Abstract
Sex-determination in C. elegans requires regulation of gene transcription and protein activity and stability. sel-10 encodes a WD40-repeat-containing F-box protein that likely mediates the ubiquitin-mediated degradation of important sex-determination factors. Loss of sel-10 results in a mild masculinization of hermaphrodites, whereas dominant alleles of sel-10, such as sel-10(n1074), cause a more severe masculinization, including a reversal of the life versus death decision in sex-specific neurons. To investigate about how sel-10 regulates sex-determination, we conducted a sel-10(n1074) suppressor screen and isolated a weak loss-of-function allele of skr-1, one of 21 Skp1-related genes in C. elegans. Skp1, Cullin, and F-box proteins, such as SEL-10, are components of the SCF E3 ubiquitin ligase complex. We present genetic evidence that the sel-10(n1074) masculinization phenotype is dependent upon skr-1 and cul-1 activity. Furthermore, we show that the SKR-1(M140I) weak loss-of-function mutation interferes with SKR-1/SEL-10 binding. Unexpectedly, we found that the G567E substitution in SEL-10 caused by the n1074 allele impairs the binding of SEL-10 to SKR-1 and the dimerization of SEL-10, which may be important for SEL-10 function. Our results suggest that SKR-1, CUL-1 and SEL-10 constitute an SCF E3 ligase complex that plays an important role in modulating sex-determination and LIN-12/Notch signaling in C. elegans.
Keywords: SCF, sex-determination, C. elegans, skr-1, sel-10, cul-1
Introduction
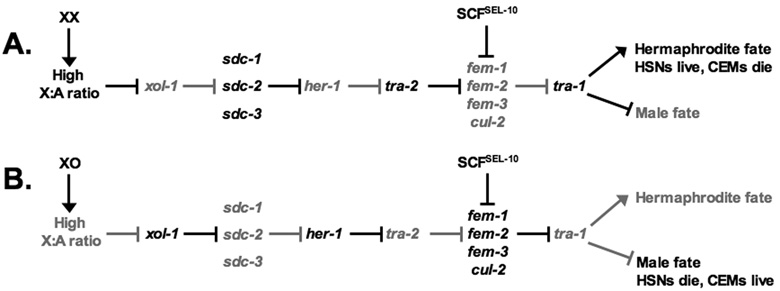
Somatic sexual differentiation in C. elegans is controlled by a negative regulatory cascade that involves signal transduction, transcriptional and translational regulation, and targeted protein degradation (Fig. 1; Zarkower, 2006). The initial determinant of sex in C. elegans is the X chromosome to autosome ratio whereby XX animals (1:1 X:A ratio) develop as hermaphrodites and XO animals (1:2 X:A ratio) develop as males (Hodgkin, 1987; Madl and Herman, 1979). This X to autosome counting mechanism (see Meyer, 2000 for review) regulates the male-specific expression of her-1 (hermaphrodization), which encodes a secreted protein that promotes male development (DeLong et al., 1993; Perry et al., 1993; Trent et al., 1991). HER-1 inhibits the activity of a transmembrane receptor TRA-2 (Transformer) in males, while TRA-2 remains active in hermaphrodites (Kuwabara et al., 1992). TRA-2 promotes hermaphrodite development by inhibiting the activity of the FEM (Feminization) protein complex composed of an ankyrin-repeat protein FEM-1 (Spence et al., 1990), a type 2C protein phosphatase FEM-2 (Chin-Sang and Spence, 1996; Pilgrim et al., 1995), a novel protein FEM-3 (Ahringer et al., 1992), and a Cullin, CUL-2 (Starostina et al., 2007). The FEM proteins and CUL-2 form a CBC (Cul2, Elongin B, Elongin C) E3 ubiquitin ligase complex referred to as CBCFEM-1, which promotes male development by targeting TRA-1 for degradation (Starostina et al., 2007). TRA-1, a DNA-binding Zinc finger protein, promotes hermaphrodite development by repressing the expression of male-specific genes in hermaphrodites (Conradt and Horvitz, 1998; Yi et al., 2000; Zarkower and Hodgkin, 1992). TRA-1 is often referred to as a master regulator of sex-determination, since loss of tra-1 results in the transformation of XX animals into fertile males (Hodgkin, 1987). Similarly, loss of any of the other core sex-determination pathway components results in a complete or near complete sexual transformation. However, there are components of the sex-determination pathway that do not act as switches between the male and hermaphrodite fate but rather “fine-tune” the pathway by targeting core pathway proteins for ubiquitin-mediated degradation.
Figure 1.
A simplified view of the sex-determination pathway in XX animals (A) and XO animals (B). Sex-determination genes depicted in black are active and genes depicted in grey are repressed (see text for detail). SCFSEL-10 is an E3 ubiquitin ligase composed of SKR-1, CUL-1, and SEL-10.
SEL-10 is an F-box protein with eight WD40-repeats and may serve as a substrate recognition component of the SCF (Skp1-Cullin-F-box) ubiquitin-ligase complex that targets substrate proteins for ubiquitin-mediated proteolysis by the proteasome (for a review on SCF complexes see Cardozo and Pagano, 2004). SEL-10 was first identified for its role in the Notch pathway and vulval development in C. elegans by targeting LIN-12/Notch and SEL-12/Presenilin for degradation (Hubbard et al., 1997; Wu et al., 1998). SEL-10 was also reported to affect sex-determination by mediating the turnover of the male-promoting proteins FEM-1 and FEM-3 (Fig. 1). sel-10(null) hermaphrodites are not visibly masculinized but exhibit a synergistic masculinization phenotype in combination with weak alleles of tra-2 (Jager et al., 2004). However, dominant mutations in sel-10, such as sel-10(n1074), cause a stronger masculinization phenotype in hermaphrodites, characterized by the reversal of the cell death fates of the sex-specific neurons: the HSNs and CEMs (Desai and Horvitz, 1989; Jager et al., 2004). The HSNs (hermaphrodite-specific neurons) control vulval muscle contraction and egg laying in hermaphrodites and are eliminated through programmed cell death (PCD) in male embryos. Mutations that cause inappropriate HSN death in hermaphrodites render the animals egg laying defective (Egl) and they become bloated with eggs (Desai et al., 1988; Desai and Horvitz, 1989; Sulston, 1976). The CEMs (cephalic male neurons), which are involved in mediating chemotaxis of males toward hermaphrodites for mating (White et al., 2007), are born in both sexes but are selectively removed through programmed cell death (PCD) in hermaphrodite embryos (Sulston and Horvitz, 1977).
n1074 and seven other dominant alleles of sel-10 cause the same single nucleotide change that results in Glycine to Glutamate substitution at amino acid 567 (G567E) in the carboxyl terminus of SEL-10, located in the eighth WD40-repeat region (Jager et al., 2004; Orlicky et al., 2003). It is unclear how SEL-10(G567E) affects the activity of the SCFSEL-10 complex to cause a dominant, masculinization defect. We thus carried out a sel-10(n1074) suppressor screen to identify components that may act with SEL-10 to regulate sex-determination.
Here we report the identification of skr-1 (Skp1-related 1) as a co-factor of sel-10. We isolated a weak loss-of-function mutation in skr-1, sm151 that specifically suppresses the masculinization defect of sel-10(n1074) animals and causes a Methionine to Isoleucine substitution at amino acid 140 (M140I). SKR-1 is a C. elegans homolog of the human Skp1 protein, a member of the SCF complex that targets substrate proteins for ubiquitin-mediated degradation by the proteasome (Bai et al., 1996; Cardozo and Pagano, 2004; Feldman et al., 1997; Skowyra et al., 1997; Zhang et al., 1995). We found that SEL-10 binds SKR-1 and this binding is compromised by the SKR-1(M140I) mutation. The SKR-1/SEL-10 binding is further reduced in the presence of both SKR-1(M140I) and SEL-10(G567E) mutations. These results suggest that SEL-10 and SKR-1 likely act in the same SCF complex and provide a mechanistic basis for the suppression of the sel-10(n1074) masculinization phenotype by skr-1(sm151). Our study also reveals an important and unexpected role for the C-terminal tail of SEL-10 in SKR-1 binding, SEL-10 dimerization, and in regulating the activity of the SCF E3 ligase complex.
Materials and methods
Strains and genetic manipulations
Strains were derived from the Bristol strain N2, grown at 20°C unless otherwise noted, and constructed using standard procedures (Brenner, 1974). Mutations used are from Brenner (1974) unless noted and are listed by linkage group (LG). LGI: dpy-5(e61), unc-29(e193), unc-75(e950), skr-1(sm151) (tm2391) (this work). LGII: smIs23[pkd-2::GFP] (Peden et al., 2007), lin-23(e1883), lin-23(ot1) (Mehta et al., 2004), tra-2(n1106) (Desai and Horvitz, 1989). LGIII: lin-12(n302) (Greenwald et al., 1983), lin-12(ar170) (Hubbard et al., 1997), cul-1(e1756), tra-1(e1488). LGIV: smIs26[pkd-2::GFP tph- 1::GFP unc-76(+)] (Peden et al., 2007), dpy-20(e1282). LGV: egl-1(n1084) (Conradt and Horvitz, 1999), sel-10(ar41) (Hubbard et al., 1997), (n1074) (Desai and Horvitz, 1989), (ok1632) (gift of the C. elegans knockout consortium, Oklahoma Medical Research Foundation), him-5(e1490), unc-76(e911). hT2[qIs48] (hereafter hT2[GFP]) (Miskowski et al., 2001) was used as a balancer for LGI and III. qIs48 is an insertion of ccEx9747 with markers: myo- 2::GFP expressed in the pharynx, pes-10::GFP expressed in embryos, and a gut promoter driving GFP in the intestine.
