Abstract
Glucocorticoid (GC) hormones are widely used in the treatment of acute lymphoblastic leukemia (ALL). Whereas a high level of GC receptor (GR) protein is associated with the sensitivity of ALL cells to steroid-mediated apoptosis, the auto-up-regulation of human (h)GR mRNA and protein is also found in hormone-sensitive ALL cell lines. We have characterized the hGR gene-proximal promoters for DNA sequences and transcription factors required for hormone responsiveness in T lymphoblasts. Sequences at −4559/−4525 and −2956/−2916, relative to the translation start site, function as strong composite GC response units (GRUs). Both GRUs include adjacent protein recognition sequences for the c-Myb transcription factor and the GR as a DNA cassette. An Ets-binding sequence overlaps the GR-binding site in the −4559/−4525 GRU, whereas an Ets-binding site present in the −2956/−2916 GRU does not overlap the GR/c-Myb-binding cassette. The Ets protein family member, PU.1, blocks hormonal activation of the −4559/−4525 GR/c-Myb-binding cassette but does not interfere with the responsiveness of the −2956/−2916 GRU. Thus, the hGR 1A GRU (described previously), the −4559/−4525 GRU, and the −2956/−2916 GRU have a similar structure and can mediate cell type-specific hormonal auto-up-regulation of hGR promoter activity in steroid-sensitive ALL cells. However, subtle differences in the GRU architecture result in differential sensitivity of the promoters to Ets family members such as PU.1. The architecture of the GRU and the spectrum of specific transcription factors present in different types of ALL might allow the development of a tailored therapy to enhance steroid sensitivity in ALL patients.
TREATMENT WITH HIGH concentrations of glucocorticoid (GC) steroid hormones triggers lysis of several lymphoid cell types, making this useful in the clinical management of lymphoid malignancies (1,2,3,4). GC-induced lysis of leukemic cells occurs via apoptosis or programmed cell death (5,6,7,8), and the steroid hormone initiates this process by binding to its intracellular binding protein, the GC receptor (GR). The GR belongs to the nuclear steroid receptor family of transcription factors. Upon binding to the GC ligand, the activated GR recognizes and binds to specific DNA sequences [GC response element (unit), GRE or GRU] in target gene promoters as a homodimer or heterodimer, followed by an alteration in target gene expression (9,10,11,12,13,14,15,16,17,18). Several DNA-binding transcription factors (c-Jun, c-Myb, Ets family proteins, etc.) functionally interact with the GR and regulate target gene expression (12,14,19,20,21,22). Further modulation of the transcriptional response involves the recruitment of additional specific coregulators that include both coactivators [(e.g. CREB-binding protein, p300, steroid receptor coactivators (SRCs)] and corepressors [such as nuclear receptor corepressor 1, receptor-interacting protein 140, Sin3, and histone deacetylase 1 (HDAC1) (23,24)].
The cellular expression level of the GR is strictly controlled and regulated in many cells and tissue types (25,26,27,28,29). Most notably, the steroid-GR complex auto-regulates GR levels at the level of GR gene expression (30,31). Whereas the GR is auto-down-regulated in most cell types (28,29,31,32,33,34,35), auto-up-regulation is characteristic of lymphoblasts that are sensitive to GC-induced apoptosis (11,12,36,37,38,39,40). The critical role of this early cellular response involving auto-up-regulation of GR expression of lymphoblasts is strongly indicated by the observation that resistant cells are converted to the hormone-sensitive phenotype by experimentally increasing intracellular GR above a certain threshold (41). Clinical analyses of acute lymphoblastic leukemia (ALL) have also revealed that a threshold cellular GR level is positively associated with a better clinical response (40,42,43,44,45,46,47).
The mechanism for auto-regulation of hGR gene expression is still not clear (11,26,27,28,29,30,31,35,37,48,49,50,51,52). There are at least 11 hGR promoters (48,49,51,52,53,54,55), but GR gene transcription is controlled primarily by three promoters (1A, 1B, and 1C) in human lymphoblasts (37,51,55). Promoters 1B and 1C exist over a span of 5 kbp upstream of the first GR coding exon (exon 2), and promoter 1A is located approximately 26 kb upstream of promoters 1B and 1C. hGR promoters 1B and 1C sit in a CpG island in the proximal promoter region and resemble promoters found in constitutively and ubiquitously expressed (housekeeping) genes (49,55). Promoter 1A does not reside in a CpG island, and its expression is regulated by a wide variety of transcription factors (11,51). In human CEM-C7 (T cell) and IM-9 (B cell) lymphoblasts, a simultaneous, coordinated auto-regulation of all three hGR gene promoters (1A, 1B, 1C) during dexamethasone (DEX) treatment is observed, although promoter 1A is much more sensitive than promoters 1B and 1C (37). No consensus-positive GREs are found in these promoters (11,12,31,48,51,56). However, we have identified a GRU in hGR promoter 1A, which is dependent upon the binding of the GR protein together with either c-Myb or a c-Ets protein family member (PU.1) at this composite sequence. This GRU mediates both up-regulation and down-regulation of promoter activity through a molecular switch mechanism (11,22). Because promoters 1B and 1C contribute the majority of GR transcripts (>90% in CEM-C7 T lymphoblasts and >95% in 697 pre-B lymphoblasts at the basal level of transcription (Ref. 37 and C.-d. Geng, unpublished), the auto-regulation of these two promoters is of significance in controlling cellular GR expression and hormone-mediated apoptosis in these lymphoblasts.
We began this study in an attempt to characterize the hGR 1B and 1C promoters. During the course of this study, new putative promoters and untranslated exon 1s were described (39,53,57). We have identified two new GRUs in the proximal promoter region. Although we are uncertain whether these sequences regulate only the immediate downstream promoters or whether they have more global effects over several promoters, the structure of these elements are strikingly similar to a GRU identified previously for the hGR 1A promoter. Thus, the distal 1A GRU and the two proximal GRUs 1) are minimal elements that retain full hormone responsiveness; and 2) contain consensus binding sites for GR, c-Myb, and Ets protein family members. These studies reveal how different hGR promoters are coordinately up-regulated in T lymphoblasts. These observations could lead to further studies aimed at identifying new therapeutic targets and strategies for improving the steroid response in T cell ALL patients.
RESULTS
GRUs that Contribute to Both Basal Expression and Auto-Up-Regulation Exist Upstream of Numerous Proximal Exon 1 Coding Sequences
Upon DEX treatment, the expression of hGR 1B transcripts is auto-up-regulated in the hormone-sensitive T lymphoblastic leukemia cell line, CEM-C7 (37). Previous studies were designed to characterize the hGR 1B promoter region, and it was concluded that it was quite complex, and could contain several transcription factor-binding elements, including three YY1 sites and four Sp1-binding sites (Fig. 1) (49,55). Because no known consensus GRE/GRU(s) have been identified in this promoter, we constructed a series of 5′- and 3′-deletions and generated luciferase reporter plasmid vectors to map the hormone-responsive sequences. While these experiments were in progress, other laboratories identified additional, new exon 1 coding sequences in this same DNA region (Fig. 1) (39,53,57). Although it has been suggested that each of these untranslated exons has its own associated specific promoter, no studies have verified this. Thus, it is not yet clear whether the sequences that we describe here are specific for a single exon 1 or have more global effects on multiple promoters. Because of this uncertainty, we have chosen to number the promoter regions relative to the A in the ATG codon for the initiator methionine in the GR-A protein as the +1 position. Sequential 5′-deletion constructs were transfected into Jurkat T-ALL cells and assayed for hormone responsiveness (Fig. 2A). Deletion of −4898/−4525 caused a drastic loss of basal promoter activity (>95%, Fig. 2A), and further deletion of the proximal promoter region, including the Sp1 and FP1 sites (−4525/−3812), nearly abolished the promoter activity and hormone responsiveness in Jurkat T cells altogether. Because constructs containing the −4525/−3609, −4307/−3609, −4122/−3609, and −4017/−3609 sequences are still able to respond to DEX treatment, it seemed possible that one or more GREs/sequences might exist between the −4017 and −3609 region. However, the contribution of this region to basal promoter activity and hormone stimulation seems limited, and maximal activity clearly requires the −4898/−4525 region.
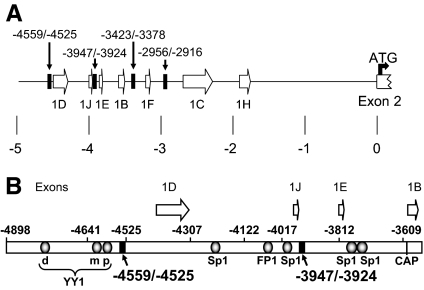
Figure 1.
Human GR Proximal Promoter and Exon 1s
A, The structure of the proximal hGR promoter. The untranslated exon 1s (1B–1H) are according to those designated in Refs. 53 and 57. The +1 position is the adenine (A) that comprises the first nucleotide in the ATG of the initiator methionine found of the hGR-A protein. The black bars designate the location of the GRUs identified in this study. B, Structure and location of transcription factor-binding sites in the upstream region of the proximal promoter. The transcription factor-binding sites indicated were determined previously (49,55). The three YY1 sites are the dYY1, mYY1, and pYY1 sites. The black bars show the location of two GRUs identified in this study. The open arrows designate the location and approximate length of the respective hGR untranslated exon 1s. CAP, Transcription initiation site for the 1B transcript.
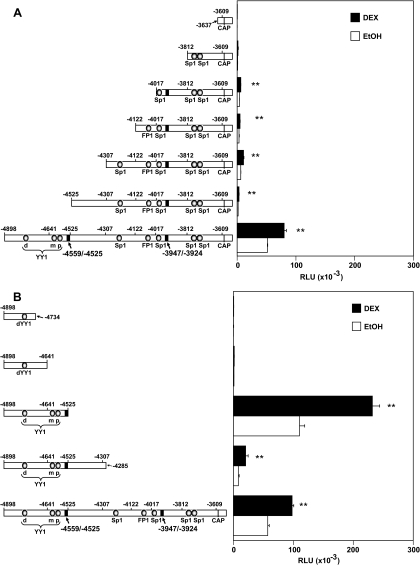
Figure 2.
Promoter Activity and Hormone Responsiveness of 5′- and 3′-End Serial Deletions of the hGR −4898/−3609 Region
5′-End or 3′-end serial deletions of the hGR-proximal promoter region. The generated luciferase reporter gene expression constructs were transfected into Jurkat cells along with 1.0 μg of the hGR expression plasmid pCYGR and 1 μg of β-gal construct (for normalizing the transfection efficiency). Parallel cultures were treated 24 h after transfection with ethanol (EtOH) vehicle (control), or 1 μm DEX. Cell lysates were made 24 h afterward, and luciferase assays were performed. A, Promoter activity and hormone responsiveness of 5′-end deletions are depicted. B, The promoter activity and hormone responsiveness of 3′-end deletions are shown. The values are the averages of three or four separate experiments, and the error bars are the sem. **, P < 0.01 for the DEX-induced values vs. their own vehicle ethanol controls. RLU, Relative light units; CAP, transcription initiation site for the 1B transcript.
