Abstract
Context: Genetic polymorphisms at the perilipin (PLIN) locus have been investigated for their potential utility as markers for obesity and metabolic syndrome (MS). We examined in obese children and adolescents (OCA) aged 7–14 yr the association of single-nucleotide polymorphisms (SNP) at the PLIN locus with anthropometric, metabolic traits, and weight loss after 20-wk multidisciplinary behavioral and nutritional treatment without medication.
Design: A total of 234 OCA [body mass index (BMI = 30.4 ± 4.4 kg/m2; BMI Z-score = 2.31 ± 0.4) were evaluated at baseline and after intervention. We genotyped four SNPs (PLIN1 6209T→C, PLIN4 11482G→A, PLIN5 13041A→G, and PLIN6 14995A→T).
Results: Allele frequencies were similar to other populations, PLIN1 and PLIN4 were in linkage disequilibrium (D′ = 0.999; P < 0.001). At baseline, no anthropometric differences were observed, but minor allele A at PLIN4 was associated with higher triglycerides (111 ± 49 vs. 94 ± 42 mg/dl; P = 0.003), lower high-density lipoprotein cholesterol (40 ± 9 vs. 44 ± 10 mg/dl; P = 0.003) and higher homeostasis model assessment for insulin resistance (4.0 ± 2.3 vs. 3.5 ± 2.1; P = 0.015). Minor allele A at PLIN4 was associated with MS risk (age and sex adjusted) hazard ratio 2.4 (95% confidence interval = 1.1–4.9) for genotype GA and 3.5 (95% confidence interval = 1.2–9.9) for AA. After intervention, subjects carrying minor allele T at PLIN6 had increased weight loss (3.3 ± 3.7 vs. 1.9 ± 3.4 kg; P = 0.002) and increased loss of the BMI Z-score (0.23 ± 0.18 vs. 0.18 ± 0.15; P = 0.003). Due to group size, risk of by-chance findings cannot be excluded.
Conclusion: The minor A allele at PLIN4 was associated with higher risk of MS at baseline, whereas the PLIN6 SNP was associated with better weight loss, suggesting that these polymorphisms may predict outcome strategies based on multidisciplinary treatment for OCA.
Perilipin single nucleotide polymorphisms are associated with higher risk of metabolic syndrome (A carriers at PLIN4 11482GA) and better weight-loss during lifestyle intervention (T carriers at PLIN6 14995AT) on obese children and adolescents.
The worldwide increasing prevalence of obesity in children and adolescents threatens to affect both their current social integration and their future adult health (1,2,3) and underscores the urgency to find efficient and successful preventive and therapeutic measures (4). The basis for this increase in obesity has been attributed to the current obesogenic environment; however, genetic factors (5) play a significant role in both the predisposition to gain as well as the resistance to lose excess body weight.
In Brazil, the prevalence of overweight experienced a 3-fold increase in children and adolescents in the period from 1974–1997, especially in urban areas and at the higher end of social class (6). Recently, the Brazilian Consumer Expenditure Survey (2002–2003) estimated that 16.7% of young people (10–19 yr) were affected by overweight, and 2.3% were affected by obesity (7), with the highest prevalence (22.4%) being detected among individuals aged 10–11 yr.
Perilipins are proteins localized at the surface of the lipid droplet in adipocytes, steroid-producing cells, and ruptured atherosclerotic plaques, and they play a key role in the cellular regulation of triglyceride deposition and mobilization (8). Perilipin facilitates the formation and storage of lipids (inhibiting lipolysis) in droplets as well as the increase in lipolysis upon stimulation with catecholamines. Perilipin knockout mouse models are resistant to diet-induced obesity, display reduced body fat, and paradoxically, develop an increased risk of glucose intolerance and peripheral insulin resistance (9). In obese women (compared with normal-weight women), basal and noradrenalin-induced rates of lipolysis were found to be 2- to 4-fold increased, whereas the perilipin content of adipocytes decreased by 50% (10).
In humans, the perilipin gene (PLIN) has been localized to chromosomal location 15q26.1 (11), within a region previously linked to obesity, hypertriglyceridemia, and diabetes (12,13). We have shown that polymorphisms in the PLIN locus were associated with obesity-related phenotypes in White, Malay, and Indian women but not in Chinese women (11,12), and none were found on a large group of French Caucasian men and women (13). Moreover the PLIN4 11482 G→A was associated with weight-loss resistance in an obese population (14). Furthermore, in patients with type 2 diabetes treated with rosiglitazone, the variant A of PLIN4 showed less increase in body weight (15). Most probably, none of the single-nucleotide polymorphisms (SNPs) examined are functional, but they may be in linkage disequilibrium (LD) with so far undetected SNPs. Taken together, the experimental and observational data support the hypothesis that PLIN plays an important role in the pathogenesis of obesity and may be involved in human lipid metabolism (16,17).
In view of the current evidence in adult populations, we investigated whether the PLIN locus was associated with anthropometric and metabolic measures and weight reduction in response to a 20-wk behavioral and nutritional recommendation treatment in a group of Brazilian obese children and adolescents (OCA).
Patients and Methods
Patients and study design
Study subjects consisted of 335 OCA aged from 7–14 yr old participating in a 20-wk lifestyle and nutritional reeducation weight-loss program with balanced diet and without weight-loss medication supported by nutritionists, physical therapists, psychologists, and endocrinologists. Of those, 75 didn’t conclude the program (22.4%), and 26 were excluded (missing results for laboratory, anthropometric, or genetic measures). Thus, statistical analyses were based on 234 OCA [78 boys and 156 girls, aged 10.7 ± 1.3 yr; body mass index (BMI) = 30.4 ± 4.5 kg/m2, BMI Z-score = 2.31 ± 0.28; 49% pubescent] randomly selected among those referred to the Children Obesity’s Outpatient Clinic of the Hospital das Clínicas of the University of São Paulo Medical School.