We sequenced the sel-10(ok1632) deletion and found a 898 base pair deletion between the 55th and 953rd base pairs after the start. There is also a 15 base pair insertion (gtattatctagtatt). The conceptual translation of sel-10(ok1632) is predicted to cause a truncation of the protein after the first 18 amino acids and thus is likely to represent a null allele.
Isolation, mapping, and cloning of skr-1(sm151)
smIs23; dpy-20(e1282); sel-10(n1074) hermaphrodites were mutagenized with 50 mM EMS and F2 non-Egl animals lacking CEMs were isolated as suppressors. From 21,800 mutagenized genomes, 5 extragenic suppressors including sm151 were isolated. sm151 was mapped to an interval on LGI based on standard three-factor mapping. sel-10(n1074) was homozygous in all mapping strains. sm151 was mapped to the right of unc-29 (genetic position 3.29) based on mapping with dpy-5(e61) unc-29(e193). 11/11 Dpy non-Unc and 0/6 Unc non-Dpy animals segregated sm151. sm151 was then mapped to position 3.63 between dpy-5 (0.0) and unc-75 (9.44). 9/28 Dpy non-Unc and 5/11 Unc non-Dpy animals segregated sm151.
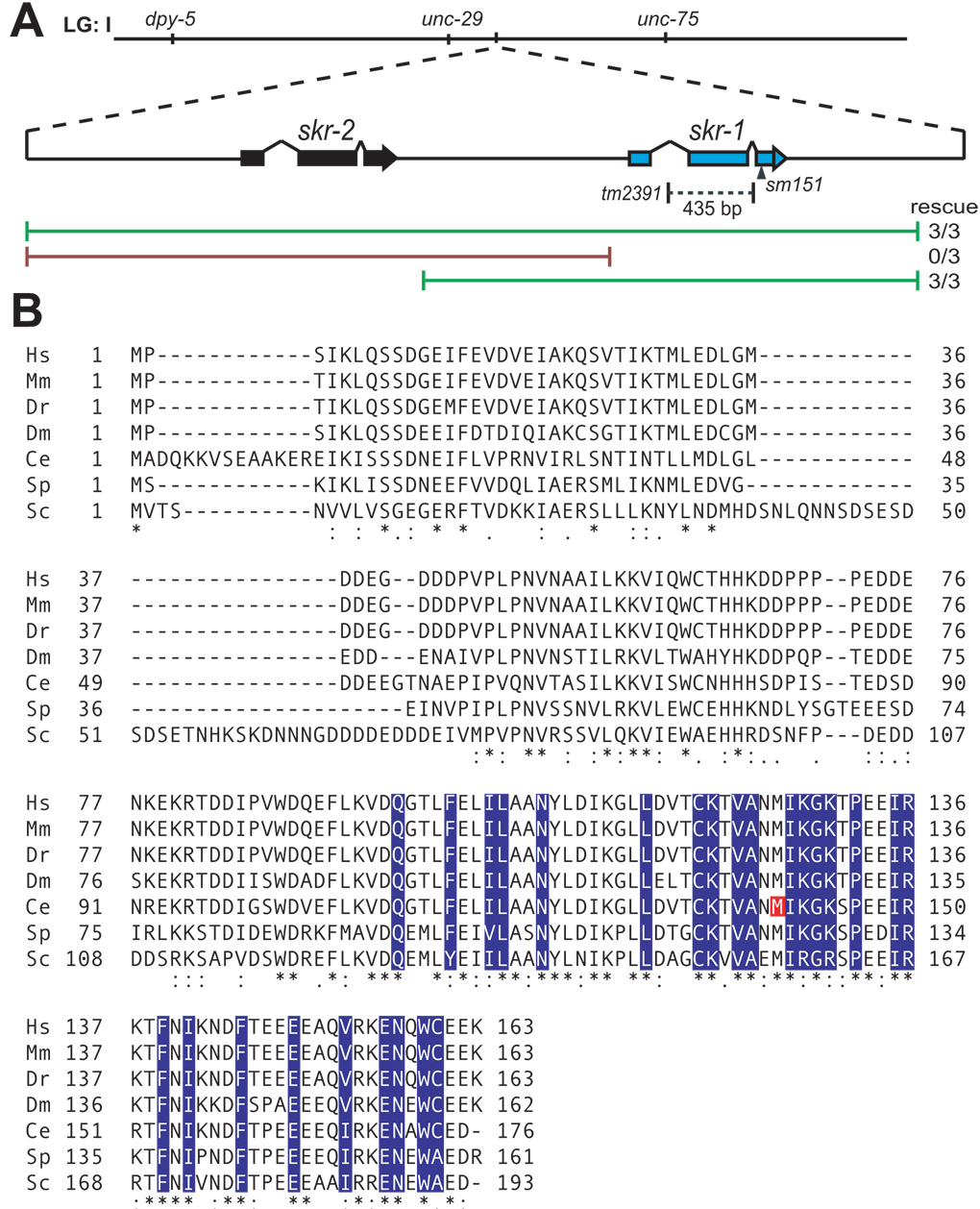
Since sm151 mapped near the skr-2 and skr-1 loci (3.77), they were tested as candidates. A 4704 base pair fragment of the skr-2 skr-1 region was amplified by PCR including 1430 base pairs upstream of the skr-2 start codon to 654 base pairs downstream of the skr-1 stop codon. sm151; smIs23; sel-10(n1074) hermaphrodites were injected with the skr-2 skr-1 PCR fragment at 20ng/ul and rol-6(su1006) at 60ng/ul. 3/3 transgenic lines exhibited rescue. To determine whether skr-2 or skr-1 rescued, two separate PCR fragments were generated and tested for rescue. A 2405 base pair fragment of skr-1 was amplified by PCR including 991 base pairs upstream of the start and 654 base pairs downstream of the stop codon. Similarly, a 3290 base pair fragment of skr-2 was amplified by PCR including 1430 base pairs upstream of the start and 1109 base pairs downstream of the stop codon. 3/3 transgenic lines carrying skr-1 rescued whereas 0/3 transgenic lines carrying skr-2 rescued (Fig. 2).
Figure 2.
(A) Schematic representation of the skr-2 and skr-1 locus of chromosome I (LG: I). The skr-1(sm151) lesion is indicated by an arrowhead and the skr-1(tm2391) deletion is indicated by a dashed line. Green lines represent PCR fragments that rescued skr-1(sm151) and the red lines represent PCR fragments that did not rescue. (B) Alignment was performed between Skp1-related proteins in human (Hs), mouse (Mm), zebrafish (Dr), fruitfly (Dm), worm (Ce), fission yeast (Sp), and budding yeast (Sc) with ClustalW2 (European Bioinformatics Institute of the European Molecular Biology Laboratory). (*) identical residues, (:) conserved residues, (.) semi-conserved residues. C. elegans SKR-1 M140 is indicated in red. F-box binding residues of human Skp1 are indicated in blue based on Schulmann et al. (2000).
The skr-1 locus was amplified by PCR from skr-1(sm151); smIs23; sel-10(n1074) animals and the entire open reading frame was sequenced. A single base pair change was identified 649 base pairs downstream of the start resulting in a codon change of ATG to ATA and an amino acid change at Methionine 140 to Isoleucine (Fig. 2).
Isolation of skr-1(tm2391)
The skr-1 deletion allele (tm2391) was isolated from pools of worms mutagenized by UV/trimethylpsoralen. A primer pair (Forward: CGCATCATACGACACACTCA; Reverse: AGGACAATGTGTGAAGTGTG) and a nested primer pair (Forward: ATCCGAGGCGGCAAAGGAAC; Reverse: GGGTAATTTAATCCTCGCAC) were used for PCR screening of the deletion allele. skr-1(tm2391) is a 435 base pair deletion that deletes from the 140th base pair through the 575th base pair after the skr-1 ATG start codon. This deletes the entire second exon and part of the first and second introns of skr-1 and is a predicted null allele (Fig. 2). To prove that this deletion allele is specific to skr-1 and does not affect skr-2, we rescued the skr-1(tm2391) mutant with an extrachromosomal array carrying skr-1(+). 2/2 lines rescued somatic phenotypes associated with skr-1(tm2391) such as larval lethality and hyperplasia of somatic gonad tissues. Sterility was not rescued as transgenes are often silenced in the germline in C. elegans (Kelly et al., 1997).
Yeast two-hybrid assays
The following cDNAs were used for yeast two-hybrid vector construction: skr-1 (yk1092h10), sel-10 (yk21f12) (gifts of Y. Kohara). cDNAs were PCR amplified with Gateway attB1 and attB2 primers and inserted into pDONR221 (Invitrogen, Carlsbad, CA) to create cDNA entry clones. Mutant cDNAs corresponding to skr-1(sm151) and sel-10(n1074) were introduced by PCR-mediated site-directed mutagenesis. Each cDNA entry clone was used to generate yeast two-hybrid vectors by Gateway reactions with pDEST22 (Gal4 Activation Domain, Trp selection) and pDEST32 (Gal4 DNA-binding domain, Leu selection; Invitrogen, Carlsbad, CA). Yeast Mav203 (MATα, leu2-3,112, trp1-190, his3Δ200, ade2-101, gal4Δ, gal80Δ, SPAL10::URA3, GAL1::lacZ, HIS3UAS GAL1::HIS3@LYS2, can1R, cyh2R; Invitrogen, Carlsbad, CA) cells were co-transformed with the relevant plasmids to be tested for interaction. Yeast strains bearing test plasmids were replica plated onto –Leu –Trp –Ura plates, –Leu –Trp plates containing 3-Amino-1,2,4-Triazole (3AT; 10mM, 25mM, 50 mM, 75 mM, and 100 mM), -Leu –Trp plates with 0.2% 5-Fluoroacetic Acid (5FOA), and –Leu –Trp plates. Strains were plated in a dilution series (0.5x) starting with OD600 = 10. A positive interaction drives expression of the lacZ, HIS3, and URA3 reporter genes that result in growth on –Ura and 3AT, and failure to grow in the presence of 5FOA. lacZ reporter expression was assayed by X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) staining of yeast transferred from –Leu –Trp plates to nitrocellulose filters and quantification of interactions was carried out by Chlorophenol red-β-D-galactopyranoside (CPRG; Invitrogen ProQuest Two-Hybrid System, Carlsbad, CA). In all assays, protein-protein interactions were tested with reciprocal Gal4 Activation domains (AD) and Gal4 DNA-binding domains (DB) and results were similar in both directions.
