resistance gene (kanR). Restriction sites in parentheses were lost during cloning. (D) Plasmid pBl-B1B contains a BamHI-BstEII fragment of the PrV-Ka genome and was used for generation of a PrV-ΔUL46-49 rescue mutant (PrV-UL46-49R). The transfer plasmids for generation of the single-gene revertants PrV-UL47R and PrV-UL48R were derived from pBl-B1B by subcloning (see text).
Extensive investigations of the UL49 and UL48 gene products of HSV-1 revealed remarkable biological properties. The UL49 protein VP22 is capable of virus-independent trafficking from cell to cell (12), and the UL48 protein VP16 stimulates transcription, especially of viral immediate-early genes (1, 6). A similar function has also been described for the UL48 homologs of bovine herpesvirus 1 (36), VZV (37), and PrV (16). The UL46 and UL47 gene products of HSV-1, VP11/12 and VP13/14, modulate VP16-mediated transactivation (33, 49, 50).
Since the UL46, UL47, UL48, and UL49 proteins represent important components of the tegument layer which fills the space between capsids and envelope membranes of mature herpesvirus particles (43), they may also play an important role during virion morphogenesis. According to widely accepted model (reviewed in reference 35), herpesvirus virions are assembled stepwise in different cell compartments. Newly replicated viral genomic DNA is encapsidated in the host cell nucleus into preformed capsids. These nucleocapsids are then translocated into the cytoplasm by sequential budding at the inner leaflet (envelopment) and fusion at the outer leaflet of the nuclear membrane (de-envelopment). In the cytoplasm, a secondary (and final) envelope containing glycosylated and nonglycosylated viral membrane proteins is acquired by the budding of nucleocapsids into trans-Golgi vesicles, which are then transported to the cell surface. Secondary envelopment has to be preceded by addition of more than 15 different tegument proteins, either to the cytoplasmic nucleocapsids or to the future budding site, which requires complex interactions between viral capsid, tegument, and membrane proteins (35).
The UL48 proteins of several alphaherpesviruses play a central role in this process, since electron-microscopic investigations of cells infected with UL48 null PrV and HSV-1 mutants revealed retention of naked nucleocapsids in the cytoplasm and a failure to release mature virions (16, 38). Consequently, productive replication of the respective PrV mutants was severely impaired (16) and no infectious progeny virus could be isolated from noncomplementing cells infected with UL48-negative HSV-1 (38, 46). In contrast, VZV and MDV, which, unlike PrV and HSV-1, are strictly cell associated in vitro, are able to productively replicate in cell culture without their UL48 homologs (7, 10).
Pull-down experiments indicated a physical interaction between the UL48 and UL49 gene products of HSV-1 (11), and in yeast two-hybrid studies the UL49 protein of PrV was shown to interact with the cytoplasmic domains of envelope glycoproteins E and M (gE and gM) (17). However, whereas the presence of at least one of these two glycoproteins is necessary for efficient formation and release of infectious PrV virions (3, 4), deletion of UL49 did not detectably affect virion morphogenesis, in vitro growth properties, or in vivo virulence of PrV (9, 17). Thus, the glycoprotein interactions of the UL49 protein may be required only for its own incorporation into virions (17, 30). Deletion of the UL49 gene products of HSV-1 and bovine herpesvirus 1 reduced viral replication efficiency but did not abolish it, indicating that these two viruses also are able to productively replicate in the absence of their UL49 protein-homologous proteins (29, 40; G. Elliott and A. Whiteley, Abstr. 26th Int. Herpesvirus Workshop, abstr. 8.09, 2001). Only the MDV UL49 protein homolog seems to be indispensable for viral growth (10).
The UL46 and UL47 gene products of HSV-1, MDV, and PrV are also nonessential proteins, although their deletion may moderately impair virus replication (10, 27, 42, 49, 50). For PrV, UL46 or UL47 null mutants exhibited slightly reduced plaque sizes in cell culture, whereas only deletion of UL47 resulted in a ca. 10-fold titer reduction, correlating with a partial retention of nonenveloped nucleocapsids in the cytoplasm of infected cells (27).
The clustered localization of the UL46 to UL49 tegument protein genes could indicate that they have evolved from a common ancestral gene in a manner similar to that proposed for the conserved glycoprotein gene cluster in the US genome regions of alphaherpesviruses (32). This would suggest that the UL46 to UL49 gene products may possess related or even redundant functions during virion morphogenesis. Thus, it appeared conceivable that deletion of one or two of the corresponding genes could be compensated for by the remaining genes of this cluster, whereas simultaneous deletion of all four genes might be lethal for the virus. Therefore, several double and triple mutants with deletions of UL46 and UL47 of HSV-1 (49), PrV (27), and MDV (10), UL48 and UL49 of PrV (16), or UL46, UL47, and UL48 of MDV (10) have been isolated and characterized. Each of these recombinants, like the corresponding single gene deletion mutants, proved to be replication competent in noncomplementing cells. However, previous attempts to delete UL46 to UL49 together from the PrV genome were not successful (9).
In the present study, a quadruple mutant lacking the entire UL46-to-UL49 gene cluster was constructed by mutagenesis of the bacterial artificial chromosome pPrV-K1 (15), which contains the genome of the wild-type strain PrV-Ka (23). Subsequently, a rescue mutant and two triple mutants lacking either UL46, UL48, and UL49 or UL46, UL47, and UL49 were generated after reinsertion of the complete deleted genome fragment or only the UL47 gene or the UL48 gene, respectively. In vitro growth properties of the obtained PrV recombinants were investigated, and virion morphogenesis was studied by electron microscopy. Viruses were propagated in porcine kidney (PSEK) cells, whereas transfection experiments, growth kinetics assays, plaque assays, and electron-microscopic analyses were performed with rabbit kidney (RK13) cells. Cells were grown in minimum essential medium supplemented with 10% fetal calf serum (Invitrogen).
