Abstract
Marek's disease virus (MDV) is an acute transforming alphaherpesvirus that causes T-cell lymphomas in chickens. We previously reported the identification of a putative oncogene, meq, that is encoded only by the oncogenic serotype of MDV. The gene product, Meq, is a latent protein that is consistently expressed in MDV-transformed lymphoblastoid cells and tumor cells. Meq has a bZIP (basic leucine zipper) structure resembling the family of Jun/Fos. The mechanism whereby Meq transforms T cells remains poorly understood. In this study, we explored the properties of Meq as a transcriptional factor. We analyzed Meq's dimerization partners and its target genes in MSB-1, an MDV-transformed T-cell line. By using in vitro assays, we first demonstrated Meq's potential to dimerize with a variety of bZIP proteins. We then identified c-Jun as the primary dimerization partner of Meq. Both are found to be colocalized in the nucleus and corecruited to promoters with AP-1 sequences. By using chromatin immunoprecipitation (ChIP), we scanned the entire MDV genome for Meq binding sites and found three regions that were enriched with Meq binding: the MDV lytic replication origin, the promoter for Meq, and the promoter for ICP4. Transactivation assays using the above promoters showed that Meq/Meq homodimers exhibited repression activity, whereas Meq/Jun heterodimers showed activation. Finally, we were able to show by ChIP that Meq is recruited to the interleukin-2 promoter in a region encompassing an AP-1 site. Thus, in addition to providing general knowledge about the transcriptional properties of Meq, our studies revealed for the first time the ability of Meq to interact with the latent MDV and host genomes. Our data suggest, therefore, a role for Meq in viral genome regulation during latency, in addition to its putative causal role in T-cell transformation.
Marek's disease (MD) is a communicable viral lymphoproliferative disease of chickens (13). The signs associated with MD are paralysis, depression, tremors, blindness, widespread T-cell lymphomas, and death. The costs of controlling MD and the associated loss make MD one of the most costly infectious diseases affecting the poultry industry. MD is caused by MD virus (MDV) (13). MDVs are composed of three antigenically related viruses, serotypes 1, 2, and 3, of which only serotype 1 MDVs are oncogenic. MDV is the only acute transforming alphaherpesvirus that causes tumors in its host. MDV was classified as an alphaherpesvirus on the basis of DNA sequence homology and genome organization (29, 55; P.Brunovskis, Z. Qian, D. Li, L. F. Lee, and H.-J. Kung, 5th Int. Symp. Marek's Dis., abstr. 44, p. 265, 1996), and its biological properties are more akin to those of gammaherpesviruses such as Epstein-Barr virus (EBV), herpesvirus saimiri, and Kaposi sarcoma-associated herpesvirus (KSHV). Like these viruses, MDV establishes latency in lymphoid cells, primarily activated or semiactivated CD4+ T cells, and causes transformation of these latently infected cells (41, 45, 51, 52).
MDV has a genome structure similar to that of herpes simplex virus, with UL and US (unique regions), each flanked, respectively, by TRL and IRL, and TRS and IRS (repeat regions). The genes residing in UL and US, mostly encoding components for genome replication and virus assembly, are highly conserved. By contrast, the genes in RL and RS are significantly divergent between MDV and herpes simplex virus, as well as among different serotypes of MDV (4, 24, 29). It was postulated that the genes in the repeat regions are likely to account for the unique lymphotropic and oncogenic properties of MDV. We first reported the identification of a gene, meq (MDV Eco Q fragment-encoded protein), as a putative oncogene (26). This gene resides in the RL region and is found only in serotype 1 MDV. It is expressed in all of the latent and tumor cells infected by MDV analyzed thus far. Meq has a structure resembling that of the Jun/Fos family of oncogenes. It is a bZIP protein with a leucine zipper domain located at the N-terminal half and proline-rich motifs in the C-terminal domain. When overexpressed in fibroblast cell lines, Meq is able to induce morphological changes, colony formation, a shortened G1 phase, and survival against a variety of extracellular stimuli (28, 33-35). Removal of Meq from transformed cells reverses the transformed phenotypes, and Meq-null mutants lack the ability to induce MD in vivo while maintaining the replicative properties (S.M.R., unpublished work). Despite the above observations implicating Meq in oncogenesis due to MDV, the mechanisms by which Meq transforms T cells remain incompletely understood. The possible role of Meq in MDV latency is even less clear.
Meq is a phosphoprotein localized to the nucleus and nucleolus. Among the kinases that can phosphorylate Meq are CDK2, PKC, and MAPK (32). The entire C-terminal domain of Meq, when linked to a heterologous DNA binding domain (e.g., Gal4), was found to be highly transactivating. Interestingly, the proline-rich repeat sequences in an isolated form have strong transcriptional repression activity, suggesting that Meq can be either a transactivator or a repressor, possibly depending on its phosphorylation status, subcellular locations, and dimerization partners.
In this study, we explored Meq's dimerization partners and its native DNA binding specificity in the MDV-transformed T-cell line MSB-1 (5). We demonstrated that Meq is able to dimerize with a variety of bZIP proteins, while different dimers bind different DNA motifs. In MSB-1 cells, Meq and c-Jun are found to be associated, colocalized in the nucleus, and corecruited to promoters with TRE/CRE sequence. By using a chromatin immunoprecipitation (ChIP)-based approach, we scanned the entire MDV genome for Meq binding sites. We found that Meq binding sites are nonrandomly distributed. Meq preferentially binds to the Meq promoter, the ICP4 promoter, and the MDV replication origin (Ori). We show that Meq homodimers bind to the MDV Ori and repress transcription from the flanking bidirectional promoters (pp38/24 and pp14). By contrast, Meq/Jun heterodimers bind to and are capable of transactivating the Meq promoter. The heterodimers were found to bind host genes as well. This suggests that Meq has the abilities to autoregulate its own expression and affect the expression of other genes. The present study not only provides information concerning Meq's function in transformed T cells but also points out the possibility that Meq may modulate MDV genome expression during latency.
MATERIALS AND METHODS
In vitro coimmunoprecipitation.
The coimmunoprecipitation experiments were performed by using a portion of Meq (amino acids [aa] 1 to 309) fused to a T7 gene 10 epitope (pT7C2bMeq/1-309). This plasmid was constructed in two steps. A BamHI fragment of MeqNco/BS (47) was first cloned into pET21b, creating p21bMeq/1-309. A blunt-ended NheI (Klenow)-, and XhoI-digested subfragment of p21bMeq/1-309 was then cloned into an MscI/XhoI-digested pCITE-2b vector (Novagen), creating pT7C2bMeq/1-309. This construct was in vitro translated with [35S]methionine (TNT system; Promega Corp., Madison, Wis.) and incubated for 20 min at 30°C alone or together with the reaction products generated from an in vitro-translated, non-epitope-tagged pC2bMeq/1-127 construct (10 μl of each). The latter was created by subcloning a BamHI-KpnI subfragment of MeqNco/BS into a BamHI-HincII-digested pCITE-2b vector. A T7-tagged version (pT7C2b/1-127) was constructed in a manner analogous to that used for pT7C2b/1-309, except that the initial pET21b clone was in this case created by subcloning a BamHI-digested, blunt-ended KpnI (with T4 DNA polymerase) subfragment of MeqNco/BS into a BamHI-digested, EcoRI-blunted (with Klenow fragment) pET21b vector.
To investigate the ability of Meq to form complexes with other bZIP members, we first used coimmunoprecipitation analysis of protein-protein interactions involving T7-Meq/1-127 and the full-length chicken c-Jun or mouse JunB protein generated by in vitro translation. T7-Meq/1-127 was translated from a supercoiled pT7C2b-Meq/1-127 plasmid (see above). Full-length chicken c-Jun was translated from an EcoRI-linearized pBSKS+c-Jun construct, which is a pBS KS+ (Stratagene, La Jolla, Calif.) subclone containing a BamHI-EcoRI subfragment of pGCJ-1 (11). Full-length mouse JunB was translated from a previously described XhoI-digested pGEM2 clone (49). This assay was performed essentially as described above for Meq-Meq. The antibodies (Abs) used were against c-Jun (Ab-2; Oncogene Science Inc.) and JunB (SC46; Santa Cruz Biotechnology, Santa Cruz, Calif.).
GST pulldown assays.
