Abstract
A new tryptophan-polyketide hybrid, codinaeopsin, was isolated from an endophytic fungus collected in Costa Rica. The structure of codinaeopsin, which was deduced from NMR and mass spectral data, contains an unusual heterocyclic unit linking indole and decalin fragments. Codinaeopsin is active against Plasmodium falciparum, the causative agent of the most lethal form of malaria (IC50 = 2.3 μg/ml or 4.7 μM).
The small molecules produced by fungi exemplify an extraordinary range of molecular diversity, a comprehensive use of biosynthetic pathways, and a remarkable ability to modulate biological processes. They represent one-fourth of all known bioactive natural products,1 and many commonly prescribed medications – including the beta-lactam antibiotics, the immunosuppressive drug cyclosporin A, and the cholesterol lowering agent lovastatin – have a fungal origin. Fungi that live in close association with other organisms, such as the endophytic fungi that live in the intercellular spaces of higher plants, seem to be especially prolific producers of biologically active small molecules with which they probably regulate their hosts and competitors.2 For the past few years, this laboratory has been part of an International Cooperative Biodiversity Group studying endophytic fungi from Costa Rica, a biodiversity hot spot on the narrow strip of land connecting the North and South American continents. A high-throughput screen for antimalarial compounds using a library of partially purified fungal extracts from Costa Rica led to the discovery of codinaeopsin (1), a previously unreported tryptophan-polyketide hybrid and the subject of this communication.
Fungal isolate CR127A was collected from a white yemeri tree (Vochysia guatemalensis) by the National Biodiversity Institute (INBio) in Costa Rica. The fungus was cultured, extracted, and the resultant extract was prefractionated at INBio by adsorbing the crude extract onto HP20 beads, washing the beads with water, and eluting fractions with ethanol/water mixtures of decreasing polarity. Weighed fractions were arrayed into 384-well plates at Harvard Medical School for high-throughput screens (HTS) in a variety of assays. A HTS live/dead assay for the most deadly malarial parasite, Plasmodium falciparum, identified strain CR127A as a potent screening positive. CR127A was cultured in a rich seed medium for five days to obtain mycelial mass, diluted into 2% malt extract broth to simulate stressful conditions, and cultured for an additional 21 days. Ethyl acetate extracts of the fermentation broth were initially partitioned by silica gel flash column chromatography using 1:1 ethyl acetate:hexanes. Fractions containing