Abstract
The Porcine Endogenous Retrovirus (PERV) Gag protein contains two late (L) domain motifs, PPPY and P(F/S)AP. Using viral release assays we demonstrate that PPPY is the dominant L domain involved in PERV release. PFAP represents a novel retroviral L domain variant and is defined by abnormal viral assembly phenotypes visualized by electron microscopy and attenuation of early PERV release as measured by viral genomes. PSAP is functionally dominant over PFAP in early PERV release. PSAP virions are 3.5-fold more infectious in vitro by TCID50 and in vivo results in more RNA positive tissues and higher levels of proviral DNA using our human PERV-A receptor (HuPAR-2) transgenic mouse model (Martina et al., 2006. Journal of Virology. 80: 3135-46). The functional hierarchies displayed by PERV L domains, demonstrates that L domain selection in viral evolution exists to promote efficient viral assembly, release and infectivity in the virus-host context.
Keywords: Porcine Endogenous Retrovirus (PERV), Late (L) domains, viral assembly, infectivity
BACKGROUND
Xenotransplantation, the use of nonhuman cells, tissues or organs for a human recipient, has the potential to alleviate the serious and growing human donor organ shortage. Pigs are presently the most suitable nonhuman donors given their physiological size and compatibility, the use of genetic engineering to remove immunological barriers (Kolber-Simonds et al., 2004), and world-wide experience with pig husbandry. A major international concern is the possibility of cross-species transmission of infectious pathogens from animal donors to immunosuppressed human transplant patients. In the case of pigs, one major concern lies with transmission of Porcine Endogenous Retrovirus (PERV) (Louz et al., 2008).
PERV is a gammaretrovirus present in 10–60 copies in all pig genomes (van der Laan et al., 2000) and is closely related to the leukemia-causing viruses, Gibbon Ape Leukemia Virus (GALV), Feline Leukemia Virus (FeLV) and Murine Leukemia Virus (MLV) (Ericsson et al., 2003). PERV envelope sequence, in vitro tropism, and receptor interference assays divides the virus into three classes: A, B and C (Akiyoshi et al., 1998; Le Tissier et al., 1997; Takeuchi et al., 1998)PERV-A and – B are able to productively infect human cell lines in vitro (Le Tissier et al., 1997; Martin et al., 2000; Wilson et al., 2000) while productive infection of the same lines has not been demonstrated for PERV-C (Wilson et al., 2000).
A key determinant in establishing a productive viral infection is the assembly and release of infectious viral particles from the host’s cells. Thus, one approach to evaluate the likelihood of cross-species transmission to human cells in xenotransplantation is to understand PERV assembly and release at the molecular level. Additionally, understanding the basis of virion assembly and infectivity for PERV, contributes to the overall knowledge of viral fitness determinants in productive retroviral infections.
Late (L) domains are one of many components contributing to efficient viral assembly and release. Three canonical Gag L domain motifs, PPXY, P(T/S)AP and YXXL, have been described for many viral families, including retroviruses (Freed, 2002). They are involved in the late stages of viral assembly and current literature documents three phenotypes for L domain mutants: 1) incomplete maturation of released particles (Gottlinger et al., 1991; Le Blanc et al., 2002), 2) reduction of viral particle release (Demirov, Orenstein, and Freed, 2002; Huang et al., 1995; Wang, Machesky, and Mansky, 2004; Yuan, Li, and Goff, 1999), and 3) Gag accumulation under the plasma membrane (Demirov and Freed, 2004; Le Blanc et al., 2002).
Failed viral assembly phenotypes have been attributed mechanistically to disruption of the viral L domain motif’s interaction with specific host cell proteins that function normally in a pathway of microvesicular and protein trafficking. Nedd4 binds the PPXY motif in Mason-Pfizer Monkey Virus (MPMV) (Gottwein et al., 2003), Human T-cell Leukemia Virus (HTLV-1) (Bouamr et al., 2003), and Rous Sarcoma Virus (RSV) (Kikonyogo et al., 2001). Tumor Supressor Gene 101 (Tsg101) binds the P(T/S)AP motif of MPMV (Gottwein et al., 2003) and Human Immunodeficiency Virus type 1 (HIV-1) (VerPlank et al., 2001). Alix/AIP1 binds the YPXL motifs of Equine Infectious Anemia Virus (Chen et al., 2005; Martin-Serrano et al., 2003; Strack et al., 2003) and MLV (Segura-Morales et al., 2005). Additional L domain sequences, interacting proteins, and functions for L domains are still being elucidated in retroviruses as well as other viral families (Licata et al., 2004; Schmitt et al., 2005; Wirblich, Bhattacharya, and Roy, 2006).
While some retroviruses have only one L domain [RSV (PPPY) (Wills et al., 1994; Xiang et al., 1996) and Equine Infectious Anemia Virus (YPDL) (Puffer et al., 1997)], others have two [MPMV (PPPY and PSAP) (Gottwein et al., 2003), HTLV-1 (PPPY and PTAP) (Heidecker et al., 2004; Wang, Machesky, and Mansky, 2004), HIV-1 (PTAP (Gottlinger et al., 1991; Huang et al., 1995) and YPDL (Strack et al., 2003))], and MLV has three [(PPPY (Yuan et al., 2000), PSAP and YPAL (Segura-Morales et al., 2005)]. Therefore, it is logical to ask why do some viruses evolve multiple L domains, and, in this situation, is there an important interaction occurring between them that determines or regulates viral assembly or release.
Mutations of the PTAP motif in HIV-1 ablate release (Gottlinger et al., 1991; Martin-Serrano, Zang, and Bieniasz, 2001). In contrast, mutation of P(T/S)AP in MPMV and HTLV impairs but does not prevent virion production (Gottwein et al., 2003; Heidecker et al., 2004; Wang, Machesky, and Mansky, 2004). Instead, PPPY is the functionally dominant domain for MPMV (Gottwein et al., 2003) and HTLV-1(Le Blanc et al., 2002; Wang, Machesky, and Mansky, 2004). Despite the secondary role played by P(T/S)AP motif in MPMV and HTLV release, its conservation suggests that its function is still selected for in viral evolution (Dorweiler et al., 2006) and our new data indicates a similar status for this domain in PERV.
We determined that Gag of a high-titer infectious PERV-A/C recombinant clone, PERV-A14/220 (Harrison et al., 2004), has two L domain motifs—a prototypic PPXY motif and a previously unidentified P(T/S)AP variant, PFAP, separated by eight residues. The variant PFAP motif is found in the two published PERV-C sequences (GenBank AAC16761.1 and AAC16763.1) while the canonical PSAP motif is found in PERV-A (GenBank AAM29192). Therefore, PFAP is the L domain sequence contributed to this A/C recombinant clone by PERV-C. PFAP resembles the previously described P(T/S)AP motif, however, its role and function as an L domain has not been previously described in PERV or any other virus.
