Abstract
Malignant gliomas are characterized by their ability to invade normal brain tissue. We have previously shown that the small GTPase Rac1 plays a role in both migration and invasion in gliomas. Here, we aim to identify Rac-activating guanine nucleotide exchange factors (GEFs) that mediate glioblastoma invasiveness. Using a brain tumor expression database, we identified three GEFs, Trio, Ect2, and Vav3, that are expressed at higher levels in glioblastoma versus low-grade glioma. The expression of these GEFs is also associated with poor patient survival. Quantitative real-time polymerase chain reaction and immunohistochemical analyses on an independent set of tumors confirmed that these GEFs are overexpressed in glioblastoma as compared with either nonneoplastic brain or low-grade gliomas. In addition, depletion of Trio, Ect2, and Vav3 by siRNA oligonucleotides suppresses glioblastoma cell migration and invasion. Depletion of either Ect2 or Trio also reduces the rate of cell proliferation. These results suggest that targeting GEFs may present novel strategies for anti-invasive therapy for malignant gliomas.
Therapeutic failures for malignant gliomas arise from the inability to treat the invading tumor cells that extend beyond the margins of the primary tumor. Invasion is a complex cellular phenomenon involving cell-cell and cell-extracellular matrix (ECM) interactions, enzymatic degradation of the ECM, and cell migration.1,2,3 Members of the Rho family of small GTPases, including RhoA, Rac1, and Cdc42, are key regulators of cell migration.4 We have previously shown that inhibition of Rac1 expression by siRNA markedly inhibits glioma cell migration and invasion.5,6,7 The closely related Rac3 GTPase is also critical for glioma invasion, but is less important for cell migration.5
Rho GTPases cycle between active GTP-bound and inactive GDP-bound forms.8 Their nucleotide state is regulated by three classes of accessory proteins: guanine nucleotide exchange factors (GEFs), which promote replacement of GDP by GTP, thereby activating the GTP-ases; Rho GTP proteins (GAPs) inactivate GTP-ases by stimulating intrinsic nucleotide hydrolysis; and the GDP dissociation inhibitors (GDIs), which essentially clamp Rho GTPases in the GDP-bound state. Once these molecular switches are activated, Rho GTPases regulate diverse cellular processes, including actin cytoskeleton organization, cell and growth survival, differentiation, oncogenic transformation, gene transcription, and migration.9
Rho GTPase GEFs (Rho GEFS) belong to either the Dbl family or the Dock family.10 All 69 Dbl proteins coded by the human genome contain a core catalytic domain termed Dbl homology (DH) domain and an adjacent Pleckstrin homology (PH) domain.11 Dock proteins, of which there are 11 coded in the human genome, contain two conserved regions, termed Dock homology region-1 and -2 (DHR1 and DHR2). In some cases, the DHR2 domain, also referred to as Docker domain, is sufficient for guanine nucleotide exchange.10 There are 80 Rho GEFS in the human genome and 22 Rho GTPases,11 of which only 15 cycle between GDP and GTP states and therefore are GEF-dependent. Moreover, most of the Rho GEFs can act on multiple Rho GTPases,11 implying that the Rho GEFs far outnumber their substrates. Thus, taking into account that for approximately a third of all Rho GEFs, the GTPase substrate specificity remains to be determined, Rac GTPases are estimated to be regulated by ∼40 different Rho GEFs.11 Very little information is currently available on the expression or activation status of any of the Rho GEFs in human tumors.11,12 In this study therefore, we used gene expression profiling to discern GEFs that act on Rac proteins and may be important for glioma invasion. Three GEFs, Trio, Ect2, and Vav3, were selected for further study because their increased expression was associated with poor patient outcome and higher tumor grade. Expression levels of these Rac GEFs were validated using quantitative real-time polymerase chain reaction (PCR) and protein levels assessed using immunohistochemistry in independent glioma specimens. Lastly, we show that depletion of the respective GEFs using RNA interference significantly inhibits the invasive behavior of glioblastoma cells. These data suggest the potential of targeting Rho GEFS as anti-invasive therapies.
Materials and Methods
Expression Profile Dataset of Rho GEFs in Human Gliomas and Nonneoplastic Brain (NB)
To identify candidate GEFs for our study we mined the expression microarray database containing 195 clinically annotated brain tumor specimens publicly available at NCBI’s Gene Expression Omnibus as dataset GSE4290. Snap-frozen specimens from epileptogenic foci (NB, n = 24) and tumor [29 low-grade astrocytomas (LGAs), 82 glioblastoma multiformes (GBMs), 11 mixed gliomas, and 49 oligodendrogliomas] with clinical information were collected at Hermelin Brain Tumor Center, Henry Ford Hospital (Detroit, MI) as previously described.13 Gene expression profiling as described previously13 was conducted on all samples using Affymetrix U133 Plus 2 GeneChips according to the manufacturer’s protocol at the Neuro-Oncology Branch at the National Cancer Institute (Bethesda, MD). Gene expression data were normalized in two ways: per chip normalization and per gene normalization. For per chip normalization, all expression data on a chip were normalized to the 50th percentile of all values on that chip. For per gene normalization, the data for a given gene were normalized to the median expression level of that gene across all samples. Gene differences were deemed statistically significant using parametric tests where variances were not assumed equal (Welch analysis of variance). Expression values were then filtered for highly variable (differentially expressed) genes (coefficient of variation >30%) across samples producing a list of 7322 genes. Principal component (PC) analysis was done to investigate the relationship between samples as previously described.13 Kaplan-Meier survival curves were developed for each cluster. One cluster had a median survival time of 401 days and the other cluster had a median survival time of 952 days. Box plots for Ect2, Trio, and Vav3 expression levels in each cluster derived from PC analysis were graphed. Significance between the two populations was tested with a two-sample t-test assuming unequal variances.
Quantitative Reverse Transcriptase-Polymerase Chain Reaction (QRT-PCR)
Total RNA from an independent cohort of frozen tissue was extracted using Trizol (Life Technologies Inc., Grand Island, NY), following standard procedures. Tumor specimens were obtained from The Nervous System Tissue Bank, University Health Network, Toronto, Canada. The following surgical specimens were obtained: nonneoplastic brain (NB, n = 4), LGA (n = 4), GBM (n = 15). All LGAs in this study have been diagnosed as grade 2 astrocytomas. The quantity and quality of RNA were assessed with a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). cDNA was synthesized from 200 ng of total RNA in a 10-μl reaction volume using the RT core kit (Eurogentec, San Diego, CA) for 10 minutes at 25°C, 30 minutes at 48°C, and then 5 minutes at 95°C. QRT-PCR was performed on 30 ng of the cDNA in a final volume of 25 μl using the Chromo4 real-time PCR detector (MJ Research, Waltham, MA) with the primers summarized in Table 1. SYBR green fluorescence was used for detection of amplification after each cycle. Negative (no template) controls were run in parallel to confirm the absence of nonspecific fluorescence in samples. PCR was done using the following protocol: 10 minutes at 95°C for activation of HotGoldStar Taq polymerase, 15 seconds at 95°C, and 1 minute at 60°C. The PCR data were analyzed with the Opticon Monitor 3.1.32 analysis software (MJ Research). Quantification was based on the number of cycles necessary to produce a detectable amount of product above background. To ensure specificity of the PCR product, the melting curves for the sample products were analyzed. The quantity is calculated relative to the sample with the lowest mean Ct value for the gene of interest. The equation for relative quantity is: relative quantity = efficiency (control Ct − unknown Ct). For each sample of interest a normalization factor (NF) is then calculated with the geometric mean of the relative quantities of all reference genes [histone H3.3 and hypoxanthine guanine phosphoribosyl transferase 1 (HPRT1)] having the same identifier as the sample of interest. The formula is the following: NF = (quantity 1 * quantity 2 … … quantity N).1/N The expression level of the sample of interest is then obtained using the equation: expression level = relative quantity/NF. Student’s t-test was used to assess statistical significance between groups.
