Abstract
Helicase activity is required for T antigen to unwind the simian virus 40 origin. We previously mapped this activity to residues 131 and 616. In this study, we generated a series of mutants with single-point substitutions in the helicase domain to discover other potential activities required for helicase function. A number of DNA unwinding-defective mutants were generated. Four of these mutants (456RA, 460ED, 462GA, and 499DA) were normal in their ability to hydrolyze ATP and were capable of associating into double hexamers in the presence of origin DNA. Furthermore, they possessed normal ability to bind to single-stranded DNA. However, they were severely impaired in unwinding origin-containing DNA fragments and in carrying out a helicase reaction with an M13 partial duplex DNA substrate. Interestingly, these mutants retained some ability to perform a helicase reaction with artificial replication forks, indicating that their intrinsic helicase activity was functional. Intriguingly, these mutants had almost completely lost their ability to bind to double-stranded DNA nonspecifically. The mutants also failed to melt the early palindrome region of the origin. Taken together, these results indicate that the mutations have destroyed a novel activity required for unwinding of the origin. This activity depends on the ability to bind to DNA nonspecifically, and in its absence, T antigen is unable to structurally distort and subsequently unwind the origin.
The large tumor (T) antigen encoded by simian virus 40 (SV40) is a phosphoprotein that has diverse roles in viral DNA replication and transformation (for reviews, see references 7, 19, 43, and 58). The initiation of DNA replication is a series of tightly regulated events. At first, T antigen binds to the 64-bp core origin of replication in a sequence-specific manner. The two pairs of GAGGC pentanucleotides, termed site II, in the core origin serve as the major binding site for T antigen (16). In the presence of ATP, individual monomers of T antigen multimerize cooperatively into a double hexamer, a bilobed structure encircling the origin (12, 28, 63, 67). Interestingly, preformed hexamers of T antigen were shown to be active in origin DNA binding and unwinding, implying that T antigen is versatile in performing these activities (62).
Formation of double hexamers is well supported by either of the two assembly units made up of pentanucleotides 1, 3, and the early palindrome (EP) region or pentanucleotides 2, 4, and the AT track (23, 24, 52). An intact origin, however, is needed for efficient unwinding (13, 33). After hexamerization, T antigen partially melts the EP region and untwists the AT track (3, 5, 6, 15, 32). At the expense of ATP hydrolysis, the distorted origin is subsequently unwound bidirectionally by the helicase activity of T-antigen double hexamers (36, 49, 54). Once unwinding is initiated, the two hexamers likely remain associated with each other, with the separated single-stranded DNA threading through the hexameric channels (50, 67). At least three other host protein factors are loaded on the helicase machine to form a functional replication initiation complex. These include replication protein A (11, 30, 69), topoisomerase I (20, 45, 46, 61), and DNA polymerase α-primase (18, 31).
Aside from its origin binding activity, T antigen is capable of binding to double-stranded and single-stranded DNA nonspecifically (8, 51). The double-stranded binding activity has been roughly mapped to two separate regions. The first region resides in the origin DNA binding domain (2, 29, 41, 42, 57). A close scrutiny of single-point substitution mutants in the origin DNA binding domain revealed that element B1 (residues 183 to 187) participates primarily in nonspecific DNA binding (48). The loss of nonspecific binding activity prevented mutant T antigens from engaging the origin DNA, suggesting that nonspecific contacts with DNA are required for origin-specific binding (44, 48). However, their DNA binding defects could also be caused by significant structural alterations resulting from mutagenesis (47). The second region is located between residues 269 and 522 (26). Although it accounts for most of the nonspecific binding (∼85%) to double-stranded DNA, full binding activity requires the cooperation between the second domain and the origin DNA binding domain (26).
Nonspecific DNA binding activity has also been observed in papillomavirus protein E1 (37, 74). During assembly over DNA, E1 undergoes a shift from an initial origin-specific binding protein to a subsequent nonspecific DNA helicase (55). Recent data from Titolo et al. (60) indicated that, in the presence of ADP-Mg, the binding specificity of T antigen was significantly reduced. These results suggest that nonspecific DNA binding is universally needed for replication initiators and is likely employed after origin-specific binding occurs. The role of nonspecific double-stranded DNA binding has been implicated previously in structural distortion of the origin (9, 38).
T antigen's single-stranded DNA binding activity requires a region between residues 301 and 627 (70). A detailed study of this activity demonstrated that it is not needed for origin-specific binding but appeared to be involved in structural distortion and unwinding of the origin-containing DNA (71). These features of single-stranded DNA binding are consistent with the fact that T antigen associates with only one strand of DNA when unwinding the replication fork (39).
As a helicase, T antigen is able to unwind DNA in a 3′-to-5′ fashion (21, 54, 68). It is capable of unwinding double-stranded DNA (66), artificial replication forks (1, 39, 49), and replication bubbles (49, 50). The region responsible for helicase activity has been mapped to residues 131 to 616 and harbors both DNA binding domains (72). As shown in the crystal structure (25), the domain by itself is organized into two closely associated tiers that possibly slide past each other to melt and unwind the origin. In the present work, we further studied the activities of the helicase domain by generating a collection of mutants with single-point substitutions. Characterization of several mutants impaired in binding nonspecifically to double-stranded DNA demonstrates that this activity is essential for T antigen to structurally distort and unwind the origin. Double-stranded DNA binding therefore participates in two critical steps during initiation of SV40 DNA replication.
MATERIALS AND METHODS
Oligonucleotide-directed mutagenesis.
Single-point substitutions of SV40 T antigen were generated as described previously (27) by annealing an oligonucleotide harboring a desired point mutation to a uridine-containing single-stranded pSK(−)SVT cDNA (26). The annealed oligonucleotide was extended with T4 DNA polymerase (New England Biolabs), and the resulting complementary strand was sealed with T4 DNA ligase. The DNA was used to transform Escherichia coli DH5α cells, and individual colonies were screened for plasmid DNA with the correct mutation by standard dideoxy DNA sequencing.
Construction of recombinant baculoviruses.
pSK(−)SVT cDNA bearing a point substitution was digested with BamHI, and the smaller fragment was ligated to a BamHI-linearized p941 baculovirus transfer vector (BD PharMingen).To make recombinant baculoviruses, p941 DNA with T-antigen inserts was cotransfected with BaculoGold (BD PharMingen) or BaculoPlatinum (Orbigen) DNA in Spodoptera frugiperda (Sf9) cells according to the manufacturer's instructions. T antigen-expressing recombinant viruses were purified by using plaque assays and screened by indirect immunofluorescence with anti-T monoclonal antibody PAb101 (22).
Protein purification.
Wild-type and mutant T antigens were purified from baculovirus-infected High 5 insect cells by immunoaffinity chromatography with monoclonal antibody PAb101 or PAb419 as described previously (28). The antibody was covalently coupled to CNBr-activated Sepharose 4B beads (Pharmacia) as described in the manufacturer's protocol. After incubating cell lysates with antibody-containing beads and washing to remove unbound proteins, T antigen was eluted with ethylene glycol buffer (50% ethylene glycol, 20 mM Tris-HCl [pH 8.5], 500 mM NaCl, 1 mM EDTA, 10% glycerol), dialyzed overnight against dialysis storage buffer (10 mM Tris-HCl [pH 8.0], 100 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 50% [vol/vol] glycerol), and stored at −20°C. The concentration of purified T antigen was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie blue or silver staining of the gel by comparison with phosphorylase B.
