Abstract
Protein ubiquitination is essential for many events linked to intracellular protein trafficking. We sought to elucidate the possible involvement of the S. cerevisiae deubiquitinating enzyme Ubp2 in transporter and receptor trafficking after we (this study) and others established that affinity purified Ubp2 interacts stably with the E3 ubiquitin ligase Rsp5 and the (ubiquitin associated) UBA domain containing protein Rup1. UBP2 interacts genetically with RSP5, while Rup1 facilitates the tethering of Ubp2 to Rsp5 via a PPPSY motif. Using the uracil permease Fur4 as a model reporter system, we establish a role for Ubp2 in membrane protein turnover. Similar to hypomorphic rsp5 alleles, cells deleted for UBP2 exhibited a temporal stabilization of Fur4 at the plasma membrane, indicative of perturbed protein trafficking. This defect was ubiquitin dependent, as a Fur4 N-terminal ubiquitin fusion construct bypassed the block and restored sorting in the mutant. Moreover, the defect was absent in conditions where recycling was absent, implicating Ubp2 in sorting at the multivesicular body. Taken together, our data suggest a previously overlooked role for Ubp2 as a positive regulator of Rsp5-mediated membrane protein trafficking subsequent to endocytosis.
Introduction
Protein ubiquitination is essential for the proper functioning of many eukaryotic cellular processes. While the covalent conjugation of polyubiquitin chains to a protein by ubiquitin ligases often leads to subsequent targeting and degradation by the 26S proteasome [1], ubiquitin attachment has also been shown to be critical for the correct execution of non-proteasomal events, ranging from the regulated methylation of histones during transcriptional elongation [2] to the intracellular trafficking of proteins [3]. For the latter, ubiquitination serves as a key signal mediating the internalization, intracellular transport and subsequent recycling or vacuolar degradation of plasma membrane-bound receptors and transporters [4]. While much progress has been made in elucidating the mechanistic basis for various steps in protein trafficking, many aspects about the core components and enzymatic steps remain unresolved.
In the budding yeast Saccharomyces cerevisiae, a single essential E3 ubiquitin ligase, Rsp5, has been implicated in the internalization of most if not all endocytosed proteins [5]. The uracil permease Fur4 is an experimentally well-characterized model substrate of Rsp5. Uracil binding, or various stress conditions such as nutrient starvation, heat shock, or the inhibition of protein synthesis trigger Fur4 phosphorylation which drives the subsequent recognition and Lys63-linked di-ubiquitination of Fur4 by Rsp5 on two specific lysine residues [6]–[10]. Similar to other membrane proteins, the ubiquitination of Fur4 results in its rapid and complete clearance from the plasma membrane into early endosomes. This is followed by the subsequent recognition of ubiquitinated Fur4 by the ESCRT-I, II, III complexes and other class E Vps proteins, followed by sorting into multivesicular bodies (MVBs) [11]. When MVB sorting is inhibited in class E vps mutants, endocytosed Fur4 recycles back up to the plasma membrane [12]. After sorting into the MVB, Fur4 is targeted to the vacuolar lumen where it is degraded [10]. Fur4 can also follow a different pathway when synthesized in the presence of its substrate, uracil. In this situation, newly synthesized Fur4 is diverted to the endosomal system directly from the Golgi apparatus [13]. It then undergoes Rsp5-dependent ubiquitination, a modification crucial for its sorting into MVBs [14], followed by vacuolar degradation after fusion of MVBs with the vacuole.
The process by which ubiquitin chains are attached to proteins is reversible. Cleavage of ubiquitin chains from ubiquitinated proteins is performed by a broad family of deubiquitinating enzymes (DUBs) which share conserved motifs, notably the Cys and His boxes, which are required for catalytic function [15]. In yeast, these DUBs fall into two major categories [16]: the ubiquitin C-terminal hydrolases (Uch), and the ubiquitin specific processing proteases (Ubp). Uchs are restricted to cleaving ubiquitin from small peptides and chemical adducts, while Ubps release ubiquitin from larger protein substrates and disassemble longer polyubiquitin chains [1]. Based on sequence analysis, the budding yeast S. cerevisiae encodes 16 Ubps, most of which have been confirmed to have general DUB activity in vitro [17].
Emerging evidence implicate members of the vesicle-based intra-organellar protein trafficking pathway as physiologically relevant targets of several of the Ubps. These include: Doa4 (Ubp4), involved in the deubiquitination of internalized endocytic cargoes such as Fur4 at the endosome immediately prior to their sorting into MVBs [18]; Ubp3, implicated in the deubiquitination of regulatory proteins in both the anterograde [19] and retrograde [20] protein transport pathways through the endoplasmic reticulum (ER) and Golgi systems, along with a possible involvement in the cytoplasm to vacuole (Cvt) trafficking pathway [21]; and Ubp1, which has been linked to the internalization and turnover of the ABC membrane transporter Ste6 via an as yet unknown mechanism [22].
Parallel studies by our group (described in this study) and another laboratory [23] have now established specific and physiologically significant physical and functional interactions between the S. cerevisiae deubiquitinating enzyme Ubp2, the E3 ligase Rsp5, along with a protein of previously unknown function, Rup1, which contains a ubiquitin associated (UBA) domain and a putative Rsp5-binding motif. Although the in vitro assays reported previously [23] have hinted at a possible role of Ubp2 in the deubiquitination of substrates of Rsp5, including the ER membrane bound transcription factor Spt23 and the two relatively uncharacterized proteins Csr2 and Ecm2 [24], physiologically significant targets of Ubp2 with respect to Rsp5-mediated trafficking remains uncertain. Here, we expand upon these initial observations and establish a critical role for Ubp2 in the ubiquitin-dependent sorting of plasma membrane proteins using Fur4 as a model substrate. Collectively, our data suggest that Ubp2 is not simply a negative regulator of Rsp5, but rather is engaged in a more complex and active regulatory relationship with Rsp5 at the MVB.
Results
Ubp2 Physically Interacts with Rsp5 and Rup1
Neither Ubp2 or Rup1 are essential for normal cell growth and deletion of either gene does not result in any obvious growth defects or overt morphological phenotypes [15], [25]. Hence, to investigate the possible role(s) of Ubp2 in yeast, we performed large-scale tandem affinity purifications (TAP) to isolate endogenous Ubp2 from S. cerevisiae strains bearing a C-terminal TAP tag [26]. Stably interacting partner proteins were resolved by SDS-PAGE, stained with silver, and identified by peptide mass fingerprinting using MALDI-TOF (matrix-assisted laser desorption/ionization-time of flight) mass spectrometry. Two polypeptides, encoding the E3 ubiquitin ligase Rsp5, and a protein of previously unknown function, YOR138c, which we have named Rup1 (Rsp5-Ubp2 interacting Protein 1), were found to reproducibly and specifically co-purify with Ubp2 (Figure 1A). Neither of these binding partners were detected in parallel purifications from an untagged negative control strain or from several hundred other TAP-tagged bait proteins [27]. These data were independently confirmed by highly-sensitive gel-free tandem mass spectrometry-based peptide shotgun sequencing (LC-MS/MS) of an aliquot of each protein mixture after in solution digestion with trypsin (http://tap.med.utoronto.ca/). Conversely, no known substrate or other previously reported Rsp5-interacting proteins [28]–[31] co-purified reproducibly with either Ubp2-TAP or Rup1-TAP (data not shown).
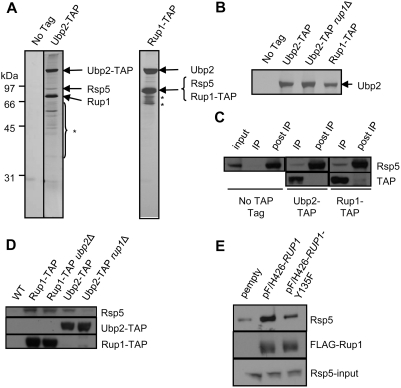
Figure 1. Ubp2, Rsp5, and Rup1 interact physically.
(A) Silver-stained SDS polyacrylamide gel showing affinity purified Ubp2-TAP and Rup1-TAP protein complexes. Arrows indicate polypeptides identified by gel band excision followed by MALDI-ToF mass spectrometry. The asterisk indicates degradation products of Ubp2 and Rup1. (B) Ubp2 recovery from Ubp2-TAP, Ubp2-TAP rup1Δ, and Rup1-TAP strains. Arrow indicates gel band identified as Ubp2 by MALDI-ToF mass spectrometry. (C) Co-immunoprecipitation of a fraction of cellular Rsp5 with Ubp2 and Rup1. Cell extracts were depleted of endogenously TAP-tagged Ubp2 or Rup1 with IgG (IP lanes), and the remaining soluble Rsp5 (post-IP lanes) and bait proteins probed by Western blot with anti-Rsp5 antibodies which also recognized the protein A portion of the TAP tag. The ‘input’ lane shows the Rsp5 level in the extract prior to IP. (D) Rup1 tethers Ubp2 to Rsp5. IgG-based immunoprecipitation of Rup1-TAP and Ubp2-TAP from WT, ubp2Δ or rup1Δ strains. The co-purification of Rsp5 with the baits was determined by Western blotting using anti-Rsp5 antibodies. (E) Plasmids expressing FLAG(F/H)-tagged wildtype Rup1 or the Y135F point mutant were transformed into cells lacking endogenous Rup1. After immunoprecipitation with anti-FLAG antibodies, Rup1 and Rsp5 were detected by Western blotting. The bottom panel show the expression levels of Rsp5 in whole cell extracts by Western blotting against Rsp5.