In different investigations, the part of the PrV genome encoding the viral dUTPase (UL50), glycoprotein N (UL49.5), four tegument proteins (UL49, UL48, UL47, and UL46), and gB (UL27) has been characterized (GenBank accession no. U38457, AJ437285, AJ010303, M17321) (5, 17, 21, 41). Assembly of the overlapping DNA sequences permitted the design of a PrV mutant possessing a 6,319-bp deletion ranging from codon 5 of UL49 to codon 636 of UL46 (Fig. 1C). For construction of this quadruple gene deletion mutant the plasmid pUC-B1K, containing a genomic 3,611-bp BamHI/KpnI fragment of PrV (16), and a 1,159-bp amplification product of the 3′ part of UL46 flanked by artificial restriction sites (UL46-PCR) (39) were used (Fig. 1C). After double digestion of pUC-B1K with SalI and EcoRI a part of the viral insert was replaced by a 278-bp SalI-EcoRI subfragment of UL46-PCR. The obtained plasmid was cleaved with SalI and XhoI, and, after Klenow treatment, the kanamycin resistance gene (kanR) gene, which had been isolated after amplification of nucleotides 1800 to 2769 of pACYC177 (GenBank accession no. X06402) by PCR with Pfx DNA polymerase (Invitrogen), was inserted. The insert fragment of the resulting plasmid was amplified by PCR with the vector-specific M13/pUC (−47) and M13/pUC reverse (−48) primers (New England Biolabs) and used for RecE- and RecT-mediated mutagenesis of pPrV-K1 in Escherichia coli, followed by selection of kanamycin-resistant clones (15). Since previous studies revealed unwanted effects of the bacterial vector sequences which are inserted at the gG gene locus of pPrV-K1 (15), they were removed by cotransfection of RK13 cells (18) with mutated bacterial artificial chromosome DNA and a plasmid containing the authentic gG gene of PrV (15). Single plaque isolates of the transfection progeny were tested for gG expression by indirect immunofluorescence, and one positive isolate was further analyzed. Thus, the genome of the resulting mutant, PrV-ΔUL46-49, should contain no other differences from that of PrV-Ka than the deletion of the four tegument protein genes (Fig. 1C).
To confirm that the phenotypic defects of PrV-ΔUL46-49 were solely caused by the introduced deletion, a revertant in which the UL46-to-UL49 gene cluster was completely restored was generated (Fig. 1D). For that purpose a 8,772-bp BamHI/BstEII subfragment of genomic BamHI fragment 1 of PrV-Ka was cloned into the vector pBluescript SK(−) (Stratagene) which had been digested with BamHI and EcoRV. After cotransfection of RK13 cells with the resulting plasmid pBl-B1B and virion DNA of PrV-ΔUL46-49, the rescue mutant PrV-UL46-49R was isolated (Fig. 1D).
Since previous studies had demonstrated that deletions of either UL47 or UL48 from the wild-type PrV genome significantly affect in vitro replication (16, 27), the corresponding ORFs, preceded by their own promoter sequences, were individually reinserted into the genome of PrV-ΔUL46-49. The single gene revertant PrV-UL47R (Fig. 1D) was obtained after transfection of RK13 cells with PrV-ΔUL46-49 DNA and a plasmid which was derived from pBl-B1B by subsequent BclI/SphI and NheI/StyI double digestions, followed by religation. Another construct, which had been obtained after insertion of a 1,602-bp PstI fragment of pBl-B1B into the same plasmid after prior digestion with SphI and StyI, was used for generation of a UL48 rescue mutant (PrV-UL48R; Fig. 1D). During plasmid cloning, all noncompatible fragment ends were blunted by Klenow treatment prior to ligation. To utilize the methylation-sensitive restriction enzyme BclI, pBl-B1B had to be propagated in the dam-negative E. coli strain GM2163 (New England Biolabs). Plaque purification of the virus revertants PrV-UL46-49R, PrV-UL47R, and PrV-UL48R was facilitated by their growth advantages compared with the parental deletion mutant, PrV-ΔUL46-49. Expression of the restored genes was verified by indirect immunofluorescence tests, and all virus recombinants were characterized by restriction analyses and Southern blot hybridization of genomic DNA (results not shown).