DNA binding assays.
Origin-containing DNA was prepared by end labeling a 112-bp HindIII-NcoI fragment of pSKori (45) with 32P in the presence of T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP. For single-stranded DNA binding, a 55-nt oligonucleotide corresponding to the bottom strand of a synthetic replication fork (39) was end labeled with 32P in the same way. For nonspecific double-stranded DNA binding, we end labeled a 400-bp non-SV40 DNA fragment (TaqI-D) derived from pSVO+ plasmid (56). All binding reactions were carried out by incubating 0.4 to 1 μg of T antigen with ∼40 fmol of 32P-labeled DNA in replication buffer (30 mM HEPES [pH 7.5], 7 mM MgCl2, 40 mM creatine phosphate, 4 mM ATP, 1 mM dithiothreitol, and 0.1 mg of bovine serum albumin per ml) for 30 min at 37°C. The DNA-protein complexes were cross-linked with 0.1% glutaraldehyde for an additional 15 min at 37°C and applied to a nondenaturing composite gel (2.5% polyacrylamide and 0.6% agarose) followed by electrophoresis in Tris-borate-EDTA (TBE) buffer at 25 mA for about 3 h at 4°C. Gels were then dried and exposed to a phosphor screen (Molecular Dynamics). Binding was quantified by scanning the screen with a PhosphorImager (Molecular Dynamics) and by using ImageQuant 5.0 software.
Origin DNA unwinding assays.
The same DNA described above in the origin DNA binding assay was also used as a substrate for origin DNA unwinding. T antigen was incubated with 32P-labeled 112-bp origin DNA in replication buffer supplemented with 50 μg of creatine phosphokinase per ml and 2.8 μg of E. coli single-stranded DNA binding protein (Pharmacia) per ml. After 1 h at 37°C, the unwinding reactions were terminated by the addition of sodium dodecyl sulfate, EDTA, and proteinase K to final concentrations of 0.5%, 20 mM, and 0.2 mg/ml, respectively, and samples were further incubated for 30 min at 37°C and then for 5 min at 65°C. The reaction mixtures were loaded on a 7% polyacrylamide gel and subjected to electrophoresis at 770 V · h under constant voltage at 3°C in TBE buffer. The gels were dried and exposed to storage phosphor screens.
ATPase assays.
To measure ATPase activity, 600 ng of T antigen was incubated with [γ-32P]ATP in ATPase buffer (25 mM 1,4-piperazinediethanesulfonic acid [pH 7.0], 0.1 mM NaCl, 5 mM MgCl2, and 0.01% Nonidet P-40) for 1 h at room temperature as described previously (10). The reaction mixtures were spotted on polyethyleneimine cellulose thin-layer plates (EM Science), air dried, and then subjected to ascending chromatography in 0.75 M KH2PO4 (pH 3.5). The plates were then air dried and exposed to X-ray film. The released inorganic phosphate and ATP were then quantitated with a Densitometer (Molecular Dynamics).
Helicase assays.
The helicase assay was carried out as described by Stahl et al. (53) with modifications. A 32-mer oligonucleotide (5′-CCAGGGTTTTCCCAGTCACGACGTTGTAAAAC-3′) was annealed to M13mp19 single-stranded DNA (Invitrogen) and elongated in the presence of dCTP and [α-32P]dATP using Klenow DNA polymerase for 20 min at room temperature. After incubation for an additional 20 min in the presence of dATP, the substrate was purified on a Centri-Spin20 column (Princeton Separations, Inc.). Four hundred nanograms of T antigen was incubated at 37°C for 30 min in helicase buffer (53) with the labeled substrate in a volume of 20 μl. The reactions were quenched by adding 5 μl of stop buffer (2% sodium dodecyl sulfate, 0.1 M EDTA, and 1 mg of proteinase K per ml). The helicase substrate and released oligonucleotide were separated by electrophoresis on a 10% native polyacrylamide gel for 360 Vh at 3°C. The synthetic replication fork DNA was assembled from two partially complementary strands as described by SenGupta and Borowiec (39). The top strand (5′-TTCTGTGACTACCTGGACGACCGGGTGACTAGCTGCGACGAGATGGGTGCACTGC-3′) was end labeled with 32P and annealed to the bottom strand (5′-GTTCTAGCACTTCGAGTCAACATGGTCGTTCCCGGTGGTCCAGGTAGTCACAGA-3′) in annealing buffer (50 mM Tris-HCl [pH 8.0] and 10 mM MgCl2) by heating the reaction mixture to 95°C and slowly cooling down to room temperature.
Similar procedures were used for making replication forks with 3′ or 5′ 30-nt single-stranded DNA overhangs. The sequences of replication fork with a 3′ single-stranded DNA tail were as follows: top, 5′ TTCTGTGACTACCTGGACGACCGGGTGACTAGCTGCGACGAGATGGGTGCACTGC-3′; bottom, 5′-TCCCGGTGGTCCAGGTAGTCACAGA-3′. The sequences of replication fork with a 5′ single-stranded DNA tail were as follows: top, 5′-TTCTGTGACTACCTGGACGACCGGG-3′; bottom, 5′-GTTCTAGCACTTCGAGTCAACATGGTCGTTCCCGGTGGTCCAGGTAGTCACAGA-3′. The annealed products were purified by gel electrophoresis. The unwinding of synthetic replication forks was conducted in a fashion similar to that for the M13mp19 helicase assay, with the exception that a 7% nondenaturing polyacrylamide gel was used, and electrophoresis was performed at 110 V for about 2.5 h at 3°C. The gels were dried, and the DNA bands were visualized and quantitated by autoradiography on a PhosphorImager.
Structural distortion assays.
The melting of the early palindrome and untwisting of the AT track (structural distortion) in the SV40 origin can be detected by KMnO4 oxidation assays as described previously by Borowiec and Hurwitz (6). In this assay, indicated amounts of T antigens were incubated with 600 ng of origin-containing pSVOΔ2792 DNA (57) in replication buffer for 20 min at 37°C. KMnO4 was then added to the reaction mixture to a final concentration of 6 mM to oxidize improperly base-paired thymine residues, followed by an immediate addition of 2-mercaptoethanol to a final concentration of 1.0 M to terminate the reaction. After the modified DNA was purified through a Centri-Spin20 column, a primer extension assay was performed by using a 32P end-labeled pBR322 EcoRI primer (New England Biolabs). The extended products were subjected to electrophoresis in a 7% sequencing gel at 1,500 V for 2.5 to 3 h. Gels were dried and exposed to a PhosphorImager screen.
RESULTS
Generation of mutations in the helicase domain.