This study included only children and adolescents with a BMI greater than the 95th percentile charts (by age and gender) from Centers for Disease Control growth charts (18). Children with diagnosed endocrine or genetic disease associated with or prone to obesity were excluded from the study. The study was approved by the Institutional Ethical Committee. The patients and their parents were given written and oral complete information about the research project, its purposes, procedures, and potential risks and had signed the consent forms.
Multidisciplinary clinical intervention
Subjects visited monthly the outpatient clinic and had a nutritional and eating behavior group meeting, a personalized nutritional orientation, a personal diet survey, a meeting with a psychologist, a group meeting (six to 10 participants per group) with a physical educator, and a consultation with an endocrinologist. Whenever necessary, the children were invited to consult the clinic psychiatrist. The nutritional course was oriented on recommendations of food-based dietary guidelines considering the dietary habits of children and families, with an average of 1800 kcal/d and no restrictive but balanced diet education program.
Dietary evaluation was based on a 3-d food intake record. In the first visit at the clinic, patients received dietary questionnaires to be filled out at home for three different days, including one weekend day. Patients were responsible for completing the food record, with their parent’s supervision. The nutrition team provided appropriate information about portion sizes using graphic examples. The notes were reviewed during treatment to correct possible errors, thereby vetting the information for use to estimate the consumption of the children using the Virtual Nutri version 1.0 for Windows software.
Anthropometric and laboratory measurements
Familial medical history was collected by questionnaire. Weight and height of patients were measured to the nearest 0.1 cm and 0.1 kg, respectively, to establish BMI and BMI Z-score according to the curves of the Centers for Disease Control. Waist circumference was measured at the umbilicus with a nonstretchable tape. Pubertal developmental stage was determined according to the method of Marshall and Tanner (19,20). Body composition was analyzed by bioelectrical impedance analysis method with body composition analyzer equipment (RJL Systems, Inc., Clinton Township, MI), using an adequately validated formula for age, weight, and height to estimate lean and fat mass (21). Sitting blood pressure was obtained using the right arm after the subject had rested quietly for at least 5 min.
To evaluate the metabolic profile, blood samples were taken via antecubital vein catheter for the measurement of glucose, insulin, total cholesterol (TC), high-density lipoprotein (HDL)-C, low-density lipoprotein (LDL)-C, very-low-density lipoprotein (VLDL)-C, and triglyceride (TG) at fasting. Glucose was determined by an automatic enzymatic colorimetric method using hexokinase (Cobas Integra; Roche, Basel, Switzerland). Insulin was determined by Auto Delfia fluoroimmunoassay (Perkin-Elmer, Turku, Finland). TC, HDL-C, and TG were analyzed by automatic enzymatic colorimetric method (Cobas Mira; F. Hoffmann-La Roche, Basel, Switzerland) using commercial kits from Roche (Mannheim, Germany). LDL-C and VLDL-C were determined by Friedewald formula VLDL-C = TG/5 and LDL-C = TC − (HDL-C + VLDL-C). The homeostasis model assessment for insulin resistance (HOMA-IR) was calculated using the following formula: insulin resistance = insulin (μU/ml) × glucose (mmol/liter)/22.5. Oral glucose tolerance test (OGTT) was performed by giving 1.75 g/kg up to a maximum of 75 g glucose solution and blood withdrawn at 0, 30, 60, 90, and 120 min afterward for the measurement of plasma glucose and insulin. Leptin and adiponectin were evaluated through ELISA kits (Linco Research, St. Louis, MO).
Metabolic syndrome (MS) was defined using the International Diabetes Foundation (IDF) criteria (22), which for the 10- to 16-yr-old age group included obesity at 90th percentile or higher (or adult cutoff if lower), assessed by waist circumference, and then two of the following: triglycerides at least 150 mg/dl, HDL-C higher than 40 mg/dl, blood pressure at least 130 mm Hg systolic or at least 85 mm Hg diastolic, plasma glucose at least 100 mg/dl (OGTT recommended), or known type 2 diabetes mellitus. We applied these criteria to all children of the study.
PLIN genotyping
The genotyping study was carried out in the Nutrition and Genomics Laboratory, at the Jean Mayer U.S. Department of Agriculture Human Nutrition Research Center on Aging. DNA extraction was performed by standard procedures. Four polymorphisms at the PLIN locus were genotyped: PLIN1 6209T→C (rs2289487, intron 2), PLIN4 11482G→A (rs894160, intron 6), PLIN5 13041A→G (rs2304795, exon 8, synonymous), and PLIN6 14995A→T (rs1052700, exon 9, untranslated region). Genotyping was carried out using the 5′-nuclease allelic discrimination TaqMan assay with ABI 7900HT system (Applied Biosystems, Foster City, CA). Standard good laboratory practices were undertaken to assure the accuracy of genotype data. Internal controls and repetitive experiments were used. Final success rate for PLIN genotyping in the study participants was 99%. This study included OCA randomly selected among those attending the Childhood Obesity’s Outpatient Unit in the city of São Paulo, Brazil. The Brazilian population is a very mixed one when considering genetics (23), and because our work is an intervention study comparing the same cohort, we didn’t consider evaluating admixture.
Statistical analysis
χ2 tests were used to verify differences between observed and expected frequencies, assuming Hardy-Weinberg equilibrium, to test LD and differences in percentages. Pairwise LD coefficients were estimated by the HELIX TREE program. D and D′ coefficients were calculated. Normal distribution for all continuous variables was tested, and some were logarithmically transformed. At baseline, independent t test for independent groups was applied to compare crude means between genotypes. In addition, to estimate and compare adjusted means, analysis of covariance was used to test the null hypotheses of no association between genetic variants and obesity-related phenotypes. For haplotypes, ANOVA and Turkey test were performed. When interactions showed a P value <0.05, post hoc analyses with Turkey tests were performed. Using the χ2 test, we investigated association between assessed categorical variables. Logistic regression models were fitted to estimate the hazard ratio with 95% confidence interval. Finally, paired-sample t tests were performed to compare variables at baseline and after intervention. All tests were conducted with an α = 0.05 and values of P < 0.05 were considered significant. Statistical analyses were performed using SPSS software. The power of the sample size was calculated through MINITAB for windows and was evaluated at 85%. The size of the group is small, and the risk of by chance findings cannot be excluded for some of the variables examined.
