Abstract
Following primary infection, KSHV establishes a lifelong persistent latent infection in the host. The mechanism of KSHV latency is not fully understood. The latent nuclear antigen (LANA or LNA) encoded by ORF73 is one of a few viral genes expressed during KSHV latency, and is consistently detected in all KSHV-related malignancies. LANA is essential for KSHV episome persistence, and regulates the expression of viral lytic genes through epigenetic silencing, and inhibition of the expression and transactivation function of the key KSHV lytic replication initiator RTA (ORF50). In this study, we used a genetic approach to examine the role of LANA in regulating KSHV lytic replication program. Deletion of LANA did not affect the expression of its adjacent genes vCyclin (ORF72) and vFLIP (ORF71). In contrast, the expression levels of viral lytic genes including immediate-early gene RTA, early genes MTA (ORF57), vIL-6 (ORF-K2) and ORF59, and late gene ORF-K8.1 were increased before and after viral lytic induction with 12-O-tetradecanoyl-phorbol-13-acetate and sodium butyrate. This enhanced expression of viral lytic genes was also observed following overexpression of RTA with or without simultaneous chemical induction. Consistent with these results, the LANA mutant cells produced more infectious virions than the wild-type virus cells did. Furthermore, genetic repair of the mutant virus reverted the phenotypes to those of wild-type virus. Together, these results have demonstrated that, in the context of viral genome, LANA contributes to KSHV latency by regulating the expression of RTA and its downstream genes.
Keywords: KSHV, LANA, viral latency, lytic replication, viral transcriptional program, reverse genetics
Introduction
Kaposi’s sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8 (HHV8), is a member of the gammaherpesvirus family. KSHV is associated with all four clinical forms of Kaposi’s sarcoma (KS), including classical KS, African endemic KS, immunosuppressive organ transplant KS, and AIDS-related KS (AIDS-KS) (Greene et al., 2007). AIDS-KS is the most common neoplasm in AIDS patients, and has evolved as an important health concern worldwide as a result of the spread of AIDS epidemics (Greene et al., 2007). KSHV is also associated with several other lymphoproliferative diseases including primary effusion lymphoma (PEL) and multicentric Castleman’s disease (MCD).
Similar to other herpesviruses, KSHV establishes a lifelong persistent latent infection in the host following primary infection. During latency, the virus is maintained as a non-integrated extrachromosomal episome, and only expresses several latent genes from the viral latent genomic locus, which includes the latent nuclear antigen (LANA or LNA) encoded by ORF73, vCyclin encoded by ORF72, vFLIP encoded by ORF-K13, and a dozen of microRNAs (Cai et al., 2005; Dittmer et al., 1998; Gao et al., 1996; Pfeffer et al., 2005; Rainbow et al., 1997; Samols et al., 2005; Sarid et al., 1998). KSHV latency can be disrupted by intracellular and extracellular signals, resulting in the expression of viral lytic genes and production of infectious virions (Deng et al., 2007). KSHV replication and transcription activator (RTA) encoded by ORF50 is necessary and sufficient for the activation of KSHV lytic replication program (Lukac et al., 1999; Lukac et al., 1998; Sun et al., 1998; Xu et al., 2005). Overexpression of RTA results in the expression of KSHV lytic genes and production of KSHV virions. A number of chemical inducers such as 12-O-tetradecanoyl-phorbol-13-acetate (TPA) and sodium butyrate (SB) are also capable of reactivating KSHV from latency (Miller et al., 1996; Moore et al., 1996).
In KS lesions, the majority of the tumor cells are latently infected by KSHV, indicating the essential role of viral latent infection in the development of the malignancy (Greene et al., 2007). However, a small subset of KS tumor cells also undergoes spontaneous lytic replication. It is believed that these lytic cells could generate infectious virions for spread to other cells, and simultaneously produce KSHV-encoded homologs of cellular cytokines (Greene et al., 2007). In addition, KSHV lytic replication and de novo infection could also induce specific cellular cytokines (Carroll et al., 2004; Qian et al., 2007; Xie et al., 2005; Ye et al., 2007). These viral and cellular cytokines could promote tumor inflammation and angiogenesis, and enhance the proliferation, invasion and dissemination of the tumor cells (Greene et al., 2007). As a result, both latent and lytic phases of viral lifecycle contribute to the pathogenesis of KSHV-induced malignancies.
In spite of intensive studies in the last decade, the mechanism of KSHV latency remains unclear (Ganem, 2006). It is postulated that some viral latent products might modulate KSHV latency. Indeed, using a reverse genetic approach, we have recently demonstrated that the viral latent protein vFLIP inhibits KSHV lytic replication and promote viral latency by repressing the AP-1 pathway (Ye et al., 2008).
LANA is another viral latent protein that has a critical role in KSHV latency. LANA is a 1,162 amino acids nuclear protein containing three distinct domains: the N-terminal proline-rich domain, the central internal repeat domain (IRD) containing several repetitive acidic residues, and the C-terminal domain with a nuclear localization signal (Gao et al., 1999; Zhang et al., 2000). Using terminal repeat (TR)-containing plasmids, it has been shown that LANA binds to the TR sequence of the viral genome to mediate its replication, and tethers the viral genome to cellular chromosomes for proper segregation into daughter cells during mitosis (Ballestas and Kaye, 2001; Cotter and Robertson, 1999; Hu et al., 2002; Krithivas et al., 2002). Transposon-mediated genetic disruption has confirmed that LANA is indeed necessary for the establishment and persistence of KSHV episome in mammalian cells (Ye et al., 2004).
In addition to episome persistence, LANA also modulates KSHV latency by regulating cell growth and survival, and the expression of viral and cellular genes. LANA targets cellular tumor suppressor p53 and pRb pathways, and oncogenic β-catenin and c-Myc pathways to promote the growth of KSHV latent cells (Cai et al., 2006; Friborg et al., 1999; Fujimuro et al., 2003; Liu et al., 2007; Radkov et al., 2000). Several studies have shown that LANA regulates the expression of cellular and viral genes through interaction with a variety of transcriptional activators or repressors such as ATF4/CREB2, Sp1, components of the mSin3 complex, and a newly identified centromeric protein KLIP1 (Krithivas et al., 2000; Lim et al., 2000; Pan et al., 2003; Verma et al., 2004). Furthermore, LANA regulates the expression of viral genes by mediating epigenetic modifications of the viral genome (Lu et al., 2006). More recently, it has been shown that LANA downregulates the expression of RTA by inhibiting its promoter activities through direct binding to the promoter and indirect binding to other transcription factors such as RBP-Jκ and Sp1, both of which also modulate the expression of RTA (Lan et al., 2005; Verma et al., 2004). In addition, LANA directly interacts with RTA to prevent its autoactivation and transactivation of other downstream viral lytic genes (Lan et al., 2005; Lan et al., 2004). Together, these studies suggest that LANA might regulate KHSV latency beyond its episome persistence function. In this study, we assessed LANA’s role in KSHV latency focusing on LANA repression of viral lytic replication program in the context of whole viral genome by generating a KSHV LANA deletion mutant BAC36ΔLANA using the KSHV BAC36 genetic system (Zhou et al., 2002). Our results revealed that deletion of LANA increased the expression of all classes of viral lytic genes including immediate-early (IE), early and late genes, and the production of KSHV infectious virions. These results suggest a role of LANA in the control of KSHV latency by repressing the expression of viral lytic genes and inhibiting the viral lytic replication program.
Results
Generation of a KSHV LANA deletion mutant BAC36ΔLANA and its revertant BAC36ΔLANArt
To examine the role of KSHV LANA in regulating viral transcriptional program and lytic replication, we generated a LANA deletion mutant BAC36ΔLANA and its revertant BAC36ΔLANArt by targeting mutagenesis using the BAC36 genetic system (Zhou et al., 2002). To delete LANA from the KSHV genome, a DNA fragment containing a kanamycin-resistance cassette (Kanr) flanked by two LoxP sites and two 50-bp DNA fragments of KSHV sequences immediately outside the LANA region was used for homologous recombination (Fig. 1A). An intermediate recombinant genome, termed BAC36ΔLANA-Kan, in which LANA was replaced with a Kanr was selected. Upon Cre-mediated recombination, the Kanr was excised resulting in the generation of a KSHV mutant with LANA deleted termed BAC36ΔLANA. This mutant virus contains a single loxP site replacing the LANA. A similar two-steps strategy was used to generate the revertant recombinant virus BAC36ΔLANArt. However, instead of a Kanr, we used a zeocin-resistance cassette (Zeor) as a selection marker (Fig. 1A). Details of the procedures were described in Materials and Methods.