One of our major aims was to understand in more detail the mechanism of helicase activity of SV40 T antigen. Therefore, 11 highly conserved residues between residues 455 and 499 in the helicase domain (residues 131 to 616) (72) were chosen and mutated. These sites were selected on the basis of high-level sequence homology among several papovavirus large T antigens. Many of the substitutions that we made represent conservative changes (e.g., Glu for Asp and Lys for Arg) when the same amino acid was present at any one site; otherwise, we introduced alanine substitutions (with the exception of position 455, where Asp was replaced with Asn). We anticipated that mild changes would minimize any deleterious effects on the structure and stability of the protein and help us identify functionally important residues. The mutant genes were cloned into p941 baculovirus transfer vector to produce recombinant baculoviruses. Mutant T antigens were then expressed and purified from baculovirus-infected insect cells by immunoaffinity chromatography (28, 40). All mutant T antigens were produced in yields and integrity similar to that for the wild-type T antigen.
Helicase activities of mutant T antigens.
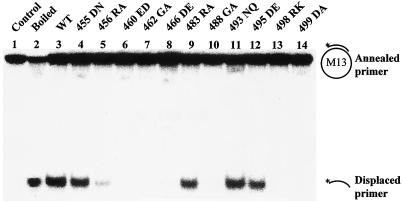
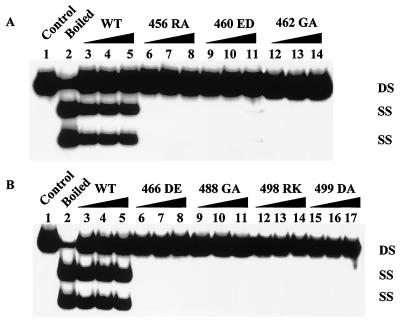
As a first test, the mutant T antigens were assessed for their ability to unwind a circular helicase substrate. Mutants 460ED, 462GA, 466DE, 488GA, 498RK, and 499DA failed to displace a short primer from the M13mp19 single-stranded DNA, and mutant 456RA exhibited a significant reduction in helicase activity (Fig. 1 and Table 1). In contrast, mutants 455DN, 483RA, 493NQ, and 495DE all had more than 50% of the wild type's helicase activity. The mutants that were deficient in helicase activity were further tested for their ability to unwind an origin-containing DNA fragment (Fig. 2). All mutant T antigens were severely defective, although mutant 460ED showed very limited unwinding when large amounts of T antigen were used (Fig. 2).
FIG. 1.
Helicase activity assays of wild-type (WT) and mutant T antigens. Four hundred nanograms of immunoaffinity-purified wild-type and mutant T antigens was incubated with an M13mp19 helicase substrate in helicase buffer for 30 min at 37°C. The reactions were stopped, and samples were analyzed on a 10% native polyacrylamide gel. The positions on the gel of the helicase substrate and product are indicated.
TABLE 1.
Summary of biochemical properties of single-point mutants
| Residue and point mutation | % Wild-type activitya
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Origin DNA binding | Single-stranded DNA binding | Nonspecific double-stranded DNA binding | ATPase | Origin DNA unwinding | Helicase
|
||||
| M13 mp19 helicase substrate | Synthetic replication fork | Replication fork with a 3′ extension | Replication fork with a 5′ extension | ||||||
| 456, Arg → Ala | 94 ± 3.5 | 85 ± 9.9 | 16 ± 0.7 | 94 | 0 | 14 ± 2.8 | 79 ± 13.7 | 76 ± 4.7 | 22 ± 6.3 |
| 460, Glu → Asp | 86 ± 10.3 | 79 ± 9.2 | 2 ± 2.3 | 100 | 0 | 0 | 65 ± 7.8 | 50 ± 9.6 | 2 ± 0.3 |
| 462, Gly → Ala | 81 ± 5.7 | 88 ± 6.5 | 3 ± 1.4 | 99 | 0 | 0 | 62 ± 12.8 | 46 ± 8.3 | 1 ± 1.1 |
| 499, Asp → Ala | 90 ± 11.4 | 74 ± 5.7 | 9 ± 5.0 | 100 | 0 | 0 | 58 ± 4.4 | 45 ± 3.5 | 4 ± 1.6 |
Quantitations were based on 400 ng of wild-type and mutant T antigens, except for the ATPase assay, for which 600 ng of proteins was used. Standard variations were determined from two to four trials of each assay.
FIG. 2.
Origin DNA unwinding by wild-type (WT) and mutant T antigens. The labeled NcoI-HindIII fragment of pSKori DNA was incubated with increasing amounts of T antigens for 1 h at 37°C in the presence of E. coli single-stranded DNA binding protein. Two displaced daughter strands were separated from the parental DNA by gel electrophoresis. (A) Four hundred (lanes 3, 6, 9, and 12), 800 (lanes 4, 7, 10, and 13), and 1,000 (lanes 5, 8, 11, and 14) ng of wild-type, 456RA, 460ED, and 462GA T antigens were assayed for their unwinding activity. (B) Four hundred (lanes 3, 6, 9, 12, and 15), 800 (lanes 4, 7, 10, 13, and 16), and 1,000 (lanes 5, 8, 11, 14, and 17) ng of wild-type, 466DE, 488GA, 498RK, and 499DA T antigens were tested. The positions of single-stranded (SS) and double-stranded (DS) DNAs are shown.
ATPase activities of mutant T antigens.
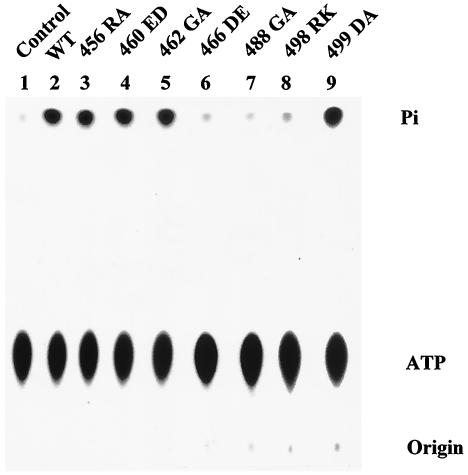
The free energy from ATP hydrolysis is required to drive T antigen to catalyze processive DNA unwinding. Thus, one explanation for the mutants' severely diminished origin DNA unwinding and helicase activities is that they were unable to hydrolyze ATP. The wild-type and mutant proteins were consequently analyzed for their ATPase activity by measuring the amounts of released phosphate. Mutants 466DE, 488GA, and 498RK had markedly reduced ATPase activity (Fig. 3, lanes 6, 7, and 8), whereas all other mutants were capable of hydrolyzing ATP at a level equivalent to that of wild-type T antigen (Fig. 3, lanes 3, 4, 5, and 9; Table 1). The addition of oligo(dT) stimulated ATPase activities of the wild type and mutants 456RA, 460ED, 462GA, and 499DA to the same extent (data not shown), further confirming that these mutant T antigens are normal in this activity. The altered ATPase activity of mutants 466DE, 488GA, and 498RK implies that these three amino acid residues play a crucial role in ATP binding and/or hydrolysis and most likely explains their undetectable helicase activity. The other four mutant T antigens (456RA, 460ED, 462GA, and 499DA) must be defective in helicase activity for some other reason.
FIG. 3.
ATPase assay of wild-type (WT) and mutant T antigens. Equal amounts of T antigen (600 ng) were incubated with 0.3 nmol of [γ-32P]ATP for 1 h at room temperature. Samples were subjected to thin-layer ascending chromatography to separate the free inorganic phosphate (Pi) from the ATP substrate. The positions of released phosphate and ATP substrate are shown.