Coprecipitation analysis of bZIP proteins of the CREB/ATF family was performed by glutathione S-transferase (GST) pulldown assays. In this assay, unlabeled bacterium-expressed control GST proteins or GST proteins fused to various bZIP domains (3) were incubated with full-length, [35S]methionine-labeled, in vitro-translated T7-Meq protein (10 ml), precipitated on glutathione-Sepharose beads, and analyzed by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) as described above. The full-length T7-Meq construct was derived from a modified pET15b subclone. The recombinant GST fusion proteins were a generous gift from N. Adya and C.-Z. Giam (Department of Medicine, Case Western Reserve University); they were purified and used in accordance with previously described methods (2).
Electrophoretic mobility shift assay (EMSA).
We expressed a six-His-tagged Meq in bacteria by nickel chelation chromatography and used it in gel retardation assays with consensus oligonucleotide probes. Meq/1-168 was derived from a pET21b subclone that has been described previously (46). The Meq product contains six-His and T7 epitope tags; bacterial expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in the Escherichia coli BL21(DE3)pLysS strain in accordance with the instructions of the manufacturer (pET system manual; Novagen). The proteins were purified under denaturing conditions and purified with Ni-nitrilotriacetic acid resin (Qiagen Inc., Valencia, Calif.) in accordance with the instructions supplied. Following elution in buffer containing 250 mM imidazole, the proteins were renatured following four changes of dialysis buffer (20 mM HEPES [pH 7.9], 100 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 1 mM dithiothreitol, 20% glycerol) overnight at 4°C as previously described (7). The protein samples were then concentrated with Centriprep-10 concentrators (Amicon Inc.) and quantified by SDS-PAGE analysis with a trypsinogen protein standard (Sigma Chemical Co., St. Louis, Mo.). Three different probes were tested: (i) a consensus cyclic AMP response element probe (CRE; CTTGGCTGACGTCAGAGAGAG; rat somatostatin promoter, −54/−34; (38); (ii) a TRE probe representing a site present in the chicken c-Jun promoter [CGGGGTGACATCATGGGCTA, −109/−90; (43); and (iii) a probe derived from sequences in the MDV origin of replication (TGCTCATTTGCATACACATCACGTGATAGT) (12). The c-Jun and c-Fos proteins were purified by nickel chelation chromatography with rat c-Jun (aa 206 to 315) and c-Fos (aa 116 to 211) bZIP constructs previously described (1). Meq, c-Jun, and c-Fos were preincubated alone or together (20 pmol of each) for 20 min at 30°C in a gel shift reaction buffer previously described (47). The bacterial proteins were then further incubated for an additional 20 min with a double-stranded probe labeled (10,000 cpm) at the 5′ ends with [32P]ATP and polynucleotide kinase (in accordance with the manufacturer's [United States Biochemical Corp., Cleveland, Ohio] instructions). The reaction products were resolved on a nondenaturing 5% polyacrylamide-Tris-glycine gel.
Cell cultures.
MDV-transformed MSB-1 chicken T cells (5) were cultured in RPMI 1640 medium supplemented with 15% heat-inactivated fetal bovine serum, 100 U of penicillin per ml, and 0.1 mg of streptomycin per ml (Invitrogen, Carlsbad, Calif.) and maintained at 37°C in 5% CO2.
RNA extraction and RT-PCR.
Cells were washed once with cold phosphate-buffered saline (PBS) and pelleted by centrifugation (500 × g, 3 min), and total RNA was extracted with the SV Total RNA Isolation System (Promega) in accordance with the manufacturer's protocol. Column-bound RNA samples were incubated with DNase (Promega) and eluted in nuclease-free water (100 μl; Promega). Reverse transcription (RT) was performed with Super-Script II reverse transcriptase (Invitrogen) in accordance with the manufacturer's protocol, with 2 μg of RNA per sample. PCR was performed with recombinant Taq polymerase (Invitrogen). Reactions were started with 94°C for 3 min, followed by 28 cycles of 94°C for 45 s, 58°C for 45 s, 72°C for 45 s, and 72°C for 10 min, and finally kept at 4°C. For primer sequences, see Table 1. For a PCR assay to confirm the authenticity of the c-fos and JunD primers, plasmid DNA (10 ng) was used as a template in the presence of the corresponding primers under the PCR conditions described above, except that the annealing temperature was 52°C.
TABLE 1.
Primers used for PCR
| Name | Type | Sequence |
|---|---|---|
| GAPDH | Forward | 5′-CAGGTCAACAACAGAGACATTGGG-3′ |
| Reverse | 5′-TACACACGGACACTTCAAGGGC-3′ | |
| c-Jun | Forward | 5′-GAGCCTACTTTCTACGAGGATGCC-3′ |
| Reverse | 5′-GTTGCTGGACTGGATGATGAGC-3′ | |
| Fra-2 | Forward | 5′-CCATCTTCCTCTTTGGTCCTTGAC-3′ |
| Reverse | 5′-GGCATTGGTCCTCACTCTCTTTC-3′ | |
| c-Fos | Forward | 5′-ACCTCCACCTTCGTCTTCACCTAC-3′ |
| Reverse | 5′-TCGTTGCTAAGTCATCAGAACACG-3′ | |
| JunD | Forward | 5′-AGGAGGATGGAAACACCCTTCTAC-3′ |
| Reverse | 5′-CCGTTGGACTGGATGATGAGG-3′ | |
| ICP4 prom. | Forward | 5′-CCTGTCGAGTCTGCGATCTTC-3′ |
| Reverse | 5′-CAAAAGGCTCTTATCATCTACTAG-3′ | |
| MDV-Ori | Forward | 5′-GATTCTTATACGGGTGGGCGTAC-3′ |
| Reverse | 5′-CATTCTGAGAGAGCATCGCGAAG-3′ | |
| Meq prom. | Forward | 5′-GCTGGAAAACCATCGTAGAAC-3′ |
| Reverse | 5′-CGTATCCACTCCCGAACCATTA-3′ | |
| gB prom. | Forward | 5′-TGGACCGTCAGTAAGTTTAGAGG-3′ |
| Reverse | 5′-CGTGGCTGTTGGTAATCTGTTC-3′ | |
| gD prom. | Forward | 5′-TCTAAGAGGCGATGGAAGAATC-3′ |
| Reverse | 5′-TCATACCGAAGCTCTAAAAGGTG-3′ | |
| IL-2 prom. AP-1 (+) | Forward | 5′-AGACTGACCTATGCACCTAAG-3′ |
| Reverse | 5′-CGAGAGGGATTTTACCTACA-3′ | |
| IL-2 prom. AP-1 (−) | Forward | 5′-AAAGCACACAGAGAGAGTGAGCG-3′ |
| Reverse | 5′-TTGTTTTTCCTCCCCGATGAC-3′ | |
| IL-2 coding | Forward | 5′-TTATCCCGTGGCTAACTAATCTGC-3′ |
| Reverse | 5′-GGCATACTGGTAAATGTTGTTGGC-3′ |
Coimmunoprecipitation assays.
MSB-1 cells were washed in cold PBS and lysed in EBC buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 0.5% NP-40, 50 mM NaF, 200 μM Na2VO4, 1 mM phenylmethylsulfonyl fluoride) with a protease inhibitor cocktail (Roche). After centrifugation at 15,000 × g for 10 min at 4°C, the clarified lysate was collected and the protein concentration was adjusted to 1 mg/ml. Twenty microliters of a protein A-protein G slurry mixture immobilized on agarose beads (Upstate) was added to the lysate, and the combination was incubated for 2 h at 4°C to reduce nonspecific binding. The cell lysate was reacted with anti-Meq polyclonal Abs (34) (1:100) overnight at 4°C with gentle rotation. The immunocomplexes were captured by the addition of 20 μl of protein A-protein G slurry mixture immobilized on agarose beads and rotated for an additional 4 h at 4°C. Beads were washed four times with EBC buffer and then boiled for 5 min in 20 μl of 2× SDS sample buffer (125 mM Tris-HCl [pH 6.8], 4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.6% bromophenol blue). Immunoprecipitates were subjected to SDS-10% PAGE and then transferred to polyvinylidene difluoride membranes (Biotechnology Systems) via semidry transfer (Amersham Pharmacia). Afterbeing blocked for 1 h at room temperature in TBST (20 mM Tris-HCl [pH 7.5], 137 mM NaCl, 0.05% Tween 20)-5% skim milk, the membranes were incubated with the primary Ab, anti-c-Jun monoclonal Ab (MAb) 610326 (1:1,000; BD Transduction Labs), overnight at 4°C, in TBST-5% skim milk. The membranes were subsequently washed with TBST three times, for 10 min each time, at room temperature. The membranes were incubated with horseradish peroxidase-conjugated Abs (ICN) for 1 h at room temperature. After incubation with the secondary antibodies, the membranes were washed three times with TBST and visualized with enhanced chemiluminescence reagents (ECL; Amersham Biosciences, United Kingdom).