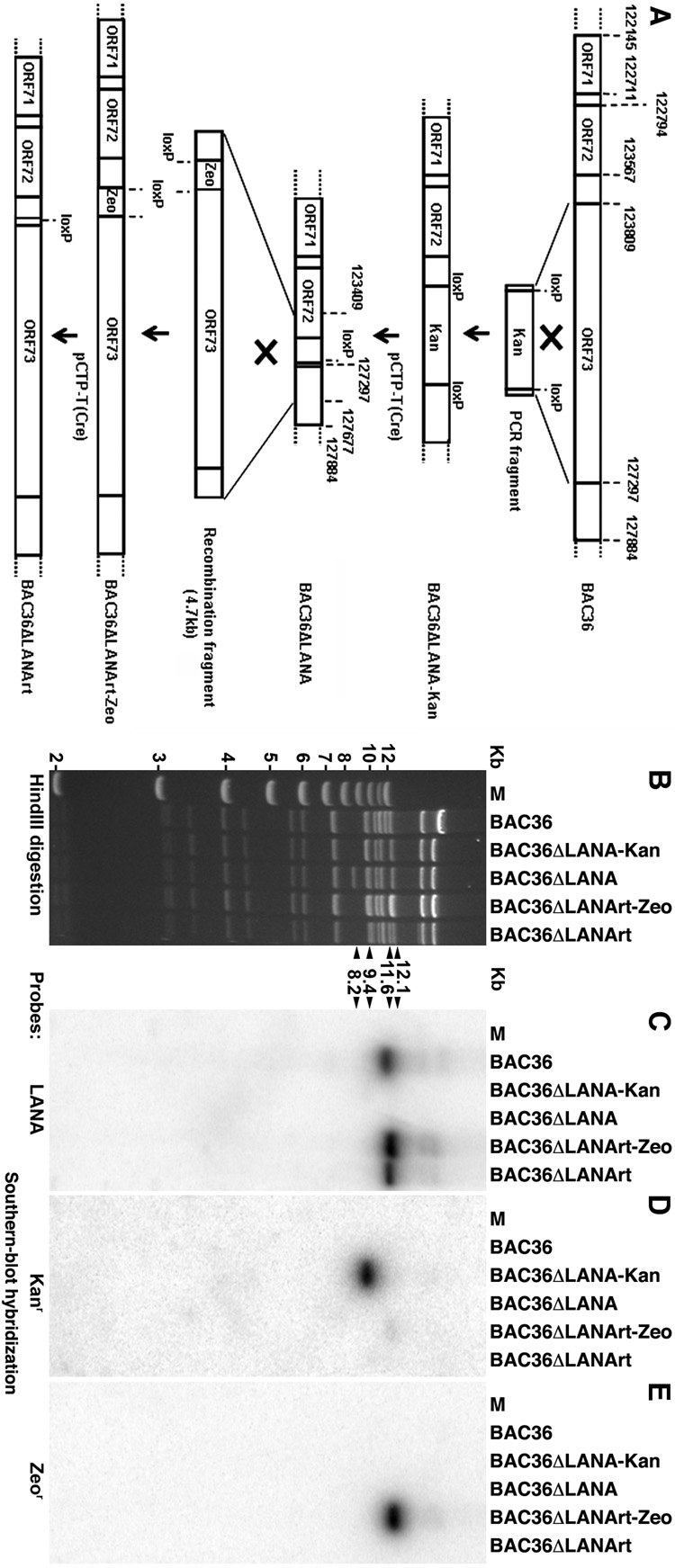
Fig. 1. Generation of a KSHV LANA deletion mutant BAC36ΔLANA and its revertant BAC36ΔLANArt.
(A) A schematic illustration showing the strategy for generating BAC36ΔLANA and BAC36ΔLANArt. A PCR fragment containing the Kanr was introduced to the BAC36 genome to replace LANA by homologous recombination resulting in the generation of an intermediate deletion mutant BAC36ΔLANA-Kan. Removal of the Kanr by Cre-mediated recombination gave rise to BAC36ΔLANA. A similar strategy was used to construct BAC36ΔLANArt using the Zeor as a selection marker. (B) HindIII restriction analysis of BAC36ΔLANA and BAC36ΔLANArt genomes, and their intermediate mutants, along with the wild-type BAC36 genome. The changes of restriction bands at the LANA locus were indicated by arrowheads and detailed in the text. (C–E) Southern-blot hybridization with LANA (C), Kanr (D) and Zeor (E) probes.
Next, we carried out genetic analysis of the LANA mutant and its revertant. Genomes of BAC36ΔLANA and BAC36ΔLANArt as well as their intermediate recombinant viruses BAC36ΔLANA-Kan and BAC36ΔLANArt-Zeo were analyzed by HindIII restriction enzyme digestion. As shown in Fig. 1B, all the recombinant genomes have restriction patterns similar to that of BAC36 except the region where LANA was manipulated. An 11.6-Kb DNA fragment containing LANA in the BAC36 lane was shifted to a 9.4-Kb band in the BAC36ΔLANA-Kan lane because of the replacement of LANA by Kanr. Of note, the new 9.4-Kb band also coincidently overlapped with another existing 9.4-Kb band. Upon excision of the Kanr, the 9.4-Kb band was shifted to an 8.2-Kb band in the BAC36ΔLANA lane. In the BAC36ΔLANArt-Zeo lane, because of the repair of LANA together with the introduction of a Zeor into the locus, the 8.2-Kb band was shifted to a 12.1-Kb band, which coincidently overlapped with another existing 12.1-Kb band. Upon excision of the Zeor, the 12.1-Kb band was shifted to an 11.6-Kb band that represented the repaired LANA band in the BAC36ΔLANArt lane. Southern-blot hybridization further confirmed the restriction patterns of different recombinant viruses (Fig. 1C–E). Indeed, the LANA probe only detected the 11.6-Kb band in the BAC36 lane, the 12.1-Kb band in the BAC36ΔLANArt-Zeo lane, and the 11.6 band in the BAC36ΔLANArt lane while no band was detected in the BAC36ΔLANA-Kan and BAC36ΔLANA lanes because of the deletion of LANA (Fig. 1C). The Kanr probe only detected the 9.4-Kb band in the BAC36ΔLANA-Kan lane (Fig. 1D) while the Zeor probe only detected the 12.1-Kb band in the BAC36ΔLANArt-Zeo lane (Fig. 1E). Together, these results demonstrated that LANA was deleted in BAC36ΔLANA but repaired in BAC36ΔLANArt, and there was no other unexpected genomic alteration in these recombinant viruses.
Reconstitution of recombinant viruses in mammalian cells
To determine the phenotypes of the recombinant viruses, we reconstituted them in HEK293 cells. Based on the percentages of GFP-positive cells at day 2 post-transfection, we estimated 40–60% transfection rates for all the recombinant viruses (Fig. 2A). While the percentages of GFP-positive cells in all the cultures remained relatively stable in the first three days, those of BAC36ΔLANA decreased significantly afterward. These results confirmed our previous study demonstrating LANA’s essential role in KSHV episome persistence using a transposon-mediated insertion LANA mutant (Ye et al., 2004).
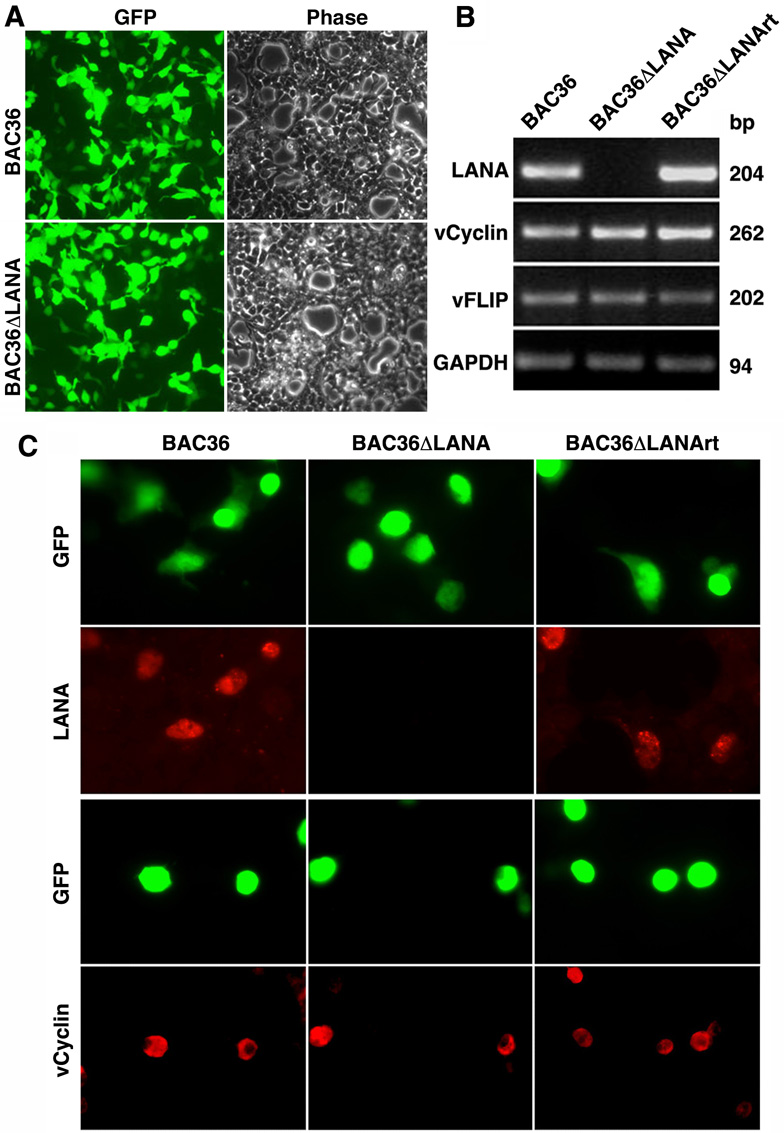
Fig. 2. Expression of LANA adjacent genes following genetic deletion of LANA from the KSHV genome.
(A) Reconstitution of recombinant viruses in HEK293 cells by transfection. Images were taken at 48 h post-transfection. (B) Expression of LANA, vCyclin and vFLIP transcripts in BAC36, BAC36ΔLANA and BAC36ΔLANArt cells analyzed by RT-PCR. GAPDH was used as a loading control. (C) Expression of LANA and vCyclin proteins in BAC36, BAC36ΔLANA and BAC36ΔLANArt cells examined by IFA using specific antibodies.
