Abstract
The endosomal sorting complex required for transport (ESCRT) proteins form multimolecular complexes that control multivesicular body formation, endosomal sorting, and transport ubiquitinated membrane proteins (including cell-surface receptors) to the endosomes for degradation. There is accumulating evidence that endosomal dysfunction is linked to neural cell degeneration in vitro, but little is known about the relationship between neural disorders and ESCRT proteins in vivo. Here we specifically deleted the hrs gene, ESCRT-0, in the neurons of mice by crossing loxP-flanked hrs mice with transgenic mice expressing the synapsin-I Cre protein (SynI-cre). Histological analyses revealed that both apoptosis and a loss of hippocampal CA3 pyramidal neurons occurred in the hrsflox/flox;SynI-cre mice. Notably, the hrsflox/flox;SynI-cre mice accumulated ubiquitinated proteins, such as glutamate receptors and an autophagy-regulating protein, p62. These molecules are particularly prominent in the hippocampal CA3 neurons and cerebral cortex with advancing age. Accordingly, we found that both locomotor activity and learning ability were severely reduced in the hrsflox/flox;SynI-cre mice. These data suggest that Hrs plays an important role in neural cell survival in vivo and provide an animal model for neurodegenerative diseases that are known to be commonly affected by the generation of proteinaceous aggregates.
The generation of proteinaceous aggregates is a common pathological feature in neurodegenerative diseases.1 Alterations in the lysosomal pathway are associated with normal brain aging, as well as with age-related neurodegenerative diseases, including Alzheimer’s and Parkinson’s. When the level of misfolded protein overwhelms the degradative pathways, cellular toxicity and neurodegeneration result.2 Cellular mechanisms for degrading misfolded protein include the ubiquitin-proteasome system, which is the main nonlysosomal degradative pathway for ubiquitinated proteins, and autophagy, a lysosome-mediated degradative pathway.3
Glutamate receptors play prominent roles in several neurodegenerative diseases.4,5,6,7 All N-methyl-d-aspartate (NMDA) receptors (NR) share one NR1 subunit and one or more NR2A-D and/or NR3 subunits, forming a heterotetrameric complex.8 Fbx2-mediated ubiquitination is required for NR1 subunit degradation.9 KEL-8, a substrate receptor for Cullin 3 ubiquitin ligases, is reported to be required for the proteolysis of the α-amino-3-hydroxy-5-methyl-isoxazolepropionic acid receptor (AMPAR) subunit GluR1.10 Thus, ubiquitination is important for the homeostatic control of glutamate receptors in neurons.
Endosomal sorting complex required for transport (ESCRT) proteins form multimolecular complexes that control multivesicular body formation and transport ubiquitinated membrane proteins to the endosomes. The ESCRTs are subdivided into four complexes.11 Ubiquitinated cargos such as epidermal growth factor receptors are initially recognized by the ESCRT-0 complex, and then sequentially handed off to ESCRT-I, -II, and -III. After these steps, the cargos are invaginated into multivesicular bodies and eventually sorted into the lysosomes. The ESCRT-0 component Hrs (also known as Hgs) plays a particularly major role in this sorting process.12 Although several studies using Hrs mutants or its deletion in mammalian cells and mice suggest that it has a role in morphogenesis and development,13,14 whether or not Hrs possesses any function in the nervous system is unknown.
Recent studies suggest that protein ubiquitination is essential for proper nervous system function.15 Ubiquitination is a key tagging process for proper protein trafficking and turnover involving proteasome- and lysosome-dependent degradation. In addition, a recent study suggested that normal multivesicular body function is essential for neural cells to avoid degeneration.16 Furthermore, ESCRT-III dysfunction is associated with a type of neurodegeneration that resembles frontotemporal dementia and other age-dependent neurodegenerative diseases.17 These findings together highlight ESCRT function as being important for maintaining neuronal homeostasis, and prompted us to investigate the in vivo role of Hrs in the central nervous system. Using the Cre-loxP system, we found that Hrs is required for the degradation of ubiquitinated proteins in the central nervous system and the survival of mouse hippocampal CA3 neurons.
Materials and Methods
Generation of Floxed Hrs Mice
To generate a neuron-specific conditional knockout of Hrs (accession no. CDB0476K; Center for Developmental Biology, Kobe, Japan), we generated a floxed hrs allele (hrsflox) using embryonic stem (ES) cell homologous recombination technology. For the targeting construct of the hrsflox/flox line, a C57BL/6J genomic clone was used to generate the hrs targeting vector, and two loxP sites were integrated, one upstream of exon 2 and one downstream of exon 4. The targeting vector was electroporated into TT2 ES cells, followed by G418 selection. Colonies surviving selection were tested for homologous recombination and incorporation of the loxP sites by Southern blot hybridization. Two clones were identified and injected into ICR 8 cell-stage embryos.18 Chimeric mice were mated to C57BL/6J mice to identify germ-line transmission of the targeted hrs allele. Removal of the neomycin selection cassette, which was surrounded by FRT (Flp recombinase target) sites, was accomplished by first mating hrsflox/flox mice to FLPeR mice19 at Riken (Kobe, Japan). All animal experiments were performed according to the guidelines laid down by the animal welfare committees of the Tohoku University Graduate School of Medicine and Riken.
Generation of hrsflox/flox;SynI-cre Mice
SynI-cre transgenic mice (a gift from Jamey Marth, University of California, San Diego, CA)20 were mated with the hrsflox/flox mice to generate hrsflox/+;SynI-cre mice. The hrsflox/+;SynI-cre mice were then mated with each other. Offspring carrying hrsflox/flox;SynI-cre and hrs+/+;SynI-cre were used for further analyses. These mice were genotyped by polymerase chain reaction (PCR) using DNA obtained from the tail.
Southern Blot Analysis
Genomic DNA from ES cells was digested with restriction enzymes, separated by electrophoresis on a 0.6% agarose gel, transferred to Hybond-N (GE Health Care, Chalfont St. Giles, UK, and hybridized with the random-primed probe.
Genotype Analysis
Genomic DNA from the mouse tail was used for PCR analysis. We genotyped the hrs flox allele using a forward primer (5′-GATGATGAGATGTTTACC-3′) and a reverse primer (5′-TTGTCCTTTACCTCTTAG-3′) that flank the 5′ loxP site. The PCR products were 354 bp for the hrsflox/flox allele and 229 bp for the wild-type allele. We amplified the hrsΔ2–4 allele using a forward (6851F: 5′-TTGTTGAATGAGTAACAAGGGTGGT-3′) and reverse primer (9100R: 5′-TGGATCCCATGAAATGGGGAACAGC-3′). The PCR products were 0.3 kbp for the hrsΔ2–4 allele and 2.3 kbp for the wild-type allele. Genotyping for the presence of the SynI-cre allele was performed using the following primer pair: forward (5′-TTACCGGTCGATGCAACGAGTGAT-3′) and reverse (5′-TTCCATGAGTGAACGAACCTGGTC-3′).
Western Blotting
Immunoblotting was performed as previously described.21 In brief, brains from mice were homogenized in lysis buffer [1% Nonidet P-40, 20 mmol/L Tris-HCl (pH 7.5), 150 mmol/L NaCl, 1 mmol/L ethylenediaminetetraacetic acid, 1 mmol/L Na3VO4, 1 mmol/L phenylmethyl sulfonyl fluoride, and 20 μg/ml aprotinin]. The lysates were precleared by centrifugation (10,000 × g) for 20 minutes at 4°C. The supernatants were then separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). After being blocked with 5% nonfat milk in Tris-buffered saline containing 0.1% Tween 20, the membranes were probed with the indicated primary antibodies. After another wash, the membranes were probed with horseradish peroxidase-conjugated secondary antibodies (Cell Signaling, Beverly, MA).