Table 1.
PCR Primer Sequences
| Primer | Sense | Anti-sense |
|---|---|---|
| ECT2 | 5′-CTCTAGGTGAGCACCCCTGT-3′ | 5′-TGTGCCGTTTTCTTGCTATCT-3′ |
| TRIO | 5′-AGTCCACCCAGAGCAACG-3′ | 5′-CGTGTAATCGTGTGTCACCA-3′ |
| VAV3 | 5′-TGAGGCACAGGACCAAAGA-3′ | 5′-TGATGTGCTTTGCTTCATTATTGT-3′ |
| RAC1 | 5′-CTGATGCAGGCCATCAAGT-3′ | 5′-CAGGAAATGCATTGGTTGTG-3′ |
| HPRT1 | 5′-TGACACTGGCAAAAACAATGCA-3′ | 5′-GGTCCTTTTCACCAGCAAGCT-3′ |
| Histone | 5′-CCACTGAACTTCTGATTCGC-3′ | 5′-GCGTGCTAGCTGGATGTCTT-3′ |
Immunohistochemistry
An independent cohort of surgical formalin-fixed, paraffin-embedded specimens was obtained (4 NB, 6 LGA, 10 GBMs) from the Department of Neuropathology, University Health Network, Toronto, Canada, for immunohistochemical validation of GEF expression. All samples were verified by a neuropathologist (Sidney Croul, University of Toronto) to confirm diagnosis. Tumor sections were primarily comprised of the tumor core. Immunohistochemistry was performed on 5-μm-thick whole tissue sections. Sections were first deparaffinized, rehydrated, treated with 1% H2O2 for 30 minutes to neutralize endogenous peroxidase activity. Heat antigen retrieval in citrate buffer was performed. Sections were then blocked in serum for 30 minutes. Whole sections were incubated with the following primary antibodies: a polyclonal rabbit anti-human Ect2 (1:100, catalog no. sc-1005; Santa Cruz Biotechnology, Santa Cruz, CA), a monoclonal mouse anti-human Rac1 (1:100, catalog no. 610651; Cell Signaling, Danvers, MA), polyclonal goat anti-human Vav3 (1:200, catalog no. ab21208, Abcam, Cambridge, MA; or 1:50, Upstate, Lake Placid, NY), or a goat anti-human Trio (1:200, catalog no. sc-6060; Santa Cruz Biotechnology) for 1 hour. Sections were then incubated with biotinylated secondary antibody. Detection was performed with avidin-biotin-horseradish peroxidase complex (ABC; Vector Laboratories, Burlingame, CA) followed by diaminobenzidine as the chromogen. Nuclei were counterstained with hematoxylin.
Semiquantification of Ect2, Trio, Vav3, and Rac1 staining was performed. Entire tissue sections were examined and a score (from 0 to 5) based on the intensity of staining of tumor cells was assigned. Because there were unequal number of cases in each category, histograms were generated by calculating a weighted score for NB, LGAs, and GBMs. For this, the number of specimens in each category was obtained (variable A). The sum of tumor cases with equal scores (from 0 to 5) in each category was calculated (variable B) and multiplied by the corresponding score (S). The total sum of the products (B*S) intensity scores in each category was obtained (C). A final weighted score was achieved according to C/A. The nonparametric Wilcoxon signed-rank test was used to determine statistical significance between groups.
Glioma Cell Lines and Culture Conditions
Human astrocytoma cell lines SNB19 and U87 (American Type Culture Collection, Manassas, VA) were maintained in minimum essential medium supplemented with 10% fetal bovine serum (Hyclone Laboratories, Inc., Logan, UT). Cells were cultured in a 37°C, 5% humidified chamber.
siRNA Preparation and Transfections
siRNA duplexes specific for Ect2, Trio, and Vav3 were designed using a web-based algorithm according to Elbashir and colleagues.14 Twenty-one nucleotide RNAs were purchased from Qiagen (Valencia, CA) in deprotected and desalted forms. Two different siRNA sequences for each GEF were used and their target sequences are summarized in Table 2. Transient transfections of siRNA were performed using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were plated at 80% confluency in Dulbecco’s modified Eagle’s medium containing 10% serum without antibiotics. Transfections were performed within 16 to 20 hours after plating. The siRNA as well as the Lipofectamine 2000 were diluted in serum-free Dulbecco’s modified Eagle’s medium. After 5 minutes, the two mixtures were combined and incubated for 20 minutes at room temperature to enable complex formation. Ect2, Trio, and Vav3 small interfering RNAs were transfected at 50 nmol/L. siRNA against the nonhuman target, luciferase, served as control. Shut-down was confirmed by QRT-PCR as described above using total RNA extracted from transfected and control cells.
Table 2.
GEF siRNA Target Sequences
| Target name | Target sequence | Start position |
|---|---|---|
| Ect2-1 | 5′-AACAAGAGTGATATTGGTTCA-3′ | 149 |
| Ect2-2 | 5′-ATAAAGCTTGGGAAAGGCGGA-3′ | 638 |
| Trio-1 | 5′-AAAAATGCCTATGTTCAACCA-3′ | 9640 |
| Trio-2 | 5′-AAAGGTCGCATTGCTGTATCA-3′ | 9514 |
| Vav3-1 | 5′-AAGAATGCCAGTCATCAGTAA-3′ | 2923 |
| Vav3-2 | 5′-AAGCAGCAGTATCTATTAATA-3′ | 3478 |
Rac Activation Assay and Western Blot Analysis
For the Rac activation assays, SNB19 cells were transfected for 24 hours with the respective GEF siRNAs or control siRNA as described above and subsequently cultured in serum for an additional 48 hours. Cells were washed twice with cold Tris-buffered saline (TBS) and lysed in 50 mmol/L Tris, pH 7.2, 0.5% Triton X-100, 10 mmol/L MgCl2 in a cocktail of protease inhibitors (Roche Diagnostics, Indianapolis, IN). Lysates were harvested by centrifugation, and equal concentrations of lysates were incubated with 20 μg of purified GST-CRIB (for Rac/Cdc42 activation) according to the manufacturer’s protocol (Pierce, Rockford, IL).
Equal amounts of protein lysates were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes. Membranes were subsequently blocked with 5% skim milk in Tris-Buffered Saline Tween 20 (TBS-T). Densitometric analysis was performed to obtain the ratio of active Rac-GTP to total Rac. Three independent experiments were performed and Student’s t-test was performed to examine statistical significance between groups. For the characterization of protein depletion, cells were lysed at days 2, 3, and 4 after transfection and Western blotting was performed using the following antibodies: a polyclonal anti-Ect2 antibody (sc-1005, Santa Cruz Biotechnology), polyclonal anti-Vav3 antibody (Cell Signaling, Beverly, MA), or a polyclonal anti-Trio antibody (CT233; gift of Richard Mains, University of Connecticut Health Center, Farmington, CT).
