Abstract
Mammalian target of rapamycin (mTOR) is a key regulator of cell growth acting via two independent targets, ribosomal protein S6 kinase 1 (S6K1) and 4EBP1. While each is known to regulate translational efficiency, the mechanism by which they control cell growth remains unclear. In addition to increased initiation of translation, the accelerated synthesis and accumulation of ribosomes are fundamental for efficient cell growth and proliferation. Using the mTOR inhibitor rapamycin, we show that mTOR is required for the rapid and sustained serum-induced activation of 45S ribosomal gene transcription (rDNA transcription), a major rate-limiting step in ribosome biogenesis and cellular growth. Expression of a constitutively active, rapamycin-insensitive mutant of S6K1 stimulated rDNA transcription in the absence of serum and rescued rapamycin repression of rDNA transcription. Moreover, overexpression of a dominant-negative S6K1 mutant repressed transcription in exponentially growing NIH 3T3 cells. Rapamycin treatment led to a rapid dephosphorylation of the carboxy-terminal activation domain of the rDNA transcription factor, UBF, which significantly reduced its ability to associate with the basal rDNA transcription factor SL-1. Rapamycin-mediated repression of rDNA transcription was rescued by purified recombinant phosphorylated UBF and endogenous UBF from exponentially growing NIH 3T3 cells but not by hypophosphorylated UBF from cells treated with rapamycin or dephosphorylated recombinant UBF. Thus, mTOR plays a critical role in the regulation of ribosome biogenesis via a mechanism that requires S6K1 activation and phosphorylation of UBF.
Cell growth (increased cell mass and size) is a prerequisite for proliferation (increased cell number), since a cell will divide only after it has reached a critical mass (38, 49, 55, 58). Thus, factors that govern cell cycle progression must also regulate growth in an interrelated fashion. Cell growth is not however, unconditionally dependent on cell cycle progression, as mutations in the budding yeast Saccharomyces cerevisiae and the fruit fly Drosophila melanogaster that block or disrupt cell division do not necessarily arrest cell growth (34, 44). Recent studies have demonstrated that cell growth and cell cycle progression in proliferating mammalian cells, like lower organisms, are also separable processes (8, 50, 63). Thus, detailed knowledge of the biochemical and molecular mechanisms governing cell size will be essential to understanding how the cell division cycle is coupled to growth and how this process is uncoupled during differentiation or is perturbed during diseases associated with deregulated growth. Our knowledge of cell cycle regulatory mechanisms has advanced considerably over the past decade. In contrast, information on the mechanisms of regulating cell growth in mammalian cells is limited.
Increased protein synthesis is one of the major anabolic events required for the growth response (28). Recent studies suggest that one of the key nodal points upon which signaling pathways converge to regulate growth is the mammalian target of rapamycin (mTOR) signaling pathway. mTOR is an important regulator of translational initiation through at least two distinct but integrated pathways (8, 10, 49, 58). One branch of this pathway controls phosphorylation of 4EBP1, releasing its inhibitory interaction with eIF4E, allowing eIF4E to associate with eIF4G to form the active eIF4F complex, a necessary component of the 40S initiation complex. eIF4E activity appears to be particularly important for the translation of mRNAs containing a highly structured 5′ untranslated region, such as transcripts encoding many proteins associated with growth and proliferation control (e.g., cyclin D1 and c-myc) (9, 10, 53, 64). The second mTOR-dependent branch leads to the phosphorylation of ribosomal protein S6 by the ribosomal S6 kinase 1 (S6K1) that stimulates the translation of mRNA with a 5′ oligopyrimidine tract (5′TOP). This class of mRNAs represents up to 30% of the total mRNA in the cell and encodes many components of the protein synthetic machinery, including ribosomal proteins (r-proteins) and translation initiation and elongation factors associated with regulating general protein initiation rates (26, 47, 56). In mammalian cells, both 4EBP1/eIF4E and S6K1 appear to be essential for efficient regulation of mammalian cell mass, and it is thought that this is largely achieved by regulating protein synthesis initiation rates, i.e., translational efficiency (8).
In addition to protein translation initiation, the rate of cell mass accumulation is dependent on the cellular level of functional ribosomes (translational capacity). Sustained increases in the rates of protein synthesis during normal processes, such as organ regeneration and hypertrophy, as well as pathological conditions, such as cardiac hypertrophy and tumor formation, all require exquisitely coordinated increases in both translation efficiency and capacity (28, 55, 57, 58). Transcription of the genes that encode the 45S rRNA precursor of the 18S, 5.8S, and 28S rRNA by RNA polymerase I (RPI) (ribosomal gene transcription [rDNA transcription]) is a major rate-limiting step in the biogenesis of ribosomes, yet the growth signaling pathways that regulate rDNA transcription and how they are coupled to those that control r-protein synthesis are poorly understood (14, 36, 40, 57, 58).
On the basis of the observations that (i) the mTOR signaling pathway promotes both growth and proliferation (49, 58) and (ii) increased rates of rRNA synthesis are necessary for accelerated rates of proliferative growth (14, 36, 57), we hypothesized that mTOR, in addition to its effects on translation, might directly regulate the function of molecules that control synthesis of rRNAs. To test this hypothesis, we have examined the role of mTOR-dependent signaling in mitogen-induced activation of rRNA synthesis. We demonstrate that mTOR activity is required for both rapid and sustained regulation of rDNA transcription in both proliferating NIH 3T3 fibroblasts and nonproliferating cardiac muscle cells. Stimulation of rDNA transcription by mTOR requires S6K1 activation and is mediated in part through the phosphorylation of the carboxy-terminal activation domain of the rDNA transcription factor, upstream binding factor (UBF). This phosphorylation promotes the interaction between UBF and the basal rDNA transcription factor SL-1. This study documents, for the first time in mammalian cells, a coordinated mechanism controlling both protein translation capacity and translational efficiency, thereby allowing for fine control of protein synthesis during growth and proliferation.
MATERIALS AND METHODS
Plasmid constructs.
The rDNA transcription reporter pSMECAT, control reporter pSMECAT-7, UBF expression vector pcDNA3-UBF1-FLAG, and UBF antisense expression vectors have been described in detail previously (2, 17, 21). The actin riboprobe vector Act-387, used to generate [32P]UTP-labeled internal standard for the in vitro transcription experiments was a gift from D. Autelitano (Cryptome Research Pty. Inc., Melbourne, Australia). This construct contains a fragment of rat β-actin (first 147 bp of exon 1 and 240 bp of exon 2) cloned into the EcoRI-BamHI site pGEM3Z which generates a transcript of 387 nucleotides. The p70 S6K1 expression constructs, PRK5 myc-tagged E2BQ (dominant negative), D3E (constitutively active), and dED3E (constitutively active, rapamycin resistant) have been described previously (16, 26, 41).
Cell culture.
NIH 3T3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 atmosphere. NIH 3T3 cells were plated at 20% confluency for 24 h and then made quiescent (serum starved) by culturing the cells for 24 h in DMEM containing 0.5% (wt/vol) bovine serum albumin (BSA). Serum-starved cells were then treated for the times indicated with 10% FBS in the presence or absence of rapamycin (final concentration of 20 nM). Neonatal cardiomyocytes were isolated from the ventricles of 1-day-old Sprague-Dawley rat pups as described previously (2, 21). Myocytes were plated at 1,250 cells/mm2 in modified Eagle's medium (MEM) containing 10% newborn calf serum and 0.1 mM bromodeoxyuridine and then allowed to attach overnight. After attachment, myocytes were maintained in serum-free defined medium (21) with KCl (50 mM) where indicated to prevent spontaneous contraction. Hypertrophic growth was initiated by adding 25 μM phenylephrine (2, 21). NISI cells were grown as previously described (18).
S6K1 activity.
Cells treated as described above were washed with phosphate-buffered saline and lysed in lysis buffer (50 mM Tris-HCl [pH 7.5], 120 mM NaCl, 1% Nonidet P-40 [NP-40], 1 mM EDTA, 50 mM NaF, 40 mM β-glycerophosphate, 0.1 mM sodium vanadate, 1 mM benzamidine, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]). Cell extracts were cleared by centrifugation at 4°C for 15 min at 12,000 × g, and aliquots of the supernatant were stored at −70°C. Endogenous or myc epitope-tagged p70 S6K1 activity was assayed after immunoprecipitation as described previously (6, 16) except 150 μM RRRRLSSLRA peptide was used as a substrate. The results are expressed in units of activity per milligram of protein lysate. One unit of activity results in the transfer of 1 pmol of γ-32Pi into peptide substrate per min.
Determination of cell growth.
Cell volume was determined by using a Sysmex CDA500 system, protein content was determined using a Bradford assay (Bio-Rad), cell DNA content was determined by Burton assay, and total RNA was determined by A260, with recovery being controlled by the addition and quantitation of a 32P-labeled RNA transcript as described previously (15, 17).
Isolation of nuclei and nuclear run-on assays.
Nuclei were harvested and resuspended in nucleus storage buffer (50 mM Tris-HCl [pH 8.3], 40% [vol/vol] glycerol, 5 mM MgCl2, 0.1 mM EDTA) as described previously (15). De novo-synthesized RNA was isolated by using nuclei normalized for DNA content. To control for recovery of the in vitro-transcribed RNA, a 3H-labeled β-actin RNA probe was added prior to extraction and the amount of 3H recovered was quantitated by liquid scintillation counting. Transcription from the rDNA promoter was measured by hybridization of in vitro-synthesized 32P-labeled run-on transcripts to slot blots of immobilized (Zetaprobe; Bio-Rad) plasmids containing the mouse rDNA promoter (positions −168 to +292 with respect to the start site of transcription initiation) and slot blots of pBluescript DNA as a control for nonspecific hybridization. Hybridization and washes were performed as described previously (15). Radioactive hybrids were detected with a Molecular Dynamics PhosphorImager, quantified using ImageQuant software, and expressed as fold increase in phosphorimager units over control values.
Preparation of nuclear extracts.
Nuclear extracts were prepared from NIH 3T3 cells (108 cells per isolation) as described previously, yielding extracts containing 6 to 10 mg of protein per ml (12). Following final dialysis, extracts were frozen in liquid N2 and stored at −80°C.
Fractionation of nuclear extract by MonoQ anion-exchange chromatography.
The standard stepwise elution from anion-exchange resin routinely used to separate RPI/SL-1 from UBF (23, 52) was refined to maximize the separation. Nuclear extracts were dialyzed (Spectra/Por 2 membrane) (molecular weight cutoff, 12,000 to 14,000) for 3 h in MonoQ buffer A [40 mM HEPES (pH 7.9), 5 mM MgCl2, 0.2 mM EDTA, 50 mM (NH4)2SO4, 0.5 mM PMSF, 0.5 mM dithiothreitol (DTT), 20% (vol/vol) glycerol]. Extracts were centrifuged at 13,000 × g for 15 min, and the supernatant (∼2 mg of protein in 1 ml) loaded onto a MonoQ anion-exchange column attached to a Pharmacia fast-performance liquid chromatography system and preequilibrated in buffer A. The column was washed for 10 min at 0.25 ml/min before elution with a linear gradient to 1 M (NH4)2SO4 in buffer A over 60 min at 0.25 ml/min. Fractions (500 μl) were collected, and the presence of RPI, SL-1, Rrn3, and UBF was detected by Western blot analysis. Fractions containing RPI (MQ340) (fractions 17 to 19, 290 to 380 mM NH4SO4) or UBF (MQ570) (fractions 25 to 27, 530 to 620 mM NH4SO4) were combined and concentrated using a Centricon-50 concentrator (Amicon). The concentrated protein was diluted 10-fold in C-20 buffer (20 mM HEPES [pH 7.9], 5 mM MgCl2, 0.2 mM EDTA, 100 mM KCl, 0.5 mM PMSF, 0.5 mM DTT, 20% [vol/vol] glycerol) and reconcentrated; this procedure was repeated once to facilitate buffer exchange. All fractions were frozen in liquid N2 and stored at −80°C. The concentration of each preparation was assessed by Western blotting, and equal amounts of each protein from either serum- or rapamycin-treated cell extracts were added to the transcription assays.