Western blot assays.
MSB-1 cells (106) were washed in cold PBS, pelleted, and lysed in 50 μl of EBC buffer. Samples were boiled for 5 min with 2× SDS sample buffer, subjected to SDS-PAGE, and then transferred and blocked in TBST-5% skim milk as described above. Membranes were incubated with primary Abs overnight at 4°C. Final dilutions of the Abs were 1:1,000 for the anti-c-Jun MAb and 1:500 for the anti-Fra-2 MAb (1600027; Geneka), in TBST-5% skim milk. The membranes were washed and incubated with horseradish peroxidase-conjugated Abs (1:3,000) for 1 h at room temperature, washed, and visualized with enhanced chemiluminescence reagents.
Immunofluorescence assay.
MSB-1 cells were fixed on slides with methanol-acetone (1:1) for 15 min at room temperature and then washed three times with PBS. After being blocked in PBS-2% bovine serum albumin (BSA) for 30 min at room temperature, cells were incubated with anti-Meq rabbit polyclonal Abs (1:500) and anti-c-Jun MAb (1:500) in 2% BSA for 1 h at 37°C. After four washes with PBS, rhodamine-conjugated anti-rabbit goat immunoglobulin G F(ab′)2 (1:1,000; ICN) and fluorescein isothiocyanate-conjugated anti-mouse sheep immunoglobulin G F(ab′)2 (1:1,000; ICN) in 2% BSA were applied as secondary Abs and allowed to react at 37°C for 1 h. Imaging was performed with a confocal microscope equipped with an argon-krypton laser (LSM510-MicroSystem; Carl Zeiss Co., Ltd.).
ChIP assay.
107 MSB-1 cells were fixed with 1% formaldehyde at room temperature for 10 min and washed with ice-cold PBS. Cells were then washed in buffer I (0.25% Triton X-100, 10 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Cell pellets were collected by centrifugation and washed in buffer II (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM HEPES, pH 6.5). Cell pellets (200 μl) were resuspended in 1 ml of lysis buffer (0.5% SDS, 10 mM EDTA, 50 mM Tris [pH 8.1], 1× protease inhibitor cocktail [Roche], 1 mg of AEBSF per ml) and sonicated four times, for 30 s each time, with 0.5-s pulses (Fisher 550 Sonic Dismembrator). Cell debris was removed by centrifugation, and the chromatin solutions were diluted 5× in dilution buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris [pH 8.1], 1× protease inhibitor cocktail). A sample of total chromatin (20 μg/ml) was collected to serve as a total-input DNA control.
Chromatin fragments were immunoprecipitated with anti-Meq polyclonal Abs (34) (1:100) or anti-c-Jun polyclonal Ab 06-225 (1:200; Upstate) overnight at 4°C. Immunocomplexes were recovered and eluted as described before (15). After reverse cross-linking at 65°C overnight, the DNA fragments were purified with a QIAquick PCR purification kit (QIAGEN) and eluted with 100 μl of 1× Tris-EDTA buffer, pH 8.0. Southern blotting was performed with cosmids (48) digested with restriction enzyme EcoRI, BamHI, or BglII overnight at 37°C and then separated on a 0.8% agarose gel. The gel was depurinated by incubation in depurination buffer (0.25 M HCl) and sequentially denatured in denaturation buffer (1.5 M NaOH, 0.5 M NaCl) for 20 min each. After denaturation, the restriction fragments were transferred to a nylon membrane (Biodyne; Pall Gelman Laboratory) by standard procedures. The DNA was immobilized on the membrane by drying at room temperature for 1 h and UV cross-linking. Immunoprecipitated DNA fragments were radiolabeled with [α32-P]dATP with a Strip-EZ DNA kit (Ambion) as recommended by the supplier. Hybridization was performed in ULTRAhyb buffer (Ambion) as recommended by the supplier.
To confirm the Southern blotting results, a PCR for viral gene promoters was performed with recombinant Taq polymerase (Invitrogen). The input DNA was either total input DNA (control) or c-Jun- or Meq-immunoprecipitated DNA fractions. Reactions were started with 94°C for 3 min, followed by 22 cycles of 94°C for 45 s, 58°C for 45 s, 72°C for 45 s, and 72°C for 10 min. For interleukin-2 (IL-2) promoter precipitation by Meq and c-Jun, the region covering the promoter AP-1 site and two control regions (one within the promoter not including AP-1 and the other in the IL-2 coding region) were chosen. For all primer sequences, see Table 1.
Dual-luciferase reporter assays.
Reporter plasmids were constructed by inserting promoter regions (Table 2) upstream of the firefly luciferase coding region (Luc) in the pGL3-Basic vector (Promega). DF1 cells were seeded in six-well plates at 10 5/well in 4 ml of Dulbecco modified Eagle medium-10% fetal bovine serum (Invitrogen) and incubated at 37°C with 5% CO2 for 24 h. For each well, 2.5 μg of plasmid DNA, including the reporters and the control expression plasmids, were transfected with the Lipofectamine reagent in accordance with the manufacturer's (Invitrogen) protocol. All wells were cotransfected with a control reporter, Renilla luciferase plasmid pRL-SV40 (Promega), which served as an internal control to normalize for variations in transfection efficiency. Cell lysates were prepared 48 h posttransfection with 1× passive lysis buffer (Promega). The dual-luciferase assay was performed in accordance with the manufacturer's protocol with a Lumat LB 9501 Luminometer (Wallac Inc.). At least three independent experiments were performed in each setting.
TABLE 2.
Flanking sequences of MDV promoters inserted into pGL3-Basic
| Name | Type | Sequence |
|---|---|---|
| meq promoter | Forward | 5′-CCACGTACTGACGAATTTAGTACC-3′ |
| Reverse | 5′-GTTGGTGCTGGAATGTTAAGAATA-3′ | |
| pp14 promoter | Forward | 5′-CTTATCCTATACCGCCGCCTCC-3′ |
| Reverse | 5′-GAGAGCATCGCGAAGAGAGAAG-3′ | |
| pp38 promoter | Forward | 5′-GAGAGCATCGCGAAGAGAGAAG-3′ |
| Reverse | 5′-CTTATCCTATACCGCCGCCTCC-3′ |
RESULTS
The dimerization potentials of Meq in vitro: c-Jun is a major partner.
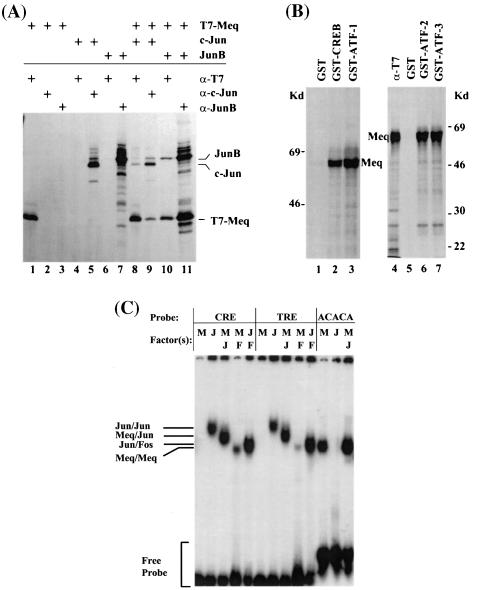
The bZIP domain of Meq has significant homology with the Jun/Fos family (26). The similarity between c-Jun and Meq in the ZIP domain extends beyond the six leucine residues to include an additional histidine (33). Given the potential of c-Jun to dimerize with a variety of bZIP proteins, we asked whether Meq might also be a promiscuous dimerization partner. Several approaches are available by which to identify the potential dimerization partners of bZIP proteins. Because of limitations of the reagents available to us, we used coprecipitation of in vitro-translated products, GST pulldown, or EMSA (47). Representative results are shown in Fig. 1. Figure 1A shows the association of in vitro-translated, T7-tagged Meq with c-Jun or JunB, with proper controls, demonstrating that the antibodies used did not cross-react with one another. Figure 1B shows GST pulldown assays involving incubations of in vitro-translated Meq and fusion proteins of GST and the CREB/ATF family members. These data show that Meq has the potential to dimerize with CREB and ATF1, -2, and -3. While neither of these approaches is highly quantitative, they revealed the potential of Meq to dimerize with a variety of bZIP proteins.
