Abstract
The transcription coactivator p300 cannot acetylate native p53 tetramers, thus revealing intrinsic conformational constraints on p300-catalyzed acetylation. Consensus site DNA is an allosteric effector that promotes acetylation of p53, suggesting that p300 has an undefined conformationally flexible interface within the p53 tetramer. To identify such conformationally responsive p300-binding sites, p300 was subjected to peptide selection from a phage-peptide display library, a technique that can define novel protein-protein interfaces. The enriched p300-binding peptides contained a proline repeat (PXXP/PXPXP) motif, and five proline repeat motifs actually reside within the p53 transactivation domain, suggesting that this region of p53 may harbor the second p300 contact site. p300 binds in vitro to PXXP-containing peptides derived from the proline repeat domain, and PXXP-containing peptides inhibit sequence-specific DNA-dependent acetylation of p53, indicating that p300 docking to both the LXXLL and contiguous PXXP motif in p53 is required for p53 acetylation. Deletion of the proline repeat motif of p53 prevents DNA-dependent acetylation of p53 by occluding p300 from the p53-DNA complex. Sequence-specific DNA places an absolute requirement for the proline repeat domain to drive p53 acetylation in vivo. Chromatin immunoprecipitation was used to show that the proline repeat deletion mutant p53 is bound to the p21 promoter in vivo, but it is not acetylated, indicating that proline-directed acetylation of p53 is a post-DNA binding event. The PXXP repeat expands the basic interface of a p300-targeted transactivation domain, and proline-directed acetylation of p53 at promoters indicates that p300-mediated acetylation can be highly constrained by substrate conformation in vivo.
The tumor suppressor protein p53 is one of the most well-studied stress-responsive eukaryotic transcription factors that function in a damage-induced cell cycle checkpoint pathway. The biochemical activity of p53 linked to its tumor suppression function is a sequence-specific DNA binding and transactivation function that controls the expression of gene products implicated in cell cycle arrest and apoptosis (39).
p53 has been dissected into functional domains that contribute to its transcription activity. The central domain of p53 contains the sequence-specific DNA binding domain that is often mutated in human cancers (31). Regulatory domains at the amino and carboxyl terminal of p53 modulate protein-protein interactions and DNA-protein interactions that affect the rate of p53-dependent transcription. The C terminus of p53 contains a domain whose phosphorylation at Ser315 in vivo by cyclin-dependent kinases (7) or at Ser392 by CK2/FACT stimulates the DNA-binding activity of p53 (23). The N-terminal domain of p53 contains the highly conserved BOX-I transactivation domain that directs the binding of p53 to the transcriptional adapter protein p300 (2). Phosphorylation of p53 in the transactivation domain at Ser15 activates p53 by an ATM-dependent pathway (37). Adjacent phosphorylation of the p53 activation domain at Thr18 or Ser20 by CHK2 activates p53 (36) by stabilizing the binding of p300 to p53 (11). Docking of p300 to the Thr18/Ser20 phosphorylated-LXXLL transactivation domain of p53 in turn promotes sequence-specific DNA-dependent acetylation in the C-terminal domain of p53, thus stabilizing the p300-p53Ac complex (12). These data highlight the complementary role of phosphorylation and acetylation in assembling a p53-p300 transcription complex.
The coactivator p300 plays a central role in signal integration with transcriptional components allowing for gene expression changes in response to a variety of stimuli (8). Tumor suppressor proteins like E2F and p53 recruit p300/CBP as their main coactivators, thus revealing these adapter polypeptides as key partners in transcription-dependent cancer control. In addition to the scaffolding role of p300/CBP, a role for the coactivator family in chromatin remodeling has been identified via an intrinsic acetyltransferase activity (24). The steady-state levels of histone acetylation mediated by p300/CBP and antagonizing histone deacetylases modulates chromatin remodeling and the rates of gene expression. Further, since the discovery that p300/CBP also acetylates nonhistone transcription factors like p53, E2F, and MyoD (18, 29, 35), most studies have demonstrated that the general role for acetylation appears to be in the stimulation of sequence-specific DNA binding.
The complex regulation and role of p53 acetylation is beginning to be unraveled (33). The original study using p53 showed that acetylation stimulates the latent DNA-binding function of p53 (18), while a later study did not show an effect of acetylation on activating the latent DNA-binding activity of p53 (14). We have started to reconstitute the stages in the assembly of the p300-p53-DNA transactivation complex in order to further clarify the regulation and function of p53 acetylation. Such studies have identified three key stages in the assembly reaction. First, phosphorylation by CHK2 at Thr18 or Ser20 in the p53 activation domain stabilizes p300 docking to the p53 activation domain (11) via the IBiD and IHD phosphopeptide binding domains of p300 (12). Second, this docking of p300 is essential for sequence-specific DNA-dependent acetylation of p53, indicating that p53 tetramer acetylation has intrinsic conformational constraints in the absence of DNA (12). Third, the function of acetylation as a post-DNA-binding event is to clamp the p300-p53AC complex into a very stable state (12). This clamping of p300-p53 after acetylation is consistent with cellular data showing that acetylation may function to recruit coactivator complexes at a promoter (4), presumably through the Bromo homology domain of p300/CBP which has the potential to bind to acetylated residues (28, 32).