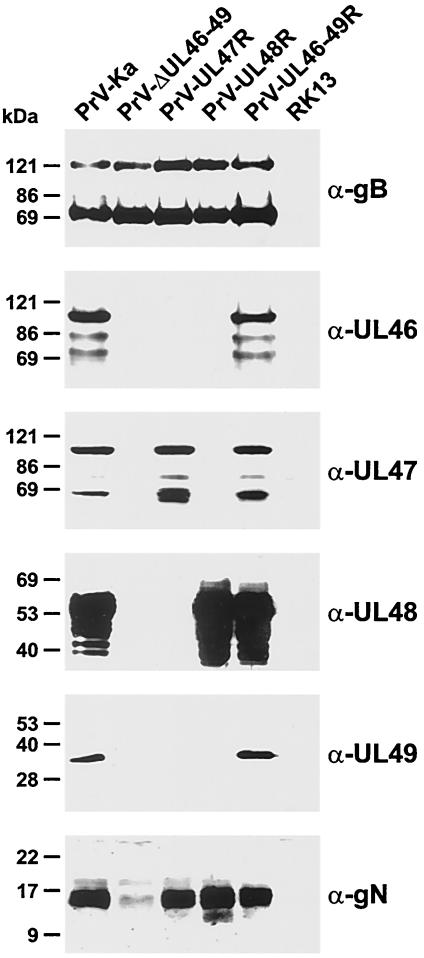
Viral protein expression was further investigated by Western blot analyses of RK13 cells which were harvested 16 h after inoculation with PrV-Ka or with the generated PrV mutants at a multiplicity of infection (MOI) of 2 (Fig. 2). Cell lysates and blots were prepared as described previously (16) and incubated with monospecific rabbit antisera against the UL46 (27), UL47 (27), UL48 (16), and UL49 (4) gene products, against gN (21), or with a gB-specific monoclonal antibody (MAb) (39). As described earlier, multiple products of the UL46, UL47, and UL48 genes could be detected in cells infected with wild-type PrV-Ka, whereas the UL49 protein, despite its differential phosphorylation (9), forms a single band with an apparent mass of ca. 33 kDa (Fig. 2). As expected, no products of the UL46, UL47, UL48, and UL49 genes were detectable in cells infected with PrV-ΔUL46-49 (Fig. 2), although expression of other viral proteins, for example, gB (Fig. 2), was unaltered. In cells infected with PrV-UL46-49R the expression of UL46 to UL49 was indistinguishable from that in PrV-Ka (Fig. 2), whereas only the UL47 gene products were detected in cells infected with PrV-UL47R and only the UL48 proteins were found in cells infected with PrV-UL48R (Fig. 2). In none of the investigated PrV mutants did we detect truncated gene products, although the first 80 codons of UL49 were preserved in the genomes of PrV-UL47R and PrV-UL48R, and short parts of the adjoining ORFs were reinserted together with the complete UL47 or UL48 gene (Fig. 1D). Whereas the resulting hybrid genes in PrV-UL48R were out of frame, a hybrid protein, consisting of amino acids 1 to 80 of the UL49 gene product and 366 to 413 of the UL48 gene product, as well as a truncated UL46 protein containing amino acids 1 to 79 and 601 to 694, might theoretically be expressed by PrV-UL47R (Fig. 1D). Apparently, however, these hypothetical proteins were either highly unstable or not sufficiently antigenic to be detected by our antisera.
FIG. 2.
Expression of viral proteins. RK13 cells were infected at a MOI of 2 with PrV-Ka, PrV-ΔUL46-49, PrV-UL47R, PrV-UL48R, or PrV-UL46-49R and incubated for 16 h at 37°C. Proteins were separated in discontinuous sodium dodecyl sulfate-polyacrylamide gels and transferred to nitrocellulose filters. Western blots of infected and noninfected cells were probed with a monoclonal antibody against gB (α-gB) or monospecific antisera against viral tegument proteins (α-UL46, α-UL47, α-UL48, and α-UL49) and gN, encoded by the UL49.5 gene (α-gN). Detected proteins were visualized by chemiluminescence reactions (Super Signal; Pierce) of peroxidase-conjugated secondary antibodies (Dianova). Molecular masses of marker proteins are indicated at the left.
Remarkably, expression of the UL49.5 gene, which encodes gN, was significantly reduced in cells infected with PrV-ΔUL46-49 (Fig. 2), although the UL49.5 open reading frame preceding the deleted gene cluster was not affected by mutagenesis (Fig. 1C). This effect was presumably caused by reduced transcription or stability of the UL49.5 mRNA. However, it appeared unlikely that it was a consequence of deletion of the common polyadenylation (poly[A]) signal of the UL49.5 and UL49 genes in PrV-ΔUL46-49, since the corresponding element of the coterminally transcribed UL48 to UL46 genes was still present (Fig. 1B and C). Significantly reduced gN expression was also observed in a previously described PrV UL49 deletion mutant and attributed to the insertion of foreign DNA sequences into the 3′-nontranslated part of the UL49.5 transcription unit (17). In PrV-ΔUL46-49 the UL49.5 mRNA may be destabilized by the bacterial kanamycin resistance gene inserted between the UL49.5 ORF and the next available polyadenylation signal. In accordance with this, wild-type-like gN expression levels were restored after replacement of the bacterial sequences by the viral UL47 or UL48 ORFs in the single-gene revertants PrV-UL47R and PrV-UL48R (Fig. 2). On the other hand, in both of these revertants the 5′-terminal part of the UL49 gene coding region, which may contain enhancer elements required for efficient transcription of the UL49.5 gene, was also restored. However, since previous studies have shown that deletion of UL49.5 has little effect on in vitro replication of PrV and that, unlike what is found for other alphaherpesviruses, gN is not required for processing or virion incorporation of gM (17, 22), a contribution of reduced gN expression to the observed phenotype of PrV-ΔUL46-49 was slight at best but cannot be totally excluded.