Sulforhodamine B (SRB) Assay
Cell growth was measured using the SRB colorimetric assay.15 Briefly, 1 day after siRNA transfection, cells were seeded at 2 × 103 cells/well in a 96-well microtiter plate. At various times, cells were fixed in 10% trichloroacetic acid for 60 minutes at 4°C, rinsed, and subsequently stained for 30 minutes at room temperature with 0.2% SRB dissolved in 1% acetic acid, followed by air-drying. The bound dye was solubilized in 100 μl of 10 mmol/L unbuffered Tris base for 30 minutes and the OD was read at 490 nm in an enzyme-linked immunosorbent assay plate reader.
Quantification of Multinucleated Cells
Glioma cells transfected with Ect2, Trio, Vav3, or luciferase-directed siRNA for 48 hours, were trypsinized and plated on coverslips overnight. The cells were subsequently washed with phosphate-buffered saline (PBS), fixed in 4% formaldehyde/PBS, permeabilized with 0.1% Triton-X 100, and incubated with 4,6-diamidino-2-phenylindole and fluorescein isothiocyanate-conjugated phalloidin (Molecular Probes, Eugene, OR) to stain for the nucleus and F-actin, respectively. Processed coverslips were mounted in 75% Vectashield mounting medium (Vector Laboratories). Images were collected using an IX70 Olympus (Center Valley, PA) inverted microscope equipped with a ×60 (1.4 NA) objective, an Orca II cooled charge-coupled device camera (Hamamatsu, Hamamatsu, Japan) and ESee (Inovision, Raleigh, NC) image analysis software. For each experimental condition, images from ∼20 fields of cells were taken in a random manner. The percentage of multinucleated cells (two or more nuclei per cell) per field was calculated as the ratio of multinucleated cells to the total number of cells per field.
Radial Cell Migration Assay
Cellular migration was quantified in a radial cell migration assay as previously described for astrocytoma cells.7,16 Glioblastoma cells were transfected with siRNA targeting luciferase, Ect2, Trio, or Vav3. After 24 hours, cells were plated onto 10-well glass slides precoated with glioma-derived ECM as previously described.17 Cell migration was determined throughout the next 24 hours.
Organotypic Brain Slice Invasion Assay
An ex vivo invasion assay into rat brain slices was performed as described previously.6,7,16,18 Briefly, 400-μm-thick vital sections were prepared from brains of Wistar rats (Charles River Laboratories, Wilmington, MA) floated on micropore membranes in culture media. Reporter cells were glioma cells stably expressing green fluorescence protein (GFP) that had been transfected with siRNA directed against Ect2, Trio, or Vav3 24 hours previously. Cells (1 × 105) were gently deposited (0.5-μl transfer volume) onto the bilateral putamen of the brain slice then incubated under standard conditions. Typically, six brain slices were used in each experiment. After 60 hours from seeding the cells, glioma cell invasion into the rat brain slices was detected using a LSM 5 Pascal laser-scanning confocal microscope (Zeiss, Thornwood, NY) to observe GFP-labeled cells in the tissue slice. Serial optical sections were obtained every 10 μm downward from the surface plane to the bottom of the slice, and for each focal plane, the area of fluorescent cells as a function of the distance from the top surface of the slice was calculated. The extent of glioma cell invasion was reported as the depth where the area of fluorescent tumor cells was half of the maximum area at the surface.6,16,19
Results
Analysis of Rac GEF Expression Profiles Using a Publicly Available Dataset
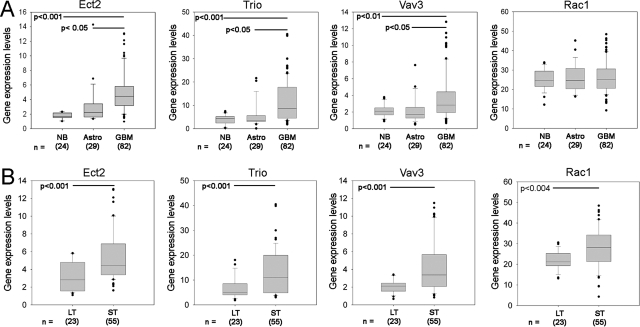
To determine the expression of Rac-activating GEFs in glioblastoma, we surveyed probes for 26 GEFs that can activate Rac11 on global expression arrays (Affymetrix) from a panel of 24 nonneoplastic and 160 brain tumor specimens (data provided by Dr. Howard Fine, Neuro-Oncology Branch, National Cancer Institute, NCBI Gene Expression Omnibus dataset GSE4290). We identified three GEFs (Ect2, Trio, and Vav3) that show significantly higher expression in glioblastoma (GBM, n = 82) than in nontumor brain (n = 24) and LGAs (n = 29) (Figure 1A).
Figure 1.
Gene expression profiling of human GEFs. A: mRNA expression levels of GEFs and Rac1 from NCBI Gene Expression Omnibus GDS1962 dataset is presented as box plot graphs. NB: nontumor brain; Astro, low-grade astrocytomas; GBM, glioblastoma microformes B: Principal component analysis of brain tumors from NCBI Gene Expression Omnibus GDS1962 dataset revealed two groups differing by their survival and were defined as short-term (ST) or long-term (LT) groups (see text). Box plot representation of mRNA expression demonstrating increased expression of Ect2, Trio, Vav3, and Rac1 in the ST group.
We applied principle component (PC) analysis13 to query the relationship between the expression of each of the three candidate GEFs and patient outcome. By PCA, the GBM patients segregated into two separate clusters displaying distinct Kaplan-Meier survival curves. The median survival of cluster 1 was 952 days (long term, LT) whereas cluster 2 had a median survival of 401 days (short term, ST).13 Examination of GEF expression in each cluster showed that GBM patients in the ST survival cluster had higher expression of Trio, Ect2, and Vav3 than GBM patients in the LT survival cluster (P < 0.001, Figure 1B). No significant change in Rac1 expression occurred across tumor grade (Figure 1A), however, Rac1 expression is elevated in GBM patients within the ST survival group compared with GBM patients in the LT group (Figure 1B).
Quantitative RT-PCR Validation of GEF Expression
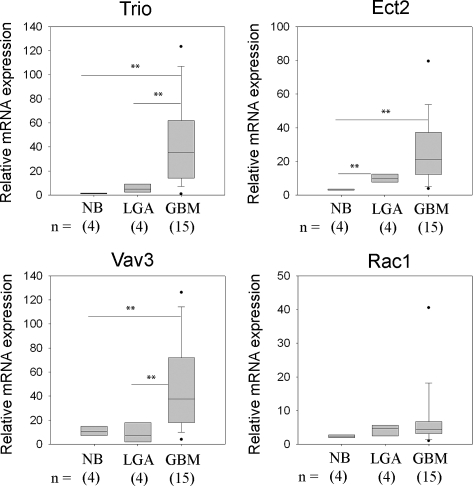
To verify the results of the expression profiling, we performed quantitative RT-PCR on independent nonneoplastic (n = 4), grade 2 LGA (n = 4), and GBM (n = 15) specimens (Figure 2). NB specimens show relatively low mRNA levels for all three GEFs examined. Expression of Ect2 in LGA was significantly higher than in NB (P < 0.005) whereas expression of Trio in LGA compared with NB did not achieve statistical significance (P = 0.053). Vav3 expression in LGA does not differ from that in NB (P = 0.697), similar to the results of the brain tumor expression profiling. In GBM specimens, the mRNA levels of the respective GEFs are significantly higher than in NB tissue (P < 0.01) (Figure 2).