Immunodepletion of UBF from MQ570.
Immunodepletion experiments were performed essentially as we have previously described (23). Antibody from UBF antiserum or preimmune serum was bound to protein A agarose beads and washed extensively in phosphate-buffered saline. Prior to immunodepletion, the control and UBF antiserum-bound beads were incubated for 30 min in C-20 buffer containing 0.05% (wt/vol) Tween 20 and then for 30 min in a solution containing 0.5 mg of BSA per ml to reduce nonspecific binding of proteins to the beads. The MQ570 fraction (UBF) was brought to a final concentration of 0.5 mg of BSA per ml and mixed with 20 μl of packed pretreated anti-UBF or control beads and incubated at 4°C for 3 h in the presence of protease and phosphatase inhibitor cocktail (catalog nos. P8340 and P5726; Sigma). The beads were removed by centrifugation, and the supernatants were used in the transcription experiments.
Baculovirus expression of Rrn3 and UBF.
FLAG-tagged mouse Rrn3 and UBF1 were expressed in Sf9 cells using the pFASTBac1 system (Life Technologies). Infected Sf9 cells were lysed in Bac-lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM DTT, 0.5 mM PMSF) and incubated with anti-FLAG M2 resin (Kodak) for 1 h at 4°C. The resin was washed extensively with Bac-lysis buffer containing 500 mM NaCl, and the bound material was eluted with 0.5 mg of FLAG peptide per ml in C-20 buffer. The purity of the eluted proteins was determined by Coomassie blue staining. To dephosphorylate UBF, FLAG-tagged UBF was bound the FLAG M2 resin and dephosphorylated by treating with alkaline phosphatase using standard conditions. After extensive washing with Bac-lysis buffer containing 500 mM NaCl, the dephosphorylated UBF was eluted with FLAG peptide as described above.
Preparation of S100 extracts and purified FLAG-tagged Rrn3.
S100 extracts for the Rrn3 activity assays were prepared from NISI cells treated with cycloheximide (CHX) (100 μg/ml) as described previously (5) or from NIH 3T3 cells treated with serum or serum plus rapamycin. NIH 3T3 cells were plated at a concentration of 106 cells per 100-mm-diameter dish and transfected 18 h later with FLAG-tagged Rrn3 (6 μg) using Lipofectamine 2000 (Invitrogen). The cells were then made quiescent (serum starved) and treated as described above. Cells were harvested in lysis buffer (50 mM Tris [pH 7.4], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100). The lysates were precleared on Sepharose CL-4B beads, and FLAG-tagged Rrn3 was purified by incubation with anti-FLAG resin and eluted with 0.5 mg of FLAG peptide per ml in C-20 buffer as described above. The activity of FLAG-tagged Rrn3 isolated from the transfected NIH 3T3 cells was compared with the activity of purified baculovirus Rrn3 (300 ng).
In vitro transcription and complementation assays.
rDNA transcription reactions were performed as previously described (4, 5). For these assays, the template DNA consisted of EcoRI-linearized plasmid pU5.1E/X containing the rat 45S rDNA promoter (positions −286 to +520) and is designed to generate a truncated transcript of 639 nucleotides. The in vitro transcription and complementation experiments utilized nuclear extracts (50 μg) or MQ fractions (equal amount of UBF or RPI) from NIH 3T3 cells and S100 extract (25 to 40 μg) from NISI cells treated with CHX, serum, or serum plus rapamycin (5). The samples were incubated for 30 min at 30°C in transcription buffer containing 0.1 μg of template, [32P]UTP, cold nucleotides, and α-amanitin (final concentration, 100 μg/ml), and the reaction was stopped by adding 5 mg of tRNA per ml in 10% (wt/vol) sodium dodecyl sulfate (SDS). An in vitro-transcribed [32P]UTP-labeled β-actin RNA riboprobe was used as an internal standard for the recovery of the RNA transcripts. RNA was extracted from the reaction mixture and redissolved in 3 μl of 0.01% (wt/vol) SDS plus 14 μl of formamide. The transcripts were resolved on 6% urea-polyacrylamide gels, which were dried and exposed to X-ray film.
Transfections and reporter assays.
NIH 3T3 cells were plated at a concentration of 0.18 × 106 cells per 60-mm-diameter dish and transfected 24 h later with the indicated constructs using Lipofectamine (Invitrogen) (21). All transfections contained equal amounts of DNA. Five hours after transfection, the culture medium was replaced with DMEM plus 0.5% BSA for a further 16 h and treated as described above. Twenty-four hours after the cardiomyocytes were plated, they were transfected using Lipofectamine 2000 (2), and hypertrophy was initiated as described above. Cell lysates were prepared and assayed for chloramphenicol acetyltransferase (CAT) or β-galactosidase activity as described previously (2, 21). To control for any differences in transfection efficiency, the data were expressed as CAT activity relative to the β-galactosidase activity, coexpressed under a cytomegalovirus (CMV) promoter. To establish the RPI dependence of the CAT assay, the essential G at position −7 in the 45S promoter was mutated in the control reporter pSMECAT-7.
SDS-PAGE and Western blot analysis.
Protein samples (10 to 20 μg) were resuspended in SDS sample buffer, boiled, size fractionated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and transferred to Immobilon P (Amersham) membranes as described previously (6, 15). The blot was subsequently probed with the following antibodies: polyclonal antibodies to S6K1 (6), UBF1/UBF2 (15), Rrn3 (5), actin, A127 subunit of RPI (15), TATA-binding protein (TBP) (Santa Cruz catalog no. 273); anti-TAFI p95 (Santa Cruz catalog no. 6569); anti-TAFI p68 (Santa Cruz catalog no. 6574); and monoclonal antibodies to the Myc epitope (41), α-tubulin, or FLAG epitope (21). The blot was developed using the appropriate secondary antibodies and enhanced chemiluminescence (Amersham).
Metabolic labeling of cells.
NIH 3T3 cells were plated at a concentration of 106 per 100-mm-diameter plate for 24 h and serum starved for 48 h in phosphate-free DMEM with 32Pi (1 mCi/5 ml) added in the last 12 h. The cells were then treated and harvested, and 32P-labeled UBF was immunoprecipitated as described previously (16).
Tryptic phosphopeptide mapping.
32P-labeled UBF was separated on an SDS-10% polyacrylamide gel, transferred to an Immobilon P membrane, exposed to autoradiography, and excised. The membrane was washed three times: with 0.5% polyvinyl pyrrolidone (molecular weight of 360,000) in 100 mM acetic acid, with water, and then with 50 mM NH4HCO3. After washing, the membrane was subjected to tryptic digestion (16, 31). Tryptic peptides were lyophilized, and phosphopeptides (5,000 cpm) were separated by two-dimensional mapping as described previously (16). Radiolabeled phosphopeptides were visualized after 24-h exposure using a PhosphorImager and ImageQuant software (Molecular Dynamics).
UBF/SL-1 immunoprecipitation.
Nuclear extracts (200 μg) from cells treated with serum or rapamycin were tumbled in C-10 buffer (C-20 buffer with 10% glycerol) containing 0.5% NP-40 and 2 μg of anti-TAFI p95 antibody at 4°C for 3 h. Washed protein A Sepharose beads (25 μl) were added, and the sample tumbled for a further 30 min. The beads were washed three times with C-10 buffer containing 0.5% NP-40, and SDS sample buffer was added. The samples were boiled, separated on SDS-8% polyacrylamide gels, transferred, and Western blotted with anti-UBF or anti-TAFI p95.
RESULTS
mTOR signaling is required for both acute and sustained activation of rDNA transcription by serum.
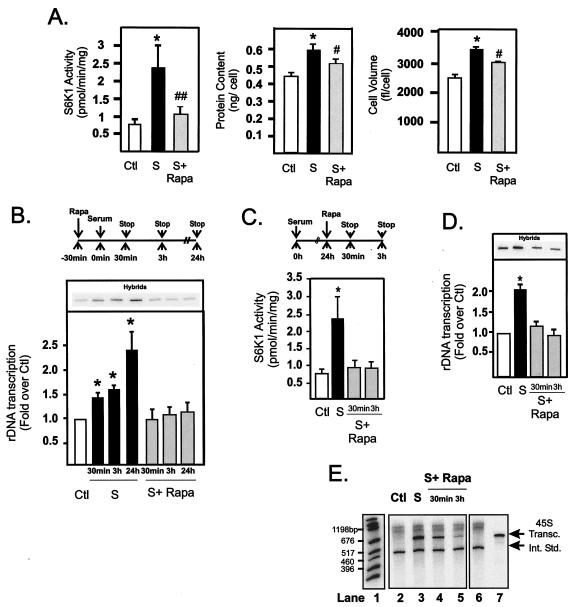
Different cell types vary markedly in their dependence on mTOR for the control of cell growth (8). To assess the suitability of NIH 3T3 cells to study mTOR-dependent regulation of rDNA transcription, we first characterized the effects of rapamycin, a potent and selective inhibitor of mTOR, on NIH 3T3 cell proliferation and growth over a 24-h time period. Quiescent cells were stimulated with DMEM containing 10% FBS in the presence or absence of 20 nM rapamycin for a further 24 h, harvested, and analyzed for activation of the mTOR-S6K1/4EBP1 pathway and parameters of cellular proliferation and growth. Treatment with serum for 24 h led to a sustained activation of mTOR-dependent signaling, with S6K1 activity elevated 2.4-fold (Fig. 1A). Rapamycin treatment blocked the increases in S6K1 activity, and this inhibition correlated with significant reductions in protein content per cell, cell volume (Fig. 1A), and proliferation (results not shown), indicating that rapamycin inhibition of mTOR leads not only to delayed cell cycle entry but also to reduced cellular growth and cell size. These results demonstrate that serum-dependent growth of NIH 3T3 cells is highly sensitive to rapamycin and support the findings of Fingar et al. (8), who demonstrated that mTOR signaling was crucial for the control of cell size in mammalian cells.
FIG. 1.
Regulation of NIH 3T3 cell growth correlates with rapid and sustained activation of rDNA transcription by mTOR. NIH 3T3 cells were plated in DMEM with 10% FBS, allowed to grow overnight, and made quiescent by incubation for 24 h in DMEM with 0.5% BSA (serum starved as a control). After serum starvation, cells were pretreated with rapamycin (20 nM) or vehicle (ethanol) for 30 min before stimulation with DMEM plus 10% FBS for 30 min, 3 h, or 24 h. Cells harvested at 24 h were assayed for S6K1 activity, protein content per cell, and cell volume (A) as described in Materials and Methods. Cells harvested at 30 min, 3 h, and 24 h were assayed for endogenous rDNA transcription rates by nuclear run-on analysis (B) as described in Materials and Methods. Alternatively, quiescent cells were stimulated with 10% FBS for 24 h before treatment with rapamycin (20 nM) for 30 min and 3 h as illustrated in the schematic (C), then harvested and assayed for S6K1 activity (C), endogenous rDNA transcription (D), and for their ability to transcribe 0.1 μg of 45S template gene in vitro (E) as described in Materials and Methods. Lane 1, markers; lanes 2 to 5, 45S transcripts (45S Transc.); lane 6, internal standard (Int. Std.); lane 7, 45S transcript. Values that are significantly different (P < 0.05) from the control values (*) and values that are significantly different (P < 0.05 [#] and P < 0.005 [##]) from the values for serum-treated cells (n = 4 to 7) are indicated. Abbreviations: Ctl, control; S, 10% FBS; S+ Rapa, 10% FBS plus 20 nM rapamycin.
