Abstract
Two template switches are necessary during plus-strand DNA synthesis of the relaxed circular (RC) form of the hepadnavirus genome. The 3′ end of the minus-strand DNA makes important contributions to both of these template switches. It acts as the donor site for the first template switch, called primer translocation, and subsequently acts as the acceptor site for the second template switch, termed circularization. A small DNA hairpin has been shown to form near the 3′ end of the minus-strand DNA overlapping the direct repeat 1 in avihepadnaviruses. Previously we showed that this hairpin is involved in discriminating between two mutually exclusive pathways for the initiation of plus-strand DNA synthesis. In its absence, the pathway leading to production of duplex linear DNA is favored, whereas primer translocation is favored in its presence, apparently through the inhibition of in situ priming. Circularization involves transfer of the nascent plus strand from the 5′ end of the minus-strand DNA to the 3′ end, where further elongation can lead to production of RC DNA. Using both genetic and biochemical approaches, we now have found that the small DNA hairpin in the duck hepatitis B virus (DHBV) makes a positive contribution to circularization. The contribution appears to be through its impact on the conformation of the acceptor site. We also identified a unique DHBV variant that can synthesize RC DNA well in the absence of the hairpin. The behavior of this variant could serve as a model for understanding the mammalian hepadnaviruses, in which an analogous hairpin does not appear to exist.
Hepadnaviruses are a family of small, enveloped DNA viruses that display a narrow host range, a tropism for the liver, and are capable of both acute and chronic infections in their hosts. The human hepatitis B virus, a member of the family, is a major worldwide health problem exposing ca. 350 million chronic carriers to an increased risk of developing hepatocellular carcinoma (11). Viral replication is necessary for maintenance of the chronic carrier state; understanding this replication is critical for limiting disease in carriers. The duck hepatitis B virus (DHBV) has proven a powerful model for studying many aspects of hepadnaviral biology, including replication (reviewed in reference 3). Although divergent at the level of nucleic acid sequence, all hepadnaviruses share a similar genetic organization and general strategy of replication.
Three template switches are integral to viral replication: one during synthesis of the minus-strand DNA and two during synthesis of plus-strand DNA during the production of the relaxed circular (RC) DNA genome. Minus-strand DNA synthesis occurs via reverse transcription of an RNA intermediate, the pregenomic RNA (23). Upon completion of the minus-strand DNA, there are two mutually exclusive pathways for plus-strand DNA synthesis. The predominant pathway gives rise to RC DNA (Fig. 1A, panels a to d), while the alternative pathway yields duplex linear (DL) DNA (Fig. 1A, panel e). Both pathways use the same RNA primer, which is capped, 18 or 19 nucleotides (nt) in length, and generated by the final RNase H cleavage during minus-strand DNA synthesis (Fig. 1A, panel a) (13, 16, 22). Regulation of the first template switch of plus-strand DNA synthesis, called primer translocation, is critical in determining whether priming initiates from direct repeat 1 (DR1) or DR2. To make RC DNA, primer translocation must occur to transfer at least the 3′ end of the primer generated at DR1 (donor site) to DR2 (acceptor site), which is near the opposite end of the minus-strand DNA (Fig. 1A, panel b) (13). Following initiation of plus-strand DNA synthesis from DR2, elongation occurs to the 5′ end of the minus-strand DNA, where it runs out of template (Fig. 1A, panel c). At this point the final template switch, termed circularization, which is facilitated in part by a small terminal redundancy (termed r) in the minus-strand DNA, allows association of the 3′ end of the nascent plus-strand DNA to the 3′ end of the minus-strand template (Fig. 1A, panel d) and, upon additional elongation, production of the RC DNA form of the genome (15). A small fraction of viruses instead make DL DNA following initiation of plus-strand DNA synthesis from DR1, a process termed in situ priming (Fig. 1A, panel e) (22). Although infection with virus containing DL DNA can lead to covalently closed circular DNA formation through a process of nonhomologous recombination, this virus is rapidly outcompeted by a virus able to efficiently synthesize RC DNA (24, 25, 27). These findings highlight the importance of efficient plus-strand template switching during reverse transcription. A mechanistic understanding of these template switches, particularly circularization, could uncover unique and important therapeutic targets, as inhibition of these steps will be detrimental to the virus.
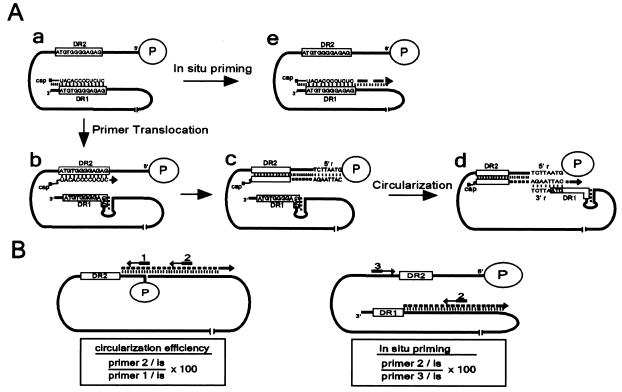
FIG. 1.
Model for plus-strand DNA replication of DHBV and depiction of primer extension analyses. (A) The full-length minus-strand DNA is shown as a solid black line, with the P protein (circle) covalently linked to the 5′ end. The two 12-nt direct repeats (DR1 and DR2) are indicated by boxes. (a) The final RNase H cleavage during minus-strand DNA synthesis generates a capped 18- or 19-nt oligoribonucleotide that serves as the primer for plus-strand DNA synthesis. The 3′ end of the primer lies within the DR1 sequence. (b) The first template switch of plus-strand DNA replication, primer translocation, involves moving at least the 3′ end of the primer from the donor site (DR1) to the distally located acceptor site (DR2), where initiation of plus-strand DNA synthesis occurs. (c) DNA synthesis proceeds to the 5′ end of the minus-strand DNA template, synthesizing approximately 50 nt of plus-strand DNA. (d) The second template switch, circularization, permits the 3′ end of the nascent plus-strand DNA to anneal to the 3′ end of the minus-strand DNA. This template switch is facilitated, in part, by a small terminal redundancy of 7 or 8 nt (5′r and 3′r). Following circularization, plus-strand DNA synthesis resumes, and elongation leads to formation of the RC form of the viral genome. (e) A small fraction of plus-strand DNA synthesis (ca. 5%) initiates from DR1 rather than DR2, a process termed in situ priming. Plus strands initiating from DR1 result in a DL form of the viral genome. In situ priming is negatively regulated, in part, by a small DNA hairpin overlapping the 5′ end of DR1. (B) Primer extension assays. Primer extension using primer 1 measures total plus-strand synthesis from DR2 at a point just prior to circularization. Primer extension using primer 2 measures plus-strand DNA that has successfully circularized, as shown at the left, and also detects priming events that occurred from DR1, as shown at the right. Primer extension with primer 3 measures the level of 5′ termini of the minus-strand DNA in the reaction. Combinations of these reactions allow for measurements of the priming events and circularization. Circularization efficiency is calculated as indicated as the percent primer 2 (DR2 priming)/primer 1. In situ priming is calculated as indicated as the percent primer 2 (DR1 priming)/primer 3 (minus-strand DNA). RNA primer utilization is an accumulation of the priming events from DR2 and DR1 relative to the amount of total minus-strand DNA (primer 1 + primer 2 [DR1 priming])/(primer 3). All reactions were normalized to a common internal standard DNA.