Figure 2.
Quantitative real-time PCR analysis of GEF and Rac1 expression. Relative mRNA expression levels of Trio, Vav3, Ect2, and Rac1 in NB, grade 2 LGAs, and GBMs were analyzed by quantitative RT-PCR using histone H3.3 and HPRT1 as reference genes, see Materials and Methods. Data are presented as box plots and asterisks indicate statistical significance between groups.
Immunohistochemical Detection of Ect2, Trio, Vav3, and Rac1 in Human Astrocytomas
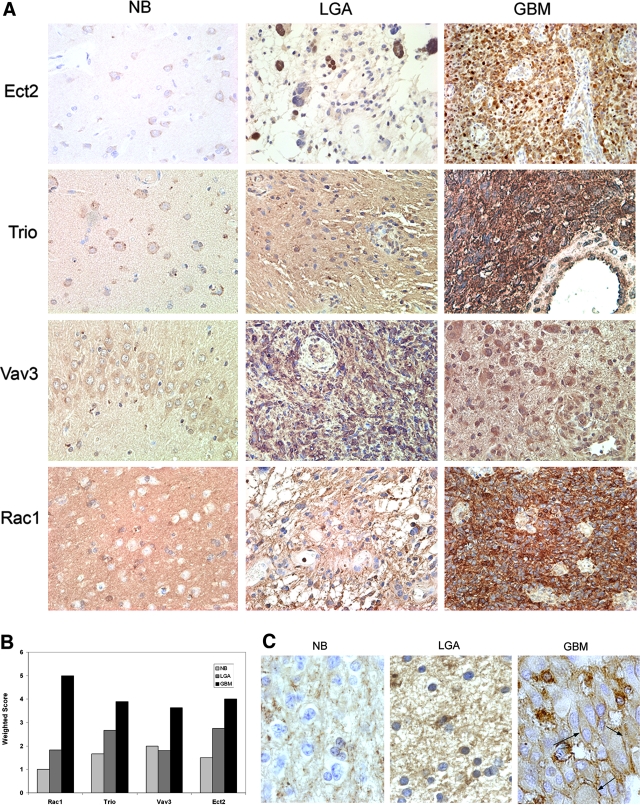
We used immunohistochemistry to examine protein expression and localization of Ect2, Trio, Vav3, and Rac1. Semiquantitative grading is represented graphically as a weighted score and shows that GBM samples expressed the highest levels of the respective GEFs and Rac1 compared to NB or LGA (all P values <0.05, Wilcoxon rank test) (Table 3; Figure 3, A and B). Ect2, Vav3, and Trio immunoreactivity was low in astrocytes, but GEF immunoreactivity in subcortical neurons was prominent (Figure 3A). Interestingly, whereas Rac1 is virtually absent in normal brain it shows a strong grade-dependent increase in protein expression, in contrast to results obtained from measuring Rac1 transcript levels.
Table 3.
Semi-Quantitative Analysis of Trio, Ect2, Vav3, and Rac1 by IHC
| Negative | Weak (score 1) | Moderate (score 2 to 3) | Strong (score 4 to 5) | |
|---|---|---|---|---|
| Rac1 | ||||
| NB | 100.00 | 0.00 | 0.00 | 0.00 |
| LGA | 0.00 | 66.67 | 16.67 | 16.67 |
| GBM | 0.00 | 0.00 | 0.00 | 100.00 |
| Ect2 | ||||
| NB | 0.00 | 50.00 | 50.00 | 0.00 |
| LGA | 0.00 | 37.50 | 25.00 | 37.50 |
| GBM | 0.00 | 0.00 | 40.00 | 60.00 |
| Trio | ||||
| NB | 0.00 | 33.33 | 66.67 | 0.00 |
| LGA | 0.00 | 25.00 | 75.00 | 0.00 |
| GBM | 0.00 | 20.00 | 10.00 | 70.00 |
| Vav3 | ||||
| NB | 0.00 | 0.00 | 100.00 | 0.00 |
| LGA | 0.00 | 0.67 | 0.33 | 0.00 |
| GBM | 0.00 | 0.00 | 0.00 | 100.00 |
Table displays percentage of cases at each staining intensity: n = 4 (NB), n = 6 (LGA), n = 10 (GBM).
Figure 3.
Immunohistochemical detection of Trio, Vav3, Ect2, and Rac1 in brain tumor specimens. A: Paraffin-embedded, formalin-fixed sections were immunostained with antibodies against Trio, Vav3, Ect2, or Rac1 and counterstained with hematoxylin. NB (n = 4), LGA (n = 6), and GBM (n = 10). Representative micrographs are shown. B: Semiquantification of immunohistochemical data was performed as described in Materials and Methods. Data represent a weighted score average. C: Higher magnification micrographs showing Rac1 staining in NB, LGA, and GBM. Black arrows point to membrane staining in GBMs. Original magnifications: ×200 (A); ×630 (C).
Whereas Trio and Vav3 primarily localize to the cytoplasm, Ect2 displays both cytoplasmic and nuclear staining. Nuclear Ect2 localization is thought to reflect the GEF in its inactive state.20 Nuclear staining is often more prominent in LGA but is also evident in GBMs (either together with cytoplasmic staining or in a mutually exclusive manner) (Figure 3A). Trio and Vav3, but not Ect2, display significant expression in endothelial cells of the tumor microvasculature (Figure 3A). Notably, whereas Rac1 is predominantly cytoplasmic in LGA, in GBM, Rac1 is both cytoplasmic and often prominently in the plasma membrane (4 of 10 GBMs examined displayed plasma membrane staining) (Figure 3C). Inactive Rac1 resides in the cytoplasm (where it is bound to GDIs), whereas activated Rac1 relocalizes to the plasma membrane.21 Thus the strong plasma membrane staining in a subset of GBMs indicates a high level of Rac activation in these tumors.