One notable feature of the p300-p53 assembly reaction is the sequence-specific DNA dependence in p53 acetylation (12), which is consistent with recent ideas that DNA can function as an allosteric effector to regulate protein-protein interactions at a promoter (26). The DNA dependence in p53 acetylation indicates that conformational restraints are placed on the p300-catalyzed acetylation reaction through conformationally flexible motifs on p53. To identify such flexible p300-docking motifs in p53, combinatorial approaches were used to show that a second, conformationally flexible p300-binding motif exists within the proline repeat domain of p53. We show here that the proline repeat domain can bind directly to p300, that the proline repeat domain responds to conformational changes mediated by DNA binding, and that p53 binding to promoter sites in vivo places an absolute requirement on the proline repeat domain for acetylation to occur. The identification of a relatively ubiquitous proline repeat p300-binding transactivation motif complementing the classic hydrophobic LXXLL motif highlights an additional layer of combinatorial regulation of core p300 protein-protein interactions at a promoter.
MATERIALS AND METHODS
Plasmids and constructs.
EGFP-PRO was constructed by ligating double-stranded oligonucleotides encoding amino acids (aa) 64 to 92 of human p53 (EGFP-PRO) into XhoI/XbaI-digested EGFP-C3 plasmid (Clontech). EGFP-NS, EGFP-BOX-I, EGFP-S20D, p21-Luc, Bax-Luc, pGL3-Basic, PG13-CAT, MG13-CAT, and CMVβ-p300 have been described previously (12). Individual PXXP deletion mutants of p53 were constructed using PCR, and the integrity of the entire reading frame was confirmed by DNA sequencing. The pCMV-p53ΔProAE plasmid (p53ΔPXXP) and baculovirus were gifts from Arnold Levine (Rockefeller University, New York, N.Y.). pcDNA3.1-p53-6KR was obtained from Ron Hay (University of St. Andrews, St. Andrews, United Kingdom). His-p300 baculovirus and all of the p300 deletion constructs used for mapping the proline repeat binding domain in p300 (see Fig. 2) were reported previously when the phospho-LXXLL-binding domains of p300 were mapped (12).
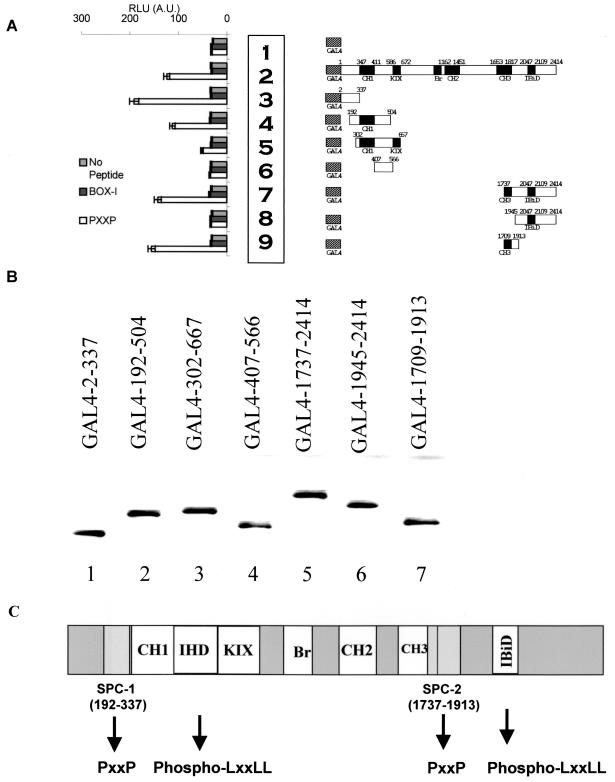
FIG. 2.