Binding of mutant T antigens to origin DNA.
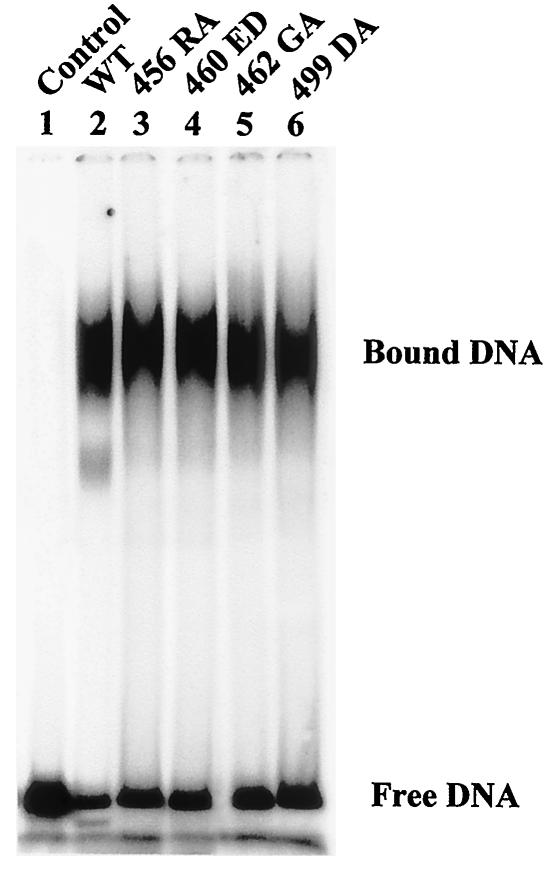
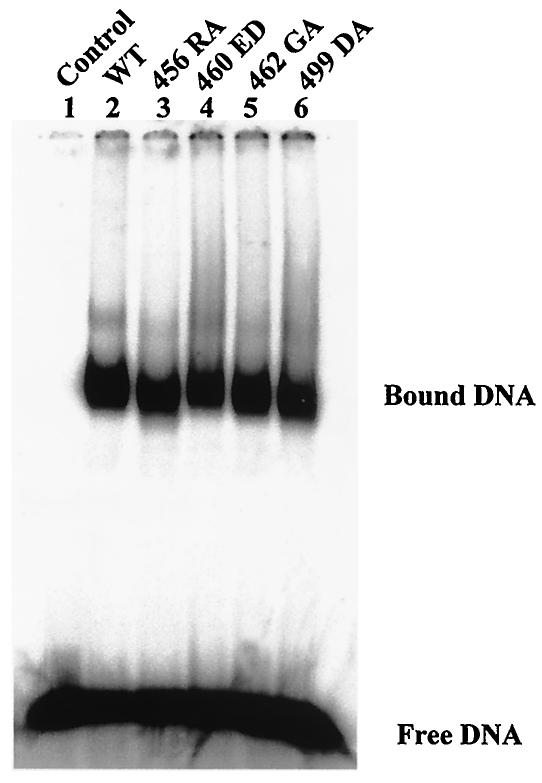
For T antigen to unwind the origin DNA, it first recognizes and binds to the core origin and then oligomerizes into a double hexamer in the presence of ATP (14, 28, 67). This sequence-specific binding is mainly mediated by the origin DNA binding domain mapping roughly from residues 131 to 259 (2, 29, 41, 42, 57). The affinity of mutants 456RA, 460ED, 462GA, and 499DA for origin DNA was measured with a gel shift assay. The results showed that all four mutants bound origin DNA almost normally (Table 1 and Fig. 4). Importantly, all those mutants oligomerized in a manner similar to that of the wild-type T antigen, as double hexamers appeared to be formed (Fig. 4). This observation indicates that the overall structure of the mutant T antigens was not seriously perturbed.
FIG. 4.
Origin DNA binding assay of wild-type (WT) and mutant T antigens. Equal amounts of T antigen (400 ng) were incubated with end-labeled NcoI-HindIII fragment of pSKori DNA for 30 min at 37°C followed by cross-linking with 0.1% glutaraldehyde for 10 min at 37°C. The DNA-protein complexes were subjected to electrophoresis on a composite gel (0.6% agarose and 2.5% acrylamide) in TBE buffer. The positions of free and bound DNAs are shown.
Binding of mutant T antigens to single-stranded DNA.
Our recent studies have revealed that T antigen employs its single-stranded DNA binding activity for unwinding origin DNA or a circular single-stranded DNA substrate (71). Single-stranded DNA binding is also required for the structural distortion of the origin. Moreover, the importance of single-stranded DNA binding in helicase activity was also demonstrated in PcrA helicase (17) and Mcm4,6,7 complexes (35). Since the single-stranded DNA binding domain has been mapped to amino acids 259 to 627 (70), mutants 456RA, 460ED, 462GA, and 499DA may have defects in single-stranded DNA binding, thus resulting in impaired helicase activity. To investigate this possibility, we carried out binding assays by using the bottom strand of an artificial replication fork as described by SenGupta and Borowiec (39). Gel shift assays showed that the affinity of mutant T antigens for the single-stranded DNA was close to normal, ranging between 74 and 88% of that of wild-type T antigen (Fig. 5 and Table 1). Additionally, all mutants oligomerized in a pattern similar to that of wild-type T antigen by forming what appears to be single hexamers (Fig. 5). Therefore, the functional single-stranded DNA binding activity of the mutants suggests that their faulty helicase activity must be due to some other defect.
FIG. 5.
The affinity of wild-type (WT) and mutant T antigens for single-stranded DNA. Equal amounts (400 ng) of wild-type and mutant T antigens were assayed for binding to a 32P end-labeled 55-nt oligonucleotide. The positions of bound and free DNAs are shown.
Nonspecific DNA binding activities of T-antigen mutants.
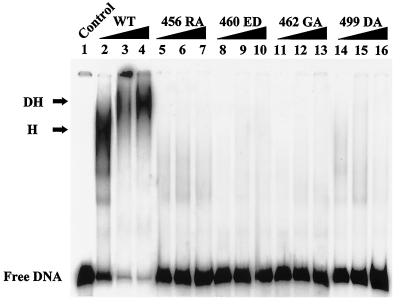
We have shown previously that full binding of T antigen to double-stranded DNA requires both the origin DNA binding domain and a poorly defined region between residues 269 and 522 (26). In addition, nonspecific DNA binding seems separable from binding to single-stranded DNA (70) and may participate in DNA unwinding since a contact with double-stranded DNA takes place (38, 39, 49). To investigate this, we tested the mutants' binding to a double-stranded DNA (TaqI fragment D of pSVO+) lacking origin sequences. As shown in Fig. 6, while the wild type was able to form hexamers at low concentrations (lane 1) or double hexamers at higher concentrations (lanes 2 and 3), mutants 460ED and 462GA displayed essentially undetectable binding activity, while mutants 456RA and 499DA possessed only about 1/10 of the wild-type activity (Table 1). Meanwhile, unlike what we observed in the binding assays with single-stranded and origin DNAs, the diffuse binding pattern of mutants 460ED and 462GA suggests that functional double hexamers were not assembled over double-stranded DNA. In contrast to the mutants' near-normal origin and single-stranded DNA binding activities, their significantly reduced binding to double-stranded DNA correlated with their completely defective origin DNA unwinding activity.