the active component (TLC: silica gel Rf=0.22, 1:1 ethyl acetate:hexanes) were pooled and further purified by reverse-phase C-18 HPLC using an acetonitrile/water gradient. Pure codinaeopsin, the active compound, was obtained with an overall yield of 18 mg/L of culture. CR127A was characterized by sequencing its internal transcribed spacer (ITS) regions of ribosomal DNA, and was found to be 98% idetical to Codinaeopsis gonytrichoides, a leaf litter fungus originally found in Japan.3 Morphological analysis of CR127A confirmed this assignment.
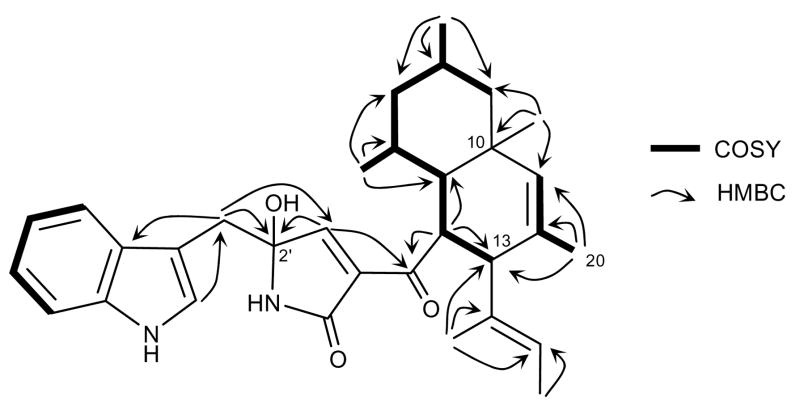
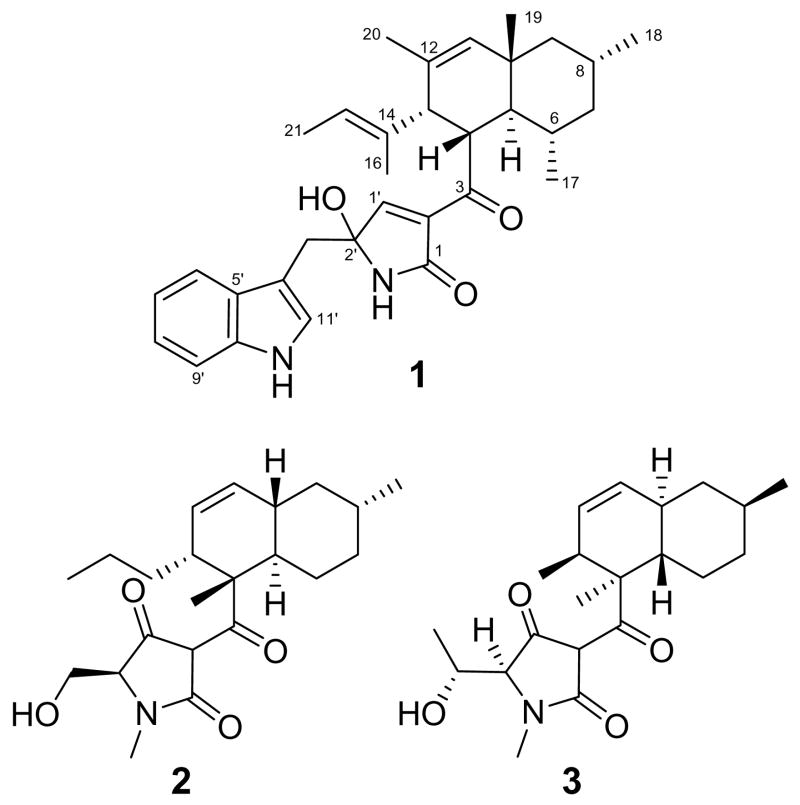
HRESIMS analysis of codinaeopsin (m/z 501.3125) indicated a molecular formula of C32H40N2O3 (calcd. m/z for [M+H]+, 501.3117). The structure of 1 was established from NMR analysis using one- and two-dimensional NMR techniques including proton, carbon, gCOSY, gHMQC, gHMBC, DEPT, and ROESY. Three distinct spin systems were detected on the basis of 1H and gCOSY experiments – an indole ring, an allylic methyl group and an aliphatic seven-carbon chain (Figure 1). The latter two were linked by a long-range HMBC correlation from the C-20 methyl to C-13, and additional HMQC and HMBC correlations adjoining these two spin systems were used to establish the decalin moiety of codinaeopsin (Figure 1). Surmising that the indole functionality arises through incorporation of tryptophan, the adjoining methylene group and heterocyclic ring were deduced, allowing for assembly of the remainder of the structure. The relative stereochemistry of the molecule was established through 1H-1H coupling constants and ROESY correlations (Supporting Information). A minor compound determined to be the C-2′ epimer of codinaeopsin was also isolated.
Figure 1.
Select COSY and HMBC correlations observed in codinaeopsin
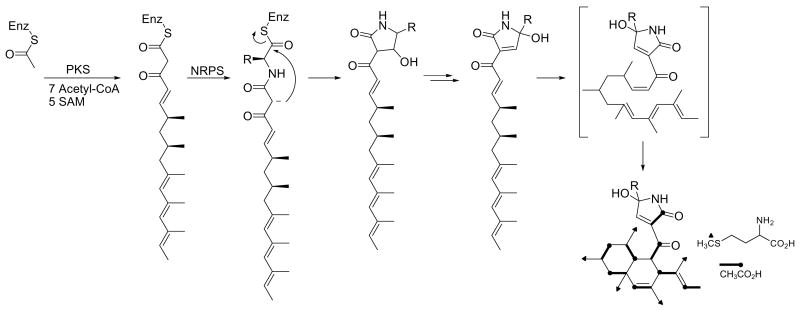
Codinaeopsin’s structure belongs to a growing family of fungal metabolites that includes the HIV integrase inhibitor equisetin4 (2), the antifungal agent cryptocin5 (3), and the telomerase inhibitor UCS1025A6, among others. These metabolites have a scaffold derived from a linear polyketide joined to an N-containing fragment. Codinaeopsin (1) has a linear C16-fragment with five methyl substituents joined to tryptophan. Equisetin (2) has a linear C14-fragment with two methyls joined to N-methyl-serine, and cryptocin (3) has a linear C12-fragment with two methyls joined to N-methyl-threonine. Earlier biosynthetic studies, including the cloning of the equisetin gene cluster7 and stable isotope labeling studies on the GKK1032-family,8 suggest a scheme for codinaeopsin’s biosynthesis that is outlined in Figure 2.
Figure 2.