Our previous studies of DHBV indicate that a small DNA hairpin in the minus-strand DNA, which overlaps DR1, contributes to limiting in situ priming (4). Here we show that the same DNA hairpin contributes positively to the circularization process. Furthermore, we provide evidence that this element provides a contribution that can be partially substituted by an alternative DNA structure in the same vicinity of the minus strand, a finding with implications for understanding replication of mammalian hepadnaviruses, which lack a similar hairpin structure.
MATERIALS AND METHODS
Molecular clones.
All molecular clones were derived from DHBV type 3 (DHBV3) (21). The molecular clones containing substitutions in the stem nucleotides of the hairpin were previously described (4). Oligonucleotide-directed mutagenesis was used to generate molecular variants with mutations in the region near DR1 (2551s4, 2551d4, 2555s4, 2555d4, s7, d7, and d7/3E; see Fig. 5B and 6A, below). The loop variants (TAA, AAA, GCA, GTA, GAC) were generated using primers each containing a randomized nucleotide incorporated at one of the loop positions, where the underline depicts the altered loop nucleotide. The AflII restriction site at position 2526 was incorporated into the upstream primers, and a primer downstream of the XbaI site at position 2662 was used to create a PCR product, which was digested with AflII and XbaI. This restriction fragment was substituted into a 0.5-mer plasmid (pD0.5G) containing the DHBV3 genome from nt 1666 to 3021. An EcoRI monomer of the genome (1.0-mer; 3,021 bp) was inserted into each 0.5-mer plasmid, generating 1.5-mer plasmids with the mutations introduced near the 5′ copy of DR1 (3′ end of the minus-strand DNA). The 1.5-mer d7/M3 and d7/3E/M3 molecular clones were generated by inserting an EcoRI monomer from pM3A (14), containing the 4-nt M3 substitution described below in Fig. 6A, into the respective 0.5-mer plasmids containing either the d7 or d7/3E mutations near the 5′ copy of DR1.
FIG. 5.
Substitutions and deletions in the hairpin have different phenotypes. (A) Schematic representation of base pairs predicted to form within the minus-strand DNA important for RC DNA synthesis. Hashed lines were used to indicate base pairing. Imperfect duplex formation between 3E-M3 and 5E-M5 is indicated by hashed lines. The hairpin described in the text is located adjacent to 3E. The donor and acceptor sites for primer translocation (DR1 and DR2) are indicated by boxes. The donor and acceptor sites for circularization are denoted (5′r and 3′r, respectively). The P protein (circle) is covalently linked to the 5′ end of the minus-strand DNA. (B) The wild-type sequence of the region near DR1 (box) in the minus strand was indicated at the top, including the sequence of the hairpin (base pairing partners indicated by arcs) and the 3E sequence (denoted by a solid line above the sequence). Substitutions were denoted by an “s” in their name, with the mutated sequences underscored. Deletions were denoted with a “d” in their name, with the mutated sequences indicated by dashes. (C) Southern blotting indicates different phenotypes for substitutions and deletions. Viral DNA was isolated from LMH cells 3 days posttransfection. The blot was hybridized with a genomic-length, minus-strand-specific RNA probe. Positions of the prominent forms of DNA replicative intermediates (RC, DL, and SS) are indicated. (D) In situ priming dramatically increased in variants containing substitutions, but not deletions, in the hairpin sequence. In situ priming was calculated as described in Materials and Methods. Samples were normalized to a wild-type reference (set to 1) and presented as the means with error bars to indicate the standard deviations. Each virus was analyzed multiple times (n = 3 to 14) from independent transfections.
FIG. 6.
Duplex formed by 3E and M3 in the presence of d7 can partially substitute for hairpin function. (A) Proposed duplex between 3E and M3 (14) with the predicted base pairing indicated by vertical hashes. The four nucleotide substitutions introduced into 3E or M3 to destabilize duplex formation in the presence of the d7 mutation are indicated by arrows (d7/3E and d7/M3, respectively). The 3E and M3 mutations were combined with the d7 mutation to generate a restoration variant (d7/3E/M3). (B) Southern blotting of viral DNA isolated from LMH cells 3 days posttransfection. The blot was hybridized with a genomic-length, minus-strand-specific RNA probe. Positions of the prominent forms of DNA replicative intermediates (RC, DL, and SS) are indicated. (C) Synthesis of RC DNA was dependent upon the duplex between 3E and M3 in the presence of the d7 mutation. RC DNA was calculated as described in Materials and Methods. Samples were normalized to a wild-type reference (set to 100) and presented as the means with error bars to indicate the standard deviations. Each virus was analyzed multiple times (n = 4 to 6) from independent transfections. (D) Modified schematic of the minus-strand DNA secondary structure (Fig. 5B) predicted to form in the presence of the d7 mutation. The imperfect duplex between 3E and M3 is now predicted to lie adjacent to DR1, where it substitutes for the hairpin as an inhibitor of in situ priming and can contribute to both template switches.
Mutations were introduced into the backgrounds of the 1.5-mer plasmids pD1.5G (wild type) and p503-3. p503-3 is a P-null version of pD1.5G (10), used to avoid expression of altered forms of the P protein, when necessary. Molecular clones constructed in the P-null background were supplied with wild-type P protein in trans by cotransfection with a P-donor plasmid, pG308-2 (4). This paper reports the results of the pD1.5G and/or p503-3 molecular clones as wild type in all analyses. All of the clones described herein express wild-type core (C) protein. All molecular clones were verified by DNA sequencing and restriction enzyme digestion analysis.
Cell culture and isolation of viral DNA.
The chicken hepatoma cell line LMH was used to replicate DHBV (2, 12). Cells were cultured as previously described (18). Calcium phosphate transfections were performed. Viral DNA was isolated from cytoplasmic capsids 3 days posttransfection as previously described (4).
Band compression analyses.
Approximately 50 ng of plasmid DNA was used as a template in dideoxy DNA sequencing reactions. Sequencing was performed with Vent exo− DNA polymerase (New England BioLabs) and a minus-sense, end-labeled oligonucleotide complementary to DHBV nucleotide coordinates 2599 to 2622, 74 nt downstream of DR1. Thermocycling parameters were 10 cycles at 95°C for 60 s, 46°C for 60 s, and 72°C for 60 s. Typically, electrophoresis was performed using 40-cm, 6% polyacrylamide gels containing 7 M urea and 1× Tris-borate-EDTA (TBE), run at 30 W. The gel used to resolve the band compression in image 1 of Fig. 3A, below, contained 7.6 M urea, 25% deionized formamide, and 1× TBE. Gels were dried, exposed to Molecular Dynamics phosphorimaging cassettes, and scanned using the Molecular Dynamics STORM.
FIG. 3.