FIG. 6.
Nonspecific double-stranded DNA binding activity of wild-type and mutant T antigens. Increasing amounts of wild-type (WT) and mutant T antigens were tested for their nonspecific binding activity to a fragment of pSVO+ (TaqI-D). Two hundred (lanes 2, 5, 8, 11, and 14), 400 (lanes 3, 6, 9, 12, and 15), and 1,000 (lanes 4, 7, 10, 13, and 16) ng of wild-type and mutant T antigens were examined. The positions of free DNA and DNA bound with the wild-type T antigen in hexamers (H) and double hexamers (DH) are shown.
Replication fork unwinding assays.
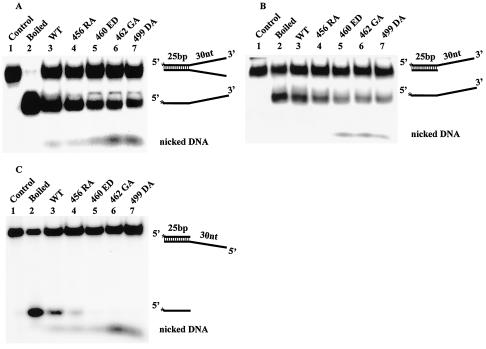
Since T antigen is naturally capable of denaturing DNA, one likely interpretation for the mutants' lack of unwinding ability is that they were intrinsically defective in helicase function regardless of the DNA substrate used. However, the unwinding of both substrates tested so far might have depended on an ability to engage with double-stranded DNA. We therefore assayed the same four mutants for helicase activity by using a synthetic replication fork DNA as the substrate. The synthetic replication fork DNA has a 25-bp double-stranded portion and two single-stranded overhangs formed by annealing two partially complementary 55-mer oligonucleotides (Fig. 7A). T antigen has been shown to unwind this synthetic replication fork efficiently in the presence of ATP (39). Since T antigen is a 3′-to-5′ helicase (68) and because the mutants can oligomerize over single-stranded DNA (Fig. 5), they may be capable of denaturing this substrate. Indeed, all four mutants were able to unwind this replication fork substrate DNA, albeit with decreased activity compared to the wild type (Fig. 7A and Table 1). Small amounts of nicked DNA were also produced, which may have been generated by a contaminating nuclease. Our data strongly suggest that mutants 456RA, 460ED, 462GA, and 499DA still bear a large portion of their intrinsic helicase activity. To demonstrate that the mutants denatured this replication fork by attaching to the free 3′ end, we generated a second substrate containing a free 3′ single-stranded tail but no 5′ tail. All four mutant T antigens denatured this substrate almost as well as they did the first replication fork substrate (Fig. 7B and Table 1). A third substrate, missing a free 3′ tail but with a free 5′ single-stranded extension, was constructed and tested (Fig. 7C). Compared to the wild type (which denatures this DNA poorly), all four mutant proteins had very low unwinding activity (Table 1) similar to their relative ability to unwind the M13 partial duplex DNA substrate (Fig. 1). Our conclusion is that the mutants' native helicase activity is at least 50% of that of the wild type and that the inability to unwind the M13 helicase substrate and origin-containing DNA correlated strongly with a defect in non-sequence-specific double-stranded DNA binding.
FIG. 7.
(A) Unwinding assay of replication forks by wild-type and mutant T antigens. Four hundred nanograms of wild-type (WT) and mutant T antigens was incubated with a synthetic replication fork DNA in helicase buffer. After 30 min at 37°C, the reactions were terminated by the addition of stop buffer, and samples were applied to a 7% nondenaturing polyacrylamide gel. The positions of the replication fork and unwound products are shown. The unwinding reactions in panel A were performed with replication forks with only a 3′ (B) or 5′ (C) single-stranded DNA extension. The positions of replication forks, unwound single strands, and nicked DNA are shown.
Structural distortion activity of mutant T antigens.
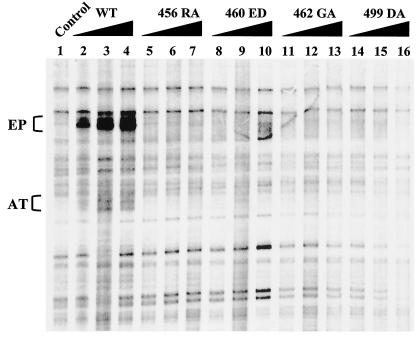
When T antigen binds to the SV40 origin, a structural distortion consisting of melting the EP region and untwisting the AT track is essential for the subsequent DNA unwinding and replication (3, 32). Structural distortion most likely involves many undefined contacts of T antigen with the sugar-phosphate backbone of the EP element and AT track (38). Although structural distortion is closely related to origin-specific binding, it has been shown previously that they are separable from each other (9, 27). Thus, it is quite conceivable that nonspecific double-stranded DNA binding plays a role in structural distortion. We therefore tested whether the nonspecific DNA binding-defective mutants could distort the structure of the origin in a KMnO4 oxidation assay as previously described (4, 5, 9, 15, 47). Figure 8 shows that all four mutants were significantly defective in inducing an EP melting signal. Under the conditions that we used, the WT's AT untwisting signal was weak, but it appeared that the mutants were defective in that activity as well. Our data support the conclusion that nonspecific DNA binding is needed for structural distortion of the origin.
FIG. 8.
Structural distortion assay of wild-type (WT) and mutant T antigens. Reaction mixtures containing 400 (lanes 2, 5, 8, 11, and 14), 800 (lanes 3, 6, 9, 12, and 15), and 1,200 (lanes 4, 7, 10, 13, 16) ng of T antigens were incubated with pSVOΔ2792 DNA in replication buffer at 37°C for 20 min followed by an immediate treatment with KMnO4. After the oxidation was terminated, a primer extension assay was performed to detect KMnO4-modified sites in the DNA, and the extended products were resolved on a 7% sequencing gel. The locations of melted EP region and untwisted AT track are indicated.
DISCUSSION
We report in this study that four mutants of SV40 T antigen with single-amino-acid substitutions in the helicase domain were extremely defective in binding nonspecifically to double-stranded DNA and in performing a helicase reaction with an M13 partial duplex DNA substrate (Table 1). In contrast, they retained a significant portion of their ability to denature artificial replication forks containing 3′ single-stranded DNA extensions (Fig. 7A and B; Table 1), implying that the mutant T antigens still serve as fairly efficient helicases. The mutant T antigens were, however, mostly incapable of unwinding a replication fork with a free 5′ single-stranded DNA extension (Fig. 7C and Table 1). These results strongly indicate that a free 3′ single-stranded DNA tail is needed for the mutant T antigens to unwind replication fork substrates. Previous footprinting data showed that T antigen protects both the single-stranded and duplex regions of replication forks or bubbles (39, 50, 54, 66, 67), suggesting a possible role of nonspecific double-stranded DNA binding in the unwinding of duplex DNA (74). In contrast, our data suggest that nonspecific double-stranded DNA binding is not strictly required for helicase activity. The failure of the mutants to unwind a replication fork with a 5′ tail (Fig. 7C) or an M13 helicase substrate that is mostly circular single-stranded DNA with a short annealed strand (Fig. 1) is probably due to their inability to properly load onto DNA that does not have a free 3′ tail. The mutant proteins can recognize and bind to all these substrates (data not shown), probably as a result of their normal single-stranded DNA binding activity (Fig. 5). We speculate from these observations that double-stranded DNA binding activity is required for T antigen to unwind DNA from a double-stranded DNA end but that this activity is mostly dispensable for melting DNA with an exposed 3′ tail.