Western blotting
10 ml cultures of yeast strains expressing AD-SKR-1 or AD-SEL-10 were grown to an OD600 of 0.5 centrifuged. Yeast cell pellets were lysed with glass beads (200 µm diameter) by vortexing and boiling in SDS buffer with 0.2% β-mercaptoethanol. Samples were centrifuged to pellet cellular debris and supernatants were run on a 10% SDS-PAGE gel. The gel was transferred to a nitrocellulose membrane at 40 volts overnight. Blots were blocked with 5% milk PBS + 0.1% Tween 20. Primary antibody was incubated for 2 hours at room temperature followed by three 10-minute washes steps and secondary antibody was incubated for 1 hour at room temperature followed by three 10-minute washes. Primary antibodies were anti-Gal4 AD (1:3000; gift of G. Odorizzi lab), and loading control was anti-phosphoglycerate kinase (PGK) monoclonal antibody (1:3000; Invitrogen, Carlsbad, CA). Secondary antibody was Goat anti-mouse HRP (1:3000; Bio-Rad, Hercules, CA). Visualization was performed with SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, Il) and X-ray film exposure.
Results
The sm151 mutation suppresses the sex-specific defects of sel-10(n1074)
We conducted a sel-10(n1074) suppressor screen (see Materials and methods) to understand how sel-10 is involved in regulating the sex-determination pathway in C. elegans. From nearly 22,000 mutagenized haploid genomes, we isolated 5 extragenic sel-10(n1074) suppressors, which suppress either the Egl phenotype, or the improperly surviving CEM phenotype in hermaphrodites, or both. One suppressor mutation, sm151, was further characterized because it is a relatively strong suppressor. Compared with sel-10(n1074) hermaphrodite animals in which 86% of CEMs improperly survive and 99% of HSNs inappropriately undergo apoptosis, sm151; sel-10(n1074) double-mutant hermaphrodites have only 14% CEM survival and HSN survival is increased to 38% (Table 1). Masculinization of sel-10(n1074) hermaphrodites is also manifested in the male-specific coelomocyte positioning (64% animals) and the male-specific B cell morphology (92% animals) (Desai and Horvitz, 1989; Sulston and Horvitz, 1977). sm151 partially suppresses both phenotypes, suggesting that its suppression of the sel-10(n1074) phenotypes is not restricted to sex-specific apoptosis (Table 1). Importantly, sm151 does not suppress the sex-specific cell death defect caused by partial loss-of-function mutations in tra-1 or tra-2, or the inappropriate HSN death phenotype in egl-1(n1084) hermaphrodites (Table 2). Furthermore, sm151 hermaphrodites and males alone do not exhibit any defects in sex-specific apoptosis or sex-determination (Table 1). Together these results suggest that sm151 is a specific suppressor of sel-10(n1074).
Table 1.
Reduction of the skr-1 activity suppresses sel-10(n1074) masculinization phenotypes
| genotype | % CEM | % HSN | % Male B cell morphology |
% Male coelomocyte position |
|---|---|---|---|---|
| + | 0 | 100 | 0 | 0 |
| + male | 100 | 0 | 100 | 100 |
| sel-10(n1074) | 86 | 1 | 92 | 64 |
| skr-1(sm151); sel-10(n1074) | 14 | 38 | 50 | 11 |
| skr-1(sm151) | 0 | 100 | 0 | 0 |
| skr-1(sm151) male3 | 100 | 0 | ND | ND |
| skr-1(sm151)/+; sel-10(n1074)4 | 62 | 3 | ND | ND |
| skr-1(tm2391)/+; sel-10(n1074)5 | 43 | 8 | ND | ND |
| skr-1(sm151)/skr-1(tm2391); sel-10(n1074)6 | 1 | 77 | ND | ND |
All strains have XX karyotype, unless otherwise indicated (XO male), and carry smIs26, which harbors Ppkd-2gfp, Ptph-1gfp, and unc-76 rescuing plasmid. n = 400 for CEMs, 100 for HSNs, and 50 for both B cell and coelomocyte scoring. Scoring of CEMs, HSNs, B cells and coelomocytes is described in Materials and Methods. ND = not determined.
Male B cell morphology: B cells in L1 larvae with enlarged nucleoli.
Male coelomocyte position: one left ventral coelomocyte located posterior to the L1 somatic gonad primordium.
Actual genotype skr-1(sm151); smIs26; him-5(e1490)
Actual genotype skr-1(sm151)/dpy-5(e61) unc-29(e193); smIs26; sel-10(n1074).
Actual genotype skr-1(tm2391)/dpy-5(e61) unc-29(e193); smIs26; sel-10(n1074).
Actual genotype skr-1(tm2391)/dpy-5(e61) skr-1(sm151); smIs26; sel-10(n1074).
Table 2.
skr-1(sm151) does not suppress tra-1, tra-2, or egl-1(gf)
| genotype | % CEM | % HSN | % Tra | n |
|---|---|---|---|---|
| + | 0 | 100 | 0 | 100 |
| + male | 100 | 0 | 100 | 100 |
| skr-1(sm151) | 0 | 100 | 0 | 100 |
| tra-1(e1488) | 99 | 0 | 100 | 100 |
| skr-1(sm151); tra-1(e1488) | 99 | 0 | 100 | 100 |
| tra-2(n1106) | 76 | 7 | 17 | 100 |
| skr-1(sm151); tra-2(n1106) | 79 | 7 | 15 | 100 |
| egl-1(n1084) | 0 | 0 | ND | 50 |
| skr-1(sm151); egl-1(n1084) | 0 | 0 | ND | 50 |
All strains carry smIs26 and have XX karyotype, unless otherwise indicated (XO male). The Tra phenotype refers to hermaphrodite animals displaying evident male tail morphology. ND = not determined.
sm151 causes a missense mutation in a C. elegans Skp1-related protein
Since sm151 is a good sel-10(n1074) suppressor and does not cause any discernable phenotypes on its own, we cloned the gene affected by sm151 to determine how it functions with sel-10 (see Materials and methods). Briefly, we mapped sm151 near the loci of two C. elegans Skp1-related genes, skr-1 and skr-2, through three-factor mapping. Since Skp1 is known to function in a complex with F-box/WD40-repeat proteins such as CDC4, the mammalian homolog of SEL-10 (Cardozo and Pagano, 2004), and since SKR-1 interacts with SEL-10 based on the yeast two-hybrid assay (Yamanaka et al., 2002), we tested whether genomic fragments containing skr-1 and skr-2 can rescue n1074 suppression by sm151 (Fig. 2A). A long PCR product containing skr-1, but not skr-2, was able to reverse the suppression of the sel-10(n1074) phenotypes by sm151. A long PCR product containing skr-2, but not skr-1, failed to reverse the suppression. We sequenced the skr-1 locus from sm151; sel-10(n1074) animals and found a single nucleotide change that results in a Methionine to Isoleucine substitution at amino acid 140. M140 is a completely conserved residue in Skp1-related proteins across diverse species (Fig. 2B). Skp1 binds to F-box proteins and M140 is adjacent to predicted F-box binding residues in mammalian Skp1 (residues shaded in blue in Fig. 2B; Schulman et al., 2000), suggesting that M140I substitution may compromise the ability of SKR-1 to bind F-box proteins.