Histology and Immunohistochemistry
Mice were perfused with 4% paraformaldehyde, and the dissected brains were postfixed for 24 hours before being embedded in paraffin. For histological analyses, 3-μm sections were stained with hematoxylin and eosin (H&E). Immunostaining was performed by the streptavidin-biotin immunoperoxidase method (Histofine SAB-PO kit; Nichirei, Tokyo, Japan) using primary antibodies. We used the antibodies at the following dilutions: anti-ubiquitin [1:200, 1B3, mouse monoclonal antibody (mAb); MBL International, Woburn, MA]; anti-ubiquitin (1:200, FK2, mAb; Biomol, Plymouth Meeting, PA); anti-GFAP (1:200, mouse mAb; Chemicon, Temecula, CA); anti-calbindin (1:200, rabbit polyclonal antibody; Chemicon). We also used anti-Hrs,22 anti-NR1,23 anti-NR2B,24 and anti-GluR1 antibodies25 as previously described. To detect mouse monoclonal antibodies, the Histofine mouse staining kit (Nichirei) was used. Immunoreactions were visualized with 3,3,-diaminobenzidine. For terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick-end labeling (TUNEL) assays, 5-μm sections were deparaffinized, and terminal transferase labeling of the fragmented DNA was performed with an in situ cell death detection kit (Fluorescein; Roche, Indianapolis, IN), according to the assay protocol of the kit.
Reverse Transcriptase (RT)-PCR
RT-PCR was performed as previously described.26 In brief, the total RNA from the brains of 8-week-old hrs+/+;SynI-cre and hrsflox/flox;SynI-cre mice was prepared using TRIzol (Invitrogen, Carlsbad, CA). PCR was performed in a 50-μl mixture consisting of 20 mmol/L Tris-HCl (pH 8.0), 2 mmol/L MgCl2,50 mmol/L KCl, 0.2 mmol/L deoxynucleotide triphosphate mixture, 1 μmol/L of various primers, 1.25 U of Ex-TaqDNA polymerase (Takara Shuzo, Kyoto, Japan), and 1 μl of the RT reaction mixture as a template. The PCR conditions were as follows: denaturation at 94°C for 2 minutes, followed by 35 cycles of 30 seconds at 94°C, 1 minute at 65°C, and 1 minute at 72°C. The following oligonucleotide primers were used: HrsE1F (5′-GAGGCAGCGGCACCTTCGAG-3′) and HrsE7R (5′-ATGGCATTCCTCAGCATCCA-3′).
In Situ Hybridization
In situ hybridization was performed as previously described.27 Mice were anesthetized and perfused with 4% paraformaldehyde. The brains were postfixed overnight at 4°C and then cryoprotected with 30% sucrose in 0.1 M phosphate buffer at 4°C. Frozen sections were cut at 20 μm on a freezing microtome and mounted on MAS-coated slides (Matsunami, Osaka, Japan). The transcription reactions were performed using a digoxigenin (DIG) RNA labeling kit (SP6/T7) (Roche). The purified plasmids were linearized and then used as templates for in vitro transcription of the DIG-labeled antisense (or sense control) RNA probes with T7 (or SP6) RNA polymerase. The transcripts were subjected to alkaline hydrolysis to reduce their size. For this, the DIG-labeled full-length cRNA was added to alkaline hydrolysis solution [40 mmol/L NaHCO3/60 mmol/L Na2CO3 (pH 10.2)] and incubated for 15 minutes at 60°C. For in situ hybridization, the sections were postfixed in 4% paraformaldehyde [freshly prepared in 0.1 mol/L phosphate buffer (pH 7.4)] for 10 minutes, washed three times with phosphate-buffered saline (PBS), treated with 0.5 μg/ml proteinase K (Sigma) for 30 minutes at 37°C, postfixed in 4% paraformaldehyde for 5 minutes, washed three times with PBS, acetylated for 10 minutes, treated 0.3% Triton X-100 for 20 minutes, and washed three times with PBS for 5 minutes each. Prehybridization was performed for 1 hour at 65°C with hybridization buffer without probe and then hybridization was done at 65°C overnight in a new hybridization buffer containing one of the DIG-labeled RNA probes. The hybridization buffer consisted of 5× saline sodium citrate (SSC; Gibco BRL/Invitrogen, Tokyo, Japan), 50% deionized formamide (Sigma, St. Louis, MO), 500 μg/ml herring sperm DNA (Roche), 5× Denhardt’s solution (Open Biosystems, Huntsville, AL), and 250 μg/ml transfer RNA (Roche). After hybridization, the sections were sequentially treated with 2× SSC/50% formamide for 30 minutes at 65°C, 2× SSC for 30 minutes at 65°C twice, and 0.2× SSC for 5 minutes at room temperature. The hybridized probe was detected with an alkaline phosphatase-conjugated anti-DIG antibody using a DIG nucleic acid detection kit (Roche) according to the manufacturer’s protocol.
Behavioral Tests
All behavioral experiments were performed with 2- to 3-month-old male mice with a mixed 129/Ola-C57BL/6 genetic background and in the light phase of their diurnal cycle, between 09:00 and 17:00 hours. All experimental protocols were approved by the Animal Care Committee of the Tohoku University School of Medicine, and all experiments were performed in compliance with the relevant laws and institutional guidelines.
Open Field Test
Mouse locomotor activity in the open field was measured using a photo-beam system (BTA-1, Muromachi-Kikai, Tokyo, Japan). The values for ambulation distance were accumulated for 30 minutes and logged onto a personal computer.
Step-Through Passive Avoidance Test
The learning ability of the mice was evaluated using a step-through passive avoidance memory test, as previously described.28 The training apparatus was a box consisting of a small lighted compartment (15 × 10 × 10 cm3) and a large dark compartment (18 × 12 × 10 cm3). A 10 × 10 cm2 guillotine door separated the two compartments. The light compartment was made of clear Plexiglas, and was illuminated by a lamp (60 W) from the outside. The dark compartment had a series of stainless-steel rods (3 mm in diameter, 1 cm apart) through which a constant electrical current could be delivered. The mice were first habituated to the box on 2 consecutive days. On the first day, they were placed in the light compartment and allowed to explore the box. The latency period for entering the dark compartment was recorded. As soon as a mouse entered the dark compartment, the door was closed, and the mouse was kept inside for 15 seconds before being returned to its cage. On the second training day, on entry into the dark compartment, the mice were given 0.5 mA of current for 5 seconds. The test session was performed 24 hours after the training session using the same paradigm, but without the foot shock. The latency period for each mouse to move into the dark compartment was recorded for up to a maximum of 300 seconds.
Wire Hanging Test
The ability of mice to hang upside down from a wire screen was tested as previously described.29 The wires were 1 mm in diameter and spaced 1 cm apart. A rectangular area of the screen was taped off to confine the mouse to an 18 × 26 cm section of the screen. After a mouse was placed on the screen, the screen was waved gently in the air three times to force the mouse to grip the wires. The screen was then immediately turned upside down, 70 cm above a large rodent housing cage. The latency period for the mouse to fall into the cage was recorded. Mice that did not fall during the 60-second trial period were removed and given a maximal score of 60 seconds.
Forced Swimming Test
Mice were individually forced to swim in an aquarium (25 × 40 × 20 cm) containing 15-cm-deep water at 25 ± 1°C; the total duration of mouse immobility was measured during a 6-minute test.30 Each mouse was judged to be immobile when it ceased struggling and remained floating motionless in the water, making only those movements necessary to keep its head above water.
Footprint Analysis
Footprint assessment was performed to detect gait abnormalities that could contribute to deficits in motor coordination on land.31 Each hindpaw was colored black and forepaw red using nontoxic dye. For each mouse, five or more consecutive strides were averaged.
Results
Expression of Hrs in Mouse Brain
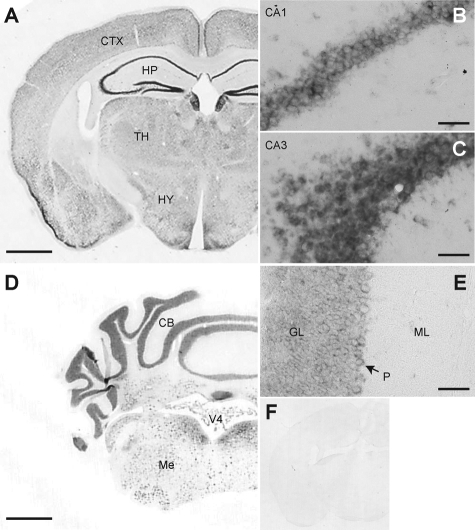
We first performed in situ hybridization to examine the expression of the hrs gene in the central nervous system because currently there is no antibody to reliably detect Hrs in brain tissue. In 5-week-old wild-type mice, hrs was ubiquitously expressed in the brain, with higher expression in the hippocampus, cerebral cortex, and hypothalamus (Figure 1, A and F). The degree of expression was higher in the CA3 than in the CA1 subfield in the hippocampus (Figure 1, B and C). Distinct expression was seen in the Purkinje cells (Figure 1, D and E).