siRNA-Mediated Depletion of Ect2, Trio, or Vav3 Suppresses Glioblastoma Cell Migration and Invasion
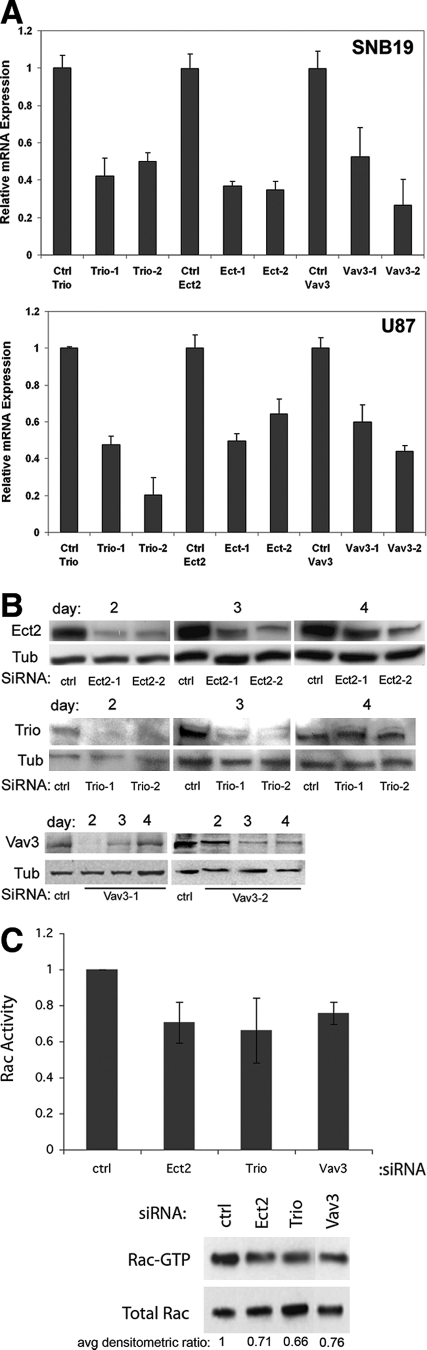
To determine the role of Trio, Ect2, and Vav3 in glioblastoma migration, we inhibited the expression of each GEF by siRNA in glioblastoma cell lines and examined the migratory behavior of the cells on glioma-derived ECM using a two-dimensional radial cell migration assay.16,17 The nontargeting siRNA sequence against the nonmammalian luciferase gene was used as a control. We obtained significant inhibition of GEF expression (40 to 80%) for all siRNA oligos in the two cell lines studied by quantitative RT-PCR (P < 0.05 for all comparisons) and Western blot analysis (Figure 4, A and B). Inhibition was maximal at days 2 to 3 after transfection and gradually declined afterward (Figure 4B). In accordance with siRNA-mediated depletion of Ect2, Trio, or Vav3, Rac activity was also diminished as demonstrated by a Rac-GTP pull-down assay in SNB19 cells (Figure 4C).
Figure 4.
Confirmation of GEF siRNA shutdown. A: Depletion of Ect2, Trio, and Vav3 was confirmed in SNB19 and U87 cells by quantitative RT-PCR. Luciferase siRNA-transfected cells were used as controls (ctrl). Expression levels were normalized first to HPRT and then to controls. Error bars represent SEM. Data shown are representative of two independent experiments. B: Time course of depletion of Ect2, Trio, and Vav3 shown by Western blotting, as detailed in Materials and Methods. Tubulin (Tub) was used as loading control. Western blots are representative of at least two independent experiments. C: Rac-GTP pull-down assay showing inhibition of Rac activity after transfection of Ect2, Trio, or Vav3 siRNA in SNB19 cells. Rac-GTP levels are normalized to total Rac in cells using densitometry. Histogram represents an average of three independent experiments. The average densitometric ratios are shown. In addition, the bars for each GEF represent an average of two different siRNA oligos. All comparisons were statistically significant (P < 0.05).
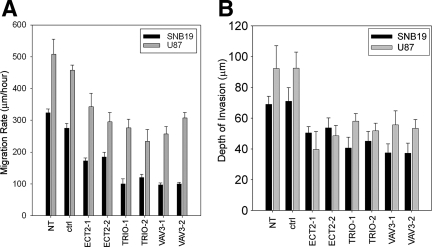
In SNB19 cells, depletion of Ect2 inhibits the migration rate by 30% whereas depletion of Trio or Vav3 inhibits the migration rate by more than 50% (Figure 5A). We obtained similar results in U87 cells (Figure 5A). To examine the role of Trio, Ect2, or Vav3 in glioblastoma cell migration in a more physiological context, we also determined the effect of depleting these GEFs in an ex vivo organotypic rat brain slice model.6,16,19 Depletion of Trio, Ect2, or Vav3 caused a 40 to 50% inhibition in invasion of SNB19 cells and a 25 to 45% inhibition in U87 cell invasion (Figure 5B).
Figure 5.
siRNA-mediated depletion of Ect2, Trio, or Vav3 expression suppresses migration in vitro and invasion ex vivo. Glioblastoma cells were transfected with siRNA targeting either luciferase (ctrl), Ect2, Trio, or Vav3. Cells not transfected (NT) with siRNA were also used as an additional control. A: After 24 hours, cells were plated onto 10-well glass slides precoated with glioma-derived ECM as previously described.17 Cell migration was determined for 24 hours. Data represent the mean ± SEM of three independent experiments. B: Glioblastoma cells stably expressing GFP were transfected with siRNA for 24 hours. Cells were subsequently implanted into the bilateral putamen on rat organotypic brain slices and observed after 48 hours. Depth of invasion was calculated as described in Materials and Methods. Data represent the mean (n = 4) ± SEM of the depth of invasion. Data shown are representative of two independent experiments.
siRNA-Mediated Depletion of Ect2 or Trio Inhibits Glioblastoma Cell Proliferation
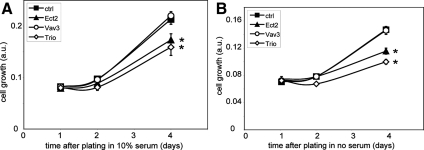
We also examined the potential role of Ect2, Trio, or Vav3 in glioblastoma cell proliferation using the colorimetric SRB assay.15,22 Whereas depletion of Vav3 only has no significant effect on cell proliferation, depletion of either Ect2 or Trio significantly inhibits cell proliferation, both in the presence and absence of serum (Figure 6, A and B). Glioblastoma cell survival however is not compromised by depleting Ect2, Trio, or Vav3, as determined by the live/dead viability assay (data not shown). It is unlikely that the decreased growth rates of cells depleted of Ect2 or Trio significantly contribute to the inhibition in cell migration and invasion, because after replating, cells display a significant lag time before resuming proliferation. Ect2 has been implicated in the control of cytokinesis.23 In line with this, we also observed that Ect2 depletion causes a strong increase in the percentage of multinucleated cells, whereas depletion of either Trio or Vav3 does not induce a multinucleated phenotype (Figure 7, A and B). Multinucleation may contribute to the inhibition in cell proliferation observed in Ect2-depleted cells.
Figure 6.
siRNA-mediated depletion of Ect2 and Trio but not Vav3 decreases the growth rate of SNB19 cells. SNB19 cells were transfected with siRNA directed against either luciferase [control, (▪)], Ect2 (▴), Vav3 (○), or Trio (224) for 1 day, and cultured on 96-well plates in the presence (A) or absence of serum (B). Cell growth was quantified by SRB staining. Data represent the mean ± SEM of four replicates and are representative of two independent experiments. *Significant (P < 0.02, one-tailed t-test) difference between GEF knockdown and control. Scale bar = 25 μm.
Figure 7.
Depletion of Ect2 induces multinucleated cells. A: SNB19 cells were transfected with the respective siRNAs for 2 days and plated on coverslips overnight. F-actin was stained with fluorescein isothiocyanate-phallodin (green) and chromatin was stained with 4,6-diamidino-2-phenylindole (blue). Nuclei in multinucleated cells are indicated by arrowheads. B: Quantification of multinucleated cells was performed as described in Materials and Methods. Data shown are the mean ± SEM from four independent experiments.