Our data also demonstrate that nonspecific double-stranded DNA binding activity is not important for T antigen to engage origin DNA. The same four mutants all showed near-normal binding to DNA containing the complete origin (Fig. 4 and Table 1). Consistent with this finding, we reported in an earlier study that inhibition of nonspecific DNA binding activity by monoclonal antibodies had only minimal effects on the origin DNA binding (26). In addition, most of the nonspecific DNA binding activity has been mapped to a region outside of the origin DNA binding domain (26, 72). Numerous studies have demonstrated that T-antigen polypeptides containing the origin DNA binding domain have poor binding to various origin-lacking DNA fragments but bind efficiently to origin-containing DNA (23, 24, 60). Our results demonstrate that the mutants form apparent double hexamers on origin DNA (Fig. 5), suggesting that binding to the origin is mediated primarily through sequence-specific binding by the origin binding domain. Overall, it is likely that nonspecific DNA binding activity is separate from origin DNA recognition and assembly into double-hexamer structures.
We previously showed that single-stranded and nonspecific double-stranded DNA binding activities reside in functional domains that partially overlap with each other (26, 70, 72). Interestingly, mutation of Glu to Ala at residue 460 abolished binding of T antigen to single-stranded DNA (71), whereas a conservative change to Asp at the same site retained the single-stranded DNA binding but resulted in a total loss of nonspecific double-stranded DNA binding (Fig. 5). These results indicate that certain residues, such as Glu 460, may be important for both binding activities. Although it has been shown that both non-sequence-specific single-stranded and double-stranded DNA binding are brought about primarily by contacts of T antigen with the sugar-phosphate backbone of DNA (39), our results indicate that these two activities are distinguishable (26, 70, 72). Structural studies revealed that either single-stranded or double-stranded DNA is encircled by the ring-like structure of T-antigen hexamers with the size of the central channel being different (25, 34, 63, 64). These observations imply possible structural changes of T-antigen hexamers in response to different DNAs (25). Therefore, the difference between single-stranded and nonspecific double-stranded DNA binding may reflect distinct conformational states of T-antigen hexamers.
Structural changes at the origin (melting of the EP region and untwisting of the AT track) are essential for DNA unwinding and replication (3, 5, 15). We demonstrate here that nonspecific DNA binding correlates well with structural distortion of the origin. Although we previously reported that origin distortion is closely linked with origin binding (47, 73), it does not rely solely on origin DNA binding since T antigen is able to weakly melt the EP region in the absence of the central palindrome in the origin (32). Additional regions or activities of T antigen are also required. These include the zinc finger region (27), the N-terminal side of the origin binding domain (9), single-stranded DNA binding (71), and nonspecific double-stranded DNA binding (this study). One possible interpretation for the mutants' failure to carry out structural distortion in the EP region is that they might have aberrant interactions with the early half of the origin. This would be consistent with the model that the sequences flanking site II are probably covered and then structurally altered by the helicase domain (25, 63). These interactions are likely to be dependent on normal nonspecific double-stranded DNA binding activity since generic protein-DNA contacts are involved (38).
The recently resolved crystal structure of the T-antigen helicase provides a structural basis for understanding the defects of mutant T antigens. All four mutated residues reside on two neighboring α-helices that lie near the central channel of the T-antigen hexamer (25). Interestingly, the side chain of residue Arg 456 protrudes into the channel, suggesting that it makes direct contacts with the accommodated DNA. On the other hand, the other three sites do not seem to directly participate in DNA binding but are probably necessary for the proper positioning of residue Arg 456 or of Lys 512 or His 513, residues that also lie in the main channel. Mutations of residues 460, 462, and 499 might abolish DNA binding by altering the local conformation of their two helices and subsequently interrupting binding of the residues in the main channel to double-stranded DNA.
The inability of the four mutants to unwind an origin-containing DNA fragment is mostly a consequence of the defect in performing the helicase reaction on double-stranded DNA. A second reason is the failure to structurally distort the origin. Hence, the mutants should be totally unable to unwind circular DNA containing the origin since both activities are required (3, 9, 14, 21, 53, 68). All of these defects appear to be due to a lack of binding double-stranded DNA. Based on these observations, we propose that the mutants are incapable of switching from a sequence-specific DNA binding protein to one that binds nonspecifically, as suggested recently for WT papilloma- virus E1 and SV40 T antigen (59, 60). Nonspecific double-stranded DNA binding is, therefore, one of the subactivities required for origin DNA unwinding in addition to origin-specific DNA binding (2, 29, 44, 48, 57), ATP binding and hydrolysis (68), oligomerization (28), interactions between hexamers (49, 65), and single-stranded DNA binding (70, 71). We conclude that non-sequence-specific double-stranded DNA binding is critical for at least two separate activities during DNA replication. First, it is needed for T antigen to melt the EP region and probably untwist the AT track. Second, this activity is required to carry out the helicase reaction from the origin.
Acknowledgments
This work was supported by grant number CA36118 from the National Cancer Institute to D.T.S.