Results obtained from 3′-sequential deletions confirmed the importance of the upstream sequences for both basal expression and hormone responsiveness. Deletion of the −4285/−3609 (FP1 and four Sp1 sites) full-length construct (−4898/−3609) yielded the −4898/−4285 construct that lost much of the basal promoter activity but was still DEX responsive (Fig. 2B). Additional deletion of −4525 to −4285 to yield construct −4898/−4525 surprisingly resulted in an increase in basal promoter activity that was about 2-fold higher than the original full-length parental DNA construct. This sequence was still DEX responsive. No known transcription factor-binding sites exist in the −4525 to −4285 sequence, but it appears that it contains an unidentified repressor sequence. Further deletion of the −4641/−4525 sequence caused a dramatic loss of promoter activity to very low levels. Because the distal hGR gene sequences (−4898/−4525) retained hormone responsiveness in T lymphoblasts, we more closely analyzed this sequence for GRE/GRU elements.
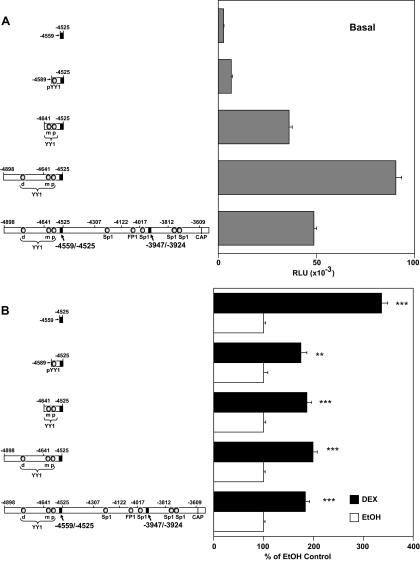
Three YY1-binding sites, distal (d), middle (m), and proximal (p), have been characterized in the −4898/−4525 hGR promoter region (49,55). dYY1 is located in the −4898/−4641 fragment, whereas mYY1 and pYY1 are close to each other and are present in the −4641/−4525 sequence. Because deletion of the −4641/−4525 region caused a nearly complete loss of all promoter activity (Fig. 2B), the role of the mYY1 and pYY1 sites in regulating hGR promoter activity and hormone responsiveness in lymphoblasts was examined. Further 5′-deletions of the dYY1, mYY1, and pYY1 binding sites causes a gradual drop of basal promoter activity (Fig. 3A). Deletion of nucleotides −4898/−4641 (which included the dYY1 site) caused about 50% loss of basal promoter activity compared with the −4898/−4525 3′-truncated sequence, although its activity was nearly the same as the full-length parental construct (Fig. 3A). The deletion of the mYY1 site to yield the −4589/−4525 fragment drastically lowers promoter activity whereas deletion of the last YY1 site (pYY1) resulted in a very low basal promoter activity (10% of full-length promoter (Fig. 3A). However, the resultant −4559/−4525 sequence shows even more robust hormone responsiveness than the full-length promoter (Fig. 3B). These data suggest that 1) the three YY1-binding sequences, especially the mYY1 and pYY1 sites, are important in contributing to the basal promoter activity; and 2) the −4559/−4525 region contains the GC-responsive sequences.
Figure 3.
Analysis of the Possible Roles for the Three YY1 Sites in the Basal Promoter Activity and Hormonal Response of the hGR-Proximal Promoter in Jurkat T Cells
A, Basal promoter activity. RLU, Relative light units. B, DEX responsiveness. Each construct is expressed as the percent of the respective ethanol treated control for the particular construct. Luciferase reporter gene assays were performed as described in the legend to Fig. 2. **, P < 0.01 and ***, P < 0.005 for the up-regulated promoter activity compared with their ethanol controls. CAP, Transcription initiation site for the 1B transcript.
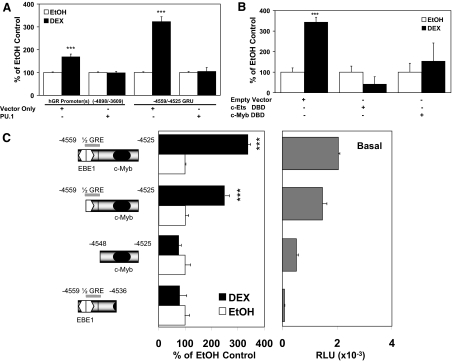
To further analyze the −4559/−4525 sequence, it was deleted in a variety of promoter contexts. Deletion of all of the sequence 3′ to the −4559/−4525 sequence did not abolish hormone responsiveness (Fig. 4A). However, the further deletion of the 34 bp from −4559 to −4525 resulted in a complete loss of hormone responsiveness. Deletion of the upstream YY1 sites has no further effect on hormone responsiveness, although basal promoter activity is lowered (data not shown).
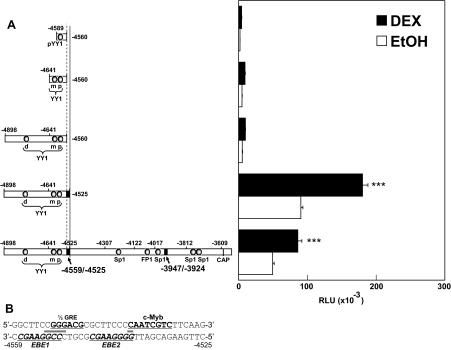
Figure 4.
Identification of the −4559/−4525 Sequence as a GRU
Deletions and luciferase reporter gene assays were done as described in Materials and Methods and the legend to Fig. 2. A, The promoter activity (open bar) and hormone responsiveness (solid bar) of the hGR constructs are shown. RLU, Relative light units. ***, P < 0.005 for DEX up-regulation of promoter activity in Jurkat cells. B, Sequence of the −4559/−4525 hGR promoter region. The half-GRE, c-Myb, and two c-Ets recognition sites (italicized) are bold and underlined. The horizontal solid bars indicate the overlapping binding recognition nucleotides between the half-GRE and EBE1 and between EBE2 and c-Myb. CAP, Transcription initiation site for the 1B transcript.
The Steroid-Dependent Auto-Up-Regulation Mediated by the −4559/−4525 DNA Sequence Depends on a Conserved GR/c-Myb Mechanism
The −4559/−4525 (35 nucleotides) DNA sequence is shown in Fig. 4B. Sequence analysis of potential protein-binding elements revealed possible recognition sites for the GR, c-Myb, and Ets family proteins. No consensus GRE is present in this sequence. However, the −4552/−4547 sequence (5′-GGGACG-3′) is homologous to a half-GRE-binding site identified previously [5′-GGGACANNNTGTCCT-3′ (58)]. Two potential Ets protein-binding sites [MVGGAWRY, with GGAW being completely conserved in all cases (59)] are found on the complementary strand of −4558/−4551 [Ets protein-binding element 1 (EBE1), with a strong, 100% homology to the consensus sequence] and on the complementary strand of −4545/−4538 (EBE2; 87% homology to the consensus sequence). Sequence analysis also identified a perfect c-Myb-binding sequence [5′-YAAYKGNH-3′ (60)] at −4538/−4531 (5′-CAATCGTC-3′, 100% homology) (Fig. 4B). The predicted weak EBE2 Ets-binding site partially overlaps with the predicted c-Myb protein recognition site, and EBE1 overlaps the half-GRE (Fig. 4B). The importance of these overlaps is described below.
We previously showed that the hGR 1A, 1B, and 1C promoters are simultaneously auto-up-regulated by DEX treatment in CEM-C7 cells, and that overexpression of a c-Myb dominant-negative DNA-binding domain (DBD) blocked the hormone response of all three promoters (12). This suggests the involvement of a c-Myb-dependent mechanism in the steroid-dependent regulation of all three promoters in lymphoblasts. Indeed, the −4559/−4525 hGR promoter sequence appears to resemble the GRU identified in the hGR 1A promoter, which contains overlapping c-Myb/Ets elements and an adjacent GR-binding sequence (FP11/12) (11,12). In the case of the 1A promoter, there appears to be a molecular switch, in which a GR/c-Myb complex may cause promoter stimulation, whereas a GR/Ets protein complex may mediate promoter repression. To test whether an Ets protein can suppress the −4559/−4525 hGR element, we focused on PU.1, an Ets protein that can act as a transcriptional repressor and is largely absent in T cells (12). Transfection of a PU.1 cDNA expression vector into Jurkat T cells completely abolishes DEX-mediated up-regulation of both the parental hGR promoter(s) sequence (−4898/−3609) and the steroid stimulation of the minimal −4559/−4525 element (Fig. 5A). In addition, hormonal responsiveness of the −4559/−4525 minimal element is completely eliminated by overexpressing a dominant-negative Ets-2 DBD (Fig. 5B). This suggests that the suppression function of Ets proteins on this promoter may depend on the physical occupancy of Ets DNA-binding sequences, one of which could block the binding of GR to the overlapping half-GRE at −4552/−4547 and the other that might block c-Myb binding to the overlapping c-Myb site at −4538/−4531 (Fig. 5B). c-Myb (present in T cells) appears to be needed for DEX up-regulation of the hGR −4559/−4525 element because overexpression of a dominant-negative c-Myb DBD also blocks DEX up-regulation of reporter gene expression (Fig. 5B). Thus, similar to the hGR 1A promoter, the −4559/−4525 element also appears to function as a molecular switch, with the combination of GR and c-Myb being auto-stimulatory, whereas an Ets protein (in this case, PU.1) acts as an inhibitory factor.
Figure 5.
The Function of the c-Myb-, Ets-, and GR-Binding Sites in the Hormone Responsiveness of the −4559/−4525 hGR Region
A, A PU.1 expression plasmid was cotransfected into Jurkat cells with the long upstream sequence (−4898/−3609) and the GRU (−4559/−4525) linked to a luciferase reporter gene, together with the GR expression plasmid and the β-gal construct, as described in the legend of Fig. 2. The hormone-stimulated promoter expression is presented as percent of its respective EtOH vehicle control. ***, P < 0.005. B, Overexpression of a dominant-negative DNA binding inhibitor of c-Ets or c-Myb blunts the hormonal response of the GRU. ***, P < 0.005 compared with the EtOH control. C, Basal promoter activity [in relative light units (RLU)] and hormonal responsiveness of the minimal GRU with the deletion of the c-Myb or EBE1/GR recognition sequences (Jurkat cells). Hormone responsiveness is plotted as percent of EtOH-treated controls for each reporter construct, respectively. ***, P < 0.005.