To further validate these results, we likewise performed reciprocal affinity purification using a strain expressing endogenous TAP-tagged Rup1 (a viable C-terminal Rsp5-TAP strain could not be generated). As expected, Rup1-TAP co-purified with a seemingly identical amount of Ubp2 and with Rsp5, albeit in an apparently sub-stoichiometric level (Figure 1A). Since immunoprecipitation of either Ubp2-TAP or Rup1-TAP resulted in a near-identical yield of Ubp2 (Figure 1B), the interaction between Ubp2 and Rup1 is seemingly complete and likely equimolar. In contrast, a substantial fraction of Rsp5 remained in solution and was not recovered with either Ubp2-TAP or Rup1-TAP after depletion of TAP-tagged proteins with IgG beads from soluble cell extracts (Figure 1C), indicating that a substantive fraction of Rsp5 exists in alternate protein complexes, or that Rsp5 binding is transient or labile. The relative proportion of cellular Rsp5 pool present in a complex with Ubp2 and Rup1 is estimated to be ∼10% of total. Since we have shown that only a modest fraction of the total cellular pool of Rsp5 is stably bound to Ubp2/Rup1, Ubp2, and by extension Rup1, may be involved in only a subset of Rsp5-dependent pathways.
To further investigate the structure-function dependencies of the interaction of Ubp2/Rup1 with Rsp5, we constructed TAP-tagged strains bearing a targeted genomic deletion in either Ubp2 or Rup1. The composition of the resulting affinity-isolated protein complexes were then examined by Western blotting. Whereas Rsp5 remained bound to Rup1-TAP in the absence of Ubp2 (Figure 1D), Ubp2-TAP did not detectably precipitate Rsp5 in the absence of Rup1. These results support a previous report [23] indicating that Rup1 likely mediates, at least in part, the interaction of Ubp2 to Rsp5. Consistent with this, we identified a putative WW domain consensus binding motif, PPPSY, encoded by amino acids 131–135 of Rup1. We generated a site-specific point mutant in which the invariant core tyrosine residue 135 was substituted with a phenylalanine (Y135F), and expressed this from an episomal plasmid in a strain lacking endogenous RUP1. This type of mutation (Y→F) has previously been shown to markedly reduce or even abolish binding of other WW domain proteins to their interacting partners bearing analogous PY motifs [32], [33]. Indeed, as expected, the interaction of this mutant variant with Rsp5 was impaired, although not abolished, relative to the wild-type control (Figure 1E), implying that the motif mediates in part the recruitment of Rsp5. Similar results were obtained after expression of the Rup1 Y135F mutant in a ubp2Δ deletion mutant background (data not shown), indicating the residual binding was not mediated by Ubp2.
UBP2 and RUP1 Interact Genetically with RSP5
To further explore the relationship of Ubp2 and Rup1 to Rsp5-related functions, we examined the effects of inducible ectopic overexpression of either Ubp2 or Rup1 in a strain harbouring a temperature-sensitive hypomorphic allele of RSP5 (rsp5-1) [34], as well as in a wild-type strain, looking for enhancement or suppression of the conditional slow-growth phenotype. As seen in the limiting dilution series shown in Figure 2A, Ubp2 overexpression was moderately toxic to the control strain as compared to an empty control vector, while the rsp5-1 mutant was found to be extremely hypersensitive. This synthetic dosage lethality was specific to UBP2, as the rsp5-1 strain did not shown any hypersensitivity to overexpression of two other unrelated deubiquitinating enzymes, UBP6 and UBP10 (Figure 2A). Indeed, although overproduction of UBP10 caused a noticeable growth defect in wildtype cells, this defect did not worsen when combined with the rsp5-1 mutation, in striking contrast to UBP2. While wildtype yeast were not detectably sensitive to the overexpression of Rup1, rsp5-1 and ubp2Δ mutants displayed modest hypersensitivity. On the other hand, a deletion of RUP1 resulted in a reduction in the growth impairment caused by the overexpression of UBP2. These data support the notion that Rup1 functions coordinately with, but not equivalently to Ubp2. To confirm that these genetic interactions were not specific to the rsp5-1 allele, another allele, rsp5-326 [35] was tested (Figure 2B). All of the above interactions were similar in this mutant background. When Rsp5 was expressed from a low copy number plasmid, pHA-RSP5 [36], in rsp5-326 strains, the growth phenotypes seen were rescued to wildtype levels, indicating that these phenotypes were specific to RSP5.
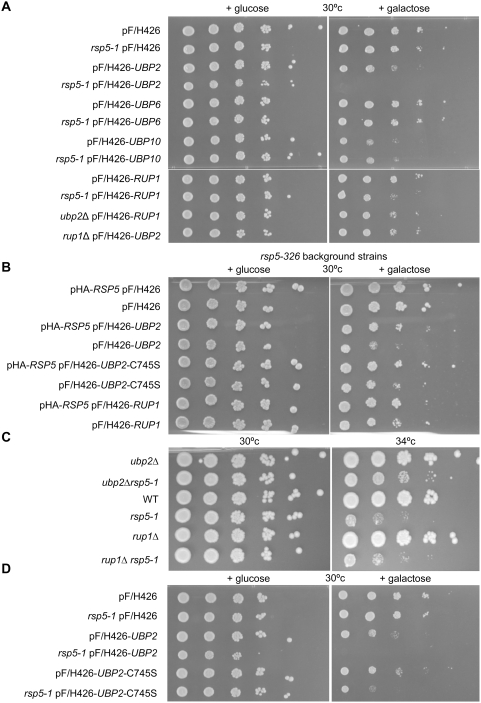
Figure 2. UBP2 and RUP1 interact genetically with RSP5.
(A, B, D) The indicated over-expression plasmids, driven by a galactose inducible promoter, were transformed into either wildtype or mutant strains. Cell cultures were then serially diluted 10-fold (starting OD600 of 0.5) and spotted onto SC-URA media containing glucose (non-inducing) or galactose (inducing). (B) Growth of rsp5-326 mutant cells bearing pHA-RSP5 (a low copy, but not galactose responsive plasmid), and/or a second overexpression plasmid as indicated. (C) Growth of cells deleted for either UBP2 and RUP1 alone, or in combination with an RSP5 temperature sensitive allele (rsp5-1), on rich media (YPD) at either 30°c (permissive for rsp5-1) or 34°c (semi-permissive for rsp5-1).
Consistent with this apparent functional antagonism (Figure 2A; [23]), deletion of the UBP2 open reading frame partially rescued the temperature-sensitivity of an rsp5-1 mutant strain (Figure 2C), although deletion of RUP1 had no observable affect. Taken together, these results are consistent with a recently proposed model [23] wherein Ubp2 directly antagonizes Rsp5 activity, and imply that Rup1 likely serves as a less critical ancillary role such as facilitating the interaction of Ubp2 with Rsp5 or possible substrates.
Ubp2 Catalytic Activity is Important, but Not Essential, for Synthetic Dosage Lethality
A key characteristic of Ubps is the presence of an evolutionarily conserved catalytic domain [15]. To test whether the catalytic activity of Ubp2 was necessary for the antagonistic interaction observed with Rsp5-deficient strains, we mutated cysteine 745, a critical residue of the core Cys box motif conserved across all members of the Ubp family, to serine, which is predicted to result in a severe or complete loss of deubiquitinating capacity [19], [37]. As expected, we observed that overexpression of this putatively inactive mutant, ubp2 C745S, led to a marked reduction in the slow-growth phenotype caused by overexpression of UBP2 in either rsp5 strain background (Figure 2, B and D), suggesting that inappropriate or excessive deubiquitination activity is largely, but not exclusively, responsible for the functional antagonism observed.
ubp2Δ Cells are Sensitive to 5-Fluorouracil (5-FU)
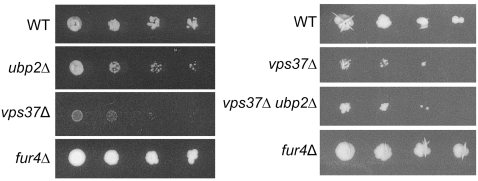
Given that Rsp5 has a well-established role in the internalization, trafficking, and subsequent vacuolar degradation of membrane bound transporters [5], we explored the possibility that Ubp2 might also modulate the efficiency or dynamics of these ubiquitination-dependent events. One well characterized substrate of Rsp5 is the uracil permease Fur4. rsp5 mutants have increased plasma membrane steady state amounts of Fur4. The same is true for mutants in class E genes that accumulate an abnormal compartment, the class E compartment, from which Fur4 recycles to plasma membrane. All these mutants display increased sensitivity to the drug 5-Fluorouracil (5-FU) [12], which is imported by Fur4. 5-FU is a toxic uracil analogue which inhibits the nucleotide synthetic enzyme thymidylate synthase and incorporates aberrantly into cellular RNA and DNA [38], leading to cell death. We checked ubp2Δ for potential sensitivity to 5-FU, using a class E mutant, vps37Δ, as a control (Figure 3). As expected, cells in which the transporter is absent (fur4Δ) are completely resistant to the drug (Figure 3; [39]). Compared to wildtype cells, ubp2Δ mutants are partially sensitive to the drug. However, they were not as sensitive as cells with a deletion in vps37Δ. The sensitivity of vps37Δ cells is not affected when combined with a mutation in UBP2, suggesting that VPS37 and UBP2 may act at the same level. These data show that Fur4 may be stabilized at the plasma membrane of ubp2Δ cells, leading to an increased sensitivity to 5-FU. The stabilization may not be complete as the sensitivity of ubp2Δ is less than that of strains defective in various sorting steps along the endocytic pathway.