Biochemical evidence for in vitro hairpin formation. The products of dideoxy sequencing reactions were electrophoresed through polyacrylamide gels containing either 7.6 M urea and 25% formamide (image 1 in panel A) or 7 M urea (the rest of the images). (A) The sequencing products for a molecular clone containing wild-type sequence are shown in panels 1 and 2, with the known sequence indicated between them as a reference. The 5′ and 3′ hairpin stem sequence is indicated by arrows next to the reference sequence. In the presence of two denaturants (image 1), the sequencing products migrated with regular spacing and the sequence was discernible. Using a single denaturant (image 2), the wild-type sequence showed anomalous migration (band compression indicated by *) upon addition of the stem-terminating nucleotides as a secondary structure formed. The band compression was resolved after introduction of a single nucleotide mutation at the DR13W position of the stem (image 3). Band compression returned upon restoring base pairing at the DR13 position (DR13R), albeit to the mutant sequence (image 4). The band compression (indicated by *) in panel 4 appeared less severe than that in panel 2. Images 2 through 4 were from the same gel. (B) Sequence of the hairpin stem influences structural stability. Band compression occurred for the wild type (indicated by *; image 1), but not for the S4C (image 2) or S4W (panel 3) variant, as expected. The restoration variant (S4R), which swapped the 5′ and 3′ sides of the stem, did not restore the band compression (image 4). All four images were from the same gel containing 7 M urea.
DNA primer extension analyses.
Primer extension was performed as described elsewhere (6), with modifications. Typically, 1 to 4 ng of viral DNA was processed for use in up to three separate primer extension reaction mixtures. Each viral DNA sample was mixed with approximately 1 ng of a DHBV-containing plasmid (pD0.5G) digested with NcoI (nt 2351) and EcoRV (nt 2650) to serve as an internal standard and allow for normalization of measurements between reactions. The DNA was subjected to alkaline hydrolysis using 0.2 N NaOH at 95°C for 5 min to remove the RNA primer from the 5′ end of the plus-strand DNA. Samples were neutralized with Tris-HCl and ethanol precipitated prior to analysis. The end-labeled oligonucleotide (primer 1) used to measure the level of plus-strand DNA that initiated from DR2 and had extended at least to the 5′ end of the minus strand (∼50 nt) was derived from nt 2537 to 2520 (minus-sense polarity; annealing temperature, 37°C). The end-labeled oligonucleotide (primer 2) used to measure the level of plus-strand DNA that initiated from DR2, successfully circularized and elongated ∼90 nt, was derived from nt 2629 to 2598 (minus-sense polarity; annealing temperature, 58°C). The end-labeled oligonucleotide (primer 3) used to measure the amount of 5′ termini of minus-strand DNA that had extended at least 112 nt was derived from nt 2425 to 2447 (plus-sense polarity; annealing temperature, 58°C). The primer extension reaction mixtures contained 1× ThermoPol buffer [10 mM KCl, 10 mM (NH4)2SO4, 20 mM Tris-HCl (pH 8.8), 2 mM MgSO4, 0.1% Triton X-100; New England BioLabs], 1 U of Vent exo− DNA polymerase (New England BioLabs), 0.2 mM (each) deoxynucleoside triphosphate, ∼0.7 pmol of end-labeled primer, internal standard plasmid DNA, and viral DNA. Ten reaction cycles were performed for each reaction, and the products were electrophoresed through 6% polyacrylamide-7.6 M urea-1× TBE buffered gels. The gels were dried and exposed to Molecular Dynamics phosphorimaging cassettes and scanned using the Molecular Dynamics STORM for visualization and quantification using Molecular Dynamics ImageQuant software.
Southern blotting analyses.
Southern blotting was performed as previously described (1) for DHBV replication intermediates using 1.25% agarose gels containing 1× TBE. Blots were probed with genomic-length, plus-strand RNA probes specific for DHBV. Membranes were exposed and scanned using Molecular Dynamics phosphorimaging cassettes and the Molecular Dynamics STORM. Visualization and quantitation were performed using Molecular Dynamics ImageQuant software. The relative levels of RC DNA and DL DNA were calculated as a percentage of all of the full-length minus-strand DNA, including those that had served as templates for plus-strand DNA synthesis.
Statistical tests.
The Wilcoxon signed rank test was used to test variants for difference in location compared with the wild-type reference virus. The largest P value was reported in all cases.
RESULTS
Description of primer extension measurements.
The template switch that circularizes the genome is a multistep process that is likely to include the following steps: the juxtaposition of the donor and acceptor sites, transfer of the nascent plus-strand DNA to the acceptor site, P protein binding, reinitiation of DNA synthesis, and elongation of the newly circularized plus-strand DNA. The primer extension reaction used herein measured the net sum of these steps. As shown in Fig. 1B, circularization efficiency was defined as the percentage of plus-strand DNA that had elongated (∼90 nt) following circularization relative to the total amount of plus-strand DNA that had initiated from DR2 and been elongated, at least, to the 5′ end of the minus-strand DNA (coordinate 2537) (15). By detecting plus-strand DNA at the latest point prior to circularization with primer 1, the analysis was not influenced by defects at prior steps in replication. The addition of a third primer (primer 3) to detect the level of minus-strand DNA 5′ termini allowed measurements of in situ priming and RNA primer utilization. In situ priming was calculated as the percentage of minus-strand DNA (measured by primer 3) that had initiated plus-strand DNA synthesis from DR1 and elongated (∼80 nt) (measured by primer 2) (Fig. 1B, right panel). In order to measure the overall efficiency of plus-strand DNA priming, RNA primer utilization was calculated as the percentage of the accumulated plus-strand DNA initiated from either DR2 (primer 1) or DR1 (primer 2; DR1 signal) relative to total minus-strand DNA (primer 3). A common internal standard was used to account for variation between the independent primer extension reactions.
The small DNA hairpin productively influences the efficiency of the circularization reaction.
A previous study showed that a DNA hairpin forms at the 3′ end of the minus-strand DNA overlapping DR1, which facilitates primer translocation, at least in part, through inhibiting in situ priming (4). In that study, a number of DHBV variants were made with mutations designed to either destabilize or completely inhibit hairpin formation. When viral DNA from those variants was analyzed by Southern blotting, in addition to higher levels of DL DNA the proportion of full-length, single-stranded (SS) DNA accumulated to higher levels. The increase in SS DNA indicated the presence of an additional defect in plus-strand DNA synthesis for these variants, but it did not discriminate between defects in RNA primer utilization and circularization. The primer extension assay described above allowed these two types of defects to be discriminated by detecting the short fragment (∼50 nt) of plus-strand DNA that has elongated from DR2 to the 5′ end of its minus-strand DNA template (Fig. 1A, panel c). The possibility that the hairpin contributed to circularization was particularly intriguing, as previous studies had suggested primer translocation and circularization may be mechanistically linked (5) and that variants containing mutations in the region of the hairpin have defects in circularization (17).