To further test this molecular switch concept, deletion analysis of the hGR −4559/−4525 sequence was performed. The entire −4559/−4525 sequence had measurable basal promoter activity and was stimulated about 3.5-fold by DEX treatment (Fig. 5C). Deletion of the EBE1 Ets-binding site from the 5′-end slightly reduced basal promoter activity (Fig. 5C). However, this construct was still hormone responsive, suggesting that the observed DEX response does not require the EBE. Further deletion of both the Ets-binding site (EBE1) and the half-GRE causes a dramatic loss of basal promoter activity and abolishes steroid up-regulation of reporter gene expression. Similarly, deletion of the c-Myb-binding site from the 3′-end results in a complete loss of basal promoter activity and hormone responsiveness in Jurkat cells (Fig. 5C).
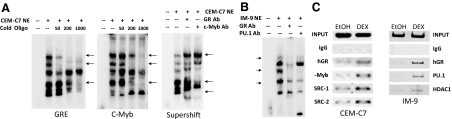
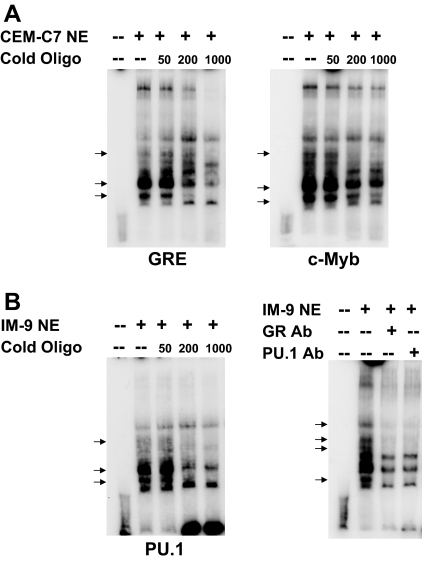
The physical binding of all of these proteins to the −4559/−4525 sequence in vitro was confirmed using an EMSA and unlabeled GRE competitor analysis (Fig. 6A). Because there may be multiple complexes formed with, for example, different coactivators, it is not possible to assign the bands present with specific protein complexes. It is clear, however, that the addition of an unlabeled GRE competitor oligonucleotide competes out some of the bands, and a number of these are also competed out by an unlabeled c-Myb competitor oligonucleotide as well. Although clear supershifted bands are not obtained using GR and c-Myb antibodies, subtle migration and intensity difference are obtained (Fig. 6A). When an IM-9 B lymphoblastoid cell nuclear extract was used, the addition of antibodies to the GR and the Ets protein family member, PU.1, causes a disruption of some of the protein-DNA complexes (Fig. 6B). Disruption of protein-DNA complexes, rather than supershifted bands, can occur when EMSAs are performed, particularly if the protein-DNA complexes are somewhat weak. Finally, to determine whether these transcription factors can form complexes in the intact cell, chromatin immunoprecipitation (ChIP) assays were performed (Fig. 6C). In CEM-C7 cells, in which auto-up-regulation of GR promoters occurs, DEX treatment causes a recruitment of the GR and c-Myb to the −4559 to −4525 GRU. Furthermore, in accord with the observed up-regulation, two known GR coactivators, SRC-1 and SRC-2 are recruited to the GRU as well. Conversely, when the GR is recruited to the GRU in IM-9 cells, which exhibit auto-down-regulation, both PU.1, and the corepressor, HDAC-1, are recruited. Thus, these data support the postulate that these transcription factors participate in regulating GR gene expression from this promoter by directly binding to the −4559/−4525 GRU sequence.
Figure 6.
Analysis of the Protein-Binding Sites in the −4559/−4525 GRU
EMSAs (panels A and B) were performed as described in Ref. 11 and ChIP assays were done as described in Ref. 12. A, EMSAs were performed using nuclear extracts of CEM-C7 cells that were treated with 1 μm DEX for 24 h. The extracts were incubated with a 32P-labeled −4559/−4525 GRU oligonucleotide, either alone or in the presence of increasing molar fold excesses of an unlabeled consensus GRE (left panel) or c-Myb consensus sequence (middle panel). Arrows indicate protein/GRU complexes that were competed out by the cold competitor oligonucleotides. In the right panel, the nuclear extract was incubated with the labeled GRU in the presence of no antibody or antibodies to the GR or c-Myb. Complexes that were altered by the antibodies are indicated by the arrows. B, EMSA supershift analysis was performed with nuclear extracts from IM-9 cells that were treated with 1 μm DEX for 24 h. Arrows indicate protein-DNA complexes that were altered by antibodies to the GR and PU.1. C, ChIP analysis. CEM-C7 (left panel) and IM-9 (right panel) cells were treated for 24 h with 1 μm DEX. ChIP analysis was performed with antibodies to the indicated proteins. Ab, Antibody; NE, nuclear extract.
Together with the results obtained in the overexpression experiments, it appears that DEX auto-up-regulation of the −4559/−4525 hGR promoter-associated element depends upon the binding of both the GR and c-Myb proteins, but not an Ets protein, whereas down-regulation involves the binding of the GR and the Ets protein, PU.1. Thus the hGR −4559/−4525 hGR promoter-associated element resembles the hGR 1A promoter (12), in that both contain a GRU in which the adjacent binding of the GR and c-Myb/Ets seems to be necessary for DEX-mediated autoregulation of GR gene expression.
A Proximal hGR GRU (−2956/−2916) is a Variation in the GR/Myb/Ets Transcription Cassette Structure that May Contribute to Cell Type-Specific Regulation of the 1C Promoter
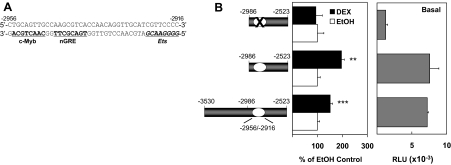
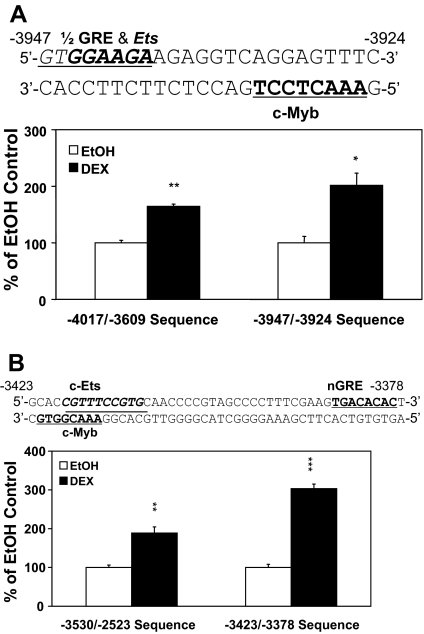
The hGR 1C promoter is in a CpG island, and most of the promoter region is GC rich (>80% GC). Although the hGR 1C promoter is auto-up-regulated by DEX in hormone- and apoptosis-sensitive CEM-C7 T-lymphoblasts (37), the mechanism is unknown. However, previous studies showed that the auto-up-regulation of the expression of hGR transcripts 1A, 1B, and 1C all depend upon the c-Myb protein in CEM-C7 and Jurkat T lymphoblasts (12). Because both the 1A promoter (12) and the GRU identified above contain a DNA cassette with neighboring GR- and c-Myb-binding sites, it seemed possible that this mechanism is conserved in the regulation of the hGR 1C promoter as well. Therefore, we searched the hGR 1C promoter region for sequences containing potential binding sites for c-Myb and the GR protein. One sequence, −2955/−2948, contains a perfect consensus c-Myb-binding sequence (5′-CAACTGCA-3′) on the complementary strand (Fig. 7A). In addition, an adjacent negative GRE (nGRE) [5′-TGACNNTN-3′ (61)] was also identified on the complementary strand at −2945/−2938 (Fig. 7A). Finally, an imperfect Ets protein-binding sequence (63% homology to the consensus site) was found, again on the complementary strand, about 14 nucleotides downstream from this c-Myb/n-GRE cassette at positions −2923/−2916 (Fig. 7A).
Figure 7.
Analysis of the hGR −2956/−2916 Sequence
A, The predicted c-Myb, nGRE, and Ets (italicized) binding sequences are bolded and underlined. B, Basal promoter activity [in relative light units (RLU)] and hormonal response of deletion constructs. Shown are 5′-end deletion of the −3530/−2523 hGR sequence, and a further internal deletion of the −2956/−2916 sequence itself. The hormonal response of each individual luciferase reporter construct in Jurkat cells is depicted as the percentage of its respective EtOH control. **, P < 0.01; ***, P < 0.005.
We next designed targeted deletions to elucidate the roles of the −2956/−2916 sequence in hGR promoter expression and hormone responsiveness in lymphoblasts. The −3530/−2987 sequences were removed to yield a −2986/−2523 reporter construct. Compared with the full-length sequence (−3530/−2523), the −2986/−2523 reporter gene containing the predicted GC-responsive sequence at −2956/−2916 retains full basal promoter activity and hormone responsiveness (Fig. 7B). Although not yet formally proven, the location of this sequence upstream of the hGR exon 1C sequences suggests that this sequence comprises or controls the hGR 1C promoter (see below). Thus, basal expression and hormone responsiveness of the hGR 1C promoter may be localized to the −2986/−2523 region of the hGR gene. Furthermore, internal deletion of the putative GRU (−2959/−2916) causes a complete loss of hormone responsiveness and a drastic reduction of promoter activity to 15% of the full-length sequence (Fig. 7B). These data suggest that the −2959/−2916 sequence in the putative hGR 1C promoter acts like the GRU in the other two hGR promoters and is responsible for hormone responsiveness of the hGR 1C promoter in lymphoblasts.