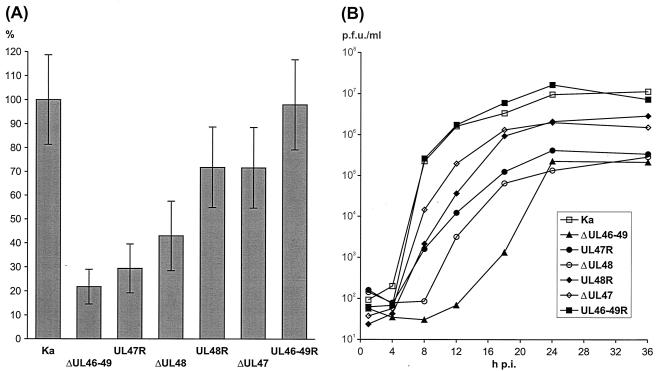
The successful isolation and propagation of the quadruple mutant PrV-ΔUL46-49 in noncomplementing RK13 and PSEK cells already demonstrated that none of the major tegument proteins encoded by UL46, UL47, UL48, and UL49 is absolutely required for productive virus replication in vitro. The infectivity of PrV-ΔUL46-49 was not strictly cell associated, since extracellular virus could be recovered after freezing and thawing infected cells, low-speed centrifugation, and filtration of the supernatant through a 0.2-μm-pore-size filter. However, the maximum titers of PrV-ΔUL46-49 were lower, and its plaques were smaller, than those of any of the described mutants lacking one or two of the four investigated tegument proteins (16, 17, 27). In the present study, plaques were visualized on infected RK13 cells which had been incubated for 48 h under semisolid medium by indirect immunofluorescence reactions of a gC-specific MAb (24). Compared to those of PrV-Ka, the average plaque diameter of PrV-ΔUL46-49 was reduced by ca. 78% (Fig. 3A). This defect of direct cell-to-cell spread was corrected in the rescue mutant PrV-UL46-49R (Fig. 3A). A moderate increase of plaque diameters from 22 to 29% of the wild-type size was observed after repair of only UL47 in PrV-UL47R (Fig. 3A). Repair of only UL48 restored cell-to-cell spread much more efficiently, since the plaques of PrV-UL48R were more than 75% of the size of PrV-Ka plaques (Fig. 3A). Obviously, the minor cell-to-cell spread defect of PrV-UL48R was predominantly caused by the absence of the UL47 protein, since plaque diameters of the single-gene deletion mutant PrV-ΔUL47 (27) were very similar (Fig. 3A).
FIG. 3.
Growth properties in RK13 cells of PrV-Ka; deletion mutants PrV-ΔUL46-49, PrV-ΔUL47, and PrV-ΔUL48; and revertants PrV-UL47R, PrV-UL48R, and PrV-UL46-49R. (A) After incubation under medium containing 6 g of methylcellulose/liter for 48 h, the infected cell monolayers were fixed with ethanol, and virus plaques were visualized by immunofluorescence reactions of a gC-specific MAb and fluorescein-conjugated secondary antibodies (Dako). The mean diameters of 30 single plaques per virus mutant were determined, and percentages of the wild-type (PrV-Ka) size were calculated. Error bars, standard deviations. (B) For analysis of one-step growth kinetics cells were infected at a MOI of 2 and incubated on ice for 1 h. Then, prewarmed medium was added, and incubation was continued at 37°C. After 1 h, nonpenetrated virus was inactivated by low-pH treatment. Immediately thereafter, as well as 4, 8, 12, 16, 24, 36, and 48 h after the temperature shift, cells were scraped into the medium and lysed by freezing and thawing. Progeny virus titers were determined by plaque assays, and the average results of two independent experiments are shown. p.i., postinfection.
One-step growth kinetics of the PrV mutants were determined as described previously (16). This revealed different contributions of the UL47 and UL48 proteins to productive replication and egress of PrV (Fig. 3B). The maximum titers of PrV-ΔUL46-49 in RK13 cells were reduced ca.100-fold compared to those of PrV-Ka and PrV-UL46-49R (Fig. 3B). The observed titer reduction of PrV-ΔUL46-49 was similar to that of the previously described single-gene deletion mutant PrV-ΔUL48 (16), and both viruses exhibited a delayed onset of replication (Fig. 3B). Virus titers were only slightly increased after restoration of UL47 gene expression in PrV-UL47R (Fig. 3B). In contrast, the maximum titer of PrV-UL48R was more than 10-fold higher than that of PrV-ΔUL46-49 and similar to that of PrV-ΔUL47 (Fig. 3B). Thus, our results confirm that the UL48 and UL47 proteins possess independent functions which are relevant for formation of mature virions and direct cell-to-cell spread of PrV. The delayed onset of replication of PrV-ΔUL48, and PrV-ΔUL46-49 might be due to the absence of the transcription-activating function of the UL48 protein (16). However, PrV-UL47R, which also lacks major parts of UL48, did not exhibit this phenotype. As mentioned above, PrV-UL47R contains an in-frame fusion gene consisting of the 5′-terminal 80 codons of UL49 and the 3′-terminal 48 codons of UL48 (Fig. 1D). Although the potential fusion protein was not detectable with the available antisera, the possibility that it is expressed and incorporated into virions and that it may induce viral gene expression early after infection cannot be ruled out. In this context, it should be mentioned that the major trans-activation domain of the HSV-1 UL48 protein has been mapped close to its C terminus (20).