Results
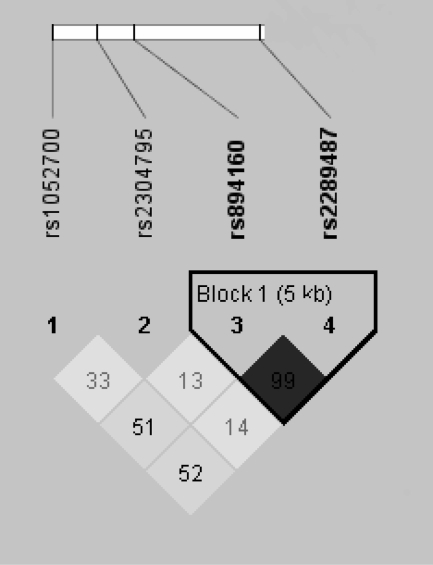
In the current study, we did not detect gender heterogeneity in the results. Therefore, data for boys and girls were analyzed together. The frequencies for the less common allele of the PLIN1, PLIN4, PLIN5, and PLIN6 polymorphisms were 0.48, 0.30, 0.38, and 0.26, respectively. Genotype distributions did not deviate from Hardy-Weinberg expectations (supplemental Table 1, published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org). Figure 1 shows the LD plot (D′) for the SNPs examined. Strong pairwise LD was observed between the PLIN1 and the PLIN4 polymorphisms (D′ = 0.99; P < 0.001). Conversely, D′ for PLIN5 and PLIN6 are much lower, and in addition, these SNPs are part of the DNA Block that includes the 3′ of the PLIN locus. Similar metabolic and anthropometric results were observed between PLIN1 and PLIN4 SNP groups due to their LD, and PLIN5 SNPs did not reveal any significant associations; therefore, we limited our study to PLIN4 and PLIN6.
Figure 1.
Pairwise (D′) plots for representative regions of PLIN gene for the four SNPs studied: PLIN1 (rs2289487), PLIN4 (rs894160), PLIN5 (rs2304795), and PLIN6 (rs1052700). Each square in the triangle plots the level of LD between a pair of sites; comparisons between neighboring sites lie along the diagonal. Darker shade indicates strong LD; lighter indicates weak LD.
Association between PLIN SNPs and baseline data
The basal anthropometric, biochemical, and clinical characteristics by genotype are shown in Table 1. No significant allele-related differences in metabolic measures were observed at baseline for PLIN6 SNPs.
Table 1.
Anthropometric and metabolic parameters at baseline and after 20 wk intervention according to PLIN4 and PLIN6 variants
|
PLIN4 11482G→A
|
PLIN6 14995A→T
|
|||||
|---|---|---|---|---|---|---|
| GG (n = 116) | GA+AA (n = 118) | P | AA (n = 134) | AT+TT (n = 100) | P | |
| Baseline | ||||||
| Age (yr) | 10.7 ± 1.3 | 10.6 ± 1.3 | 0.91 | 10.7 ± 1.4 | 10.6 ± 1.2 | 0.83 |
| Weight (kg) | 66.8 ± 14.1 | 69.8 ± 16.5 | 0.18 | 69.1 ± 16.3 | 67.3 ± 14.1 | 0.43 |
| BMI (kg/m2) | 30.1 ± 3.9 | 30.6 ± 5.0 | 0.53 | 30.7 ± 4.7 | 30.0 ± 4.1 | 0.28 |
| BMI Z-score | 2.30 ± 0.3 | 2.32 ± 0.3 | 0.88 | 2.32 ± 0.3 | 2.29 ± 0.3 | 0.46 |
| Waist circumference (cm) | 95.7 ± 9.5 | 97.1 ± 11.7 | 0.39 | 97.1 ± 11.4 | 95.6 ± 9.6 | 0.33 |
| Blood pressure systolic (mm Hg) | 111 ± 13 | 113 ± 16 | 0.42 | 112 ± 14 | 112 ± 15 | 0.99 |
| Blood pressure diastolic (mm Hg) | 70 ± 12 | 70 ± 12 | 0.80 | 70 ± 12 | 71 ± 11 | 0.35 |
| Fasting glycemia (mg/dl) | 85 ± 7 | 86 ± 8 | 0.16 | 85 ± 8 | 86 ± 7 | 0.37 |
| Fasting insulin (μU/ml) | 16.5 ± 9.8 | 18.6 ± 9.8 | 0.03a | 18.3 ± 10.3 | 16.5 ± 9.1 | 0.20 |
| HOMA-IR | 3.5 ± 2.1 | 4.0 ± 2.3 | 0.02a | 3.9 ± 2.2 | 3.6 ± 2.2 | 0.21 |
| TC (mg/dl) | 158.3 ± 31.9 | 159.5 ± 32.0 | 0.75 | 159.4 ± 34.1 | 158.3 ± 28.7 | 0.98 |
| HDL-C (mg/dl) | 43.6 ± 10.1 | 40.0 ± 9.0 | 0.003a | 42.4 ± 9.9 | 40.9 ± 9.4 | 0.27 |
| LDL-C (mg/dl) | 95.6 ± 28.1 | 97.5 ± 29.3 | 0.64 | 96.3 ± 29.9 | 97.0 ± 27.0 | 0.76 |
| TG (mg/dl) | 93.9 ± 42.5 | 110.7 ± 49.1 | 0.003a | 102.9 ± 46.9 | 101.6 ± 46.5 | 0.98 |
| Diet (kcal/d) | 1951 ± 648 | 2001 ± 672 | 0.69 | 1946 ± 671 | 2031 ± 639 | 0.51 |
| After intervention (20 wk) | ||||||
| Weight (kg) | 64.5 ± 14.5 | 67.1 ± 15.7 | 0.19 | 67.2 ± 16.2 | 64.0 ± 13.4 | 0.43 |
| Weight loss (kg) | 2.4 ± 3.7 | 2.7 ± 3.6 | 0.51 | 1.9 ± 3.5 | 3.3 ± 3.7 | 0.00a |
| BMI (kg/m2) | 28.3 ± 4.3 | 28.8 ± 4.8 | 0.51 | 29.1 ± 4.9 | 27.8 ± 3.9 | 0.04a |
| BMI Z-score | 2.10 ± 0.3 | 2.12 ± 0.4 | 0.65 | 2.14 ± 0.4 | 2.06 ± 0.3 | 0.11 |