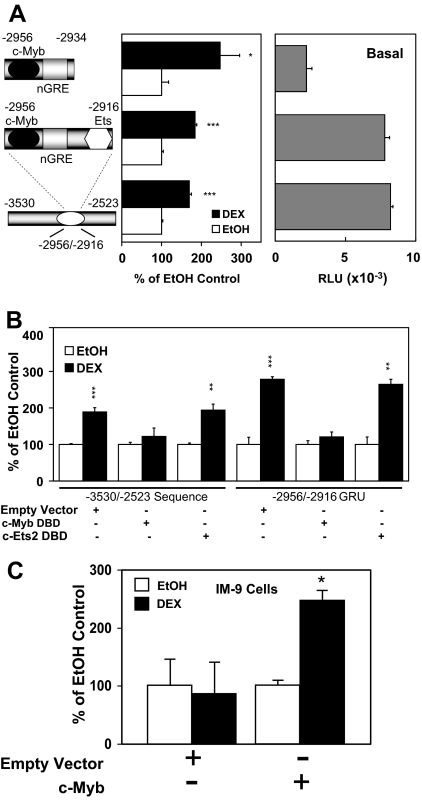
The Hormonal Activation of the −2956/−2916 Sequence by DEX in Lymphoblasts Depends upon Both the GR- and c-Myb-Binding Sites
The −2956/−2916 sequence contains predicted binding sites for c-Myb, the GR, and an Ets protein. This was confirmed in vitro using EMSAs with CEM-C7 and IM-9 cell nuclear extracts (see below). To examine the putative roles for the GR and c-Myb proteins in the hormonal responsiveness of this element, we examined the deletion variant, −2956/−2934, in which the 3′-end containing the Ets-binding sequence was removed. There was a clear hormonal response for the −2956/−2934 sequence containing only the c-Myb- and GR-binding sites, although the Ets-binding sequence is important for basal activity, because the removal of this site causes about 60% loss of basal promoter activity (Fig. 8A). Internal deletion of this Ets-binding sequence (data not shown) likewise results in reduced basal promoter activity (∼50%) but retention of full hormonal responsiveness compared with the −2986/−2523 sequence. Thus, it appears that the −2923/−2916 Ets-binding site is important for basal promoter activity, but not directly for steroid-mediated up-regulation. This is borne out by experiments involving dominant-negative inhibitors (Fig. 8B). Overexpression of a c-Myb DBD dominant-negative inhibitor causes a complete loss of DEX stimulation for both the full-length sequence (−3530/−2523) and the minimal −2956/−2916 sequence, whereas overexpression of an Ets2 DBD dominant-negative protein does not interfere with steroid stimulation of either sequence in Jurkat cells. If the c-Myb binding site in the −2956/−2916 GRU is functional, then the overexpression of c-Myb in IM-9 cells (which lack c-Myb) should reverse the hormonal regulation from a down-regulation to an up-regulation. Transient transfection of a c-Myb cDNA expression vector with a promoter 1C-luciferase reporter gene did, indeed, switch the autoregulation from negative to positive (Fig. 8C). Thus, it appears that hormonal stimulation of the −2956/−2916 sequence requires both the GR and c-Myb proteins.
Figure 8.
Analysis of the c-Ets and c-Myb Protein-Binding Sites in the −2956/−2916 hGR Promoter Region
A, The basal promoter activity [in relative light units (RLU)] and hormone responsiveness of the minimal GRU is shown compared with the long construct. A further deletion of the c-Ets sequence from the GRU was also performed. The DEX response is displayed as the percent of the respective EtOH controls. *, P < 0.05; ***, P < 0.005. B, Dominant-negative DBD inhibitors of c-Myb or c-Ets were expressed along with the luciferase reporter gene constructs for both the long sequence (left) and the minimal GRU (right). The DEX responsiveness is shown as the percent of the respective EtOH control. **, P < 0.01; ***, P < 0.005. C, IM-9 cells were transfected with an empty vector or a c-Myb cDNA expression vector, a full-length (−3530/−2523) hGR promoter/luciferase construct, and cytomegalovirus-β-gal (normalization control). Luciferase assays were performed as described in the legend to Fig. 2. *, P < 0.05.
EMSAs were performed to determine whether the GR, c-Myb, and an Ets protein bind to the hGR −2956/−2916 GRU (Fig. 9). Competition experiments using nuclear extracts from DEX-treated CEM-C7 cells showed that, in general, the same bands were competed out using unlabeled GRE and c-Myb consensus oligonucleotides (Fig. 9A). For nuclear extracts obtained from IM-9 cells that were treated with DEX (Fig. 9B), a cold PU.1 oligonucleotide was able to specifically compete out some of the bands. Furthermore, the addition of antibodies against the GR and PU.1 resulted in the same altered gel shift pattern, consistent with the disruption of a subset of the protein-DNA complexes. We have attempted to perform ChIP analysis on the −2956/−2916 sequence. Unfortunately, the extremely high GC content of this region prohibited these studies. The EMSA results, along with the reporter gene analyses, strongly suggest that the GR, c-Myb, and PU.1 can bind to the hGR −2956/−2916 promoter sequence.
Figure 9.
EMSA of the −2956/−2916 GRU
EMSAs were performed as described in Ref. 11. A, EMSAs were performed using nuclear extracts of CEM-C7 cells that were treated with 1 μm DEX for 24 h. The extracts were incubated with a 32P-labeled −2956/−2916 GRU oligonucleotide, either alone or in the presence of increasing molar fold excesses of an unlabeled consensus GRE (left panel) or c-Myb consensus sequence (right panel). Arrows indicate protein/GRU complexes that were competed out by the cold competitor oligonucleotides. B, EMSAs were performed using nuclear extracts of IM-9 cells that were treated with 1 μm DEX for 24 h. The extracts were incubated with a 32P-labeled −2956/−2916 GRU oligonucleotide, either alone or in the presence of increasing molar fold excesses of an unlabeled PU.1 consensus sequence (left panel). Arrows indicate protein/GRU complexes that were competed out by the cold competitor oligonucleotides. EMSA supershift analysis (right panel) was performed with nuclear extracts from IM-9 cells that were treated with 1 μm DEX for 24 h. Arrows indicate protein-DNA complexes that were altered by antibodies to the GR and PU.1. Ab, Antibody; NE, nuclear extract.
Two Additional Potential GRUs May Exist in Other GR Promoters
Preliminary experiments have been performed on two other regions of the hGR promoter to determine whether the GRU motif may be conserved even further. Serial deletion reporter gene assays (Fig. 2A) and computer analysis revealed a sequence, −3947/−3924, which contains a half-GRE, an overlapping Ets site, and a c-Myb recognition sequence (Fig. 10A). This architecture is remarkably similar to the arrangement of these transcription factor-binding sites in the −4559/−4525 GRU described above. Furthermore, the −3947/−3924 sequence alone conferred about the same amount of DEX responsiveness to a reporter gene as did the longer −4017/−3609 fragment (Fig. 10A). The potential role of this −3947/−3924 GRU, if any, is discussed below (Discussion). Another potential GRU was noted at positions −3423/−3378 (Fig. 10B). There was an overlap of a putative c-Myb sequence and a putative c-Ets sequence; however, the putative GR-binding element (a potential nGRE) was located substantially downstream from the c-Myb/c-Ets sites. To determine whether this region was hormone responsive and acted as a GRU, we compared the hormonal regulation of the long −3530/−2523 DNA sequence that contains both this site and the previously identified −2956/−2916 GRU to a reporter gene that contains only the −3423/−3378 potential GRU (Fig. 10B). The −3423/−3378 element was indeed hormone responsive. Thus, we have identified, so far, four apparent GRU-containing DNA sequences in the proximal promoter region that extends almost 5 kbp upstream of the initiator ATG for the GR protein. These occur at positions −4559/−4525, −3947/−3924, −3423/−3378, and −2956/−2916 (Fig. 1).
Figure 10.
Analysis of Minor GRUs
A, The DNA sequences of the predicted half-GRE, and c-Myb-binding sequences in the −3947/−3924 hGR gene GRU are displayed in bold, whereas the putative c-Ets site is italicized and underlined with a thin underline. The hormone responsiveness of the GRU-like sequence was analyzed in the context of a long construct (−4017/−3609) as well as for the minimal GRU (−3947/−3924). The assay was done in Jurkat cells as described earlier. The up-regulation of promoter activity is shown as percent over the respective EtOH control. *, P < 0.05; **, P < 0.01. B, The putative nGRE, c-Myb, and c-Ets (italicized) sequences of the hGR −3423/−3378 GRU are indicated. The hormone responsiveness of the GRU-like sequences in Jurkat cells in the context of an extended sequence (left) and for the minimal GRU (right) was determined. The results are given as the percentage of DEX induction vs. the respective EtOH control. **, P < 0.01; ***, P < 0.005.
The GRUs May Form a Common Theme with Potentially Important Variations
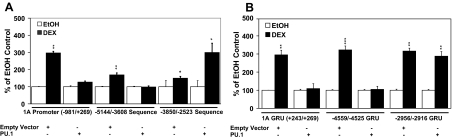
Previously, we had identified a strong GRU (+243/+269) in the hGR 1A promoter region (12). For the hGR 1A promoter/exon, we have proposed a molecular switch mechanism, in which binding of the GR and c-Myb proximal to each other would stimulate hGR gene transcription (auto-up-regulation), whereas binding of the hGR proximal to an Ets protein (with PU.1 being the strongest Ets protein candidate) would inhibit hGR gene transcription (auto-down-regulation). An inspection of the architecture of the three strongest GRUs (1A, −4559/−4525, and −2956/−2916) led to a prediction. The Ets-binding site overlaps with the c-Myb-binding site in the hGR 1A GRU (+243/+269), and it overlaps with the GR- or c-Myb-binding site in the −4559/−4525 GRU (Fig. 4B). Thus, exogenous expression of an Ets protein should block hGR auto-up-regulation from these two GRUs in T lymphoblasts. In contrast, the Ets-binding site for the −2956/−2916 GRU does not overlap with the GR- or c-Myb-binding sites (Fig. 7A). Thus, if the hypothesis is correct, exogenous overexpression of an Ets protein should not block hormone-mediated up-regulation from this GRU in T lymphoblasts. We used long sequences that contained these GRU elements (Fig. 11A) and the minimal GRUs themselves (Fig. 11B) to test this hypothesis. As predicted, overexpression of the Ets protein, PU.1, in Jurkat cells (that normally lack PU.1) blocked up-regulation from the GRUs in which the EBE overlapped with the c-Myb- and/or GR-binding site [hGR 1A −981/+269 (Fig. 11A) and +243/+269 (Fig. 11B), and the −5144/−3608 (Fig. 11A) and −4559/−4525 (Fig. 11B) sequences], but not those constructs, either long (−3850/+85; Fig. 11A) or short (−2956/−2916; Fig. 11B), in which the Ets-binding site does not overlap with the other protein-binding sites in the GRU.
Figure 11.
Comparative Analysis of the Multiple hGR Gene Promoter GRUs
A, Comparison of the long sequences for the hGR 1A promoter (−981/+269), the sequence that contains the two most 5′-GRUs in the proximal GR promoter (−5144/−3608), and the sequence that contains the most 3′ two GRUs in the proximal GRU promoter (−3850/−2523), and the effect of PU.1 overexpression in Jurkat cells. B, Comparison of the minimal GRUs for the hGR 1A promoter (+243/+269), the strongest 5′-upstream GRU (1D?; −4559/−4525) in the proximal GR promoter, and the strongest 3′-GRU (1C?; −2956/−2916) in the proximal GRU promoter, and the effect of PU.1 overexpression in Jurkat cells. The results are displayed as DEX induced percent over the EtOH control. *, P < 0.05; **, P < 0.01; ***, P < 0.005.