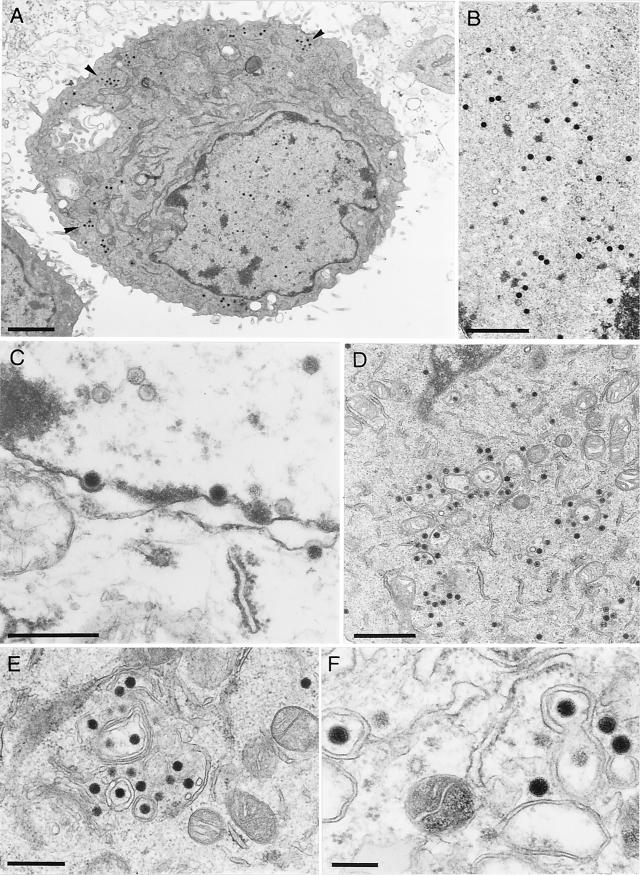
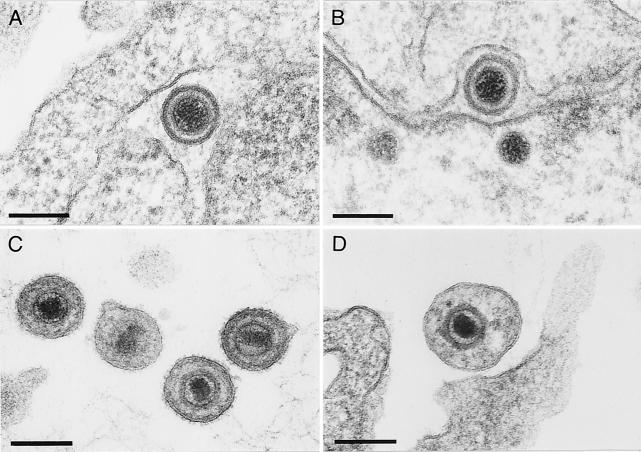
To investigate the functions of the UL46 to UL49 gene products of PrV in more detail, RK13 cells were fixed 12 h after infection with PrV-ΔUL46-49 (Fig. 4) or PrV-UL47R or PrV-UL48R (data not shown) and analyzed by electron microscopy as previously described (19). Apparently, the four tegument proteins are not required for the early steps of virion formation, since no obvious difference in intranuclear virion maturation was detectable in cells infected with PrV-ΔUL46-49 (Fig. 4A and B). DNA-filled capsids were also transported through the nuclear membrane (Fig. 4C), and de-enveloped nucleocapsids were observed in the cytoplasm (Fig. 4A, D, and E). However, secondary envelopment in the trans-Golgi region, as well as release of mature virions, was rarely detected (Fig. 4A). Only in a few infected cells were cytoplasmic nucleocapsids localized in the vicinity of proliferated membranes (Fig. 4E), and budding events could be observed (Fig. 4F). Most of the few extracellular virus particles of PrV-ΔUL46-49 were abnormally shaped (Fig. 5D). These virions contained neither the electron-dense tegument layer nor visible glycoprotein spikes at the envelope, which are both typical features of mature wild-type PrV virions (Fig. 5C) (19). In contrast, perinuclear virions of PrV-Ka (Fig. 5A) and PrV-ΔUL46-49 (Fig. 5B) had similar appearances.
FIG. 4.
Virion morphogenesis of PrV-ΔUL46-49. Electron microscopy of RK13 cells which had been fixed 12 h after infection at a MOI of 1 showed different stages of intranuclear capsid maturation (A and B), transit through the nuclear membrane (C), and accumulation of nonenveloped cytoplasmic nucleocapsids (A [arrowheads], D, and E). Occasionally, budding events were detectable in the cytoplasm (F). Bars, 2 (A), 1 (B and D), 0.5 (C and E), or 0.2 μm (F).
FIG. 5.
Structure of perinuclear (A and B) and extracellular (C and D) virions of PrV-Ka (A and C) and PrV-ΔUL46-49 (B and D). Ultrathin sections were prepared 12 h after infection of RK13 cells at a MOI of 1, counterstained with uranyl acetate, and examined by electron microscopy. Bars, 150 nm.
Correlating with the minor effects of reinsertion of the UL47 gene on one-step growth and plaque formation (see above), the defects in virion morphogenesis of PrV-UL47R were similar to those observed with PrV-ΔUL46-49. In contrast, PrV-UL48R, which expressed UL48 but which lacked UL46, UL47, and UL49, exhibited almost wild-type-like properties in electron-microscopic analyses, including efficient release of mature virions (data not shown).
To our knowledge PrV-ΔUL46-49 is the first described alphaherpesvirus with a deletion of the entire UL46-to-UL49 tegument protein gene cluster. Despite exhibiting a severe replication defect and drastically reduced viral titers, the obtained virus mutant could be propagated in noncomplementing cells. Thus, even if the UL46 to UL49 gene products of PrV possess redundant or overlapping functions, they are either nonessential for virus replication or can be compensated for by other viral proteins. For MDV and HSV-1, generation of similar mutants was hampered by the essential character of UL49 and UL48, respectively (10, 46). For PrV, deletion of the UL48 gene has also been shown to have the most profound effect on viral replication (16) while deletion of UL46, UL47, or UL49 had only slight (27) or no effects on viral replication (9, 17). Like many of its homologs, the PrV UL48 protein enhances viral immediate-early gene expression in infected cells (1, 6, 16, 36, 37). The HSV-1 UL48 protein was further shown to prevent rapid degradation of viral mRNA by modulation of the virion host shutoff activity of the UL41 protein (28). A similar function of the UL48 protein of PrV is conceivable, although yeast two-hybrid studies and immunoprecipitation experiments provided no evidence for physical interactions between the homologous UL48 and UL41 gene products of PrV (data not shown). Although the onset of progeny virus formation was delayed in cells infected with UL48-negative PrV mutants, electron-microscopic studies did not reveal an inhibition of the early steps in virion formation (16). Growth kinetics of the quadruple mutant PrV-ΔUL46-49 were even more delayed, indicating that the UL46, UL47, and UL49 proteins of PrV, like their HSV-1 homologs (11, 33, 49, 50), might be involved in trans-activation of viral gene expression early after infection. However, nucleocapsid formation in the nucleus and egress from the nucleus in cells infected with wild-type PrV and in cells infected with PrV-ΔUL46-49 appeared very similar. Thus, although the UL47, UL48, and UL49 gene products of PrV were detectable in both compartments, the host cell nucleus and the cytoplasm (9, 16, 27), they are apparently not crucial for the nuclear steps of virion formation.