Figure 3. Loss of UBP2 results in hypersensitivity to 5-fluorouracil (5-FU).
Wildtype, ubp2Δ, vps37Δ, fur4Δ single mutants, and vps37Δubp2Δ double mutant cells were transformed with a URA3 plasmid and subsequently tested for sensitivity to 5-FU. Exponentially growing cells were serially diluted 10-fold (starting OD600 ∼0.7) and spotted onto SD plates with 5 µM 5-FU. Plates were imaged either after 4 days (left panel) or 7 days (right panel).
ubp2Δ Cells Have Increased Steady State Amounts of Plasma Membrane Localized Fur4
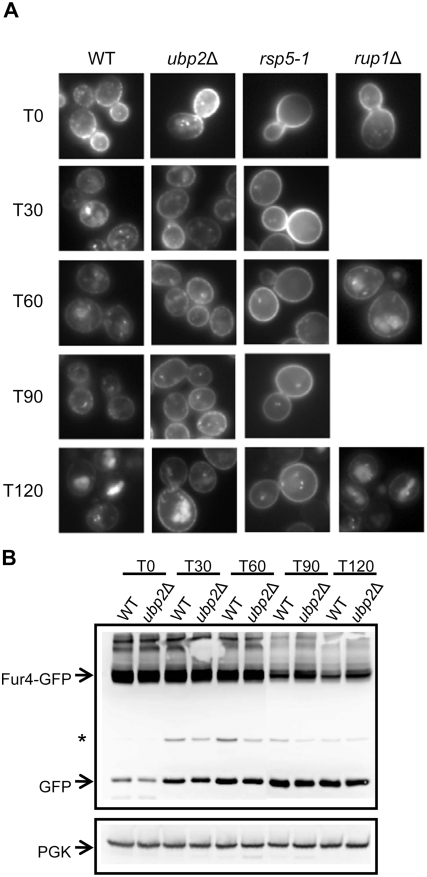
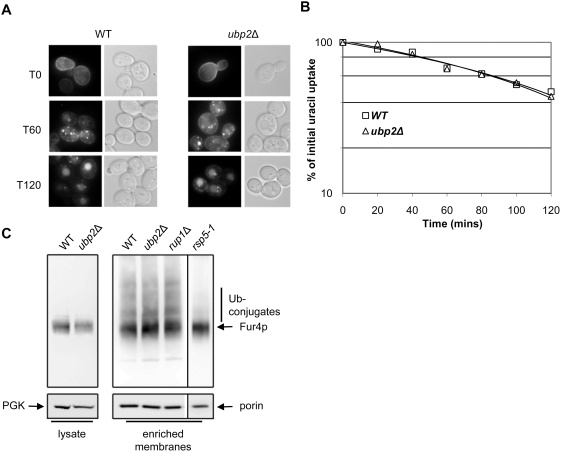
In order to define whether the 5-FU sensitivity of ubp2Δ cells indeed comes from defects in trafficking, or merely from impaired RNA metabolism [40], we followed the fate of a GFP-tagged version of Fur4 in ubp2Δ and rup1Δ cells, using as control rsp5 mutant cells (Figure 4, A and B). A galactose-inducible version of Fur4-GFP was similarly targeted to the plasma membrane after galactose induction in all the strains. Glucose was then added to stop Fur4-GFP synthesis and chase to the plasma membrane any Fur4-GFP still in the secretory pathway. Uracil was then added to trigger Fur4-GFP endocytosis. Cells were harvested at various time points after the addition of uracil, and subjected to whole-cell imaging (Figure 4A). Total protein extracts were also prepared in parallel for Western blotting against Fur4-GFP in the case of wildtype and ubp2Δ cells (Figure 4B). Uracil triggered a progressive loss of plasma membrane GFP fluorescence in wildtype cells, a transient apparition of punctuate intracellular fluorescent dots (likely endosomes), followed by apparition of luminal vacuolar fluorescence (corresponding to free GFP, not immediately degraded by vacuolar proteases). Fur4-GFP displayed almost the same fate in rup1Δ cells. In contrast, in rsp5-1 cells, even at a permissive temperature, plasma membrane fluorescence was still detectable after two hours of uracil treatment, together with very faint vacuolar fluorescence. In ubp2Δ cells, the situation was intermediate between that observed in wildtype and rsp5 cells. Plasma membrane staining was still observed after 90 min of uracil treatment, and after two hours, both plasma membrane and vacuolar fluorescence were evidenced. In agreement with this observation, Fur4-GFP degradation was delayed in ubp2Δ compared to wildtype cells (Figure 4B).
Figure 4. Uracil-induced Fur4 sorting is perturbed in ubp2Δ mutant cells.
A FUR4-GFP fusion reporter was transformed into cells, which were then grown in raffinose overnight. Galactose was added (OD600 = 0.6) for 2 hours to induce synthesis, followed by glucose for 10 min to chase Fur4-GFP to the plasma membrane, and then uracil (40 µg/ml) to trigger transporter internalization. (A) Fur4-GFP signal viewed by fluorescence microscopy at the indicated time points (min) after uracil addition. T0 indicates cells immediately before addition. (B) Fur4-GFP protein levels as measured by Western blotting. Total protein extracts were generated from cells harvested at the indicated time points following uracil addition. Fur4-GFP was probed with anti-GFP antibodies. 3-phosphoglycerate kinase (PGK) was used as a loading control. The asterisk indicates an unknown background band.
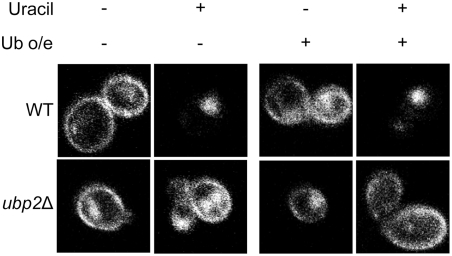
Deubiquitinating enzymes such as Ubp4 (Doa4) have previously been linked to the overall recycling of ubiquitin prior to the proteasomal or vacuolar degradation of substrates [18], [41]. Depletion of the free ubiquitin pools in a yeast cell has the potential to lead to defects in protein sorting from the plasma membrane [6], presumably by indirectly reducing the efficiency of Rsp5-mediated ubiquitination at various steps of the sorting pathway. To exclude the possibility that the Fur4-GFP sorting defect observed in ubp2Δ mutants was an artifact due to a general reduction in free ubiquitin levels, we assessed whether the phenotype could be suppressed by the forced overexpression of ubiquitin from a plasmid using a strongly inducible copper promoter. Impaired sorting in the ubp2 deletion mutant was not restored (Figure 5). This data, along with other previously reported data showing that ubp2 mutants exhibit wildtype levels of free ubiquitin [17], indicate that the impaired kinetics of Fur4-GFP sorting in ubp2Δ mutants are indeed likely a direct result of improper regulation of the sorting pathway, and not due to a deficit in cellular ubiquitin levels.
Figure 5. The overexpression of ubiquitin does not rescue the sorting defect in ubp2Δ cells.
pFUR4-GFP and a plasmid overexpressing ubiquitin (+) or an empty plasmid (−) were transformed into cells. Strains were grown in sucrose, and diluted to OD600 = 0.5 in media containing galactose and 0.1 mM copper sulphate (to overexpress ubiquitin) and grown for 4 hours. Fur4 transcription was stopped by adding glucose for 1 hour to chase Fur4 to the plasma membrane. Uracil (40 µg/ml) was then added to induce Fur4 internalization. The GFP signal was viewed by fluorescence confocal microscopy before (−) and at 60 min after (+) uracil addition. Note that the left panel contains the same images as Figure 7, as the experiments were done in parallel.