Serum treatment of quiescent cells results in rapid and sustained activation of mTOR signaling (13, 30). To determine the role of ribosomal gene transcription in mTOR-dependent activation of growth, we examined rDNA transcription rates in NIH 3T3 cells 30 min, 3 h, and 24 h after stimulation with serum in the presence or absence of pretreatment with rapamycin (Fig. 1B, schematic). Serum stimulation of quiescent cells activated S6K1 activity 3.5-fold within 30 min, and this activity was abolished by rapamycin (results not shown). Concomitantly, serum rapidly stimulated rDNA transcription, as measured by nuclear run-on transcription from the 45S pro-moter, to 40% of the maximal value within 30 min (Fig. 1B). While activation remained constant for 3 h, maximal activation required a further 18 to 24 h of stimulation, suggesting the existence of both acute and sustained mechanisms regulating rDNA transcription in response to serum. Incubation of the cells with rapamycin for 30 min prior to serum treatment completely inhibited both the rapid and sustained induction of rDNA transcription (Fig. 1B). Together, these data demonstrate that activated mTOR is required for both the rapid and maximal activation of rDNA transcription by serum. Importantly, since the number of nuclei assayed by nuclear run-on, per time point, was normalized for DNA content, an increased gene copy number prior to cell division could not account for the sustained response.
To determine whether increased mTOR activity was also required to maintain a stimulated level of transcription, we examined how rapidly rapamycin could inhibit rDNA transcription in exponentially growing cells (Fig. 1C, schematic). Rapamycin treatment of exponentially growing cells rapidly inhibited S6K1 activity (Fig. 1C) with a corresponding inhibition of rDNA transcription rates (Fig. 1D). The genome of eukaryotic cells contain between 200 and 400 copies of the 45S ribosomal gene. Nuclear run-on transcription measures the level of active RPI loaded onto the ribosomal genes. However, it does not indicate whether the increased transcription is due to accelerated initiation-elongation rates on a fixed number of genes or whether additional ribosomal genes are being transcribed.
To clarify the mechanism of increased transcription, we measured the effect of rapamycin on RPI activity in a cell-free transcription assay (Fig. 1E). Modified nuclear extracts were prepared from NIH 3T3 cells and examined for their ability to support RPI transcription from a linearized 45S rDNA template (see Materials and Methods). Serum treatment (24-h treatment) significantly increased the activity of RPI transcription compared to that of serum-starved (control) cells (Fig. 1E, compare lanes 2 and 3). This activation was rapidly inhibited by rapamycin (within 30 min), and inhibition was maximal after 3 h (lanes 4 and 5). These results indicate that the major effect of mTOR to activate rDNA transcription is to activate one or more of the RPI transcription components. Treatment of primary rat fibroblasts with rapamycin also resulted in a rapid inhibition of rDNA transcription (results not shown), confirming that our results were not restricted to immortalized cell lines. Thus, in mammalian cells, sustained mTOR activation is required to maintain serum activation of rDNA transcription.
S6K1 activation is necessary and sufficient to activate rDNA transcription in mammalian cells.
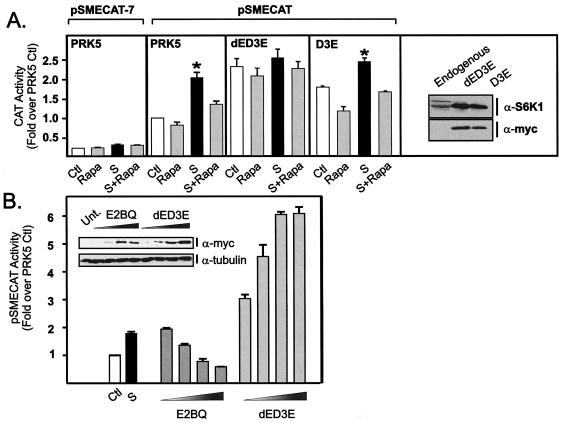
Given the role of S6K1 in the regulation of r-protein synthesis, we hypothesized that this major target of mTOR signaling may mediate rapamycin-sensitive stimulation of rDNA transcription, allowing coordinate regulation of ribosome biogenesis. To examine this possibility, we performed cotransfection experiments with mutants of S6K1 and a well-characterized 45S gene reporter system, pSMECAT (2, 17, 21, 41, 54). pSMECAT contains the mouse rDNA promoter (positions −152 to +60) upstream of an internal ribosomal entry site to drive the expression of CAT. We have previously shown by S1 nuclease analysis that the site of transcription initiation for pSMECAT is that predicted for RNA transcripts from RPI (16). Moreover, mutational inactivation of the 45S promoter by mutating the G at position −7 which is essential for transcription by RPI (pSMECAT-7) did not reveal the presence of cryptic RPII or RPIII initiation sites (21). Similarly, in these studies, the level of CAT activity for pSMCAT-7 was <15% of that for pSMECAT and did not significantly respond to serum or rapamycin treatment (Fig. 2A). On the other hand, serum induced pSMECAT activity to a level similar to that observed for the endogenous ribosomal genes (compare Fig. 2A, panel pSMECAT PRK5, and Fig. 1B). Rapamycin significantly inhibited the serum activation of pSMECAT and to a lesser extent inhibited basal levels. Importantly, rapamycin repression of pSMECAT was rescued by forced expression of a constitutively active, partially rapamycin-insensitive version of S6K1 (dED3E) (41) (Fig. 2A), suggesting that the major pathway downstream of mTOR responsible for regulating rDNA transcription was S6K1. In contrast, overexpression of a constitutively active version of S6K1 that retains full rapamycin sensitivity (D3E) (41) (Fig. 2A) was unable to rescue rapamycin inhibition of rDNA transcription. Thus, S6K1 activity is sufficient to restore rDNA transcription in the presence of rapamycin. Interestingly, equivalent forced expression of either D3E or dED3E in quiescent cells led to a significant increase in pSMECAT expression (Fig. 2A), indicating that elevated S6K1 activity alone is sufficient to stimulate rDNA transcription in the absence of serum or growth factors.
FIG. 2.
S6K1 is necessary and sufficient to regulate rDNA transcription in mammalian cells. (A) NIH 3T3 cells (0.18 × 106 cells/well) were transfected 24 h after plating with the following plasmid constructs: a reporter construct for rDNA transcription (pSMECAT) (0.45 μg), a control vector in which an essential G at position −7 in the 45S promoter was mutated in the control reporter (pSMECAT-7), empty vector PRK5 (0.45 μg), or the indicated S6K1 mutants, dED3E or D3E (0.45 μg). The cells were then transferred into DMEM containing 0.5% BSA. After 24 h of serum starvation, the cells were pretreated with rapamycin (20 nM) or vehicle (ethanol) for 30 min and then stimulated with DMEM plus 10% FBS for a further 24 h before being assayed for CAT activity. Samples were analyzed in parallel for endogenous and recombinant S6K1 expression with an antibody directed to the carboxy-terminal region of S6K1 or the myc tag. (B) Cells were plated and transfected as described above with pSMECAT (0.45 μg) and increasing amounts of the dominant-negative construct E2BQ (0.45, 0.9, 1.8, and 3.6 μg) or dED3E (0.45, 0.9, 1.8, and 3.6 μg) and then transferred into DMEM containing 10% BSA. Total plasmid DNA per transfection was equalized with the empty vector, PRK5. After 48 h, the cells were assayed for CAT activity. Samples were analyzed in parallel for recombinant S6K1 expression (myc) and tubulin as a control for loading (see insert). Values that are significantly different (P < 0.05) from the control values (serum-starved cells) (n = 5) are indicated (*). Abbreviations: Ctl, control; Rapa, 20 nM rapamycin; S, 10% FBS; S+Rapa, 10% FBS plus 20 nM rapamycin; α-S6K1, anti-S6K1; Unt., untransfected.
To test whether S6K1 activity is required for rDNA transcription, pSMECAT was cotransfected with the interfering mutant of S6K1 (E2BQ) which acts in a dominant-negative fashion when overexpressed (26). Dominant-negative S6K1 repressed serum-induced rDNA transcription in a dose-dependent fashion (Fig. 2B), confirming the requirement of S6K1 activity for serum stimulation of rDNA transcription. Expression of dED3E to levels similar to those observed for E2BQ (Fig. 2B, insert) gave a dose-dependent activation of rDNA transcription, consistent with the results presented in Fig. 2A, indicating that high levels of plasmid DNA do not inhibit pSMECAT transcription. These observations demonstrate a novel link between S6K1 activity and rDNA transcription in mammalian cells and are consistent with the increased cell size noted with S6K1 transfection (8).
mTOR-dependent activation of rDNA transcription is independent of proliferation.
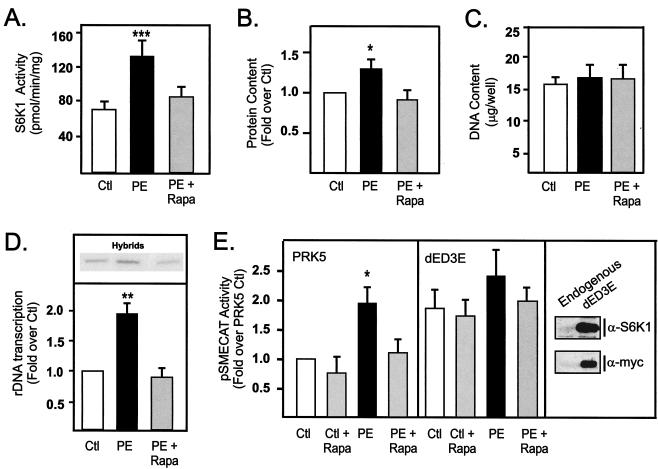
The ability of mTOR to rapidly regulate rDNA transcription suggests a direct mechanism of action. However, we have also shown that maximal activation of rDNA transcription by serum requires 24 h, which correlates with the documented effects of S6K1 on the transition from the G1 phase to the S phase of the cell cycle (30, 42). To examine the possibility that sustained effects of mTOR on rDNA transcription might be the indirect consequence of regulation of cell cycle progression, we examined the ability of rapamycin to inhibit rDNA transcription in primary cultures of postmitotic neonatal cardiomyocytes. These cells do not proliferate or undergo DNA synthesis in response to growth factor stimulation; instead, they undergo hypertrophic growth (19). Treatment of primary cultures of neonatal cardiomyocytes with the α1-adrenergic receptor agonist phenylephrine stimulated S6K1 activity twofold (Fig. 3A) and hypertrophic growth, as defined by an increase in protein content without a change in DNA content (Fig. 3B and C). This correlated with a twofold activation of endogenous rDNA transcription (Fig. 3D) with no change in cell number (data not shown), which is consistent with previous studies (2, 19, 21). Rapamycin treatment blocked these increases (Fig. 3A to D). In cotransfection experiments, phenylephrine stimulated pSMECAT activity twofold, and this activation was abolished by rapamycin (Fig. 3E, PRK5 panel). Enforced expression of the constitutively active rapamycin-resistant S6K1 mutant (dED3E) rescued rapamycin-mediated repression of the reporter for rDNA transcription pSMECAT. Thus, the ability of mTOR to regulate rRNA synthesis is independent of its effects on cell cycle and is mediated, at least in part, through S6K1. These findings are consistent with the observation that mTOR-dependent growth of fibroblasts continues despite a blockade of cell cycle progression (8).