In contrast, the cytoplasmic steps of virion morphogenesis were substantially impaired in cells infected with PrV-ΔUL46-49, leading to an inhibition of secondary nucleocapsid envelopment in the trans-Golgi region. Similar defects have been observed with a PrV UL48 null mutant and, although much less pronounced, also with a PrV UL47 deletion mutant (16, 27). In all cases, these defects detected by electron microscopy correlated with reduced plaque sizes and virus titers, which were most severely affected in cells infected with PrV-ΔUL46-49. After reinsertion of the entire UL46-to-UL49 gene cluster into the genome of PrV-ΔUL46-49, wild-type growth properties were fully restored. Single-gene repair of UL47 had only moderate stimulatory effects on virus replication, whereas plaque sizes and maximum virus titers of the UL48 rescue mutant PrV-UL48R were significantly increased compared to those of the parental mutant PrV-ΔUL46-49. However, wild-type-like growth properties were not fully achieved, and the remaining replication defect of PrV-UL48R was very similar to that of previously described UL47-negative PrV mutants (27). The additional absence of UL46 and UL49 from the genome of PrV-UL48R had no detectable effects, which is in agreement with the almost wild-type-like growth properties of UL46- or UL49-negative PrV mutants (9, 17, 27). Thus, our results indicate that the contributions of UL48 and UL47 to both direct cell-to-cell spread and release of infectious virions are additive and presumably independent of each other.
A remarkable finding of electron-microscopic analyses of previously described UL48 and UL48/UL49 null PrV mutants was the presence of high numbers of capsidless particles in and around infected cells which contained the UL46, UL47, and, if expressed, UL49 tegument proteins (16). This finding indicated that UL48 is required for secondary envelopment of cytoplasmic nucleocapsids and for prevention of premature budding at membranes in the trans-Golgi region (16). Moderate amounts of capsidless particles were also produced by wild-type PrV and many described recombinants but were not detectable with PrV mutants lacking the nonessential envelope glycoproteins gE, gI, and gM concomitantly (3, 4). Interestingly, capsidless particles were also not produced in or released from cells infected with PrV-ΔUL46-49 (Fig. 4A), PrV-UL47R, or PrV-UL48R (data not shown), indicating that interactions of UL46 or UL49 with gE or gM might be a prerequisite for efficient budding in the trans-Golgi region. Yeast two-hybrid studies revealed physical interactions of the UL49 gene product with gE and gM (17). Although capsid-containing and capsidless particles were also formed in the absence of UL49 (16), it would be interesting to test whether reinsertion of the UL49 gene into the genome of PrV-ΔUL46-49 enhances secondary envelopment.
PrV-ΔUL46-49 did not produce capsidless particles, but budding events involving nucleocapsids were occasionally observed in the cytoplasm of infected cells, and the resulting particles were also detected in the extracellular space (Fig. 5D). These “virions” apparently contained neither the electron-dense tegument material nor the typical glycoprotein projections on their envelopes, as observed in mature virions of PrV and other herpesviruses (Fig. 5C) (19, 43). The absence of a pronounced tegument layer was not surprising, since four major tegument proteins had been deleted in PrV-ΔUL46-49. However, considering the apparent absence of glycoprotein spikes, it appears questionable whether these abnormal PrV-ΔUL46-49 particles represent infectious virions.
Nevertheless, PrV-ΔUL46-49 was shown to be able to productively replicate in porcine and rabbit cells. The progeny virus was not strictly cell associated since it could be filtered and was resistant to freezing and thawing. Similar levels of infectious virus were also obtained after infection of cells with other PrV mutants that exhibit a defect in secondary envelopment due to deletions of gE, gI, and gM (3, 4), the UL37 tegument protein (26), or the UL3.5 gene product (14). These findings suggest that, besides the described tegumentation and envelopment pathway (35), a second but less efficient pathway of virus egress may exist. In the past, a direct release of primary enveloped virions from the perinuclear space was proposed for HSV-1 (45). However, UL34 or UL31 null PrV mutants which are blocked prior to primary envelopment were also demonstrated to retain a low level of infectivity in noncomplementing cells (15, 25). Thus, possibly nucleocapsids or other immature virus particles released during host cell disintegration represent the infectious units of PrV-ΔUL46-49 and the other mentioned deletion mutants. On the other hand, the possibility that PrV-ΔUL46-49 forms small amounts of regular mature virions which were undetectable in the electron microscope cannot be excluded.