A deletion in ubp2 could result in abnormal vesicle morphology leading to indirect defects in Fur4 sorting. To determine if this was the case, wildtype and ubp2Δ cells were labelled with the fluorescent lipid-binding dye FM4-64, washed, and endocytosis of the dye was examined. Compared to wildtype cells, ubp2Δ cells displayed a normal rate of sorting of the dye to endosomes and eventually to the vacuolar membrane (Figure S1). Normal endosome and vacuolar morphologies were also seen, although subtle differences may not be detected with this assay. Taken together, these data implicate Ubp2 in the intracellular sorting of Fur4, specifically at a step in the trafficking of Fur4-GFP from the plasma membrane to the vacuole.
ubp2Δ Cells Display Normal Plasma Membrane Fur4 Ubiquitination and Internalization
Plasma membrane stabilization of Fur4 subsequent to conditions normally triggering its endocytosis can be observed in two types of situations: in mutants impaired in the internalization step of endocytosis [10], [13], and in class E mutants with impaired MVB sorting. In the latter case, Fur4 recycles from the endosomal class E compartment back to plasma membrane. In order to decipher the origin of plasma membrane stabilization of Fur4-GFP in ubp2Δ cells, we first searched, using vps class E mutants, for conditions preventing any recycling.
Fur4-GFP was expressed in wildtype and vps37Δ cells, and cells were subjected to carbon starvation (CS) to induce internalization of Fur4-GFP from the plasma membrane. Glucose (CA), was then added, to induce recycling of Fur4 back up to the plasma membrane, in the presence or absence of an inhibitor of protein synthesis, cycloheximide (CHX). Fur4-GFP localization was followed by both fluorescence microscopy (Figure S2A) and by monitoring uracil uptake (Figure S2B). In both WT and vps37Δ cells, Fur4-GFP was internalized completely after carbon starvation as expected (Figure S2A, [12]). Fur4-GFP was rapidly targeted back up to the plasma membrane in vps37Δ cells after the addition of glucose but not upon addition of glucose+CHX. Recycling only occurred in vps37Δ cells, as the mutation blocked the entry of Fur4-GFP into the vacuolar lumen; in wildtype cells all Fur4-GFP was targeted to vacuolar lumen, where it was degraded [12]. As expected, uracil uptake increased over 60 minutes post-glucose addition in vps37Δ cells in the absence of CHX (Figure S2B). Taken together, these data show that recycling of Fur4-GFP cannot be detected in the presence of CHX, although the exact mechanism is unknown. Thus, triggering endocytosis of Fur4-GFP in the presence of CHX was a good condition in which to define the level of action of Ubp2.
After two hours of galactose induction of Fur4-GFP followed by glucose, CHX was added to wildtype and ubp2Δ cells. The fate of Fur4-GFP was then followed by fluorescence imaging and uracil uptake measurement (Figure 6, A and B). Plasma membrane fluorescence disappeared in a similar way in wildtype and ubp2Δ cells (Figure 6A), and the loss of uracil uptake occurred with identical rate in both type of cells (Figure 6B). Endocytic internalization of Fur4 is thus normal in ubp2Δ cells. In agreement with the observation of a normal rate of internalization, the pattern of ubiquitination (regularly spaced Fur4 species evidenced above the main Fur4-GFP signal) on Western blots of plasma membrane enriched fractions was identical in wildtype and ubp2Δ cells (Figure 6C). This is in comparison to rsp5-1 cells, which showed a distinct lack of Fur4-ubiquitin conjugates as expected. This data implies that Ubp2 is likely not involved in the maintenance or deubiquitination of Fur4 at the plasma membrane.
Figure 6. UBP2 is not necessary for cycloheximide (CHX) triggered internalization of Fur4.
(A) pFUR4-GFP bearing cells were grown in raffinose overnight. Galactose was added for 2 hours to induce synthesis, and glucose was then added for 10 min to chase Fur4-GFP to the plasma membrane. CHX (0.1mg/ml) was added and GFP signal examined by fluorescence microscopy and Nomarski optics. Time refers to the time after addition of CHX, with 0 min as pre-induction. (B) Uracil uptake was measured at different times after the addition of CHX in WT and ubp2Δ strains. Results are expressed as a percentage of the initial uracil uptake, and plotted on a log scale. (C) Fur4 ubiquitin profile at the plasma membrane is unchanged in ubp2Δ mutants. pFUR4 was transformed into WT, ubp2Δ, rup1Δ, and rsp5-1, and cells, which were grown in raffinose overnight. Expression from pFUR4 was induced for 90 min with galactose before adding glucose for 15min to chase Fur4 to the plasma membrane. Total protein extracts (lysate) and enriched membrane fractions were collected and analyzed by Western blotting to visualize Fur4 and Fur4-ubiquitin conjugates. 3-phosphoglycerate kinase (PGK) and porin, a mitochondrial membrane protein, were used as loading controls.
Ubp2 is Involved in Efficient Fur4-GFP Ubiquitination in the Late Steps of Endocytosis
If Ubp2 is not required for plasma membrane ubiquitination and internalization, it must therefore be required for subsequent steps of trafficking. In a survey of all membrane bound Fur4 in the cell, the ubiquitin status of Fur4 appears to be unaffected in ubp2Δ cells (data not shown). As any subtle and rapid ubiquitination/deubiquitination events may have been missed, and in order to more closely decipher the origin of the Fur4 trafficking defect in ubp2Δ cells, we investigated the role of ubiquitination in uracil-induced endocytosis.
Defects in Rsp5-dependent, ubiquitin-dependent trafficking of a number of membrane-bound proteins, either at plasma membrane, or at MVB, can be bypassed using an in frame fusion of ubiquitin to these various cargoes. This is notably the case for Fur4 [13]. Hence, we examined whether expression of the Fur4-GFP reporter construct bearing ubiquitin as an N-terminal fusion (pUb-FUR4-GFP) could likewise suppress the sorting defect following the loss of Ubp2 activity. Control experiments (Figure 7) confirmed that, as expected, the fusion construct rescued, at least partially, the internalization defect seen in rsp5-1 cells at 60 minutes post-uracil addition. Likewise, the Ub-Fur4-GFP fusion protein was also efficiently cleared from the plasma membrane in the ubp2 deletion mutants, as the fluorescence signal detected at the plasma membrane was virtually absent one hour after uracil induction. The concomitant increase in the background signal intensity detected in the vacuoles of ubp2Δ cells was expected due to missorting of nascent Ub-Fur4-GFP to the vacuole directly from the Golgi [13]. Taken together, these data suggest, albeit somewhat paradoxically, that the loss of Ubp2 activity impairs proper Rsp5-mediated ubiquitination of Fur4-GFP, likely at the level of MVB sorting. This sorting step was restored with Ub-Fur4-GFP, and the protein, correctly sorted to MVB, did not recycle to the plasma membrane.
Figure 7. Ub-Fur4-GFP is correctly sorted at the MVB in ubp2Δ mutant cells.
pFUR4-GFP or pUb-FUR4-GFP expression plasmids were transformed into WT and ubp2Δ cells. Strains were grown in sucrose, diluted (OD600 = 0.5) in media containing galactose and Fur4 synthesis induced for 4 hours. Transcription was stopped by adding glucose for 1 hour to chase Fur4 fusion reporters to the plasma membrane. Uracil (40 µg/ml) was then added to trigger internalization. GFP signal was viewed by fluorescence confocal microscopy both before (T0) and 60 min after uracil addition. (Note: cells in the left panel were also transformed with a control vector (YEp46Δ) and incubated with 0.1 mM copper sulphate, as the experiment was done in parallel to that shown in Figure 5.)
Ubp2 May Modulate the Sorting of Ste2
The yeast α-factor receptor, Ste2, is another well-characterized substrate of Rsp5 [42]. Similar to Fur4, Ste2 is ubiquitinated, internalized, and subsequently degraded in the vacuole upon substrate-binding [42]–[44]. Defects in the sorting and degradation of Ste2 result in hypersensitivity to α-factor [45], as the presence of α-factor triggers cell cycle arrest in MATa recipient cells [46]. When a disk of α-factor is spotted on a lawn of MATa cells, a gradient zone of growth inhibition forms, termed a halo. ubp2 deletion mutants exhibit hypersensitivity to α-factor by halo assay as compared to a WT control strain (Figure 8), suggesting a defect in clearing Ste2 from the plasma membrane of these cells. Surprisingly, the sensitivity of ubp2Δ cells to α-factor was slightly greater than in end3 deletion mutants lacking an endocytic machinery component, which was previously found to be sensitive to α-factor [45], and rup1Δ cells have a sensitivity similar to ubp2Δ cells. These data suggest that Ubp2 modulates the ubiquitination and sorting of other plasma membrane associated proteins other than Fur4, but this effect may happen with slightly different kinetics depending on the cargo. The possible involvement of Rup1 in Ste2 transport is currently unclear and will be subject to further study.
Figure 8. ubp2Δ cells are sensitive to alpha-factor consistent with a defect in Ste2 receptor sorting.
α-factor (5 ul of 100 µM, 40 µM or water alone) was spotted onto sterile filter disks, and placed onto solid media containing WT, ubp2Δ, rup1Δ, or end3Δ MATa cells. Plates were then incubated at 30°C for one day and imaged. Halo diameters (mm) were measured and averaged from five replicates of 100 µM alpha factor spots.