DISCUSSION
These studies were initiated to characterize the GC-responsive elements controlling expression of the hGR gene 1B and 1C promoters. However, in addition to the five hGR transcripts (1A1, 1A2, 1A3, 1B, and 1C) that we described previously in lymphoblasts (51), recent studies (published after the inception of this work) have identified several new hGR exon 1 transcript variants (1D, 1E, 1F, 1H, 1I, and 1J), presumably driven by their own individual respective promoters (Fig. 1) (39,53,57). Promoter/exon 1D, 1J, and 1E sequences are located in the DNA region that was previously described as part of the 1B promoter, whereas promoter/exon 1F sequences are found in the original 1C promoter region. Thus, it is possible that results ascribed to the hGR 1B (55) and 1C promoters may involve other promoters as well, and multiple promoters may even exist in a single DNA sequence that is being analyzed. For example, we originally thought that the −4559/−4525 and −3947/−3924 GRUs were associated with the hGR 1B promoter/exon, whereas we ascribed the −3423/−3378 and −2956/−2916 GRUs to the hGR 1C promoter. It is now possible that the −4559/−4525 GRU actually controls the 1D promoter, which is up-regulated by DEX treatment in both CEM-C7 T lymphoblasts and the steroid-sensitive, pre-B ALL cell line, 697 (39). Likewise, the −3947/−3924 GRU may regulate expression of the hGR 1E promoter/exon. Exon 1E expression is biased toward the immune system (57), although its responsiveness to steroid hormone has not been studied (39). Similarly, the −3423/−3378 GRU may be associated with expression of the hGR 1F promoter/exon, which is also biased to the immune system (57) and is auto-up-regulated by GC treatment in both CEM-C7 T lymphoblasts and 697 B lymphoblasts (39), rather than the hGR 1C promoter/exon, as we had originally thought. Finally, the −2956/−2916 GRU is 5′ to the hGR 1C promoter/exon, and it may actually control the expression of the hGR 1C-containing transcripts. However, a note of caution must be raised. Although it seems clear that there are multiple (at least 11) hGR exon 1s, the promoters that control their regulation have been neither identified nor characterized. Individual promoters have not been proven for each transcript, and the processing of the primary transcripts has not been studied. Furthermore, the GRUs could act as enhancers and control the expression of other transcripts that are not immediately downstream of the GRU. With regard to the present study, the precise transcript that is regulated by the GRU is not critical. What is important is the seemingly common molecular mechanism that is involved, and the significance of a certain variation on the common theme.
What was striking in previous studies was that the expression of three transcripts (1A3, 1B, and 1C) occurred in an apparent coordinate fashion (37). Thus, all three transcripts (plus the minor 1A1 and 1A2 transcripts) are down-regulated by steroid treatment in IM-9 B lymphoblasts and up-regulated in CEM-C7 T lymphoblasts. This suggested a common molecular mechanism for the regulation of these transcripts. The identification of a hGR/c-Myb/Ets GRU in the hGR 1A promoter (12) and a molecular switch mechanism for autoregulation of gene expression led us to search for similar motifs in the regulation of other GR transcripts. Interestingly, we have identified three additional GRUs (−4559/−4525, −3947/−3924, and −3423/−3378) that we believe act in a fashion similar to the hGR 1A GRU, and a fourth GRU (−2956/−2916) that is a biologically important variation on this theme. We propose that all four involve the direct binding of the GR, c-Myb, and Ets proteins. Although the overall scheme appears to be conserved, the precise architecture of the GRUs varies.
Whereas the hGR binds to its own site in the hGR 1A GRU, there are overlapping adjacent c-Myb- and Ets-binding sites. Thus, we propose that either c-Myb or an Ets protein, but not both, can bind adjacent to the hGR. C-Myb and Ets protein binding are mutually exclusive. We have proposed that the interaction of the hGR and c-Myb forms a complex that stimulates transcription of the hGR 1A transcript, whereas an hGR/Ets protein complex inhibits gene transcription (12). For the −4559/−4525 (1D promoter?) sequence, there are nearby (though not adjacent) hGR- and c-Myb-binding sites (Fig. 4B). Two potential Ets protein-binding sequences are present, one that overlaps with the putative GR-binding sequence and one that overlaps, by only one nucleotide, with the c-Myb site. Thus, we propose that, like the hGR 1A promoter, a combination of the hGR and c-Myb results in auto-up-regulation. The positive role of c-Myb is indicated by the suppression of hormone-dependent up-regulation when a dominant-negative c-Myb DBD mutant is expressed (Fig. 5B). The inhibitory role of an Ets protein [PU.1 (Fig. 5A) or a dominant-negative Ets DBD mutant (Fig. 5B)] suggests that binding of an Ets protein blocks hGR and/or c-Myb binding to this GRU, thus blunting steroid-mediated up-regulation. Thus, as for the hGR 1A promoter, a molecular switch mechanism may occur for the −4559/−4525 (1D promoter?) sequence.
A similar molecular switch mechanism may function for the −3947/−3924 (1E promoter?) sequence (Fig. 10A). Because a potential Ets-binding site overlaps the putative GR-binding site, we predict that the expression of an Ets protein would block auto-up-regulation from this sequence. The −3423/−3378 sequence (Fig. 10B; 1F promoter?) has the GR-binding site at a considerable distance for the c-Myb-binding site. However, once again, an Ets-binding site overlaps with the c-Myb-binding site, reminiscent of the molecular switch seen for the hGR 1A GRU. Thus, we would predict that an Ets protein would block hormone-mediated up-regulation of the −3423/−3378 sequence (1F promoter?). Additional experiments are required to test these predictions.
Finally, the −2956/−2916 sequence (1C promoter?) displays an interesting variation on the GRU theme. It also has apparent binding sites for the hGR, c-Myb, and Ets proteins. However, there is no overlap in these sequences (Fig. 7A). Thus, if the molecular switch model were true, we would predict that a dominant-negative c-Myb DBD mutant would block steroid-induced auto-up-regulation, but that the expression of an Ets protein would not do so in cells (such as T lymphoblasts) that contain c-Myb. Indeed, a c-Myb DBD blocked up-regulation (Fig. 8B), whereas neither a c-Ets DBD (Fig. 8B) nor an inhibitory Ets protein like PU.1 (Fig. 11B) did so. This has potentially important biological implications (discussed below).
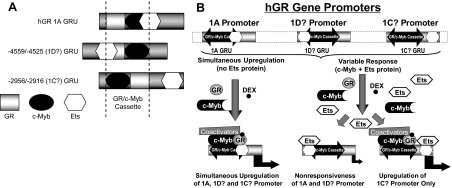
The architecture of the promoter 1A, −4559/−4525 (promoter 1D?), and −2956/−2916 (promoter 1C?) GRUs is shown in Fig. 12A. We propose that a GR/c-Myb cassette is the fundamental unit responsible for hGR promoter auto-up-regulation. An Ets-binding site that overlaps either element (GR or c-Myb) can blunt auto-up-regulation and perhaps even reverse it to auto-down-regulation. The mode of regulation depends not only on the promoter elements, but also on the transcription factors present in the cell lineage. Figure 12B summarizes the way in which the GRUs that we have identified are similar and can differ.
Figure 12.
Diagrammatic Representative of GRU Architecture in the GR Promoter and the Effects that Varied Architecture May Have on Hormonal Regulation and Cell Type Specificity
A, Linear arrangement of the hGR 1A GRU, the −4559/−4525 (1D?) GRU, and the −2956/−2916 (1C?) GRU. Whereas the proposed negative regulatory c-Ets sites overlap with positive regulatory GR (1A) or c-Myb (−4559/−4525)-binding sites, the c-Ets site does not overlap with a positive regulatory element in the −2956/−2916 GRU. B, Proposed model of how the lineage specific expression of a negative regulatory c-Ets protein could suppress GC-mediated up-regulation of the hGR 1A and 1D? promoters but have no effect on auto-up-regulation of the 1C? promoter because of a lack of transcriptional interference in the latter case.
The question that remains is: what is the role of this common GRU mechanism, and what is the importance of any variations in it? Because there may be multiple promoters for the GR, it seems logical that a common mechanism for the regulation of (at least some) GR transcripts should exist. This would allow a coordinated, concerted regulation of GR gene expression in processes (programmed cell death?) that require a similar mode of gene regulation. However, fine tuning of the response (GR gene expression) may be required under certain physiological states, so that subtle, although perhaps important, variations on a theme are required. There appear to be at least two ways that this can be achieved for the regulation of GR gene expression. First, the architecture of the GR can be varied to allow transcription factors to either compete for overlapping DNA-binding sites (e.g. c-Myb and c-Ets protein on the −4559/−4525, −3947/−3924, −3423/−3378, and hGR 1A +243/+269 GRUs), or the binding sites can be physically separated such that transcription factor interference does not occur (e.g. the −2956/−2916GRU). The second mechanism to allow versatility is the spectrum of transcription factors that are expressed in a certain cell type or lineage. For example, T cell ALL and early B cell ALL lymphoblasts contain the c-Myb transcription factor (Ref. 12 and Geng, C.-d., J. R. Schwartz, and W. V. Vedeckis, in preparation). However, the 697 early B cell ALL line contains the negative regulatory c-Ets protein, PU.1, whereas the CEM-C7 T-ALL line does not (12). Thus, although the 1A3 and 1C transcripts are up-regulated in CEM-C7 cells in a coordinated manner, the 1A3 transcript is down-regulated in 697 cells whereas the 1C transcript is up-regulated (Geng, C.-d., J. R. Schwartz, and W. V. Vedeckis, in preparation). Presumably both transcripts are up-regulated in CEM-C7 cells because they do not contain the opposing PU.1 transcription factor. However, in 697 cells, it appears that the expressed PU.1 suppresses up-regulation of the 1A3 transcription (because of the interference in the binding of c-Myb to the composite, overlapping GRU in the hGR 1A promoter), whereas PU.1 does not suppress up-regulation of the 1C promoter because the c-Ets (PU.1)-binding site does not overlap with the c-Myb (stimulatory)-binding site in the −2956/−2916 GRU.
In summary, a conserved element, the GR promoter GRU, which is controlled by the GR protein itself, c-Myb, and a c-Ets protein member, has been identified. This element may allow for the coordinate regulation (positive or negative) in certain cell types that contain a specific repertoire of transcription factors, whereas slight variations in the architecture of the GRU can allow for the independent regulation of certain GR transcripts in cell lineages containing a different composition of transcription factors. The similarity of the GRUs and the variations on this theme allow for fine tuning of the control of GR gene expression that is tailored for the cell type, the cellular response to hormone, and the physiological status of the cell.
MATERIALS AND METHODS
Cell Lines and Cell Culture
The human Jurkat (T-ALL) and IM-9 (B lymphoblastoid) cell lines (from American Type Culture Collection, Manassas, VA) were grown in RPMI 1640 supplemented with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Human CEM-C7 (T-ALL) cells (a gift from Dr. E Brad Thompson, University of Texas Medical Branch, Galveston, TX) were maintained in RPMI 1640 with 10% dialyzed fetal bovine serum (FBS) (Invitrogen). Cells were grown in a 5% CO2 incubator at 37 C. To treat the cells, a final concentration of 1 μm DEX (Sigma, St. Louis, MO; from a 1 mm of stock solution in ethanol) was added to cultures whereas an equivalent volume of ethanol vehicle alone was added to the controls.