To investigate the role of the hairpin in circularization, many of the same DHBV variants previously used to establish the hairpin as an inhibitor of in situ priming were analyzed (Fig. 2A) (4). Five independent sets of variants were used initially to reduce and subsequently restore the number of Watson-Crick base pairs within the stem of the hairpin through either substitution of a single base-pairing partner (DR10, DR11, DR12, DR13 series) or by substituting the entire 4-nt stem sequence (S4 series). Molecular clones designed to interfere with hairpin formation and/or stability contained substitutions in either the 3′ half (referred to as W variants) or the 5′ half (C variants) of the stem. These mutations were also combined to restore Watson-Crick base-pairing potential, albeit to mutant sequences (R variants) (Fig. 2A). If the hairpin contributed to circularization, there were two predictions. First, the relative circularization efficiency of the W and C variants should have been lower than that of the wild-type reference virus, due to destabilizing effects of the substitutions. Second, the relative circularization efficiency of the double mutants (R variants) should have increased with respect to either of the W or C variants due to the restoration of base pairing.
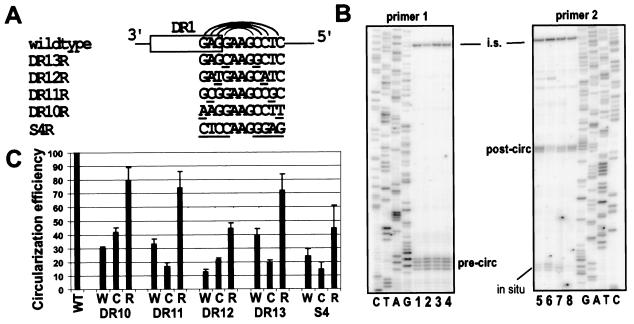
FIG. 2.
Primer extension analyses of variants indicated that the presence of the hairpin increases circularization efficiency. (A) Substitutions were introduced into the stem nucleotides of the hairpin to initially reduce (W and C variants) and subsequently restore (R variants) to mutant sequence the potential for Watson-Crick base pairing. The R variant for each set of mutations is shown, with the substituted nucleotides in both sides of the stem underscored. The W and C variants contained the same substitutions as the R variants introduced into either the 3′ or 5′ half of the stem, respectively. Sequence is indicated in the minus-strand orientation. (B) Primer extension analyses on DNA replication intermediates isolated from LMH cells using primers 1 and 2 with samples from the DR13 series. Samples included internal standard DNA. Primer 1 detected plus-strand DNA that initiated from DR2 and elongated, at least, to the 5′ end of the minus-strand DNA (pre-circ). Primer 2 detected the fraction of plus-strand DNA detected by primer 1 that successfully circularized and elongated (post-circ). Primer 1 also detected plus strands initiating from DR1 (in situ). Detection of the internal standard (i.s.) is shown near the top of both gel images. The sequencing ladders were generated using the same primers as in the respective primer extension reactions. Lanes 1 and 8, wild-type reference; lanes 2 and 7, DR13W; lanes 3 and 6, DR13C; lanes 4 and 5, DR13R. (C) The circularization efficiency for each variant was calculated as described in the legend for Fig. 1B. Samples were normalized to a wild-type reference (set to 100) and are presented as means with error bars to indicate the standard deviations. Each virus was analyzed multiple times (n = 2 to 7) from independent transfections.
Molecular clones encoding each of the variants were expressed in the LMH cell line via transient transfection. The intracellular replication intermediates were isolated for analysis of circularization efficiency by primer extension (Fig. 2B). The relative circularization efficiency was calculated as described for Fig. 1B. For each of the five sets of variants, the W and C variants had decreased circularization efficiencies compared with the wild-type reference virus (Fig. 2C). These results were consistent with the idea that the W and C variants led to destabilization of the hairpin and thus loss of circularization function. More significantly, the second prediction was met, as the circularization efficiency of each R variant was higher than that of either of the respective W or C variants. We interpret these findings as compelling evidence that the presence of the small DNA hairpin is important to the efficient circularization of DHBV. Furthermore, the contribution of the hairpin sequence to productive circularization appeared to be significant in magnitude, as the circularization efficiencies of variants were reduced to levels as low as 10% of that for the wild-type reference virus in some variants (Fig. 2C, DR12W and S4C).
When the ability of the restoration variants to inhibit in situ priming was previously measured, it was found that none of the restoration variants were able to suppress in situ priming as well as the wild-type reference virus (4). Similarly, none of the restoration variants were able to support the circularization process as well as the wild-type reference (Fig. 2C, R variants compared to WT). Thus, just having Watson-Crick base-pairing potential in a hairpin stem at this location is not sufficient for complete functional activity. Moreover, there is a correlation between the hairpin's ability to regulate in situ priming and promote circularization, suggesting a mechanistic link between these processes. This linkage could be explained if the sequence of the hairpin contributed to its own stability or the stability of a nearby DNA structure that was important for both processes. Although some of the variants also exhibited slight changes in their total RNA primer utilization, in general, the level of plus-strand priming was similar to that in the wild-type reference virus for these variants (data not shown).
The phenotypes described were not due to changes in the amino acid sequence of any of the virally encoded proteins. Cotransfection analysis indicated that viruses containing mutations in the hairpin sequence retained their phenotypes, yet did not affect the phenotype of the cotransfected virus, expressing wild-type viral proteins (Southern blotting [data not shown]). Therefore, the effects described were cis-acting in nature. Although the W and R variants contained changes in DR2 to retain sequence identity between DR1 and DR2, variants containing only the DR2 mutations had little, if any, influence on the circularization reaction when measured using primer extension (data not shown).
Biochemical evidence indicates the hairpin can form a stable secondary structure.
Anomalous mobility of DNA molecules occurs during electrophoresis in both native and denaturing polyacrylamide gels upon formation of secondary structures (9). Hirao and colleagues showed that it is possible to establish a correlation between anomalous mobility during electrophoresis and the melting temperature (Tm) of a DNA secondary structure (9). A similar biochemical analysis was performed on our hairpin variants by using electrophoresis of dideoxy sequencing products. Secondary structures present at the 3′ end of SS DNA molecules can cause them to migrate electrophoretically faster than DNA molecules of the same size. This faster migration is detected in gel electrophoresis as a compression of bands in the sequencing ladder (band compression) at the point of secondary structure formation, thus making the sequence indiscernible. A consequence is an unusually large and irregular band spacing directly above the band compression due to the void left by the faster-migrating species that contain the secondary structure. Changing the electrophoresis conditions, in particular the denaturing conditions of the gel, can influence the ability of the secondary structure to form. Using dideoxy sequencing reactions made it possible to visualize the impact each additional nucleotide of the structure has upon its electrophoretic mobility.