We compared the functional importance of both L domain motif pairs, PPPY-PFAP and PPPY-PSAP, in PERV release and infectivity. Our results demonstrate that PPPY is the primary L domain required for PERV release, as observed at the level of viral RNA genomes quantified by RT-qPCR and at the level of Gag protein quantified by fluorescent-based Western blotting. Mutation of the P(F/S)AP domains produces a viral release arrest phenotype observed by electron microscopy, defining their role as functional L domains. We also show that the PSAP domain compared to PFAP enhances both early viral release and infectivity in vitro, and in vivo infection of human PERV receptor transgenic mice. The functional hierarchy of PSAP over PFAP in early release and infectivity suggests that the prevalence of the PSAP motif in gammaretroviruses as the secondary L domain is due to a selection for optimal virion fitness.
RESULTS
The PPPY motif is the dominant L domain for PERV release
A series of PERV mutants was constructed (Figure 1) to determine the individual contributions of: i) PPPY and P(F/S)AP and ii) PFAP and PSAP L domain motifs in PERV-A14/220 release. Viral release was measured in the supernatants at the relatively late time point of 72 hours post-transfection. Protein levels were quantified by densitometry of fluorescent-based Gag Western blots using a monoclonal anti-PERV Gag antibody and based on the 24kDa form of processed Gag (Figure 2A). We also measured cellular levels of Gag precursor (Figure 2B). By normalizing the amount of gag detected in the supernatant to the total amount of Gag protein detected in both cells and supernatants, we created a metric that we call “average release efficiency” (Figure 2D) (adapted from the literature (Gottwein and Krausslich, 2005)). In parallel, PERV pol RT-qPCR was used to quantify viral genomes in both cells and supernatants. In this testing, average release efficiency was determined by normalizing supernatant PERV pol cDNA copy numbers to the total PERV pol cDNA copy numbers of both cells and supernatants (Figure 2E).
FIGURE 1.

Schematic of the 524 a.a. Gag protein (bp 4231-5805) present in the high infectious titer proviral clone PERV-A14/220 (Harrison et al., 2004). Processed Gag components [Matrix (MA), Capsid (CA) and Nucleocapsid (NC)] are delineated based on similarity to MLV Gag sequences. The two L domain motifs, PPPY and PFAP, reside in p12 and are underlined. Mutations were introduced into the L domains by site-directed mutagenesis. Resulting amino acid sequences are shown for each construct used to evaluate the role of the L domains.
FIGURE 2.
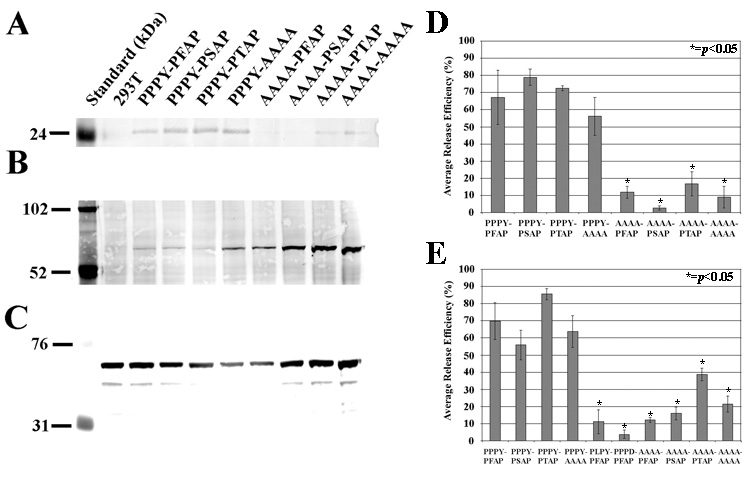
Average release efficiency of PERV L domain constructs. Gag protein levels were determined by densitometry of fluorescence-based Western blots: (A) p24 Gag detected in supernatant virion pellets and (B) precursor Gag detected in cellular extracts with mouse monoclonal anti-Gag39, and (C) actin detected in cellular extracts with anti-actin. Actin is a loading control to normalize cellular Gag protein levels. Viral genome quantification was done by PERV pol RT-qPCR. A minimum of three experiments was averaged to obtain the Average Release Efficiency (%) for Gag protein (D) or viral genomes (E). Release efficiency is an expression of viral protein (or genomic RNA) present in the supernatant relative to the total Gag protein (or viral genomic RNA) present in both the supernatant and the cell. Student’s t-tests were applied to determine if the release of mutant constructs were statistically significant from the wild-type, PPPY-PFAP (*=p<0.05).
The first conclusion is that PPPY is functionally dominant in PERV release at the level of both Gag protein and viral genome metrics. Mutation of all four residues of PPPY to AAAA, resulted in significantly lower average release efficiencies (p<0.05) compared to PPPY-PFAP for both Gag protein and viral genomes (Figures 2D and 2E). Single amino acid changes, PLPY and PPPD, also exhibited similar reductions (Figure 2E). Additionally, the average release efficiencies of PPPY-P(F/S)AP and PPPY-AAAA are equivalent. Thus, P(F/S)AP, alone, is not able to support PERV release. Moreover, in the absence of the PPPY domain, PTAP, the dominant functional L domain in HIV-1 release, does not rescue PERV virion release. We also observed that the average release efficiencies of all the second L domains (PFAP, PSAP and PTAP) are equivalent at 72 hours (Figures 2D and 2E).
The P(F/S)AP motif displays a functional hierarchy early in PPPY-dominant release
We tested the possibility that at an early time point after transfection that differences in the viral release efficiencies of the second L domains might be observed. Figure 3 shows that both PPPY-PSAP and PPPY-PTAP have significantly higher average release efficiencies than PPPY-PFAP at 24 hours post-transfection. In addition, PPPY-PTAP has a significant higher average release efficiency (p<0.03) compared to PPPY-PSAP. This establishes a functional hierarchy for PERV release - PTAP>PSAP>PFAP.
FIGURE 3.