Figure 1.
In situ hybridization of 5-week-old wild-type mouse brain probed for hrs mRNA. A: Coronal section through the cerebrum (hippocampus) showing the ubiquitous expression of hrs mRNA. Stronger hrs expression was seen in the CA3 (C) than the CA1 (B) subfield. D and E: Coronal section through the cerebellum showing strong expression of hrs in the granular cell layers and Purkinje cells. F: In situ hybridization of the section consecutive to A with the sense probe as a negative control. CTX, cerebral cortex; HP, hippocampus; TH, thalamus; HY, hypothalamus; CB, cerebellum; V4, fourth ventricle; Me, medulla oblongata; GL, granular layer; P, Purkinje cell; ML, molecular layer. Scale bars: 1 mm (A, D); 50 μm (B, C, E). Original magnifications: ×40 (A, D); ×400 (B, C, E).
Deletion of Hrs in the Central Nervous System
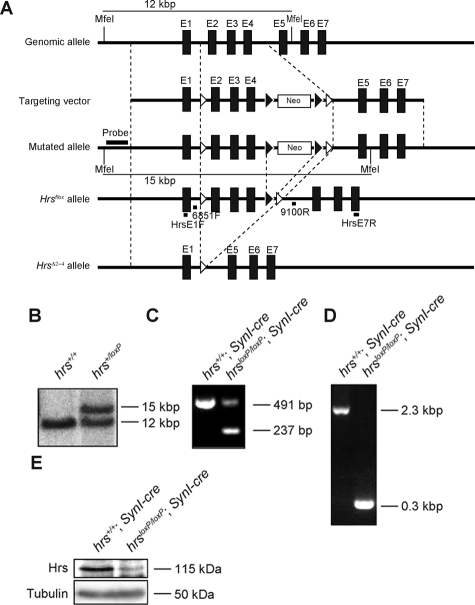
An hrs flox mouse was generated with loxP sites flanking exons 2 to 4 of the mouse hrs locus (Figure 2A). Heterozygous hrs flox ES cells were identified by Southern blot analysis (Figure 2B). To delete hrs specifically in the mouse brain, homozygous hrs flox mice (hrsloxP/loxP) were crossed with SynI-cre transgenic mice. To assess whether exons 2 to 4 were deleted, we performed RT-PCR analysis of the brains from hrsloxP/loxP;SynI-cre, and hrs+/+;SynI-cre mice. A pair of primers spanning exons 1 and 7, respectively, amplified a 491-bp fragment from the hrs+/+;SynI-cre brain and a 237-bp fragment from the hrsloxP/loxP;SynI-cre brain (Figure 2C). Sequence analyses of the RT-PCR fragments showed that the hrs mRNA transcript from the hrsloxP/loxP;SynI-cre brain contained a deletion of nucleotides 74 through 327, resulting in a frameshift and a stop codon in exon 5 (data not shown). PCR analyses of the genomic DNA from these brains revealed 0.3-kbp fragments (Figure 2D). Moreover, immunoblotting analysis with an anti-Hrs mAb revealed that ∼60% of the Hrs expression was suppressed in the hrsloxP/loxP;SynI-cre brain (Figure 2E). Because SynI-cre transgenic mice specifically express Cre recombinase in differentiated neurons, and not in astroglia,20 these data suggest that both neurons and glial cells express Hrs. We conclude that the hrs flox allele represents a functional conditional allele.
Figure 2.
Generation of floxed hrs mice. A: Schematic representation of the hrs genomic locus, targeting vector, and hrs mutated locus. The targeting vector was designed to replace exon 2 (E2) to E4, which encode Hrs amino acids 74 to 327. The expected fragments generated by MfeI digestion were 12 and 15 kb for the wild-type and mutated alleles, respectively. Open and closed arrowheads denote the positions of the loxP and FRT sequences, respectively. B: Southern blot analysis of the hrs mutation in ES cell clones. DNA was digested with MfeI, and the blot was probed with the flanking 5′ probe as shown in A. Lines indicate the positions of the DNA fragments corresponding to the wild-type and mutated alleles. C: RT-PCR analysis of the total RNA from the brains of hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice. The primers used were HrsE1F and HrsE7R (see Materials and Methods). D: Genomic PCR analysis of the hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre brains. The primers used were 6851F and 9100R (see Materials and Methods). E: Western blot analysis for Hrs. Lysates from hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre brains were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and blotted with an anti-Hrs antibody.
Loss of Weight in Hrs Mutant Mice
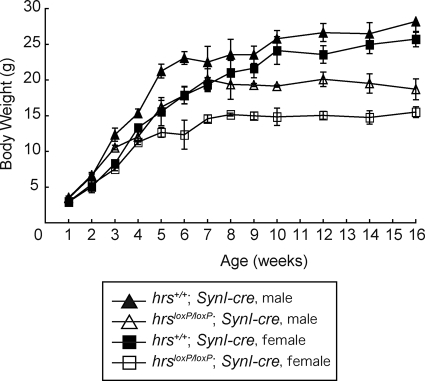
HrsloxP/loxP;SynI-cre mice were morphologically indistinguishable from their littermates at birth. Hrs+/+;SynI-cre, hrs+/loxP;SynI-cre, and hrsloxP/loxP;SynI-cre mice were obtained at the expected Mendelian ratios and were viable at least for several months. Hrs+/loxP;SynI-cre mice did not differ in growth or behavior from their hrs+/+;SynI-cre littermates. However, growth retardation of the hrsloxP/loxP;SynI-cre mice became detectable by 3 weeks of age and gradually worsened; most of the hrsloxP/loxP;SynI-cre mice did not show any increase in their body weight after they reached 8 weeks of age (Figure 3). Moreover, the hrsloxP/loxP;SynI-cre mice were infertile.
Figure 3.
Phenotypes of the hrsloxP/loxP;SynI-cre mice. Growth curves for hrs+/+;SynI-cre (males, n = 12; females, n = 9) and hrsloxP/loxP;SynI-cre (males, n = 10; females, n = 9) mice. Error bars indicate SE.