Using plasmid DNA as a template, dideoxy DNA sequencing reactions were performed with a minus-sense, end-labeled primer annealing 74 nt downstream of DR1. Electrophoresis of the DNA sequencing products was performed using either polyacrylamide gels containing a single denaturant (urea) or a combination of two denaturants (urea and formamide). As shown in Fig. 3A (image 1), when electrophoresis of the products of the wild-type sequencing reaction was performed in the presence of two strong denaturants, uniform band spacing was seen and the sequence was clearly discernible. The known nucleotide sequence is provided as a reference. However, electrophoresis of the products of a sequencing reaction generated using the same wild-type template on a sequencing gel containing a single denaturant resulted in a band compression at the site of the predicted hairpin (Fig. 3A, image 2). The band pattern appeared uniform and clearly discernible, as the sequence is read 5′ to 3′ until the position of the hairpin. Following addition of two stem-forming nucleotides, anomalous, faster-migrating species were present, making the sequence indiscernible through this region (Fig. 3A, compare the band pattern to the known sequence provided at the left). In particular, anomalous migration was apparent after addition of only the second Watson-Crick base-pairing partner in the 3′ half of the stem (DR12 position) but became more pronounced as additional stem nucleotides were added. Evidence for the interpretation that this was a result of secondary structure formation was based upon the second denaturant, formamide, inhibiting band compression (Fig. 3A, image 1). Thus, the results from this analysis provide biochemical evidence that the wild-type hairpin sequence (5′-CTCCGAAGGAG-3′) can form a stable secondary structure.
Next, the effects single-nucleotide W and C substitutions had upon the band compression were investigated. Electrophoresis of the dideoxy sequencing products for the DR13W variant is shown in image 3 of Fig. 3A. Electrophoresis of the products of the sequencing reaction for the DR13W molecular clone was performed on the same gel as wild type in image 2 using a single denaturant, urea. In this case, there was no indication of band compression, as band spacing appeared uniform and the pattern indicated the expected sequence, which is shown to the left. A similar result was obtained upon analyzing the DR13C molecular clone, as well as the molecular clones of DR12W and DR12C (data not shown). In fact, these mutations were sufficient to inhibit the band compression under native electrophoresis conditions (data not shown). The results can be interpreted to indicate that base pairing between the stem nucleotides at the DR12 and DR13 positions of the stem make a significant contribution to the stability of the hairpin. Band compression lost following introduction of the W or C substitutions returned when the base-pairing potential at each respective position was restored (R variants). As shown in image 4 of Fig. 3A, the band compression was less severe for the DR13R molecular clone when compared with the wild-type sequencing product, which was run on the same gel (shown in image 2). This was also true of the DR12R analysis (data not shown). This may indicate that the hairpins of the restoration variants were less stable than the wild-type hairpin in the presence of additional 3′ sequence. Thus, the biochemical evidence provided corroborating evidence that the wild-type hairpin sequence was capable of forming a stable secondary structure, and that single nucleotide mutations could significantly alter its stability. Furthermore, the severity of the band compression for the DR12R and DR13R molecular variants was reduced compared with that of the wild-type sequence, suggesting an explanation for the inability to completely restore functional activity, be it suppression of in situ priming and/or promotion of circularization, during virus replication in cell cultures.
The large magnitude of the defect in circularization of the S4C and S4W variants in cell culture was not surprising considering that these mutations were expected to completely eliminate hairpin formation. What was surprising was the low magnitude of restoration of the S4R variant in terms of promoting circularization efficiency (Fig. 2C) and in regulating in situ priming (4). In the S4R variant, the overall G+C content was not changed, nor was the relative position of the G+C base pairs, as each side of the stem was simply replaced with its genetic complement. When the S4 series was analyzed using the band compression assay, there was no indication of anomalous migration of the sequencing products from either the S4W or S4C molecular clones as expected (Fig. 3B, images 2 and 3, respectively). Interestingly, band compression was also absent for the S4R sequencing product (Fig. 3B, image 4). This finding highlights the importance of stem sequence on small DNA hairpin stability.
The loop-terminating nucleotides contribute to function.
The genetic and biochemical analyses of the base-pairing partners in the hairpin stem were consistent with the hairpin forming and contributing to both plus-strand template switches (this work and reference 4). To gain insight into the mechanism by which the secondary structure contributes to circularization, the contribution of the loop to function, if any, was evaluated. To this end, molecular variants were analyzed that contained single nucleotide substitutions at each of the positions within the loop without altering the sequence of the stem. A set of base substitutions, introduced randomly, were chosen for analysis. In addition to the wild-type loop sequence (5′-GAA-3′), variants containing the loop sequences TAA, AAA, GCA, GTA, and GAC were analyzed. Primer extension was performed to measure circularization efficiency, and both primer extension and Southern blotting were used to measure the proportion of in situ priming for each of these variants.
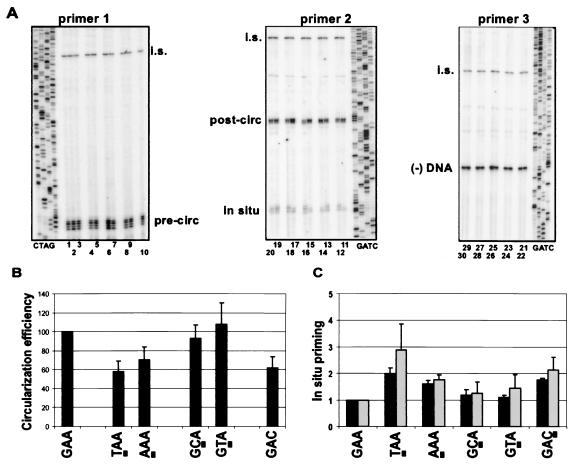
As described earlier, a quantitative primer extension assay was used to measure the levels of circularization and in situ priming (Fig. 1B and 4A). Relative to the wild-type reference, circularization efficiency and the level of in situ priming were unaffected by changes in the central nucleotide of the loop, as indicated by the variants 5′-GTA-3′ and 5′-GCA-3′ (Fig. 4B and C, respectively). However, the circularization efficiency was reduced to a similar level (∼60% of wild type) in each of the variants, with a change in one of the loop-terminating nucleotides (TAA, AAA, GAC; P < 0.03) (Fig. 4B). Similarly, when these variants were assayed for their ability to negatively regulate in situ priming, the variants that altered the loop-terminating nucleotides had 1.5- to 2-fold higher levels of in situ priming when measured by both primer extension and Southern blotting (Fig. 4C) (P < 0.03). Clearly, the sequence of the loop contributed to function; however, the contribution was not equal for all loop positions and, rather, it appeared limited to the loop-terminating nucleotides.
FIG. 4.
Loop-terminating nucleotides contribute to function. (A) Primer extension was performed to measure in situ priming and circularization efficiency of the loop variants. Primer extension with primers 1 and 2 were as described in the legend for Fig. 2B. Primer 3 was used to detect the 5′ termini of minus-strand DNA [(−) DNA]. The internal standard (i.s.) for each reaction is shown near the top of each gel. Sequencing ladders were generated using the same primers as the respective primer extension reactions. Each virus is indicated by its loop sequence (5′ to 3′ in the minus-strand orientation). Lanes 1, 11, and 21, wild-type reference (GAA); lanes 2, 12, and 22 and lanes 3, 13, and 23, two independent molecular isolates of GAC; lanes 4, 14, and 24 and lanes 5, 15, and 25, two independent molecular isolates of GCA; lanes 6, 16, and 26, not germane; lanes 7, 17, and 27, GTA; lanes 8, 18, and 28 and lanes 9, 19, and 29, two independent molecular isolates of TAA; lanes 10, 20, and 30, AAA. (B) The circularization process is sensitive to mutations in the loop-terminating nucleotides. Circularization efficiency was described in the legend for Fig. 1B. Samples were normalized to a wild-type reference (set to 100) and presented as the means with error bars to indicate the standard deviations. Each virus was analyzed multiple times (n = 4 to 6) from independent transfections. (C) The level of in situ priming is also sensitive to mutations in the loop-terminating nucleotides. In situ priming was calculated using both primer extension (black bars) and Southern blotting (gray bars; gels not shown); calculations were described in the legend for Fig. 1B and Materials and Methods, respectively. Samples were normalized to a wild-type reference (set to 1) and presented as the means with error bars to indicate the standard deviations. Each virus was analyzed multiple times (n = 4 to 8) from independent transfections.