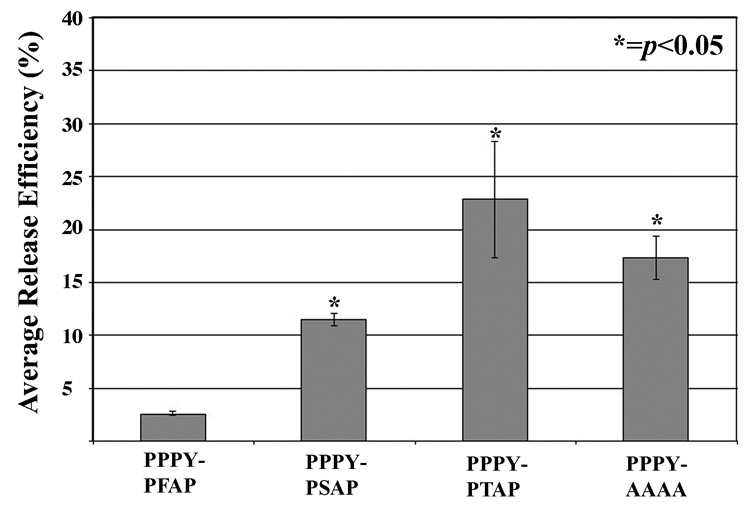
Average release efficiencies of second domain variants in early viral release. Release efficiencies were determined by viral genome quantification 24 hours post-transfection in both supernatants and cells as described in Figure 2. There is a functional hierarchy for early viral release in the presence of the dominant PPPY motif: PTAP>PSAP>PFAP.
An unexpected result was that the presence of the second L domain appears to attenuate early PERV release. There is significantly less viral release efficiency comparing either PPPY-PFAP or PPPY-PSAP to a construct with the second domain replaced by alanines, PPPY-AAAA (Figure 3). Attenuation is only observed at the 24-hour time point, as the average release efficiencies of PPPY-PFAP, PPPY-PSAP and PPPY-AAAA are equivalent at 72 hours post-transfection. Therefore, the second L domain attenuates early PERV release, PFAP attenuating release more than PSAP, consistent with the observed functional hierarchy.
Aberrant assembly and release phenotypes are observed in both PPPY and PFAP mutants by electron microscopy
Thin-section electron microscopy was used to visualize 293T cells 48 hours after transfection with either wild-type PPPY-PFAP (Figure 4A and 4D) or the L domain mutants, PPPD-PFAP (Figure 4B and 4E) and PPPY-AAAA (Figure 4C). All sections were reviewed and photographed by an independent expert (MW) blinded to the identities of the samples. PPPD-PFAP shows thickenings under the plasma membrane at curved surfaces, which are presumably sites of Gag accumulation prior to virion budding and consistent with an early budding arrest phenotype (Le Blanc et al., 2002). Higher magnification reveals viral cores that do not resemble those of the wild-type PERV (compare 4D and 4E). Nonetheless, budded virions are visualized, consistent with the results in Figure 2E for the presence of some viral genomes in the supernatant. Knocking out the second L domain entirely (PPPY-AAAA), results in a spectrum of late stage arrest phenotypes including grape-like clusters of virions at the cell surface (Figure 4C) as well as tethered viral particles at the budding sites (see middle arrow). Thus, both L domains of PERV are involved in virion assembly and release.
FIGURE 4.
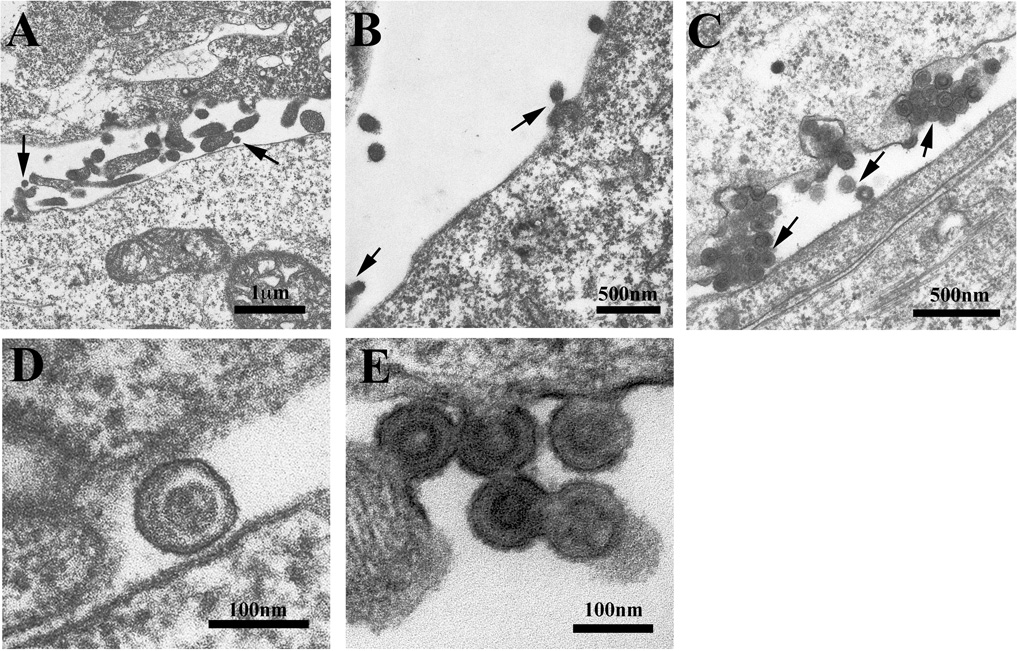
Electron micrographs of 293T cells transfected with the following PERV provirus constructs: (A and D) PPPY-PFAP, (B and E) PPPD-PFAP and (C) PPPY-AAAA. Arrows indicate normal virion budding and phenotypes in (A), tethered viral particles at the cell membrane in (B) and dysmorphic grape-like clusters incompletely budded from the cell surface with tethered viral particles (middle arrow) in (C). Higher magnification images reveal aberrant PPPY-mutant cores compared to PPPY-PFAP (D and E). Micrographs are representative samples of multiple images obtained for each of the L domain mutant phenotypes.
PERV L domains enhance, but are not required for viral release and infectivity
Alanine mutations of both motifs (AAAA-AAAA) results in significantly lower average release efficiency (p<0.01) compared to PPPY-PFAP (Figures 2D and 2E). Nonetheless, we measured an average of 6.87 × 104 ± 3.15 × 10 4 (standard error) viral genomes of AAAA-AAAA in these supernatants demonstrating that viral release occurs even in the absence of both L domains. However, detecting viral genomes in the supernatant does not directly correlate with infectious virus. Therefore, we tested supernatants containing AAAA-AAAA virions for infection of permissive 293T cells. PERV pol genomic DNA copy numbers were measured at 72 hours and 2 weeks post infection. At 72 hours, PERV pol copy numbers in 500 ng of genomic DNA for AAAA-AAAA were readily detected at 4.8 × 104 ± 2.66 × 104. At two weeks the levels increased significantly, consistent with productive infection (2.1 × 105 ± 5.5 × 104). Thus, the L domains of PERV are not absolutely required for release of an infectious virion and a productive infection.
PSAP virions are more infectious than PFAP virions in vitro
Given a hierarchy for PERV release, PSAP>PFAP, we questioned whether there was any difference in the infectivity of these virions. We tested this possibility with supernatants harvested at 72 hours, a time at which virion release is equivalent by both protein and genome quantification. TCID50 values were determined for PPPY-PFAP, PPPY-PSAP and PPPY-AAAA in stably infected 293T producer lines (Table 1; n=3 with 6 replicates for each dilution). PPPY-PSAP virions are 3.5-fold and 3.8-fold more infectious than PPPY-PFAP and PPPY-AAAA, respectively.