Discussion
Despite the rapidly expanding interest in DUBs as likely critical regulators of core cellular processes such as protein trafficking [47], [48], the physiological roles and targets of most of the known or predicted deubiquitinating enzymes remain unknown. In this report, we sought to confirm and extend the observations of previous studies regarding the molecular associations and putative functions of Ubp2 [23], [24], [49]. In principle, most of the genetic and physical interaction data reported in this study and previously by another group [23] point to an antagonistic, and presumably regulatory relationship with Ubp2/Rup1 counteracting Rsp5. In light of this perspective, the finding that ubp2Δ mutants have a defect in Rsp5-mediated transporter and receptor sorting reported here is especially surprising.
The accumulation of Fur4-GFP seen at the plasma membrane under uracil induced conditions could be due to a defect in internalization or an increase in recycling of Fur4 to the plasma membrane. We see that the internalization step itself does not seem to be impaired. Since the defect in Fur4-GFP trafficking appears only in recycling permissive conditions in ubp2Δ cells, our data imply that the accumulation of Fur4-GFP at the plasma membrane is most likely due to a more efficient recycling of the permease. An aberrant accumulation of membrane proteins at the plasma membrane can result from a defect in MVB formation, as has been reported with ESCRT mutants [12]. Therefore, it is likely that ubp2Δ mutants have a defect in internalization into MVBs. This defect is not complete, however, as Fur4-GFP eventually reaches the vacuolar lumen following a temporal lag (i.e. after 120 minutes post uracil addition versus 30 minutes in WT cells). Further evidence that places Ubp2 at the MVB sorting step is the fact that sensitivity to 5-FU does not seem to worsen in a vps37Δubp2Δ double mutant when compared to a vps37Δ single mutant alone. In addition, Ubp2, Rup1, and Rsp5 have been shown in other studies to be present at the MVB. Rsp5 was found to be located at various sites within the endocytic pathway, including plasma membrane invaginations, late endosomes, and MVBs by colocalization with Pep12, an endosomal marker, and Vps32, a component of an ESCRT complex located at the MVB [50]. Rsp5, Rup1 and Ubp2 have also been shown to interact with Hse1 [49], a component of the ESCRT-0 complex located at the MVB. In addition to Fur4, we see a role for Ubp2 in the proper clearance of another membrane-bound protein, Ste2, from the plasma membrane, suggesting Ubp2 is generally required for efficient sorting.
Consistent with these results, Ubp2 has recently been shown to be important for the Rsp5-mediated trafficking of a vacuolar protease, carboxypeptidase S (Cps), which is sorted to the vacuolar lumen from the Golgi via the MVB pathway [49]. In addition, a role for Ubp2 in properly sorting the membrane bound amino acid permease Gap1 directly from the Golgi to the MVB has also been reported [49]. In this study, we establish, at least in the case of Fur4 (and possibly for Ste2), the additional involvement of Ubp2 in sorting of endocytic cargo from the plasma membrane to the MVB. It seems, therefore, that Ubp2 has a role in modulating Rsp5-mediated trafficking of various types of membrane associated cargo.
What then, specifically, is the role of Ubp2? Although Ubp2 has been shown to deubiquitinate a non-essential ER-membrane bound transcription factor, Spt23, in vitro [23], there is as yet no evidence for the physiological processing of Spt23 in vivo (M.L.; unpublished observations). Indeed, counterintuitive to expectation, our observation that a ub-Fur4 fusion can rescue the sorting defect in ubp2Δ mutants suggests inefficient or improper ubiquitination of the internalized transporter.
One possibility is that Ubp2 may be involved in modulating the efficiency of Rsp5 ubiquitination, by deubiquitinating auto-ubiquitinated Rsp5, or by sequestering Rsp5 to a specific cellular compartment or substrate. Deubiquitination of Rsp5 would lead to the subsequent stabilization of the E3 ub-ligase as is the case for many previously identified E3-DUB physically interacting pairs [51], [52]. However, this appears unlikely, as no detectable decrease in Rsp5 levels was detected in a ubp2Δ mutants (M.L.; unpublished observations), and Rsp5 has not been reported to be (auto)ubiquitinated in vivo.
A second possibility is that Ubp2 may be involved in ubiquitin chain length editing of internalized receptors and transporters at the MVB. Most substrates of the endocytic pathway are covalently attached with single ubiquitin moieties or short K63-linked ubiquitin chains [6], [53]–[55], although a Vps pathway substrate with a K63-linked polyubiquitin chain has been recently reported [56]. The recent finding that Ubp2 can efficiently and preferentially deubiquitinate K63 linked chains [24] suggests a possible role for Ubp2 in editing Rsp5-catalyzed polyubiquitin chains down to a length suitable for recognition by various proteins in the endocytic/ESCRT machinery. Although a difference in the ubiquitin profile of Fur4 in a WT versus ubp2Δ mutants was not detected, our Western blot assay represented the sum of all membrane compartments in the cell, and may not be sensitive enough to visualize a subtle kinetic defect at the MVB. It would be interesting and informative to look at the ubiquitination of Fur4 in conditions where more subtle differences in ubiquitination levels at the MVB could be detected.
A third, and more likely (although currently speculative) possibility is a role of Ubp2 in the deubiquitination of the cargo (e.g. Fur4) right at the MVB, which is then followed by a second, subsequent re-ubiquitination by Rsp5 for efficient sorting into the MVB. In support of this model, it was previously reported that Rsp5-dependent ubiquitination of cargo must occur at the MVB stage for proper sorting into MVBs [57]. Further support for this model is our data showing a physical interaction between Rsp5 and Ubp2 through tethering by Rup1, allowing for a close physical proximity between the Ub ligase and the DUB at the MVB. Another DUB, Doa4, also appears to play a somewhat related, but different, role at the MVB, as deubiquitination by Doa4 itself is not sufficient for the proper re-ubiquitination of Fur4. Although it has not yet been shown conclusively that deubiquitination must occur prior to cargo re-ubiquitination, this will be an interesting area to address in the future. In addition to cargo deubiquitination, we cannot currently rule out an (additional) indirect role of Ubp2 on the sorting machinery itself, adding another possible level of complexity in the regulation of trafficking. We are also unable to conclusively exclude a role for Ubp2 at the plasma membrane, as there might have been subtle defects in ubp2Δ cells at the internalization step of endocytosis that remained undetected in our experimental conditions.
The role of Rup1 also remains enigmatic. Residual Ubp2-Rsp5 binding was detected in the absence of Rup1, implying that Ubp2 and Rsp5 can interact, either directly, or possibly through another unknown adaptor protein. This could explain why Rup1 is not required for proper MVB sorting, although a minor sorting defect not apparent in the assay conditions tested cannot be ruled out. Aspects of our genetic data imply that Rup1 has a positive role in Ubp2 activity. For example, deletion of RUP1 suppressed, albeit only partially, the temperature-sensitive phenotype of rsp5-1 mutants. The presence of a UBA domain in Rup1 is also suggestive. Ubiquitin-binding motifs, such as UBA or UIM (ubiquitin interacting motif) domains, are present in a sizeable fraction of proteins involved in intracellular protein trafficking [58], and have been suggested to facilitate the “passing-off” of ubiquitinated endocytic cargo or components from one functional module in the processing machinery to another [58], [59]. Hence, Rup1 could play a complex role at the MVB, in both recruiting ubiquitinated proteins to the ternary complex and tethering Ubp2 to Rsp5.
Materials and Methods
Strains and Growth Media
All transformations of yeast strains were done using a standard protocol [60], [61]. All yeast strains were grown in YP (yeast extract and peptone)+2% glucose and at 30°C unless otherwise specified. Relevant genotypes of strains are detailed in Table 1. TAP-tagged deletion mutant strains were constructed by direct transformation or by mating a relevant deletion strain with a suitable TAP tagged strain. Tetrads were pulled, and haploids containing both HIS3 (TAP) and NAT-resistance (deletion) markers were selected. The presence of the deletion and the expression of the TAP-tagged protein were confirmed by PCR and Western blotting, respectively. ubp2Δ::NatMX and rup1Δ::NatMX strains were constructed as described in [62] by transformation with EcorRI-cut p4339 into cells or by PCR of the NatMX cassette from p4339 followed by transformation. For Figure 2, wildtype strains in 2A and 2D, and rup1Δ in 2C, are derived from FY56 which is isogenic to rsp5-1. For other figures, wildtype strains are BY4741. ubp2Δrsp5-1 and rup1Δrsp5-1 were constructed either from direct transformation of the deletion cassette into rsp5-1, or by mating of haploids, tetrad dissection, and selection of haploids with the required genotype. vps37Δ was constructed as described in [63] and vps37Δubp2Δ was constructed by crossing single mutant strains.