| BMI loss Z-score | 0.21 ± 0.2 | 0.19 ± 0.1 | 0.43 | 0.18 ± 0.1 | 0.23 ± 0.2 | 0.01a |
| Waist circumference (cm) | 93.1 ± 9.5 | 94.5 ± 10.6 | 0.51 | 95.5 ± 10.5 | 91.4 ± 9.0 | 0.05a |
| Fasting glycemia (mg/dl) | 80 ± 7 | 80 ± 8 | 0.75 | 80 ± 8 | 80 ± 7 | 0.56 |
| Fasting insulin (μU/ml) | 14.1 ± 7.5 | 15.0 ± 8.0 | 0.51 | 15.5 ± 7.8 | 13.2 ± 7.6 | 0.02a |
| HOMA-IR | 2.7 ± 1.4 | 3.0 ± 1.7 | 0.43 | 3.1 ± 1.5 | 2.6 ± 1.6 | 0.01a |
| TC (mg/dl) | 156.9 ± 29.0 | 158.7 ± 33.2 | 0.79 | 156.6 ± 32.6 | 159.3 ± 29.1 | 0.45 |
| HDL-C (mg/dl) | 47.0 ± 10.7 | 43.6 ± 9.6 | 0.02a | 45.6 ± 10.4 | 44.8 ± 10.1 | 0.64 |
| LDL-C (mg/dl) | 90.4 ± 27.1 | 94.6 ± 31.0 | 0.35 | 91.2 ± 30.7 | 94.3 ± 26.9 | 0.31 |
| TG (mg/dl) | 98.7 ± 46.2 | 102.4 ± 52.1 | 0.71 | 100.2 ± 53.9 | 100.9 ± 42.2 | 0.60 |
| Diet (kcal/d) | 1637 ± 543 | 1493 ± 549 | 0.21 | 1618 ± 551 | 1474 ± 540 | 0.25 |
n = 234.
P < 0.05, independent t test.
However, there were significant baseline differences for PLIN4 with higher TG (P = 0.003) and lower HDL-C (P = 0.003) for the minor A allele. We also observed that the minor A allele of PLIN4 was associated with higher insulin concentrations (P = 0.034) and HOMA-IR measures (P = 0.015). No significant associations were observed between these two SNPs and glucose, TC, leptin, and adiponectin concentrations or for body composition results. None of the PLIN SNPs showed any association with baseline daily caloric intake (Table 1).
Using the IDF criteria in children, we observed that 44 (19%) of the 232 children and adolescents were classified as having MS (two didn’t have all needed measurements available for classification) (Table 2).
Table 2.
Prevalence of MS per polymorphisms PLIN4 and PLIN6
| MS (IDF criteria) (n = 232) |
PLIN4 11482 G→A
|
PLIN6 14995 A→T
|
||
|---|---|---|---|---|
| GG (%) | GA+AA (%) | AA (%) | AT+TT (%) | |
| Waist circumference ≥90th percentile | 99.1 | 99.1 | 99.2 | 99.0 |
| Blood pressure ≥130 SBP or ≥85 DBP (mm Hg) | 12.6 | 22.8 | 18.0 | 17.5 |
| Glucose ≥100 mg/dl | 2.6 | 4.3 | 3.8 | 3.0 |
| HDL <40 mg/dl | 40.5 | 51.7 | 43.3 | 50.0 |
| TG ≥150 mg/dl | 10.3 | 17.8 | 16.4 | 11.0 |
| Total (n = 44, 19% have MS) | 12.1 | 25.9 | 18.2 | 20.0 |
DBP, Diastolic blood pressure; IDF, International Diabetes Federation; SBP, systolic blood pressure.
At baseline, subjects carrying the minor A allele at PLIN4 had a significantly higher prevalence of MS (25.9%) compared with 12.1% for the GG genotype.
Hazard ratios for the presence of MS using as reference the wild-type GG for PLIN4 11482G→A was evaluated using logistic regression after adjusting for sex, age, and pubertal stage. The MS hazard ratio for the genotype GA was 2.38 (95% confidence interval = 1.1–4.9) and for AA 3.46 (95% confidence interval = 1.2–9.9). There was no difference in MS prevalence for SNP PLIN6. These results were confirmed on the whole group at baseline when including children who didn’t finish the intervention.
OGTT
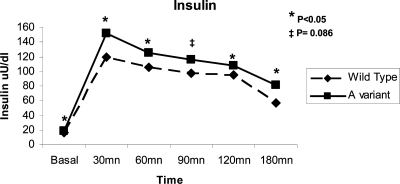
An OGTT was performed in 229 of the 234 obese children. We didn’t observe allele-related differences for glucose; however, the average area under the curve for insulin was significantly higher for carriers of the minor A allele at PLIN4 (P = 0.005). Specifically, A allele carriers had significant or borderline significantly higher insulin levels at baseline (P = 0.066) and 30 (P = 0.011), 60 (P = 0.026), 90 (P = 0.086), and 120 (P = 0.035) min after the glucose load (Fig. 2). No significant difference for the insulin response was observed for PLIN6.