Discussion
In this study we used gene expression profiling, quantitative PCR, and immunohistochemistry to identify GEFs that are overexpressed in glioblastoma. We found three RacGEFs, Ect2, Trio, and Vav3, that are overexpressed in glioblastoma compared to LGAs or NB and show that each of these GEFs contributes to the invasive behavior of these tumors. All three GEFs that we identified in this study can act on multiple GTPases. Ect2 can catalyze nucleotide exchange on Rac1, Cdc42, and RhoA.23,24 Trio has two DH domains; one that can act on RhoG and to a lesser extent on Rac1 and the other DH domain can act on RhoA.25,26 Vav3 can act on RhoA, RhoG, and to a lesser extent on Rac1.27 Thus, it is conceivable that in addition to Rac proteins, other Rho family members contribute to glioblastoma invasion by virtue of these respective GEFs. Rho GEFs are thought to mediate the activation of Rho GTPases by various stimuli, including cell adhesion, growth factor receptor activation, and cytokines.28,29 For the vast majority of these GEFs, the distinct stimuli that lead to their activation remain to be identified. It is likely however that Ect2, Trio, and Vav3 mediate downstream effects that are initiated by distinct signals that stimulate glioma migration and invasion.
The best characterized functions of Ect2 are in the regulation of chromosome attachment to spindle microtubules and cytokinesis.23,30,31,32,33 In keeping with this, we observed that depletion of Ect2 causes the formation of large multinucleated glioblastoma cells, which is likely to contribute to the inhibitory effect of Ect2 depletion on the migratory behavior of glioblastoma cells. However, Ect2 also has been shown to interact with the Par6/Par3/atypical protein kinase C polarity complex,34 which has been implicated in astrocyte polarization and migration downstream of Cdc42.35 These considerations suggest that defects in Ect2-regulated cell polarity may also contribute to glioblastoma cell migration.36 In line with our findings, it was recently reported that Ect2 is overexpressed in glioblastoma and correlates with poor prognosis in glioma patients.37 This study also showed that siRNA-mediated depletion of Ect2 inhibits glioblastoma cell invasion and proliferation.
Trio has been shown to play an essential role in neuronal migration and axon guidance in a number of model systems.38 More recently, Trio also has been implicated in the migration of distal tip cells in the nematode Caenorhabditis elegans.39 Interestingly, both in this system and in the phagocytic clearance of apoptotic cells in C. elegans, the action of Trio is mediated by RhoG that in turn activates Rac proteins via the Dock1/ELMO bipartite GEF.40,41 Notably, Dock1/ELMO has also recently been shown to be critical for glioblastoma migration.42 However, the identity of the signaling events that regulate Trio still primarily remains elusive.
There are three mammalian Vav proteins that are structurally highly homologous. Vav1 is predominantly expressed in hematopoietic tissues, whereas Vav2 and Vav3 are broadly expressed.43,44 Vav proteins have several protein-protein interaction domains including a Src homology 2 (SH2) and two Src homology 3 (SH3) domains that can, respectively, interact with a large variety of tyrosine kinase receptors or adaptor proteins.43 A number of the receptors that can recruit Vav proteins, including EGFR, PDGFR, IGF1R, and TrkA are abundantly expressed in malignant glioma,45,46,47,48 providing a potential mechanism for the role of Vav3 in glioblastoma migration and invasion.
Rho proteins have been found to be overexpressed in a number of human tumors.12,49 In our study we show that Rac1 mRNA is not altered in astrocytomas, but that Rac1 protein levels are strongly increased in a grade-dependent manner. These data are consistent with studies showing that Rac1 protein levels are increased in high-grade (85%) versus low-grade (20%) gliomas and correlate with poor survival.50 Thus, our findings support the contention that Rac1 protein levels in glioblastoma are posttranscriptionally regulated, either by an increase in RNA stability, translation efficiency, and/or protein stability.
Importantly, our immunohistochemical analysis demonstrating plasma membrane association of Rac1, suggests that Rac1 is activated in GBM versus LGA. Although the antibody that we used in these immunohistochemistry studies also recognizes Rac2 and Rac3, it is likely that the staining primarily reflects Rac1 protein levels, because Rac2 is primarily expressed in hematopoietic cells and Rac3 levels in glioblastoma cells are typically lower than those of Rac1.5 The activation of Rac1 in GBM is in line with our finding that Ect2, Trio, and Vav3 are overexpressed in these tumors, although it is likely that other events also contribute, including decreased expression of GAP proteins that act on Rac1.51
The overexpression of Trio, Ect2, and Vav3 in GBM tissue and their role in glioblastoma cell invasion suggests that these GEFs present novel potential drug targets for therapeutic intervention. Ultimately, it will be important to carefully compare the expression of these GEFs in the invasive edge and tumor core and these studies are ongoing in our laboratories. There is a burgeoning literature indicating that targeting GEFs by small molecule inhibitors is indeed feasible.52 Whether it will be possible to specifically inhibit a single GEF or a small subset of GEFs remains to be seen however. The window of opportunity for therapeutic intervention in glioblastoma also needs to be carefully assessed given that there is some expression of these GEFs in NB, including peritumoral brain.
Rho GEFs are thought to be primarily regulated in a posttranslational manner, either by phosphorylation or interaction with phosphatidylinositol (PI) lipids and other proteins.11,29 Kinases that can phosphorylate and activate Rho GEFs include EGFR and PDGFR,28 two receptors that are, respectively, amplified or overexpressed during glioma progression.48 The PIP3 phosphatase PTEN is lost in more than 30% of primary GBMs, leading to an increase in plasma membrane PIP3 levels, which in turn can recruit and/or activate Rho GEFs through the PI3K pathway.53,54 Thus, additional GEFs that are not deregulated at the expression level may still be hyperactive in high-grade gliomas. Overall, the knowledge that Rac1 and its regulators are essential for glioma migration and invasion underlines the notion of inhibiting Rac1-mediated signaling as a therapeutic avenue for malignant glioma.
Acknowledgments
We thank Satoko Nakada (Department of Human Pathology, Kanazawa University Graduate School of Medicine, Kanazawa, Japan) for assistance with immunohistochemical analysis, Sidney Croul (University Health Network, University of Toronto, Toronto, Canada) for assistance with the diagnosis of the brain tumor and nonneoplastic brain specimens used in this study, and Richard Mains (University of Connecticut Health Center, Farmington, CT) for the generous gift of anti-Trio antibody.
Footnotes
Address reprint requests to James T. Rutka, M.D., Ph.D., Division of Neurosurgery, Suite 1503, the Hospital for Sick Children, 555 University Ave., Toronto, Ontario Canada M5G 1X8; or Marc Symons, Ph.D., Center for Oncology and Cell Biology, The Feinstein Institute for Medical Research at North Shore-LIJ, 350 Community Dr. Manhasset, NY 11030. E-mail: james.rutka@sickkids.ca and msymons@nshs.edu.