FIG. 1.
The in vitro binding partners of Meq. (A) Coimmunoprecipitation of T7-tagged Meq with full-length chicken c-Jun or mouse JunB proteins. The proteins were in vitro translated in the presence of [35S]methionine and incubated with antibodies (anti-T7 [α-T7], anti-c-Jun [α-c-Jun], and anti-JunB [α-JunB]) as shown. Lanes 1 to 7 contain reaction mixtures involving a single protein and a single antibody to demonstrate the specificity of the antibody. Lanes 8 to 11 contain reaction mixtures of proteins in pairs with the individual antibodies. (B) GST pulldown assays of Meq by CREB family members. GST fusion proteins made with CREB and ATF-1, -2, and -3 (lanes 2, 3, 6, and 7, respectively) were incubated with [35S]methionine-labeled T7-Meq and precipitated on glutathione-Sepharose beads, and the eluates were analyzed by SDS-PAGE. GST was used as a negative control (lanes 1 and 5). T7 antibody (α-T7) was used as a control to immunoprecipitate and identify T7-Meq (lane 4). (C). EMSA analysis of Meq dimmers. Bacterium-expressed and nickel chelation column-purified Meq (M), c-Jun (J), and c-Fos (F) were incubated alone or in pairs with radiolabeled oligonucleotide probes, followed by gel retardation analysis. The CRE and TRE probes were derived, respectively, from collagen and c-Jun promoters. The ACACA probe was derived from the MDV origin of replication. Details are described in Materials and Methods. Kd, kilodaltons.
To determine the functional significance of these protein interactions in terms of the DNA sequences bound, we used EMSA to compare the relative affinity of homodimers and heterodimers of Meq toward the previously identified Meq binding motifs. Previously, by the CASTing approach, we defined two high-affinity motifs that bind, respectively, the Meq/Meq homodimer and the Meq/Jun heterodimer (46). The Meq/Meq dimer primarily binds to the core sequence ACACA. However, ACACA alone is not sufficient for high-affinity binding and requires a secondary structure (such as DNA bending), molded by the surrounding sequence. As we pointed out before, one such motif was found in the replication origin of the MDV genome (MDV-Ori). By contrast, Meq/Jun heterodimers bind canonical AP-1 sequences, i.e., either a TRE (TGACTCA) or a CRE (TGACGTCA) consensus (47). In this study, we tested the abilities of Meq/Meq, Meq/Jun, and Meq/Fos to bind these motifs found in MDV and cellular promoters. The ACACA oligonucleotide was derived from MDV-Ori, which resides in a region that also serves as a bidirectional promoter for the transcription of pp38/pp24 and pp14 (17, 22, 46).
The AP-1 motifs were derived, respectively, from collagen (CRE) and c-Jun (TRE) promoters. We previously showed that the bZIP domain of Meq is necessary and sufficient for binding to these motifs, and the C-terminal domain has no effect on binding, per se (47). In order to distinguish c-Jun, c-Fos, and Meq by size, we used a Meq construct that carries the N-terminal half of Meq, which includes the complete bZIP domain. As shown in Fig. 1C, Meq/Meq homodimers bind strongly to MDV-Ori but only weakly, if at all, to TRE or CRE. By contrast, Meq/Jun binds strongly to TRE and CRE. In fact, in an equimolar mixture of Meq and c-Jun, the form that binds to TRE and CRE is almost exclusively Meq/Jun. Fos does not form dimers itself (unpublished data; 44). Meq and Fos, when mixed together do form Meq/Fos heterodimers, which bind to TRE and CRE, but the affinity seems to be less than that of Meq/Jun (Fig. 1C). This suggests that the form that binds (TRE or CRE) is primarily the Meq/Jun heterodimer. The situation is completely different for the ACACA motif represented by MDV-Ori. Here, neither Jun/Jun nor Meq/Jun forms tight complexes with the promoter. The species that binds most avidly is the Meq homodimer. These data suggest that depending on the dimerization partner, Meq can transcriptionally regulate different sets of genes.
The potential dimerization partners of Meq in MSB-1 cells: c-Jun and Fra-2.
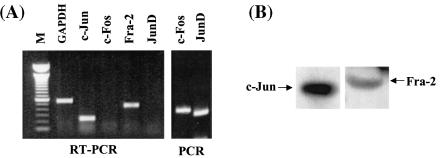
The above data demonstrated the potential of Meq to dimerize in vitro with multiple members of the Jun/Fos family of bZIP proteins and the ability of heterodimers and homodimers to bind distinct enhancer motifs. A key question, then, is what will the functional dimerization partner(s) of Meq inside the tumor cells be? To address this, we employed MSB-1 cells, an MDV-transformed T-lymphoblastoid cell line, which has been widely used to study viral latency, reactivation, and transformation (5). Since not all available antibodies of cellular bZIP proteins (mostly raised against mammalian proteins) react with chicken counterparts, we first used an RT-PCR assay to ascertain their expressions in MSB-1. To design primers for chicken bZIP genes, we surveyed the National Center for Biotechnology Information GenBank database and were able to identify sequences for chicken c-Jun, JunD, Fos, and Fra-2 (Fig. 2A). The RT-PCR results positively identified the expression of c-Jun and Fra-2 but not other species. The primers for JunD and Fos, however, produced positive PCR products on the appropriate plasmids carrying the chicken junD and c-fos genes, indicating that the lack of expression is not due to faulty primers. Western blot analysis confirmed the expression of c-Jun and Fra-2 in MSB-1 (Fig. 2B).
FIG. 2.
Expression of bZIP genes and proteins in MSB-1 cells. (A) RT-PCR detection of the Jun/Fos family of proteins. RT-PCR was used to analyze the expression of c-Fos, c-Jun, Fra-2, and JunD in MSB-1 cells. Primers for the chicken bZIP genes were designed by surveying the National Center for Biotechnology Information GenBank database. Lane M, 100-bp DNA ladder. The rightmost two lanes are positive controls for the primer sets for junD and c-fos. The templates used in these PCRs are plasmids RCAS-JD (a gift of M Aoki and P. Vogt), and RCAS-cfos (a gift of M. Castellazzi). (B) Western blot analysis of c-Jun and Fra-2. Antibodies against c-Jun and Fra-2 were used to probe MSB-1 protein extracts. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Association and colocalization of Meq and c-Jun in MSB-1 cells.
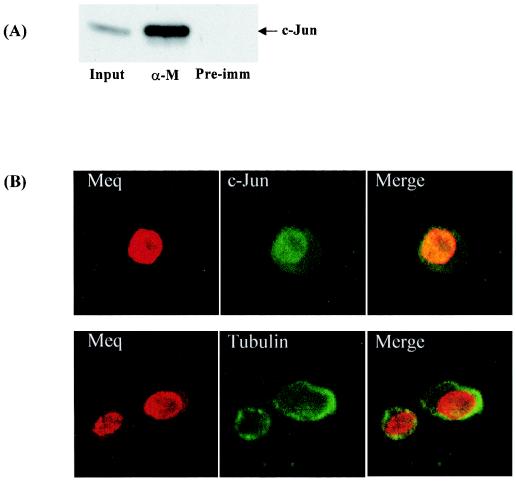
The presence of c-Jun prompted us to investigate whether c-Jun is a natural partner of Meq. We first studied their association by coimmunoprecipitation with MSB-1 cell extracts. Figure 3A shows the coprecipitation of Meq and c-Jun, with antibodies against Meq. A colocalization experiment was then carried out with differentially labeled antibodies against Meq and c-Jun. Importantly, colocalization of Meq and c-Jun in the nucleus is evident on the basis of the merged images (Fig. 3B).
FIG. 3.