To investigate the effects of separate and concomitant deletion of UL46, UL47, UL48, and UL49 on the virulence of PrV, animal experiments have to be performed. Up to now, only a UL49 single-gene deletion mutant has been characterized in vivo in a rat model and shown to be indistinguishable from wild-type PrV (9). It will be of particular interest to study also the function of the other tegument proteins of PrV during infection of the central nervous system, since in neurons the cytoplasmic steps of virion assembly and egress may be distinct from those observed in other cell types (13).
Acknowledgments
This study was supported by a grant from the Deutsche Forschungsgemeinschaft (Me 854/5-2).
We thank B. G. Klupp and M. Kopp for making antisera and virus mutants available and G. A. Smith and P. Ioannou for providing the plasmids required for cloning and mutagenesis of the PrV genome in E. coli. The technical assistance and photographic help of C. Ehrlich, P. Meyer, E. Zorn. and H. Stephan are greatly appreciated.
REFERENCES
- 1.Batterson, W., and B. Roizman. 1983. Characterization of the herpes simplex virion-associated factor responsible for the induction of alpha genes. J. Virol. 46:371-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ben-Porat, T., R. A. Veach, and S. Ihara. 1983. Localization of the regions of homology between the genomes of herpes simplex virus type 1 and pseudorabies virus. Virology 127:194-204. [DOI] [PubMed] [Google Scholar]
- 3.Brack, A. R., J. M. Dijkstra, H. Granzow, B. G. Klupp, and T. C. Mettenleiter. 1999. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J. Virol. 73:5364-5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack, A. R., B. G. Klupp, H. Granzow, R. Tirabassi, L. W. Enquist, and T. C. Mettenleiter. 2000. Role of the cytoplasmic tail of pseudorabies virus glycoprotein E in virion formation. J. Virol. 74:4004-4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bras, F., S. Dezelee, B. Simonet, X. Nguyen, P. Vende, A. Flamand, and M. J. Masse. 1999. The left border of the genomic inversion of pseudorabies virus contains genes homologous to the UL46 and UL47 genes of herpes simplex virus type 1, but no UL45 gene. Virus Res. 60:29-40. [DOI] [PubMed] [Google Scholar]
- 6.Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, J. I., and K. Seidel. 1994. Varicella-zoster virus (VZV) open reading frame 10 protein, the homolog of the essential herpes simplex virus protein VP16, is dispensable for VZV replication in vitro. J. Virol. 68:7850-7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davison, A. J., and J. E. Scott. 1986. The complete DNA sequence of varicella-zoster virus. J. Gen. Virol. 67:1759-1816. [DOI] [PubMed] [Google Scholar]
- 9.del Rio, T., H. C. Werner, and L. W. Enquist. 2002. The pseudorabies virus VP22 homologue (UL49) is dispensable for virus growth in vitro and has no effect on virulence and neuronal spread in rodents. J. Virol. 76:774-782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorange, F., B. K. Tischer, J. F. Vautherot, and N. Osterrieder. 2002. Characterization of Marek's disease virus serotype 1 (MDV-1) deletion mutants that lack UL46 to UL49 genes: MDV-1 UL49, encoding VP22, is indispensable for virus growth. J. Virol. 76:1959-1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elliott, G., G. Mouzakitis, and P. O'Hare. 1995. VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J. Virol. 69:7932-7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elliott, G., and P. O'Hare. 1997. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell 88:223-233. [DOI] [PubMed] [Google Scholar]
- 13.Enquist, L. W., P. J. Husak, B. W. Banfield, and G. A. Smith. 1999. Infection and spread of alphaherpesviruses in the nervous system. Adv. Virus Res. 51:237-347. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs, W., B. G. Klupp, H. Granzow, H. J. Rziha, and T. C. Mettenleiter. 1996. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J. Virol. 70:3517-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fuchs, W., B. G. Klupp, H. Granzow, N. Osterrieder, and T. C. Mettenleiter. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs, W., H. Granzow, B. G. Klupp, M. Kopp, and T. C. Mettenleiter. 2002. The UL48 tegument protein of pseudorabies virus is critical for intracytoplasmic assembly of infectious virions. J. Virol. 76:6729-6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuchs, W., B. G. Klupp, H. Granzow, A. Mundt, C. Hengartner, L. W. Enquist, and T. C. Mettenleiter. 2002. Physical interaction between envelope glycoproteins E and M of pseudorabies virus and the major tegument protein UL49. J. Virol. 76:8208-8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, F. L., and A. J. van der Eb. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456-467. [DOI] [PubMed] [Google Scholar]
- 19.Granzow, H., B. G. Klupp, W. Fuchs, J. Veits, N. Osterrieder, and T. C. Mettenleiter. 2001. Egress of alphaherpesviruses: comparative ultrastructural study. J. Virol. 75:3675-3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greaves, R., and P. O'Hare. 1989. Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J. Virol. 63:1641-1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jöns, A., H. Granzow, R. Kuchling, and T. C. Mettenleiter. 1996. The UL49.5 gene of pseudorabies virus codes for an O-glycosylated structural protein of the viral envelope. J. Virol. 70:1237-1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jöns, A., J. M. Dijkstra, and T. C. Mettenleiter. 1998. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J. Virol. 72:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaplan, A. S., and A. Vatter. 1959. A comparison of herpes simplex and pseudorabies virus. Virology 7:394-407. [DOI] [PubMed] [Google Scholar]