Loss of Pyramidal Neurons in the Hippocampal CA3 Subfield in Hrs Mutant Mice
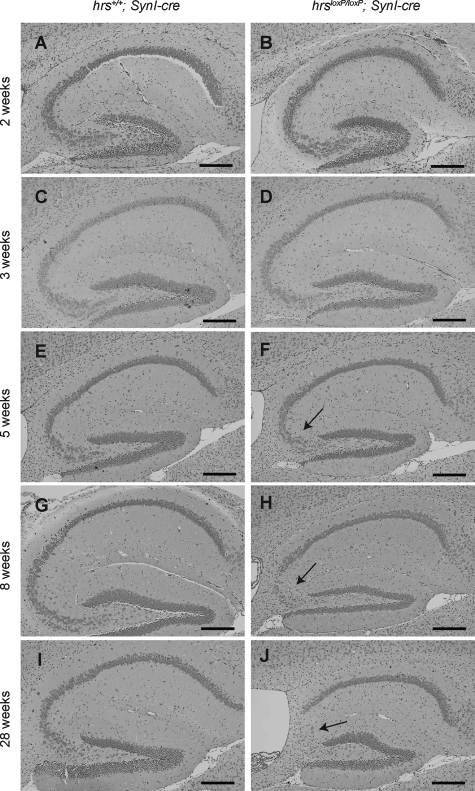
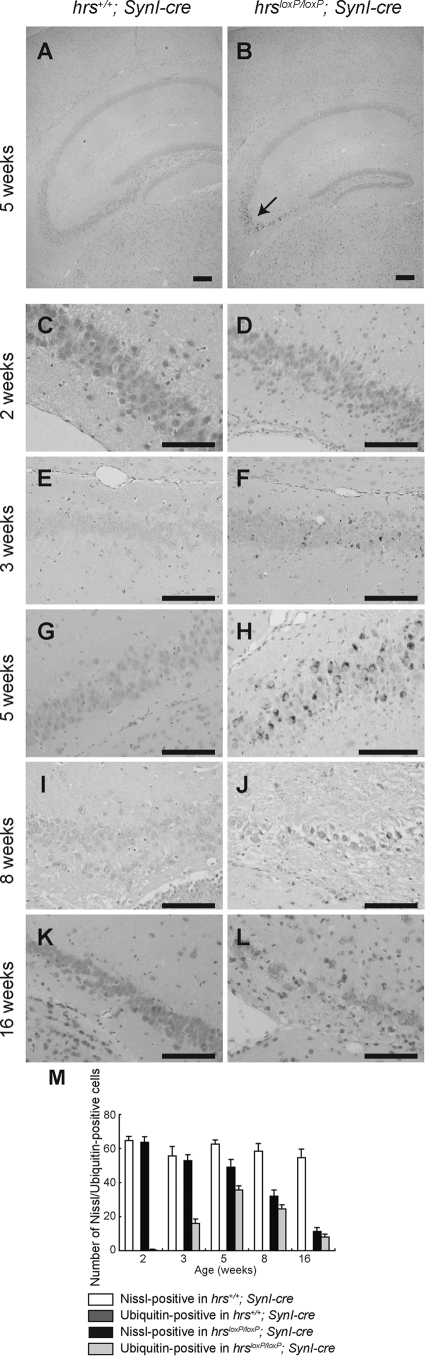
We next performed histopathological examinations of the hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice by Nissl staining with cresyl violet. The gross anatomy of the Hrs mutant brain was normal. In the 2- and 3-week old brains, no histopathological difference was observed between the hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice (Figure 4, A–D). However, the number of pyramidal neurons in the hippocampal CA3 subfield was reduced in the 5-week-old hrsloxP/loxP;SynI-cre brain (Figure 4, E and F), and progressive decreases were observed in the 8- and 28-week-old hrsloxP/loxP;SynI-cre brains (Figure 4, G–J). In contrast, we could not detect any decrease in the pyramidal neurons in CA1. No difference was observed in any other regions, including the cerebral cortex, substantia nigra, striatum, cerebellum, or hypothalamus by Nissl staining or H&E staining (data not shown).
Figure 4.
Abnormalities in the hippocampal CA3 subfield of hrsloxP/loxP;SynI-cre mice. Nissl staining of anterior coronal hippocampus sections of mice. Mice were hrs+/+;SynI-cre (A, C, E, G, I) and hrsloxP/loxP;SynI-cre (B, D, F, H, J) and were 2 weeks (A, B), 3 weeks (C, D), 5 weeks (E, F), 8 weeks (G, H), and 28 weeks (I, J) of age. Note the loss of pyramidal cells in the CA3 subfield in the hrsloxP/loxP;SynI-cre hippocampus (arrows). Scale bars = 250 μm. Original magnifications, ×100.
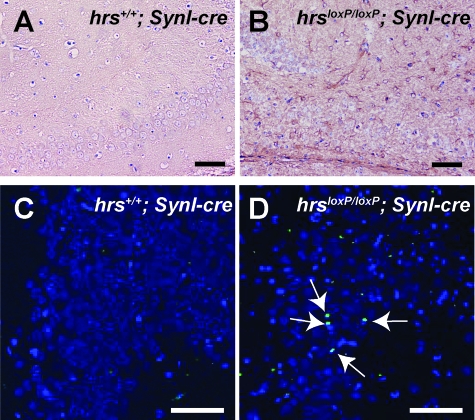
Immunostaining for the glial marker GFAP (glial fibrillary acidic protein) showed increased GFAP in the hippocampal CA3 subfield of the hrsloxP/loxP;SynI-cre mice, suggesting the presence of neural damage in this region (Figure 5, A and B). To determine whether the reduced number of neurons observed in the hrsloxP/loxP;SynI-cre mouse brain was caused by cell death, we performed TUNEL staining, which detects the DNA fragmentation in dying cells. Several TUNEL-positive cells were detected in the hippocampal CA3 subfield of 5-week-old hrsloxP/loxP;SynI-cre mice, but no TUNEL-positive cells were detected in the hrs+/+;SynI-cre hippocampus (Figure 5, C and D). These results suggest that neural cell death occurs in a specific brain region of the hrsloxP/loxP;SynI-cre mice, the hippocampal CA3 subfield.
Figure 5.
Neural cell death occurs in the hippocampal CA3 subfield of hrsloxP/loxP;SynI-cre mice. A and B: Anti-GFAP antibody staining. Brain sections from hrs+/+;SynI-cre (A) and hrsloxP/loxP;SynI-cre (B) mice were immunostained with a GFAP-specific antibody. The GFAP signal was increased in the hippocampus of hrsloxP/loxP;SynI-cre mice. C and D: TUNEL staining of the hippocampal CA3 subfield. Hippocampus sections of 5-week-old hrs+/+;SynI-cre (C) and hrsloxP/loxP;SynI-cre (D) mice were stained for TUNEL. Arrows indicate positive staining for TUNEL. Original magnifications, ×400. Scale bars = 50 μm.
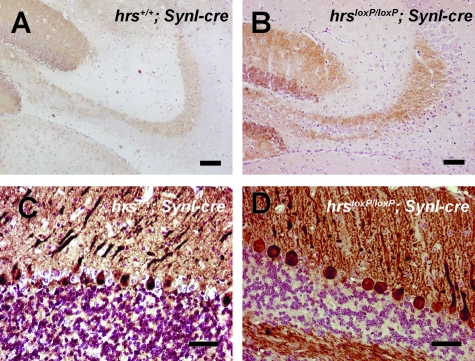
To examine the mossy fiber pathway that connects granule cells to CA3 pyramidal cells, we performed immunostaining assays for calbindin, which selectively stains neurons in the dentate gyrus containing the mossy fiber pathway and Purkinje cells. Despite the profound reduction of pyramidal cells in the hippocampal CA3 subfield of hrsloxP/loxP;SynI-cre mice, the staining pattern of calbindin was not significantly different between the hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice (Figure 6, A and B). Immunohistochemical staining with an anti-calbindin antibody demonstrated no difference in the Purkinje cell numbers between the hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice (Figure 6, C and D).
Figure 6.
A and B: Calbindin immunostaining for mossy fibers. Sections were prepared from the brains of 8-week-old hrs+/+;SynI-cre (A) and hrsloxP/loxP;SynI-cre (B) mice. C and D: Calbindin immunostaining for Purkinje cells. The sections were prepared from the brains of 8-week-old hrs+/+;SynI-cre (C) and hrsloxP/loxP;SynI-cre (D) mice. Scale bars: 0.25 mm (A, B); 25 μm (C, D). Original magnifications: ×200 (A, B); ×400 (C, D).