Coimmunoprecipitation and colocalization of Meq and Jun in MSB-1 cells. (A) Coprecipitation of Meq and c-Jun. MSB-1 cell extracts were precipitated by antibodies against Meq (α-M) or by preimmune serum (Pre-imm) and then subjected to Western blotting with a c-Jun monoclonal antibody. The input consisted of 10% of the total cell lysate before precipitation. (B) Nuclear colocalization of c-Jun and Meq. Cells were permeabilized and stained with rabbit anti-Meq and mouse anti-c-Jun antibodies. Secondary staining was performed with rhodamine-conjugated anti-rabbit and fluorescein isothiocyanate-conjugated anti-mouse antibodies. Mouse anti-tubulin antibodies were used to delineate the cytoplasm from the nucleus.
Chromosomal Meq binding sites in the MDV genome.
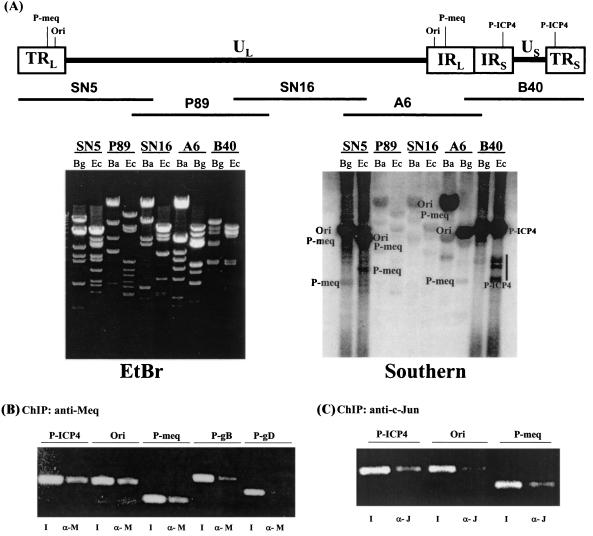
From our previous studies on Meq recognition sequences, we showed that Meq/Meq homodimers bind the ACACA motif and Meq/Jun heterodimers bind TRE and CRE sites. Numerous such motifs and sites are distributed along the chicken genomes and the MDV latent genome. It is difficult to predict a priori the binding sites of Meq. We therefore decided to globally define the binding sites on the latent MDV chromosome by the ChIP approach (15, 36). The complete knowledge of the MDV genome sequences and the restriction sites (29, 55) made this task possible. MSB-1 cells were cross-linked with formaldehyde and then sonicated to shear the chromatin into an average size of 500 bp. Meq antibodies were used to precipitate the chromatin region where Meq resides. After removal of the cross-linking agent, the coprecipitated DNA fragments were isolated. To effectively scan the chromosomal sites associated with Meq, we radiolabeled the isolated DNA by random priming and used it to probe a Southern filter containing overlapping cosmid clones of the MDV genome which were digested with restriction enzyme BglII, BamHI, or EcoRI (19). Five overlapping cosmid clones, which span the entire MDV genome, were used (48). As shown in Fig. 4A, right side, while there is a general background of hybridization signals, the relative intensities of various bands are clearly nonuniform. We interpret this to mean that there is a general low-affinity binding of Meq at numerous sites along the MDV genome but that there are also high-affinity binding sites. On the basis of the restriction enzyme map, we were able to narrow down the high-intensity hybridization regions to three sites, MDV-Ori, the Meq promoter, and the ICP4 promoter. In agreement with the predicted Meq recognition sequences, the MDV replication origin contains an ACACA motif and the promoters of Meq and ICP4 contain AP-1 or AP-1-like motifs (6, 46, 47).
FIG. 4.
Identification of the Meq and Jun binding sites on the MDV chromosome by the ChIP approach. (A) Chromosomal binding sites of Meq. The top panel depicts the MDV genome structure, with the MDV origin of replication (Ori) and the promoters for Meq (P-Meq) and ICP4 (P-ICP4) marked. The five cosmids (SN5, P89, SN16, A6, and B40) spanning the entire MDV genome are indicated. Cosmids were digested with the restriction enzyme EcoRI (Ec), BglII (Bg), or BamHI (Ba) and separated on an agarose gel. The gel was stained with ethidium bromide (EtBr; left panel) and Southern blotted with radiolabeled probes derived from ChIP (Southern; right panel). The DNA associated with Meq chromatin was radiolabeled as described in Materials and Methods. (B) PCR verification of Meq binding sites. PCR primers were designed for the Meq-precipitated (α-M) regions (Ori, P-ICP4, and P-meq). Other MDV promoters served as controls (P-gB, glycoprotein B promoter; P-gD, glycoprotein D promoter). For each primer set, a PCR with the total input DNA (I) before precipitation was carried out. (C) PCR identification of c-Jun binding sites. A ChIP assay was performed for c-Jun (α-J), and the precipitated DNA was assayed by PCR.
To verify the Southern blot data, we then designed specific PCR primers for the Meq-precipitated chromatin regions. Primers for other MDV promoters were also designed to serve as controls. For each primer set, a PCR with the input DNA (total genomic DNA) was carried out to ensure that the priming efficiency is comparable. These primers were then used to produce PCR products with the Meq-associated chromatin DNA. As shown in Fig. 4B, amplicons for MDV-Ori, the Meq promoter, and the ICP4 promoter are readily detected. Amplicons for other MDV promoters, such as the promoters for gB and gD, which contain AP-1 sites, showed much less intensity. Thus, there seems to be a general agreement between the presence of Meq binding motifs and the ChIP results. Since there are numerous potential Meq binding elements in the MDV genome, the selectivity toward certain sites is likely to be governed by other presently unknown factors, such as local chromatin structure or adjacent transcriptional binding sites.
We then asked whether c-Jun is also recruited to these high-affinity binding sites, presumably in the form of Meq/Jun heterodimers. We performed the ChIP assay for c-Jun and assayed for the presence of MDV-Ori and the Meq promoter. Figure 4C shows that c-Jun is recruited to the Meq promoter and the ICP4 promoter, both carrying AP-1 sites, but, interestingly, not to the MDV replication origin, confirming our observations with the gel shift experiment described earlier (Fig. 1C).
Differential transactivation and repression activities of Meq on different promoters.
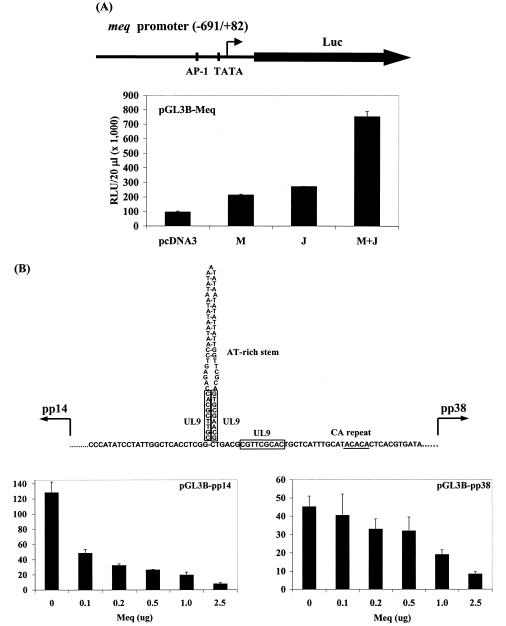
To study the functional significance of Meq binding to these sites, we first cloned the Meq promoter and linked it to a luciferase reporter gene. We tested the transactivation potential of Meq on the Meq promoter, either alone or in combination with c-Jun. As shown in Fig. 5A, the Meq/Meq homodimer has a modest effect on the transcriptional strength of the Meq promoter, in agreement with our previous report (47). Jun/Jun homodimers also failed to transactivate the Meq promoter to any significant extent. By contrast, the Meq/Jun heterodimer is a potent transactivation complex. This supports the findings our ChIP experiment, in which Meq and c-Jun were found to be present on this promoter, poised to transcribe the gene for Meq.
FIG. 5.
Regulation of MDV gene promoter activity by Meq and c-Jun. (A) Activation of the meq promoter. The top diagram illustrates the meq promoter-driven luciferase (Luc) reporter in the pGL3-Basic vector. Luciferase activation (bottom panel) by transient expression of Meq (M) and/or c-Jun (J) constructs in transfected cells. RLU, relative luciferase units. (B) Repression of the bidirectional pp14 and pp38 promoters by Meq. The top diagram depicts the MDV origin of replication and the flanking bidirectional promoters of pp14 and pp38. Boxed areas indicate UL9 binding sites. The underlined sequence is the CA repeat binding site for Meq. The dose-dependent repression of the pp14 and pp38 promoter-driven luciferase activities by transient expression of Meq constructs in transfected cells is shown in the bottom left and right panels, respectively.