skr-1(sm151) is a weak loss-of-function allele of skr-1
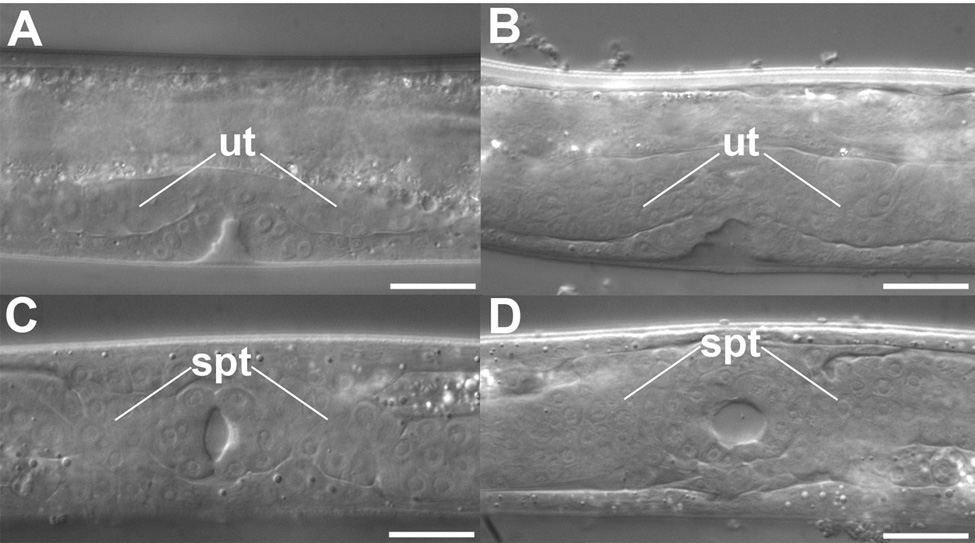
To determine the nature of the skr-1(sm151) mutation, we conducted a genetic analysis of skr-1. skr-1(sm151) does not have any discernable phenotypes on its own but RNAi-mediated knockdown of skr-1 causes lethality (Nayak et al., 2002; Yamanaka et al., 2002), suggesting that sm151 might be a weak allele. However, given the high sequence homology between skr-1 and skr-2, it is possible that skr-1(RNAi) may cause a knockdown of both skr genes, which leads to lethality. We thus screened for and obtained a deletion allele (tm2391) of skr-1 (see Materials and methods). tm2391 removes the entire second exon of skr-1 as well as part of the first and second introns and is a predicted null allele. This deletion does not disrupt the neighboring skr-2 gene (Fig. 2A). skr-1(tm2391) animals can not be maintained as a homozygous strain due to embryonic and larval lethality (data not shown). Rare skr-1(tm2391) homozygous escapers become uncoordinated sterile adults with hyperplasia of several tissues including the uterus and the spermatheca of the somatic gonad (Fig. 3). The lethality and hyperplasia phenotypes of skr-1(tm2391) animals are similar to those reported for animals treated with skr-1/skr-2 RNAi (Nayak et al., 2002). However, since skr-1(tm2391) disrupts skr-1 but not skr-2, we conclude that skr-1 and skr-2 do not have redundant functions and that loss of skr-1 is responsible for the observed lethality, hyperplasia, and sterility defects of skr-1/skr-2 RNAi treated animals. The skr-1(tm2391) hyperplasia phenotype is similar to that seen in animals deficient in cul-1 or lin-23, which encodes a C. elegans Cullin and an F-box protein with WD40-repeats, respectively (Kipreos et al., 2000; Kipreos et al., 1996). Hyperplasia of post-embryonic tissues in lin-23 and cul-1 mutants is due to a requirement of SCFLIN-23 to degrade cell cycle regulators in response to developmental cues (Kipreos et al., 2000; Kipreos et al., 1996). It has been suggested that lin-23 and cul-1 function together with skr-1 and/or skr-2 to promote degradation of cell cycle regulators (Nayak et al., 2002). Therefore, our results suggest that SKR-1, but not SKR-2, likely plays the major role in mediating the functions of the SCFLIN-23 ubiquitin ligase complex. Indeed, skr-2 is expressed in intestine and skr-1 is ubiquitously expressed (Yamanaka et al., 2002).
Figure 3.
DIC lateral views of (A) N2 normal uterine development and (B) skr-1(tm2391) uterine hyperplasia (ut = uterus). DIC ventral views of (C) N2 normal somatic gonad development and (D) skr- 1(tm2391) somatic gonad hyperplasia (sg = somatic gonad). Bars = 25 µm.
The stronger defects seen in the skr-1(tm2391) mutant suggests that skr-1(sm151) is a weak allele. However, since sm151/+ weakly suppresses sel-10(n1074) masculinization phenotypes (Table 1), it remains possible that this semi-dominant suppression is the result of a gain-of-function or a neomorphic function of skr-1. We thus compared the extent of sel- 10(n1074) suppression in sm151/sm151, sm151/tm2391, and +/tm2391 backgrounds. sel-10(n1074) phenotypes are very sensitive to the dose of skr-1: +/tm2391 suppresses to a greater extent than +/sm151 and sm151/tm2391 is a better suppressor than sm151/sm151 (Table 1). Therefore, we conclude that sm151 is a weak loss-of-function allele and that sel-10(n1074) masculinization is dependent on skr-1(+) activity. We were not able to assess the masculinization phenotypes of skr-1(tm2391); sel-10(n1074) animals due to the high penetrance of embryonic and larval lethality. Given that there are 21 skr genes in C. elegans (Nayak et al., 2002; Yamanaka et al., 2002), we cannot rule out the possibility that sel-10(n1074) masculinization phenotypes are also dependent on another skr gene..
Since skr-1 encodes a member of the SCF complex (Nayak et al., 2002; Yamanaka et al., 2002), we surmised that loss of other SCF components might also suppress the sel-10(n1074) phenotypes. cul-1 encodes a Cullin component of the SCF complex (Kipreos et al., 1996). CUL-1 also binds to SKR-1 (as well as other SKR proteins) in a yeast two-hybrid assay (Nayak et al., 2002; Yamanaka et al., 2002). Therefore we tested whether loss of cul-1 could suppress sel-10(n1074) masculinization. Due to the general sickness of cul-1(e1756) animals, we found that the presence or absence of the HSNs could not be reliably scored. However, the CEMs could be easily detected with a Ppkd-2::GFP reporter. The percentage of improperly surviving CEMs was significantly less in cul-1(e1756); sel-10(n1074) hermaphrodites than in sel-10(n1074) hermaphrodite animals (Table 3). This suggests that sel-10(n1074) masculinization is dependent upon an SCF complex that is composed of SKR-1, CUL-1 and SEL-10. The observation that the cul-1(e1756) strong loss-of-function allele did not completely suppress sel-10(n1074) masculinization likely reflects the fact that cul-1(e1756) homozygotes from heterozygous mothers exhibit partial cul-1(+) maternal rescue (Kipreos et al., 1996).
Table 3.
Loss of cul-1 activity suppresses sel-10(n1074) masculinization phenotypes
| genotype | % CEM | n |
|---|---|---|
| + | 0 | 100 |
| + male | 100 | 100 |
| cul-1(e1756) | 0 | 100 |
| sel-10(n1074) | 76 | 100 |
| cul-1(e1756); sel-10(n1074) | 35 | 44 |
| cul-1(e1756) male | 100 | 20 |
All strains have XX karyotype, unless otherwise indicated (XO male), and carry smIs23, which harbors Ppkd-2gfp. cul-1(e1756) animals were the non-GFP progeny from cul-1(e1756)/hT2[GFP] parents.
skr-1(sm151) genetically interacts with sel-10 in lin-12 activity assays
sel-10 was first identified as a regulator of LIN-12/Notch signaling (Hubbard et al., 1997), which is required for the anchor cell/ventral uterine precursor (AC/VU) cell fate decision (Greenwald et al., 1983; Kimble and Hirsh, 1979). Loss of lin-12 causes two initially equivalent cells, Z1.ppp and Z4.aaa, to adopt an AC fate and is thus called a 2AC phenotype (Greenwald et al., 1983). LIN-12/Notch is also required for the vulval precursor cells (VPCs) to adopt proper fates (1° or 2° vulval fates, or a 3° non-vulval fate). Hyperactive lin-12 activity results in all VPCs adopting 2° fates and thus over-induction of the vulval tissue, which is known as a Multivulva (Muv) phenotype (Sundaram and Greenwald, 1993).
Since SEL-10 is suggested to target LIN-12 for ubiquitin-mediated degradation, loss of sel-10 would result in an increase in the level of LIN-12. Phenotypically, loss of sel-10 suppresses the 2AC phenotype associated with lin-12 reduction-of-function (rf) alleles and enhances the Muv phenotype of lin-12 gain-of-function (gf) alleles (Hubbard et al., 1997). Although sel-10(n1074) has a dominant effect on sex-determination, sel-10(n1074) is a recessive reduction-of-function mutation with respect to lin-12 activity assays (Jager et al., 2004). Unlike sel-10(n1074), which suppresses the lin-12(rf) 2AC phenotype from 80% to 25%, skr-1(sm151) did not suppress the 2AC phenotypes of the lin-12(rf) animals on its own, but further reduced the 2AC phenotype of lin-12(rf); sel-10(n1074) animals from 25% 2AC to 18% 2AC (Table 4). Importantly, skr-1(sm151) did not confer additional suppression of the lin-12(rf) 2AC phenotype in a sel-10(ok1632) null background (see Materials and methods), which has 19% 2AC. The effect of skr-1(sm151) in the lin-12(rf) assay is small and can only be detected in the n1074 background. Similarly, we did not observe an enhancement of the lin-12(gf) Muv phenotype by skr-1(sm151). However, we found that skr-1(sm151) greatly enhances the Muv phenotype of lin-12(gf); sel- 10(n1074) animals from 31% to 88% and does not enhance the Muv phenotype of lin-12(gf); sel-10(ok1632) (Table 5).
Table 4.
skr-1(sm151) suppresses lin-12(rf) in a sel-10(n1074) background
| genotype | % 2 AC phenotype (25°C) | n |
|---|---|---|
| skr-1(sm151) | 0 | 100 |
| skr-1(sm151); sel-10(n1074) | 0 | 100 |
| skr-1(sm151); sel-10(ok1632) | 0 | 100 |
| lin-12(ar170) | 80 | 100 |
| skr-1(sm151); lin-12(ar170) | 77 | 100 |
| lin-12(ar170); sel-10(n1074) | 25* | 200 |
| lin-12(ar170); sel-10(ok1632) | 19 | 100 |
| skr-1(sm151); lin-12(ar170); sel-10(n1074) | 18* | 200 |
| skr-1(sm151); lin-12(ar170); sel-10(ok1632) | 18 | 100 |
| skr-1(sm151); unc-76 | 0 | 100 |
| skr-1(sm151); sel-10(ar41) unc-76 | 0 | 100 |
| lin-12(ar170); unc-76 | 79 | 100 |
| skr-1(sm151); lin-12(ar170); unc-76 | 81 | 100 |
| lin-12(ar170); sel-10(ar41) unc-76 | 21 | 100 |
| skr-1(sm151); lin-12(ar170); sel-10(ar41) unc-76 | 19 | 100 |
ACs were scored with DIC (100x) at the late L3/early L4 stage.