REFERENCES
- 1.Alexandrov, A. I., M. R. Botchan, and N. R. Cozzarelli. 2002. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J. Biol. Chem. 277:44886-44897. [DOI] [PubMed] [Google Scholar]
- 2.Arthur, A. K., A. Höss, and E. Fanning. 1988. Expression of simian virus 40 T antigen in Escherichia coli: localization of T-antigen origin DNA-binding domain to within 129 amino acids. J. Virol. 62:1999-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borowiec, J. A. 1992. Inhibition of structural changes in the simian virus 40 core origin of replication by mutation of essential origin sequences. J. Virol. 66:5248-5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding: how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. [DOI] [PubMed] [Google Scholar]
- 5.Borowiec, J. A., F. B. Dean, and J. Hurwitz. 1991. Differential induction of structural changes in the simian virus 40 origin of replication by T antigen. J. Virol. 65:1228-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borowiec, J. A., and J. Hurwitz. 1988. Localized melting and structural changes in the SV40 origin of replication induced by T-antigen. EMBO J. 7:3149-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bullock, P. A. 1997. The initiation of simian virus 40 DNA replication in vitro. Crit. Rev. Biochem. Mol. Biol. 32:503-568. [DOI] [PubMed] [Google Scholar]
- 8.Carroll, R. B., L. Hager, and R. Dulbecco. 1974. Simian virus 40 T antigen binds to DNA. Proc. Natl. Acad. Sci. USA 71:3754-3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, L., W. S. Joo, P. A. Bullock, and D. T. Simmons. 1997. The N-terminal side of the origin-binding domain of simian virus 40 large T antigen is involved in A/T untwisting. J. Virol. 71:8743-8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark, R., D. P. Lane, and R. Tjian. 1981. Use of monoclonal antibodies as probes of simian virus 40 T antigen ATPase activity. J. Biol. Chem. 256:11854-11858. [PubMed] [Google Scholar]
- 11.Collins, K. L., and T. J. Kelly. 1991. Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol. Cell. Biol. 11:2108-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dean, F. B., J. A. Borowiec, T. Eki, and J. Hurwitz. 1992. The simian virus 40 T antigen double hexamer assembles around the DNA at the replication origin. J. Biol. Chem. 267:14129-14137. [PubMed] [Google Scholar]
- 13.Dean, F. B., J. A. Borowiec, Y. Ishimi, S. Deb, P. Tegtmeyer, and J. Hurwitz. 1987. Simian virus 40 large tumor antigen requires three core replication origin domains for DNA unwinding and replication in vitro. Proc. Natl. Acad. Sci. USA 84:8267-8271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dean, F. B., P. Bullock, Y. Murakami, C. R. Wobbe, L. Weissbach, and J. Hurwitz. 1987. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc. Natl. Acad. Sci. USA 84:16-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean, F. B., and J. Hurwitz. 1991. Simian virus 40 large T antigen untwists DNA at the origin of DNA replication. J. Biol. Chem. 266:5062-5071. [PubMed] [Google Scholar]
- 16.Deb, S., S. Tsui, A. Koff, A. L. DeLucia, R. Parsons, and P. Tegtmeyer. 1987. The T-antigen-binding domain of the simian virus 40 core origin of replication. J. Virol. 61:2143-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dillingham, M. S., P. Soultanas, P. Wiley, M. R. Webb, and D. B. Wigley. 2001. Defining the roles of individual residues in the single-stranded DNA binding site of PcrA helicase. Proc. Natl. Acad. Sci. USA 98:8381-8387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dornreiter, I., W. C. Copeland, and T. S. Wang. 1993. Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol. Cell. Biol. 13:809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fanning, E., and R. Knippers. 1992. Structure and function of simian virus 40 large tumor antigen. Annu. Rev. Biochem. 61:55-85. [DOI] [PubMed] [Google Scholar]
- 20.Gai, D., R. Roy, C. Wu, and D. T. Simmons. 2000. Topoisomerase I associates specifically with simian virus 40 large-T-antigen double hexamer-origin complexes. J. Virol. 74:5224-5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goetz, G. S., F. B. Dean, J. Hurwitz, and S. W. Matson. 1988. The unwinding of duplex regions in DNA by the simian virus 40 large tumor antigen-associated DNA helicase activity. J. Biol. Chem. 263:383-392. [PubMed] [Google Scholar]
- 22.Gurney, E. G., R. O. Harrison, and J. Fenno. 1980. Monoclonal antibodies against simian virus 40 T antigens: evidence for distinct subclasses of large T antigen and for similarities among nonviral T antigens. J. Virol. 34:752-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joo, W. S., H. Y. Kim, J. D. Purviance, K. R. Sreekumar, and P. A. Bullock. 1998. Assembly of T-antigen double hexamers on the simian virus 40 core origin requires only a subset of the available binding sites. Mol. Cell. Biol. 18:2677-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim, H. Y., B. A. Barbaro, W. S. Joo, A. E. Prack, K. R. Sreekumar, and P. A. Bullock. 1999. Sequence requirements for the assembly of simian virus 40 T antigen and the T-antigen origin binding domain on the viral core origin of replication. J. Virol. 73:7543-7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, D., R. Zhao, W. Lilyestrom, D. Gai, R. Zhang, J. A. DeCaprio, E. Fanning, A. Jochimiak, G. Szakonyi, and X. S. Chen. 2003. Structure of the replicative helicase of the oncoprotein SV40 large tumour antigen. Nature 423:512-518. [DOI] [PubMed] [Google Scholar]
- 26.Lin, H.-J. L., R. Upson, and D. T. Simmons. 1992. Nonspecific DNA binding activity of simian virus 40 large T antigen: evidence for the cooperation of two regions for full activity. J. Virol. 66:5443-5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loeber, G., J. E. Stenger, S. Ray, R. E. Parsons, M. E. Anderson, and P. Tegtmeyer. 1991. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J. Virol. 65:3167-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mastrangelo, I. A., P. V. C. Hough, J. S. Wall, M. Dodson, F. B. Dean, and J. Hurwitz. 1989. ATP-dependent assembly of double hexamers of SV40 T antigen at the viral origin of DNA replication. Nature 338:658-662. [DOI] [PubMed] [Google Scholar]
- 29.McVey, D., M. Strauss, and Y. Gluzman. 1989. Properties of the DNA-binding domain of the simian virus 40 large T antigen. Mol. Cell. Biol. 9:5525-5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melendy, T., and B. Stillman. 1993. An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 268:3389-3395. [PubMed] [Google Scholar]
- 31.Murakami, Y., and J. Hurwitz. 1993. Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J. Biol. Chem. 268:11008-11017. [PubMed] [Google Scholar]
- 32.Parsons, R., M. E. Anderson, and P. Tegtmeyer. 1990. Three domains in the simian virus 40 core origin orchestrate the binding, melting, and DNA helicase activities of T antigen. J. Virol. 64:509-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parsons, R., and P. Tegtmeyer. 1992. Spacing is crucial for coordination of domain functions within the simian virus 40 core origin of replication. J. Virol. 66:1933-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.San Martin, M. C., C. Gruss, and J. M. Carazo. 1997. Six molecules of SV40 large T antigen assemble in a propeller-shaped particle around a channel. J. Mol. Biol. 268:15-20. [DOI] [PubMed] [Google Scholar]
- 35.Sato, M., T. Gotow, Z. You, Y. Komamura-Kohno, Y. Uchiyama, N. Yabuta, H. Nojima, and Y. Ishimi. 2000. Electron microscopic observation and single-stranded DNA binding activity of the Mcm4,6,7 complex. J. Mol. Biol. 300:421-431. [DOI] [PubMed] [Google Scholar]
- 36.Scheffner, M., R. Wessel, and H. Stahl. 1989. SV40 T antigen catalyzed duplex DNA unwinding. Curr. Top. Microbiol. Immunol. 144:37-45. [DOI] [PubMed] [Google Scholar]
- 37.Sedman, J., and A. Stenlund. 1996. The initiator protein E1 binds to the bovine papillomavirus origin of replication as a trimeric ring-like structure. EMBO J. 15:5085-5092. [PMC free article] [PubMed] [Google Scholar]