FIG. 3.
Rapamycin inhibits rDNA transcription independent of its effect on proliferation. Cultures of primary neonatal cardiomyocytes maintained in defined serum-free medium were stimulated with vehicle (ethanol) or the hypertrophic agent phenylephrine (25 μM) in the presence or absence of rapamycin (20 nM). After 48 h, the cells were harvested and assayed for S6K1 activity (A), protein content (B), DNA content (C), or rDNA transcription (D) as described in Materials and Methods. Alternatively, cultures of primary neonatal cardiomyocytes were transfected with pSMECAT (0.45 μg), S6K1 mutant dED3E (0.45 μg), or empty vector PRK5 (0.45 μg) and then transferred to defined serum-free medium. After 24 h, the cells were stimulated as described above, harvested 24 h later, and then assayed for CAT activity (E). Expression of endogenous and recombinant S6K1 was determined by Western blotting with an antibody directed to the carboxy-terminal region of S6K1 (α-S6K1) or the myc tag, respectively. Values that are significantly different from the control values (n = 5 to 7) are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.005. Abbreviations: Ctl, control; PE, 25 μM phenylephrine; PE + Rapa, PE plus 20 nM rapamycin.
mTOR regulates the level of the rDNA transcription factor UBF.
UBF is a key factor implicated in the regulation of rDNA transcription whose activity is subject to regulation at multiple levels, including growth factor-induced expression and phosphorylation (reviewed in references 14 and 36). UBF levels have been demonstrated to be functionally limiting for rDNA transcription rates in NIH 3T3 cells (2, 17, 21). Thus, we examined whether mTOR-dependent activation of rDNA transcription might be mediated through changes in the levels of UBF.
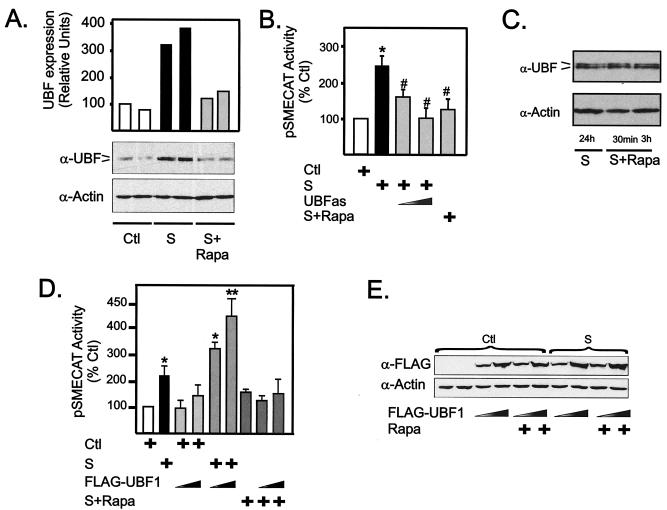
Stimulation of quiescent NIH 3T3 fibroblasts with serum for 24 h led to a significant increase in UBF protein expression (Fig. 4A), consistent with data from NIH 3T6 fibroblasts (11). Growth factor induction of UBF expression is independent of effects on proliferation and DNA content, since UBF expression is stimulated by growth factors in terminally differentiated cardiomyocytes (19, 21). Rapamycin completely blocked the induction of UBF protein (Fig. 4A), demonstrating that serum-induced UBF expression requires activation of mTOR. Expression of UBF antisense RNA in NIH 3T3 cells significantly inhibited the activity of a cotransfected reporter for rDNA transcription, pSMECAT, in a dose-dependent manner to the level observed with rapamycin treatment (Fig. 4B). These results demonstrate that serum induction of rDNA transcription is dependent on increased levels of UBF, consistent with previous data (2). Together, these data indicate that mTOR can regulate rDNA transcription by regulating the accumulation of UBF protein.
FIG. 4.
mTOR regulates UBF expression. (A) NIH 3T3 cells were made quiescent, pretreated with 20 nM rapamycin or vehicle (ethanol) for 30 min, stimulated with serum (by growing in DMEM containing 10% FBS for 24 h), and harvested. Equal amounts of protein were analyzed for UBF expression by Western blotting using polyclonal antibodies that recognize both forms of UBF (UBF1 and UBF2) (α-UBF) and an anti-actin antibody (α-Actin) to verify loading. Samples from duplicate cultures are shown. (B) NIH 3T3 cells were transfected with pSMECAT (0.45 μg) and where indicated with increasing amounts (0.9 and 1.8 μg) of a vector driving expression of UBF antisense RNA (pCMV5-UBFas) or the empty vector (pCMV5) and then transferred to DMEM containing 0.5% BSA. After 24 h of serum starvation, cells were pretreated with rapamycin (20 nM) or vehicle (ethanol) for 30 min before stimulation with DMEM containing 10% FBS for a further 24 h. The cells were harvested and assayed for CAT activity. (C) NIH 3T3 cells growing exponentially in the presence of DMEM containing 10% FBS were treated with rapamycin (20 nM) for 30 min or 3 h, harvested, and assayed by Western blotting for UBF expression as described above for panel A. (D and E) NIH 3T3 cells were transfected with pSMECAT (0.45 μg) and where indicated, with increasing amounts of a construct driving the expression of an epitope-tagged version of UBF1 (FLAG-tagged UBF-1 [FLAG-UBF1]) (0.9 and 1.8 μg) or the empty vector (pCDNA3) and then transferred to DMEM containing 0.5% BSA. After 24 h of serum starvation, cells were pretreated with rapamycin (20 nM) or vehicle (ethanol) for 30 min before stimulation with DMEM containing 10% FBS for a further 24 h as indicated. The cells were then harvested and assayed for CAT activity (D) or expression of recombinant UBF using an anti-FLAG antibody; actin was used as a loading control (E). Values that are significantly different from the control values (*, P < 0.05; **, P < 0.01) and values that are significantly different (P < 0.05) from the values for serum-treated cells (n = 5) (#) are indicated. Abbreviations: Ctl, control (serum starved); S, 10% FBS; S+Rapa, 10% FBS plus 20 nM rapamycin.
Acute treatment of exponentially growing cells with rapamycin significantly inhibited rDNA transcription (Fig. 1D and E). However, no effect of rapamycin on endogenous UBF protein levels was observed in this time frame (Fig. 4C), indicating that additional levels of regulation are required. Moreover, overexpression of UBF1 was insufficient to rescue repression of serum-induced rDNA transcription by rapamycin (Fig. 4D). Control Western blots demonstrated that rapamycin had no effect on the expression of ectopic UBF (Fig. 4E). Together, these data indicate that an mTOR-dependent mechanism in addition to elevated expression of UBF is required to regulate rDNA transcription.
Rapid activation of rDNA transcription by mTOR is mediated by phosphorylation of the carboxy-terminal activation domain of UBF.
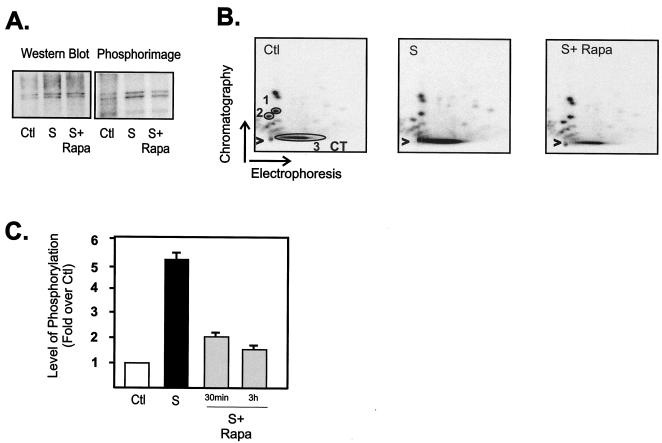
In addition to changes in expression, maximal transcriptional activity of UBF requires phosphorylation at multiple sites within the carboxy-terminal acidic tail region (amino acids 675 to 765) (14, 27, 61, 62). This carboxy-terminal phosphorylation facilitates the efficient interaction of UBF with the basal rDNA transcription factor SL-1 (23, 27, 60) and is essential for transactivation (62). Since rapid phosphorylation of this region is regulated by serum and growth factors (24, 27, 32, 39, 62), we examined whether the rapid inhibition of rDNA transcription by rapamycin (30 min or 3 h) might be mediated in part through a reduction in the level of phosphorylation of the UBF carboxy-terminal domain. NIH 3T3 cells were metabolically labeled with 32Pi, and UBF was immunopurified and subjected to two-dimensional tryptic phosphopeptide mapping. Western blotting of a typical immunoprecipitate revealed increased UBF levels in response to serum treatment and no effect of short-term rapamycin treatment on the level of UBF (Fig. 5A), consistent with the data presented in Fig. 4C. Exposure of the membrane to the phosphorimager revealed the characteristic [32P]-labeled doublet (Fig. 5A) that was excised for phosphopeptide mapping. Consistent with previous reports (27, 61, 62), a number of phosphopeptides were detected. Upon serum stimulation, the majority of the 32P label was incorporated into a large tryptic fragment which did not migrate significantly in the chromatography phase (Fig. 5B, CT) and which was previously identified as the carboxy-terminal activation domain of UBF (27, 61, 62). Rapamycin treatment prevented serum-stimulated phosphorylation of the activation domain (Fig. 5B). To quantitate these effects and account for any variations in loading, the ratio of carboxy-terminal domain phosphorylation over the intensity of two invariant peptides (peptides 1 and 2) was determined using ImageQuant software. Pooled data of separate experiments were plotted in Fig. 5C. Phosphorylation of the carboxy-terminal domain was stimulated fivefold by serum treatment, and rapamycin treatment for both 30 min and 3 h significantly reduced the phosphorylation (Fig. 5C). These studies suggest that the acute regulation of rDNA transcription by mTOR is dependent, at least in part, on the phosphorylation of the carboxy-terminal activation domain of UBF.
FIG. 5.
mTOR regulates phosphorylation of the UBF carboxy-terminal activation domain. NIH 3T3 cells were made quiescent (serum starved as a control), stimulated with serum (grown in DMEM containing 10% FBS for 24 h), and then treated with rapamycin (20 nM) or vehicle (ethanol) for 30 min or 3 h. The cells were also labeled with 1 mCi of 32Pi per plate 12 h after serum was added. 32P-labeled UBF was immunoprecipitated, separated by SDS-PAGE, and transferred to Immobilon P membranes. The membranes were then subjected to Western blot analysis using anti-UBF antibodies and phosphorimager analysis (A). Alternatively, 32P-labeled UBF was excised and digested with trypsin, and the resultant peptides were separated by electrophoresis and chromatography as described in Materials and Methods. Radiolabeled phosphopeptides were visualized after 24-h exposure using a PhosphorImager (B). Panels A and B show typical results for a 3-h rapamycin treatment. (C) The results from two or three separate experiments incorporating 30-min and 3-h treatment with rapamycin were quantitated using ImageQuant software (Molecular Dynamics), and the level of carboxy-terminal domain (CT) phosphorylation was expressed relative to the phosphorylation of peptides 1 and 2 (circled 1 and 2 in panel B). Abbreviations: Ctl, control; S, 10% FBS; S+ Rapa, 10% FBS plus 20 nM rapamycin.