Table 1. Yeast Strains Used in this Study.
| Strain/Plasmid | Relevant Genotype | Origin |
| WT | BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) | [68] |
| Ubp2-TAP | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 UBP2::TAP-HIS3 | [69] |
| Rup1-TAP | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 RUP1::TAP-HIS3 | [69] |
| ubp2 Δ | ubp2Δ::NATMX, derived from Y3656 (MATα can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0) | This study, [70] |
| ubp2 Δ | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 ubp2Δ::KanMX3 | [25] |
| rup1 Δ | rup1Δ::NATMX, derived from Y3656 (MATα can1Δ::MFA1pr-HIS3-MFα1pr-LEU2 his3Δ leu2Δ0 ura3Δ0 met15Δ0 lys2Δ0) | This study, [70] |
| rup1 Δ | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 rup1Δ::KanMX3 | [25] |
| rup1 Δ | rup1Δ::NATMX, derived from FY56 (MATα his4-912 ΔR5 lys2-128Δ ura3-52) | This study, [71] |
| Ubp2-TAP rup1 Δ | MATa UBP2::TAP-HIS3, rup1Δ::NATMX | This study |
| Rup1-TAP ubp2 Δ | MATa RUP1::TAP-HIS3, ubp2Δ::NATMX | This study |
| rsp5-1 | FW1808 (MATα his4-912 ΔR5 lys2-128Δ ura3-52 rsp5-1) | [71] |
| rsp5-326 | MATa leu2-3,112 ura3-52 his3-Δ200 trp1-Δ901 ade2-101 suc2-Δ9 | [35] |
| ubp2 Δ rsp5-1 | ubp2Δ::KanMX3 rsp5-1 | This study, [71] |
| rup1 Δ rsp5-1 | rup1Δ::KanMX3 rsp5-1 | This study, [71] |
| fur4 Δ | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 fur4Δ::KanMX3 | [25] |
| end3 Δ | MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0 end3Δ::KanMX3 | [25] |
| vps37 Δ | vps37::HIS | This study |
| vps37 Δ ubp2 Δ | vps37::HIS ubp2Δ::KanMX3 | This study |
Plasmids
Plasmids allowing for inducible overexpression of N-terminal affinity tagged proteins were constructed by recombineering essentially as previously described [64]. Briefly, full-length ORFs were amplified from yeast genomic DNA using primers similar to the Yeast Genepairs Primers (Research Genetics) bearing homology to the 5′ and 3′ ends of each ORF. The product from this first PCR reaction was re-amplified using primers which contain sequences homologous to those flanking a SmaI cloning site downstream of a galactose inducible promoter in a high copy yeast expression plasmid with a selectable URA3 marker gene (pF/H426). The PCR product was integrated into the parental plasmid, linearized with SmaI, by co-transformation and in vivo homologous recombination, resulting in an in-frame fusion to a triple FLAG/His10/HA2 (F/H) epitope affinity tag immediately upstream of the ORF. Correct expression of the fusion protein was confirmed by Western blotting. The mutant constructs pF/H426-UBP2-C745S and pF/H426-RUP1-Y135F, bearing site specific point mutations were constructed using the QuikChange II Site-Directed Mutagenesis kit (Stratagene). Mutants were confirmed by dideoxy DNA sequencing. pHA-RSP5 (p[HA-RSP5, CEN, LEU2]) was from [36]. The galactose inducible expression plasmids pFUR4 (pFL38GalFUR4), pFUR4-GFP (pFL38GalFUR4-GFP), and ubiquitin-fusion construct pUb-FUR4-GFP (pFL38GalUb-FUR4-GFP; containing a variant ubiquitin with all its relevant lysine residues mutated to arginine to ensure that no additional ubiquitin can be added to the ubiquitin moiety, and glycine 76 replaced with a valine, to prevent proteolytic cleavage) were from [8], [13], [65]. The chromosome encoded uracil permease is produced in very small amounts and cells that expressed the permease under the Gal1 promoter from a centromeric plasmid were used for accurate measurement of the permease activity and for the immunodetection of the protein [7] The cooper-inducible ubiquitin expression plasmid pCUP1-Ub-TRP1 (YEp96) and the control vector (YEp46Δ) are from [66].
TAP-tagged Protein Purification and Mass Spectrometry
Large-scale TAP-tagged protein purifications were separated on 10% polyacrylamide gels, and silver stained using a standard protocol. Subsequent analysis of the mixtures of purified proteins by both gel-free tandem mass spectrometry and gel-based MALDI-ToF mass spectrometry were performed essentially as previously described [27].
Cell Lysis, MiniGel Electrophoresis and Western Blotting
Cells were harvested and lysed by glass bead beating in YEB (Yeast Extraction Buffer: 245 mM KCl, 1 mM EDTA, 5 mM EGTA, 50 mM Hepes-KOH pH 8.0, 10% glycerol, 2 mM DTT) containing an EDTA-free protease inhibitor cocktail (Complete; Roche) or YEB containing 350 mM KCl to decrease non-specific binding to antibody beads (Rup1 Y135F binding experiments) and centrifuged at 14 krpm at 4°C for 10 minutes to pellet insoluble debris. For the immunoprecipitation experiments, approximately 6 mg of total soluble protein extract (∼10 mg/ml) was then incubated for 1 hour at 4°C with anti-FLAG M2 Agarose affinity beads (Sigma-Aldrich) for F/H tagged strains, or with IgG Sepharose 6 Fast Flow beads (Amersham Pharmacia) for TAP-tagged strains. The beads were washed extensively, and tightly bound proteins were eluted with 15 ul of SDS sample buffer by heating at 90°C for 5 minutes. The samples were separated by electrophoresis on a 4–12% Bis-Tris polyacrylamide gel (Invitrogen), and the proteins transferred from the gel onto nitrocellulose membrane (Trans-Blot Transfer Medium; BioRad). The membranes were then extensively pre-blocked with a 5% milk protein solution, followed by overnight incubation with mouse monoclonal anti-HA antibody (12CA5 cell line; a kind gift from Mike Tyers, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Ontario) to detect the TAP tag, rabbit anti-Rsp5 antibody (a kind gift from Linda Hicke [67]), or mouse anti-FLAG antibody (Sigma-Aldrich). Detection was performed by ECL (SuperSignal West Pico, Pierce) using suitable secondary antibodies conjugated to horseradish peroxidase. For the Fur4 immunoblots, whole cell lysates (Figure 6) were made by cell disruption followed by centrifugation at 3000 g, and total protein extracts (Figure 4B) were prepared by the NaOH-TCA lysis technique as previously described [10]. To better detect ubiquitinated species of Fur4, membrane enriched fractions of cells producing the lower molecular weight Fur4, and not the Fur4-GFP fusion were analyzed. Membrane-enriched protein fractions were prepared as previously described [18] except that cells were broken in a “One Shot” Cell Disrupter (Constant Systems LDT) at maximum pressure. Proteins in sample buffer were heated at 37°C, resolved by SDS-polyacrylamide gel electrophoresis in 10% acrylamide gels using tricine buffer, and transferred to nitrocellulose membranes. The membranes were probed with monoclonal anti-GFP antiserum (Roche), anti-PGK (Molecular Probes), rabbit anti-Fur4 antibody [10], or monoclonal antibodies against porin (Molecular Probes). Horseradish peroxidase-conjugated anti-mouse or anti-rabbit immunoglobulin G was used as the secondary antibody (Sigma) and was detected by enhanced chemiluminescence (ECL).
Serial Dilutions
Strains were grown overnight to saturation in SC-URA or YP media containing 2% glucose, and diluted with sterile water to OD600 = 0.5. 10-fold dilutions with sterile water were then made, and 3 µl of the cell suspension spotted onto media plates containing rich (YP) or selective (SC-URA) media+2% glucose or 2% sucrose/1% galactose. Plates were incubated for 2–4 days at 30°C or 34°C and imaged. For 5-FU plates, cells were grown to OD600 ∼0.7. 10-fold dilutions were then made, and 20 µl of the cell suspension spotted onto media plates containing SD+5 µM 5-FU (Sigma). Plates were incubated for 4 or 7 days and imaged.
Uracil uptake assay
Uracil uptake was measured as previously described [10], by incubating a 1 ml culture of exponentially growing cells with 5×10−6 M radiolabeled uracil for 20 s at 30°C. Cells were quickly filtered through Whatman GF/C filters, which were then washed twice with ice-cold water and counted for radioactivity. Fur4p activity was measured various times after the addition of cycloheximide (100 µg/ml; Sigma) as previously described [10].
Microscopy
Cells grown to exponential growth phase in YNB medium was concentrated by a factor of ten by centrifugation. Cells were viewed immediately, without fixation, under a fluorescence microscope (type BY61, Olympus, Tokyo, Japan) and images captured with a digital camera. For Figures 5 and 7, fluorescence images of live yeast cells at a 512×512 pixel resolution were generated with a Leica DM IRBE confocal microscope using a 100× objective with immersion oil. Images were processed using Leica TCS software.
α-factor Halo Assay
MATa cells from an overnight culture were added to pre-warmed (55°C) sterile 0.5% agar, vigorously mixed, and poured evenly as a thin layer onto plates containing solid YP+2% glucose media. After solidifying, sterile filter disks soaked with 5 ul of various concentrations of α-factor (Pepceuticals) were gently overlaid onto the cell lawn, followed by incubation at 30°C for one day prior to scoring.