Deletion of LANA did not affect the expression of latent genes vCyclin and vFLIP
Both vCyclin and vFLIP are adjacent to LANA in the KSHV genome. These three genes are transcribed from the latent locus as three transcripts: two tricistronic transcripts of 5.8-Kb and 5.4-Kb encoding all three genes, and a 1.7-Kb biscistronic transcript encoding vCyclin and vFLIP (Dittmer et al., 1998; Sarid et al., 1999). We used reverse transcription PCR (RT-PCR) to detect the expression of LANA, vCyclin and vFLIP transcripts. As shown in Fig. 2B, while LANA was detected in the BAC36 and BAC36ΔLANArt cells, they were absent in the BAC36ΔLANA cells. In contrast, vCyclin and vFLIP were detected in cells of all three recombinant viruses at similar expression levels. We further examined the expression of LANA protein by immunofluorescence antibody assay (IFA). As shown in Fig. 2C, while we detected LANA-specific speckle patterns in all GFP-positive BAC36 and BAC36ΔLANArt cells, we did not detect the same speckle patterns in any BAC36ΔLANA cells. In contrast to LANA, vCyclin protein was detected in GFP-positive cells of all three recombinant viruses (Fig. 2C). Together, these results confirmed that LANA was deleted in BAC36ΔLANA but repaired in BAC36ΔLANArt, and indicated that the genetic manipulation procedures did not disrupt the expression of vCyclin and vFLIP.
Deletion of LANA enhanced the expression of KSHV lytic genes
To assess the effect of LANA deletion on KSHV transcription program, we examined the expression of a set of KSHV genes representing different classes of viral genes including one latent gene vCyclin, one IE gene RTA, three early genes MTA (ORF57), ORF59 and vIL-6 (ORF-K2), and one late gene ORF-K8.1 by reverse-transcription real-time quantitative PCR (RT-qPCR). As shown in Fig. 3A, deletion of LANA, in general, had marginal effect on the expression of vCyclin transcripts with or without induction with TPA and sodium butyrate (T/B), thus further confirming the results of RT-PCR (Fig. 2B). Nevertheless, RT-qPCR did detect a 30–35% reduction of vCyclin transcripts in the BAC36 and BAC36ΔLANArt cells at 48 h post-induction (hpi) albeit no change was observed for the BAC36ΔLANA cells under the same condition.
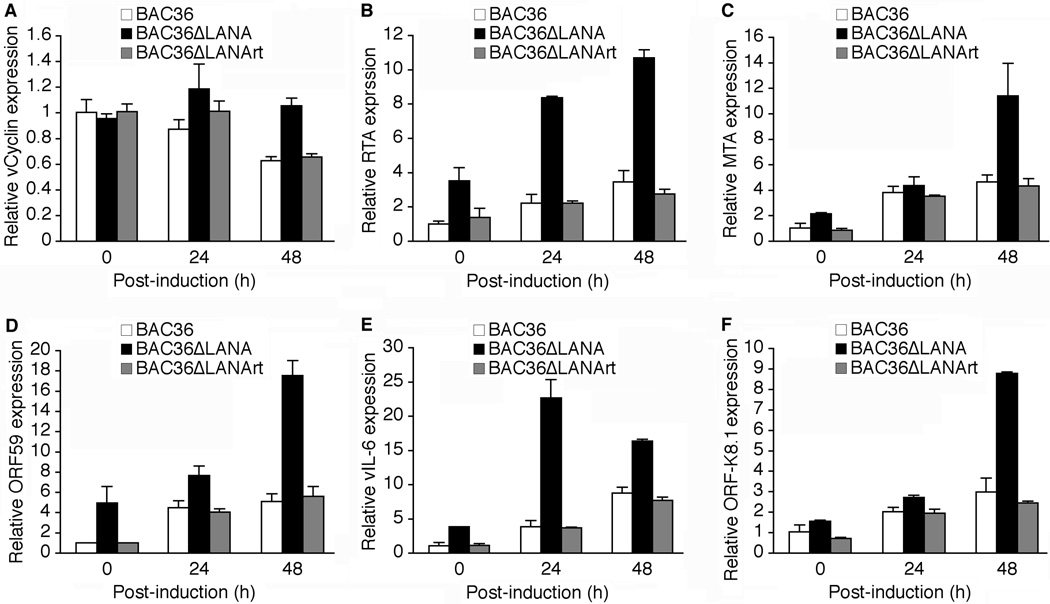
Fig. 3.
Effect of LANA deletion on KSHV transcriptional program. mRNA levels of different KSHV genes in BAC36, BAC36ΔLANA and BAC36ΔLANArt cells following induction with T/B for 0, 24 and 48 h were examined by RT-qPCR. The viral genes were: (A) vCyclin, (B) RTA, (C) MTA, (D) ORF59, (E) vIL-6 and (F) ORF-K8.1. The mRNA levels of KSHV genes in BAC36 cells at 0 hpi were used as the baselines (1). The relative mRNA levels were first normalized to those of GAPDH, and then to the percentages of GFP-positive cells in the cultures.
RTA is a critical transactivator in KSHV switch from latency to lytic replication. As shown in Fig. 3B, the expression of RTA transcripts in BAC36ΔLANA cells was 3.5-fold higher than that of BAC36 cells. Upon induction with T/B, the expression of RTA transcripts at 48 hpi was increased 3.5- and 3.0-fold in BAC36 and BAC36ΔLANA cells, respectively. Thus, the expression of RTA transcripts remained 3.1-fold higher in BAC36ΔLANA cells than in BAC36 cells following induction with T/B. The expression patterns of RTA transcripts in BAC36ΔLANArt cells were the same as those of BAC36 cells. These results indicated that deletion of LANA enhanced the expression of RTA transcripts with or without chemical induction.
We next examined the expression of other KSHV lytic genes. MTA, a functional homologue of the Epstein-Barr virus (EBV) MTA gene, is essential for viral lytic replication (Majerciak et al., 2007). Co-expression of MTA and RTA synergistically activates some promoters of KSHV early and late genes (Kirshner et al., 2000; Malik et al., 2004; Palmeri et al., 2007). In uninduced cells, the expression level of MTA transcripts was 2.1-fold higher in BAC36ΔLANA cells than in BAC36 cells (Fig. 3C). While we detected minimal differences of MTA transcripts between BAC36 and BAC36ΔLANA cells at 24 hpi, we found that the expression of MTA transcripts at 48 hpi was increased 4.7-fold in BAC36 cells while that of BAC36ΔLANA cells was increased 5.4-fold compared to uninduced cells. Thus, the expression level of MTA transcripts was 2.4-fold higher in BAC36ΔLANA cells than in BAC36 cells at 48 hpi. Again, we found that the expression patterns of MTA transcripts in BAC36ΔLANArt cells were the same as those of BAC36 cells (Fig. 3C).
The expression levels of ORF59, vIL-6 and ORF-K8.1 transcripts were also higher in the BAC36ΔLANA cells than in the BAC36 cells with or without chemical induction though they had different expression patterns (Fig. 3D–F). Both ORF59 and ORF-K8.1 are commonly used as KSHV lytic markers because of the availability of specific antibodies (Chandran et al., 1998; Majerciak et al., 2006; Zoeteweij et al., 1999). ORF59 encodes a viral accessory protein expressed in the early phase of viral replication, whereas ORF-K8.1 encodes a glycoprotein expressed in the late phase viral lytic replication (Chan and Chandran, 2000; Chandran et al., 1998). vIL-6 is one of the several well-defined KSHV genes containing the RTA-responsive element (RRE) in their promoter regions (Deng et al., 2002). As shown in Fig. 3D–F, the expression patterns of ORF-K8.1 transcripts were similar to those of MTA transcripts while those of ORF59 and vIL-6 transcripts resembled to the RTA transcripts albeit the vIL-6 transcripts peaked at 24 hpi rather than at 48 hpi. The basal expression levels of ORF59, vIL-6 and ORF-K8.1 transcripts in BAC36ΔLANA cells were 5.0-, 3.8- and 1.6-fold higher, respectively, than those in BAC36 cells. In BAC36 and BAC36ΔLANA cells, ORF59 transcripts were induced 5.1- and 3.5-fold while ORF-K8.1 transcripts were induced 3.0- and 5.6-fold, respectively, at 48 hpi. Upon induction with T/B, vIL-6 transcripts were expressed at a faster kinetics in BAC36ΔLANA cells than in BAC36 cells. At 24 hpi, the expression levels of vIL-6 transcripts were increased 3.8-fold in BAC36 cells, while those in BAC36ΔLANA cells were further increased 6.0-fold on top of their higher baseline levels. Thus, the expression levels of vIL-6 transcripts at 24 hpi remained at 6.0-fold higher in BAC36ΔLANA cells than in BAC36 cells. Interestingly, while the expression levels of vIL-6 transcripts at 48 hpi in BAC36ΔLANA cells decreased slightly from the previous time point, those of BAC36 cells were further increased 2.3-fold. Nevertheless, the expression levels of vIL-6 transcripts at 48 hpi in BAC36ΔLANA cells remained at higher levels than those of BAC36 cells (Fig. 3E). As expected, the expression patterns of ORF59, vIL-6 and ORF-K8.1 transcripts in BAC36ΔLANArt cells were similar to those of BAC36ΔLANA cells (Fig. 3D–F). Together, these results indicated that deletion of LANA also enhanced the expression of RTA downstream lytic genes. However, the expression patterns of these genes varied according to the individual viral lytic genes, possibly reflecting their differential expression kinetics.