Supported by the Canadian Institutes of Health Research (grant MUP-76410 to J.T.R. and Canada Graduate Schlorship to B.S.); the Ruth L. Kirschstein National Research Service (award F32 CA112986-01 to N.L.T.), the Russell Becker-American Brain Tumor Association (grant to N.L.T.), the National Institutes of Health (grants NS-42262 to M.E.B. and NS-060023 to M.S.), the Project to Cure Foundation, the Little Louie Foundation (to M.S.), Brainchild, the Laurie Berman Fund, and the Wiley Fund (to J.T.R.).
References
- Christofori G. New signals from the invasive front. Nature. 2006;441:444–450. doi: 10.1038/nature04872. [DOI] [PubMed] [Google Scholar]
- Demuth T, Reavie LB, Rennert JL, Nakada M, Nakada S, Hoelzinger DB, Beaudry CE, Henrichs AN, Anderson EM, Berens ME. MAP-ing glioma invasion: mitogen-activated protein kinase kinase 3 and p38 drive glioma invasion and progression and predict patient survival. Mol Cancer Ther. 2007;6:1212–1222. doi: 10.1158/1535-7163.MCT-06-0711. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, Wyckoff J, Condeelis J. Cell migration in tumors. Curr Opin Cell Biol. 2005;17:559–564. doi: 10.1016/j.ceb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- Chan AY, Coniglio SJ, Chuang YY, Michaelson D, Knaus UG, Philips MR, Symons M. Roles of the Rac1 and Rac3 GTPases in human tumor cell invasion. Oncogene. 2005;24:7821–7829. doi: 10.1038/sj.onc.1208909. [DOI] [PubMed] [Google Scholar]
- Chuang YY, Tran NL, Rusk N, Nakada M, Berens ME, Symons M. Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res. 2004;64:8271–8275. doi: 10.1158/0008-5472.CAN-04-2097. [DOI] [PubMed] [Google Scholar]
- Salhia B, Rutten F, Nakada M, Beaudry C, Berens M, Kwan A, Rutka JT. Inhibition of Rho-kinase affects astrocytoma morphology, motility, and invasion through activation of Rac1. Cancer Res. 2005;65:8792–8800. doi: 10.1158/0008-5472.CAN-05-0160. [DOI] [PubMed] [Google Scholar]
- Symons M, Settleman J. Rho family GTPases: more than simple switches. Trends Cell Biol. 2000;10:415–419. doi: 10.1016/s0962-8924(00)01832-8. [DOI] [PubMed] [Google Scholar]
- Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- Lu M, Kinchen JM, Rossman KL, Grimsley C, Hall M, Sondek J, Hengartner MO, Yajnik V, Ravichandran KS. A steric-inhibition model for regulation of nucleotide exchange via the Dock180 family of GEFs. Curr Biol. 2005;15:371–377. doi: 10.1016/j.cub.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- Gómez del Pulgar T, Benitah SA, Valeron PF, Espina C, Lacal JC. Rho GTPase expression in tumourigenesis: evidence for a significant link. Bioessays. 2005;27:602–613. doi: 10.1002/bies.20238. [DOI] [PubMed] [Google Scholar]
- Tran NL, McDonough WS, Savitch BA, Fortin SP, Winkles JA, Symons M, Nakada M, Cunliffe HE, Hostetter G, Hoelzinger DB, Rennert JL, Michaelson JS, Burkly LC, Lipinski CA, Loftus JC, Mariani L, Berens ME. Increased fibroblast growth factor-inducible 14 expression levels promote glioma cell invasion via Rac1 and nuclear factor-{kappa}B and correlate with poor patient outcome. Cancer Res. 2006;66:9535–9542. doi: 10.1158/0008-5472.CAN-06-0418. [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren JT, Bokesch H, Kenney S, Boyd MR. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- Valster A, Tran NL, Nakada M, Berens ME, Chan AY, Symons M. Cell migration and invasion assays. Methods. 2005;37:208–215. doi: 10.1016/j.ymeth.2005.08.001. [DOI] [PubMed] [Google Scholar]
- Berens M, Rief M, Loo M, Giese A. The role of extracellular matrix in human astrocytoma migration and proliferation studied in a microliter scale assay. Clin Exp Metastasis. 1994;12:405–415. doi: 10.1007/BF01755884. [DOI] [PubMed] [Google Scholar]
- Jung S, Ackerley C, Ivanchuk S, Mondal S, Becker LE, Rutka JT. Tracking the invasiveness of human astrocytoma cells by using green fluorescent protein in an organotypical brain slice model. J Neurosurg. 2001;94:80–89. doi: 10.3171/jns.2001.94.1.0080. [DOI] [PubMed] [Google Scholar]
- Jung S, Hinek A, Tsugu A, Hubbard SL, Ackerley C, Becker LE, Rutka JT. Astrocytoma cell interaction with elastin substrates: implications for astrocytoma invasive potential. Glia. 1999;25:179–189. doi: 10.1002/(sici)1098-1136(19990115)25:2<179::aid-glia8>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Saito S, Liu XF, Kamijo K, Raziuddin R, Tatsumoto T, Okamoto I, Chen X, Lee CC, Lorenzi MV, Ohara N, Miki T. Deregulation and mislocalization of the cytokinesis regulator ECT2 activate the Rho signaling pathways leading to malignant transformation. J Biol Chem. 2004;279:7169–7179. doi: 10.1074/jbc.M306725200. [DOI] [PubMed] [Google Scholar]
- Quinn MT, Evans T, Loetterle LR, Jesaitis AJ, Bokoch GM. Translocation of Rac correlates with NADPH oxidase activation. Evidence for equimolar translocation of oxidase components. J Biol Chem. 1993;268:20983–20987. [PubMed] [Google Scholar]
- Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc. 2006;1:1112–1116. doi: 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- Tatsumoto T, Xie X, Blumenthal R, Okamoto I, Miki T. Human ECT2 is an exchange factor for Rho GTPases, phosphorylated in G2/M phases, and involved in cytokinesis. J Cell Biol. 1999;147:921–928. doi: 10.1083/jcb.147.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solski PA, Wilder RS, Rossman KL, Sondek J, Cox AD, Campbell SL, Der CJ. Requirement for C-terminal sequences in regulation of Ect2 guanine nucleotide exchange specificity and transformation. J Biol Chem. 2004;279:25226–25233. doi: 10.1074/jbc.M313792200. [DOI] [PubMed] [Google Scholar]
- Blangy A, Vignal E, Schmidt S, Debant A, Gauthier-Rouviere C, Fort P. TrioGEF1 controls Rac- and Cdc42-dependent cell structures through the direct activation of RhoG. J Cell Sci. 2000;113:729–739. doi: 10.1242/jcs.113.4.729. [DOI] [PubMed] [Google Scholar]
- Debant A, Serra-Pages C, Seipel K, O'Brien S, Tang M, Park SH, Streuli M. The multidomain protein Trio binds the LAR transmembrane tyrosine phosphatase, contains a protein kinase domain, and has separate Rac-specific and Rho-specific guanine nucleotide exchange factor domains. Proc Natl Acad Sci USA. 1996;93:5466–5471. doi: 10.1073/pnas.93.11.5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Movilla N, Dosil M, Zheng Y, Bustelo XR. How Vav proteins discriminate the GTPases Rac1 and RhoA from Cdc42. Oncogene. 2001;20:8057–8065. doi: 10.1038/sj.onc.1205000. [DOI] [PubMed] [Google Scholar]