Figure 2.
Line plots of insulin response to OGTT according to PLIN4 polymorphism: genotype GG compared with carriers of one or two A allele at PLIN4.
Associations between PLIN and relevant traits after weight-loss intervention
After 20 wk clinical intervention, 226 children and adolescents (96.6%) lowered their BMI Z-score (−0.20 ± 0.16) and their body weight (mean −2.53 kg). Simultaneously, a number of metabolic variables experienced significant improvement, including lowering of insulin and HOMA-IR and increase of HDL in the entire group as well as when considering groups separated per genotype (P < 0.0001). We didn’t find any allele-related differences for daily caloric intake evaluated at the end of the intervention period (Table 1). Despite this, carriers of the minor T PLIN6 allele showed a significantly greater weight loss (P = 0.003) and loss of BMI Z-score (P = 0.014), accompanied by a significant lower waist circumference (P = 0.05), lower fasting insulin (P = 0.02), and lower HOMA-IR (P = 0.012) compared with wild type. Carriers of the A allele at PLIN4 maintained significantly lower HDL-C levels (P = 0.005) when comparing with wild type and had a significant decrease in TG concentrations (P = 0.022).
Haplotype analysis
Two different patterns were found associated with these PLIN SNPs in this obese population. Carriers of the minor allele PLIN4 had a higher-risk metabolic status. Conversely, the SNP PLIN6 was not associated with a different metabolic status but to a better response to weight loss during the multidisciplinary program. To gain further insight, we divided the group of 234 OCA in four haplotypes groups using the two SNPs PLIN4 11482G→A and PLIN6 14995A→T. The four groups are 1.1 (GA) (35.9%), 1.2 (GT) (13.7%), 2.1 (AA) (21.4%), and 2.2 (AT) (29%) (1 codes the common and 2 codes the presence of minor allele). The anthropometric, biochemical, and clinical characteristics of the study subjects according to the haplotypes are presented in Table 3 for baseline and after intervention.
Table 3.
Anthropometric and metabolic parameters (baseline and after intervention) for haplotypes PLIN4 11482G→A/PLIN6 14995A→T
| Haplotype PLIN4/PLIN6 | GA 1.1 (n = 84, 35.9%) | GT 1.2 (n = 32, 13.7%) | AA 2.1 (n = 50, 21.4%) | AT 2.2 (n = 68, 29%) |
|---|---|---|---|---|
| Baseline | ||||
| Age (yr) | 10.7 ± 1.3 | 10.5 ± 1.4 | 10.6 ± 1.5 | 10.7 ± 1.2 |
| Weight (kg) | 68 ± 14.6 | 63.6 ± 12.2 | 70.9 ± 18.7 | 69 ± 14.7 |
| BMI (kg/m2) | 30.5 ± 4.1 | 29.3 ± 3.2 | 31.1 ± 5.7 | 30.3 ± 4.4 |
| BMI Z-score | 2.31 ± 0.3 | 2.28 ± 0.2 | 2.34 ± 0.3 | 2.30 ± 0.3 |
| Waist circumference (cm) | 96.7 ± 9.7 | 93.2 ± 8.3 | 97.8 ± 13.8 | 96.7 ± 10.1 |
| Blood pressure systolic (mm Hg) | 111 ± 13 | 112 ± 13 | 114 ± 16 | 112 ± 16 |
| Blood pressure diastolic (mm Hg) | 70 ± 13.0 | 72 ± 10 | 70 ± 11 | 70 ± 12 |
| Fasting glycemia (μU/dl) | 84 ± 7.4 | 87 ± 6.7 | 87 ± 7.5 | 86 ± 7.6 |
| Fasting insulin (μU/dl) | 17.8 ± 10.5 | 13.3 ± 6.7 | 19.2 ± 10.1 | 18.1 ± 9.7 |
| HOMA-IR | 3.73 ± 2.2 | 2.85 ± 1.5 | 4.15 ± 2.0 | 3.90 ± 2.4 |
| TC (mg/dl) | 158.5 ± 33.4 | 157.9 ± 28 | 160.9 ± 35.7 | 158.5 ± 29.3 |
| HDL-C (mg/dl) | 43.55 ± 10.3 | 43.66 ± 9.8 | 40.40 ± 9.0 | 39.66 ± 9.0 |
| LDL-C (mg/dl) | 94.83 ± 28.9 | 97.75 ± 26.2 | 98.72 ± 31.7 | 96.57 ± 27.5 |
| TG (mg/dl) | 98.45 ± 46.2 | 81.81 ± 28.2 | 110.38 ± 47.6 | 110.96 ± 50.5 |
| Diet (kcal/d) | 1935 ± 587 | 2024 ± 923 | 1965 ± 811 | 2033 ± 539 |
| After intervention (20 wk) | ||||
| Weight (kg) | 66.2 ± 14.7 | 60 ± 13.0 | 68.9 ± 18.5 | 65.8 ± 13.3 |
| Weight loss (kg) | 1.90 ± 3.7 | 3.59 ± 3.6 | 1.98 ± 3.1 | 3.20 ± 3.8 |
| BMI (kg/m2) | 28.9 ± 4.4 | 26.9 ± 3.6 | 29.4 ± 5.6 | 28.3 ± 4.0 |
| BMI Z-score | 2.13 ± 0.3 | 2.00 ± 0.3 | 2.16 ± 0.4 | 2.10 ± 0.4 |
| BMI loss Z-score | 0.18 ± 0.1 | 0.29 ± 0.2 | 0.18 ± 0.1 | 0.20 ± 0.1 |
| Waist circumference (cm) | 94.2 ± 9.0 | 88.8 ± 10.9 | 98.4 ± 12.9 | 92.3 ± 8.4 |
| Fasting glycemia (μU/dl) | 80 ± 7.4 | 81 ± 7.4 | 82 ± 7.9 | 80 ± 7.4 |
| Fasting insulin (μU/dl) | 14.4 ± 7.3 | 12.9 ± 8.1 | 17.2 ± 8.4 | 13.3 ± 7.4 |
| HOMA-IR | 2.80 ± 1.3 | 2.42 ± 1.5 | 3.49 ± 1.8 | 2.64 ± 1.6 |
| TC (mg/dl) | 156.2 ± 29.4 | 158.8 ± 28.5 | 157.5 ± 38 | 159.5 ± 29.6 |