Endogenous UBF from serum-stimulated cells and recombinant baculovirus UBF rescue rapamycin-mediated repression of RPI transcription.
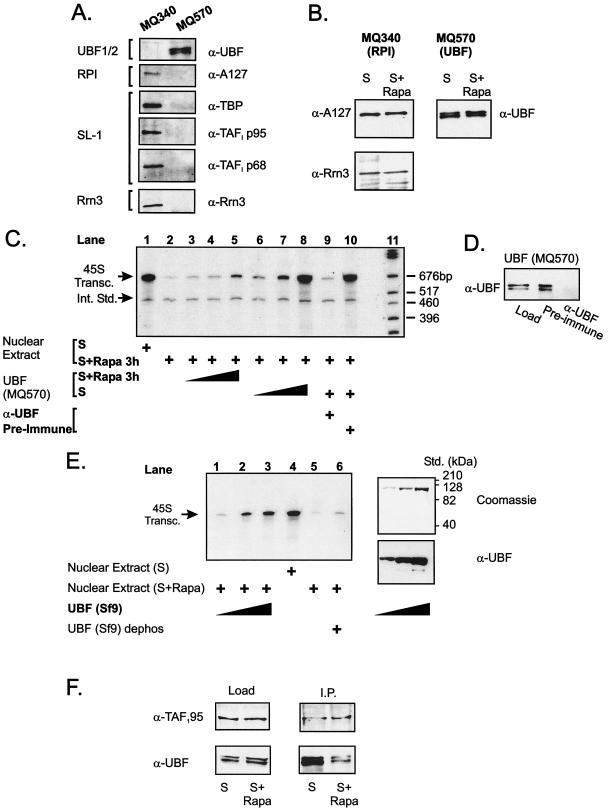
To further explore the role of UBF phosphorylation in the regulation of RPI transcription, we examined the relative ability of partially purified UBF from serum- or rapamycin-treated NIH 3T3 cells to rescue transcription in nuclear extracts from rapamycin-treated cells. UBF was separated from RPI/SL-1 by MonoQ anion-exchange chromatography of nuclear extracts as described previously (48). Western blot analysis with antibodies to the A127 subunit of RPI or UBF revealed effective separation, with RPI eluting at ∼340 mM NH4SO4 and UBF eluting at ∼570 mM NH4SO4. The presence of SL-1 in the fractions containing RPI and their absence from the fractions containing UBF was confirmed by Western blot analysis using antibodies to three subunits of SL-1, TBP, TAFI p95, and TAFI p68. Rrn3 (also called transcription initiation factor 1A or TIF-1A) has been implicated in the regulation of rDNA transcription in response to mitogens by recruiting the RPI complex to the core rDNA transcription factors on the rDNA promoter (5, 67). The presence of Rrn3 in the RPI/SL-1 complex but not the UBF-containing fractions was also confirmed by Western blotting (Fig. 6A ). Each pool was concentrated, and the volume was adjusted to ensure that an equal amount of each factor purified from cells treated with either serum or rapamycin could be added to the transcription reaction mixtures (Fig. 6B).
FIG. 6.
UBF from serum-stimulated cells and recombinant baculovirus UBF rescues rapamycin-inhibited RPI activity. NIH 3T3 cells were made quiescent, stimulated with serum (in DMEM plus 10% FBS) for 24 h, and treated with rapamycin (20 nM) or vehicle (ethanol) for 3 h. Nuclear extracts were prepared and fractionated as described in Materials and Methods. (A) Fractions containing RPI/SL-1 (MQ340) and UBF (MQ570) were pooled and analyzed by Western blotting for expression of RPI (anti-A127), SL-1 (anti-TBP, anti-TAFI p95, and anti-TAFI p68), Rrn3 (anti-Rrn3), or UBF (anti-UBF). (B) The RPI/SL-1 and UBF fractions were concentrated, and the volume was adjusted to ensure that an equal amount of each factor purified could be added to the transcription reaction mixtures. In addition, the level of Rrn3 was determined asillustrated by Western blotting. (C) Nuclear extracts (50 μg) were assayed for their ability to transcribe 0.1 μg of the 45S template gene in vitro and respond to partially purified UBF (MQ570). Lanes 3 to 5 and 6 to 8 contain 1, 3, and 10 μl of UBF (MQ570) from rapamycin- or serum-treated extracts, respectively. In lanes 9 and 10, the UBF fraction was precleared with anti-UBF or preimmune serum before addition to the transcription reaction mixture. Lane 11 contains molecular size markers. (D) Western blot illustrates effective immunodepletion of UBF from MQ570 fraction. (E) Nuclear extracts (50 μg) were assayed for their ability to transcribe 0.1 μg of the 45S template gene in vitro and respond to purified baculovirus-expressed UBF. Lanes 1 to 3 contain 10, 25, and 50 ng of UBF added to a rapamycin-treated nuclear extract. Lane 4 contains serum-treated nuclear extract. Lanes 5 to 6 illustrates the effect of dephosphorylating the baculovirus UBF on transcription in a rapamycin-treated nuclear extract. The purity of the baculovirus-expressed UBF is illustrated by the Coomassie blue and Western blots. (F) Nuclear extracts (200 μg) from serum- or rapamycin treated cells were incubated with anti-TAF1 p95 antibodies (2 μg) in C-10 buffer containing 0.5% NP-40 at 4°C for 3 h, and then washed protein A Sepharose beads (25 μl) were added. The sample was tumbled for 30 min, and the beads were washed three times with C-10 buffer containing 0.5% NP-40. Sample loading buffer was added, and the samples were boiled and separated on SDS-8% polyacrylamide gels, transferred, and Western blotted with anti-UBF or anti-TAF1 p95 antibodies. Abbreviations: α-UBF, anti-UBF; UBF1/2, UBF1 and UBF2; S, 10% FBS; S+ Rapa, 10% FBS plus 20 nM rapamycin; 45S Transc., 45S transcripts; Int. Std., internal standard; Std., standards; dephos, dephosphorylated; I.P., immunoprecipitation.
Nuclear extracts from serum-stimulated cells supported robust transcription that was severely repressed by rapamycin (Fig. 6C, compare lanes 1 and 2). Addition of increasing amounts of hypophosphorylated UBF (MQ570) from rapamycin-treated cells only marginally activated transcription (lanes 3 to 5). In contrast, phosphorylated UBF from serum-treated cells activated transcription in a dose-dependent manner (lanes 6 to 8), with the highest level of UBF restoring transcription to the level observed in serum-treated extracts (compare lanes 1 and 8). Immunodepletion of UBF inhibited the ability of MQ570 fractions from serum-stimulated cells to rescue transcription (lane 9), whereas preimmune serum had no effect (lane 10). Western blot analysis confirmed that UBF was quantitatively and specifically cleared from the MQ570 fraction by the anti-UBF antiserum (Fig. 6D). To confirm that UBF was the effective component in the MQ570 fraction, we examined the ability of recombinant phosphorylated UBF to rescue transcription in nuclear extracts from rapamycin-treated cells. As shown in Fig. 6E, lanes 1 to 3, highly purified recombinant UBF was able to rescue transcription in rapamycin-treated extracts almost back to the level observed for serum-treated cells (compare lanes 3 and 4). Importantly, the ability of the recombinant UBF to rescue transcription was dependent on phosphorylation, since phosphatase-treated UBF was unable to rescue transcription of extracts from rapamycin-treated cells (Fig. 6E, lane 6).
Phosphorylation of the carboxy-terminal activation domain of UBF regulates its ability to recruit the SL-1 complex to the rDNA promoter (27, 60). To confirm that rapamycin-dependent inhibition of UBF phosphorylation correlated with a reduced ability of UBF to interact with SL-1, we performed coimmunoprecipitation experiments. SL-1 was immunoprecipitated from serum- and rapamycin-treated NIH 3T3 cell extracts using antibodies to TAFI p95, and the amount of coimmunoprecipitated UBF was detected by Western blotting (Fig. 6F). The extracts contained equal levels of UBF, and although UBF coimmunoprecipitated with SL-1 in extracts from serum-treated cells, this was reduced by approximately 80% in rapamycin-treated extracts (Fig. 6F). Thus, rapamycin treatment leads to dephosphorylation of the carboxy-terminal activation domain of UBF and its dissociation from SL-1.
Rrn3 (TIF-1A), RPI, and SL-1 are not inhibited by rapamycin.
Rrn3 is an essential factor for mammalian rDNA transcription (5, 36). In response to mitogen stimulation, Rrn3 is phosphorylated and activated by the ERK and RSK kinases, suggesting that this factor plays a central role in coupling extracellular growth stimuli to ribosome biogenesis (67). Accordingly, we examined whether Rrn3 activity might also be regulated by mTOR/S6K1 signaling.
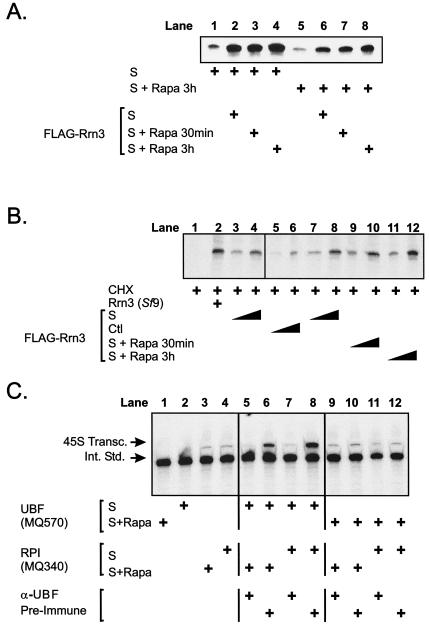
FLAG-tagged Rrn3 was affinity purified from transfected NIH 3T3 cells treated with serum or rapamycin for 30 min or 3 h, and its ability to rescue in vitro transcription in extracts from NIH 3T3 cells treated with serum or rapamycin (3 h) was measured. Rrn3 purified from serum-treated NIH 3T3 cells added to extracts from serum-treated NIH 3T3 cells (Fig. 7A, lane 2) was able to stimulate transcription, demonstrating that Rrn3 is a rate-limiting factor for rDNA transcription, even under these optimal growth conditions (compare lanes 1 and 2). Rrn3 purified from cells treated with rapamycin for 30 min (lane 3) or 3 h (lane 4) stimulated transcription to a level equivalent to that of Rrn3 from serum-treated extracts (compare lanes 3 and 4 with lane 2).
FIG. 7.