Table 1.
| TCID50/mL ± SDa (n=3) | |
|---|---|
| PPPY-PFAP | 8.93×103 ± 3.62×103 |
| PPPY-PSAP | 3.13×104 ± 6.9×103b |
| PPPY-AAAA | 8.17×103 ± 6.10×103b |
SD = Standard Deviation
p<0.03; two-tailed Student’s t-test
The greater infectivity of PPPY-PSAP was also established in single-round infection experiments by qPCR measuring PERV pol copy numbers present in genomic DNA. We tested both 293T cells and HeLa 72 hours after infection. PERV pol copy numbers detected in 250 ng of genomic DNA for PPPY-PSAP are significantly higher compared to PPPY-PFAP and PPPY-AAAA ( p<0.05; Figure 5). These results further support the greater infectivity of PPPY-PSAP compared to PPPY-PFAP and demonstrate that it is not 293T cell-specific.
FIGURE 5.
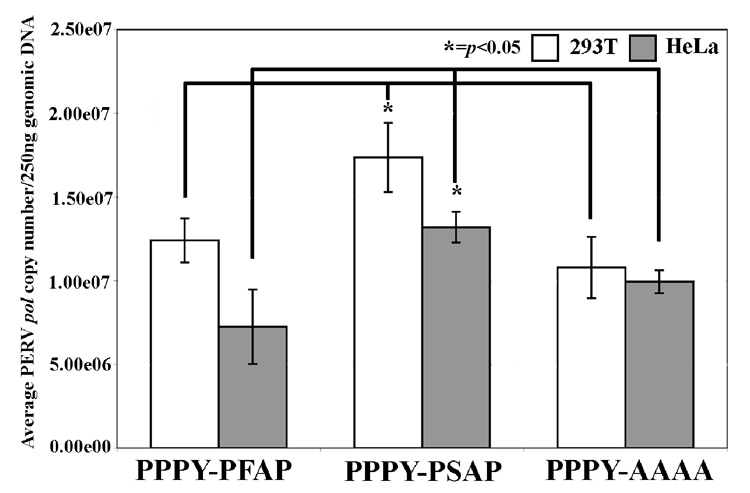
PERV pol copies present in 250ng genomic DNA of 293T and HeLa cells 72 hours after infection with cell-free supernatants from PPPY-PFAP, PPPY-PSAP or PPPY-AAAA PERV. The graph represents the average of three independent experiments. A two-tailed Student’s t-test was applied to determine if PERV pol copy numbers present in PPPY-PSAP were statistically different compared to PPPY-PFAP. PERV pol copy numbers of PPPY-PSAP were 1.4-fold and 1.8-fold more than PPPY-PFAP in 293T and HeLa cells, respectively. PPPY-PSAP PERV pol copy numbers were 1.6-fold and 1.3-fold more than PPPY-PFAP in 293T and HeLa cells, respectively (* = p<0.05).
PSAP virions are more infectious than PFAP virions in vivo
PPPY-PFAP and PPPY-PSAP infectivity was evaluated in vivo using our human PERV-A receptor 2 (HuPAR-2) transgenic mouse model for productive PERV infection (Martina et al., 2006). The HuPAR2 mouse model tests the L domain motifs in an environment akin to a natural host infection where multiple rounds of both viral release and infectivity are required for a productive infection. Cell-free supernatants from stable PPPY-PFAP and PPPY-PSAP producer cell lines were used to infect transgenic and non-transgenic littermates. Mice were injected intraperitoneally and intravenously with a dose of 104 infectious units of either PPPY-PFAP or PPPY-PSAP, normalized based on the TCID50 values determined immediately prior to injections. At 4 and 8 weeks after infection, the mice were sacrificed and multiple tissue compartments were harvested. PERV pol copy numbers were determined in genomic DNA and cellular RNA in the various tissues by qPCR and RT-qPCR, respectively (Table 2).
Table 2.
| DNAa |
RNAb |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| 4 weeks |
4 weeks |
||||||||
| PPPY-PFAP |
PPPY-PSAP |
PPPY-PFAP |
PPPY-PSAP |
||||||
| Tissue | PERV positive animalsc | Avg PERV pol copy number ± SEd | PERV positive animals | Avg PERV pol copy number ± SE | Tissue | PERV positive animals | Avg PERV pol copy number ± SE | PERV positive animals | Avg PERV pol copy number ± SE |
| Liver | 3/7 | 596 ± 98.1 | 6/8 | 1,644 ± 404 | Liver | 2/7 | 327 ± 199 | 5/8 | 692 ± 96.2 |
| Lung | 0/7 | 4/7 | 40.0 ± 6.9 | Lung | N/Ae | 0/7 | |||
| Spleen | 0/8 | 7/8 | 48.0 ± 13.8 | Spleen | N/A | 6/8 | 766 ± 498 | ||
| Small Bowel | 0/8 | 6/8 | 363 ± 103 | Small Bowel | N/A | 6/8 | 1,351 ± 535.6 | ||
| Kidney | 8/8 | 42.5 ± 9.12 | 6/8 | 90.0 ± 26.3 | Kidney | 4/8 | 71 ± 8 | 3/8 | 43 ± 10.4 |
| Heart | 0/8 | 0/8 | Heart | N/A | N/A | ||||
| Brain | 8/8 | 986 ± 109 | 7/8 | 1,043 ± 523.5 | Brain | 5/8 | 845 ± 429 | 6/8 | 512 ± 165 |
| Bone marrow | 6/8 | 17,200 ± 3,079 | 7/8 | 26,741± 3,984 | Bone marrow | 6/8 | 194 ± 46.1 | 6/8 | 246 ± 38.9 |
|
8 weeks |
8 weeks |
||||||||
| Liver | 1/9 | 219 | 7/9 | 2,844 ± 527.3 | Liver | 0/9 | 5/9 | 1,393 ± 719.1 | |
| Lung | 1/9 | 18.7 | 6/9 | 29 ± 19 | Lung | 0/9 | 5/9 | 78 ± 24 | |
| Spleen | 0/9 | 6/9 | 367 ± 207 | Spleen | N/A | 0/9 | |||
| Small Bowel | 3/9 | 197 ± 89.5 | 4/9 | 375 ± 63.5 | Small Bowel | 3/9 | 438 ± 342 | 4/9 | 415 ± 222 |
| Kidney | 7/9 | 765 ± 505 | 8/9 | 1,574 ± 490.7 | Kidney | 3/9 | 184 ± 42.3 | 4/9 | 225 ± 36.0 |
| Heart | 0/9 | 6/9 | 178 ± 64.1 | Heart | N/A | N/A | |||
| Brain | 5/9 | 547 ± 106 | 6/9 | 1,305 ± 336 | Brain | 2/9 | 298 ± 192 | 6/9 | 803 ± 168 |
| Bone marrow | 5/9 | 1,642 ± 381.8 | 7/9 | 29,947 ± 16,999 | Bone marrow | 5/9 | 553 ± 132 | 6/9 | 1,576 ± 293 |
Data are expressed as copy number per 2 µg of genomic DNA
Data are expressed as copy number per 1 µg of total RNA
An animal was designated “PERV positive” if any result was >10 copies in 2 µg of DNA or >10 copies in 1 µg of total RNA (after background subtraction)
SE = standard error
N/A = tissue not analyzed because DNA was demonstrated as negative
PERV pol was not detected in the tissues of the non-transgenic littermates (data not shown). As early as 4 weeks after infection, PERV pol copy numbers are evident in both DNA and RNA of the majority of animals and tissues of the PPPY-PSAP animals at levels consistently higher than the PPPY-PFAP group. These differences remain clear at 8 weeks. The in vitro infectivity results are therefore recapitulated in vivo, indicating that there is a real infectivity advantage for PPPY-PSAP PERV based on selection of the second L domain.