Deletion of LANA enhanced the expression of KSHV lytic proteins
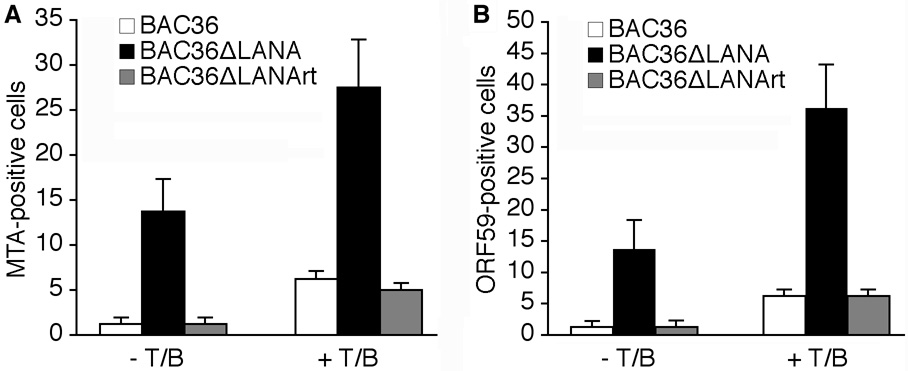
Since we had observed enhanced expression levels of KSHV lytic transcripts following deletion of LANA from the KSHV genome, we further determined whether these results could be translated into protein levels. We carried out IFA staining for MTA and ORF59 proteins using specific antibodies in uninduced or T/B-induced cells. We detected low numbers of MTA- and ORF59-positive cells at about 1.0-1.5% in uninduced BAC36 cells, indicating that the majority of these cells were at tight latency. In contrast, BAC36ΔLANA cells were more leaky for the expression of viral lytic proteins, and had about 12–14.0% MTA- and ORF59-positive cells, respectively, indicating a reduced level of repression of viral lytic transcriptional program in these cells (Fig. 4A and B). The enhanced expression levels of KSHV lytic proteins in BAC36ΔLANA cells were also observed following induction with T/B. As shown in Fig. 4A and B, the percentages of both MTA- and ORF59-positive cells were increased 5.0-fold in BAC36 cells, and 2.0- and 2.7-fold, respectively, in BAC36ΔLANA cells following induction with T/B. Thus, BAC36ΔLANA cells had 4.4- and 5.8-fold more MTA- and ORF59-positive cells, respectively, than BAC36 cells following induction with T/B. The patterns of MTA- and ORF59-positive cells in BAC36ΔLANA cells were almost the same as the BAC36 cells (Fig. 4). These results demonstrated that deletion of LANA enhanced the expression of viral lytic proteins with and without induction with T/B.
Fig. 4. Effect of LANA deletion on the expression of KSHV lytic proteins.
(A–B) Percentages of actual MTA- (A) and ORF59-positive cells (B) detected by immunofluorescence antibody assay in uninduced and T/B-induced BAC36, BAC36ΔLANA and BAC36ΔLANArt cells.
LANA repressed multiple stages of KSHV lytic replication
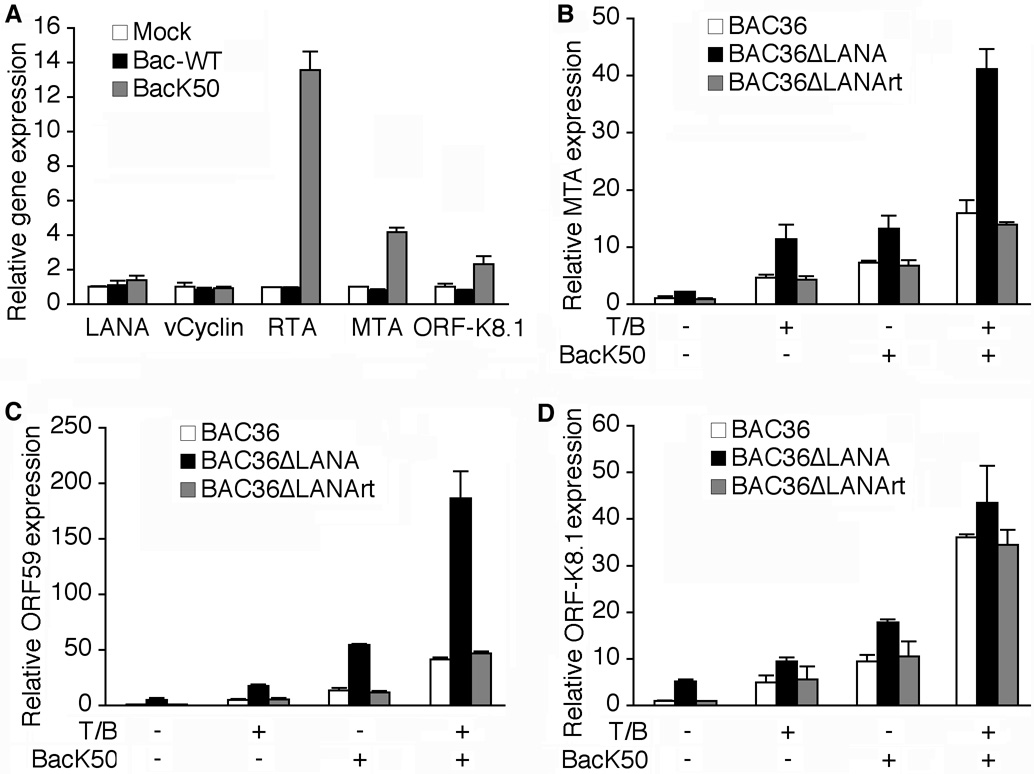
Our results indicated that LANA regulated the expression of RTA during both KSHV latency and lytic replication (Fig. 3B). Previous studies have shown that during T/B-induction of KSHV lytic replication, a series of viral and cellular events occur, including epigenetic remodeling of KSHV genome, and induction of cellular transcriptional factors and expression of RTA, that synergistically culminate in the activation of KSHV lytic cascade (Chen et al., 2001; Sun et al., 1999; Wang et al., 2004; Xie et al., 2007). Since RTA is necessary and sufficient for activating KSHV lytic transcription program, we directly examined the effect of LANA deletion on RTA transactivation of KSHV lytic genes. We induced KSHV lytic replication by infecting the cells with BacK50, a RTA baculovirus, known to effectively reactivate KSHV from latency (Vieira and O'Hearn, 2004). As shown in Fig. 5A, overexpression of RTA increased the level of RTA transcripts by 13.6-fold while simultaneously induced MTA and ORF-K8.1 transcripts by 4.2- and 2.3-fold, respectively. The expression of LANA and vCyclin transcripts was not induced following RTA expression. In contrast to the RTA baculovirus, infection with a wild-type baculovirus (Bac-WT) without containing RTA had no effect on the expression of any viral transcripts, indicating that the observed induction of MTA and ORF-K8.1 transcripts was caused by the overexpression of RTA rather than any unspecific effects caused by baculovirus infection. As shown in Fig. 5B–D, overexpression of RTA increased the expression of MTA, ORF59 and ORF-K8.1 transcripts by 7.2-, 13.4- and 9.4-fold, respectively, in BAC36 cells. As expected, the expression levels of MTA, ORF59 and ORF-K8.1 transcripts without any induction were already higher in BAC36ΔLANA cells than in BAC36 cells by 2.1-, 5.0- and 5.2-fold, respectively. Following induction with RTA, the expression levels of these transcripts in BAC36ΔLANA cells were further increased by 6.2-, 11.0- and 3.5-fold, respectively. Overall, the expression levels of MTA, ORF59 and ORF-K8.1 transcripts were 1.8-, 4.1- and 1.9-fold higher in BAC36ΔLANA cells than BAC36 cells, respectively, following induction with RTA. It was noted that both ORF59 and ORF-K8.1 transcripts had higher expression levels following induction with RTA than with T/B in both BAC36 and BAC36ΔLANA cells while the MTA transcripts had similar expression levels following induction with both methods in these cells. These results were consistent with those reported in another study (Vieira and O'Hearn, 2004), and suggested that the initial induction of RTA might be a “limiting factor” during T/B-induced KSHV lytic replication. Indeed, results of RT-qPCR indicated that the induction of RTA transcripts was not very efficient following T/B treatment (Fig. 3B). When we combined the induction methods of T/B and RTA, the expression levels of MTA, ORF59 and ORF-K8.1 transcripts were increased by 15.9-, 41.5-, and 36.1-fold, respectively, in BAC36 cells, and 41.1-, 186.5- and 43.4-fold, respectively, in BAC36ΔLANA cells, using uninduced BAC36 cells as the baseline, suggesting a synergistic effect of the two induction methods. While the expression levels of MTA and ORF59 transcripts remained substantially higher in BAC36ΔLANA cells than in BAC36 cells, the difference for the ORF-K8.1 transcripts became minimal following the combined induction of T/B and RTA (Fig. 5D). Thus, the expression of ORF-K8.1 appeared to have reached the maximum limit following combined induction with these two methods. Again, we found that results from the BAC36ΔLANArt cells were similar to those of BAC36 cells, indicating that the observed enhanced expression of viral genes were indeed due to the deletion of LANA but not because of any alterations in other parts of the viral genome. Together, these results indicated that LANA also repressed the expression of RTA downstream viral lytic genes.
Fig. 5. Expression of KSHV lytic genes following different induction methods of KSHV lytic replication program.
(A) BacK50 but not control Bac-WT induced the expression of KSHV lytic MTA and ORF-K8.1 transcripts in BAC36 cells. No induction was observed with the LANA and vCyclin transcripts. Viral transcripts were analyzed at 48 h after infection with the baculoviruses. The mRNA levels of KSHV genes in the mock-infected cells were used as the baselines (1). (B–D) mRNA levels of different KSHV lytic genes in BAC36, BAC36ΔLANA and BAC36ΔLANArt cells following induction with T/B, BacK50, or combination of both methods for 48 h were examined by RT-qPCR. The viral genes were: (B) MTA, (C) ORF59, and (D) ORF-K8.1. The mRNA levels of KSHV genes in BAC36 cells at 0 hpi were used as the baselines (1). In all the experiments, the relative mRNA levels were first normalized to those of GAPDH, and then to the percentages of GFP-positive cells in the cultures.