Band compression analysis was performed on each of these variants. It was evident that the loop-terminating nucleotides contributed to the stability of the hairpin, as they inhibited the band compression at the site of the hairpin, whereas the compression remained for the variants that contained changes in the central nucleotide of the loop (data not shown). Taken together, these results provide evidence that the trinucleotide loop sequence of the DHBV hairpin had a sequence requirement of 5′-GNA-3′ for both functional activity during replication and structural stability in vitro. Our results were consistent with the studies of others that showed a structural advantage for small DNA hairpins with trinucleotide loops that contained this loop sequence (7-9). The contribution of the loop-terminating nucleotides was subsequently shown to be through a sheared base pair between the 5′ G and 3′ A in the loop (26). Our findings indicate that the contribution of the loop sequence is likely providing stability to the hairpin, although other contributions cannot be ruled out.
Deletions and substitutions in the hairpin sequence result in different phenotypes.
Substitutions designed to decrease base pairing in the stem resulted in an increase in the proportion of in situ priming, consistent with the hairpin acting as an inhibitor of in situ priming (4). However, a previously described variant that removes the 11 nt adjacent to DR1 (d11) (Fig. 5B), which would remove the hairpin, did not have a significant increase in the level of in situ priming (6). As the phenotype of this variant was not consistent with the hairpin being the sole contributor to the inhibition of in situ priming, we were interested in understanding how the hairpin could be removed without increasing in situ priming. One possibility was that the phenotype was linked to the mutation being a deletion rather than a substitution.
A previous study showed that base-pairing interactions between distally located regions of the minus-strand DNA (Fig. 5A; 3E and M3, 5E and M5) are important for both template switches during plus-strand DNA synthesis of DHBV (14). The duplex formed by 3E and M3, adjacent to the hairpin described herein (Fig. 5A), would also be affected by the deletion in the d11 variant (Fig. 5B). In order to understand better the phenotype of the d11 variant, the region encompassed by this deletion was mutated further, comparing substitutions with deletions. Two sets of 4-nt mutations, one overlapping 3E and the other within the hairpin, were initially generated (Fig. 5B). The first set of mutations either substituted (2555s4) or deleted (2555d4) the 4 nt adjacent to the hairpin in 3E. The phenotypes of the 2555s4 and 2555d4 mutants were very similar when determined by both Southern blotting (Fig. 5C) and primer extension (data not shown). These variants had phenotypes similar to those of previously described 3E variants, which had defects in both RNA primer utilization and circularization (14). A second set of mutations was designed to either substitute (2551s4, referred to as S4C previously) or delete (2551d4) the 4 nt comprising the 5′ half of the hairpin's stem (Fig. 5B). Both of these mutations were expected to inhibit hairpin formation. Unlike the 2555s4 and 2555d4 variants within 3E, the comparable deletion and substitution within the hairpin resulted in distinct phenotypes (Fig. 5C). Whereas a 4-nt substitution in the hairpin stem (2551s4) increased in situ priming by sixfold compared to the wild-type reference, deletion of the same nucleotides (2551d4) only increased in situ priming by twofold (Fig. 5D). The 2551d4 variant was able to synthesize some RC DNA, although not to levels comparable with those of the wild-type reference (Fig. 5C). Thus, deletions and substitutions within the hairpin sequence did not have the same consequence upon replication.
To gain insight into the phenotype of the 2551d4 variant, two more variants were analyzed with mutations within the hairpin sequence. This set either substituted or deleted the first 7 nt following DR1 (s7 or d7, respectively) (Fig. 5B). Again, both mutations were expected to inhibit hairpin formation and not alter the potential for 3E to base pair with M3. The s7 variant synthesized no detectable RC DNA and high levels of DL DNA (Fig. 5C and D). The phenotype of the s7 variant was similar to that of other substitution variants that disrupt base pairing within the hairpin, including the 2551s4 variant (Fig. 5C and D). Surprisingly, the d7 variant had a phenotype distinct from the previous hairpin substitutions and deletions. This variant produced high levels of RC DNA, although less than the wild-type reference (Fig. 5C and 6D). The level of in situ priming by this variant was as low, or lower, than that in the wild-type reference (Fig. 5D and Table 1). Thus, this variant was able to perform both plus-strand template switches quite well despite deletion of 7 nt within the hairpin. Overall, these analyses indicated that substitutions and deletions lead to significantly different phenotypes. Substitutions behaved as if the hairpin were a negative regulator of in situ priming. Deletions within the hairpin behaved as if one or more other determinants could contribute to the regulation of in situ priming and circularization, at least in the absence of the hairpin.
TABLE 1.
Efficiency of template switches as determined by primer extensiona
| Virus | Level relative to wild type
|
||
|---|---|---|---|
| In situ priming | Circularization | RNA primer utilization | |
| Wild type | 1 | 100 | 100 |
| d7 | 0.23 ± 0.13 | 80.1 ± 5.4 | 87.7 ± 16.9 |
| d7/3E | 3.70 ± 0.61 | 0 ± 0 | 39.5 ± 5.0 |
| d7/M3 | 3.06 ± 0.70 | 0 ± 0 | 32.7 ± 6.9 |
| d7/3E/M3 | 0.33 ± 0.09 | 76.7 ± 9.3 | 71.6 ± 6.3 |
| s7 | 10.7 ± 2.6 | 5.1 ± 3.0 | 128 ± 38 |
Calculations were performed as described in the text. The values for wild type were set to either 1 (in situ priming) or 100 (circularization and RNA primer utilization). Values reported are the means ± standard deviations.
The function of the hairpin can be partially replaced by a displaced 3E/M3 duplex.
It was possible that the different phenotypes of the substitution and deletion variants were due to their effect upon the conformation of the 3′ end of the minus-strand DNA, by changing the location of the 3E/M3 duplex with respect to DR1. That is, the substitution variants had disrupted formation of the hairpin and in doing so had removed an inhibitor of in situ priming; whereas, in the variants containing deletions of the hairpin sequence, the relative location of the 3E/M3 duplex with respect to DR1 changed and may have contributed to the replication phenotype (compare Fig. 5A and 6B). The d7 variant was interesting, as it replicated well in the absence of the hairpin. It was possible the 3E/M3 duplex in the presence of the d7 mutation acted to suppress in situ priming and promote production of RC DNA.