DNA Constructs
The human promoter 1A-, 1B-, and 1C-luciferase reporter constructs, pXP1–1A −981/+269, pXP-1A +241/+269 (FP11/12, 1A GRU), pXP 1B+1C, pXP1–1B and pXP1−1 C, have been described previously (11,49,51,55). Because of the discovery of new untranslated exons in the proximal promoter region (previously referred to by us as the 1B+1C region), we have chosen to modify our terminology to make it consistent with that used by the laboratories that discovered these new exon 1s. Specifically, we will use the convention of designating the A in the ATG codon for the initiator methionine of the GR-A form (53,57) of the GRα protein as the +1 position. All upstream sequences used in our constructs will be designated with a minus numbering system from this +1 site. The relationship of the elements that were characterized in this study to the structure of the hGR-proximal promoter region is shown in Fig. 1. PCR amplification was used to make the 5′- and 3′- sequential deletion of proximal promoter-reporter constructs, as well as the internal deletion constructs pXP −4898/−4525 (Δ−4559/−4525) and pXP −2983/−2523 (Δ−2958/−2914). The synthesized PCR primers (Integrated DNA Technologies, Coralville, IA) used for sequential promoter deletions and mutagenesis are listed in Table 1. The PCR-amplified fragments were digested with restriction enzymes BamHI/HindIII, and inserted into the pXP1 vector digested with the same enzymes. To generate the pXP1 −4559/−4525 and pXP1 −2958/−2914 GRU luciferase reporter vectors, the sense and antisense −4559/−4525 or −2958/−2914 oligonucleotides with attached BamHI and HindIII adaptor sequences (Table 1) were synthesized (Integrated DNA Technologies) and annealed to form double-stranded DNA. The double-stranded DNA oligonucleotides were inserted into the empty pXP-1 vector to generate the reporter constructs. All the constructs obtained were confirmed by DNA sequencing for correct sequences inserted in the vector. The pCYGR plasmid was a gift from Dr. John A. Cidlowski (National Institute of Environmental Health Sciences, Research Triangle Park, NC). The human c-Myb expression construct, pcDNA3-c-MybHA and c-Myb DBD expression construct, pcDNA3-c-Myb DBD, were kindly provided by Dr. Giuseppe Raschellà (Ente Nuove Tecnologie Energia Ambiente, Rome, Italy). pFN Ets 2 DBD was obtained from Dr. Craig A. Hauser (The Burnham Institute, La Jolla, CA). PU.1/Spi1 expression constructs are gifts from Dr. Françoise Moreau-Gachelin (Laboratoire de Transduction du Signal et Oncogenèse, Section de Recherche-Institut Curie, Paris, France).
Table 1.
Sequences Used to Generate Constructs
| Promoter Sequences | Forward Primers/Oligos | Reverse Primers/Oligos | Note | |
|---|---|---|---|---|
| 5′-Upstream Promoter Region | −4898/−3609 | (−4898) 5′-TTG GGA TCC GGA GTT GCT CAG CGC CCT-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | PCR |
| −4526/−3609 | (−4526) 5′-GTC GGA TCC AGA TGT CAG AGC AGG GGG AGC-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | ||
| −4307/−3609 | (−4307) 5′-AGA GGA TCC GTG CCG CAC AAG GTA GGA GGC TCG GTC G-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | ||
| −4122/−3609 | (−4122) 5′-AAC GGA TCC ATC TGT AGG GAG TTG AAC GCT G-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | ||
| −4017/−3609 | (−4017) 5′-CAC GGA TCC GAG GGC AGC AAA TGT CAA GAT TCG-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | ||
| −3812/−3609 | (−3812) 5′-AAG GGA TCC AGC TCG CTG GAG GTT TTG-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | ||
| −3637/−3609 | (−3637) 5′-GGT GGA TCC TGC TTT GCA ACT TCT CTC CCA GTG-3′ | 5′-CGC AAG CTT CTC GCA CTG GGA GAG AAG TTG CAA AG-3′ (−3609) | ||
| −4898/−4284 | (−4898) 5′-TTG GGA TCC GGA GTT GCT CAG CGC CCT-3′ | 5′-TGC AAG CTT CGA GCC TCC TAC CTT GTG CAG −3′ (−4284) | ||
| −4898/−4525 | (−4898) 5′-TTG GGA TCC GGA GTT GCT CAG CGC CCT-3′ | 5′-CGC AAG CTT GAA GAC GAT TGG GGA AGC GCG T-3′ (−4525) | ||
| −4898/−4642 | (−4898) 5′-TTG GGA TCC GGA GTT GCT CAG CGC CCT-3′ | 5′-TCG AAG CTT GCC TAA GCC GTC CTT CCT TGG −3′ (−4642) | ||
| −4898/−4734 | (−4898) 5′-TTG GGA TCC GGA GTT GCT CAG CGC CCT-3′ | 5′-AAG AAG CTT GTC CGG AGA CGA TCT ACG GG-3′ (−4734) | ||
| −4641/−4525 | (−4641) 5′-TTA GGA TCC CCG CGA TCG CGA ACC TTT GCC AAG ATG G-3′ | 5′-TGA AAG CTT GAA GAC GAT TGG GGA AGC GCG T-3′ (−4525) | ||
| −4589/−4525 | (−4589) 5′-GGG GGA TCC ACA CTG TAC CCT ACC AAG ATG-3′ | 5′-TGA AAG CTT GAA GAC GAT TGG GGA AGC GCG T-3′ (−4525) | ||
| −4898/−4560 | (−4898) 5′-TTG GGA TCC GGA GTT GCT CAG CGC CCT-3′ | 5′-TGA AAG CTT GCC CGC CGC CAT CTT GGT AGG GTA C-3′ (−4560) | ||
| −4641/−4560 | (−4641) 5′-TTA GGA TCC CCG CGA TCG CGA ACC TTT GCC AAG ATG G-3′ | 5′-TGA AAG CTT GCC CGC CGC CAT CTT GGT AGG GTA C-3′ (−4560) | ||
| −4560/−4525 | (−4560) 5′-GAT CCG GCT TCC GGG ACG CGC TTC CCC AAT CGT CTT CA-3′ | 5′-AGC TTG AAG ACG ATT GGG GAA GCG CGT CCC GGA AGC CG-3′(−4525) | Ds DNA Annealing | |
| −4554/−4525 | (−4554) 5′-GAT CCC GGG ACG CGC TTC CCC AAT CGT CTT CA-3′ | 5′-AGC TTG AAG ACG ATT GGG GAA GCG CGT CCC GG-3′(−4525) | ||
| −4548/−4525 | (−4548) 5′-GAT CCG CGC TTC CCC AAT CGT CTT CA-3′ | 5′-AGC TTG AAG ACG ATT GGG GAA GCG CG-3′ (−4525) | ||
| −4560/−4536 | (−4560) 5′-GAT CCG GCT TCC GGG ACG CGC TTC CCC AA-3′ | 5′-AGC TTT GGG GAA GCG CGT CCC GGA AGC CG-3′ (−4536) | ||
| −3947/−3924 | (−3947) 5′-GAT CCG TGG AAG AAG AGG TCA GGA GTT TCA-3′ | 5′-AGC TTG AAA CTC CTG ACC TCT TCT TCC ACG-3′ (−3924) | ||
| (Continued) | ||||
Table 1A.
Continued
| Promoter Sequences | Forward Primers/Oligos | Reverse Primers/Oligos | Note | |
|---|---|---|---|---|
| 3′-Downstream Promoter Region | −2986/−2523 | (−2986) 5′-AAC GGA TCC AGG GCG TGG GGG CAG GGA CCG CGG GCG CCC CTG CAG TTG CCA AGC GTC ACC-3′ | 5′-GGA AAG CTT CTA TTT AAG AAA GTC TCC CAT TGC CCA GCT G-3′ (−2523) | PCR |
| −2986/−2523 | (−2986) 5′-AAC GGA TCC AGG GCG TGG GGG CAG GGA CCG CGG GCG CCC GGC CGC CGC GCG GCC CCT CGG GCG GGG A-3′ | 5′-GGA AAG CTT CTA TTT AAG AAA GTC TCC CAT TGC CCA GCT G-3′ (−2523) | ΔGRU | |
| −2958/−2917 | (−2958) 5′-GAT CCC TGC AGT TGC CAA GCG TCA CCA ACA GGT TGC ATC GTT CCC A-3′ | 5′-AG CTT GGG AAC GAT GCA ACC TGT TGG TGA CGC TTG GCA ACT GCA GG-3′ (−2917) | ΔEts | |
| −2958/−2925 | (−2958) 5′-GAT CCC CTG CAG TTG CCA AGC GTC ACC AAC AGG TTG CA-3′ | 5′-AG CTT GCA ACC TGT TGG TGA CGC TTG GCA ACT GCA GGG GA-3′ (−2925) | Ds DNA Annealing | |
| −3425/−3377 | (−3425) 5′-GAT CCG GCA CCG TTT CCG TGC AAC CCC GTA GCC CCT TTC GAA GTG ACA CAC TTA-3′ | 5′-AG CTT AAG TGT GTC ACT TCG AAA GGG GCT ACG GGG TTG CAC GGA AAC GGT GCC G-3′ (−3377) | ||
Ds DNA, Double-stranded DNA.
Transient Transfection and Luciferase Reporter Gene Assay
For transient transfection of reporter expression vectors, Jurkat cells cultured in six-well plates (5 × 105 cells/ml) were transfected with plasmid vectors using the Superfect transfection reagent (QIAGEN, Valencia, CA). Cells were treated by EtOH (vehicle control) or DEX (1 μm) 24 h after transfection, and this was followed by an additional 24 h of incubation. Cells were harvested, washed with PBS, lysed, and assayed for firefly luciferase and β-galactosidase (β-gal) activities on an Ascent Luminoskan luminometer (Thermo Electron, Franklin, MA) as previously described (11,12,51). To transfect IM-9 B cells, electroporation was used based on the minor modification of an unpublished protocol (kindly provided by Dr. E. Brad Thompson). Cells growing at log phase were collected by centrifugation and washed with PBS. The cells were resuspended at 1 × 107 cells/ml in serum-free RPMI 1640 containing 1.25% dimethylsulfoxide (DMSO). Cell aliquots (400 μl; 2.5 × 106 cells) were mixed with the promoter expression vector plus 1.5 μg of a β-gal expression vector (normalization control). The mixture was transferred into 0.4-cm gap electroporation cuvettes (Bio-Rad Laboratories, Inc., Hercules, CA). Then cuvettes were electroporated using 975 μF and 275 V with a Gene Pulser II (Bio-Rad) Electroporator and then cooled on ice for 5 min. Electroporated cells from each cuvette were diluted into 5 ml of RPMI 1640 supplemented with 10% FBS and 1.25% DMSO and cultured overnight. Cells were then washed two times with RPMI 1640 containing 10% FBS to remove DMSO. The cell pellets were resuspended in 6 ml fresh RPMI 1640 (with 10% FBS) and split into 3-ml aliquots. The cells were cultured for another 4 h before ethanol vehicle (control) or DEX (1 μm final concentration) was added. After 24 h of treatment, cells were collected, washed, and analyzed for luciferase and β-gal activity. Results from three separate experiments were used for statistical analysis.