Deletion of LANA increased the production of KSHV infectious virions
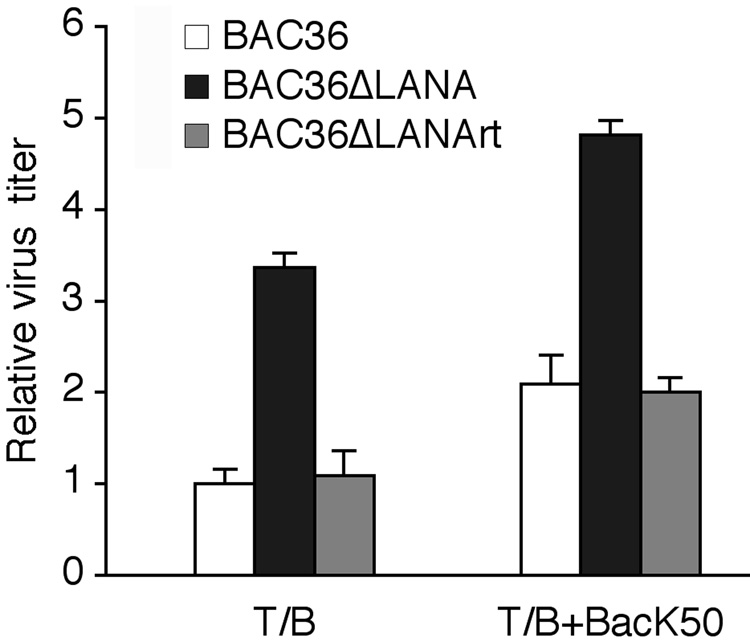
The above experiments demonstrated that deletion of LANA from the KSHV genome increased the expression of KSHV lytic genes. We further determined whether this enhanced viral lytic replication program resulted in the increased production of KSHV infectious virions. We induced the cells with T/B alone or together with BacK50. At 96 hpi, the cultured supernatants were collected and titrated for infectious virions by infected 293T cells. BAC36 cells typically generated 104 GFP-positive cells units (GFU) following induction with T/B (Zhou et al., 2002). As shown in Fig. 6, BAC36ΔLANA cells produced 3.4-fold more infectious virions than BAC36 cells following induction with T/B. Consistent with the gene expression results (Fig. 6), induction with both RTA and T/B further increased the yields to 2.1- and 4.8-fold in BAC36 and BAC36ΔLANA cells, respectively, when we used the T/B-induced BAC36 cells as the baseline (Fig. 6). BAC36ΔLANArt cells produced similar amounts of infectious virions as the BAC36 cells under both induction conditions. Thus, deletion of LANA increased the production of KSHV infectious virions following induction of viral lytic replication. These results also demonstrated that LANA was not an essential gene for viral lytic replication and virion production.
Fig. 6.
Effect of LANA deletion on the production of KSHV infectious virions. BAC36, BAC36ΔLANA and BAC36ΔLANArt cells were induced with T/B or RTA alone, or together for 96 h. Supernatants were then titrated for infectious virions by infecting HEK293 cells. The relative virus titers were calculated using T/B-induced BAC36 cells as the baseline (1).
Discussion
Extensive studies have shown that the major KSHV latent protein LANA is a multifunctional protein regulating different phases of viral lifecycle. LANA is essential for KSHV latency as it plays a critical role in KSHV episome persistence by mediating episome replication, and episome segregation into daughter cells during mitosis (Ballestas and Kaye, 2001; Cotter and Robertson, 1999; Hu et al., 2002; Krithivas et al., 2002). LANA also regulates cell growth and survival by targeting multiple cellular tumor suppressor and oncogenic pathways, including p53, pRb, β-catenin and c-Myc pathways, and thus further contributes to KSHV latency (Cai et al., 2006; Friborg et al., 1999; Fujimuro et al., 2003; Liu et al., 2007; Radkov et al., 2000). Recently, several studies have shown that LANA might control KSHV latency by repressing KSHV lytic transcriptional program (Lan et al., 2005; Lan et al., 2004; Verma et al., 2004). While these studies were mostly carried out by overexpressing the viral gene, their results have suggested a novel function of LANA in regulating KSHV latency. In the current study, we have examined the role of LANA in regulating KSHV lytic transcriptional program in the context of whole viral genome. We used a genetic approach to delete LANA from the KHSV genome, and assessed the effect on viral replication. Our results showed that deletion of LANA from the viral genome indeed increased the expression of KSHV lytic genes during both viral latency and lytic replication. As a result, LANA deletion mutant cells produced more KSHV infectious virions. These results have confirmed that, in addition to regulation of KSHV episome persistence, and cell growth and survival, LANA also controls KSHV latency by repressing viral lytic transcriptional program (Lan et al., 2004).
During KSHV latency, only a handful of KSHV genes, including LANA, vCyclin and vFLIP, and the recently described viral microRNAs are expressed while viral lytic genes are generally silent (Greene et al., 2007). It is believed that such tight control of expression of KSHV lytic genes is regulated by one or more of the viral products expressed during viral latency (Ye et al., 2008). Indeed, by means of the same genetic approach used in this study, we have recently shown that KSHV latent gene vFLIP controls viral latency by repressing KSHV lytic transcriptional program (Ye et al., 2008). vFLIP represses the expression of KSHV lytic genes, including the key lytic switch gene RTA, by inhibiting the activation of AP-1, which is an essential cellular transcriptional factor for activating the expression of several viral lytic genes and viral lytic replication program (Pan et al., 2006; Wang et al., 2004; Xie et al., 2007). In the current study, we have shown that deletion of LANA also increases the expression of KSHV lytic genes including RTA in uninduced KSHV-infected cells. These results confirmed those obtained in previous studies using single viral gene (Lan et al., 2005; Lan et al., 2004), and indicated that, similar to vFLIP, LANA also controls viral latency by repressing KSHV lytic transcriptional program. Thus, KSHV at least uses two viral latent proteins LANA and vFLIP to promote viral latency by controlling viral lytic transcriptional program. It remains to be determined whether these two viral proteins carry out these similar functions independently or synergistically at different phases of viral lifecycle.
While the mechanism of LANA controls KSHV latency might be complex, previous studies have shown that LANA represses the expression of KSHV lytic genes through epigenetic silencing of KSHV genome, and inhibition of the expression and transactivation function of RTA (Lan et al., 2005; Lan et al., 2004; Lu et al., 2006; Verma et al., 2004). Interestingly, a LANA-deficient murine gammaherpesvirus 68 had impaired viral replication in murine fibroblasts albeit an enhanced kinetics of viral gene expression because of p53 induction and cell death (Forrest et al., 2007). Thus, the detailed mechanism of LANA repression of KSHV lytic transcriptional program during viral latency remains to be further elucidated. Of particular interest is the comparison of specific epigenetic modifications of KSHV genome and promoters of specific viral latent and lytic genes in the LANA deletion mutant and wild-type virus cells. Such information could provide further insights into the mechanism of LANA regulation of cellular genes during KSHV latency.
KSHV lytic replication is a multistep process usually initiated with epigenetic remodeling of the viral genome, induction of specific cellular transcriptional factors and RTA (Chen et al., 2001; Sun et al., 1999; Wang et al., 2004; Xie et al., 2007). Treatment of cells harboring wild-type virus with T/B induced KSHV lytic replication, indicating that these chemical agents were at least sufficient to partially release different blocks of viral lytic replication, including the repression effects of vFLIP and LANA, and initiate the viral lytic replication cascade. Nevertheless, deletion of LANA from the KSHV genome further enhanced the viral lytic transcriptional program upon chemical induction, indicating that LANA might exert its repression effects at various stages of viral lytic replication, and treatment with T/B was not sufficient to completely release these repression effects. Indeed, in the T/B induction experiments, we showed that LANA regulated the expression of RTA while in the RTA induction experiments, we showed that LANA also regulated the transactivation function of RTA. Furthermore, a synergistic effect was observed when the cells were induced with both T/B and RTA. Together, these results confirm the existence of multiple blocks in KSHV lytic replication, and suggest that induction with neither method is sufficient to completely overcome these blocks. Interestingly, KSHV lytic genes remained expressed at higher levels in the LANA deletion mutant even when the cells were induced with both T/B and RTA, indicating that induction with both methods was also not sufficient to completely overcome the blocks imposed by LANA alone in the wild-type virus. Thus, we conclude that the mechanism of KSHV regulation of latency and lytic replication is rather complex, and the optimal combination for inducing KSHV lytic replication remains to be uncovered.
Our results have also shown that individual KSHV lytic genes are differentially regulated by LANA. It was interesting that deletion of LANA accelerated the expression kinetics of vIL-6 following induction with T/B. At 24 hpi, vIL-6 was induced to much higher levels in BAC36ΔLANA cells than BAC36 cells. vIL-6 was also induced to a higher level in BAC36ΔLANA cells than other KSHV lytic genes including RTA in cells of both mutant and wild-type viruses. We postulated that the expression of vIL-6 could also be regulated at a RTA-independent mechanism, at least at the early stage of viral lytic replication, and this mode of expression were highly regulated by LANA. In fact, an RTA-independent activation mechanism has been described for regulating the expression of vIL-6 (Chatterjee et al., 2002). Further examination of LANA and RTA transcriptional and functional regulation of KSHV genes at the whole genome scale should promise to reveal insights into the expression control of viral genes, and the mechanism of KSHV latency and reactivation.