| HDL-C (mg/dl) | 47.21 ± 10.4 | 46.50 ± 11.5 | 42.91 ± 10.0 | 44.05 ± 9.3 |
| LDL-C (mg/dl) | 89.53 ± 27.7 | 92.68 ± 26.0 | 93.98 ± 35.5 | 95.03 ± 27.5 |
| TG (mg/dl) | 98.85 ± 47.0 | 98.25 ± 44.8 | 102.63 ± 64.6 | 102.20 ± 41.2 |
| Diet (kcal/d) | 1664 ± 585 | 1551 ± 406 | 1550 ± 505.5 | 1439 ± 597 |
At baseline
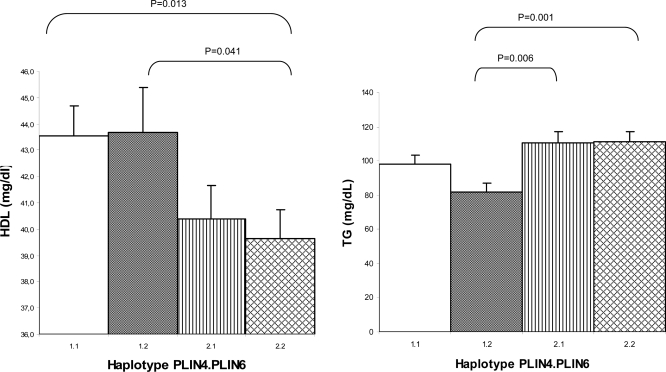
We didn’t observe significant differences either for anthropometric measures or for daily caloric ingestion between the four groups. Statistically significant differences were observed for insulin, HOMA-IR, HDL-C (Fig. 3), and TG (Fig. 3).
Figure 3.
Mean measures for HDL and TG at baseline for haplotypes PLIN4 11482G→A/PLIN6 14995A→T: haplotypes GA = 1.1, GT = 1.2, AA = 2.1, and AT = 2.2. Independent t test, P < 0.05.
The 1.2 haplotype (carrying exclusively the T allele for PLIN6) showed a significantly lower HOMA-IR than the three other groups: 1.1 (P = 0.045), 2.1 (P = 0.001), and 2.2 (P = 0.009).
The highest prevalence of MS was 25% for haplotypes 2.1 (AA) and 26.5% for 2.2 (AT), both carrying the minor allele A for PLIN4. The two other groups showed a lower prevalence for MS: 14.3% for 1.1 (GA) and a remarkably low 6.3% for 1.2 (GT). The high prevalence of MS of group 2.1 (carrying exclusively the A allele of PLIN4) was due to hypertriglyceridemia (20% had TG ≥150 mg/dl) and hypertension (27.7%). We also observed in this group 2.1 a higher prevalence of glucose ≥100 mg/dl, which was doubled compared with the other three groups. We observed that no children from the 1.2 (GT) haplotype had TG levels of 150 mg/dl or higher.
After intervention (Table 3)
The 1.2 haplotype had the best response to weight-loss intervention (loss of BMI Z-score 0.29 ± 0.2 and mean loss of weight 3.59 kg). The 1.2 group had a significantly better loss of BMI Z-score than the three other groups: 1.1 (P = 0.009), 2.1 (P = 0.011), and 2.2 (P = 0.038).
The haplotype 2.1 remained with a significantly higher HOMA-IR than the three other groups: 1.1 (P = 0.035), 1.2 (P = 0.016), and 2.2 (P = 0.018).
Discussion
Our data show that in OCA, the PLIN locus is associated with MS risk and with response to a weight-loss program based on dietary and behavioral advice. Specifically, carriers of variant A of PLIN4 11482G→A may be at higher risk for MS, whereas being carrier of variant T of PLIN6 14995A→T predicts a better weight-loss response to a multidisciplinary behavioral intervention.
For the SNPs studied, there were in our population no significant differences at baseline for either anthropometric measures or daily caloric ingestion. However, carriers of allele A of PLIN4 showed a significantly higher hazard ratio for MS, primarily from hypertriglyceridemia and hypertension. This appears to be in contrast with previous reports showing that the minor A allele at PLIN4 has been associated with lower risk of obesity (11,12). However, in the presence of obesity, our data support that this allele was associated with a worse metabolic profile consisting of higher TG, lower HDL-C, higher insulin response to the oral glucose load, and higher insulin resistance (HOMA-IR). The influence of PLIN4 SNP in MS is confirmed by the haplotypes of the SNPs PLIN4 11482G→A/PLIN6 14995A→T, as shown by the 4-fold prevalence of MS observed among groups 2.1 (AA) and 2.2 (AT), both containing minor allele A at PLIN4, as compared with haplotype group 1.2 (GT), containing exclusively the minor allele T at PLIN6. We observed that group 1.2 had the lowest prevalence of MS. Previous evidence shows that obese women homozygous for the minor A allele at PLIN4 had a higher basal and noradrenaline-induced lipolysis and reduced perilipin (10) as compared with wild type. We propose that in our population, despite the potential for a higher lipolysis associated with this allele, the very sedentary life and bad dieting, probably overcame the protection offered by increased fatty acid release from the adipocyte and brought up the metabolic unbalance resulting from nonesterified fatty acids flooding the blood circulation. Therefore, being obese combined with higher lipolysis induces first insulin resistance (24) and followed probably by early type 2 diabetes (25).