Accumulation of Ubiquitinated Proteins in the Hrs Mutant Brain
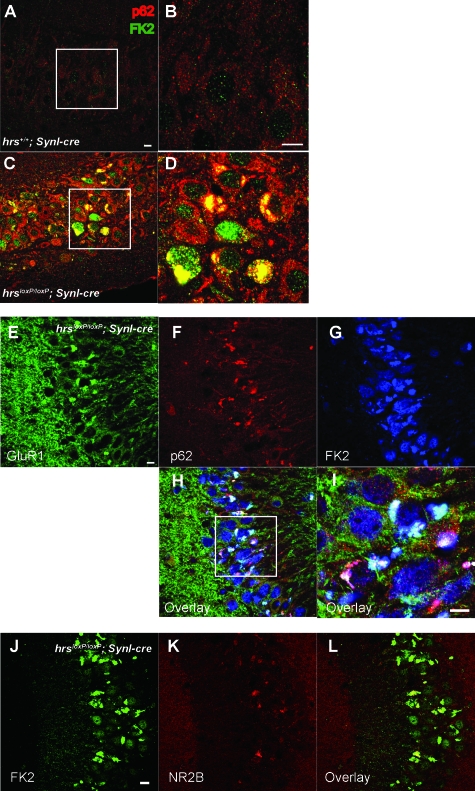
Because Hrs has an essential role in the endocytic sorting of ubiquitinated proteins, we investigated whether ubiquitinated proteins accumulated in the Hrs mutant brains by immunohistochemical analysis. In the hrsloxP/loxP; SynI-cre mouse brain, numerous granules stained by an anti-ubiquitin antibody appeared in the CA3 subfield, cerebral cortex, hypothalamus, and less frequently, in Purkinje cells (Figures 7 and 8). In the 16-week-old hrsloxP/loxP;SynI-cre mouse brain, there were fewer ubiquitin-positive pyramidal cells, because most of the pyramidal cells were already lost (Figure 7, K–M). Interestingly, ubiquitin-positive aggregates were also observed in the 3-week-old hrsloxP/loxP;SynI-cre mouse brain, which did not show the loss of hippocampal CA3 pyramidal neurons (Figure 4, C and D; and Figure 7, E, F, and M). The number of ubiquitin-positive neurons in the cerebral cortex gradually increased with age (Figure 8, A–J and O). On the other hand, we could not detect any ubiquitin-positive aggregates in the hrsloxP/loxP;MX1-cre mouse liver, which was sufficiently knocked-out by the injection of polyriboinosinic polyribocytidylic acid (data not shown). These data suggest that Hrs plays a crucial role in the degradation of ubiquitinated proteins in neural cells.
Figure 7.
Ubiquitin-positive inclusions in the hrsloxP/loxP;SynI-cre brain. Immunohistochemistry of hippocampal CA3 sections from hrs+/+;SynI-cre (A, C, E, G, I, K) and hrsloxP/loxP;SynI-cre (B, D, F, H, J, L) mice at various weeks of age, stained with an anti-ubiquitin antibody (1B3). A and B: Five weeks old, low magnification. C and D: Two weeks old. E and F: Three weeks old. G and H: Five weeks old. I and J: Eight weeks old. K and L: Sixteen weeks old. Arrow indicates ubiquitin-positive cells. M: Nissl- or ubiquitin-positive cells in the hippocampal CA3 subfield were counted in comparable areas for hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice. Data represent the mean ± SE of three mice. Scale bars = 50 μm. Original magnifications: ×100 (A, B); ×400 (C–L).
Figure 8.
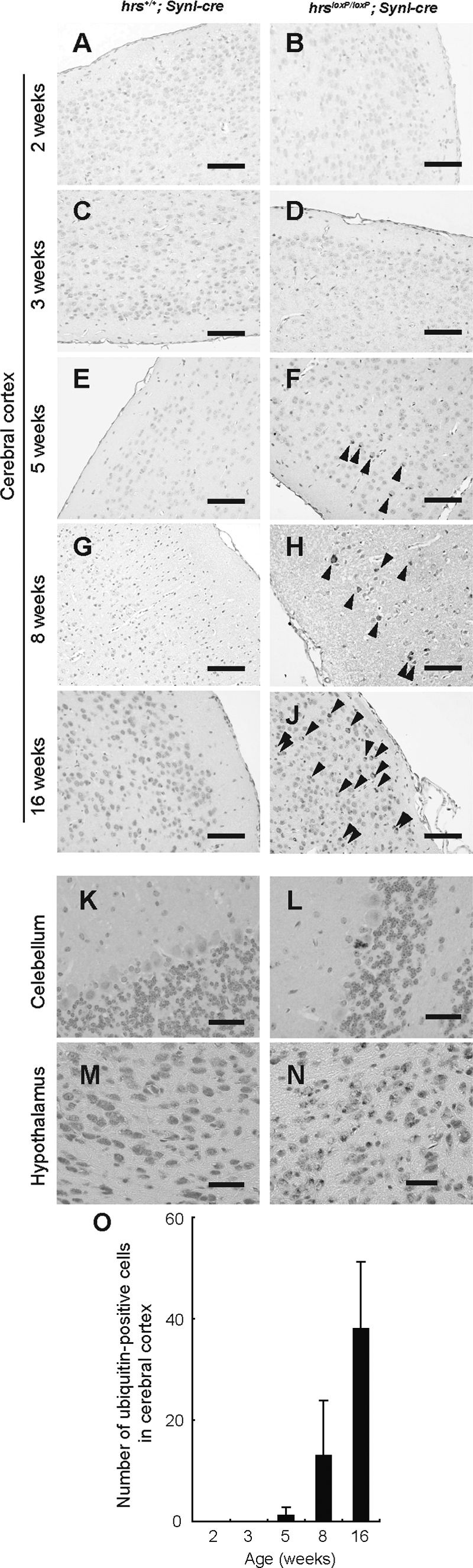
Ubiquitin-positive inclusions in the hrsloxP/loxP;SynI-cre brain. Immunohistochemistry of cerebral cortex (A–J), Purkinje cells (K, L), and hypothalamus (M, N) sections from hrs+/+;SynI-cre (A, C, E, G, I, K, M) and hrsloxP/loxP;SynI-cre (B, D, F, H, J, L, N) mice at various weeks of age, stained with an anti-ubiquitin antibody (1B3). A and B: Two weeks old. C and D: Three weeks old. E, F, K, and L: Five weeks old. G and H: Eight weeks old. I, J, M, and N: Sixteen weeks old. Arrowheads indicate ubiquitin-positive cells. O: Ubiquitin-positive cells in the cerebral cortex were counted in comparable areas for each hrsloxP/loxP;SynI-cre mouse, and five fields were counted in each area for each mouse. Data represent the mean ± SE of three mice. Scale bars = 100 μm. Original magnifications, ×400.
Expression Pattern of Glutamate Receptors, PSD-95, and p62
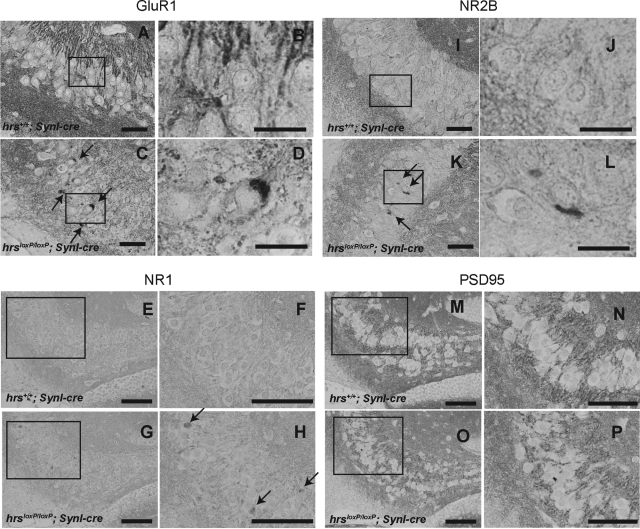
Because Hrs binds to ubiquitinated receptors and sorts them into lysosomes through multivesicular bodies, we performed immunohistochemical analyses in the hippocampus for NR1 and NR2B, the major subunits of NR, and for GluR1, the major subunit of AMPAR. In hrsloxP/loxP;SynI-cre mice, NR1-, NR2B-, and GluR1-positive aggregates were observed in the perikarya of CA3 pyramidal cells (Figure 9, A–L). We suspected that the PSD-95 that is abundant in virtually all mature excitatory glutaminergic synapses might also be involved in the glutamate receptor accumulation. Previous studies indicate that Hrs binds to PSD-95β32 and that PSD-95 controls glutaminergic synapse function.33 Nevertheless, PSD-95-positive aggregates were not detected in the hrsloxP/loxP;SynI-cre mice (Figure 9, M–P). These data suggest that Hrs affects the degradation of NR and AMPAR without affecting PSD-95.
Figure 9.