- 24.Klupp, B. G., and T. C. Mettenleiter. 1999. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J. Virol. 73:3014-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp, B. G., H. Granzow, and T. C. Mettenleiter. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063-10073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp, B. G., H. Granzow, E. Mundt, and T. C. Mettenleiter. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927-8936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kopp, M., B. G. Klupp, H. Granzow, W. Fuchs, and T. C. Mettenleiter. 2002. Identification and characterization of the pseudorabies virus tegument proteins UL46 and UL47: role for UL47 in virion morphogenesis in the cytoplasm. J. Virol. 76:8820-8833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lam, Q., C. A. Smibert, K. E. Koop, C. Lavery, J. P. Capone, S. P. Weinheimer, and J. R. Smiley. 1996. Herpes simplex virus VP16 rescues viral mRNA from destruction by the virion host shutoff function. EMBO J. 15:2575-2581. [PMC free article] [PubMed] [Google Scholar]
- 29.Liang, X., B. Chow, Y. Li, C. Raggio, D. Yoo, S. Attah-Poku, and L. A. Babiuk. 1995. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus UL49 homolog is dispensable for virus growth. J. Virol. 69:3863-3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyman, M. G., G. L. Demmin, and B. W. Banfield. 2003. The attenuated pseudorabies virus strain Bartha fails to package the tegument proteins Us3 and VP22. J. Virol. 77:1403-1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McGeoch, D. J., M. A. Dalrymple, A. J. Davison, A. Dolan, M. C. Frame, D. McNab, L. J. Perry, J. E. Scott, and P. Taylor. 1988. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J. Gen. Virol. 69:1531-1574. [DOI] [PubMed] [Google Scholar]
- 32.McGeoch, D. J. 1990. Evolutionary relationships of glycoprotein genes in the S regions of alphaherpesvirus genomes. J. Gen. Virol. 71:2361-2367. [DOI] [PubMed] [Google Scholar]
- 33.McKnight, J., P. Pellett, F. Jenkins, and B. Roizman. 1987. Characterization and nucleotide sequence of two herpes simplex virus 1 genes whose products modulate α-trans-inducing factor-dependent activation of α genes. J. Virol. 61:992-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mettenleiter, T. C. 2000. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet. Res. 31:99-115. [DOI] [PubMed] [Google Scholar]
- 35.Mettenleiter, T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misra, V., A. C. Bratanich, D. Carpenter, and P. O'Hare. 1994. Protein and DNA elements involved in transactivation of the promoter of the bovine herpesvirus (BHV) 1 IE-1 transcription unit by the BHV α gene trans-inducing factor. J. Virol. 68:4898-4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moriuchi, H., M. Moriuchi, S. E. Straus, and J. I. Cohen. 1993. Varicella-zoster virus open reading frame 10 protein, the herpes simplex virus VP16 homolog, transactivates herpesvirus immediate-early gene promoters. J. Virol. 67:2739-2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mossman, K. L., R. Sherburne, C. Lavery, J. Duncan, and J. R. Smiley. 2000. Evidence that herpes simplex virus VP16 is required for viral egress downstream of the initial envelopment event. J. Virol. 74:6287-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nixdorf, R., B. G. Klupp, A. Karger, and T. C. Mettenleiter. 2000. Effects of truncation of the carboxy terminus of pseudorabies virus glycoprotein B on infectivity. J. Virol. 74:7137-7145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pomeranz, L. E., and J. A. Blaho. 2000. Assembly of infectious herpes simplex virus type 1 virions in the absence of full-length VP22. J. Virol. 74:10041-10054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robbins, A. K., D. J. Dorney, M. W. Wathen, M. E. Whealy, C. Gold, R. J. Watson, L. E. Holland, S. D. West, M. Levine, J. C. Glorioso, and L. W. Enquist. 1987. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J. Virol. 61:2691-2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roizman, B., and D. M. Knipe. 2001. Herpes simplex viruses and their replication, p. 2399-2459. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 43.Roizman, B., and P. E. Pellet. 2001. The family herpesviridae: A brief introduction, p. 2381-2397. In D. M. Knipe, P. M. Howley, D. E. Griffin, R. A. Lamb, M. A. Martin, B. Roizman, and S. E. Straus (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, Pa.
- 44.Telford, E. A., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 45.Torrisi, R. M., C. DiLazzaro, A. Pavan, L. Pereira, and G. Campadelli-Fiume. 1992. Herpes simplex virus envelopment and maturation studied by fracture label. J. Virol. 66:554-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weinheimer, S. P., B. A. Boyd, S. K. Durham, J. L. Resnick, and D. R. O'Boyle. 1992. Deletion of the VP16 open reading frame of herpes simplex virus type 1. J. Virol. 66:258-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wild, M. A., S. Cook, and M. Cochran. 1996. A genomic map of infectious laryngotracheitis virus and the sequence and organization of genes present in the unique short and flanking regions. Virus Genes 12:107-116. [DOI] [PubMed] [Google Scholar]
- 48.Yanagida, N., S. Yoshida, K. Nazerian, and L. F. Lee. 1993. Nucleotide andpredicted amino acid sequences of Marek's disease virus homologues of herpes simplex virus major tegument proteins. J. Gen. Virol. 74:1837-1845. [DOI] [PubMed] [Google Scholar]
- 49.Zhang, Y., D. A. Sirko, and J. L. McKnight. 1991. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J. Virol. 65:829-841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, Y., and J. McKnight. 1993. Herpes simplex virus type 1 UL46 and UL47 deletion mutants lack VP11 and VP12 or VP13 and VP14, respectively, and exhibit altered viral thymidine kinase expression. J. Virol. 67:1482-1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziemann, K., T. C. Mettenleiter, and W. Fuchs. 1998. Gene arrangement within the unique long genome region of infectious laryngotracheitis virus is distinct from that of other alphaherpesviruses. J. Virol. 72:847-852. [DOI] [PMC free article] [PubMed] [Google Scholar]