After the 20-wk dietary and behavioral intervention, a large variability in responses was observed, and our initial hypothesis was that variability at the PLIN locus may be, in part, involved in this differential response. Indeed, the results from our study support the notion that after intervention, carriers of the minor T allele for the PLIN6 had a higher mean weight loss (3.3 vs. 1.9 kg) when compared with the wild type and a greater success reducing obesity as measured by the loss of BMI Z-score (10 vs. 7.8%). Moreover, using haplotype analysis, we compared the groups 1.2 (carrying exclusively the T allele of PLIN6) and 1.1 (none of the minor alleles) to evaluate the influence of the presence of PLIN6 variant, and our data show that the baseline MS prevalence in the 1.2 group (6%) was less than half as compared with the 1.1 group (14%). Likewise, BMI Z score after intervention experienced a greater decrease in 1.2 (12.7%) as compared with 1.1 subjects (7.8%). This study is the first to report that PLIN6 variant has a better response to weight loss in multidisciplinary intervention in obese children, and this observation needs to be validated in an independent sample to verify whether it applies to other populations. Even though PLIN6 variant was associated with obesity risk in some populations (12,26), we confirm the findings that this variant was not associated with other MS risk factors. PLIN6 is located in the 3′ region where alternative splicing occurs, altering the transcription product resulting in perilipin isoform (12) with different lipolytic efficiency (27).
A multidisciplinary program of lifestyle and dietary change (28) is described as the best intervention for childhood obesity and successful changes are noted in the majority of the children of our group after 5 months. However, under the common feature of obesity, children differ considerably in their metabolic status, which may drive the diversity in responses observed in this and other multidisciplinary programs. We have shown in this study that the genetic variability at the PLIN locus may have a significant effect on response to weight loss.
It should be taken into consideration that the A variant of PLIN4, shown as being a protective variant (12,13,29) against obesity, could become an obstacle to regain metabolic homeostasis once obesity has been established. This observation is in agreement with a recent finding suggesting that obesity may modify the associations between PLIN variations and diabetes risk (30). Whereas our sample size could be considered small, our intervention study is highly controlled, and small sample sizes are often sufficient in this design. However, we need to be very cautious before extrapolating our conclusions to other populations, and replication of our findings is essential.
In conclusion, these findings show that these common PLIN polymorphisms may predict outcome strategies based on multidisciplinary treatment for obesity in children and should be considered on a prevention/treatment basis and that preventive counseling with lifestyle changes should be emphasized in overweight children with this PLIN4 A variant.
Supplementary Material
Acknowledgments
We sincerely thank research subjects who participated in the study. We also thank the support of Eliana Frazzatto, Mariana Rodrigues, and Barbara Lourenço.
Footnotes
This work was supported by Grants CAPES (Coordenação de Aperfeiçoamento de Pessoa de Nivel Superior) (Brazil), NIH/NHLBI HL54776 and NIH/NIDDK DK075030, and by contract 58-1950-9-001 from the U.S. Department of Agriculture Research Service.
Disclosure Statement: The authors have nothing to disclose.
First Published Online September 23, 2008
Abbreviations: BMI, Body mass index; HDL, high-density lipoprotein; HOMA-IR, homeostasis model assessment for insulin resistance; LD, linkage disequilibrium; LDL, low-density lipoprotein; MS, metabolic syndrome; OCA, obese children and adolescents; OGTT, oral glucose tolerance test; SNP, single-nucleotide polymorphism; TC, total cholesterol; TG, triglyceride; VLDL, very-low-density lipoprotein.
References
- Kiess W, Reich A, Muller G, Meyer K, Galler A, Bennek J, Kratzsch J 2001 Clinical aspects of obesity in childhood and adolescence: diagnosis, treatment and prevention. Int J Obes Relat Metab Disord 25(Suppl 1):S75–S79 [DOI] [PubMed] [Google Scholar]
- Ebbeling CA, Pawlak DB, Ludwig DS 2002 Childhood obesity: public-health crisis, common sense cure. Lancet 360:473–482 [DOI] [PubMed] [Google Scholar]
- Gortmaker SL, Must A, Perrin JM, Sobol AM, Dietz WH 1993 Social and economic consequences of overweight in adolescence and young adulthood. N Engl J Med 329:1008–1012 [DOI] [PubMed] [Google Scholar]
- Barlow SE, Dietz WH 1998 Obesity evaluation and treatment: Expert Committee recommendations. The Maternal and Child Health Bureau, Health Resources and Services Administration and the Department of Health and Human Sciences. Pediatrics 102:1–11 [DOI] [PubMed] [Google Scholar]
- Mutch DM, Clément K 2006 Genetics of human obesity. Best Pract Res Clin Endocrinol Metab 20:647–664 [DOI] [PubMed] [Google Scholar]
- Wang Y, Monteiro C, Popkin BM 2002 Trends of obesity and underweight in older children and adolescents in the United States, Brazil, China, and Russia. Am J Clin Nutr 75:971–977 [DOI] [PubMed] [Google Scholar]
- IBGE 2006 Consumer Expenditure Survey, 2002–2003: Rio de Janeiro. http://www.ibge.gov.br/home/estatistica/populacao/condicaodevida/pof/2003medidas/pof2003medidas.pdf (accessed 20 June 2007) [Google Scholar]