Rrn3, RPI, and SL-1 activities are not inhibited by rapamycin. (A) The ability of purified Rrn3 to complement in vitro transcription of 0.1 μg of the 45S template gene in S100 extracts from NIH 3T3 cells (40 μg) treated with serum or rapamycin (3 h) was assayed. Lanes 1 to 4 contain S100 extracts from serum-treated cells complemented with equal amounts (50 ng) of affinity-purified FLAG-tagged Rrn3 from cells treated with serum or rapamycin (30 min or 3 h). Lanes 5 to 8 contain S100 extracts from cells treated with rapamycin (3 h) complemented with purified Rrn3 from cells treated with serum or rapamycin (30 min or 3 h). The amount of affinity-purified Rrn3 was determined by silver-stained polyacrylamide gels and Western blotting against recombinant Rrn3 from Sf9 cells (5, 25; data not shown). (B) The ability of purified Rrn3 to complement in vitro transcription of 0.1 μg of the 45S template gene in S100 extracts from CHX-treated NISI cells (40 μg) was assayed. Lane 2 contains purified baculovirus-expressed Rrn3, and lanes 3 and 4 contain active FLAG-tagged Rrn3 complementing transcription from the CHX-treated S100 cell extracts. Lanes 5 to 12 contain increasing doses (10 and 25 ng) of FLAG-tagged Rrn3 purified from serum-starved cells (lanes 5 and 6), serum-treated cells (lanes 7 and 8), and cells treated with rapamycin for 30 min (lanes 9 and 10) or 3 h (lanes 11 and 12). (C) UBF (MQ570 [lanes 1 and 2]) and RPI/SL-1 (MQ340 [lanes 3 and 4]) fractions from cells treated with serum or rapamycin (3 h) were assayed for their ability to transcribe 0.1 μg of the 45S template gene in vitro. Alternatively, RPI/SL-1 fractions precleared with anti-UBF or preimmune sera were assayed in the presence (+) of UBF from cells treated with serum (lanes 5 to 8) or rapamycin (lanes 9 to 12). The amount of affinity-purified FLAG-tagged Rrn3 was determined by silver-stained polyacrylamide gels and Western blotting against recombinant Rrn3 from Sf9 cells (5, 25; data not shown). Abbreviations: S, 10% FBS; S + Rapa, 10% FBS plus 20 nM rapamycin; FLAG-Rrn3, FLAG-tagged Rrn3, Ctl, control; 45S Transc., 45S transcripts; Int. Std., internal standard; α-UBF, anti-UBF.
To confirm that the apparent lack of rapamycin effect on Rrn3 activity was not due to Rrn3 being reactivated by the serum-treated cell extracts, we also examined the ability of Rrn3 to activate transcription in extracts from rapamycin-treated cells. As shown previously, rapamycin repressed rDNA transcription (Fig. 7A, compare lanes 1 and 5). Rrn3 purified from serum-treated cells activated transcription in rapamycin-treated extracts to levels greater than those observed for serum-treated cells (compare lanes 6 and 1). Thus, Rrn3 is a limiting factor for transcription in extracts from both serum- and rapamycin-treated cells. Importantly, however, the addition of Rrn3 purified from cells treated with rapamycin for 30 min (lane 7) and 3 h (lane 8) to extracts from rapamycin-treated cells stimulated transcription to the level observed for active Rrn3 purified from serum-treated cells (compare lanes 7 and 8 with lane 6). Thus, rapamycin does not inhibit Rrn3 activity, suggesting that alterations in Rrn3 activity are unlikely to contribute to the repression of rDNA transcription observed in response to inhibition of mTOR/S6K signaling. Notably, Rrn3 purified from either serum- or rapamycin-treated cells is not able to activate transcription in rapamycin-treated extracts (lanes 6 to 8) to the same extent as in serum-treated extracts (lanes 2 to 4), which is also consistent with the difference between these two extracts being due to the rapamycin-dependent repression of UBF demonstrated above.
To further confirm that Rrn3 activity was not repressed after inhibition of mTOR/S6K1 signaling, we performed the standard assay for Rrn3 activity based on the observation that Rrn3 is rapidly inhibited by CHX and that purified Rrn3 can fully complement RPI transcription in CHX-treated cell extracts (5). FLAG-tagged Rrn3 was affinity purified from transfected NIH 3T3 cells treated with rapamycin for 30 min or 3 h, and its ability to rescue in vitro transcription in CHX-treated NISI cell extracts was measured in the standard CHX assay (5, 25). For positive controls, recombinant Rrn3 purified from Sf9 cells reconstituted transcription from CHX-treated NISI S100 extracts (Fig. 7B, compare lanes 1 and 2), as did increasing amounts of FLAG-tagged Rrn3 purified from serum-treated cells (Fig. 7B, compare lanes 3 and 4), demonstrating that the transcription system was responsive to Rrn3 as previously reported (1, 5). Rrn3 purified from exponentially growing NIH 3T3 cells stimulated transcription to a greater extent than Rrn3 from serum-starved cells (compare lanes 5 and 6 with lanes 7 and 8), consistent with serum-mediated regulation of Rrn3 shown in previous studies (67). Importantly, treatment of serum-stimulated cells with rapamycin for 30 min (lanes 9 and 10) or 3 h (lanes 11 and 12) had no significant effect on the ability of Rrn3 to rescue transcription compared to the Rrn3 activity from serum-treated cells (lanes 7 and 8). Taken together, these experimental results demonstrate that acute rapamycin-mediated repression of rDNA transcription does not appear to be mediated by regulation of Rrn3 activity.
While our studies show that UBF is a major target for inactivation by rapamycin, the possibility that RPI/SL-1 was independently regulated was tested. The UBF fractions (MQ570) did not support transcription, consistent with their depletion of RPI and SL-1 (Fig. 7C, lanes 1 and 2). The RPI/SL-1 (MQ340) fractions supported rDNA transcription (lanes 3 and 4), consistent with previous studies demonstrating that nuclear extracts depleted of UBF are still able to initiate basal rDNA transcription in vitro (51). Importantly, there was no significant difference in transcriptional activity between RPI/SL-1 extracts from serum- and rapamycin-treated cells. As these extracts contain equivalent levels of Rrn3 (Fig. 6B), these data are consistent with Rrn3 playing no major role in rapamycin inhibition of rDNA transcription. Furthermore, the RPI/SL-1 fractions from cells treated with serum (lane 6) and rapamycin (lane 7) were significantly activated by UBF purified from serum-treated cells, and this was prevented by depleting the extracts of UBF with anti-UBF antibodies (lanes 5 and 7). Finally, transcription from either RPI/SL-1 fraction (serum or rapamycin treated) was not activated by UBF purified from rapamycin-treated extracts (lanes 9 to 12).
Taken together, these data demonstrate that Rrn3, RPI, and SL-1 are not significantly affected by inhibition of mTOR/S6K signaling. Instead, rapamycin repression of rDNA transcription is mediated by inactivation of UBF via a mechanism that involves decreased phosphorylation of the carboxy-terminal activation domain, leading to reduced association between UBF and SL-1.
DISCUSSION
Regulated cell growth is absolutely required for faithful cell division and hence proliferation. Understanding how these responses are coordinated has been the focus of much recent attention (38, 55, 57). A key issue has been to elucidate which mitogen-activated intracellular signaling pathways also mediate cell growth. Recent studies have clearly demonstrated that mTOR signaling is a primary mechanism by which the size and growth of mammalian cells are regulated (8, 10, 49, 58). The exact mechanism by which this is achieved is equivocal but appears to require the coordinate activation of two downstream targets of mTOR, 4EBP1, and S6K1 (8). Here, we provide evidence that at least part of the mechanism by which mTOR regulates cell growth is by modulating the rate of transcription of ribosomal genes, an important limiting step in the synthesis of ribosomes and hence of translational capacity. Moreover, we demonstrate that the stimulation of rDNA transcription by mTOR requires S6K1 activity and is mediated largely through the phosphorylation of the carboxy-terminal activation domain of the rDNA transcription factor UBF.
Two independent observations led us to conclude that S6K1 is the major pathway downstream of mTOR that mediates serum activation of rDNA transcription. First, overexpression of a constitutively active, partially rapamycin-insensitive mutant of S6K1 signaling was shown to be sufficient to overcome the inhibition of rDNA transcription by rapamycin. Second, a dominant interfering mutant of S6K1 abolished serum-stimulated rDNA transcription. Given the well-established role for S6K1 in regulating r-protein synthesis, our study provides evidence for a novel mechanism for the coordinated regulation of two of the major steps in ribosome biogenesis by a single pathway in mammalian cells. Interestingly, studies from S. cerevisiae demonstrate that mTOR exerts control over general translation initiation and r-protein gene transcription as well as synthesis and processing of the 35S and 5S precursor RNA (44, 66). Thus, taken with the current data, mTOR represents a critical step in linking translational efficiency and capacity, and this function is conserved in yeast and mammalian cells.
These findings contrast with a previous study in mammalian cells that found only limited (50%) inhibition of rDNA transcription in lymphosarcoma P1798 cells after 24- to 48-h treatments with 1 μM rapamycin, a 50-fold-higher concentration than is required to inhibit mTOR (33). The reason for the discrepancy is unclear, but recent studies on the ability of rapamycin to reduce cell size and protein content in a variety of cell lines clearly demonstrate that different cell types may possess various degrees of dependence on mTOR in the control of cell growth (8, 49, 58). Nonetheless, in addition to NIH 3T3 cells, we observed potent inhibition of rDNA transcription in primary fibroblasts and cardiomyocytes, reinforcing mTOR-dependent regulation of rDNA transcription as a general phenomenon.
It should be noted that our studies do not preclude a contribution from the recently identified S6K1 homologue, S6K2 (50), in the regulation of rDNA transcription induced by serum or growth factors. This highly homologous kinase has all the known S6K1 regulatory sites conserved, is similarly dependent on mTOR signaling, and is inhibited by the dominant interfering mutant of S6K1, as indicated by the ability of this mutant to inhibit the translational regulation of 5′TOP messages (26). Conversely, these messages are unaffected in S6K1 knockout mice containing redundant signaling from S6K2 (50). Interestingly, the S6K1 knockout mice are viable and fertile but significantly smaller, suggesting that S6K2 cannot completely complement all the S6K1 growth-related functions (50).
Consistent with the observation that mTOR-dependent regulation of cell growth is readily separated from its regulation of cellular proliferation (8), we demonstrated that mTOR-S6K1 signaling exhibits tight control over rDNA transcription in nonproliferating primary cardiomyocytes. Moreover, we have found that rapamycin is able to effectively block rDNA transcription in NIH 3T3 cells in which the Rb family had been functionally inactivated by retrovirus-mediated expression of the viral oncoprotein 16E7 (G. Poortinga, K. M. Hannan, R. B. Pearson and G. A. McArthur, unpublished data). Thus, despite the demonstrated role for S6K1 in the G1-to-S phase transition (30, 42) S6K1-dependent regulation of rDNA transcription is independent of cell cycle progression.
Our data indicate that mTOR activity is required for the rapid activation of rDNA transcription in response to serum stimulation. It also appears to be required to maintain stimulated transcription, since rRNA synthesis in exponentially growing cells was significantly reduced within 30 min of treatment with rapamycin. Indeed, the rapidity of the rapamycin inhibition of transcription suggested that mTOR must modulate a component of the rDNA transcription apparatus or an immediate upstream regulator whose amount or activity is acutely regulated. Consistent with this, we found that rapamycin treatment led to the rapid dephosphorylation of the carboxy-terminal activation domain of UBF. Phosphorylation of this domain is thought to activate UBF transcriptional activity by promoting its interaction with the TBP component of SL-1, thereby facilitating recruitment of RPI (60). The key role of UBF phosphorylation in mTOR-dependent regulation of rDNA transcription was supported by experiments showing that both partially purified phosphorylated UBF from serum-stimulated cells and purified recombinant UBF were able to reconstitute the RPI transcriptional activity of both nuclear extracts and partially purified RPI/SL-1 fractions from rapamycin-treated cells. Conversely, both hypophosphorylated UBF from rapamycin-treated cells and dephosphorylated, recombinant UBF failed to elevate RPI activity. Consistent with these findings, coimmunoprecipitation studies revealed endogenous UBF and RPI/SL-1 interacted less tightly in extracts from rapamycin-treated cells than exponentially growing cells. Thus, we conclude that mTOR regulates rDNA transcription by phosphorylation of the UBF carboxy-terminal activation domain, thereby promoting the interaction between UBF and SL-1 and the formation of a stable initiation complex at the rDNA promoter.