Materials and methods
Cell lines and cell culture
Human embryonic kidney 293 (HEK293) cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 50 µg/ml of gentamicin. The Spodoptera frugiperda pupal ovarian cell line Sf9 was grown at 27 °C in complete BaculoGold TNM-FH insect medium (BD Biosciences, San Jose, CA).
Generation of KSHV LANA deletion mutant
The strategy for generating the KSHV LANA deletion mutant and its revertant was illustrated in Fig. 1A. The procedures were essentially carried out as previously described (Zhou et al., 2002). Briefly, a linear DNA fragment containing a Kanr flanked by loxP sites was PCR-amplified with primers LANAK-F and LANAK-R that carry the homologous recombination sequences immediately outside the LANA locus (Table 1). The purified PCR fragment was electroporated into competent E. coli containing the BAC36 genome and the pKD46 plasmid to facilitate homologous recombination (Datsenko and Wanner, 2000). After the first round of homologous recombination, the kanamycin- and chloramphenicol-resistant transformants containing the intermediate recombinant KSHV LANA deletion mutant, termed BAC36ΔLANA-Kan, were isolated and analyzed by HindIII digestion (Fig. 1B). The Kanr was removed from the intermediate mutant by Cre-mediated recombination upon transfection of the plasmid pCTP-T expressing the Cre recombinase into the BAC36ΔLANA-Kan E. coli (Saeki et al., 2001). The resulting recombinant KSHV LANA deletion mutant, termed BAC36ΔLANA, was further verified by HindIII restriction digestion and Southern-blot hybridization (Fig. 1C–E).
Table 1.
Primers used for genetic manipulation, RT-qPCR and RT-PCR
| Gene | Application | Primer |
|---|---|---|
| Kanr | Mutagenesis | LANAK-F: 5’-accgcctccataatttactttggttgtcagaccagatttcccgaggatggtgtaggctggagctgcttc-3’ |
| Kanr | Mutagenesis | LANAK-R: 5’-cggcgacggtggcttctagggaggaaaaagggggagaggtgtggcttttacatatgaatatcctccttag-3’ |
| LANA HindIII-PacI fragment | Mutagenesis | LANAF: 5’-tttaagcttcatgggttattggccgtttctgtttct-3’ |
| LANA HindIII-PacI fragment | Mutagenesis | PacI-LANAR: 5’tttttaattaagtcatttcctgtggagagtccccaggacct-3’ |
| LANA EcoRI-PacI fragment | Mutagenesis | PacI-LANAL: 5’tttttaattaaaagccacacctctccccctttttcctc-3’ |
| LANA EcoRI-PacI fragment | Mutagenesis | EcoRI-LANAL: 5’-tttgaattctaacttacgcatatgcgaagtaagaga-3’ |
| vCyclin | RT-qPCR | vCyclin-F: 5′-gctgataatagaggcgggcaatgag-3′ |
| vCyclin | RT-qPCR | vCyclin-R: 5’-gttggcgtggcgaacagaggcagtc-3’ |
| RTA | RT-qPCR | ORF50-F: 5′-cacaaaaatggcgcaagatga-3′ |
| RTA | RT-qPCR | ORF50-R: 5′-tggtagagttgggccttcagtt-3′ |
| MTA | RT-qPCR | ORF57-F: 5′-acgaatcgagggacgacg-3′ |
| MTA | RT-qPCR | ORF57-R: 5′-cgggttcggacaattgct-3′ |
| vIL-6 | RT-qPCR | ORF-K2-F: 5′-acccttgcagatgccgg-3′ |
| vIL-6 | RT-qPCR | ORF-K2-R: 5′-ggatgctatgggtgatcgatg-3′ |
| ORF59 | RT-qPCR | ORF59-F: 5′-cgagtcttcgcaaaaggttc-3′ |
| ORF59 | RT-qPCR | ORF59-R: 5′-aagggaccaactggtgtgag-3′ |
| ORF-K8.1 | RT-qPCR | ORF-K8.1-F: 5′-aaagcgtccaggccaccacaga-3′ |
| ORF-K8.1 | RT-qPCR | ORF-K8.1-R: 5′-ggcagaaaatggcacacggttac-3′ |
| GAPDH | RT-qPCR | GAPDH-F1: 5′-cccctggccaaggtcatcca-3′ |
| GAPDH | RT-qPCR | GAPDH-R1: 5′-acagccttggcagcgccagt-3′ |
| vCyclin | RT-PCR | vCyclin-F: 5’-gactgcctctgttcgccacgcc-3’ |
| vCyclin | RT-PCR | vCyclin-R: 5’-gccaggaatacaacctagaacc-3’ |
| vFLIP | RT-PCR | vFLIP-F: 5’-cgtctacgtggagaacagtgagctg-3’ |
| vFLIP | RT-PCR | vFLIP-R: 5’-ctgggcacggatgacagggaagtg-3’ |
| LANA | RT-PCR | LANA-F: 5′-tacggttggcgaagtcacatc-3′ |
| LANA | RT-PCR | LANA-R: 5′-cctcgcagcagactacacctccac-3′ |
| GAPDH | RT-PCR | GAPDH-F2: 5′-gaaggtgaaggtcggagtc-3′ |
| GAPDH | RT-PCR | GAPDH-R2: 5′-gaagatggtgatgggatttc-3′ |
The BAC36ΔLANA revertant was constructed with similar strategy (Fig. 1A). For this purpose, a 4.7-Kb HindIII-EcoRI DNA fragment containing the entire LANA-coding frame and a Zeor was generated. Two PCR reactions were carried out using BAC36 as a template: a HindIII-PacI DNA fragment amplified with primers LANAF and PacI-LANAR, and an EcoRI-PacI fragment amplified using primers PacI-LANAL and EcoRI-LANAL (Table 1). The two PCR fragments were purified, ligated and cloned into the HindIII and EcoRI sites of pBluescript II KS (Stratagene, La Jolla, CA). The resulting plasmid DNA was digested with PacI, and a Zeor was then cloned into this site to generate plasmid pBSK-HZeoE. Following HindIII and EcoRI digestion of the pBSK-HZeoE DNA, the 4.7-Kb HindIII-EcoRI fragment was gel purified, and electroporated into E. coli containing the BAC36ΔLANA genome and the pKD46 plasmid. Upon homologous recombination, zeocin- and chloramphenicol-resistant colonies containing an intermediate revertant recombinant, termed BAC36ΔLANArt-Zeo, was selected. The Zeor was then removed by Cre-mediated recombination. The resulting mutant revertant construct, termed BAC36ΔLANArt, was verified by HindIII restriction digestion and Southern-blot hybridization (Fig. 1B–E).
Restriction analysis and Southern-blot hybridization
DNA preparations of different recombinant viruses were digested with HindIII restriction enzyme, separated on 0.8% agarose gels by electrophoresis, and transferred to Zeta-Probe GT membranes (Zhou et al., 2002). The membranes were hybridized with 32P-labeled LANA, Kanr and Zeor probes, and specific signals were detected with a Typhoon™ 9410 Imaging System (GE Healthcare Bio-Sciences Corp., Piscataway, NJ).
Reconstitution of recombinant viruses in mammalian cells
HEK293 cells were plated in T-75 flasks one day before transfection. Cells at 70–80% confluency were transfected with 25 µg of a fresh DNA preparation of a recombinant virus using Targefect-293 F-2 Transfectio Reagent according to the instructions of the manufacturer (Targeting Systems, El Cajon, CA). All DNA preparations of different recombinant viruses were verified for integrity by restriction analysis and Southern-blot hybridization (Fig. 1). At 24 h post-transfection, the transfected cells were split into either T-25 flasks or coverslips in 24-well plates at 70–80% confluency. The transfection efficiency was monitored by counting the percentages of GFP-positive cells at 48 h post-transfection.
Baculovirus
Recombinant baculovirus BacK50 expressing KSHV RTA was generously provided by Dr. Jeffrey Vieira at the University of Washington, Seattle, WA and propagated in Sf9 cells (Vieira and O'Hearn, 2004). Virus was collected by centrifugation to remove the cell debris when the majority of the cells underwent lysis. The resulting supernatants containing infectious BacK50 were aliquoted and stored at −80 °C as stocks. A wild-type baculovirus without RTA Bac-WT was used as a control.
Induction of viral lytic replication
To induce viral lytic replication, cells of different recombinant viruses were treated with ng/ml of TPA and 3 mM sodium butyrate. To examine LANA’s effect on RTA transactivation of other downstream viral lytic genes, cells were also infected with BacK50 or Bac-WT with or without T/B. For the later experiments, cells were infected with BacK50 for 2 h before the addition of T/B. To examine the expression of viral genes, cells were collected at different hpi and examined for the expression of viral transcripts and proteins. To examine the production of virions, supernatants collected from the cultures at 96 hpi were centrifuged to remove cell debris, and used to inoculate HEK293 in 24-well plates in triplicates. The numbers of GFP-positive cells generated from 1 ml of supernatant reflecting the relative viral titers (GFU) were determined as previously described (Gao et al., 2003; Pan et al., 2006).
Reverse-transcription real-time quantitative PCR (RT-qPCR) and reverse transcription PCR (RT-PCR)
Total RNA samples from triplicates were prepared using the SV total RNA Isolation System (Promega Corporation, Madison, WI). cDNA samples were reverse-transcribed from total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, Carlsbad, CA), and primed with random hexamers. Real-time PCR reactions were carried out for RTA, MTA, vIL-6, ORF59, ORF-K8.1, and GAPDH as described previously (Yoo et al., 2005). The primers are listed in Table 1. Briefly, each real-time PCR (qPCR) reaction was carried out in a total reaction volume of 20 µl consisting of SYBR-PCR master mix, the cDNA template and gene-specific primers in a DNA Engine Opticon 2 Continuous Fluorescence Detector (Bio-Rad Laboratories, Hercules, CA). PCR reactions were carried out in triplicates for each sample. The relative mRNA levels were first normalized to those of GAPDH, and then to the percentages of GFP-positive cells in the cultures.