- Schiller MR. Coupling receptor tyrosine kinases to Rho GTPases—GEFs what’s the link. Cell Signal. 2006;18:1834–1843. doi: 10.1016/j.cellsig.2006.01.022. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Hall A. Guanine nucleotide exchange factors for Rho GTPases: turning on the switch. Genes Dev. 2002;16:1587–1609. doi: 10.1101/gad.1003302. [DOI] [PubMed] [Google Scholar]
- Oceguera-Yanez F, Kimura K, Yasuda S, Higashida C, Kitamura T, Hiraoka Y, Haraguchi T, Narumiya S. Ect2 and MgcRacGAP regulate the activation and function of Cdc42 in mitosis. J Cell Biol. 2005;168:221–232. doi: 10.1083/jcb.200408085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piekny A, Werner M, Glotzer M. Cytokinesis: welcome to the Rho zone. Trends Cell Biol. 2005;15:651–658. doi: 10.1016/j.tcb.2005.10.006. [DOI] [PubMed] [Google Scholar]
- Yüce O, Piekny A, Glotzer M. An ECT2-central spindlin complex regulates the localization and function of RhoA. J Cell Biol. 2005;170:571–582. doi: 10.1083/jcb.200501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S, Tatsumoto T, Lorenzi MV, Chedid M, Kapoor V, Sakata H, Rubin J, Miki T. Rho exchange factor ECT2 is induced by growth factors and regulates cytokinesis through the N-terminal cell cycle regulator-related domains. J Cell Biochem. 2003;90:819–836. doi: 10.1002/jcb.10688. [DOI] [PubMed] [Google Scholar]
- Liu XF, Ishida H, Raziuddin R, Miki T. Nucleotide exchange factor ECT2 interacts with the polarity protein complex Par6/Par3/protein kinase Czeta (PKCzeta) and regulates PKCzeta activity. Mol Cell Biol. 2004;24:6665–6675. doi: 10.1128/MCB.24.15.6665-6675.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etienne-Manneville S, Hall A. Integrin-mediated activation of Cdc42 controls cell polarity in migrating astrocytes through PKCzeta. Cell. 2001;106:489–498. doi: 10.1016/s0092-8674(01)00471-8. [DOI] [PubMed] [Google Scholar]
- Liu XF, Ohno S, Miki T. Nucleotide exchange factor ECT2 regulates epithelial cell polarity. Cell Signal. 2006;18:1604–1615. doi: 10.1016/j.cellsig.2006.01.007. [DOI] [PubMed] [Google Scholar]
- Sano M, Genkai N, Yajima N, Tsuchiya N, Homma J, Tanaka R, Miki T, Yamanaka R. Expression level of ECT2 proto-oncogene correlates with prognosis in glioma patients. Oncol Rep. 2006;16:1093–1098. [PubMed] [Google Scholar]
- Bateman J, Van Vactor D. The Trio family of guanine-nucleotide-exchange factors: regulators of axon guidance. J Cell Sci. 2001;114:1973–1980. doi: 10.1242/jcs.114.11.1973. [DOI] [PubMed] [Google Scholar]
- deBakker CD, Haney LB, Kinchen JM, Grimsley C, Lu M, Klingele D, Hsu PK, Chou BK, Cheng LC, Blangy A, Sondek J, Hengartner MO, Wu YC, Ravichandran KS. Phagocytosis of apoptotic cells is regulated by a UNC-73/TRIO-MIG-2/RhoG signaling module and armadillo repeats of CED-12/ELMO. Curr Biol. 2004;14:2208–2216. doi: 10.1016/j.cub.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Henson PM. Engulfment: ingestion and migration with Rac, Rho and TRIO. Curr Biol. 2005;15:R29–R30. doi: 10.1016/j.cub.2004.12.017. [DOI] [PubMed] [Google Scholar]
- Lundquist EA, Reddien PW, Hartwieg E, Horvitz HR, Bargmann CI. Three C. elegans Rac proteins and several alternative Rac regulators control axon guidance, cell migration and apoptotic cell phagocytosis. Development. 2001;128:4475–4488. doi: 10.1242/dev.128.22.4475. [DOI] [PubMed] [Google Scholar]
- Jarzynka MJ, Hu B, Hui KM, Bar-Joseph I, Gu W, Hirose T, Haney LB, Ravichandran KS, Nishikawa R, Cheng SY. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo XR. Regulatory and signaling properties of the Vav family. Mol Cell Biol. 2000;20:1461–1477. doi: 10.1128/mcb.20.5.1461-1477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo XR. Vav proteins, adaptors and cell signaling. Oncogene. 2001;20:6372–6381. doi: 10.1038/sj.onc.1204780. [DOI] [PubMed] [Google Scholar]
- Akbasak A, Sunar-Akbasak B. Oncogenes: cause or consequence in the development of glial tumors. J Neurol Sci. 1992;111:119–133. doi: 10.1016/0022-510x(92)90060-x. [DOI] [PubMed] [Google Scholar]
- Louis DN, Gusella JF. A tiger behind many doors: multiple genetic pathways to malignant glioma. Trends Genet. 1995;11:412–415. doi: 10.1016/s0168-9525(00)89125-8. [DOI] [PubMed] [Google Scholar]
- Louis DN, Seizinger BR. Genetic basis of neurological tumours. Baillieres Clin Neurol. 1994;3:335–352. [PubMed] [Google Scholar]
- Zhu Y, Parada LF. The molecular and genetic basis of neurological tumours. Nat Rev Cancer. 2002;2:616–626. doi: 10.1038/nrc866. [DOI] [PubMed] [Google Scholar]
- Fritz G, Just I, Kaina B. Rho GTPases are over-expressed in human tumors. Int J Cancer. 1999;81:682–687. doi: 10.1002/(sici)1097-0215(19990531)81:5<682::aid-ijc2>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Iwadate Y, Sakaida T, Saegusa T, Hiwasa T, Takiguchi M, Fujimoto S, Yamaura A. Proteome-based identification of molecular markers predicting chemosensitivity to each category of anticancer agents in human gliomas. Int J Oncol. 2005;26:993–998. [PubMed] [Google Scholar]
- Yuan S, Miller DW, Barnett GH, Hahn JF, Williams BR. Identification and characterization of human beta 2-chimaerin: association with malignant transformation in astrocytoma. Cancer Res. 1995;55:3456–3461. [PubMed] [Google Scholar]
- Bos JL, Rehmann H, Wittinghofer A. GEFs and GAPs: critical elements in the control of small G proteins. Cell. 2007;129:865–877. doi: 10.1016/j.cell.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Fleming IN, Batty IH, Prescott AR, Gray A, Kular GS, Stewart H, Downes CP. Inositol phospholipids regulate the guanine-nucleotide-exchange factor Tiam1 by facilitating its binding to the plasma membrane and regulating GDP/GTP exchange on Rac1. Biochem J. 2004;382:857–865. doi: 10.1042/BJ20040916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Luby-Phelps K, Das B, Shu X, Xia Y, Mosteller RD, Krishna UM, Falck JR, White MA, Broek D. Role of substrates and products of PI 3-kinase in regulating activation of Rac-related guanosine triphosphatases by Vav. Science. 1998;279:558–560. doi: 10.1126/science.279.5350.558. [DOI] [PubMed] [Google Scholar]