The carboxy terminus of UBF contains no consensus substrate recognition motifs for S6K1, and recombinant and immunopurified S6K1 failed to phosphorylate UBF in vitro, suggesting that UBF is not a S6K1 substrate (R. B. Pearson, K. M. Hannan, R. D. Hannan, and R. B. Pearson, unpublished data). Moreover, conditional deletion of the S6 gene does not block synthesis of the 45S rRNA precursor in mammalian cells (63), indicating that UBF activation is not mediated via S6K1-dependent phosphorylation of the ribosomal S6 protein. Thus, it is likely that S6K1 regulates rDNA transcription via the phosphorylation of a novel target(s), upstream of UBF. This is consistent with recent studies demonstrating that S6K1 has targets in addition to S6, including the transcription factor CREM-τ (7), the translation elongation factor CBP80 (65), the translation elongation regulator eEF2 kinase (3), and the proapoptotic protein Bad-1 (22).
Casein kinase 2 (CK2) has been shown to phosphorylate the carboxy-terminal activation domain of UBF in vitro and can be coimmunoprecipitated with UBF from whole-cell extracts, suggesting that UBF might be a bona fide substrate of CK2 in vivo (37, 40, 62). However, there have been only limited reports that CK2 is regulated in a growth-dependent manner. Indeed, we found rapamycin treatment of NIH 3T3 cells had no effect on CK2 activity (55.05 ± 17 versus 51.12 ± 17.2 pmol/min/mg for serum- and rapamycin-treated samples, respectively) (59). Alternatively, it is possible that mTOR/S6K1 modulates UBF phosphorylation by restraining a phosphatase. This would be consistent with the following observations: (i) the UBF carboxy-terminal activation domain is rapidly dephosphorylated following inhibition of mTOR with rapamycin, and (ii) rapamycin increases the activity of protein phosphatase 2A in certain cell types (43). Further studies are required to identify the molecular mechanism by which mTOR regulates UBF phosphorylation.
Interestingly, our data also demonstrate that rapamycin blocks the serum induction of UBF protein accumulation (11). Previous studies and experiments herein demonstrate that UBF is limiting for transcription, despite its apparent abundance compared to the other RPI transcription components (2, 17). Thus, maximal activation of rDNA transcription by mTOR might require increases in both UBF expression and UBF phosphorylation. The mechanism responsible for the regulation of UBF expression is not clear and may be mediated either via S6K1 or 4EBP1/eIF4E or indeed both mTOR-dependent pathways. UBF mRNA does not contain a 5′TOP (L. Rothblum and R. D. Hannan, unpublished data), seemingly precluding regulation of UBF by ribosomal S6 protein-dependent mechanisms. On the other hand, 4EBP1/eIF4E signaling has been shown to regulate the translation of proteins containing a complex 5′ untranslated region (10) similar to that found in the UBF mRNA. Our studies do not exclude the possibility that the increase in UBF expression is a consequence of an increase in gene copy number as the cells progress from the G1 phase to the S phase in response to mTOR activation. However, it has been demonstrated that UBF expression is up regulated by growth stimuli in postmitotic cardiomyocytes, which are unable to undergo DNA synthesis (19, 20), suggesting that mitogens can stimulate UBF expression independent of changes in DNA content. Importantly, however, overexpression of recombinant UBF was not sufficient to stimulate rDNA transcription in serum-starved cells, and it did not overcome rapamycin-induced inhibition of rDNA transcription during serum stimulation. Thus, increased expression of UBF in the absence of some mitogenic signaling is insufficient to up regulate rDNA transcription.
In addition to mTOR-dependent signaling, the Ras/MAPK (mitogen-activated protein kinase) and phosphatidylinositol 3-kinase (PI3K) pathways have been identified as critical effectors of growth (28), the former largely acting via c-myc and the latter via a complex network involving PDK1, PKB, TSC1/TSC2, and S6K1 (28, 45, 46). Surprisingly, given the critical role that ribosome biogenesis occupies in growth, direct activation of rDNA transcription by these signaling pathways has been to a large extent overlooked. Indeed, the first credible evidence in mammalian cells for a direct link from extracellular growth factor signaling to rDNA transcription came from a recent study demonstrating that the epidermal growth factor could cause immediate up regulation of rDNA transcription via ERK directly phosphorylating UBF (54). ERK-dependent phosphorylation of amino acids 117 and 201 within HMG boxes 1/2 of UBF was shown to prevent their binding to DNA (54) suggesting that cyclic phosphorylation of UBF on these residues might be required for rDNA transcription. More recently, it has been shown that mitogenic activation also leads to the ERK/RSK-mediated phosphorylation of the mammalian homologue of yeast RPI initiation factor Rrn3P (1, 35). Rrn3 was originally identified as a factor whose activity was required for the full complementation of RPI-dependent transcription in extracts from quiescent or CHX-treated cells (29). Interestingly, we were unable to demonstrate acute regulation of Rrn3 by mTOR, despite the important role this factor is thought to play in growth factor-dependent regulation of rDNA transcription (5, 67).
Clearly, the data presented here along with the recent studies describing ERK-dependent regulation of ribosome biogenesis demonstrate that core components of the RPI transcription apparatus are direct targets for activation by mitogenic signaling pathways. The critical challenge in the future will be to determine how the complex array of signaling cascade initiated by growth factors is integrated at the level of the rDNA transcription components to coordinate rRNA synthesis with growth and proliferation.
Acknowledgments
R.D.H. and R.B.P. contributed equally to this study.
This work was supported in part by grants from the National Health and Medical Research Council of Australia to R.D.H (NHMRC grants 166900 and 251688), G.A.M (NHMRC grant 208908), and R.B.P. (NHMRC grant 251688).
We thank Affrica Jenkins and Katarzyna Jastrzebski for technical assistance and Walter Thomas and David Bowtell for critical reading of the manuscript.
This article is dedicated to the memory of Nicole Lundie, an outstanding Ph.D. student in Richard Pearson's laboratory, who passed away tragically on 1 August 2003.
REFERENCES
- 1.Bodem, J., G. Dobreva, U. Hoffmann-Rohrer, S. Iben, H. Zentgraf, H. Delius, M. Vingron, and I. Grummt. 2000. TIF-IA, the factor mediating growth-dependent control of ribosomal RNA synthesis, is the mammalian homolog of yeast Rrn3p. EMBO Rep. 1:171-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brandenburger, Y., A. Jenkins, D. J. Autelitano, and R. D. Hannan. 2001. Increased expression of UBF is a critical determinant for rRNA synthesis and hypertrophic growth of cardiac myocytes. FASEB J. 15:2051-2053. [DOI] [PubMed] [Google Scholar]
- 3.Brennan, P., J. W. Babbage, G. Thomas, and D. Cantrell. 1999. p70s6k integrates phosphatidylinositol 3-kinase and rapamycin-regulated signals for E2F regulation in T lymphocytes. Mol. Cell. Biol. 19:4729-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cassidy, B. G., H. F. Yang-Yen, and L. I. Rothblum. 1986. Transcriptional role for the nontranscribed spacer of rat ribosomal DNA. Mol. Cell. Biol. 6:2766-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavanaugh, A. H., I. Hirschler-Laszkiewicz, Q. Hu, M. Dundr, T. Smink, T. Misteli, and L. I. Rothblum. 2002. Rrn3 phosphorylation is a regulatory checkpoint for ribosome biogenesis. J. Biol. Chem. 277:27423-27432. [DOI] [PubMed] [Google Scholar]
- 6.Conus, N. M., B. A. Hemmings, and R. B. Pearson. 1998. Differential regulation by calcium reveals distinct signaling requirements for the activation of Akt and p70S6k. J. Biol. Chem. 273:4776-4782. [DOI] [PubMed] [Google Scholar]
- 7.de Groot, R. P., L. M. Ballou, and P. Sassone-Corsi. 1994. Positive regulation of the cAMP-responsive activator CREM by the p70 S6 kinase: an alternative route to mitogen-induced gene expression. Cell 79:81-91. [DOI] [PubMed] [Google Scholar]
- 8.Fingar, D. C., S. Salama, C. Tsou, E. Harlow, and J. Blenis. 2002. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 16:1472-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Control of translation by the target of rapamycin proteins. Prog. Mol. Subcell. Biol. 27:143-174. [DOI] [PubMed] [Google Scholar]
- 10.Gingras, A. C., B. Raught, and N. Sonenberg. 2001. Regulation of translation initiation by FRAP/mTOR. Genes Dev. 15:807-826. [DOI] [PubMed] [Google Scholar]
- 11.Glibetic, M., L. Taylor, D. Larson, R. Hannan, B. Sells, and L. Rothblum. 1995. The RNA polymerase I transcription factor UBF is the product of a primary response gene. J. Biol. Chem. 270:4209-4212. [DOI] [PubMed] [Google Scholar]
- 12.Haglund, R. E., and L. I. Rothblum. 1987. Isolation, fractionation and reconstitution of a nuclear extract capable of transcribing ribosomal DNA. Mol. Cell. Biochem. 73:11-20. [DOI] [PubMed] [Google Scholar]
- 13.Han, J. W., R. B. Pearson, P. B. Dennis, and G. Thomas. 1995. Rapamycin, wortmannin, and the methylxanthine SQ20006 inactivate p70s6k by inducing dephosphorylation of the same subset of sites. J. Biol. Chem. 270:21396-21403. [DOI] [PubMed] [Google Scholar]
- 14.Hannan, K. M., R. D. Hannan, and L. I. Rothblum. 1998. Transcription by RNA polymerase I. Front. Biosci. 3:d376-d398. [DOI] [PubMed] [Google Scholar]
- 15.Hannan, K. M., B. K. Kennedy, A. H. Cavanaugh, R. D. Hannan, I. Hirschler-Laszkiewicz, L. S. Jefferson, and L. I. Rothblum. 2000. RNA polymerase I transcription in confluent cells: Rb downregulates rDNA transcription during confluence-induced cell cycle arrest. Oncogene 19:3487-3497. [DOI] [PubMed] [Google Scholar]
- 16.Hannan, K. M., G. Thomas, and R. B. Pearson. 2003. Activation of S6K1 (p70 ribosomal protein S6 kinase 1) requires an initial calcium-dependent priming event involving formation of a high-molecular-mass signalling complex. Biochem. J. 370:469-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hannan, R., V. Stefanovsky, T. Arino, L. Rothblum, and T. Moss. 1999. Cellular regulation of ribosomal DNA transcription: both rat and Xenopus UBF1 stimulate rDNA transcription in 3T3 fibroblasts. Nucleic Acids Res. 27:1205-1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hannan, R. D., W. M. Hempel, A. Cavanaugh, T. Arino, S. I. Dimitrov, T. Moss, and L. Rothblum. 1998. Affinity purification of mammalian RNA polymerase I. Identification of an associated kinase. J. Biol. Chem. 273:1257-1267. [DOI] [PubMed] [Google Scholar]
- 19.Hannan, R. D., J. Luyken, and L. I. Rothblum. 1995. Regulation of rDNA transcription factors during cardiomyocyte hypertrophy induced by adrenergic agents. J. Biol. Chem. 270:8290-8297. [DOI] [PubMed] [Google Scholar]