Materials and Methods for Figures S1 and S2 are described in Supporting Information file Materials and Methods S1.
Supporting Information
Endosome and Vacuolar morphology is normal in ubp2Δ cells. Cells were incubated with the lipid-binding fluorescent dye FM4-64 and washed. Endocytosis of the dye and endosome/vacuolar morphology were monitored by fluorescence microscopy and Nomarski optics at the indicated time points (min) after staining.
(1.96 MB TIF)
Recycling of Fur4-GFP cannot be detected in the presence of cycloheximide. WT and vps37Δ cells transformed with pFur4-GFP were cultured at 30°C and Fur4-GFP synthesis was induced for 90 min by adding galactose. Glucose was added to block Fur4-GFP synthesis (CC). 20 min later, cells were subjected to carbon starvation (CS) to trigger endocytosis of the permease for 60 minutes. Cultures were divided in two equal fractions. Glucose (CA) or glucose+cycloheximide (CA+CHX 0.1 mg/ml) was then added. (A) Cells were visualized by fluorescence microscopy at t = 0 (0'CC), after 60 minutes of carbon starvation (60'CS) and 60 minutes after the addition of carbon (60'CA) or carbon+cycloheximide (60'CA+CHX). Note that Fur4-GFP was efficiently internalized in all the strains upon CS. (B) Uracil uptake was measured 3, 10, 30 and 60 min after the addition of glucose. Results are expressed as a percentage of the initial uracil uptake measured immediately before carbon starvation, and plotted on a linear scale.
(0.46 MB TIF)
(0.04 MB DOC)
Acknowledgments
We thank Erin O'Shea, Jonathan Weissman, Steve Hanes, Scott Emr, Teresa Zoladek, Daniel Finley, and Charles Boone for strains and plasmids; and Linda Hicke and Mike Tyers for antibodies. We also acknowledge Maanasa Raghavan, Janet N.Y. Chan, Jennifer Rilstone, Peter Wong, Michael Davey, Oxana Pogoutse and Grace Guo for technical assistance; David Jansma for support to M.H.Y.L. with the confocal microscopy; and Elizabeth Conibear and members of the Emili Laboratory for constructive discussions.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by funds from the Protein Engineering Network Centre of Excellence of Canada (PENCE) to A.E.. Ontario Graduate Scholarships and University of Toronto Open Fellowships to M.H.Y.L. Work in R.H.T.'s lab was supported by the Centre National de la Recherche Scientifique and the Universities Paris 6 and Paris 7, by a grant from the Association pour la Recherche sur le Cancer (ARC) (grant no. 3298), by a special grant from the CNRS (ACI BCMS), and by a grant from the European Program 6 (FP6) (RUBICON, contrat no. 018683). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Hochstrasser M. Ubiquitin-dependent protein degradation. Annu Rev Genet. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 2.Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- 3.Hicke L. Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. Faseb J. 1997;11:1215–1226. doi: 10.1096/fasebj.11.14.9409540. [DOI] [PubMed] [Google Scholar]
- 4.Dupre S, Urban-Grimal D, Haguenauer-Tsapis R. Ubiquitin and endocytic internalization in yeast and animal cells. Biochim Biophys Acta. 2004;1695:89–111. doi: 10.1016/j.bbamcr.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 5.Horak J. The role of ubiquitin in down-regulation and intracellular sorting of membrane proteins: insights from yeast. Biochim Biophys Acta. 2003;1614:139–155. doi: 10.1016/s0005-2736(03)00195-0. [DOI] [PubMed] [Google Scholar]
- 6.Galan JM, Haguenauer-Tsapis R. Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. Embo J. 1997;16:5847–5854. doi: 10.1093/emboj/16.19.5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. A PEST-like sequence mediates phosphorylation and efficient ubiquitination of yeast uracil permease. Mol Cell Biol. 1998;18:314–321. doi: 10.1128/mcb.18.1.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seron K, Blondel MO, Haguenauer-Tsapis R, Volland C. Uracil-induced down-regulation of the yeast uracil permease. J Bacteriol. 1999;181:1793–1800. doi: 10.1128/jb.181.6.1793-1800.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchal C, Haguenauer-Tsapis R, Urban-Grimal D. Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J Biol Chem. 2000;275:23608–23614. doi: 10.1074/jbc.M001735200. [DOI] [PubMed] [Google Scholar]
- 10.Volland C, Urban-Grimal D, Geraud G, Haguenauer-Tsapis R. Endocytosis and degradation of the yeast uracil permease under adverse conditions. J Biol Chem. 1994;269:9833–9841. [PubMed] [Google Scholar]
- 11.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 12.Bugnicourt A, Froissard M, Sereti K, Ulrich HD, Haguenauer-Tsapis R, et al. Antagonistic roles of ESCRT and Vps class C/HOPS complexes in the recycling of yeast membrane proteins. Mol Biol Cell. 2004;15:4203–4214. doi: 10.1091/mbc.E04-05-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blondel MO, Morvan J, Dupre S, Urban-Grimal D, Haguenauer-Tsapis R, et al. Direct sorting of the yeast uracil permease to the endosomal system is controlled by uracil binding and Rsp5p-dependent ubiquitylation. Mol Biol Cell. 2004;15:883–895. doi: 10.1091/mbc.E03-04-0202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morvan J, Froissard M, Haguenauer-Tsapis R, Urban-Grimal D. The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic. 2004;5:383–392. doi: 10.1111/j.1398-9219.2004.00183.x. [DOI] [PubMed] [Google Scholar]
- 15.Baker RT, Tobias JW, Varshavsky A. Ubiquitin-specific proteases of Saccharomyces cerevisiae. Cloning of UBP2 and UBP3, and functional analysis of the UBP gene family. J Biol Chem. 1992;267:23364–23375. [PubMed] [Google Scholar]
- 16.Wilkinson KD. Ubiquitination and deubiquitination: targeting of proteins for degradation by the proteasome. Semin Cell Dev Biol. 2000;11:141–148. doi: 10.1006/scdb.2000.0164. [DOI] [PubMed] [Google Scholar]
- 17.Amerik AY, Li SJ, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- 18.Dupre S, Haguenauer-Tsapis R. Deubiquitination step in the endocytic pathway of yeast plasma membrane proteins: crucial role of Doa4p ubiquitin isopeptidase. Mol Cell Biol. 2001;21:4482–4494. doi: 10.1128/MCB.21.14.4482-4494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen M, Stutz F, Belgareh N, Haguenauer-Tsapis R, Dargemont C. Ubp3 requires a cofactor, Bre5, to specifically de-ubiquitinate the COPII protein, Sec23. Nat Cell Biol. 2003;5:661–667. doi: 10.1038/ncb1003. [DOI] [PubMed] [Google Scholar]
- 20.Cohen M, Stutz F, Dargemont C. Deubiquitination, a new player in Golgi to endoplasmic reticulum retrograde transport. J Biol Chem. 2003;278:51989–51992. doi: 10.1074/jbc.C300451200. [DOI] [PubMed] [Google Scholar]
- 21.Baxter BK, Abeliovich H, Zhang X, Stirling AG, Burlingame AL, et al. Atg19p ubiquitination and the cytoplasm to vacuole trafficking pathway in yeast. J Biol Chem. 2005;280:39067–39076. doi: 10.1074/jbc.M508064200. [DOI] [PubMed] [Google Scholar]
- 22.Schmitz C, Kinner A, Kolling R. The deubiquitinating enzyme Ubp1 affects sorting of the ATP-binding cassette-transporter Ste6 in the endocytic pathway. Mol Biol Cell. 2005;16:1319–1329. doi: 10.1091/mbc.E04-05-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kee Y, Lyon N, Huibregtse JM. The Rsp5 ubiquitin ligase is coupled to and antagonized by the Ubp2 deubiquitinating enzyme. Embo J. 2005;24:2414–2424. doi: 10.1038/sj.emboj.7600710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kee Y, Munoz W, Lyon N, Huibregtse JM. The deubiquitinating enzyme Ubp2 modulates Rsp5-dependent Lys63-linked polyubiquitin conjugates in Saccharomyces cerevisiae. J Biol Chem. 2006;281:36724–36731. doi: 10.1074/jbc.M608756200. [DOI] [PubMed] [Google Scholar]
- 25.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 26.Rigaut G, Shevchenko A, Rutz B, Wilm M, Mann M, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 27.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 28.Hein C, Springael JY, Volland C, Haguenauer-Tsapis R, Andre B. NPl1, an essential yeast gene involved in induced degradation of Gap1 and Fur4 permeases, encodes the Rsp5 ubiquitin-protein ligase. Mol Microbiol. 1995;18:77–87. doi: 10.1111/j.1365-2958.1995.mmi_18010077.x. [DOI] [PubMed] [Google Scholar]
- 29.Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, et al. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- 30.Dunn R, Klos DA, Adler AS, Hicke L. The C2 domain of the Rsp5 ubiquitin ligase binds membrane phosphoinositides and directs ubiquitination of endosomal cargo. J Cell Biol. 2004;165:135–144. doi: 10.1083/jcb.200309026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gwizdek C, Hobeika M, Kus B, Ossareh-Nazari B, Dargemont C, et al. The mRNA nuclear export factor Hpr1 is regulated by Rsp5-mediated ubiquitylation. J Biol Chem. 2005;280:13401–13405. doi: 10.1074/jbc.C500040200. [DOI] [PubMed] [Google Scholar]