These values are statistically different based on Chi square analysis, P < 0.03. unc-76(e911) is linked to sel-10(ar41) and was included in relevant control strains.
Table 5.
skr-1(sm151) enhances lin-12(gf) in a sel-10(n1074) background
| genotype | % Muv (20°C) | n |
|---|---|---|
| skr-1(sm151) | 0 | 100 |
| skr-1(sm151); sel-10(n1074) | 0 | 100 |
| skr-1(sm151); sel-10(ok1632) | 0 | 100 |
| lin-12(n379) | 10 | 100 |
| skr-1(sm151); lin-12(n379) | 12 | 100 |
| lin-12(n379); sel-10(n1074) | 31 | 100 |
| lin-12(n379); sel-10(ok1632) | 96 | 100 |
| skr-1(sm151); lin-12(n379); sel-10(n1074) | 88 | 100 |
| skr-1(sm151); lin-12(n379); sel-10(ok1632) | 97 | 100 |
| skr-1(sm151); unc-76 | 0 | 100 |
| skr-1(sm151); sel-10(ar41) unc-76 | 0 | 100 |
| lin-12(n379); unc-76 | 11 | 100 |
| skr-1(sm151); lin-12(n379); unc-76 | 12 | 100 |
| lin-12(n379); sel-10(ar41) unc-76 | 59 | 100 |
| skr-1(sm151); lin-12(n379); sel-10(ar41) unc-76 | 57 | 100 |
The Muv phenotype was scored in adult animals using DIC (40x). unc-76(e911) is linked to sel-10(ar41) and was included in relevant control strains.
Since skr-1(sm151) had no effect on the lin-12(rf) or lin-12(gf) phenotype in sel-10(+) genetic backgrounds, skr-1(sm151) may be a very weak allele and the phenotypic assays are not sensitive enough to detect a difference. Alternatively, skr-1(sm151) could be allele-specific to sel-10(n1074). To distinguish between these two possibilities, we tested the effect of skr-1(sm151) on the lin-12(rf) or lin-12(gf) phenotype in another sel-10 mutant background, sel-10(ar41). sel-10(ar41) contains a stop codon before the WD40-repeats but does not behave genetically as a null mutation (Hubbard et al., 1997; Table 5), possibly due to read through of the stop codon. In the lin-12(gf) phenotypic assay, sel-10(ar41) does not enhance the Muv phenotype as well as sel-10(ok1632null) (Table 5), again indicating that sel-10(ar41) probably is not a null allele.
In these lin-12 activity assays, skr-1(sm151) did not have any significant effect on the lin-12(rf) or lin-12(gf) phenotypes in a sel-10(ar41) background. Although there is little room for further suppression of the lin-12(rf) 2AC phenotype by skr-1(sm151) in lin-12(rf); sel-10(ar41) animals compared with lin-12(rf); sel-10(ok1632null) animals (21% to 19%; Table 4), there is a significant difference in the percentage of the Muv phenotype between lin-12(gf); sel-10(ar41) (59%) and lin-12(gf); sel-10(ok1632) (96%) animals, indicating that the Muv phenotype of lin-12(gf); sel-10(ar41) can be enhanced (Table 5). The fact that skr-1(sm151) can enhance the effect of sel-10(n1074) but not that of sel-10(ar41) in the lin-12(rf) or lin-12(gf) assay is consistent with skr-1(sm151) being an allele-specific suppressor/enhancer of sel-10(n1074). However, additional non-null alleles of sel-10 are not available to further test this hypothesis.
skr-1(sm151) does not genetically interact with lin-23
There are many F-box proteins in C. elegans, including SEL-10 and LIN-23, both of which also contain WD40 repeats. The observation that skr-1(tm2391) phenocopies lin-23(null) mutants (Fig. 3) prompted us to test whether skr-1(sm151) could enhance the defect caused by lin-23 mutations. Since there are no partial loss-of-function lin-23 alleles that cause hyperplasia, we tested instead whether skr-1(sm151) could enhance the defect of lin-23(ot1) animals, which are deficient in AVL GABAergic neuron axon outgrowth as assayed by the unc-47::GFP reporter (Mehta et al., 2004). No enhancement of lin-23(ot1) AVL axonal outgrowth defect was seen, suggesting that skr-1(sm151) activity is sufficient for lin-23. However, lin-23(ot1) is almost null for the AVL axon defects and leaves little room for enhancement. Therefore we tested whether skr-1(sm151) could cause an AVL axon defect in a lin-23(ot1)/+ genetic background. We did not see any effect of skr-1(sm151) in this background either. Taken together, skr-1(sm151) does not have a detectable effect on the processes regulated by lin-23.
SKR-1(M140I) and SEL-10(G567E) reduce SKR-1/SEL-10 binding
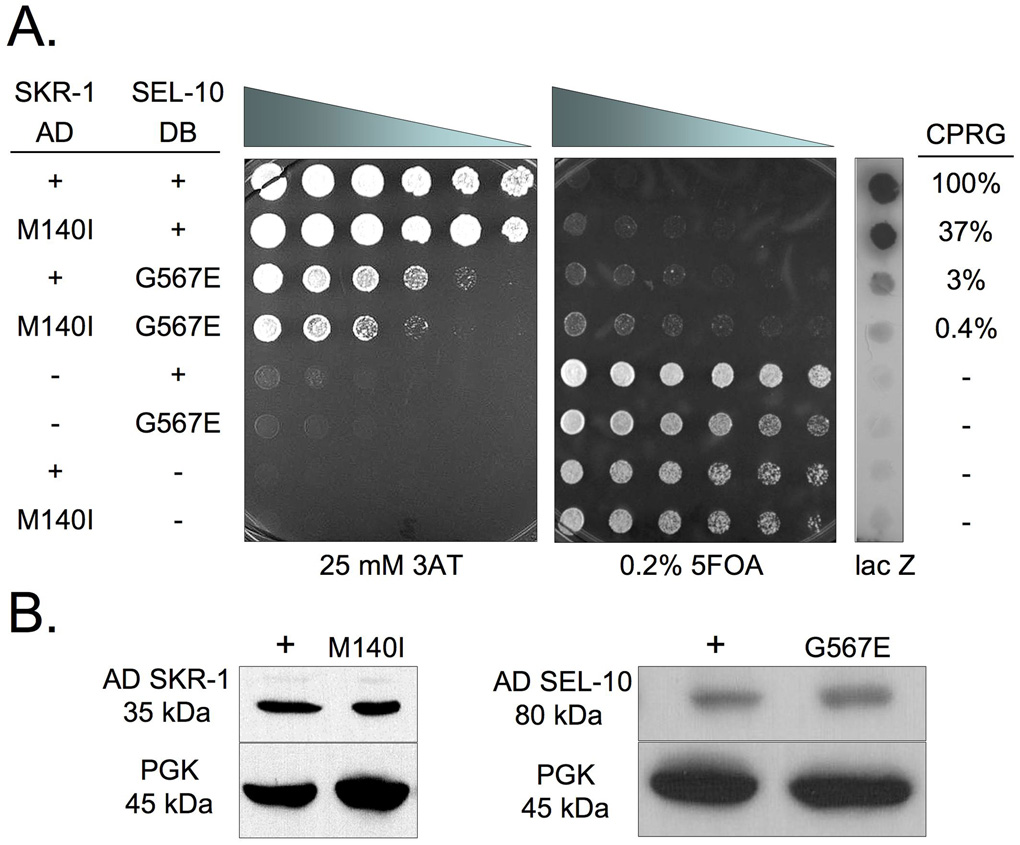
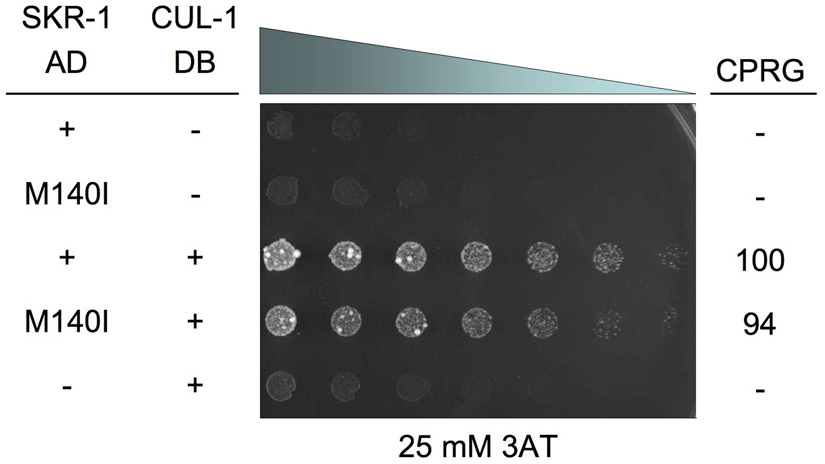
Since SKR-1(M140I) alters a residue that is within the F-box interaction region of SKR-1, SKR-1(M140I) may affect SKR-1 binding to SEL-10. We used the yeast two-hybrid assay to assess the binding of the SKR-1 proteins (wild-type and M140I) to SEL-10 as described previously by Yamanaka et al. (2002). We found a strong protein interaction between GAL4 activation domain (AD) SKR-1 fusion (AD-SKR-1) and GAL4 DNA binding domain (DB) SEL-10 fusion (DB-SEL-10) based on growth on 3-Amino-1,2,4-Triazole (3AT), inhibition of growth on 0.2% 5-fluoroorotic acid (5FOA), and lacZ staining (see Materials and methods; Fig. 4A). DB-SKR-1 and AD-SEL-10 also interact strongly as revealed by the same assays but the interaction is slightly weaker in all assays, particularly in growth inhibition on 0.2% 5FOA and lacZ staining (data not shown). A quantitative measurement of the β-galactosidase activity was performed with a chlorophenolred-β-D-galactopyranoside (CPRG) assay (see Materials and methods) and we found that the SKR-1(M140I)/SEL-10 interaction is 37% as strong as SKR-1/SEL-10 in this assay (Fig. 4A). This suggests that SKR-1(M140I) reduces SKR-1 binding to SEL-10.