DISCUSSION
Efficient viral assembly, release and infectivity govern virion fitness. Here we provide the first report for the relative contributions of the L domains in PERV Gag to virion fitness. PERV contains two functional L domains, PPPY and P(F/S)AP, that both contribute to viral assembly and release. Mutation of the PFAP domain results in classic L domain cell-surface arrest and abnormal particle assembly phenotypes as observed by electron microscopy; therefore, we define PFAP as a novel L domain motif variant of P(T/S)AP. Mutation of the PPPY domain results in aberrant viral assembly phenotypes, indicating PPPY is a functional PERV L domain. However, the two motifs differentially contribute to early and late viral release and late particle infectivity. One functional hierarchy is represented by PPPY over P(F/S)AP for relatively late viral release (72 hours post-transfection). Another functional hierarchy is PSAP over PFAP for early viral release (24 hours post-transfection). Yet despite the significant reduction in viral genomes released, double L domain mutant virions, AAAA-AAAA are still infectious. Interestingly, we also demonstrated that PSAP virions are more infectious as compared to PFAP and AAAA. Therefore, in the context of viral evolution, L domains are selected for enhancing the efficiency of virion release and infectivity, but are not the sole determinants.
PPPY is the dominant PERV L domain in viral release. Mutation of the entire domain (AAAA-PFAP) resulted in a significant reduction of viral particle release measured at the viral genome and the protein level. Additionally, single residue mutations (PLPY-PFAP and PPPD-PFAP) also resulted in significant reductions of viral particle release. Our results for PERV are consistent with the functional dominance of PPPY in other retroviruses containing more than one L domain. Decreased viral particle release due to single or multiple residue PPPY mutants has been demonstrated for HTLV-1 (Heidecker et al., 2004; Le Blanc et al., 2002; Wang, Machesky, and Mansky, 2004) and MPMV (Gottwein et al., 2003; Yasuda and Hunter, 1998). Deletion of the PPPY motif in M-MuLV, a gammaretrovirus like PERV that also contains PSAP and a third L domain, YPDL (Table 4), reduced release by 80% as measured by supernatant RT activity (Yuan et al., 2000). The functional hierarchy of PPPY over additional L domain sequence in viral release is therefore not limited to PERV. In fact, all the gammaretroviral Gag sequences in the public domain contain the PPPY motif (Table 4).
The PPPY motif is also conserved for the deltaretroviruses and is the dominant L domain required for both Bovine Leukemia Virus (Wang, Norris, and Mansky, 2002) and HTLV-1 (Bouamr et al., 2003; Heidecker et al., 2004; Le Blanc et al., 2002; Wang, Machesky, and Mansky, 2004). Given the documented dominance of the PPPY motif in viruses with more than one functional L domain across multiple viral families, we propose that this has led to PPPY’s conservation in viral evolution. Further support for this hypothesis is the recent data suggesting that one step in the endogenization of GALV in the Koala’s genome involved the loss of viral fitness through mutation of its PPIY motif (Oliveira et al., 2007). However, the secondary domain in a PPPY-dominant context must also have a function in order to be carried through the course of viral evolution.
PERV presents two natural variants for the second L domain sequence, PFAP and PSAP. We have demonstrated a functional hierarchy of these two domains in early PPPY-dominant release and late particle infectivity. PPPY-PSAP was significantly greater than PPPY-PFAP in early release (24 hours) at the level of viral genomes. The enhanced efficiency of PSAP over PFAP could be attributed to the chemical similarity of the serine side-chain of PSAP to that of threonine in PTAP while the phenylalanine of PFAP lacks the hydroxyl group. This may influence the binding affinity of viral-host factors involved in viral release. Consistent with this hypothesis is that binding of Tsg101 to PTAP has a 7-fold higher affinity than to PSAP (Pornillos et al., 2003). However, we also have shown that whatever early gains in efficiency are observed for PSAP over PFAP, that by 72 hours, virion release is equivalent.
An unexpected finding was that PPPY-PSAP is ~3.5-fold more infectious than PPPY-PFAP by TCID50. This infectivity difference is not due to differences in RT activity, as assays of both PFAP and PSAP virions demonstrated equivalent amounts (data not shown). Moreover, both 293T and HeLa cells showed the same relative increase for PPPY-PSAP infection, demonstrating that the difference is not cell type-dependent. We also demonstrated that infectivity is greater for PSAP than PFAP in vivo using our HuPAR-2 transgenic mouse model for PERV infection (Martina et al., 2006).
Furthermore, PPPY-AAAA demonstrated in vitro infectivity similar to that of PPPY-PFAP. These results indicate that the second domain is not required for infectivity. Thus, it is PSAP that appears to be responsible for the increased infectious virion fitness demonstrated in vitro and in vivo. Recently, the mutation of PSAP in Vesicular Stomatitis Virus (VSV) was shown to affect viral pathogenesis. Mutation of the serine to threonine, yielding an HIV-1-like PTAP motif, attenuated infection both in vitro and in vivo (Irie et al., 2007). These data support our observation that PSAP is playing a role in viral infectivity and suggest that it is an inherent property of that particular motif sequence. Excluding PERV, four out of eight known gammaretroviral Gag sequences have PSAP in addition to PPPY and two others have the related motif, PSGP (Table 4). From our experiments, it would be hypothesized that the prevalence and evolutionary survival of PSAP as a secondary motif is due to its role in particle infectivity.