Mapping of the proline repeat binding domain of p300 (SPC-1/2). (A) HCT116 p53−/− cells were transfected with 5 μg of GAL4 control (row 1), GAL4-p300 (row 2), or the indicated GAL4-p300 miniproteins derived from the N and C termini of p300 (rows 3 to 9). Lysates were captured onto the solid phase with an anti-GAL4 antibody, and the indicated biotinylated peptide (1 ng) (PXXP peptide [from p53 aa 55 to 74], unphosphorylated LXXLL motif [from p53], or no peptide) was added and the amount of peptide bound to p300 protein was quantitated as RLU by using peroxidase linked to streptavidin. (B) Quantitation of GAL4 fusion protein levels. HCT116 p53−/− cells were transfected with 5 μg of GAL4 control (lane 1), GAL4-p300 (lane 2), or the indicated GAL4-p300 miniproteins derived from the N and C termini of p300 (lanes 3 to 9). Lysates were immunoblotted with an anti-GAL4 antibody. (C) Schematic diagram illustrating the two minimal sites for proline contact (SPC-1 and SPC-2) relative to other p300 subdomains including C/H1, IHD, C/H3, KIX, Bromo, C/H2, and IBiD. The minimal N-terminal SPC-1 domain (aa 192 to 337) is distinct from the phospho-LXXLL p53-binding domain IHD. The minimal C-terminal SPC-2 domain (aa 1737 to 1913) is distinct from the IBiD domain and partially overlaps with the C/H3 domain (aa 1653 to 1817).
Cell culture, transfections, phage display, ELISAs, and Western blots.
Transient transfections into A375 and Saos-2 cells and two-site enzyme-linked immunosorbent assays (ELISAs) to measure p53 acetylation, GAL-p300 peptide binding, and p300-p53 complex stability were carried out as previously described (12). Full-length p300 and MDM2 were purified as described previously (11). Sf9-expressed wild-type p53 and p53ΔProAE (p53ΔPXXP) tetramers were purified by heparin-Sepharose chromatography as described previously (22). The liposome-mediated method of transfection was carried out using Lipofectamine 2000 based on the manufacturer's recommendations. Luciferase activity was quantified using a luminometer (Fluoroskan Ascent FL), and β-galactosidase activity was quantified using a DIAS microplate reader (Dynatech) at a wavelength of 420 nm. Primary antibodies included anti-p300 (N15), anti-GAL4DBD (RK5C1), anti-Bax (N20), anti-hBrg1 (H-88), and anti-TRRAP (T-17) (Santa Cruz Biotechnology, Inc.). Anti-HisG was supplied by Invitrogen. Anti-MDM2 (2A10), anti-p53 (DO-1), and anti-p53 (ICA-9) were described previously (21). Anti-acetyl p53 K373/382, anti-acetyl p53 K373, anti-acetylhistone H4 (chromatin immunoprecipitation [ChIP] grade), and anti-acetyllysine were supplied by Upstate Biotechnology Incorporated. Anti-histone was supplied by Roche Diagnostics. Anti-enhanced green fluorescent protein (EGFP) was supplied by Clontech. Anti-p21 (Ab-1) was supplied by Calbiochem. Horseradish peroxidase-conjugated secondary antibodies were supplied by DAKO. The phage-peptide display and biopanning procedure using pure p300 or MDM2 was performed as previously described (38). Immunoprecipitation of protein complexes was performed as described previously (12).
ChIP.
HCT116 (p53−/−) cells (107) were transfected with the indicated plasmids, and after 48 h, the cells were treated with 1% formaldehyde for 30 min at room temperature on a rocker. The cells were then washed and scraped into 10 ml of phosphate-buffered saline with protease inhibitors. Cells were collected by a 5-min spin at 1,500 × g at 4°C, resuspended in 200 μl of ChIP lysis buffer A (5 mM HEPES [pH 7.9], 85 mM KCl, 0.5% [vol/vol] NP-40, 1× protease inhibitor mixture [PIM]), and incubated on ice for 10 min, giving a crude nuclear fraction. The pellet was then resuspended in 400 μl of ChIP lysis buffer B (1% [wt/vol] sodium dodecyl sulfate [SDS], 10 mM EDTA, 50 mM Tris-HCl [pH 8.1]) and incubated on ice for 10 min. The DNA was then sonicated by using a W-385 sonicator (Ultrasonics Inc.) with a microtip and five 10-s bursts at 40% maximum power to give fragments of approximately 600 bp. Initially, optimum conditions for sonication were determined by agarose gel analysis after reversing cross-links and precipitated DNA. Cell debris was removed by spinning at 14,000 × g for 10 min at 4°C, and supernatant was transferred to a 15-ml Falcon tube. Approximately 10% of the lysate was retained as an input control. The remaining lysate was then diluted 10-fold in 3.6 ml of ChIP IP buffer (0.01% [wt/vol] SDS, 1% [vol/vol] Triton X-100, 1.2 mM EDTA, 167 mM NaCl, 16.7 mM Tris-HCl [pH 8.0], 5 μM trichostatin A, 1× PIM), giving a chromatin solution which