Figure 4.
(A) A yeast two-hybrid assay with AD-SKR-1, DB-SEL-10, and the respective M140I and G567E mutants or empty vectors (−). Yeast were plated in a 0.5x dilution series from left to right starting at OD600 = 10. Positive interactions exhibit increased growth on 3AT and decreased growth on 5FOA relative to negative controls (lanes 5–8). Positive interactions are shown by lacZ staining. ®-galactosidase activity based on the CPRG assay is shown as a percentage of the activity observed in yeast expressing SKR-1(+)/SEL-10(+). Yeast were grown for two days before imaging. (B) Western blot analysis of yeast lysates containing AD-SKR-1 and AD-SKR-1(M140I) (left) and AD-SEL-10 and AD-SEL-10(G567E) (right). Phosphoglycerate kinase (PGK) was used as a loading control. See Materials and Methods for additional details.
We also tested the interaction between SKR-1(WT or M140I) and SEL-10(G567E) proteins and found that SKR-1(M140I) had significantly reduced binding to SEL-10(G567E) compared to the SKR-1/SEL-10(G567E) interaction in all three yeast two-hybrid assays (Fig. 4A). Surprisingly, SEL-10(G567E) binds only weakly to SKR-1 compared to the SEL-10/SKR-1 interaction (3% of the SEL-10/SKR-1 interaction; Fig. 4A). The F-box motif mediates the interaction between F-box proteins and the Skp1 proteins (Bai et al., 1996). Intriguingly, the F-box of SEL-10 is located at the amino terminal region, whereas the G567E mutation locates at the SEL-10 carboxyl terminal tail. Based on the structures of SCF complexes containing the mammalian SEL-10 homologues (Hao et al., 2007; Orlicky et al., 2003), the C-terminal tails of F-box proteins do not contact Skp1. We thus examined if the G567E mutation may affect the stability of SEL-10 in yeast by performing western blot analysis (see Materials and methods) on yeast lysates to determine the expression levels of SEL-10, SEL-10(G567E), SKR-1, and SKR-1(M140I). We did not detect any significant difference in protein expression of the wild-type versus the mutant forms of SEL-10 and SKR-1 (Fig. 4B). Therefore we conclude that the M140I mutation in SKR-1 and the G567E mutation in SEL-10 both reduce SKR-1/SEL-10 binding. Our data suggest that the C-terminus of SEL-10 is important for binding to SKR-1, which is an unexpected finding that has not been reported previously. Despite greatly reduced binding between SKR-1(M140I) and SEL-10(G567E) (0.4% of the SKR-1/SEL-10 interaction), SKR-1(M140I)/SEL-10(G567E) still interact at a level well above background in the yeast two-hybrid system and may retain some activity in promoting degradation of their target protein(s) in vivo (Fig. 4A).
We also tested if the M140I mutation disrupts SKR-1 binding to another SCF component, CUL-1 (Yamanaka et al., 2002; Nayak et al., 2002). We found no significant difference between the binding of SKR-1/CUL-1 and SKR-1(M140I)/CUL-1 in all yeast two-hybrid assays (Fig. 5 and data not shown). We conclude that the SKR-1 M140I mutation specifically interferes with the binding to SEL-10, especially the binding to the SEL-10(G567E) protein, which underlies the strong suppression of the sel-10(n1074) defects by skr-1(sm151).
Figure 5.
A yeast two-hybrid assay with AD-SKR-1 or AD-SKR-1(M140) and DB-CUL-1 or empty vectors (−).Yeast were plated in a 0.5x dilution series from left to right starting at OD600 = 10. Positive interactions exhibit increased growth on 3AT relative to negative controls (lanes 1, 2, and 5). ®-galactosidase activity based on CPRG assay is shown as a percentage of the activity observed in yeast expressing SKR-1(+)/CUL-1(+). Yeast were grown for two days before imaging. See Materials and Methods for additional details.
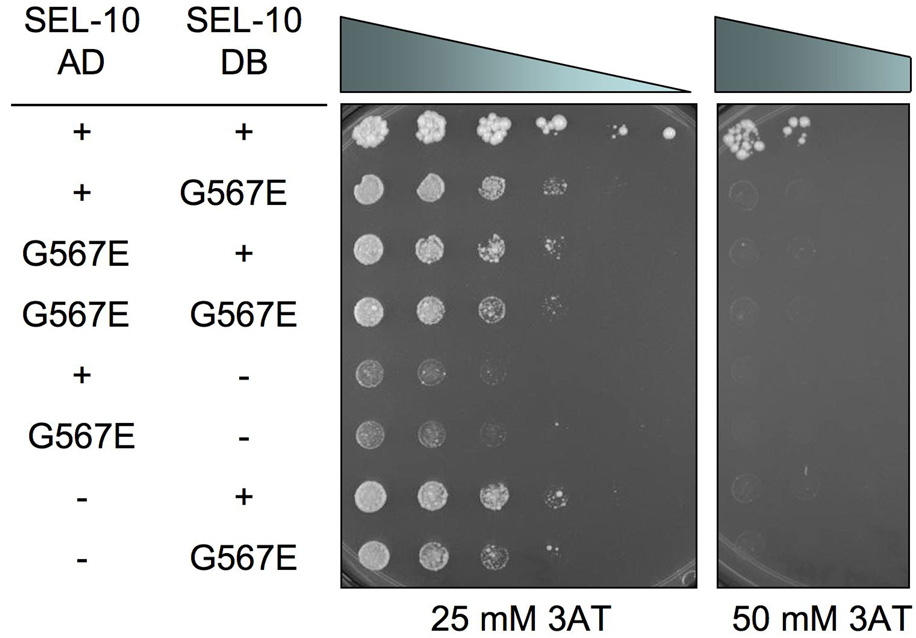
The SEL-10 G567E mutation disrupts the formation of SEL-10 homodimers
The yeast SCFCDC4 and mammalian SCFFbw7 complexes form dimers that are important for the function of the complex as an E3 ubiquitin ligase (Hao et al., 2007; Tang et al., 2007). The WD40-repeat F-box proteins CDC4, human β-TrCP, and C. elegans SEL-10 all contain a conserved D-domain, just N-terminal to the F-box domain, that is required for dimerization (Tang et al., 2007). We thus tested if SEL-10 interacts with itself by the yeast two-hybrid analysis. We found that SEL-10 does self interact, possibly by forming homodimers, and that the G567E mutation disrupts SEL-10 dimerization (Fig. 6). Therefore, sel-10(n1074) may cause reduction of sel-10 activity and the activity of the SCFSEL-10 complex by disrupting SEL-10 dimerization.
Figure 6.
A yeast two-hybrid assay with AD-SEL-10 and DB-SEL-10. Yeast were plated in a 0.5x dilution series from left to right starting at OD600 = 10. Positive interactions exhibit increased growth on 3AT relative to negative controls (lanes 5–8). Yeast were grown for seven days before imaging. See Materials and Methods for additional details.
Discussion
SKR-1 is a critical component of the C. elegans SCFSEL-10 and SCFLIN-23 complexes
The SCF complex is a multi-subunit E3 ubiquitin ligase, conserved from yeast to humans, that attaches polyubiquitin chains to its protein substrates and targets them for degradation by the proteasome. In yeast and humans, Skp1, Rbx, and Cul1 are the common components and the variable F-box proteins provide substrate specificity (Kipreos and Pagano, 2000). Interestingly, in yeast and humans, there is a single Skp1 gene but in C. elegans there are 21 Skp1-related (skr) genes (Nayak et al., 2002; Yamanaka et al., 2002). The reason for so many skr genes in C. elegans is not clear.
In this study, we present genetic evidence suggesting that SKR-1 is the Skp1 protein for two major C. elegans SCF complexes, SCFSEL-10 and SCFLIN-23, and that it is not redundant with other SKR proteins. The F-box protein SEL-10 is involved in the ubiquitin-mediated degradation of LIN-12 in the AC/VU decision and vulval induction (Hubbard et al., 1997) and has been proposed to promote degradation of FEM-1 and FEM-3 in sex determination (Jager et al., 2004). We show that SKR-1 is a critical component of the SCFSEL-10 complex that regulates the activity of these pathways. First, SEL-10 and SKR-1 physically interact in yeast 2-hyrid assays, whereas other SKR proteins do not interact with SEL-10 (Yamanaka et al., 2002). Second, our genetic analysis suggests that the role of sel-10 in sex determination is dependent on skr-1(+) activity: skr-1(sm151) or skr-1(sm151)/skr-1(tm2391) suppresses masculinization of hermaphrodites caused by sel-10(n1074) (Table 1). In addition, skr-1(sm151) can further suppress the 2AC defect of the lin-12(rf); sel-10(n1074) mutant and enhance the Muv defect of the lin-12(gf); sel-10(n1074) mutant (Table 4). It remains possible that the SCFSEL-10 complex uses SKR-1 in some cells and other SKR proteins in other cell types. However, we find this unlikely since SKR-1::GFP is widely expressed in C. elegans (Yamanaka et al., 2002) and our genetic analysis of skr-1 and sel-10 interactions includes several different cell types such as HSN and CEM neurons, the AC/VU cells, and P.np cells, which become either vulval tissue or hypodermis. Since a loss-of-function mutation in cul-1 suppresses masculinization of hermaphrodites caused by sel-10(n1074) (Table 3), this result suggests that CUL-1 is also a component of the SCFSEL-10 complex.