Interestingly, in the presence of the dominant PPPY, both the PSAP and PFAP second L domains appear to attenuate early PERV release (24 hours; Figure 3). These results suggest that another function of the second L domains may be to regulate the pace of viral assembly or release at the membrane to insure the completion of the process and release of fully functional virions. Mechanistically, this regulation might represent a competition or an interplay of host cell factors binding the PPPY domain (e.g. NEDD4) and the P(F/S)AP domain (e.g. Tsg101). Therefore, future studies could address these questions by studying which host cell factors are binding PERV L domains and mapping their potential interactions.
It is important to note that viral genomes are detected in the supernatant of the double L domain alanine mutant (AAAA-AAAA), albeit at significantly reduced levels. We demonstrated that these virions are infectious and the levels increase through several passages of the infected target cells proving that these are productive infections. Therefore, L domain-independent mechanisms or presently unknown motifs must contribute to PERV release under these conditions. The double L domain mutants of MPMV, HTLV-1 and Ebola Virus VP40 demonstrated a similar phenomenon of low but measurable viral release levels (Gottwein et al., 2003; Heidecker et al., 2004; Neumann et al., 2005). Even HIV-1 lacking the p6 protein, which contains PTAP, replicates in some T-cell lines and primary PBMCs, albeit with a delay (Demirov, Orenstein, and Freed, 2002). Thus, the evidence at present indicates that L domains have evolved and are selected to enhance the efficiency of viral assembly and release but they are not absolutely required.
In conclusion, we have demonstrated a hierarchy of functional L domains for PERV virion release and infectivity. The dominant role of PERV PPPY in the presence of an additional L domain motif is consistent with published results for other multiple L domain-containing viruses. Our results represent the first functional studies of viral release and infectivity for the second L domains, P(F/S)AP, in gammaretroviruses. We demonstrate that these are functional in release and impact on infectivity in vitro and in vivo. Thus, both L domains have a role in determining viral fitness and selective pressures for maintaining a productive viral life cycle, explaining the conservation of multiple L domain motifs in viruses during viral evolution.
METHODS
Cell lines and plasmid constructs
293T and HeLa cells were maintained in DMEM (Gibco, Carlsbad, CA) supplemented with 10% Fetal Bovine Serum (HyClone, Logan, UT), 5% 1M Hepes (Gibco), 5% 100mM sodium pyruvate (Gibco) and 5% 100X Penicillin-Streptomycin-Glutamine (Gibco). Cells were incubated at 37°C in the presence of 7% CO2. The wild-type PERV refers to the high infectious titer PERV-A14/220 clone (PERV A/C recombinant; GenBank AY570980) (Harrison et al., 2004) that was a kind gift from Dr. Y. Takeuchi (University College London). Mutations were introduced using site-directed mutagenesis with the QuikChange Mutagenesis kit (Stratagene, Foster City, CA) according to the manufacturer’s protocol.
Viral release assay by RNA genomes
3×106 293T cells were transfected with 10µg of wild-type or mutant PERV-A14/220 proviral plasmid using ProFection Mammalian Transfection System (Promega, Madison, WI). Either 24 or 72 hours after transfection, 8mL of supernatant was filtered (0.45µm) and ultracentrifuged (23,000rpm × 2 hours, 4°C). Pelleted viral genomic RNA was extracted (QIAamp Viral RNA Mini Kit; Qiagen, Valencia, CA) and purified using the RNeasy Mini Kit (Qiagen), including on-column DNaseI digestion. 8µl of supernatant viral genomic RNA was used for pol-specific cDNA synthesis with the SuperScript First Strand Synthesis kit (Invitrogen; Carlsbad, CA). Cellular RNA was also extracted using the RNeasy Mini Kit (Qiagen). 2.5 µg of purified cellular RNA was used for pol-specific cDNA synthesis. A volumetric equivalent of 53.3µl viral supernatant and a mass equivalent of 11.25ng of cellular RNA were used for PERV pol detection by TaqMan qPCR (Ericsson et al., 2003) on the Applied Biosystems 7900HT Fast Real Time PCR System (Foster City, CA). Copy numbers from untransfected 293T cells (negative control) and supernatants (typically <100 copies) were subtracted from the experimental values of PERV pol copy numbers. Average Release Efficiency was calculated for the RT-qPCR data using the following equation: (copy # supernatant PERV pol RNA / copy # supernatant PERV pol RNA + copy # cellular PERV pol RNA).
Western blot for virion release
3×106 293T cells were transfected with 10µg of wild-type or mutant PERV-A14/220 proviral plasmids using ProFection Mammalian Transfection System (Promega). 72 hours after transfection, 8mL of supernatant was filtered (0.45µm) and ultracentrifuged (23,000rpm × 2 hours, 4°C). Pelleted virions were resuspended in 100µl 5× Lamelli loading buffer. 2×106 transfected 293T cells were resuspended in 400µl 5× Lamelli loading buffer. 40µl of the resuspended virion pellet and 25µl of the cell preparation in loading buffer was run on a 10% NuPAGE Bis-Tris gel (Invitrogen). Protein was transferred to Hybond-LFP PVDF membrane (GE Healthcare, Piscataway, NJ) and immunoblotted 1:1000 with the primary mouse anti-Gag39 monoclonal antibody (kind gift of Dr. G. Byrne (Xu et al., 2003)) in addition to 1:3000 rabbit anti-actin (Sigma) for cellular protein blots. Secondary antibodies were ECL Plex goat anti-mouse Cy5 and goat anti-rabbit Cy3 (GE Healthcare) used at 1:1250. Blots were visualized using the TyphoonTrio Imager (GE Healthcare) and analyzed using ImageQuant 5.1(GE Healthcare) in terms of average volume density. We used these results to calculate Average Release Efficiency in terms of viral protein detected in the supernatants and cells by replacing the copy # term in the equation described above with the average volume density.
Electron microscopy
As previously described (Gilula, Epstein, and Beers, 1978), cells were grown in 35mm2 dishes, fixed for 30 minutes on ice in 2.5% glutaraldehyde in 0.1M Na cacodylate buffer (pH7.3). Following treatment with 1% osmium tetroxide for 1 hour at room temperature, each sample was incubated in 0.5% tannic acid in 0.05M cacodylate buffer for 30 minutes then rinsed in 1% Na2SO4 for 10 min and then rinsed in 0.1M cacodylate buffer. The cells were dehydrated in a graded ethanol series then cleared in HPMA (2-hydroxypropyl methacrylate) twice for 15 minutes each and embedded in LX-112 (Ladd, Burlington VT). After polymerization at 60°C for 24 hours, the plastic petri dish was broken away and the thin resin disk was cut up into small pieces and attached to blank blocks with Superglue. Thin sections were cut using a diamond knife, mounted on copper slot grids, stained with uranyl acetate and lead citrate and examined on a Philips CM-100 electron microscope. Specific images were recorded on Kodak SO-163 film and negatives were scanned with a Fujifilm FineScan 2750xl.