Proposed biosynthesis of codinaeopsin. Completed structure shows labeling patterns observed in feeding experiments. PKS, polyketide synthase; SAM, S-adenosyl methionine; NRPS, nonribosomal peptide synthase (condensation-adenylation-thiolation).
Early steps assemble the polyketide unit shown. Methyl branches may arise from incorporation of propionate instead of acetate, or from transfer of methyl groups to an acetate-derived polyketide. Stable isotope labeling studies using [1-13C]-acetate and [S-13CH3]-L-methionine resulted in the enrichments shown in Table 1. Enrichment of carbons 1, 3, 5, 7, 9, 11, 13, and 15 with C-1 of acetate and of pendant methyl groups with the S-methyl group of methionine conform to the acetate-methionine pathway normally observed for fungi.9 These steps are likely catalyzed by an iterative type I polyketide synthase (PKS), which uses each active site in the multifunctional enzyme repeatedly during assembly.10 These PKS modules also contain the methyltransferase domains. A nonribosomal peptide synthetase (NRPS) module then introduces an L-tryptophan, and subsequent intramolecular condensation yields the central heterocyclic ring characteristic of this family.11 Most frequently, this ring is a tetramic acid (2,4-pyrrolidindione) as seen in equisetin (2) and cryptocin (3). In codinaeopsin, the ring differs by having a reduced carboxylate carbon and an oxidized Cα of tryptophan. This formal oxidation-reduction could also be accomplished by a series of tautomeric shifts involving enol and imine intermediates within the ring. This latter mechanism is consistent with finding both C-2′ epimers in the culture supernatant. However, these and other questions about the biosynthesis of codinaeopsin will require analyzing the PKS-NRPS cluster involved in its biosynthesis. The linear PKS-assembled unit is then cyclized, possibly by an intramolecular Diels-Alder-like addition similar to those which have been established for solanapyrone12 and lovastatin.13
Table 1.
NMR assignments of codinaeopsin in CD3OD (600MHz), and 13C abundance after administration of labeled precursors
| position | δC | δH | [1-13C]-acetate enrichmenta | [S-13CH3]-L-Met enrichmenta |
|---|---|---|---|---|
| 1 | 170.5 | 3.3 | ||
| 2 | 137.1 | |||
| 3 | 199.8 | 4.1 | ||
| 4 | 49.6 | 3.43 (dd, J = 12.4, 7.0 Hz) | ||
| 5 | 47.2 | 1.50 (dd, J = 12.4 Hz) | 5.6 | |
| 6 | 33.4 | 1.20 (m) | ||
| 7 | 48.3 | 0.61, 1.49 | 5.9 | |
| 8 | 28.7 | 1.72 (m) | ||
| 9 | 49.9 | 0.85, 1.41 | 4.3 | |
| 10 | 37.7 | |||
| 11 | 136.8 | 5.21 (s) | 5.4 | |
| 12 | 130.5 | |||
| 13 | 53.1 | 2.47 (d, J = 7.0 Hz) | 6.0 | |
| 14 | 137.3 | |||
| 15 | 124.5 | 4.51 (q, J = 6.5 Hz) | 5.6 | |
| 16 | 13.5 | 1.29 (3H, d, J = 6.5 Hz) | ||
| 17 | 25.4 | 0.22 (3H, d, J = 6.7 Hz) | 46.9 | |
| 18 | 23.1 | 0.82 (3H, d, J = 7.5 Hz) | 41.2 | |
| 19 | 22.6 | 0.82 (3H, s) | 44.3 | |
| 20 | 22.7 | 1.37 (3H, s) | 39.4 | |
| 21 | 14.2 | 1.30 (3H) | 43.2 | |
| 1′ | 157.0 | 7.34 (s) | ||
| 2′ | 88.5 | |||
| 3′ | 34.5 | 3.32 (d), 3.44 (d, J = 14.0 Hz) | ||
| 4′ | 109.0 | |||
| 5′ | 129.3 | |||
| 6′ | 119.3 | 7.57 (d, J = 7.8 Hz) | ||
| 7′ | 120.2 | 7.01 (dd, J = 7.5 Hz) | ||
| 8′ | 122.3 | 7.06 (dd, J = 7.5 Hz) | ||
| 9′ | 112.4 | 7.29 (d, J = 8.0 Hz) | ||
| 10′ | 137.7 | |||
| 11′ | 125.4 | 7.10 (s) |
The initial antimalarial activity of codinaeopsin was discovered using the high-throughput whole parasite screen previously described,14 and the same screen was used to follow biological activity during the isolation. Pure codinaeopsin has an IC50 of 4.66 μM (2.33 μg/ml) in this assay against the 3D7 strain of P. falciparum, the common drug-sensitive laboratory strain. It has essentially the same activity against the pyrimethamine- and mefloquine-resistant HB3 strain and the chloroquine-pyrimethamine- and mefloquine-resistant Dd2 (Supporting Information). The practical utility of these observations will require further studies.