RT-PCR reactions were also carried out to detect the expression of vCyclin, vFLIP, and LANA transcripts in HEK293 cells harboring BAC36, BAC36ΔLANA and BAC36ΔLANArt as previously described (Ye et al., 2004). The primers were listed in Table 1. The specific PCR products were examined on 2% agarose gels.
Immunofluorescence antibody assay (IFA)
IFA was carried out as previously described (Ye et al., 2004). Briefly, cells on glass coverslips were first fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) at 4 °C for 20 min, and then permeabilized with 0.5% Triton X-100 in PBS at 4 °C for 10 min. The cells were blocked with 3% bovine serum albumin in PBS at 37 °C for 1 h followed by incubation with a primary antibody at 37 °C for 1 h. Specific signal was detected by incubating the cells with a secondary antibody at 37 °C for another 45 min followed by examination with an Axiovert Fluorescence Microscope (Carl Zeiss, Maple Grove, MN). Rabbit polyclonal antibodies were used to detect vCyclin and MTA (a gift of Dr. Zhiming Zheng). A goat anti-rabbit IgG Rhodamine conjugate was used to reveal the specific signals (Santa Cruz Biotechnology, Santa Cruz, CA). A mouse monoclonal antibody was used to detect ORF59 protein (a gift of Dr. Bala Chandran), and specific signals were revealed with a goat anti-mouse IgG Alexia Fluor 568 conjugate (Invitrogen). LANA was detected with a rat monoclonal antibody (ABI, New York, NY) and the specific signal was revealed with a goat anti-rat IgG Alexia Fluor 568 conjugate (Invitrogen).
Acknowledgements
This work was supported by grants from the National Institute of Health (CA096512, CA124332, CA119889 and DE017333) and an American Cancer Society Research Scholar Grant (#RSG-04-195) to S-J Gao. We thank Dr. Bala Chandran at Rosalind Franklin University of Medicine and Science and Dr. Zhiming Zheng at National Institute of Health for providing the antibodies, Dr. Dr. Jeffrey Vieira at the University of Washington, Seattle, WA for recombinant baculovirus BacK50, Dr. B. L. Wanner at Purdue University, West Lafayette, Indiana for plasmid pKD46, and Dr. E. A. Chiocca at Harvard Medical School for plasmid pCTP-T.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ballestas ME, Kaye KM. Kaposi's sarcoma-associated herpesvirus latency-associated nuclear antigen 1 mediates episome persistence through cis-acting terminal repeat (TR) sequence and specifically binds TR DNA. J. Virol. 2001;75(7):3250–3258. doi: 10.1128/JVI.75.7.3250-3258.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai QL, Knight JS, Verma SC, Zald P, Robertson ES. EC5S ubiquitin complex is recruited by KSHV latent antigen LANA for degradation of the VHL and p53 tumor suppressors. PLoS Pathog. 2006;2(10):e116. doi: 10.1371/journal.ppat.0020116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi's sarcoma-associated herpesvirus expresses an array of viral microRNAs in latently infected cells. Proc. Natl. Acad. Sci. USA. 2005;102(15):5570–5575. doi: 10.1073/pnas.0408192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll PA, Brazeau E, Lagunoff M. Kaposi's sarcoma-associated herpesvirus infection of blood endothelial cells induces lymphatic differentiation. Virology. 2004;328(1):7–18. doi: 10.1016/j.virol.2004.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SR, Chandran B. Characterization of human herpesvirus 8 ORF59 protein (PF-8) and mapping of the processivity and viral DNA polymerase-interacting domains. J. Virol. 2000;74(23):10920–10929. doi: 10.1128/jvi.74.23.10920-10929.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran B, Bloomer C, Chan SR, Zhu L, Goldstein E, Horvat R. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology. 1998;249(1):140–149. doi: 10.1006/viro.1998.9316. [DOI] [PubMed] [Google Scholar]
- Chatterjee M, Osborne J, Bestetti G, Chang Y, Moore PS. Viral IL-6-induced cell proliferation and immune evasion of interferon activity. Science. 2002;298(5597):1432–1435. doi: 10.1126/science.1074883. [DOI] [PubMed] [Google Scholar]
- Chen J, Ueda K, Sakakibara S, Okuno T, Parravicini C, Corbellino M, Yamanishi K. Activation of latent Kaposi's sarcoma-associated herpesvirus by demethylation of the promoter of the lytic transactivator. Proc. Natl. Acad. Sci. USA. 2001;98(7):4119–4124. doi: 10.1073/pnas.051004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotter MA, Robertson ES. The latency-associated nuclear antigen tethers the Kaposi's sarcoma-associated herpesvirus genome to host chromosomes in body cavity-based lymphoma cells. Virology. 1999;264(2):254–264. doi: 10.1006/viro.1999.9999. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Liang Y, Sun R. Regulation of KSHV lytic gene expression. Curr. Top. Microbiol. Immunol. 2007;312:157–183. doi: 10.1007/978-3-540-34344-8_6. [DOI] [PubMed] [Google Scholar]
- Deng H, Song MJ, Chu JT, Sun R. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus) J. Virol. 2002;76(16):8252–8264. doi: 10.1128/JVI.76.16.8252-8264.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer D, Lagunoff M, Renne R, Staskus K, Haase A, Ganem D. A cluster of latently expressed genes in Kaposi's sarcoma-associated herpesvirus. J. Virol. 1998;72(10):8309–8315. doi: 10.1128/jvi.72.10.8309-8315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest JC, Paden CR, Allen RD, III, Collins J, Speck SH. ORF73-null murine gammaherpesvirus 68 reveals roles for mLANA and p53 in virus replication. J. Virol. 2007;81(21):11957–11971. doi: 10.1128/JVI.00111-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friborg J, Jr, Kong W, Hottiger MO, Nabel GJ. p53 inhibition by the LANA protein of KSHV protects against cell death. Nature. 1999;402(6764):889–894. doi: 10.1038/47266. [DOI] [PubMed] [Google Scholar]
- Fujimuro M, Wu FY, ApRhys C, Kajumbula H, Young DB, Hayward GS, Hayward SD. A novel viral mechanism for dysregulation of beta-catenin in Kaposi's sarcoma-associated herpesvirus latency. Nat. Med. 2003;9(3):300–306. doi: 10.1038/nm829. [DOI] [PubMed] [Google Scholar]
- Ganem D. KSHV infection and the pathogenesis of Kaposi's sarcoma. Annu. Rev. Pathol. 2006;1:273–296. doi: 10.1146/annurev.pathol.1.110304.100133. [DOI] [PubMed] [Google Scholar]
- Gao SJ, Deng JH, Zhou FC. Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 2003;77(18):9738–9749. doi: 10.1128/JVI.77.18.9738-9749.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SJ, Kingsley L, Hoover DR, Spira TJ, Rinaldo CR, Saah A, Phair J, Detels R, Parry P, Chang Y, Moore PS. Seroconversion to antibodies against Kaposi's sarcoma-associated herpesvirus-related latent nuclear antigens before the development of Kaposi's sarcoma. N. Engl. J. Med. 1996;335(4):233–241. doi: 10.1056/NEJM199607253350403. [DOI] [PubMed] [Google Scholar]
- Gao SJ, Zhang YJ, Deng JH, Rabkin CS, Flore O, Jenson HB. Molecular polymorphism of Kaposi's sarcoma-associated herpesvirus (Human herpesvirus 8) latent nuclear antigen: evidence for a large repertoire of viral genotypes and dual infection with different viral genotypes. J. Infect. Dis. 1999;180(5):1466–1476. doi: 10.1086/315098. [DOI] [PubMed] [Google Scholar]
- Greene W, Kuhne K, Ye F, Chen J, Zhou F, Lei X, Gao SJ. Molecular biology of KSHV in relation to AIDS-associated oncogenesis. Cancer Treat. Res. 2007;133:69–127. doi: 10.1007/978-0-387-46816-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Garber AC, Renne R. The latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus supports latent DNA replication in dividing cells. J. Virol. 2002;76(22):11677–11687. doi: 10.1128/JVI.76.22.11677-11687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirshner JR, Lukac DM, Chang J, Ganem D. Kaposi's sarcoma-associated herpesvirus open reading frame 57 encodes a posttranscriptional regulator with multiple distinct activities. J. Virol. 2000;74(8):3586–3597. doi: 10.1128/jvi.74.8.3586-3597.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 2002;76(22):11596–11604. doi: 10.1128/JVI.76.22.11596-11604.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krithivas A, Young DB, Liao G, Greene D, Hayward SD. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 2000;74(20):9637–9645. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan K, Kuppers DA, Robertson ES. Kaposi's sarcoma-associated herpesvirus reactivation is regulated by interaction of latency-associated nuclear antigen with recombination signal sequence-binding protein Jkappa, the major downstream effector of the Notch signaling pathway. J. Virol. 2005;79(6):3468–3478. doi: 10.1128/JVI.79.6.3468-3478.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan K, Kuppers DA, Verma SC, Robertson ES. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 2004;78(12):6585–6594. doi: 10.1128/JVI.78.12.6585-6594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Sohn H, Gwack Y, Choe J. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) binds ATF4/CREB2 and inhibits its transcriptional activation activity. J. Gen. Virol. 2000;81(Pt 11):2645–2652. doi: 10.1099/0022-1317-81-11-2645. [DOI] [PubMed] [Google Scholar]