Determination of TCID50
293T (104 cells/well) were seeded in 48-well plates. Cells were infected with 4-fold serial dilutions (undiluted to 1:16,384; 6 wells/dilution) of cell-free supernatant from PERV-infected, stable cell lines. Genomic DNA was extracted after 72 hours with DirectPCR lysis reagent (Viagen, Los Angeles, CA) and analyzed by PCR for PERV-A/C env using the following: fwd primer 5’-ATGTCTGCCTTCGATCAGTAATCCC-3’ and rev primer 5’-CTCAAACCACCCTTGAGTAGTTTCC-3’. TCID50/ml was calculated using the Karber formula.
Assessment of PERV infection by DNA PCR
2×105 293T or HeLa cells were seeded in 6-well plates. Eight hours later, cells were infected with 1mL of cell-free supernatant in the presence of 4µg/mL polybrene and left at 37°C overnight. After sixteen hours, the supernatant was replaced with 2mL fresh media. Genomic DNA was isolated 72 hours later and PERV pol copies in 500ng were determined by PERV pol qPCR.
In vivo infectivity
FVB/Nj mice transgenic for the human PAR-2 (HuPAR-2) receptor (Martina et al., 2006) were injected both intraperitoneally and intravenously with cell-free supernatants of either PERV wild type (PFAP) or L domain variant (PSAP). The amount of infectious virus injected was normalized based on TCID50 determinations. Organs were harvested at 4 and 8 weeks post-exposure. 2µg of extracted DNA and 1µg of extracted RNA were analyzed for the presence of PERV pol by qPCR. HuPAR-2 negative littermates injected with wild-type PERV were used as controls.
Table 3.
| VIRUS | ACCESSION NUMBER | L DOMAIN TYPE | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
PPXY |
P(T/S)AP |
YXXL |
|||||||
| PERV-A | AAF65926 | 149PPPY151 | 161PSAP164 | ||||||
| PERV-C | AAC16761 | 149PPPY152 | 161PFAP164 | ||||||
| PERV-A 14/220a | AAT77166 | 149PPPY152 | 161PFAP164 | ||||||
| GALV (Gibbon Ape Leukemia Virus) | P21416 | 161PPPY164 | 122PPIY125 | 115PSAP118 | |||||
| MLV (Murine Leukemia Virus) | AAB03090 | 161PPPY164 | 109PTAP112 | 115PSGP118 | 129YPAL132 | ||||
| MuLV (Friend Murine Leukemia Virus) | NP_040332 | 162PPPY165 | 111PSAP114 | 131YPAL134 | |||||
| M-MuLV (Moloney Murine Leukemia Virus) | AAB59942 | 162PPPYb165 | 111PSAP114 | 131YPAL135 | 333YQRL336 | ||||
| FeLV (Feline Leukemia Virus) | AAA43055 | 157PPPY160 | 168PSGP171 | ||||||
| FSV (Feline Sarcoma Virus) | AAA43046 | 157PPPY160 | 168PSGP171 | ||||||
| ARV (Avian Reticuloendotheliosis Virus) | AAZ57417 | 154PPPY157 | 127PSAP131 | ||||||
PPPY L domain has been documented in the published literature as functional for viral release and assembly (Yuan, Li, and Goff, 1999)
Present Manuscript
ACKNOWLEDGEMENTS
We would like to thank Dr. Malcom Wood of the TSRI Microscopy Core for preparation of the EM samples and image acquisition. This work was supported by NIH grant R01 AI52349, an AHA Fellowship Award to KTM (0615044Y) and the Molly Baber Research Fund.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Akiyoshi DE, Denaro M, Zhu H, Greenstein JL, Banerjee P, Fishman JA. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J Virol. 1998;72(5):4503–4507. doi: 10.1128/jvi.72.5.4503-4507.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouamr F, Melillo JA, Wang MQ, Nagashima K, de Los Santos M, Rein A, Goff SP. PPPYVEPTAP motif is the late domain of human T-cell leukemia virus type 1 Gag and mediates its functional interaction with cellular proteins Nedd4 and Tsg101 [corrected] J Virol. 2003;77(22):11882–11895. doi: 10.1128/JVI.77.22.11882-11895.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280(49):40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Freed EO. Retrovirus budding. Virus Res. 2004;106(2):87–102. doi: 10.1016/j.virusres.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Demirov DG, Orenstein JM, Freed EO. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J Virol. 2002;76(1):105–117. doi: 10.1128/JVI.76.1.105-117.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorweiler IJ, Ruone SJ, Wang H, Burry RW, Mansky LM. Role of the human T-cell leukemia virus type 1 PTAP motif in Gag targeting and particle release. J Virol. 2006;80(7):3634–3643. doi: 10.1128/JVI.80.7.3634-3643.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericsson TA, Takeuchi Y, Templin C, Quinn G, Farhadian SF, Wood JC, Oldmixon BA, Suling KM, Ishii JK, Kitagawa Y, Miyazawa T, Salomon DR, Weiss RA, Patience C. Identification of receptors for pig endogenous retrovirus. Proc Natl Acad Sci U S A. 2003;100(11):6759–6764. doi: 10.1073/pnas.1138025100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. Viral late domains. J Virol. 2002;76(10):4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilula NB, Epstein ML, Beers WH. Cell-to-cell communication and ovulation. A study of the cumulus-oocyte complex. J Cell Biol. 1978;78(1):58–75. doi: 10.1083/jcb.78.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of mutations affecting the p6 gag protein on human immunodeficiency virus particle release. Proc Natl Acad Sci U S A. 1991;88(8):3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Bodem J, Muller B, Schmechel A, Zentgraf H, Krausslich HG. The Mason-Pfizer monkey virus PPPY and PSAP motifs both contribute to virus release. J Virol. 2003;77(17):9474–9485. doi: 10.1128/JVI.77.17.9474-9485.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwein E, Krausslich HG. Analysis of human immunodeficiency virus type 1 Gag ubiquitination. J Virol. 2005;79(14):9134–9144. doi: 10.1128/JVI.79.14.9134-9144.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison I, Takeuchi Y, Bartosch B, Stoye JP. Determinants of high titer in recombinant porcine endogenous retroviruses. J Virol. 2004;78(24):13871–13879. doi: 10.1128/JVI.78.24.13871-13879.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidecker G, Lloyd PA, Fox K, Nagashima K, Derse D. Late assembly motifs of human T-cell leukemia virus type 1 and their relative roles in particle release. J Virol. 2004;78(12):6636–6648. doi: 10.1128/JVI.78.12.6636-6648.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Orenstein JM, Martin MA, Freed EO. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69(11):6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie T, Carnero E, Okumura A, Garcia-Sastre A, Harty RN. Modifications of the PSAP region of the matrix protein lead to attenuation of vesicular stomatitis virus in vitro and in vivo. J Gen Virol. 2007;88(Pt 9):2559–2567. doi: 10.1099/vir.0.83096-0. [DOI] [PubMed] [Google Scholar]