- 32.Chen HI, Einbond A, Kwak SJ, Linn H, Koepf E, et al. Characterization of the WW domain of human yes-associated protein and its polyproline-containing ligands. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 33.Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, et al. Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem. 2001;276:15164–15173. doi: 10.1074/jbc.M010484200. [DOI] [PubMed] [Google Scholar]
- 34.Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katzmann DJ, Sarkar S, Chu T, Audhya A, Emr SD. Multivesicular body sorting: ubiquitin ligase Rsp5 is required for the modification and sorting of carboxypeptidase S. Mol Biol Cell. 2004;15:468–480. doi: 10.1091/mbc.E03-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaminska J, Gajewska B, Hopper AK, Zoladek T. Rsp5p, a new link between the actin cytoskeleton and endocytosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2002;22:6946–6948. doi: 10.1128/MCB.22.20.6946-6958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- 38.Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 39.Jund R, Lacroute F. Genetic and physiological aspects of resistance to 5-fluoropyrimidines in Saccharomyces cerevisiae. J Bacteriol. 1970;102:607–615. doi: 10.1128/jb.102.3.607-615.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, et al. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci U S A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Swaminathan S, Amerik AY, Hochstrasser M. The Doa4 deubiquitinating enzyme is required for ubiquitin homeostasis in yeast. Mol Biol Cell. 1999;10:2583–2594. doi: 10.1091/mbc.10.8.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dunn R, Hicke L. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Mol Biol Cell. 2001;12:421–435. doi: 10.1091/mbc.12.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reneke JE, Blumer KJ, Courchesne WE, Thorner J. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell. 1988;55:221–234. doi: 10.1016/0092-8674(88)90045-1. [DOI] [PubMed] [Google Scholar]
- 44.Schandel KA, Jenness DD. Direct evidence for ligand-induced internalization of the yeast alpha-factor pheromone receptor. Mol Cell Biol. 1994;14:7245–7255. doi: 10.1128/mcb.14.11.7245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raths S, Rohrer J, Crausaz F, Riezman H. end3 and end4: two mutants defective in receptor-mediated and fluid-phase endocytosis in Saccharomyces cerevisiae. J Cell Biol. 1993;120:55–65. doi: 10.1083/jcb.120.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bucking-Throm E, Duntze W, Hartwell LH, Manney TR. Reversible arrest of haploid yeast cells in the initiation of DNA synthesis by a diffusible sex factor. Exp Cell Res. 1973;76:99–110. doi: 10.1016/0014-4827(73)90424-2. [DOI] [PubMed] [Google Scholar]
- 47.Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695:189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 48.Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 49.Ren J, Kee Y, Huibregtse JM, Piper RC. Hse1, a Component of the Yeast Hrs-STAM Ubiquitin-sorting Complex, Associates with Ubiquitin Peptidases and a Ligase to Control Sorting Efficiency into Multivesicular Bodies. Mol Biol Cell. 2007;18:324–335. doi: 10.1091/mbc.E06-06-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang G, McCaffery JM, Wendland B, Dupre S, Haguenauer-Tsapis R, et al. Localization of the Rsp5p ubiquitin-protein ligase at multiple sites within the endocytic pathway. Mol Cell Biol. 2001;21:3564–3575. doi: 10.1128/MCB.21.10.3564-3575.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- 52.Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, et al. The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 2006;281:38738–38747. doi: 10.1074/jbc.M605959200. [DOI] [PubMed] [Google Scholar]
- 53.Terrell J, Shih S, Dunn R, Hicke L. A function for monoubiquitination in the internalization of a G protein-coupled receptor. Mol Cell. 1998;1:193–202. doi: 10.1016/s1097-2765(00)80020-9. [DOI] [PubMed] [Google Scholar]
- 54.Springael JY, Galan JM, Haguenauer-Tsapis R, Andre B. NH4+-induced down-regulation of the Saccharomyces cerevisiae Gap1p permease involves its ubiquitination with lysine-63-linked chains. J Cell Sci. 1999;112(Pt 9):1375–1383. doi: 10.1242/jcs.112.9.1375. [DOI] [PubMed] [Google Scholar]
- 55.Gitan RS, Eide DJ. Zinc-regulated ubiquitin conjugation signals endocytosis of the yeast ZRT1 zinc transporter. Biochem J. 2000;346 Pt 2:329–336. [PMC free article] [PubMed] [Google Scholar]
- 56.Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, et al. Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation. Traffic. 2007;8:1280–1296. doi: 10.1111/j.1600-0854.2007.00610.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leon S, Erpapazoglou Z, Haguenauer-Tsapis R. Ear1p and ssh4p are new adaptors of the ubiquitin ligase rsp5p for cargo ubiquitylation and sorting at multivesicular bodies. Mol Biol Cell. 2008;19:2379–2388. doi: 10.1091/mbc.E08-01-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hicke L, Dunn R. Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol. 2003;19:141–172. doi: 10.1146/annurev.cellbio.19.110701.154617. [DOI] [PubMed] [Google Scholar]
- 59.Swanson KA, Kang RS, Stamenova SD, Hicke L, Radhakrishnan I. Solution structure of Vps27 UIM-ubiquitin complex important for endosomal sorting and receptor downregulation. Embo J. 2003;22:4597–4606. doi: 10.1093/emboj/cdg471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gietz D, St Jean A, Woods RA, Schiestl RH. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soni R, Carmichael JP, Murray JA. Parameters affecting lithium acetate-mediated transformation of Saccharomyces cerevisiae and development of a rapid and simplified procedure. Curr Genet. 1993;24:455–459. doi: 10.1007/BF00351857. [DOI] [PubMed] [Google Scholar]
- 62.Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 63.Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, et al. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 64.Emili A, Schieltz DM, Yates JR, 3rd, Hartwell LH. Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell. 2001;7:13–20. doi: 10.1016/s1097-2765(01)00150-2. [DOI] [PubMed] [Google Scholar]
- 65.Marchal C, Dupre S, Urban-Grimal D. Casein kinase I controls a late step in the endocytic trafficking of yeast uracil permease. J Cell Sci. 2002;115:217–226. doi: 10.1242/jcs.115.1.217. [DOI] [PubMed] [Google Scholar]
- 66.Hanna J, Leggett DS, Finley D. Ubiquitin depletion as a key mediator of toxicity by translational inhibitors. Mol Cell Biol. 2003;23:9251–9261. doi: 10.1128/MCB.23.24.9251-9261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stamenova SD, Dunn R, Adler AS, Hicke L. The Rsp5 ubiquitin ligase binds to and ubiquitinates members of the yeast CIN85-endophilin complex, Sla1-Rvs167. J Biol Chem. 2004;279:16017–16025. doi: 10.1074/jbc.M313479200. [DOI] [PubMed] [Google Scholar]
- 68.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 69.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 70.Tong AH, Lesage G, Bader GD, Ding H, Xu H, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 71.Wu X, Chang A, Sudol M, Hanes SD. Genetic interactions between the ESS1 prolyl-isomerase and the RSP5 ubiquitin ligase reveal opposing effects on RNA polymerase II function. Curr Genet. 2001;40:234–242. doi: 10.1007/s00294-001-0257-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Endosome and Vacuolar morphology is normal in ubp2Δ cells. Cells were incubated with the lipid-binding fluorescent dye FM4-64 and washed. Endocytosis of the dye and endosome/vacuolar morphology were monitored by fluorescence microscopy and Nomarski optics at the indicated time points (min) after staining.
(1.96 MB TIF)
Recycling of Fur4-GFP cannot be detected in the presence of cycloheximide. WT and vps37Δ cells transformed with pFur4-GFP were cultured at 30°C and Fur4-GFP synthesis was induced for 90 min by adding galactose. Glucose was added to block Fur4-GFP synthesis (CC). 20 min later, cells were subjected to carbon starvation (CS) to trigger endocytosis of the permease for 60 minutes. Cultures were divided in two equal fractions. Glucose (CA) or glucose+cycloheximide (CA+CHX 0.1 mg/ml) was then added. (A) Cells were visualized by fluorescence microscopy at t = 0 (0'CC), after 60 minutes of carbon starvation (60'CS) and 60 minutes after the addition of carbon (60'CA) or carbon+cycloheximide (60'CA+CHX). Note that Fur4-GFP was efficiently internalized in all the strains upon CS. (B) Uracil uptake was measured 3, 10, 30 and 60 min after the addition of glucose. Results are expressed as a percentage of the initial uracil uptake measured immediately before carbon starvation, and plotted on a linear scale.
(0.46 MB TIF)
(0.04 MB DOC)