Western Blotting
Cells were collected and lysed with Laemmli sodium dodecyl sulfate sample buffer containing a protease inhibitor cocktail. Proteins resolved on SDS-PAGE (8%) were transferred to Immobilon-Nitrocellulose membranes (Millipore Corp., Bedford, MA). The membranes were blocked by 5% nonfat milk and developed using an Enhanced Chemiluminescence kit (Amersham Pharmacia Biotech, Arlington Heights, IL). Rabbit polyclonal antiactin, antitubulin, anti-hGR (H300), anti-c-Ets 1/2, anti-c-Myb, anti-Spi-B, and anti-PU.1/Spi1 antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA).
EMSAs
EMSAs were performed exactly as described previously (11), using nuclear extracts from CEM-C7 and/or IM-9 cells treated with 1 μm DEX or EtOH vehicle. For the supershift assays, antibodies to hGR (GR H300, sc-8992), c-Myb (c-Myb H141, sc-7874), and PU.1 (PU.1 T21, sc-352) were from Santa Cruz Biotechnology. Antibodies (2 μg) were incubated with the binding reaction mixture for 45 min at room temperature before the labeled oligonucleotides were added. The reactions were then incubated for an additional 15 min before electrophoresis. For the competition gel shift assays, double-stranded consensus DNA oligos that are recognized by the GR, c-Myb, or PU.1 were added to the binding reaction at 50, 200, and 1000 fold concentration over the isotopically labeled GRU probes. Synthesized GRU oligonucleotides from hGR gene promoters were as follows: GRU −4559/−4525, sense 5′-GGCTTCCGGGACGCGCTTCCCCAATCGTCTTC-3′, antisense 5′-GAAGACGATTGGGGAAGCGCGTCCCGGAAGCC-3′; and GRU −2956/−2916, sense 5′-GCGGGGAACGATGCAACCTGTTGGTGACGCTTGGCAACTGCAGG-3′ and antisense 5′-CCTGCAGTTGCCAAGCGTCACCAACAGGTTGCATCGTTCCC CGC-3′. The consensus oligonucleotides for the GRE (sc-2545, 5′-AGAGGATCTGTACAGGATGTTCTAGAT-3′), c-Myb (sc-2583, 5′-TACAGGCATAACGGTTCCGTAGTGA-3′), and Ets (PU.1) (sc-2549, 5′-GGGCTGCTTGAGGAAGTATAAGAAT-3′), were purchased from Santa Cruz Biotechnology. The binding reactions were resolved via 5% nondenaturing PAGE and visualized by autoradiography after being dried on a gel dryer.
ChIP Assays
Chromatin immunoprecipitation assays were performed as described previously (12). Antibodies used were: GR H300 (sc-8992); c-Myb H141 (sc-7874); PU.1 T-21(sc-352); SRC-1 (sc-8995); and SRC-2 (sc-8996) (all from Santa Cruz Biotechnology). Anti-HDAC1 (2E10) was obtained from Upstate Biotechnology (Lake Placid, NY; catalog no. 05-614). Cells (1 × 107) were cross linked with 1% formaldehyde and lysed to release nuclei. The chromatin released from the nuclei was sonicated. One portion was saved and used to determine the input. The sonicated chromatin was precleared with normal IgG (Santa Cruz Biotechnology). For immunoprecipitation, 6 μg of protein-specific antibodies or normal IgG (control) were added to the chromatin and incubated overnight at 4 C. The immune complexes were collected using salmon sperm DNA/protein A-agarose beads, followed by extensive washing. Then the chromatin DNA-protein-antibody complexes were eluted and the DNA-protein formaldehyde cross-links were reversed. DNA was purified using a Qiaquick PCR purification kit (QIAGEN) and used as the PCR template. The primers used for PCR amplification were as follows: forward primer, 5′-CCGTAGATCGTCTCCGGACAAGA-3′; and reverse primer, 5′-CTCCCCCTGCTCTGACATCTTGAA-3′, which spans −4755/−4507 region of the hGR gene promoter containing −4559/−4525 GRU sequence. PCR mixtures were resolved on 1.5% agarose gels and visualized by EtBr staining after 28–35 cycles of amplification.
Acknowledgments
We thank Dr. John A. Cidlowski (National Institute of Environmental Health Sciences, Research Triangle Park, NC) for the pCYGR GR protein expression construct; Dr. Giuseppe Raschellà (Ente Nuove Tecnologie Energia Ambiente, Rome, Italy) for the pcDNA3-c-MybHA and pcDNA3-c-Myb DBD expression constructs, Dr. Craig A. Hauser (The Burnham Institute, La Jolla, CA) for the pFN Ets 2 DBD expression vector, and Dr. Françoise Moreau-Gachelin (Laboratoire de Transduction du Signal et Oncogenèse, Section de Recherche-Institut Curie, Paris, France) for the PU.1/Spi1 expression construct.
Footnotes
This work was supported by the research support fund from Department of Biochemistry and Molecular Biology (to W.V.V. and C.-d. G.), a Hurricane Katrina Recovery Fund from the Louisiana Cancer Research Consortium (to W.V.V.), a Pilot Incentive Award from the Louisiana Cancer Research Consortium (to C.-d. G.), and National Cancer Institute Grant CA116042 (to W.V.V.).
Disclosure Statement: The authors have no disclosures.
First Published Online October 22, 2008
Abbreviations: ALL, Acute lymphoblastic leukemia; ChIP, chromatin immunoprecipitation; DBD, DNA-binding domain; DEX, dexamethasone; DMSO, dimethylsulfoxide; dYY1, distal YY1; EBE, Ets protein-binding element; FBS, fetal bovine serum; β-gal, β-galactosidase; GC, glucocorticoid; GR, glucocorticoid receptor; GRE, GC response element; GRU, GC response unit; HDAC, histone deacetylase; mYY1, middle YY1; pYY1, proximal YY1; SRC, steroid receptor coactivator.
References
- Pearson OH, Eliel LP, Rawson RW, Dobriner K, Rhoads CP 1949 ACTH- and cortisone-induced regression of lymphoid tumors in man. A preliminary report. Cancer 2:943–945 [DOI] [PubMed] [Google Scholar]
- Baxter JD, Harris AW, Tomkins GM, Cohn M 1971 Glucocorticoid receptors in lymphoma cells in culture: relationship to glucocorticoid killing activity. Science 171:189–191 [DOI] [PubMed] [Google Scholar]
- Norman MR, Thompson EB 1977 Characterization of a glucocorticoid-sensitive human lymphoid cell line. Cancer Res 37:3785–3791 [PubMed] [Google Scholar]
- Ploner C, Schmidt S, Presul E, Renner K, Schrocksnadel K, Rainer J, Riml S, Kofler R 2005 Glucocorticoid- induced apoptosis and glucocorticoid resistance in acute lymphoblastic leukemia. J Steroid Biochem Mol Biol 93:153–160 [DOI] [PubMed] [Google Scholar]
- Robertson AM, Bird CC, Waddell AW, Currie AR 1978 Morphological aspects of glucocorticoid-induced cell death in human lymphoblastoid cells. J Pathol 126:181–187 [DOI] [PubMed] [Google Scholar]
- Wyllie AH 1980 Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature 284:555–556 [DOI] [PubMed] [Google Scholar]
- Smets LA, Salomons G, van den Berg J 1999 Glucocorticoid induced apoptosis in leukemia. Adv Exp Med Biol 457:607–614 [DOI] [PubMed] [Google Scholar]
- Gaynon PS, Carrel AL 1999 Glucocorticosteroid therapy in childhood acute lymphoblastic leukemia. Adv Exp Med Biol 457:593–605 [DOI] [PubMed] [Google Scholar]
- Cato AC, Ponta H, Herrlich P 1992 Regulation of gene expression by steroid hormones. Prog Nucleic Acid Res Mol Biol 43:1–36 [DOI] [PubMed] [Google Scholar]
- Beato M, Chalepakis G, Schauer M, Slater E 1989 DNA regulatory elements for steroid hormones. J Steroid Biochem 32:737–748 [DOI] [PubMed] [Google Scholar]
- Geng C-d, Vedeckis WV 2004 Steroid-responsive sequences in the human glucocorticoid receptor gene 1A promoter. Mol Endocrinol 18:912–924 [DOI] [PubMed] [Google Scholar]
- Geng C-D, Vedeckis WV 2005 c-Myb and members of the c-Ets family of transcription factors act as molecular switches to mediate opposite steroid regulation of the human glucocorticoid receptor 1A promoter. J Biol Chem 280:43264–43271 [DOI] [PubMed] [Google Scholar]
- Morin B, Woodcock GR, Nichols LA, Holland LJ 2001 Heterodimerization between the glucocorticoid receptor and the unrelated DNA-binding protein, Xenopus glucocorticoid receptor accessory factor. Mol Endocrinol 15:458–466 [DOI] [PubMed] [Google Scholar]
- Mullick J, Anandatheerthavarada HK, Amuthan G, Bhagwat SV, Biswas G, Camasamudram V, Bhat NK, Reddy SEP, Rao V, Avadhani NG 2001 Physical interaction and functional synergy between glucocorticoid receptor and Ets2 proteins for transcription activation of the rat cytochrome P-450c27 promoter. J Biol Chem 276:18007–18017 [DOI] [PubMed] [Google Scholar]
- Widen C, Zilliacus J, Gustafsson JA, Wikstrom AC 2000 Glucocorticoid receptor interaction with 14–3-3 and Raf-1, a proposed mechanism for cross-talk of two signal transduction pathways. J Biol Chem 275:39296–39301 [DOI] [PubMed] [Google Scholar]
- Morin B, Zhu C, Woodcock GR, Li M, Woodward RN, Nichols LA, Holland LJ 2000 The binding site for Xenopus glucocorticoid receptor accessory factor and a single adjacent half-GRE form an independent glucocorticoid response unit. Biochemistry 39:12234–12242 [DOI] [PubMed] [Google Scholar]
- Drouin J, Sun YL, Tremblay S, Lavender P, Schmidt TJ, De Léan A, Nemer M 1992 Homodimer formation is rate-limiting for high affinity DNA binding by glucocorticoid receptor. Mol Endocrinol 6:1299–1309 [DOI] [PubMed] [Google Scholar]
- Wang JC, Stromstedt PE, Sugiyama T, Granner DK 1999 The phosphoenolpyruvate carboxykinase gene glucocorticoid response unit: identification of the functional domains of accessory factors HNF3 β (hepatic nuclear factor-3β) and HNF4 and the necessity of proper alignment of their cognate binding sites. Mol Endocrinol 13:608–618 [DOI] [PubMed] [Google Scholar]