- Liu J, Martin HJ, Liao G, Hayward SD. The Kaposi's sarcoma-associated herpesvirus LANA protein stabilizes and activates c-Myc. J. Virol. 2007;19(81):10451–10459. doi: 10.1128/JVI.00804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Day L, Gao SJ, Lieberman PM. Acetylation of the latency-associated nuclear antigen regulates repression of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 2006;80(11):5273–5282. doi: 10.1128/JVI.02541-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Kirshner JR, Ganem D. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 1999;73(11):9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac DM, Renne R, Kirshner JR, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252(2):304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- Majerciak V, Pripuzova N, McCoy JP, Gao SJ, Zheng ZM. Targeted disruption of Kaposi's sarcoma-associated herpesvirus ORF57 in the viral genome is detrimental for the expression of ORF59, K8alpha, and K8.1 and the production of infectious virus. J. Virol. 2007;81(3):1062–1071. doi: 10.1128/JVI.01558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerciak V, Yamanegi K, Zheng ZM. Gene structure and expression of Kaposi's sarcoma-associated herpesvirus ORF56, ORF57, ORF58, and ORF59. J. Virol. 2006;80(24):11968–11981. doi: 10.1128/JVI.01394-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik P, Blackbourn DJ, Cheng MF, Hayward GS, Clements JB. Functional co-operation between the Kaposi's sarcoma-associated herpesvirus ORF57 and ORF50 regulatory proteins. J. Gen. Virol. 2004;85(Pt 8):2155–2166. doi: 10.1099/vir.0.79784-0. [DOI] [PubMed] [Google Scholar]
- Miller G, Rigsby MO, Heston L, Grogan E, Sun R, Metroka C, Levy JA, Gao SJ, Chang Y, Moore P. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 1996;334(20):1292–1297. doi: 10.1056/NEJM199605163342003. [DOI] [PubMed] [Google Scholar]
- Moore PS, Gao SJ, Dominguez G, Cesarman E, Lungu O, Knowles DM, Garber R, Pellett PE, McGeoch DJ, Chang Y. Primary characterization of a herpesvirus agent associated with Kaposi' s sarcoma. J. Virol. 1996;70(1):549–558. doi: 10.1128/jvi.70.1.549-558.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D, Spadavecchia S, Carroll KD, Lukac DM. Promoter- and cell-specific transcriptional transactivation by the Kaposi's sarcoma-associated herpesvirus ORF57/Mta protein. J. Virol. 2007;24(81):13299–13314. doi: 10.1128/JVI.00732-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H, Xie J, Ye F, Gao SJ. Modulation of Kaposi's sarcoma-associated herpesvirus infection and replication by MEK/ERK, JNK, and p38 multiple mitogen-activated protein kinase pathways during primary infection. J. Virol. 2006;80(11):5371–5382. doi: 10.1128/JVI.02299-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan HY, Zhang YJ, Wang XP, Deng JH, Zhou FC, Gao SJ. Identification of a novel cellular transcriptional repressor interacting with the latent nuclear antigen of Kaposi's sarcoma-associated herpesvirus. J. Virol. 2003;77(18):9758–9768. doi: 10.1128/JVI.77.18.9758-9768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, Russo JJ, Ju J, Randall G, Lindenbach BD, Rice CM, Simon V, Ho DD, Zavolan M, Tuschl T. Identification of microRNAs of the herpesvirus family. Nat. Methods. 2005;2(4):269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Qian LW, Xie J, Ye F, Gao SJ. Kaposi's sarcoma-associated herpesvirus infection promotes invasion of primary human umbilical vein endothelial cells by inducing matrix metalloproteinases. J. Virol. 2007;81(13):7001–7010. doi: 10.1128/JVI.00016-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radkov SA, Kellam P, Boshoff C. The latent nuclear antigen of Kaposi sarcoma-associated herpesvirus targets the retinoblastoma-E2F pathway and with the oncogene Hras transforms primary rat cells. Nat. Med. 2000;6(10):1121–1127. doi: 10.1038/80459. [DOI] [PubMed] [Google Scholar]
- Rainbow L, Platt GM, Simpson GR, Sarid R, Gao SJ, Stoiber H, Herrington CS, Moore PS, Schulz TF. The 222- to 234-kilodalton latent nuclear protein (LNA) of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) is encoded by orf73 and is a component of the latency-associated nuclear antigen. J. Virol. 1997;71(8):5915–5921. doi: 10.1128/jvi.71.8.5915-5921.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y, Fraefel C, Ichikawa T, Breakefield XO, Chiocca EA. Improved helper virus-free packaging system for HSV amplicon vectors using an ICP27-deleted, oversized HSV-1 DNA in a bacterial artificial chromosome. Mol. Ther. 2001;3(4):591–601. doi: 10.1006/mthe.2001.0294. [DOI] [PubMed] [Google Scholar]
- Samols MA, Hu J, Skalsky RL, Renne R. Cloning and identification of a microRNA cluster within the latency-associated region of Kaposi' sarcoma-associated herpesvirus. J. Virol. 2005;79(14):9301–9305. doi: 10.1128/JVI.79.14.9301-9305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid R, Flore O, Bohenzky RA, Chang Y, Moore PS. Transcription mapping of the Kaposi' sarcoma-associated herpesvirus (human herpesvirus 8) genome in a body cavity-based lymphoma cell line (BC-1) J. Virol. 1998;72(2):1005–1012. doi: 10.1128/jvi.72.2.1005-1012.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarid R, Wiezorek JS, Moore PS, Chang Y. Characterization and cell cycle regulation of the major Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) latent genes and their promoter. J. Virol. 1999;73(2):1438–1446. doi: 10.1128/jvi.73.2.1438-1446.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. USA. 1998;95(18):10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R, Lin SF, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J. Virol. 1999;73(3):2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma SC, Borah S, Robertson ES. Latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus up-regulates transcription of human telomerase reverse transcriptase promoter through interaction with transcription factor Sp1. J. Virol. 2004;78(19):10348–10359. doi: 10.1128/JVI.78.19.10348-10359.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J, O'Hearn PM. Use of the red fluorescent protein as a marker of Kaposi's sarcoma-associated herpesvirus lytic gene expression. Virology. 2004;325(2):225–240. doi: 10.1016/j.virol.2004.03.049. [DOI] [PubMed] [Google Scholar]
- Wang SE, Wu FY, Chen H, Shamay M, Zheng Q, Hayward GS. Early activation of the Kaposi's sarcoma-associated herpesvirus RTA, RAP, and MTA promoters by the tetradecanoyl phorbol acetate-induced AP1 pathway. J. Virol. 2004;78(8):4248–4267. doi: 10.1128/JVI.78.8.4248-4267.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Ajibade AO, Ye F, Kuhne K, Gao SJ. Reactivation of Kaposi's sarcoma-associated herpesvirus from latency requires MEK/ERK, JNK and p38 multiple mitogen-activated protein kinase pathways. Virology. 2007;371(1):139–154. doi: 10.1016/j.virol.2007.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Pan H, Yoo S, Gao SJ. Kaposi's sarcoma-associated herpesvirus induction of AP-1 and interleukin 6 during primary infection mediated by multiple mitogen-activated protein kinase pathways. J. Virol. 2005;79(24):15027–15037. doi: 10.1128/JVI.79.24.15027-15037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, AuCoin DP, Huete AR, Cei SA, Hanson LJ, Pari GS. A Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 ORF50 deletion mutant is defective for reactivation of latent virus and DNA replication. J. Virol. 2005;79(6):3479–3487. doi: 10.1128/JVI.79.6.3479-3487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Blackbourn DJ, Mengel M, Xie JP, Qian LW, Greene W, Yeh IT, Graham D, Gao SJ. Kaposi's sarcoma-associated herpesvirus promotes angiogenesis by inducing angiopoietin-2 expression via AP-1 and Ets1. J. Virol. 2007;81(8):3980–3991. doi: 10.1128/JVI.02089-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Xie JP, Kang T, Greene W, Kuhne K, Lei XF, Li QH, Gao S-J. Kaposi’s sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-κB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 2008;82(9):4235–4249. doi: 10.1128/JVI.02370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye FC, Zhou FC, Yoo SM, Xie JP, Browning PJ, Gao SJ. Disruption of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen leads to abortive episome persistence. J. Virol. 2004;78(20):11121–11129. doi: 10.1128/JVI.78.20.11121-11129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SM, Zhou FC, Ye FC, Pan HY, Gao SJ. Early and sustained expression of latent and host modulating genes in coordinated transcriptional program of KSHV productive primary infection of human primary endothelial cells. Virology. 2005;343(1):47–64. doi: 10.1016/j.virol.2005.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Deng JH, Rabkin C, Gao SJ. Hot-spot variations of Kaposi's sarcoma-associated herpesvirus latent nuclear antigen and application in genotyping by PCR-RFLP. J. Gen. Virol. 2000;81(Pt 8):2049–2058. doi: 10.1099/0022-1317-81-8-2049. [DOI] [PubMed] [Google Scholar]
- Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 2002;76(12):6185–6196. doi: 10.1128/JVI.76.12.6185-6196.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoeteweij JP, Eyes ST, Orenstein JM, Kawamura T, Wu L, Chandran B, Forghani B, Blauvelt A. Identification and rapid quantification of early- and late-lytic human herpesvirus 8 infection in single cells by flow cytometric analysis: characterization of antiherpesvirus agents. J. Virol. 1999;73(7):5894–5902. doi: 10.1128/jvi.73.7.5894-5902.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]