We also cloned the MDV-Ori, which contains the flanking bidirectional promoters for the pp14 and pp38/24 genes (46). The segment we cloned carries the entire sequence between the transcriptional start sites of pp14 and pp38. Two reporter constructs were made; in one of them, the luciferase reporter gene is linked to the start site of pp38, and in the other, it is linked to pp14. We tested the potential significance of Meq/Meq homodimer binding to the transcription of these two genes. We found that the Meq/Meq homodimer has a generally suppressive effect on the transcription of both pp38 and pp14, and this suppression occurred in a dose-dependent manner (Fig. 5B). We have also tested the effect of Meq and C-Jun on the transcription of either the pp38 or the pp14 promoter and found that addition of Jun did not increase the transactivation potential (data not shown), in agreement with the notion that the Meq/Meq homodimer is the principal form that binds to this region. These data provide the first evidence that Meq/Meq homodimers and Meq/Jun heterodimers have distinct functional activities.
Corecruitment of Meq and c-Jun to the chicken IL-2 promoter.
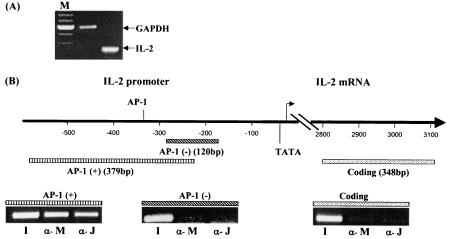
Having demonstrated Meq's ability to bind and activate viral promoters, we were curious about whether Meq is also anchored on the promoter of cellular genes. We have previously shown that Meq alone has the ability to transform rodent fibroblasts, presumably because of its interaction with the host genes (35). Although the database for the chicken promoters is limited, there are a few chicken promoters with AP-1 sites present, among which we selected the IL-2 promoter because of the implication of IL-2 in T-cell transformation (18, 25, 40) and the expression of IL-2 in the MSB-1 cell line (Fig. 6A). ChIP assays with Meq and c-Jun antibodies were carried out as before. As shown in Fig. 6B, both Meq and c-Jun are recruited to a region encompassing an AP-1 site in the promoter, but not a region without such a site or a region representing the coding sequence of IL-2. This suggests that Meq/Jun may also mediate the transcription of host genes.
FIG. 6.
Corecruitment of Meq and Jun to the chicken IL-2 promoter. (A) Expression of IL-2 in MSB-1 cells. An RT-PCR assay for IL-2 transcripts with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a control was carried out as described in Materials and Methods. Lane M, 100-bp DNA ladder. (B) ChIP analysis of Meq and c-Jun on the IL-2 promoter. The top diagram shows the chicken gene for IL-2 and its promoter. An AP-1 site is located 350 bp upstream of the transcriptional initiation site (GenBank accession no. AJ224516). Three sets of PCR primers were designed to cover the regions marked by bars (see Table 1 for primer sequences). The PCR products corresponding to the AP-1 (+) region, the AP-1 (−) region, and the coding region are shown. Lane I (input) on each panel shows the PCR product resulting from the total DNA before ChIP. Anti-Meq (α-M) and anti-c-Jun (α-J) antibodies were used in the ChIP assay as before.
DISCUSSION
In this study, we extended our previous observations, showing that Meq behaves like a prototypic transcriptional factor equipped with the ability to homodimerize with itself and heterodimerize with c-Jun. Significantly, we provide data, for the first time, pertaining to the in vivo Meq dimerization partners inside transformed cells. By using a variety of approaches, we first showed that Meq can dimerize with c-Jun, JunB, Fos, CREB, and ATF1, -2, and -3 in vitro. On the basis of the charge neutralization rules (10, 28, 54), we previously predicted that Meq is likely to be a promiscuous dimerization partner. Because of the availability of the clones, the dimerization partners we tested are not all of chicken origin and the approaches used are not particularly quantitative. Notwithstanding, our results showed that Meq, in a manner similar to that of c-Jun, has the potential to dimerize with a variety of cellular bZIP proteins. The biological effects and the target genes Meq modulates therefore critically depend on the types of partner molecules expressed in particular cell types.
The present results also confirmed our previous report that the preferred binding motif of Meq/Meq homodimers differs from that of Meq/Jun heterodimers. We found that Meq/Meq does not bind to TRE or CRE present in the c-Jun or somatostatin promoter, whereas Meq/Jun and Meq/Fos heterodimers do. By contrast, Meq/Meq, but not Meq/Jun, binds strongly to the ACACA motif present in the MDV replication origin. These results suggest that Meq homodimers and heterodimers are involved in the regulation of different sets of genes. A similar situation has been reported for c-Jun, where Jun/Fos targets TRE sites, whereas Jun/ATF preferentially binds to CRE sites (56, 57). Not only are the binding sites for Meq homodimers and heterodimers different, but the transactivation ability also differs. We found that Meq/Jun is a strong transactivator, whereas Meq/Meq acts as a repressor in the promoters tested. To identify the direct targets of Meq, we used ChIP to scan the entire MDV chromosome for Meq binding sites. This was made possible by the relatively small size of the MDV genome and the availability of overlapping cosmid clones (48). Antibodies against Meq were used to precipitate the chromatin and DNA associated with Meq. The DNA was radiolabeled to probe a Southern blot of restriction enzyme-digested cosmids. Despite the general background often associated with ChIP experiments and the presence of multiple potential Meq binding sites throughout the genome, our data showed that the hybridization signals are nonrandom, with intensity clustered at certain regions. On the basis of the restriction enzyme maps, the more intense signals are located in MDV-Ori, the Meq promoter, and the ICP4 promoter. Both the Meq and ICP4 promoters carry AP-1-like sequences, and MDV-Ori contains an ACACA sequence. However, they are not the only promoters that carry such sequences; additional factors contributing to the high-affinity binding of Meq to these sites must exist. These factors include the local chromatin structure, the posttranslational modification status of the histones, the proximity to other enhancers, etc. It was estimated that in mammalian cells, there are about 3,000 c-Jun molecules per cell and about 1 million AP-1 sites on host chromosomes (58). The high-affinity binding sites for c-Jun are influenced by the presence of other enhancers such as neighboring ETS binding sites, suggesting synergistic effects between c-Jun and other transcriptional factors. Whether this is also the case for Meq is not clear and requires the identification of more Meq binding sites on the MDV chromosome, as well as the host genome.
In this study, we chose to focus on the Meq promoter and the MDV-Ori region for more detailed analysis. PCR results validated our Southern blot data, showing that Meq was recruited to these sites. A ChIP assay with an antibody against c-Jun revealed, interestingly, that c-Jun was recruited to the Meq promoter, but not to MDV-Ori, consistent with our gel shift data showing that the MDV-Ori oligonucleotide binds only Meq homodimers. The finding that Meq binds the MDV chromosome is interesting and suggests that, in addition to being a transforming protein, Meq may participate in the regulation of MDV genes and, hence, the replication process. Considering its overwhelming presence in the latent state as opposed to the lytic viral phases, Meq is more likely to be involved in inhibition of replication or latency establishment. Transactivation assays showed that Meq/Jun transactivates the Meq promoter strongly, whereas Meq homodimers repress the divergent promoters overlapping MDV-Ori. These findings implicate Meq in the autoregulation of its own transcription and the suppression of lytic genes such as that for pp38. Given that the Meq binding site is in proximity to the UL9 (origin binding protein) consensus binding site (59, 60), one could further speculate that Meq binding may influence MDV DNA replication by either direct contact with or modulation of the chromatin conformation recognized by UL9 (8). Increasing evidence suggests that transcriptional factors binding to the replication origin in viruses is one way in which the initiation of DNA replication is controlled (for examples, see references 9, 42, and 53). It is interesting that other oncogenic herpesviruses, such as EBV and KSHV, also express bZIP proteins. EBV encodes Zta (also known as ZEBRA or BZLF1), and KSHV encodes K-bZIP (also known as K8) (16, 21, 31). Unlike Meq, they are neither in the immediate family of Jun/Fos nor able to heterodimerize with the Jun/Fos proteins. But, like Meq, they form homodimers and bind to sites close to the viral lytic replication origins (20, 30, 50). Thus, there seems to be a common theme of the bZIP proteins encoded by oncogenic herpesviruses in that they may serve dual roles in viral transcription, as well as DNA replication.