To determine if the base pairing between 3E and M3 occurred in the d7 variant, four nucleotide substitutions were introduced into either the 3E or M3 sequence (Fig. 6A) combined with the d7 mutation to generate the d7/3E and d7/M3 variants, respectively. Very little RC DNA was detected by Southern blotting for either of these variants (Fig. 6C and D). However, when the 3E and M3 substitutions were combined with the d7 mutation (d7/3E/M3), restoring base-pairing potential between 3E and M3, albeit with mutant sequence, the ability to synthesize RC DNA was restored to a level similar to that of the d7 variant (Fig. 6C and D). These findings indicated that base pairing occurred between 3E and M3 in the presence of the d7 mutation and that it was necessary for the ability of this virus to synthesize RC DNA and suppress in situ priming.
In both the d7 and d7/3E/M3 variants, the proportion of RC DNA production was slightly lower than that of the wild-type reference virus, although the proportion of DL DNA did not increase (Fig. 6D and Table 1). Next, the primer extension assay, described earlier, was used to measure the level of RNA primer utilization (total priming from DR1 and DR2) and the circularization efficiency for each of these variants. When compared to the wild-type reference, the d7 and d7/3E/M3 variants were slightly deficient for both RNA primer utilization and circularization (Table 1). Thus, the reduction in RC production in these two variants seems to be an accumulation of two defects, albeit mild in degree. Unlike the d7 and d7/3E/M3 variants, the proportion of in situ priming increased in the d7/3E and d7/M3 single mutants (Table 1). We interpreted these results to indicate that in the presence of the d7 mutation, the duplex between 3E and M3 may, at least partially, act as an inhibitor of in situ priming. The proportion of in situ priming detected for the d7/3E and d7/M3 variants was lower than that for the s7 variant. The reason for this is not completely clear, but it should be noted that RNA primer utilization was significantly lower in the d7/3E and d7/M3 variants than in the wild-type reference (Table 1).
Thus, juxtaposition of DR1 and the duplex formed by 3E and M3 (Fig. 6B) can functionally substitute, to a large extent, for the hairpin in promoting both plus-strand template switches. In addition, we interpret the intermediate phenotype of the 2551d4 variant to indicate that the plus-strand template switches are sensitive to the precise conformation of the 3′ end of the minus-strand DNA determined, in part, by the relative juxtaposition of DR1 and the 3E/M3 duplex.
DISCUSSION
We have provided evidence based upon viral replication in cell culture that a small DNA hairpin overlapping DR1 in the minus-strand DNA acts not only in regulating in situ priming and primer translocation (4), but also in promoting efficient circularization. Corroborating evidence for hairpin formation was provided using an in vitro assay (Fig. 3). Use of this assay not only supported the notion that the predicted hairpin could form a stable secondary structure, it also provided insight into the sequence contributions of the stem and loop to function. Previous studies of small DNA hairpins with unusually high thermostability showed both the stem and loop sequences contributed to stability (7-9). In particular, trinucleotide loops containing the sequence 5′-GNA-3′ were shown by nuclear magnetic resonance to be particularly stable with a sheared base pair forming between the G and A in the loop (26). Our results indicate the DHBV stem-loop likely falls into this class of structures. Recently, Bevilacqua and colleagues (20) identified the 5′-cGNABg-3′ class of DNA hairpin loops through a process that selects for stable DNA secondary structures. Interestingly, they proposed that this tetraloop sequence is a variation of the 5′-GNA-3′ loop, in which the fourth loop nucleotide is bulged out while sheared base pairing still occurs between the G and A bases. This is interesting, as the hairpin loop in the avian hepadnaviruses exists in two forms: as a trinucleotide 5′-GAA-3′ loop described herein or a 5′-GAAT-3′ tetraloop, such as in the heron hepatitis B virus.
It was previously noted that mutations in the cis-acting elements 3E, M (M3 and M5), and 5E affect both primer translocation-utilization and circularization, suggesting the template switches may be mechanistically linked (5). The mechanistic linkage, in that case, is likely through effects upon the conformation of the minus-strand DNA, providing a relative juxtaposition of the donor and acceptor sites for both plus-strand template switches (14). Here, we have extended our previous findings to show that the hairpin at DR1 also contributes to both template switches (4). The argument for mechanistic linkage can be extended even further upon consideration of the d7 and d7/3E/M3 variants. Although these variants appear to substitute functionally for the hairpin, they do not replicate to the level of the wild-type reference. They have mild defects in both primer translocation-utilization and circularization, and those defects are distributed between the two processes (Table 1). Perhaps it is not surprising that the two template switches continue to show examples of being mechanistically linked, as the 3′ end of the minus-strand DNA is involved in both template switches, first as the donor site for primer translocation and then as the acceptor site for circularization.
The specific contribution made by the hairpin to circularization remains to be defined, although we favor the second of two general models. The first model is based upon the premise that efficient circularization of the genome depends upon a precise juxtaposition of the donor and acceptor sites (Fig. 1A, panel d, 5′r and 3′r). Although the base pairing among 3E, M3, M5, and 5E depicted in Fig. 5A predicts a means to localize the donor and acceptor sites, it does not necessarily provide a precise juxtaposition. However, if the hairpin were to act as a recognition element for the P protein, or another component associated with the donor site, it would facilitate precise juxtaposition of the two ends. Although very intriguing, this model is difficult to reconcile with the phenotypes of the d7 and d7/3E/M3 variants, which lack the hairpin but circularize quite well in its absence (80% of wild-type [Table 1]). Alternatively, the precise juxtaposition of the donor and acceptor sites may be inherent in a higher-order conformation not apparent from the base pairing depicted in Fig. 5A or, perhaps, another element acts as the recognition site, such as the duplex between 3E and M3. Our results are more consistent with a model in which the hairpin influences the conformation of the acceptor site. The hairpin may act by imparting a specific conformation on the acceptor site. For instance, the presence of double-stranded DNA in the hairpin, or in the 3E-M3 duplex in the presence of the d7 mutation, may result in an extension of a helical nature to the nearby SS DNA, contributing to its ability to associate with either the dissociated nascent plus-strand DNA or the intact duplex between the nascent plus strand and the 5′ end of the minus-strand DNA. There is precedence for a small DNA hairpin near the 3′ end of the acceptor strand to enhance hybridization between two strands of nucleic acid (19). Whether circularization occurs through a triple-stranded intermediate including 5′r, 3′r, and the nascent plus-strand DNA or involves displacement and transfer of the nascent plus strand is not known. In either case, it would not be surprising that additional structural components in the template, such as those described herein, may exist to facilitate the circularization process.
RNA primer utilization defects have been reported for variants that interfere with the 3E-M3 or M5-5E interactions (6, 14). A simple rationale for that finding was that in the absence of these interactions the hairpin still formed and inhibited in situ priming. Interestingly, similar mutations in 3E and M3, when introduced into the d7 background, also led to RNA primer utilization defects in a background lacking the hairpin (Table 1). This suggests that an alternative explanation may be necessary to understand these priming defects. One possibility is that base-pairing interactions between 3E-M3 and M5-5E, in addition to localizing the donor and acceptor sites, also contribute either directly or indirectly to the general plus-strand priming reaction at DR1 and/or DR2. Alternatively, plus-strand priming may actually be occurring in these variants but is failing to elongate to a position capable of being detected in the primer extension assay. For instance, a secondary structure may form in the absence of the 3E-M3 interaction, such as a minus-strand equivalent to epsilon, causing elongation from DR1 to be aborted and RNA primer utilization to be underrepresented in our analysis.