- 20.Hannan, R. D., J. Luyken, and L. I. Rothblum. 1996. Regulation of ribosomal DNA transcription during contraction-induced hypertrophy of neonatal cardiomyocytes. J. Biol. Chem. 271:3213-3220. [DOI] [PubMed] [Google Scholar]
- 21.Hannan, R. D., V. Stefanovsky, L. Taylor, T. Moss, and L. I. Rothblum. 1996. Overexpression of the transcription factor UBF1 is sufficient to increase ribosomal DNA transcription in neonatal cardiomyocytes: implications for cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 93:8750-8755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada, H., J. S. Andersen, M. Mann, N. Terada, and S. J. Korsmeyer. 2001. p70S6 kinase signals cell survival as well as growth, inactivating the pro-apoptotic molecule BAD. Proc. Natl. Acad. Sci. USA 98:9666-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hempel, W. M., A. H. Cavanaugh, R. D. Hannan, L. Taylor, and L. I. Rothblum. 1996. The species-specific RNA polymerase I transcription factor SL-1 binds to upstream binding factor. Mol. Cell. Biol. 16:557-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershey, J. C., M. Hautmann, M. M. Thompson, L. I. Rothblum, T. A. Haystead, and G. K. Owens. 1995. Angiotensin II-induced hypertrophy of rat vascular smooth muscle is associated with increased 18 S rRNA synthesis and phosphorylation of the rRNA transcription factor, upstream binding factor. J. Biol. Chem. 270:25096-25101. [DOI] [PubMed] [Google Scholar]
- 25.Hirschler-Laszkiewicz, I., A. H. Cavanaugh, A. Mirza, M. Lun, Q. Hu, T. Smink, and L. I. Rothblum. 2003. Rrn3 becomes inactivated in the process of ribosomal DNA transcription. J. Biol. Chem. 278:18953-18959. [DOI] [PubMed] [Google Scholar]
- 26.Jefferies, H. B., S. Fumagalli, P. B. Dennis, C. Reinhard, R. B. Pearson, and G. Thomas. 1997. Rapamycin suppresses 5′TOP mRNA translation through inhibition of p70s6k. EMBO J. 16:3693-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kihm, A. J., J. C. Hershey, T. A. J. Haystead, C. S. Madsen, and G. K. Owens. 1998. Phosphorylation of the rRNA transcription factor upstream binding factor promotes its association with TATA binding protein. Proc. Natl. Acad. Sci. USA 95:14816-14820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kozma, S. C., and G. Thomas. 2002. Regulation of cell size in growth, development and human disease: PI3K, PKB and S6K. Bioessays 24:65-71. [DOI] [PubMed] [Google Scholar]
- 29.Lampert, A., and P. Feigelson. 1974. A short lived polypeptide component of one of two discrete functional pools of hepatic nuclear alpha-amanitin resistant RNA polymerases. Biochem. Biophys. Res. Commun. 58:1030-1038. [DOI] [PubMed] [Google Scholar]
- 30.Lane, H. A., A. Fernandez, N. J. Lamb, and G. Thomas. 1993. p70s6k function is essential for G1 progression. Nature 363:170-172. [DOI] [PubMed] [Google Scholar]
- 31.Luo, K. X., T. R. Hurley, and B. M. Sefton. 1991. Cyanogen bromide cleavage and proteolytic peptide mapping of proteins immobilized to membranes. Methods Enzymol. 201:149-152. [DOI] [PubMed] [Google Scholar]
- 32.Luyken, J., R. D. Hannan, J. Y. Cheung, and L. I. Rothblum. 1996. Regulation of rDNA transcription during endothelin-1-induced hypertrophy of neonatal cardiomyocytes. Hyperphosphorylation of upstream binding factor, an rDNA transcription factor. Circ. Res. 78:354-361. [DOI] [PubMed] [Google Scholar]
- 33.Mahajan, P. B. 1994. Modulation of transcription of rRNA genes by rapamycin. Int. J. Immunopharmacol. 16:711-721. [DOI] [PubMed] [Google Scholar]
- 34.Montagne, J., M. J. Stewart, H. Stocker, E. Hafen, S. C. Kozma, and G. Thomas. 1999. Drosophila S6 kinase: a regulator of cell size. Science 285:2126-2129. [DOI] [PubMed] [Google Scholar]
- 35.Moorefield, B., E. A. Greene, and R. H. Reeder. 2000. RNA polymerase I transcription factor Rrn3 is functionally conserved between yeast and human. Proc. Natl. Acad. Sci. USA 97:4724-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moss, T., and V. Y. Stefanovsky. 2002. At the center of eukaryotic life. Cell 109:545-548. [DOI] [PubMed] [Google Scholar]
- 37.Moss, T., and V. Y. Stefanovsky. 1995. Promotion and regulation of ribosomal transcription in eukaryotes by RNA polymerase I. Prog. Nucleic Acid Res. Mol. Biol. 50:25-66. [DOI] [PubMed] [Google Scholar]
- 38.Neufeld, T. P., and B. A. Edgar. 1998. Connections between growth and the cell cycle. Curr. Opin. Cell Biol. 10:784-790. [DOI] [PubMed] [Google Scholar]
- 39.O'Mahony, D. J., W. Q. Xie, S. D. Smith, H. A. Singer, and L. I. Rothblum. 1992. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J. Biol. Chem. 267:35-38. [PubMed] [Google Scholar]
- 40.Paule, M. R. 1998. Transcription of ribosomal genes by eukaryotic RNA polymerase 1. Landes Bioscience, Austin, Tex.
- 41.Pearson, R. B., P. B. Dennis, J. W. Han, N. A. Williamson, S. C. Kozma, R. E. Wettenhall, and G. Thomas. 1995. The principal target of rapamycin-induced p70s6k inactivation is a novel phosphorylation site within a conserved hydrophobic domain. EMBO J. 14:5279-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pearson, R. B., and G. Thomas. 1995. Regulation of p70s6k/p85s6k and its role in the cell cycle. Prog. Cell Cycle Res. 1:21-32. [DOI] [PubMed] [Google Scholar]
- 43.Peterson, R. T., B. N. Desai, J. S. Hardwick, and S. L. Schreiber. 1999. Protein phosphatase 2A interacts with the 70-kDa S6 kinase and is activated by inhibition of FKBP12-rapamycin-associated protein. Proc. Natl. Acad. Sci. USA 96:4438-4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Powers, T., and P. Walter. 1999. Regulation of ribosome biogenesis by the rapamycin-sensitive TOR-signaling pathway in Saccharomyces cerevisiae. Mol. Biol. Cell 10:987-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Radimerski, T., J. Montagne, M. Hemmings-Mieszczak, and G. Thomas. 2002. Lethality of Drosophila lacking TSC tumor suppressor function rescued by reducing dS6K signaling. Genes Dev. 16:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Radimerski, T., J. Montagne, F. Rintelen, H. Stocker, J. van Der Kaay, C. P. Downes, E. Hafen, and G. Thomas. 2002. dS6K-regulated cell growth is dPKB/dPI3K-independent, but requires dPDK1. Nat. Cell Biol. 4:251-255. [DOI] [PubMed] [Google Scholar]
- 47.Raught, B., A. C. Gingras, and N. Sonenberg. 2001. The target of rapamycin (TOR) proteins. Proc. Natl. Acad. Sci. USA 98:7037-7044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rothblum, L. I., R. Reddy, and B. Cassidy. 1982. Transcription initiation site of rat ribosomal DNA. Nucleic Acids Res. 10:7345-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmelzle, T., and M. N. Hall. 2000. TOR, a central controller of cell growth. Cell 103:253-262. [DOI] [PubMed] [Google Scholar]
- 50.Shima, H., M. Pende, Y. Chen, S. Fumagalli, G. Thomas, and S. C. Kozma. 1998. Disruption of the p70s6k/p85s6k gene reveals a small mouse phenotype and a new functional S6 kinase. EMBO J. 17:6649-6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Smith, S. D., D. J. O'Mahony, B. T. Kinsella, and L. I. Rothblum. 1993. Transcription from the rat 45S ribosomal DNA promoter does not require the factor UBF. Gene Expr. 3:229-236. [PMC free article] [PubMed] [Google Scholar]
- 52.Smith, S. D., E. Oriahi, D. Lowe, H. F. Yang-Yen, D. O'Mahony, K. Rose, K. Chen, and L. I. Rothblum. 1990. Characterization of factors that direct transcription of rat ribosomal DNA. Mol. Cell. Biol. 10:3105-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sonenberg, N., and A. C. Gingras. 1998. The mRNA 5′ cap-binding protein eIF4E and control of cell growth. Curr. Opin. Cell Biol. 10:268-275. [DOI] [PubMed] [Google Scholar]
- 54.Stefanovsky, V. Y., G. Pelletier, R. Hannan, T. Gagnon-Kugler, L. I. Rothblum, and T. Moss. 2001. An immediate response of ribosomal transcription to growth factor stimulation in mammals is mediated by ERK phosphorylation of UBF. Mol. Cell 8:1063-1073. [DOI] [PubMed] [Google Scholar]
- 55.Su, T. T., and P. H. O'Farrell. 1998. Size control: cell proliferation does not equal growth. Curr. Biol. 8:R687-R689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang, H., E. Hornstein, M. Stolovich, G. Levy, M. Livingstone, D. Templeton, J. Avruch, and O. Meyuhas. 2001. Amino acid-induced translation of TOP mRNAs is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of S6K1 and rpS6 phosphorylation. Mol. Cell. Biol. 21:8671-8683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas, G. 2000. An encore for ribosome biogenesis in the control of cell proliferation. Nat. Cell Biol. 2:E71-E72. [DOI] [PubMed]
- 58.Thomas, G., and M. N. Hall. 1997. TOR signalling and control of cell growth. Curr. Opin. Cell Biol. 9:782-787. [DOI] [PubMed] [Google Scholar]
- 59.Tiganis, T., C. M. House, and B. E. Kemp. 1996. Protein kinase CK2: biphasic kinetics with peptide substrates. Arch. Biochem. Biophys. 325:289-294. [DOI] [PubMed] [Google Scholar]
- 60.Tuan, J. C., W. Zhai, and L. Comai. 1999. Recruitment of TATA-binding protein-TAFI complex SL1 to the human ribosomal DNA promoter is mediated by the carboxy-terminal activation domain of upstream binding factor (UBF) and is regulated by UBF phosphorylation. Mol. Cell. Biol. 19:2872-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Voit, R., A. Kuhn, E. E. Sander, and I. Grummt. 1995. Activation of mammalian ribosomal gene transcription requires phosphorylation of the nucleolar transcription factor UBF. Nucleic Acids Res. 23:2593-2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Voit, R., A. Schnapp, A. Kuhn, H. Rosenbauer, P. Hirschmann, H. G. Stunnenberg, and I. Grummt. 1992. The nucleolar transcription factor mUBF is phosphorylated by casein kinase II in the C-terminal hyperacidic tail which is essential for transactivation. EMBO J. 11:2211-2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Volarevic, S., M. J. Stewart, B. Ledermann, F. Zilberman, L. Terracciano, E. Montini, M. Grompe, S. C. Kozma, and G. Thomas. 2000. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science 288:2045-2047. [DOI] [PubMed] [Google Scholar]
- 64.West, M. J., M. Stoneley, and A. E. Willis. 1998. Translational induction of the c-myc oncogene via activation of the FRAP/TOR signalling pathway. Oncogene 17:769-780. [DOI] [PubMed] [Google Scholar]
- 65.Wilson, K. F., W. J. Wu, and R. A. Cerione. 2000. Cdc42 stimulates RNA splicing via the S6 kinase and a novel S6 kinase target, the nuclear cap-binding complex. J. Biol. Chem. 275:37307-37310. [DOI] [PubMed] [Google Scholar]
- 66.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao, J., X. Yuan, M. Frodin, and I. Grummt. 2003. ERK-dependent phosphorylation of the transcription initiation factor TIF-IA is required for RNA polymerase I transcription and cell growth. Mol. Cell 11:405-413. [DOI] [PubMed] [Google Scholar]