As a transforming protein, Meq is likely to interact with host genes as well. The gene for v-Jun, in the form of a mutated version of the gene for c-Jun, is known to be a potent retrovirus oncogene and AP-1 transcriptional factor (27, 57, 58). It is also known that AP-1 activation is generally observed in T-cell lymphomas (23, 39) and that one of the target genes is that for IL-2, which is a critical cytokine involved in T-cell proliferation (14, 37, 61). This prompted us to determine whether Meq is involved in IL-2 transcription in the MSB-1 cell line. We have data showing that both Meq and c-Jun are recruited to the IL-2 promoter. While we do not know whether an IL-2 autocrine loop contributes to the transformation of MSB-1, the present findings provide an interesting lead for future research in this direction.
In summary, we report here the in vitro dimerization potentials, the in vivo functional partner, and the chromosomal binding sites of Meq in a naturally MDV-infected T-cell line. We found that Meq/Meq homodimers and Meq/Jun heterodimers bind to different DNA motifs with different transactivation capacities. In addition to advancing our understanding of how Meq functions in T cells, this report provides new leads to the possible roles of Meq in MDV replication.
Acknowledgments
A.M.L., Y.I., and L.X. contributed equally to the data presented in this publication.
We acknowledge the support from the National Institutes of Health (CA46613, CA91574, USDA 2001-02390, and USDA 2002-35204-11621) to H.-J.K. and L.L. A.M.L. was supported by Vaadia-BARD Postdoctoral Award FI-323-2001 from BARD, The United States-Israel Binational Agricultural Research and Development Fund.
We thank Ling-Yu Chen for assistance with preparation of the anti-Meq antibody. H.-J.K. acknowledges the original contribution of P.B., who initiated this work.
REFERENCES
- 1.Abate, C., D. Luk, R. Gentz, F. J. Rauscher III, and T. Curran. 1990. Expression and purification of the leucine zipper and DNA-binding domains of Fos and Jun: both Fos and Jun contact DNA directly. Proc. Natl. Acad. Sci. USA 87:1032-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adya, N., and C.-Z. Giam. 1995. Distinct regions in human T-cell lymphotropic virus type I Tax mediate interactions with activator protein CREB and basal transcription factors. J. Virol. 69:1834-1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adya, N., L. J. Zhao, W. Huang, I. Boros, and C.-Z. Giam. 1994. Expansion of CREB's DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at positions 282 to 284 near the conserved DNA-binding domain of CREB. Proc. Natl. Acad. Sci. USA 91:5642-5646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afonso, C. L., E. R. Tulman, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2001. The genome of turkey herpesvirus. J. Virol. 75:971-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akiyama, Y., and S. Kato. 1974. Two cell lines from lymphomas of Marek's disease. Biken J. 17:105-116. [PubMed] [Google Scholar]
- 6.Anderson, A. S., A. Francesconi, and R. W. Morgan. 1992. Complete nucleotide sequence of the Marek's disease virus ICP4 gene. Virology 189:657-667. [DOI] [PubMed] [Google Scholar]
- 7.Angel, P., E. A. Allegretto, S. T. Okino, K. Hattori, W. J. Boyle, T. Hunter, and M. Karin. 1988. Oncogene jun encodes a sequence-specific trans-activator similar to AP-1. Nature 332:166-171. [DOI] [PubMed] [Google Scholar]
- 8.Aslani, A., S. Simonsson, and P. Elias. 2000. A novel conformation of the herpes simplex virus origin of DNA replication recognized by the origin binding protein. J. Biol. Chem. 275:5880-5887. [DOI] [PubMed] [Google Scholar]
- 9.Baumann, M., R. Feederle, E. Kremmer, and W. Hammerschmidt. 1999. Cellular transcription factors recruit viral replication proteins to activate the Epstein-Barr virus origin of lytic DNA replication, oriLyt. EMBO J. 18:6095-6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baxevanis, A., and C. Vinson. 1993. Interactions of coiled coils in transcription factors: where is the specificity? Curr. Opin. Genet. Dev. 3:278-285. [DOI] [PubMed] [Google Scholar]
- 11.Bos, T. J., F. J. Rauscher III, T. Curran, and P. K. Vogt. 1989. The carboxy terminus of the viral Jun oncoprotein is required for complex formation with the cellular Fos protein. Oncogene 4:123-126. [PubMed] [Google Scholar]
- 12.Bradley, G., M. Hayashi, G. Lancz, A. Tanaka, and M. Nonoyama. 1989. Structure of the Marek's disease virus BamHI-H gene family: genes of putative importance for tumor induction. J. Virol. 63:2534-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calnek, B. W., and R. L. Witter. 1997. Marek's disease, 10th ed. Iowa State University Press, Ames.
- 14.Cantrell, D. A., and K. A. Smith. 1984. The interleukin-2 T-cell system: a new cell growth model. Science 224:1312-1316. [DOI] [PubMed] [Google Scholar]
- 15.Chen, H., R. J. Lin, W. Xie, D. Wilpitz, and R. M. Evans. 1999. Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell 98:675-686. [DOI] [PubMed] [Google Scholar]
- 16.Countryman, J., H. Jenson, R. Seibl, H. Wolf, and G. Miller. 1987. Polymorphic proteins encoded within BZLF1 of defective and standard Epstein-Barr viruses disrupt latency. J. Virol. 61:3672-3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui, Z. Z., L. F. Lee, J.-L. Liu, and H.-J. Kung. 1991. Structural analysis and transcriptional mapping of the Marek's disease virus gene encoding pp38, an antigen associated with transformed cells. J. Virol. 65:6509-6515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Duprez, V., G. Lenoir, and A. Dautry-Varsat. 1985. Autocrine growth stimulation of a human T-cell lymphoma line by interleukin 2. Proc. Natl. Acad. Sci. USA 82:6932-6936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feinberg, A., and B. Vogelstein. 1983. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 132:6-13. [DOI] [PubMed]
- 20.Gao, Z., A. Krithivas, J. E. Finan, O. J. Semmes, S. Zhou, Y. Wang, and S. D. Hayward. 1998. The Epstein-Barr virus lytic transactivator Zta interacts with the helicase-primase replication proteins. J. Virol. 72:8559-8567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruffat, H., S. Portes-Sentis, A. Sergeant, and E. Manet. 1999. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J. Gen. Virol. 80:557-561. [DOI] [PubMed] [Google Scholar]
- 22.Hong, Y., and P. M. Coussens. 1994. Identification of an immediate-early gene in the Marek's disease virus long internal repeat region which encodes a unique 14-kilodalton polypeptide. J. Virol. 68:3593-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwai, K., N. Mori, M. Oie, N. Yamamoto, and M. Fujii. 2001. Human T-cell leukemia virus type 1 tax protein activates transcription through AP-1 site by inducing DNA binding activity in T cells. Virology 279:38-46. [DOI] [PubMed] [Google Scholar]
- 24.Izumiya, Y., H. Jang, M. Ono, and T. Mikami. 2001. A complete genomic DNA sequence of Marek's disease virus type 2, strain HPRS24. Curr. Top. Microbiol. Immunol. 255:191-221. [DOI] [PubMed]
- 25.Jain, J., V. E. Valge-Archer, and A. Rao. 1992. Analysis of the AP-1 sites in the IL-2 promoter. J. Immunol. 148:1240-1250. [PubMed] [Google Scholar]
- 26.Jones, D., L. Lee, J.-L. Liu, H.-J. Kung, and J. K. Tillotson. 1992. Marek disease virus encodes a basic-leucine zipper gene resembling the fos/jun oncogenes that is highly expressed in lymphoblastoid tumors. Proc. Natl. Acad. Sci. USA 89:4042-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karin, M., Z. Liu, and E. Zandi. 1997. AP-1 function and regulation. Curr. Opin. Cell Biol. 9:240-246. [DOI] [PubMed] [Google Scholar]