Immunohistochemistry of GluR1 (A--D), NR1 (E–H), NR2B (I–L), and PSD-95 (M–P) in the hippocampus CA3 subfield. Hippocampus sections from hrs+/+;SynI-cre (A, C, E, G, I, K, M, O) and hrsloxP/loxP;SynI-cre (B, D, F, H, J, L, N, P) 5-week-old mice were stained with GluR1-, NR1-, NR2B-, and PSD-95-specific antibodies. Arrows indicate protein aggregates. Insets show a higher magnification. Scale bars: 50 μm (A, C, E–H, I, K, M–P); 20 μm (B, D, J, L). Original magnifications, ×400.
Because Hrs is involved in the autophagic pathway,34 it was possible that insufficient autophagy-dependent protein degradation was the reason for the aggregation of ubiquitinated protein in the CA3 region. We therefore examined the expression of two proteins: LC3, a specific marker of autophagosomes, and p62, which is regulated by autophagy and thus accumulates when autophagy is insufficient.35 Although LC3-positive vesicles could not be detected in either the control or Hrs mutant mouse brain (data not shown), aggregated p62 was clearly observed in the pyramidal cell perikarya in the CA3 subfield of the Hrs mutant (Figure 10, A–D). Consistent with data in Figure 7, signals detected by FK2, including mono-and polyubiquitinated proteins, were also observed in hrsloxP/loxP;SynI-cre mice. Notably, p62, GluR1, and NR2B were co-localized with each other, as well as with the ubiquitinated proteins (Figure 10, E–L).
Figure 10.
Immunohistochemistry of p62, FK2, GluR1, and NR2B in the hippocampus CA3 subfield. A–D: Aggregated p62 (red) and ubiquitinated proteins (green) were observed in the hrsloxP/loxP;SynI-cre mice (C, D), but not in the hrs+/+;SynI-cre mice (A, B). p62 was co-localized with ubiquitinated proteins (yellow). E–L: GluR1 and NR2B were also co-localized with p62 and FK2 in the hrsloxP/loxP;SynI-cre mice. Scale bars = 10 μm. Original magnifications, ×400.
Learning Ability and Locomotor Activity Impairments in Hrs Mutant Mice
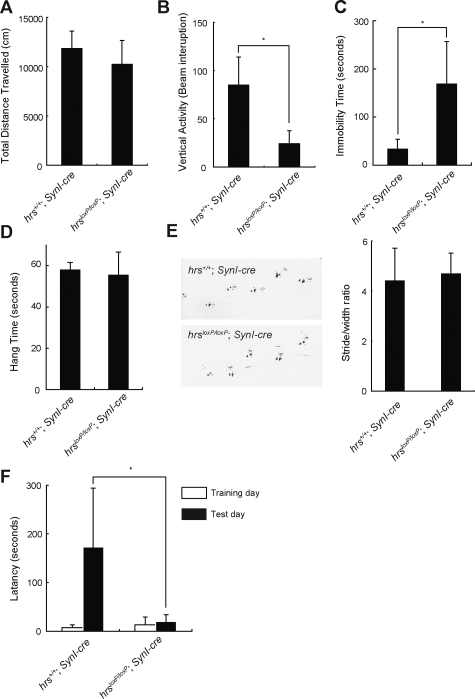
To investigate the effect of ubiquitinated protein accumulation in neural cells, behavioral analyses were performed with hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice. First, in an open field test, there were significant differences in the vertical (rearing) activity, but not in the horizontal activity, between the two groups (Figure 11, A and B). These results indicate that locomotor activity was impaired in the hrsloxP/loxP;SynI-cre mice.
Figure 11.
Behavioral examinations. A and B: The open-field activity test. Data are averages ± standard errors (n = 10) for the total distance (A) and vertical activity (B) of the hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice for 30 minutes. C: Increase of immobility time in the forced swimming test in the hrsloxP/loxP;SynI-cre mice. Values are expressed as means ± standard errors (n = 10). D: Wire hanging test. Hrs+/+;SynI-cre and hrsloxP/loxP;SynI-cre mice were tested (n = 10). E: Left, paw placement records of 12-week-old mice; right, stride lengths corrected for paw base widths (stride/width ratio) of hrsloxP/loxP;SynI-cre and hrs+/+;SynI-cre littermate mice. Values are expressed as means ± standard errors of three mice. F: Loss of memory function of the hrsloxP/loxP;SynI-cre mice in the step-through passive avoidance test. Data represent means ± standard errors of the step-through latency period (n = 10). Statistical differences between groups were determined by Student’s t-test. *P < 0.05.
Next, because ubiquitinated proteins accumulated in the hypothalamus of the hrsloxP/loxP;SynI-cre mice, we investigated the mental status of the two groups of mice by the forced swimming test, which is used to evaluate depression in rodents. In this test, a depressed state is induced in mice by forcing them to swim in an aquarium from which they cannot escape. The hrsloxP/loxP;SynI-cre mice showed a significantly longer duration of immobility (Figure 11C). To test whether hrsloxP/loxP;SynI-cre mice had weaker muscles, we performed a wire-hanging test. No significant difference was observed between the two groups of mice in this test (Figure 11D). Footprint analysis also showed no difference between the two groups (Figure 11E). Collectively, these results indicate that the hrsloxP/loxP;SynI-cre mice were in a depressive state.
Finally, we tested whether the loss of hippocampal CA3 neurons in the hrsloxP/loxP;SynI-cre mice affected their learning ability. Mice were examined using a passive avoidance task. Twenty-four hours after training, the latency period for mice to enter the dark box was significantly shorter for the hrsloxP/loxP;SynI-cre mice than for the hrs+/+;SynI-cre mice (Figure 11F). This result is compatible with the observation of hippocampal neuron loss in the hrsloxP/loxP;SynI-cre mice.
Discussion
In the present study, we detected the obvious accumulation of ubiquitinated proteins in 5-week-old hrsloxP/loxP;SynI-cre brain, although neurodegeneration was not apparent in the hippocampus until after 8 weeks, suggesting that the aggregation of ubiquitinated proteins precedes neurodegeneration in these mice. We also found that ubiquitinated proteins in the cerebral cortex of hrsloxP/loxP;SynI-cre mice increased with age, and that their learning ability was impaired, in accordance with the increase in ubiquitinated proteins. These findings reveal the importance of understanding Hrs’ role in the mechanism of neurodegenerative diseases because some ubiquitin-positive inclusions, such as phosphorylated tau and TDP-43, accumulate in the cerebral cortex, which results in the progressive loss of cognitive ability.36 In this study, we could not detect the expression of phosphorylated tau in the cerebral cortex of hrsloxP/loxP;SynI-cre mice using an anti-phosphorylated tau antibody (data not shown). Further study is required to reveal the relationship between the ESCRT proteins and neurodegenerative disease.
In neurons, NMDA receptor proteins usually concentrate at the postsynaptic density (PSD), a specialized apparatus beneath synapses that consists of receptors, scaffolding molecules, and signal-transduction enzymes. Synaptic transmission modulates the composition of the PSD, in part by the activity-dependent ubiquitination and degradation of PSD components.37 The Mdm2-mediated ubiquitination of PSD-95 is critical for regulating the cell-surface expression of AMPA receptors in synaptic plasticity.38 The overexpression of Hrs blocks the postsynaptic targeting of PSD-95β, which instead accumulates on large endosomal vesicles.32 In this study, we found AMPAR- and NMDAR-positive aggregates in the hippocampus of hrsloxP/loxP;SynI-cre mice, although their expression levels, except in the aggregates, were not significantly different between the hrsloxP/loxP;SynI-cre and control mice. These observations suggest that the loss of hippocampal CA3 pyramidal neurons is not caused by excitotoxicity, but by protein aggregate-induced cellular toxicity. Generally, ESCRTs including Hrs were thought to regulate an endosomal pathway, not a proteasomal pathway.11 Thus, we conclude that the loss of Hrs impairs the lysosome-dependent degradative pathway, thereby advancing the accumulation of ubiquitinated proteins, including glutamate receptors, resulting in neurodegeneration. However, we cannot exclude the possibility that we failed to detect altered CA3 pyramidal neurons in the hrsloxP/loxP;SynI-cre mice.