- 38.SenGupta, D. J., and J. A. Borowiec. 1994. Strand and face: the topography of interactions between the SV40 origin of replication and T-antigen during the initiation of replication. EMBO J. 13:982-992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.SenGupta, D. J., and J. A. Borowiec. 1992. Strand-specific recognition of a synthetic DNA replication fork by the SV40 large tumor antigen. Science 256:1656-1661. [DOI] [PubMed] [Google Scholar]
- 40.Simanis, V., and D. P. Lane. 1985. An immunoaffinity purification procedure for SV40 large T antigen. Virology 144:88-100. [DOI] [PubMed] [Google Scholar]
- 41.Simmons, D. T. 1986. DNA-binding region of the simian virus 40 tumor antigen. J. Virol. 57:776-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Simmons, D. T. 1988. Geometry of the simian virus 40 large tumor antigen-DNA complex as probed by protease digestion. Proc. Natl. Acad. Sci. USA 85:2086-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simmons, D. T. 2000. SV40 large T antigen: functions in DNA replication and transformation. Adv. Virus Res. 55:75-134. [DOI] [PubMed] [Google Scholar]
- 44.Simmons, D. T., G. Loeber, and P. Tegtmeyer. 1990. Four major sequence elements of simian virus 40 large T antigen coordinate its specific and nonspecific DNA binding. J. Virol. 64:1973-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simmons, D. T., R. Roy, L. Chen, D. Gai, and P. W. Trowbridge. 1998. The activity of topoisomerase I is modulated by large T antigen during unwinding of the SV40 origin. J. Biol. Chem. 273:20390-20396. [DOI] [PubMed] [Google Scholar]
- 46.Simmons, D. T., P. W. Trowbridge, and R. Roy. 1998. Topoisomerase I stimulates SV40 T antigen-mediated DNA replication and inhibits T antigen's ability to unwind DNA at nonorigin sites. Virology 242:435-443. [DOI] [PubMed] [Google Scholar]
- 47.Simmons, D. T., R. Upson, K. Wun-Kim, and W. Young. 1993. Biochemical analysis of mutants with changes in the origin-binding domain of simian virus 40 tumor antigen. J. Virol. 67:4227-4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simmons, D. T., K. Wun-Kim, and W. Young. 1990. Identification of simian virus 40 T antigen residues important for specific and nonspecific binding to DNA and for helicase activity. J. Virol. 64:4858-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smelkova, N. V., and J. A. Borowiec. 1997. Dimerization of simian virus 40 T-antigen hexamers activates T-antigen DNA helicase activity. J. Virol. 71:8766-8773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smelkova, N. V., and J. A. Borowiec. 1998. Synthetic DNA replication bubbles bound and unwound with twofold symmetry by a simian virus 40 T-antigen double hexamer. J. Virol. 72:8676-8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spillman, T., D. Giacherio, and L. P. Hager. 1979. Single strand DNA binding of simian virus 40 tumor antigen. J. Biol. Chem. 254:3100-3104. [PubMed] [Google Scholar]
- 52.Sreekumar, K. R., A. E. Prack, D. R. Winters, B. A. Barbaro, and P. A. Bullock. 2000. The simian virus 40 core origin contains two separate sequence modules that support T-antigen double-hexamer assembly. J. Virol. 74:8589-8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stahl, H., P. Droge, and R. Knippers. 1986. DNA helicase activity of SV40 large tumor antigen. EMBO J. 5:1939-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stahl, H., M. Scheffner, M. Wiekowski, and R. Knippers. 1988. DNA unwinding function of the SV40 large tumor antigen. Cancer Cells 6:105-112. [Google Scholar]
- 55.Stenlund, A. 2003. E1 initiator DNA binding specificity is unmasked by selective inhibition of non-specific DNA binding. EMBO J. 22:954-963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stillman, B. W., and Y. Gluzman. 1985. Replication and supercoiling of simian virus 40 DNA in cell extracts from human cells. Mol. Cell. Biol. 5:2051-2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strauss, M., P. Argani, I. J. Mohr, and Y. Gluzman. 1987. Studies on the origin-specific DNA-binding domain of simian virus 40 large T antigen. J. Virol. 61:3326-3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sullivan, C. S., and J. M. Pipas. 2002. T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis. Microbiol. Mol. Biol. Rev. 66:179-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Titolo, S., K. Brault, J. Majewski, P. W. White, and J. Archambault. 2003. Characterization of the minimal DNA binding domain of the human papillomavirus E1 helicase: fluorescence anisotropy studies and characterization of a dimerization-defective mutant protein. J. Virol. 77:5178-5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Titolo, S., E. Welchner, P. W. White, and J. Archambault. 2003. Characterization of the DNA-binding properties of the origin-binding domain of simian virus 40 large T antigen by fluorescence anisotropy. J. Virol. 77:5512-5518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trowbridge, P. W., R. Roy, and D. T. Simmons. 1999. Human topoisomerase I promotes initiation of simian virus 40 DNA replication in vitro. Mol. Cell. Biol. 19:1686-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uhlmann-Schiffler, H., S. Seinsoth, and H. Stahl. 2002. Preformed hexamers of SV40 T antigen are active in RNA and origin-DNA unwinding. Nucleic Acids Res. 30:3192-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Valle, M., C. Gruss, L. Halmer, J. M. Carazo, and L. E. Donate. 2000. Large T-antigen double hexamers imaged at the simian virus 40 origin of replication. Mol. Cell. Biol. 20:34-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.VanLoock, M. S., A. Alexandrov, X. Yu, N. R. Cozzarelli, and E. H. Egelman. 2002. SV40 large T antigen hexamer structure: domain organization and DNA-induced conformational changes. Curr. Biol. 12:472-476. [DOI] [PubMed] [Google Scholar]
- 65.Weisshart, K., P. Taneja, A. Jenne, U. Herbig, D. T. Simmons, and E. Fanning. 1999. Two regions of simian virus 40 T antigen determine cooperativity of double-hexamer assembly on the viral origin of DNA replication and promote hexamer interactions during bidirectional origin DNA unwinding. J. Virol. 73:2201-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wessel, R., U. Ramsperger, H. Stahl, and R. Knippers. 1992. The interaction of SV40 large T antigen with unspecific double-stranded DNA: an electron microscopic study. Virology 189:293-303. [DOI] [PubMed] [Google Scholar]
- 67.Wessel, R., J. Schweizer, and H. Stahl. 1992. Simian virus 40 T-antigen DNA helicase is a hexamer which forms a binary complex during bidirectional unwinding from the viral origin of DNA replication. J. Virol. 66:804-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wiekowski, M., M. W. Schwarz, and H. Stahl. 1988. Simian virus 40 large T antigen DNA helicase. Characterization of the ATPase-dependent DNA unwinding activity and its substrate requirements. J. Biol. Chem. 263:436-442. [PubMed] [Google Scholar]
- 69.Wold, M. S., and T. Kelly. 1988. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. USA 85:2523-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu, C., D. Edgil, and D. T. Simmons. 1998. The origin DNA-binding and single-stranded DNA-binding domains of simian virus 40 large T antigen are distinct. J. Virol. 72:10256-10259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu, C., R. Roy, and D. T. Simmons. 2001. Role of single-stranded DNA binding activity of T antigen in simian virus 40 DNA replication. J. Virol. 75:2839-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wun-Kim, K., and D. T. Simmons. 1990. Mapping of helicase and helicase substrate binding domains on simian virus 40 large T antigen. J. Virol. 64:2014-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wun-Kim, K., R. Upson, W. Young, T. Melendy, B. Stillman, and D. T. Simmons. 1993. The DNA-binding domain of simian virus 40 tumor antigen has multiple functions. J. Virol. 67:7608-7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang, L., I. Mohr, E. Fouts, D. A. Lim, M. Nohaile, and M. Botchan. 1993. The E1 protein of bovine papilloma virus 1 is an ATP-dependent DNA helicase. Proc. Natl. Acad. Sci. USA 90:5086-5090. [DOI] [PMC free article] [PubMed] [Google Scholar]