In addition to SEL-10, LIN-23 is another well-described F-box protein in C. elegans. lin-23 promotes cell cycle exit during C. elegans development and appears to work in conjunction with cul-1 based on their similar loss-of-function phenotypes such as hyperplasia of several tissues (Kipreos et al., 2000). It has been suggested that both skr-1 and skr-2 may work with lin-23 and cul-1 to regulate cell cycle exit (Nayak et al., 2002), since both SKR-1 and SKR-2 bind CUL-1 in a yeast 2-hybrid assay. However, the strong sequence similarity between skr-1 and skr-2 prevents dissection of their respective contributions by RNAi, which is predicted to knockdown both genes (Nayak et al., 2002). Our observation that a null allele of skr-1 results in hyperplasia of several tissues similar to that seen in lin-23 and cul-1 loss-of-function mutants (Fig. 3) establishes that SKR-2 could not substitute for SKR-1 in the SCFLIN-23 complex and that SKR-1 likely is the major, if not the only, SKR component for the SCFLIN-23 complex.
skr-1(sm151) is a specific suppressor/enhancer of sel-10(n1074)
Although skr-1(sm151) was isolated as a strong suppressor of the masculinization phenotype caused by the semi-dominant sel-10(n1074) mutation, skr-1(sm151) does not suppress the masculinization phenotype caused by loss-of-function mutations in tra-1 or tra-2. The skr-1(sm151) mutant by itself is superficially wild-type and does not display any detectable defects. Interestingly, skr-1(sm151) specifically enhances the Muv defect of the lin-12(gf); sel-10(n1074) mutant but not that of the lin-12(gf); sel-10(ar41) mutant, which contains a sel-10 strong loss-of-function allele, but not a null allele (Table 5). In addition, skr-1(sm151) does not enhance or suppress the AVL neuron axonal outgrowth defect caused by a loss-of-function mutation in another closely related F-box gene, lin-23. These results together suggest that skr-1(sm151) is likely a mutation that specifically suppresses/enhances the defect of sel-10(n1074) animals in sex-determination and lin-12 activity assays. There are no other non-null sel-10 alleles available for testing whether skr-1(sm151) is truly an allele-specific suppressor/enhancer of sel-10(n1074).
Paradoxically, complete loss of the sel-10 activity also results in weak masculinization of hermaphrodites (Jager et al., 2004), suggesting that the stronger masculinization phenotype caused by sel-10(n1074) is unlikely a result of increased SEL-10 activity but rather a gain of new function for the SEL-10(G567E) protein. One hypothesis is that there are redundant F-box proteins that target FEM-1 and FEM-3 for degradation (Jager et al., 2004), although there are no other known F-box proteins except SEL-10 that affect sex-determination. Loss of sel-10 is thus compensated for by other F-box genes, which would explain why loss of sel-10 results in a weak masculinization phenotype. On the other hand, the sel-10(n1074) mutation may result in the formation of a stable but non-functional SCFSEL-10(G567E) complex such that other redundant F-box proteins do not gain access to FEM-1 or FEM-3 (Jager et al., 2004). As a result, sel-10(n1074) causes a stronger masculinization defect. Since we isolated skr-1(sm151) as a suppressor of the sel-10(n1074) masculinization defect and the sm151 (M140I) mutation greatly reduces SKR-1 binding to SEL-10(G567E), it is possible that the SKR-1(M140I)-containing SCFSEL-10(G567E) complex may now allow other F-box proteins to gain access to the complex and degrade FEM-1 and FEM-3. However, given that SEL-10(G567E) binds wild-type SKR-1 very poorly (Fig. 4A) and fails to form SEL-10 homodimers, it seems unlikely that SEL-10(G567E) would result in the formation of a stable, dominant-negative SCFSEL-10(G567E) complex. In addition, we find that neither skr-1(sm151) nor cul-1(e1756) can enhance the mild masculinization defect caused by sel-10(ok1632null) (Table 7), suggesting that there are no other F-box proteins forming E3 ligase complexes with SKR-1 and CUL-1 and acting redundantly with SEL-10 to regulate sex-determination. In fact, our results are more consistent with sel-10(n1074) causing the formation of an unstable, compromised SCFSEL-10(G567E) complex with reduced E3 ligase activity. Indeed, sel-10(n1074) behaves as a loss-of-function mutation with respect to the lin-12 signaling (Table 4 and Table 5) (Jager et al., 2004). Similarly, the SCFSEL-10(G567E) complex may have reduced E3 ligase activity towards all of its substrates, including FEM-1 and FEM-3.
Table 7.
skr-1(sm151) and cul-1(e1756) do not enhance the masculinization defect caused by sel-10(ok1632null)
| genotype | % CEM | % HSN | n |
|---|---|---|---|
| sel-10(ok1632) | 1 | 97 | 100 |
| skr-1(sm151) | 0 | 100 | 100 |
| skr-1(sm151); sel-10(ok1632) | <1 | 97 | 100 |
| cul-1(e1756) | 0 | ND | 31 |
| cul-1(e1756); sel-10(ok1632) | 0 | ND | 26 |
All strains carry smIs26, except for strains bearing cul-1(e1756), which carry smIs23. ND = not determined.
The SEL-10 G567E mutation is located at the C-terminus inside the eighth WD40-repeat region, which is a site involved in substrate binding in homologous SCF complexes (Hao et al., 2007; Jager et al., 2004; Orlicky et al., 2003). It is possible that the G567E substitution alters SEL-10 substrate binding in a way that SCFSEL-10(G567E) targets a protein for degradation that normally is not recognized by SCFSEL-10. In this case, such a neomorphic activity of the SCFSEL-10(G567E) complex may target a hermaphrodite-promoting, sex-determination factor for degradation. If this factor normally is not targeted for degradation, then even a small decrease in steady-state level of this protein may have a large impact on sex-determination. We note that even if the SCFSEL-10(G567E) complex is less stable due to reduced SKR-1/SEL-10 binding caused by the G567E mutation (Fig. 4A), any such degradation of its neomorphic target could have a significant effect in sex determination and could be suppressed by the SKR-1(M140I) mutation that further destabilizes the SCF complex.
The C-terminus of SEL-10
The SEL-10 G567E mutation is located at the C-terminus inside the eighth WD40-repeat region, a site that is predicted to be involved in substrate binding based on the structure of the Skp1/CDC4 complex and the structure of the Skp1/Fbw7 complex (Hao et al., 2007; Jager et al., 2004; Orlicky et al., 2003). The C-terminal tails of the F-box proteins appear to be away from Skp1 in these structures. It is thus surprising that the G567E mutation greatly reduces SEL-10 binding to SKR-1. Since G567E also disrupts SEL-10 dimerization, which has been shown to be important for the SCF activity (Tang et al., 2007), it is possible that SEL-10 dimerization is important for the SEL-10/SKR-1 interaction and that G567E impairs the SEL-10/SKR-1 binding by disrupting SEL-10 dimerization. The F-box of SEL-10 is presumed to confer SKR-1 binding and we confirmed that an F-box deletion prevents SKR-1/SEL-10 interaction (data not shown). The C-terminus of SEL-10 is thus required for efficient SKR-1 binding but is not sufficient for mediating SKR-1 binding in the absence of the F-box. Our study thus reveals a previously unreported role of the C-terminus of SEL-10 in stabilizing the interaction between a F-box protein and a Skp1 protein. It will be interesting to see if the C-terminus of other F-box proteins influences Skp1 binding.
Table 6.
skr-1(sm151) does not show genetic interactions with lin-23
| genotype | % AVL defects | n |
|---|---|---|
| + | 0 | 100 |
| skr-1(sm151) | 1 | 100 |
| lin-23(ot1) | 93 | 100 |
| skr-1(sm151); lin-23(ot1) | 99 | 100 |
| lin-23(ot1)/+ | 0 | 50 |
| lin-23(ot1)/+; skr-1(sm151)/+ | 0 | 50 |
| lin-23(ot1)/+; skr-1(sm151) | 0 | 50 |
lin-23(ot1) animals show defects in AVL axon outgrowth as assayed by unc-47::GFP. All strains were scored as L4s with oxIs12[unc-47::GFP].