Endosomes are major targets of genetic and epigenetic pathogenic factors in many neurodegenerative diseases.39 In Huntington’s disease, the expansion of a polyglutamate tract in huntingtin (Htt) causes neuronal loss in the striatum and cortex.40 Mutant Htt is believed to promote cell death in several ways that involve endosome dysfunction, and Htt normally associates with HIP1, a protein that co-localizes on early endosomes with its ligand Hrs.41 Recent studies indicate that ESCRTs not only facilitate the trafficking of ubiquitylated proteins from endosomes to lysosomes, but play a critical role in autophagy; ESCRT III dysfunction causes autophagosome accumulation and neurodegeneration because of an abnormal fusion process between autophagosomes and endosomal compartments or lysosomes.17,42 Moreover, autophagy is essential for the survival of neural cells, and its impairment is implicated in the pathogenesis of neurodegenerative disorders involving ubiquitin-containing inclusion bodies.17,43,44 In hrsloxP/loxP;SynI-cre mice, we found that ubiquitinated proteins accumulated in the hippocampus, which is similar to the finding for autophagy-deficient mice.43,44 In this context, we previously demonstrated that Hrs plays a crucial role in autophagosome maturation.34 Although we have not yet obtained direct evidence for it, it appears that the autophagic pathway is impaired in the hippocampal CA3 subfields of the hrsloxP/loxP;SynI-cre mice, because p62, which is degraded by the autophagic pathway,44 was among the accumulated proteins. Further study will be required to elucidate the function of Hrs in autophagy by using other organ-specific Cre transgenic mice.
It was striking that the loss of pyramidal neurons by Hrs depletion occurred only in the hippocampal CA3 subfield. It was also interesting that NR accumulated only in the CA3, and not in the CA1 subfield, even though NR1, a common subunit of NR,45 is distributed in both CA1 and CA3. We expect that one of the following ideas will explain this selective phenotype: First, the expression of hrs is higher in the CA3 subfield than in CA1 (Figure 1, B and C). Second, certain cargos that are critical for neural survival and sensitive to Hrs-dependent sorting and degradation may be restricted to CA3. Further study will be required to elucidate the reason for this specificity of Hrs-dependent neurodegeneration.
In severely depressed patients, emotional arousal, cognitive abnormality, and vulnerability to psychotic episodes are linked to hypothalamic-pituitary-adrenal axis activity.46 Excessive stimulation of the axis is implicated in depression, and hyperactivity of the hypothalamic-pituitary-adrenal axis is observed in the majority of patients with depression.47 We found that hrsloxP/loxP;SynI-cre mice showed more immobility than controls in a forced swimming test, and that ubiquitinated proteins accumulated in the hypothalamus of these mice, although hypothalamic neurons were not lost. These observations suggest that ubiquitinated proteins in the neural cells of hrsloxP/loxP;SynI-cre mice might impair hypothalamic function and the hypothalamic-pituitary-adrenal axis. We also found that the loss of Hrs markedly impaired the retention of passive avoidance behavior 24 hours after the training trial. A previous study showed that CA3 NMDAR function is absent during memory formation in CA3-NR1 KO mice,48 which is similar to the phenotype of Hrs mutant mice. Our data are compatible with this study.
The present study indicates that Hrs plays a pivotal role in the survival of neural cells through its involvement in the degradation pathway for ubiquitinated proteins. Although it is still unknown whether Hrs selectively recognizes harmful gene products associated with neurodegenerative disorders such as Huntington’s, Parkinson’s, and Alzheimer’s disease, our Hrs mutant mice provide an excellent animal model system for studying the molecular mechanisms of neurodegenerative diseases.
Acknowledgments
We thank Dr. Jamey D. Marth for the Synapsin-I Cre transgenic mice; Kazuhiko Yanai and Eiko Sakurai for their help with the behavioral examinations; Drs. Hiromu Yawo, Yasuto Itoyama, Ichiro Sora, and Toshio Watanabe for discussion; and Ms. Rie Ito for technical assistance.
Footnotes
Address reprint requests to Nobuyuki Tanaka, Department of Microbiology and Immunology, Tohoku University Graduate School of Medicine, 2-1 Seiryo-machi, Aoba-ku, Sendai, 980-8575 Japan. E-mail: n-tanaka@m.tains.tohoku.ac.jp.
Supported in part by the Ministry of Education, Culture, Sports, Science, and Technology of Japan (grants-in-aid for Scientific Research on Priority Areas 19041067 and 20012053 to N.T. and 19059001 to K.S.); the Japan Society for the Promotion of Science [grants-in-aid for Scientific Research (C) 19590484 to N.T. and 19590488 to K.M. and a grant-in-aid for Exploratory Research 19659108 to K.S.]; and the Naito Research Foundation.
References
- Ballatore C, Lee VM, Trojanowski JQ. Tau-mediated neurodegeneration in Alzheimer’s disease and related disorders. Nat Rev Neurosci. 2007;8:663–672. doi: 10.1038/nrn2194. [DOI] [PubMed] [Google Scholar]
- Trojanowski JQ, Lee VM. “Fatal attractions” of proteins: a comprehensive hypothetical mechanism underlying Alzheimer’s disease and other neurodegenerative disorders. Ann N Y Acad Sci. 2000;924:62–67. doi: 10.1111/j.1749-6632.2000.tb05561.x. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Aarts MM, Tymianski M. Molecular mechanisms underlying specificity of excitotoxic signaling in neurons. Curr Mol Med. 2004;4:137–147. doi: 10.2174/1566524043479202. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Excitotoxic and excitoprotective mechanisms: abundant targets for the prevention and treatment of neurodegenerative disorders. Neuromolecular Med. 2003;3:65–94. doi: 10.1385/NMM:3:2:65. [DOI] [PubMed] [Google Scholar]
- Moghaddam B. Bringing order to the glutamate chaos in schizophrenia. Neuron. 2003;40:881–884. doi: 10.1016/s0896-6273(03)00757-8. [DOI] [PubMed] [Google Scholar]
- Tzschentke TM. Glutamatergic mechanisms in different disease states: overview and therapeutical implications—an introduction. Amino Acids. 2002;23:147–152. doi: 10.1007/s00726-001-0120-8. [DOI] [PubMed] [Google Scholar]
- Mori H, Mishina M. Structure and function of the NMDA receptor channel. Neuropharmacology. 1995;34:1219–1237. doi: 10.1016/0028-3908(95)00109-j. [DOI] [PubMed] [Google Scholar]
- Kato A, Rouach N, Nicoll RA, Bredt DS. Activity-dependent NMDA receptor degradation mediated by retrotranslocation and ubiquitination. Proc Natl Acad Sci USA. 2005;102:5600–5605. doi: 10.1073/pnas.0501769102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer H, Rongo C. KEL-8 is a substrate receptor for CUL3-dependent ubiquitin ligase that regulates synaptic glutamate receptor turnover. Mol Biol Cell. 2006;17:1250–1260. doi: 10.1091/mbc.E05-08-0794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M. A protein’s final ESCRT. Traffic. 2005;6:2–9. doi: 10.1111/j.1600-0854.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- Raiborg C, Rusten TE, Stenmark H. Protein sorting into multivesicular endosomes. Curr Opin Cell Biol. 2003;15:446–455. doi: 10.1016/s0955-0674(03)00080-2. [DOI] [PubMed] [Google Scholar]