- Kikonyogo A, Bouamr F, Vana ML, Xiang Y, Aiyar A, Carter C, Leis J. Proteins related to the Nedd4 family of ubiquitin protein ligases interact with the L domain of Rous sarcoma virus and are required for gag budding from cells. Proc Natl Acad Sci U S A. 2001;98(20):11199–11204. doi: 10.1073/pnas.201268998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolber-Simonds D, Lai L, Watt SR, Denaro M, Arn S, Augenstein ML, Betthauser J, Carter DB, Greenstein JL, Hao Y, Im GS, Liu Z, Mell GD, Murphy CN, Park KW, Rieke A, Ryan DJ, Sachs DH, Forsberg EJ, Prather RS, Hawley RJ. Production of alpha-1,3-galactosyltransferase null pigs by means of nuclear transfer with fibroblasts bearing loss of heterozygosity mutations. Proc Natl Acad Sci U S A. 2004;101(19):7335–7340. doi: 10.1073/pnas.0307819101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Blanc I, Prevost MC, Dokhelar MC, Rosenberg AR. The PPPY motif of human T-cell leukemia virus type 1 Gag protein is required early in the budding process. J Virol. 2002;76(19):10024–10029. doi: 10.1128/JVI.76.19.10024-10029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Tissier P, Stoye JP, Takeuchi Y, Patience C, Weiss RA. Two sets of human-tropic pig retrovirus. Nature. 1997;389(6652):681–682. doi: 10.1038/39489. [DOI] [PubMed] [Google Scholar]
- Licata JM, Johnson RF, Han Z, Harty RN. Contribution of ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J Virol. 2004;78(14):7344–7351. doi: 10.1128/JVI.78.14.7344-7351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louz D, Bergmans HE, Loos BP, Hoeben RC. Reappraisal of biosafety risks posed by PERVs in xenotransplantation. Rev Med Virol. 2008;18(1):53–65. doi: 10.1002/rmv.559. [DOI] [PubMed] [Google Scholar]
- Martin U, Winkler ME, Id M, Radeke H, Arseniev L, Takeuchi Y, Simon AR, Patience C, Haverich A, Steinhoff G. Productive infection of primary human endothelial cells by pig endogenous retrovirus (PERV) Xenotransplantation. 2000;7(2):138–142. doi: 10.1034/j.1399-3089.2000.00052.x. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003;100(21):12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Zang T, Bieniasz PD. HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001;7(12):1313–1319. doi: 10.1038/nm1201-1313. [DOI] [PubMed] [Google Scholar]
- Martina Y, Marcucci KT, Cherqui S, Szabo A, Drysdale T, Srinivisan U, Wilson CA, Patience C, Salomon DR. Mice transgenic for a human porcine endogenous retrovirus receptor are susceptible to productive viral infection. J Virol. 2006;80(7):3135–3146. doi: 10.1128/JVI.80.7.3135-3146.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann G, Ebihara H, Takada A, Noda T, Kobasa D, Jasenosky LD, Watanabe S, Kim JH, Feldmann H, Kawaoka Y. Ebola virus VP40 late domains are not essential for viral replication in cell culture. J Virol. 2005;79(16):10300–10307. doi: 10.1128/JVI.79.16.10300-10307.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira NM, Satija H, Kouwenhoven IA, Eiden MV. Changes in viral protein function that accompany retroviral endogenization. Proc Natl Acad Sci U S A. 2007;104(44):17506–17511. doi: 10.1073/pnas.0704313104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J Cell Biol. 2003;162(3):425–434. doi: 10.1083/jcb.200302138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puffer BA, Parent LJ, Wills JW, Montelaro RC. Equine infectious anemia virus utilizes a YXXL motif within the late assembly domain of the Gag p9 protein. J Virol. 1997;71(9):6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79(5):2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Morales C, Pescia C, Chatellard-Causse C, Sadoul R, Bertrand E, Basyuk E. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. J Biol Chem. 2005;280(29):27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114(6):689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Patience C, Magre S, Weiss RA, Banerjee PT, Le Tissier P, Stoye JP. Host range and interference studies of three classes of pig endogenous retrovirus. J Virol. 1998;72(12):9986–9991. doi: 10.1128/jvi.72.12.9986-9991.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Laan LJ, Lockey C, Griffeth BC, Frasier FS, Wilson CA, Onions DE, Hering BJ, Long Z, Otto E, Torbett BE, Salomon DR. Infection by porcine endogenous retrovirus after islet xenotransplantation in SCID mice. Nature. 2000;407(6800):90–94. doi: 10.1038/35024089. [DOI] [PubMed] [Google Scholar]
- VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, Carter CA. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag) Proc Natl Acad Sci U S A. 2001;98(14):7724–7729. doi: 10.1073/pnas.131059198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Machesky NJ, Mansky LM. Both the PPPY and PTAP motifs are involved in human T-cell leukemia virus type 1 particle release. J Virol. 2004;78(3):1503–1512. doi: 10.1128/JVI.78.3.1503-1512.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Norris KM, Mansky LM. Analysis of bovine leukemia virus gag membrane targeting and late domain function. J Virol. 2002;76(16):8485–8493. doi: 10.1128/JVI.76.16.8485-8493.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills JW, Cameron CE, Wilson CB, Xiang Y, Bennett RP, Leis J. An assembly domain of the Rous sarcoma virus Gag protein required late in budding. J Virol. 1994;68(10):6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, Wong S, VanBrocklin M, Federspiel MJ. Extended analysis of the in vitro tropism of porcine endogenous retrovirus. J Virol. 2000;74(1):49–56. doi: 10.1128/jvi.74.1.49-56.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirblich C, Bhattacharya B, Roy P. Nonstructural protein 3 of bluetongue virus assists virus release by recruiting ESCRT-I protein Tsg101. J Virol. 2006;80(1):460–473. doi: 10.1128/JVI.80.1.460-473.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Cameron CE, Wills JW, Leis J. Fine mapping and characterization of the Rous sarcoma virus Pr76gag late assembly domain. J Virol. 1996;70(8):5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Sharma A, Okabe J, Cui C, Huang L, Wei YY, Wan H, Lei Y, Logan JS, Levy MF, Byrne GW. Serologic analysis of anti-porcine endogenous retroviruses immune responses in humans after ex vivo transgenic pig liver perfusion. Asaio J. 2003;49(4):407–416. [PubMed] [Google Scholar]
- Yasuda J, Hunter E. A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release. J Virol. 1998;72(5):4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74(16):7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Li X, Goff SP. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. Embo J. 1999;18(17):4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]