In summary, codinaeopsin (1), a stereochemically rich tryptophan-polyketide hybrid with an unusual heterocyclic link, has been isolated from an endophytic fungus from Costa Rica. Codinaeopsin belongs to a family of small molecules with a broad range of biological activities, and further studies in other assays are in progress.
Supplementary Material
Experimental details, data obtained in antimalarial screens, NMR spectra, sequence information. This material is available free of charge via the Internet at http://pubs.acs.org.
Scheme 1.
Acknowledgments
This work was supported by 5UO1TW007404 (NIH-FIC, JC). We thank Giselle Tamayo and the staff of the Instituto Nacional de Biodiversidad (INBio, Costa Rica) for their assistance with collecting fungi and performing morphological analysis, and Mary Lynn Baniecki (Harvard Medical School) for performing the malarial screens.
References
- 1.Henkel T, Brunne RM, Müller H, Reichel F. Angew Chem Int Ed. 1999;38:643. doi: 10.1002/(SICI)1521-3773(19990301)38:5<643::AID-ANIE643>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 2.(a) Schulz B, Boyle C. Mycol Res. 2005;109:661. doi: 10.1017/s095375620500273x. [DOI] [PubMed] [Google Scholar]; (b) Strobel GA. Critical Reviews in Biotechnology. 2002;22:315. doi: 10.1080/07388550290789531. [DOI] [PubMed] [Google Scholar]
- 3.Yokoyama T. IFO Res Commun. 1975;7:114. [Google Scholar]
- 4.Vesonder RF, Tjarks LW, Rohwedder WK, Burmeister HR, Laugal JA. J Antibiot (Tokyo) 1979;32:759. doi: 10.7164/antibiotics.32.759. [DOI] [PubMed] [Google Scholar]
- 5.Li JY, Strobel G, Harper J, Lobkovsky E, Clardy J. Org Lett. 2000;2:767. doi: 10.1021/ol000008d. [DOI] [PubMed] [Google Scholar]
- 6.Agatsuma T, Akama T, Nara S, Matsumiya S, Nakai R, Ogawa H, Otaki S, Ikeda S, Saitoh Y, Kanda Y. Org Lett. 2002;4:4387. doi: 10.1021/ol026923b. [DOI] [PubMed] [Google Scholar]
- 7.Sims JW, Fillmore JP, Warner DD, Schmidt EW. Chem Commun (Camb) 2005:186. doi: 10.1039/b413523g. [DOI] [PubMed] [Google Scholar]
- 8.Oikawa H. J Org Chem. 2003;68:3552. doi: 10.1021/jo0267596. [DOI] [PubMed] [Google Scholar]
- 9.O’Hagan D. Nat Prod Rep. 1993;10:593. doi: 10.1039/np9931000593. [DOI] [PubMed] [Google Scholar]
- 10.Crawford JM, Thomas PM, Scheerer JR, Vagstad AL, Kelleher NL, Townsend CA. Science. 2008;320:243. doi: 10.1126/science.1154711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schobert R, Schlenk A. Bioorg Med Chem. 2008;16:4203. doi: 10.1016/j.bmc.2008.02.069. [DOI] [PubMed] [Google Scholar]
- 12.(a) Oikawa H, Katayama K, Suzuki Y, Ichihara A. J Chem Soc, Chem Commun. 1995:1321. [Google Scholar]; (b) Oikawa H, Suzuki Y, Naya A, Katayama K, Ichihara A. J Am Chem Soc. 1994;116:3605. [Google Scholar]; (c) Katayama K, Kobayashi T, Oikawa H, Honma M, Ichihara A. Biochim Biophys Acta. 1998;1384:387. doi: 10.1016/s0167-4838(98)00040-5. [DOI] [PubMed] [Google Scholar]
- 13.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. J Am Chem Soc. 2000;122:11519. [Google Scholar]
- 14.Baniecki ML, Wirth DF, Clardy J. Antimicrob Agents Chemother. 2007;51:716. doi: 10.1128/AAC.01144-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Experimental details, data obtained in antimalarial screens, NMR spectra, sequence information. This material is available free of charge via the Internet at http://pubs.acs.org.