- Komada M, Soriano P. Hrs, a FYVE finger protein localized to early endosomes, is implicated in vesicular traffic and required for ventral folding morphogenesis. Genes Dev. 1999;13:1475–1485. doi: 10.1101/gad.13.11.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura S, Takeshita T, Asao H, Kimura Y, Murata K, Sasaki Y, Hanai JI, Beppu H, Tsukazaki T, Wrana JL, Miyazono K, Sugamura K. Hgs (Hrs), a FYVE domain protein, is involved in Smad signaling through cooperation with SARA. Mol Cell Biol. 2000;20:9346–9355. doi: 10.1128/mcb.20.24.9346-9355.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Deconstructing the axon: Wallerian degeneration and the ubiquitin-proteasome system. Trends Neurosci. 2004;27:3–6. doi: 10.1016/j.tins.2003.10.015. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JA, Beigneux A, Ahmad ST, Young SG, Gao FB. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Curr Biol. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- Yagi T, Tokunaga T, Furuta Y, Nada S, Yoshida M, Tsukada T, Saga Y, Takeda N, Ikawa Y, Aizawa S. A novel ES cell line, TT2, with high germline-differentiating potency. Anal Biochem. 1993;214:70–76. doi: 10.1006/abio.1993.1458. [DOI] [PubMed] [Google Scholar]
- Dymecki SM. Flp recombinase promotes site-specific DNA recombination in embryonic stem cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:6191–6196. doi: 10.1073/pnas.93.12.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Kaneko K, Asao H, Kasai H, Endo Y, Fujita T, Takeshita T, Sugamura K. Possible involvement of a novel STAM-associated molecule “AMSH” in intracellular signal transduction mediated by cytokines. J Biol Chem. 1999;274:19129–19135. doi: 10.1074/jbc.274.27.19129. [DOI] [PubMed] [Google Scholar]
- Asao H, Sasaki Y, Arita T, Tanaka N, Endo K, Kasai H, Takeshita T, Endo Y, Fujita T, Sugamura K. Hrs is associated with STAM, a signal-transducing adaptor molecule. Its suppressive effect on cytokine-induced cell growth. J Biol Chem. 1997;272:32785–32791. doi: 10.1074/jbc.272.52.32785. [DOI] [PubMed] [Google Scholar]
- Abe M, Fukaya M, Yagi T, Mishina M, Watanabe M, Sakimura K. NMDA receptor GluRepsilon/NR2 subunits are essential for postsynaptic localization and protein stability of GluRzeta1/NR1 subunit. J Neurosci. 2004;24:7292–7304. doi: 10.1523/JNEUROSCI.1261-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukaya M, Kato A, Lovett C, Tonegawa S, Watanabe M. Retention of NMDA receptor NR2 subunits in the lumen of endoplasmic reticulum in targeted NR1 knockout mice. Proc Natl Acad Sci USA. 2003;100:4855–4860. doi: 10.1073/pnas.0830996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy GG, Al-Ayyan M, Andrew D, Fukaya M, Watanabe M, Todd AJ. Widespread expression of the AMPA receptor GluR2 subunit at glutamatergic synapses in the rat spinal cord and phosphorylation of GluR1 in response to noxious stimulation revealed with an antigen-unmasking method. J Neurosci. 2004;24:5766–5777. doi: 10.1523/JNEUROSCI.1237-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Takeshita T, Miura S, Murata K, Kimura Y, Ishii N, Nose M, Sakagami H, Kondo H, Tashiro F, Miyazaki JI, Sasaki H, Sugamura K. Loss of hippocampal CA3 pyramidal neurons in mice lacking STAM1. Mol Cell Biol. 2001;21:3807–3819. doi: 10.1128/MCB.21.11.3807-3819.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeren-Wiemers N, Gerfin-Moser A. A single protocol to detect transcripts of various types and expression levels in neural tissue and cultured cells: in situ hybridization using digoxigenin-labelled cRNA probes. Histochemistry. 1993;100:431–440. doi: 10.1007/BF00267823. [DOI] [PubMed] [Google Scholar]
- Chen Z, Sakurai E, Hu W, Jin C, Kiso Y, Kato M, Watanabe T, Wei E, Yanai K. Pharmacological effects of carcinine on histaminergic neurons in the brain. Br J Pharmacol. 2004;143:573–580. doi: 10.1038/sj.bjp.0705978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sango K, McDonald MP, Crawley JN, Mack ML, Tifft CJ, Skop E, Starr CM, Hoffmann A, Sandhoff K, Suzuki K, Proia RL. Mice lacking both subunits of lysosomal beta-hexosaminidase display gangliosidosis and mucopolysaccharidosis. Nat Genet. 1996;14:348–352. doi: 10.1038/ng1196-348. [DOI] [PubMed] [Google Scholar]
- Zomkowski AD, Santos AR, Rodrigues AL. Putrescine produces antidepressant-like effects in the forced swimming test and in the tail suspension test in mice. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:1419–1425. doi: 10.1016/j.pnpbp.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Klapdor K, Dulfer BG, Hammann A, Van der Staay FJ. A low-cost method to analyse footprint patterns. J Neurosci Methods. 1997;75:49–54. doi: 10.1016/s0165-0270(97)00042-3. [DOI] [PubMed] [Google Scholar]
- Chetkovich DM, Bunn RC, Kuo SH, Kawasaki Y, Kohwi M, Bredt DS. Postsynaptic targeting of alternative postsynaptic density-95 isoforms by distinct mechanisms. J Neurosci. 2002;22:6415–6425. doi: 10.1523/JNEUROSCI.22-15-06415.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Béique JC, Lin DT, Kang MG, Aizawa H, Takamiya K, Huganir RL. Synapse-specific regulation of AMPA receptor function by PSD-95. Proc Natl Acad Sci USA. 2006;103:19535–19540. doi: 10.1073/pnas.0608492103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamai K, Tanaka N, Nara A, Yamamoto A, Nakagawa I, Yoshimori T, Ueno Y, Shimosegawa T, Sugamura K. Role of Hrs in maturation of autophagosomes in mammalian cells. Biochem Biophys Res Commun. 2007;360:721–727. doi: 10.1016/j.bbrc.2007.06.105. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Koike M, Sou YS, Ueno T, Hara T, Mizushima N, Iwata J, Ezaki J, Murata S, Hamazaki J, Nishito Y, Iemura S, Natsume T, Yanagawa T, Uwayama J, Warabi E, Yoshida H, Ishii T, Kobayashi A, Yamamoto M, Yue Z, Uchiyama Y, Kominami E, Tanaka K. Homeostatic levels of p62 control cytoplasmic inclusion body formation in autophagy-deficient mice. Cell. 2007;131:1149–1163. doi: 10.1016/j.cell.2007.10.035. [DOI] [PubMed] [Google Scholar]
- Forman MS, Trojanowski JQ, Lee VM. Neurodegenerative diseases: a decade of discoveries paves the way for therapeutic breakthroughs. Nat Med. 2004;10:1055–1063. doi: 10.1038/nm1113. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Colledge M, Snyder EM, Crozier RA, Soderling JA, Jin Y, Langeberg LK, Lu H, Bear MF, Scott JD. Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron. 2003;40:595–607. doi: 10.1016/s0896-6273(03)00687-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA. Endosome function and dysfunction in Alzheimer’s disease and other neurodegenerative diseases. Neurobiol Aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Metzler M, Li B, Gan L, Georgiou J, Gutekunst CA, Wang Y, Torre E, Devon RS, Oh R, Legendre-Guillemin V, Rich M, Alvarez C, Gertsenstein M, McPherson PS, Nagy A, Wang YT, Roder JC, Raymond LA, Hayden MR. Disruption of the endocytic protein HIP1 results in neurological deficits and decreased AMPA receptor trafficking. EMBO J. 2003;22:3254–3266. doi: 10.1093/emboj/cdg334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, Stenmark H. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, Yokoyama M, Mishima K, Saito I, Okano H, Mizushima N. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, Tanaka K. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- Köhr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–446. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- de Kloet ER, Derijk RH, Meijer OC. Therapy insight: is there an imbalanced response of mineralocorticoid and glucocorticoid receptors in depression? Nat Clin Pract Endocrinol Metab. 2007;3:168–179. doi: 10.1038/ncpendmet0403. [DOI] [PubMed] [Google Scholar]
- Berton O, Nestler EJ. New approaches to antidepressant drug discovery: beyond monoamines. Nat Rev Neurosci. 2006;7:137–151. doi: 10.1038/nrn1846. [DOI] [PubMed] [Google Scholar]
- Nakazawa K, Quirk MC, Chitwood RA, Watanabe M, Yeckel MF, Sun LD, Kato A, Carr CA, Johnston D, Wilson MA, Tonegawa S. Requirement for hippocampal CA3 NMDA receptors in associative memory recall. Science. 2002;297:211–218. doi: 10.1126/science.1071795. [DOI] [PMC free article] [PubMed] [Google Scholar]