- Ray D, Bosselut R, Ghysdael J, Mattei MG, Tavitian A, Moreau-Gachelin F 1992 Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU. 1. Mol Cell Biol 12:4297–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier JM, Bourachot B, Doucas V, Yaniv M, Moreau-Gachelin F 1993 Functional interference between the Spi-1/PU.1 oncoprotein and steroid hormone or vitamin receptors. EMBO J 12:5089–5096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brogan IJ, Murray IA, Cerillo G, Needham M, White A, Davis JRE 1999 Interaction of glucocorticoid receptor isoforms with transcription factors AP-1 and NF-κB: lack of effect of glucocorticoid receptor β. Mol Cell Endocrinol 157:95–104 [DOI] [PubMed] [Google Scholar]
- Geng CD, Pedersen KB, Nunez BS, Vedeckis WV 2005 Human glucocorticoid receptor α transcript splice variants with exon 2 deletions: evidence for tissue- and cell type-specific functions. Biochemistry 44:7395–7405 [DOI] [PubMed] [Google Scholar]
- Robyr D, Wolffe AP, Wahli W 2000 Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol Endocrinol 14:329–347 [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O'Malley BW 1999 Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol 69:3–12 [DOI] [PubMed] [Google Scholar]
- Freeman AI, Munn HL, Lyons V, Dammermann A, Seckl JR, Chapman KE 2004 Glucocorticoid down-regulation of rat glucocorticoid receptor does not involve differential promoter regulation. J Endocrinol 183:365–374 [DOI] [PubMed] [Google Scholar]
- Okret S, Poellinger L, Dong Y, Gustafsson JA 1986 Down-regulation of glucocorticoid receptor mRNA by glucocorticoid hormones and recognition by the receptor of a specific binding sequence within a receptor cDNA clone. Proc Natl Acad Sci USA 83:5899–5903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinyak JE, Dorin RI, Hoffman AR, Perlman AJ 1987 Tissue-specific regulation of glucocorticoid receptor mRNA by dexamethasone. J Biol Chem 262:10441–10444 [PubMed] [Google Scholar]
- Rosewicz S, McDonald AR, Maddux BA, Goldfine ID, Miesfeld RL, Logsdon CD 1988 Mechanism of glucocorticoid receptor down-regulation by glucocorticoids. J Biol Chem 263:2581–2584 [PubMed] [Google Scholar]
- Dong Y, Poellinger L, Gustafsson JA, Okret S 1988 Regulation of glucocorticoid receptor expression: evidence for transcriptional and posttranslational mechanisms. Mol Endocrinol 2:1256–1264 [DOI] [PubMed] [Google Scholar]
- Burnstein KL, Bellingham DL, Jewell CM, Powell-Oliver FE, Cidlowski JA 1991 Autoregulation of glucocorticoid receptor gene expression. Steroids 56:52–58 [DOI] [PubMed] [Google Scholar]
- Govindan MV, Pothier F, Leclerc S, Palaniswami R, Xie B 1991 Human glucocorticoid receptor gene promotor-homologous down regulation. J Steroid Biochem Mol Biol 40:317–323 [DOI] [PubMed] [Google Scholar]
- Hoeck W, Rusconi S, Groner B 1989 Down-regulation and phosphorylation of glucocorticoid receptors in cultured cells. Investigations with a monospecific antiserum against a bacterially expressed receptor fragment. J Biol Chem 264:14396–14402 [PubMed] [Google Scholar]
- Vedeckis WV, Ali M, Allen HR 1989 Regulation of glucocorticoid receptor protein and mRNA levels. Cancer Res (Suppl) 49:2295s–2302s [PubMed] [Google Scholar]
- Alksnis M, Barkhem T, Strömstedt PE, Ahola H, Kutoh E, Gustafsson JA, Poellinger L, Nilsson S 1991 High level expression of functional full length and truncated glucocorticoid receptor in Chinese hamster ovary cells. Demonstration of ligand-induced down-regulation of expressed receptor mRNA and protein. J Biol Chem 266:10078–10085 [PubMed] [Google Scholar]
- Burnstein KL, Jewell CM, Cidlowski JA 1990 Human glucocorticoid receptor cDNA contains sequences sufficient for receptor down-regulation. J Biol Chem 265:7284–7291 [PubMed] [Google Scholar]
- Eisen LP, Elsasser MS, Harmon JM 1988 Positive regulation of the glucocorticoid receptor in human T-cells sensitive to the cytolytic effects of glucocorticoids. J Biol Chem 263:12044–12048 [PubMed] [Google Scholar]
- Pedersen KB, Vedeckis WV 2003 Quantification and glucocorticoid regulation of glucocorticoid receptor transcripts in two human leukemic cell lines. Biochemistry 42:10978–10990 [DOI] [PubMed] [Google Scholar]
- Pedersen KB, Geng C-d, Vedeckis WV 2004 Three mechanisms are involved in glucocorticoid receptor auto-regulation in a human T-lymphoblast cell line. Biochemistry 43:10851–10858 [DOI] [PubMed] [Google Scholar]
- Presul E, Schmidt S, Kofler R, Helmberg A 2007 Identification, tissue expression, and glucocorticoid responsiveness of alternative first exons of the human glucocorticoid receptor. J Mol Endocrinol 38:79–90 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Irving JA, Minto L, Matheson E, Nicholson L, Ploner A, Parson W, Kofler A, Amort M, Erdel M, Hall A, Kofler R 2006 Glucocorticoid resistance in two key models of acute lymphoblastic leukemia occurs at the level of the glucocorticoid receptor. FASEB J 20:2600–2612 [DOI] [PubMed] [Google Scholar]
- Ramdas J, Liu W, Harmon JM 1999 Glucocorticoid- induced cell death requires autoinduction of glucocorticoid receptor expression in human leukemic T cells. Cancer Res 59:1378–1385 [PubMed] [Google Scholar]
- Kato GJ, Quddus FF, Shuster JJ, Boyett J, Pullen JD, Borowitz MJ, Whitehead VM, Crist WM, Leventhal BG 1993 High glucocorticoid receptor content of leukemic blasts is a favorable prognostic factor in childhood acute lymphoblastic leukemia. Blood 82:2304–2309 [PubMed] [Google Scholar]
- Gomi M, Moriwaki K, Katagiri S, Kurata Y, Thompson EB 1990 Glucocorticoid effects on myeloma cells in culture: correlation of growth inhibition with induction of glucocorticoid receptor messenger RNA. Cancer Res 50:1873–1878 [PubMed] [Google Scholar]
- Nanni P, Nicoletti G, Prodi G, Galli MC, De Giovanni C, Grilli S, Lollini PL, Gobbi M, Cavo M, Tura S 1982 Glucocorticoid receptor and in vitro sensitivity to steroid hormones in human lymphoproliferative diseases and myeloid leukemia. Cancer 49:623–632 [DOI] [PubMed] [Google Scholar]
- Leventhal BG 1981 Glucocorticoid receptors in lymphoid tumors. Cancer Res 41:4861–4862 [PubMed] [Google Scholar]
- Bloomfield CD, Peterson BA, Zaleskas J, Frizzera G, Smith KA, Hildebrandt L, Gail-Peczalska KJ, Munck A 1980 In vitro glucocorticoid studies for predicting response to glucocorticoid therapy in adults with malignant lymphoma. Lancet 1:1952–1956 [DOI] [PubMed] [Google Scholar]
- Schmidt S, Rainer J, Riml S, Ploner C, Jesacher S, Achmuller C, Presul E, Skvortsov S, Crazzolara R, Fiegl M, Raivio T, Janne OA, Geley S, Meister B, Kofler R 2006 Identification of glucocorticoid-response genes in children with acute lymphoblastic leukemia. Blood 107:2061–2069 [DOI] [PubMed] [Google Scholar]
- Zong J, Ashraf J, Thompson EB 1990 The promoter and first, untranslated exon of the human glucocorticoid receptor gene are GC rich but lack consensus glucocorticoid receptor element sites. Mol Cell Biol 10:5580–5585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breslin MB, Vedeckis WV 1998 The human glucocorticoid receptor promoter upstream sequences contain binding sites for the ubiquitous transcription factor Yin Yang 1. J Steroid Biochem Mol Biol 67:369–381 [DOI] [PubMed] [Google Scholar]
- Breslin MB, Vedeckis WV 1996 The glucocorticoid receptor and c-jun promoters contain AP-1 sites that bind different AP-1 transcription factors. Endocrine 5:15–22 [DOI] [PubMed] [Google Scholar]
- Breslin MB, Geng C-D, Vedeckis WV 2001 Multiple promoters exist in the human GR gene, one of which is activated by glucocorticoids. Mol Endocrinol 15:1381–1395 [DOI] [PubMed] [Google Scholar]
- Breslin M, Vedeckis WV, Identification of alternative promoter usage for the regulation of the human glucocorticoid receptor gene. Program of the 80th Annual Meeting of The Endocrine Society, New Orleans, LA, 1998, p 133 [Google Scholar]
- Turner JD, Muller CP 2005 Structure of the glucocorticoid receptor (NR3C1) gene 5′ untranslated region: identification, and tissue distribution of multiple new human exon 1. J Mol Endocrinol 35:283–292 [DOI] [PubMed] [Google Scholar]
- Wei P, Inamdar N, Vedeckis WV 1998 Transrepression of c-jun gene expression by the glucocorticoid receptor requires both AP-1 sites in the c-jun promoter. Mol Endocrinol 12:1322–1333 [DOI] [PubMed] [Google Scholar]
- Nunez BS, Vedeckis WV 2002 Characterization of promoter 1B in the human glucocorticoid receptor gene. Mol Cell Endocrinol 189:191–199 [DOI] [PubMed] [Google Scholar]
- Encio IJ, Detera-Wadleigh SD 1991 The genomic structure of the human glucocorticoid receptor. J Biol Chem 266:7182–7188 [PubMed] [Google Scholar]
- Turner JD, Schote AB, Macedo JA, Pelascini LPL, Muller CP 2006 Tissue specific glucocorticoid receptor expression, a role for alternative first exon usage? Biochem Pharmacol 72:1529–1537 [DOI] [PubMed] [Google Scholar]
- Truss M, Beato M 1993 Steroid hormone receptors: interaction with deoxyribonucleic acid and transcription factors. Endocr Rev 14:459–479 [DOI] [PubMed] [Google Scholar]
- Oikawa T, Yamada T 2003 Molecular biology of the Ets family of transcription factors. Gene 303:11–34 [DOI] [PubMed] [Google Scholar]
- Deng QL, Ishii S, Sarai A 1996 Binding site analysis of c-Myb: screening of potential binding sites by using the mutation matrix derived from systematic binding affinity measurements. Nucleic Acids Res 24: 766–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warriar N, Pagé N, Govindan MV 1996 Expression of human glucocorticoid receptor gene and interaction of nuclear proteins with the transcriptional control element. J Biol Chem 271:18662–18671 [DOI] [PubMed] [Google Scholar]