- Tai ES, Ordovas JM 2007 The role of perilipin in human obesity and insulin resistance. Curr Opin Lipidol 18:152–156 [DOI] [PubMed] [Google Scholar]
- Tansey JT, Sztalryd C, Gruía-Gray J, Roush DL, Zee JV, Gravilova O, Reitman ML, Deng CX, Li C, Kimmel AR, Londos C 2001 Perilipin ablation results in a lean mouse with aberrant adipocyte lipolysis, enhanced leptin production, and resistance to diet-induced obesity. Proc Natl Acad Sci USA 98:6494–6499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottagui-Tabar S, Ryden M, Lofgren P, Faulds G, Hoffstedt J, Brookes AJ, Andersson I, Arner P 2003 Evidence for an important role of perilipin in the regulation of human adipocyte lipolysis. Diabetologia 46:789–797 [DOI] [PubMed] [Google Scholar]
- Qi L, Corella D, Sorli JV, Portoles O, Shen H, Coltell O, Godoy D, Greenberg AS, Ordovas JM 2004 Genetic variation at the perilipin (PLIN) locus is associated with obesity-related phenotypes in white women. Clin Genet 66:299–310 [DOI] [PubMed] [Google Scholar]
- Qi L, Shen H, Larson I, Schaefer EJ, Greenberg AS, Tregouet DA, Corella D, Ordovas JM 2004 Gender-specific association of a perilipin gene haplotype with obesity risk in a white population. Obes Res 12:1758–1765 [DOI] [PubMed] [Google Scholar]
- Meirhaeghe A, Thomas S, Ancot F, Cottel D, Arveiler D, Ferrieres J, Amouyel P 2006 Study of the impact of perilipin polymorphisms in a French population. J Negat Results Biomed 5:10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corella D, Qi L, Sorli JV, Godoy D, Portoles O, Coltell O, Greenberg AS, Ordovas JM 2005 Obese subjects carrying the 11482G>A polymorphism at the perilipin locus are resistant to weight loss after dietary energy restriction. J Clin Endocrinol Metab 90:5121–5126 [DOI] [PubMed] [Google Scholar]
- Kang S, Cha B, Kim H, Kim S, Hur K, Lee H, Shim W, Ahn C, Lee HC 2006 The 11482G>A polymorphism in the perilipin gene is associated with weight gain with rosiglitazone treatment in type 2 diabetes. Diabetes Care 29:1320–1324 [DOI] [PubMed] [Google Scholar]
- Yan W, Chen S, Huang J, Shen Y, Qiang B, Gu D 2004 Polymorphisms in PLIN and hypertension combined with obesity and lipid profiles in Han Chinese. Obes Res 12:1733–1737 [DOI] [PubMed] [Google Scholar]
- Perez-Martinez P, Lopez-Miranda J, Perez-Jimenez F, Ordovas JM 2008 Influence of genetic factors in the modulation of postprandial lipemia. Atheroscler Suppl 9:49–55 [DOI] [PubMed] [Google Scholar]
- US Department of Health and Human Services 2000 CDC growth charts: United States. http://www.cdc.gov/growthcharts/ [Google Scholar]
- Marshall WA, Tanner JM 1969 Variations in the pattern of pubertal changes in girls. Arch Dis Child 44:291–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WA, Tanner JM 1970 Variations in the pattern of pubertal changes in boys. Arch Dis Child 45:13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecoraro P, Guida B, Caroli M, Trio R, Falconi C, Principato S, Pietrobelli A 2003 Body mass index and skinfold thickness versus bioimpedance analysis: fat mass prediction in children. Acta Diabetol 40(Suppl 1):S278–S281 [DOI] [PubMed] [Google Scholar]
- Zimmet P, Alberti G, Kaufman F, Tajima N, Silink M, Arslanian S, Wong G, Bennett P, Shaw J, Caprio S; International Diabetes Federation Task Force on Epidemiology and Prevention of Diabetes 2007 The metabolic syndrome in children and adolescents. Lancet 369: 2059–2061 [DOI] [PubMed] [Google Scholar]
- Pimenta JR, Zuccherato LW, Debes AA, Maselli L, Soares RP, Moura-Neto RS, Rocha J, Bydlowski SP, Pena SD 2006 Color and genomic ancestry in Brazilians: a study with forensic microsatellites. Hum Hered 62:190–195 [DOI] [PubMed] [Google Scholar]
- Avramoglu RK, Basciano H, Adeli K 2006 Lipid and lipoprotein dysregulation in insulin resistant states. Clin Chim Acta 368:1–19 [DOI] [PubMed] [Google Scholar]
- Ten S, Maclaren N 2004 Insulin resistance syndrome in children. J Clin Endocrinol Metab 89:2526–2539 [DOI] [PubMed] [Google Scholar]
- Qi L, Tai ES, Tan CE, Shen H, Chew SK, Greenberg AS, Corella D, Ordovas JM 2005 Intragenic linkage disequilibrium structure of the human perilipin gene (PLIN) and haplotype association with increased obesity risk in a multiethnic Asian population. J Mol Med 83: 448–456 [DOI] [PubMed] [Google Scholar]
- Jang Y, Kim OY, Lee JH, Koh SJ, Chae JS, Kim JY, Park S, Cho H, Lee JE, Ordovas JM 2006 Genetic variation at the perilipin locus is associated with changes in serum free fatty acids and abdominal fat following mild weight loss. Int J Obes (Lond) 30:1601–1608 [DOI] [PubMed] [Google Scholar]
- Suskind RM, Blecker U, Udall Jr JN, Von Almen TK, Schumacher HD, Carlisle L, Sothern MS 2000 Recent advances in the treatment of childhood obesity. Pediatr Diabetes 1:23–33 [DOI] [PubMed] [Google Scholar]
- Perez-Martinez P, Yiannakouris N, Lopez-Miranda J, Arnett D, Tsai M, Galan E, Straka R, Delgado-Lista J, Province M, Ruano J, Borecki I, Hixson J, Garcia-Bailo B, Perez-Jimenez F, Ordovas JM 2008 Postprandial triacylglycerol metabolism is modified by the presence of genetic variation at the perilipin (PLIN) locus in 2 white populations. Am J Clin Nutr Mar 87:744–752 [DOI] [PubMed] [Google Scholar]
- Qi L, Zhang C, Greenberg A, Hu FB 2008 Common variation in perilipin gene, central obesity, and risk of type 2 diabetes in US women. Obesity (Silver Spring) 16:1061–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.