- 28.Kung, H.-J., L. Xia, P. Brunovskis, D. Li, J.-L. Liu, and L. F. Lee. 2001. Meq: an MDV-specific bZIP transactivator with transforming properties. Curr. Top. Microbiol. Immunol. 255:245-260. [DOI] [PubMed] [Google Scholar]
- 29.Lee, L. F., P. Wu, D. Sui, D. Ren, J. Kamil, H.-J. Kung, and R. L. Witter. 2000. The complete unique long sequence and the overall genomic organization of the GA strain of Marek's disease virus. Proc. Natl. Acad. Sci. USA 97:6091-6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, C. L., H. Li, Y. Wang, F. X. Zhu, S. Kudchodkar, and Y. Yuan. 2003. Kaposi's sarcoma-associated herpesvirus lytic origin (ori-Lyt)-dependent DNA replication: identification of the ori-Lyt and association of K8 bZip protein with the origin. J. Virol. 77:5578-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin, S. F., D. R. Robinson, G. Miller, and H.-J. Kung. 1999. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J. Virol. 73:1909-1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu, J., M. D. Hebert, Y. Ye, D. J. Templeton, H. Kung, and A. G. Matera. 2000. Cell cycle-dependent localization of the CDK2-cyclin E complex in Cajal (coiled) bodies. J. Cell Sci. 113:1543-1552. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J.-L., and H.-J. Kung. 2000. Marek's disease herpesvirus transforming protein MEQ: a c-Jun analogue with an alternative life style. Virus Genes 21:51-64. [PubMed] [Google Scholar]
- 34.Liu, J.-L., L. F. Lee, Y. Ye, Z. Qian, and H.-J. Kung. 1997. Nucleolar and nuclear localization properties of a herpesvirus bZIP oncoprotein, MEQ. J. Virol. 71:3188-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, J.-L., Y. Ye, L. F. Lee, and H.-J. Kung. 1998. Transforming potential of the herpesvirus oncoprotein MEQ: morphological transformation, serum-independent growth, and inhibition of apoptosis. J. Virol. 72:388-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Louie, M. C., H. Q. Yang, A. H. Ma, W. Xu, J. X. Zou, H.-J. Kung, and H. W. Chen. 2003. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA 100:2226-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merlo, J. J., and A. Y. Tsygankov. 2001. Herpesvirus saimiri oncoproteins Tip and StpC synergistically stimulate NF-κB activity and interleukin-2 gene expression. Virology 279:325-338. [DOI] [PubMed] [Google Scholar]
- 38.Montminy, M. R., K. A. Sevarino, J. A. Wagner, G. Mandel, and R. H. Goodman. 1986. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc. Natl. Acad. Sci. USA 83:6682-6686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori, N., M. Fujii, K. Iwai, S. Ikeda, Y. Yamasaki, T. Hata, Y. Yamada, Y. Tanaka, M. Tomonaga, and N. Yamamoto. 2000. Constitutive activation of transcription factor AP-1 in primary adult T-cell leukemia cells. Blood 95:3915-3921. [PubMed] [Google Scholar]
- 40.Nagarkatti, M., M. Hassuneh, A. Seth, K. Manickasundari, and P. S. Nagarkatti. 1994. Constitutive activation of the interleukin 2 gene in the induction of spontaneous in vitro transformation and tumorigenicity of T cells. Proc. Natl. Acad. Sci. USA 91:7638-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nazerian, K., and J. M. Sharma. 1975. Detection of T-cell surface antigens in a Marek's disease lymphoblastoid cell line. J. Natl. Cancer Inst. 54:277-279. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen-Huynh, A. T., and P. A. Schaffer. 1998. Cellular transcription factors enhance herpes simplex virus type 1 oriS-dependent DNA replication. J. Virol. 72:3635-3645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nishimura, T., and P. K. Vogt. 1988. The avian cellular homolog of the oncogene jun. Oncogene 3:659-663. [PubMed] [Google Scholar]
- 44.O'Shea, E. K., R. Rutkowski, W. F. Stafford III, and P. S. Kim. 1989. Preferential heterodimer formation by isolated leucine zippers from fos and jun. Science 245:646-648. [DOI] [PubMed] [Google Scholar]
- 45.Parcells, M. S., R. L. Dienglewicz, A. S. Anderson, and R. W. Morgan. 1999. Recombinant Marek's disease virus (MDV)-derived lymphoblastoid cell lines: regulation of a marker gene within the context of the MDV genome. J. Virol. 73:1362-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qian, Z., P. Brunovskis, L. Lee, P. K. Vogt, and H.-J. Kung. 1996. Novel DNA binding specificities of a putative herpesvirus bZIP oncoprotein. J. Virol. 70:7161-7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian, Z., P. Brunovskis, F. Rauscher III, L. Lee, and H.-J. Kung. 1995. Transactivation activity of Meq, a Marek's disease herpesvirus bZIP protein persistently expressed in latently infected transformed T cells. J. Virol. 69:4037-4044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reddy, S. M., B. Lupiani, I. M. Gimeno, R. F. Silva, L. F. Lee, and R. L. Witter. 2002. Rescue of a pathogenic Marek's disease virus with overlapping cosmid DNAs: use of a pp38 mutant to validate the technology for the study of gene function. Proc. Natl. Acad. Sci. USA 99:7054-7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryder, K., L. F. Lau, and D. Nathans. 1988. A gene activated by growth factors is related to the oncogene v-jun. Proc. Natl. Acad. Sci. USA 85:1487-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarisky, R. T., Z. Gao, P. M. Lieberman, E. D. Fixman, G. S. Hayward, and S. D. Hayward. 1996. A replication function associated with the activation domain of the Epstein-Barr virus Zta transactivator. J. Virol. 70:8340-8347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schat, K. A., C. L. Chen, B. W. Calnek, and D. Char. 1991. Transformation of T-lymphocyte subsets by Marek's disease herpesvirus. J. Virol. 65:1408-1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schat, K. A., C. L. Chen, W. R. Shek, and B. W. Calnek. 1982. Surface antigens on Marek's disease lymphoblastoid tumor cell lines. J. Natl. Cancer Inst. 69:715-720. [PubMed] [Google Scholar]
- 53.Schepers, A., D. Pich, and W. Hammerschmidt. 1996. Activation of oriLyt, the lytic origin of DNA replication of Epstein-Barr virus, by BZLF1. Virology 220:367-376. [DOI] [PubMed] [Google Scholar]
- 54.Schuermann, M., J. Hunter, G. Hennig, and R. Muller. 1991. Non-leucine residues in the leucine repeats of Fos and Jun contribute to the stability and determine the specificity of dimerization. Nucleic Acids Res. 19:739-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tulman, E. R., C. L. Afonso, Z. Lu, L. Zsak, D. L. Rock, and G. F. Kutish. 2000. The genome of a very virulent Marek's disease virus. J. Virol. 74:7980-7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Dam, H., and M. Castellazzi. 2001. Distinct roles of Jun:Fos and Jun:ATF dimers in oncogenesis. Oncogene 20:2453-2464. [DOI] [PubMed] [Google Scholar]
- 57.Vogt, P. K. 2001. Jun, the oncoprotein. Oncogene 20:2365-2377. [DOI] [PubMed] [Google Scholar]
- 58.Wisdom, R. 1999. AP-1: one switch for many signals. Exp. Cell Res. 253:180-185. [DOI] [PubMed] [Google Scholar]
- 59.Wu, T. F., H. H. Chen, and H. Wu. 2001. Functional characterization of Marek's disease virus (MDV) origin-binding protein (OBP): analysis of its origin-binding properties. Virus Genes 23:227-239. [DOI] [PubMed] [Google Scholar]
- 60.Wu, T. F., W. Sun, M. Boussaha, R. Southwick, and P. M. Coussens. 1996. Cloning and sequence analysis of a Marek's disease virus origin binding protein (OBP) reveals strict conservation of structural motifs among OBPs of divergent alphaherpesviruses. Virus Genes 13:143-157. [DOI] [PubMed] [Google Scholar]
- 61.Yamada, G., Y. Kitamura, H. Sonoda, H. Harada, S. Taki, R. C. Mulligan, H. Osawa, T. Diamantstein, S. Yokoyama, and T. Taniguchi. 1987. Retroviral expression of the human IL-2 gene in a murine T cell line results in cell growth autonomy and tumorigenicity. EMBO J. 6:2705-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]