The results obtained from the d7 variants are particularly interesting in light of the predicted absence of a similarly placed hairpin in the mammalian hepadnaviruses. We speculate that the replication strategy used by the mammalian hepadnaviruses is similar to the d7 variants in DHBV. The findings reported herein further emphasize the contributions made by the DHBV nucleic acid template to its own replication and suggest this may be a common feature of all hepadnaviruses, if not all elements that undergo reverse transcription.
Acknowledgments
We are grateful to the members of the Loeb lab for helpful discussions. We thank Jesse Summers, Bill Sugden, Peter Angeletti, and Paul Ahlquist for critically reviewing the manuscript.
This work was supported by National Institutes of Health grants PO1 CA22443, P30 CA07175, and T32 CA09135.
REFERENCES
- 1.Calvert, J., and J. Summers. 1994. Two regions of an avian hepadnavirus RNA pregenome are required in cis for encapsidation. J. Virol. 68:2084-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Condreay, L. D., C. E. Aldrich, L. Coates, W. S. Mason, and T. T. Wu. 1990. Efficient duck hepatitis B virus production by an avian liver tumor cell line. J. Virol. 64:3249-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ganem, D. 1996. Hepadnaviridae and their replication, p. 2703-2737. In B. N. Fields, D. M. Knipe, P. M. Howley, et al. (ed.), Fields virology, 3rd ed. Lippincott-Raven, Philadelphia, Pa.
- 4.Habig, J. W., and D. D. Loeb. 2002. Small DNA hairpin negatively regulates in situ priming during duck hepatitis B virus reverse transcription. J. Virol. 76:980-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Havert, M. B., L. Ji, and D. D. Loeb. 2002. Analysis of duck hepatitis B virus reverse transcription indicates a common mechanism for the two template switches during plus-strand DNA synthesis. J. Virol. 76:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havert, M. B., and D. D. Loeb. 1997. cis-acting sequences in addition to donor and acceptor sites are required for template switching during synthesis of plus-strand DNA for duck hepatitis B virus. J. Virol. 71:5336-5344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirao, I., G. Kawai, S. Yoshizawa, Y. Nishimura, Y. Ishido, K. Watanabe, and K. Miura. 1994. Most compact hairpin-turn structure exerted by a short DNA fragment, d(GCGAAGC) in solution: an extraordinarily stable structure resistant to nucleases and heat. Nucleic Acids Res. 22:576-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirao, I., G. Kawai, S. Yoshizawa, Y. Nishimura, Y. Ishido, K. Watanabe, and K. Miura. 1993. Structural features and properties of an extraordinarily stable hairpin-turn structure of d(GCGAAGC). Nucleic Acids Symp. Ser. 29:205-206. [PubMed] [Google Scholar]
- 9.Hirao, I., Y. Nishimura, Y. Tagawa, K. Watanabe, and K. Miura. 1992. Extraordinarily stable mini-hairpins: electrophoretical and thermal properties of the various sequence variants of d(GCGAAAGC) and their effect on DNA sequencing. Nucleic Acids Res. 20:3891-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch, R. C., J. E. Lavine, L. J. Chang, H. E. Varmus, and D. Ganem. 1990. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature 344:552-555. [DOI] [PubMed] [Google Scholar]
- 11.Kane, M. A. 1998. Status of hepatitis B immunization programmes in 1998. Vaccine 16(Suppl.):S104-S108. [DOI] [PubMed] [Google Scholar]
- 12.Kawaguchi, T., K. Nomura, Y. Hirayama, and T. Kitagawa. 1987. Establishment and characterization of a chicken hepatocellular carcinoma cell line, LMH. Cancer Res. 47:4460-4464. [PubMed] [Google Scholar]
- 13.Lien, J. M., C. E. Aldrich, and W. S. Mason. 1986. Evidence that a capped oligoribonucleotide is the primer for duck hepatitis B virus plus-strand DNA synthesis. J. Virol. 57:229-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu, N., R. Tian, and D. D. Loeb. 2003. Base pairing among three cis-acting sequences contributes to template switching during hepadnavirus reverse transcription. Proc. Natl. Acad. Sci. USA 100:1984-1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loeb, D. D., K. J. Gulya, and R. Tian. 1997. Sequence identity of the terminal redundancies on the minus-strand DNA template is necessary but not sufficient for the template switch during hepadnavirus plus-strand DNA synthesis. J. Virol. 71:152-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loeb, D. D., R. C. Hirsch, and D. Ganem. 1991. Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: implications for other reverse transcribing elements. EMBO J. 10:3533-3540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Loeb, D. D., and R. Tian. 2001. Mutations that increase in situ priming also decrease circularization for duck hepatitis B virus. J. Virol. 75:6492-6497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeb, D. D., and R. Tian. 1995. Transfer of the minus strand of DNA during hepadnavirus replication is not invariable but prefers a specific location. J. Virol. 69:6886-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mir, K. U., and E. M. Southern. 1999. Determining the influence of structure on hybridization using oligonucleotide arrays. Nat. Biotechnol. 17:788-792. [DOI] [PubMed] [Google Scholar]
- 20.Nakano, M., E. M. Moody, J. Liang, and P. C. Bevilacqua. 2002. Selection for thermodynamically stable DNA tetraloops using temperature gradient gel electrophoresis reveals four motifs: d(cGNNAg), d(cGNABg), d(cCNNGg), and d(gCNNGc). Biochemistry 41:14281-14292. [DOI] [PubMed] [Google Scholar]
- 21.Sprengel, R., C. Kuhn, H. Will, and H. Schaller. 1985. Comparative sequence analysis of duck and human hepatitis B virus genomes. J. Med. Virol. 15:323-333. [DOI] [PubMed] [Google Scholar]
- 22.Staprans, S., D. D. Loeb, and D. Ganem. 1991. Mutations affecting hepadnavirus plus-strand DNA synthesis dissociate primer cleavage from translocation and reveal the origin of linear viral DNA. J. Virol. 65:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Summers, J., and W. S. Mason. 1982. Replication of the genome of a hepatitis B-like virus by reverse transcription of an RNA intermediate. Cell 29:403-415. [DOI] [PubMed] [Google Scholar]
- 24.Yang, W., and J. Summers. 1995. Illegitimate replication of linear hepadnavirus DNA through nonhomologous recombination. J. Virol. 69:4029-4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang, W., and J. Summers. 1998. Infection of ducklings with virus particles containing linear double-stranded duck hepatitis B virus DNA: illegitimate replication and reversion. J. Virol. 72:8710-8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshizawa, S., G. Kawai, K. Watanabe, K. Miura, and I. Hirao. 1997. GNA trinucleotide loop sequences producing extraordinarily stable DNA minihairpins. Biochemistry 36:4761-4767. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, Y. Y., and J. Summers. 2000. Low dynamic state of viral competition in a chronic avian hepadnavirus infection. J. Virol. 74:5257-5265. [DOI] [PMC free article] [PubMed] [Google Scholar]