Acknowledgements
We gratefully acknowledge Ning Zheng and Xuedong Liu for advice and discussion, the technical assistance of Eric Griffiths, the Caenorhabditis Genetics Stock Center for strains, and Xue lab members for discussions and comments on the manuscript. This work is supported by a NIH NRSA postdoctoral fellowship (5 F32 GM075612) to D.J.K., a Burroughs Wellcome Fund Career Award and NIH R01 grants (GM66262 and GM59083) to D.X., and a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.M.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ahringer J, Rosenquist TA, Lawson DN, Kimble J. The Caenorhabditis elegans sex determining gene fem-3 is regulated post-transcriptionally. Embo. J. 1992;11:2303–2310. doi: 10.1002/j.1460-2075.1992.tb05289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, Elledge SJ. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chin-Sang ID, Spence AM. Caenorhabditis elegans sex-determining protein FEM-2 is a protein phosphatase that promotes male development and interacts directly with FEM-3. Genes. Dev. 1996;10:2314–2325. doi: 10.1101/gad.10.18.2314. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell. 1998;93:519–529. doi: 10.1016/s0092-8674(00)81182-4. [DOI] [PubMed] [Google Scholar]
- Conradt B, Horvitz HR. The TRA-1A sex determination protein of C. elegans regulates sexually dimorphic cell deaths by repressing the egl-1 cell death activator gene. Cell. 1999;98:317–327. doi: 10.1016/s0092-8674(00)81961-3. [DOI] [PubMed] [Google Scholar]
- DeLong L, Plenefisch JD, Klein RD, Meyer BJ. Feedback control of sex determination by dosage compensation revealed through Caenorhabditis elegans sdc-3 mutations. Genetics. 1993;133:875–896. doi: 10.1093/genetics/133.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai C, Garriga G, McIntire SL, Horvitz HR. A genetic pathway for the development of the Caenorhabditis elegans HSN motor neurons. Nature. 1988;336:638–646. doi: 10.1038/336638a0. [DOI] [PubMed] [Google Scholar]
- Desai C, Horvitz HR. Caenorhabditis elegans mutants defective in the functioning of the motor neurons responsible for egg laying. Genetics. 1989;121:703–721. doi: 10.1093/genetics/121.4.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RM, Correll CC, Kaplan KB, Deshaies RJ. A complex of Cdc4p, Skp1p, and Cdc53p/cullin catalyzes ubiquitination of the phosphorylated CDK inhibitor Sic1p. Cell. 1997;91:221–230. doi: 10.1016/s0092-8674(00)80404-3. [DOI] [PubMed] [Google Scholar]
- Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- Hao B, Oehlmann S, Sowa ME, Harper JW, Pavletich NP. Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell. 2007;26:131–143. doi: 10.1016/j.molcel.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Hodgkin J. A genetic analysis of the sex-determining gene, tra-1, in the nematode Caenorhabditis elegans. Genes. Dev. 1987;1:731–745. doi: 10.1101/gad.1.7.731. [DOI] [PubMed] [Google Scholar]
- Hubbard EJ, Wu G, Kitajewski J, Greenwald I. sel-10, a negative regulator of lin-12 activity in Caenorhabditis elegans, encodes a member of the CDC4 family of proteins. Genes. Dev. 1997;11:3182–3193. doi: 10.1101/gad.11.23.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Schwartz HT, Horvitz HR, Conradt B. The Caenorhabditis elegans F-box protein SEL-10 promotes female development and may target FEM-1 and FEM-3 for degradation by the proteasome. Proc. Natl. Acad. Sci. U S A. 2004;101:12549–12554. doi: 10.1073/pnas.0405087101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly WG, et al. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble J, Hirsh D. The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 1979;70:396–417. doi: 10.1016/0012-1606(79)90035-6. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Gohel SP, Hedgecock EM. The C. elegans F-box/WD-repeat protein LIN-23 functions to limit cell division during development. Development. 2000;127:5071–5082. doi: 10.1242/dev.127.23.5071. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Lander LE, Wing JP, He WW, Hedgecock EM. cul-1 is required for cell cycle exit in C. elegans and identifies a novel gene family. Cell. 1996;85:829–839. doi: 10.1016/s0092-8674(00)81267-2. [DOI] [PubMed] [Google Scholar]
- Kipreos ET, Pagano M. The F-box protein family. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-5-reviews3002. REVIEWS3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwabara PE, Okkema PG, Kimble J. tra-2 encodes a membrane protein and may mediate cell communication in the Caenorhabditis elegans sex determination pathway. Mol. Biol. Cell. 1992;3:461–473. doi: 10.1091/mbc.3.4.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madl JE, Herman RK. Polyploids and sex determination in Caenorhabditis elegans. Genetics. 1979;93:393–402. doi: 10.1093/genetics/93.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta N, Loria PM, Hobert O. A genetic screen for neurite outgrowth mutants in Caenorhabditis elegans reveals a new function for the F-box ubiquitin ligase component LIN-23. Genetics. 2004;166:1253–1267. doi: 10.1534/genetics.166.3.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer BJ. Sex in the worm counting and compensating X-chromosome dose. Trends in Genetics. 2000;16:247–253. doi: 10.1016/s0168-9525(00)02004-7. [DOI] [PubMed] [Google Scholar]
- Miskowski J, Li Y, Kimble J. The sys-1 gene and sexual dimorphism during gonadogenesis in Caenorhabditis elegans. Dev. Biol. 2001;230:61–73. doi: 10.1006/dbio.2000.9998. [DOI] [PubMed] [Google Scholar]
- Nayak S, Santiago FE, Jin H, Lin D, Schedl T, Kipreos ET. The Caenorhabditis elegans Skp1-related gene family: diverse functions in cell proliferation, morphogenesis, and meiosis. Curr. Biol. 2002;12:277–287. doi: 10.1016/s0960-9822(02)00682-6. [DOI] [PubMed] [Google Scholar]
- Orlicky S, Tang X, Willems A, Tyers M, Sicheri F. Structural basis for phosphodependent substrate selection and orientation by the SCFCdc4 ubiquitin ligase. Cell. 2003;112:243–256. doi: 10.1016/s0092-8674(03)00034-5. [DOI] [PubMed] [Google Scholar]
- Peden E, Kimberly E, Gengyo-Ando K, Mitani S, Xue D. Control of sex-specific apoptosis in C. elegans by the BarH homeodomain protein CEH-30 and the transcriptional repressor UNC-37/Groucho. Genes. Dev. 2007;21:3195–3207. doi: 10.1101/gad.1607807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry MD, Li W, Trent C, Robertson B, Fire A, Hageman JM, Wood WB. Molecular characterization of the her-1 gene suggests a direct role in cell signaling during Caenorhabditis elegans sex determination. Genes. Dev. 1993;7:216–228. doi: 10.1101/gad.7.2.216. [DOI] [PubMed] [Google Scholar]
- Pilgrim D, McGregor A, Jackle P, Johnson T, Hansen D. The C. elegans sex-determining gene fem-2 encodes a putative protein phosphatase. Mol. Biol. Cell. 1995;6:1159–1171. doi: 10.1091/mbc.6.9.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman BA, Carrano AC, Jeffrey PD, Bowen Z, Kinnucan ER, Finnin MS, Elledge SJ, Harper JW, Pagano M, Pavletich NP. Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature. 2000;408:381–386. doi: 10.1038/35042620. [DOI] [PubMed] [Google Scholar]
- Skowyra D, Craig KL, Tyers M, Elledge SJ, Harper JW. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- Spence AM, Coulson A, Hodgkin J. The product of fem-1, a nematode sex-determining gene, contains a motif found in cell cycle control proteins and receptors for cell-cell interactions. Cell. 1990;60:981–990. doi: 10.1016/0092-8674(90)90346-g. [DOI] [PubMed] [Google Scholar]
- Starostina NG, Lim JM, Schvarzstein M, Wells L, Spence AM, Kipreos ET. A CUL-2 ubiquitin ligase containing three FEM proteins degrades TRA-1 to regulate C. elegans sex determination. Dev. Cell. 2007;13:127–139. doi: 10.1016/j.devcel.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE. Post-embryonic development in the ventral cord of Caenorhabditis elegans. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 1976;275:287–297. doi: 10.1098/rstb.1976.0084. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Sundaram M, Greenwald I. Genetic and phenotypic studies of hypomorphic lin-12 mutants in Caenorhabditis elegans. Genetics. 1993;135:755–763. doi: 10.1093/genetics/135.3.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Orlicky S, Lin Z, Willems A, Neculai D, Ceccarelli D, Mercurio F, Shilton BH, Sicheri F, Tyers M. Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell. 2007;129:1165–1176. doi: 10.1016/j.cell.2007.04.042. [DOI] [PubMed] [Google Scholar]
- Trent C, Purnell B, Gavinski S, Hageman J, Chamblin C, Wood WB. Sex-specific transcriptional regulation of the C. elegans sex-determining gene her-1. Mech. Dev. 1991;34:43–55. doi: 10.1016/0925-4773(91)90090-s. [DOI] [PubMed] [Google Scholar]
- White JQ, Nicholas TJ, Gritton J, Truong L, Davidson ER, Jorgensen EM. The sensory circuitry for sexual attraction in C. elegans males. Curr. Biol. 2007;17:1847–1857. doi: 10.1016/j.cub.2007.09.011. [DOI] [PubMed] [Google Scholar]
- Wu G, Hubbard EJ, Kitajewski JK, Greenwald I. Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein, and SEL-12 presenilin. Proc. Natl. Acad. Sci. U S A. 1998;95:15787–15791. doi: 10.1073/pnas.95.26.15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka A, Yada M, Imaki H, Koga M, Ohshima Y, Nakayama K. Multiple Skp1-related proteins in Caenorhabditis elegans: diverse patterns of interaction with Cullins and F-box proteins. Curr. Biol. 2002;12:267–275. doi: 10.1016/s0960-9822(02)00657-7. [DOI] [PubMed] [Google Scholar]
- Yi W, Ross JM, Zarkower D. Mab-3 is a direct tra-1 target gene regulating diverse aspects of C. elegans male sexual development and behavior. Development. 2000;127:4469–4480. doi: 10.1242/dev.127.20.4469. [DOI] [PubMed] [Google Scholar]
- Zarkower D. Somatic sex determination. WormBook. 2006:1–12. doi: 10.1895/wormbook.1.84.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkower D, Hodgkin J. Molecular analysis of the C. elegans sex-determining gene tra-1: a gene encoding two zinc finger proteins. Cell. 1992;70:237–249. doi: 10.1016/0092-8674(92)90099-x. [DOI] [PubMed] [Google Scholar]
- Zhang H, Kobayashi R, Galaktionov K, Beach D. p19Skp1 and p45Skp2 are essential elements of the cyclin A-CDK2 S phase kinase. Cell. 1995;82:915–925. doi: 10.1016/0092-8674(95)90271-6. [DOI] [PubMed] [Google Scholar]