was then precleared with 100 μl of protein G beads, which had been preadsorbed with sonicated salmon sperm DNA (20 μg of DNA in Tris-EDTA, 1-mg/ml bovine serum albumin per 100 μl of beads), for 45 min at 45°C on a rotary mixer. Beads were then collected by centrifugation at 2,000 × g, and chromatin solution was transferred to a fresh Falcon tube. At this stage, the solution was split into three different microcentrifuge tubes to allow different antibody immunoprecipitations. Each tube was then incubated with 2 μg of the desired antibody overnight at 4°C on a rotating wheel. The immune complexes were then captured with 40 μl of protein G beads, prepared as described above, for 1 h at 4°C on a rotating wheel. The beads were then washed with 1 ml of ChIP wash buffer 1 (0.1% [wt/vol] SDS, 1% [vol/vol] Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 150 mM NaCl), ChIP wash buffer 2 (0.1% [wt/vol] SDS, 1% [vol/vol] Triton X-100, 2 mM EDTA, 20 mM Tris-HCl [pH 8.1], 500 mM NaCl), ChIP wash buffer 3 (0.25 M LiCl, 1% [vol/vol] NP-40, 1% [wt/vol] deoxycholate, 1 mM EDTA, 10 mM Tris-HCl [pH 8.1]), and finally twice with Tris-EDTA (10 mM Tris.HCl [pH 8]-1 mM EDTA). The protein-DNA complexes were then eluted by adding 250 μl of ChIP elution buffer (1% SDS-0.1 M NaHCO3) to the beads and vortexing before incubating at room temperature for 15 min. The eluate was transferred to a fresh tube, and the elution process was repeated with the beads. The eluates were then combined, and the cross-links were reversed by adding 20 μl of 5 M NaCl and incubating for 4 h at 65°C. After incubation, 10 μl of 0.5 M EDTA (pH 8.0), 20 μl of 1 M Tris-HCl (pH 6.8), and 10 μl of proteinase K (2 mg/ml) were added and incubated for 1 h at 45°C. The DNA was then recovered by phenol-chloroform extraction. PCRs were carried out according to the recommendations of the manufacturer (Qiagen) by using HotStar Taq DNA polymerase. To each 50-μl reaction mixture, 350 ng of each primer was incorporated, and titration of template (1, 2, and 5 μl) was carried out to verify linearity range. An input DNA sample was also incorporated to act as a control for the PCRs. For p21 ChIP and GAPDH PCRs, the cycle conditions were as follows: an initial 10-min Taq activation step at 95°C followed by 30 cycles of 1 min at 95°C, 1 min at 55°C, and 2 min at 72°C, followed by a final 10-min extension at 72°C. The PCR products were then run on a 1% agarose gel, and PCR products were quantified using a GeneGnome bioimager and accompanying software. The oligonucleotides for ChIP included the following: GAPDH (forward, AAAAGCGGGGAGAAAGTAGG; reverse, CTAGCCTCCCGGGTTTCTCT) and p21 (forward, CCAGCCCTTGGATGGTTT; reverse, GCCTCCTTTCTGTCCTGA).
RESULTS
P300 binds proline repeat polypeptides.
The phage-peptide display technique is a relatively powerful method to identify novel protein-binding domains on a polypeptide and to select for high-affinity peptide ligands under carefully controlled in vitro conditions. For example, using this approach Shimizu et al. previously identified a novel MDM2-binding site in the core domain of p53 that is required for p53 ubiquitination (38), and an independent study selected peptides from a phage-peptide library that only bind to the activated conformation of protein kinase C (1). The enrichment procedure we used involves presentation of approximately 10 ng of native protein to a phage-peptide library, and the selection for high-affinity peptides is usually extended to three rounds of binding and elution in order to develop a peptide consensus site. Two rounds of selection do not enrich enough to acquire peptides of a similar sequence, and four rounds of selection give only one sequence of the highest affinity.
To define the flexible contact site in p53 that mediates DNA-dependent acetylation by p300, peptides that bind to full-length p300 protein were selected from a phage-peptide library. Selection of high-affinity binding phage yielded four types of related proline repeat peptide consensus motifs: PXXPXXP, PXXP, PXPXP, and PRLP (where P is proline, R is arginine, L is leucine, and X is any amino acid) (representative peptides are shown in Fig. 1A). The sequence ITFPAKPTNYPY was selected three times, the sequence NFMESLPRLPMH was selected twice, and the PRLP motif was enriched in four peptides. The remaining peptides were enriched only once, and this proportion of duplicate to unique peptides with a common consensus motif indicates that the three rounds of selection were optimal for this enrichment of p300-binding peptides.
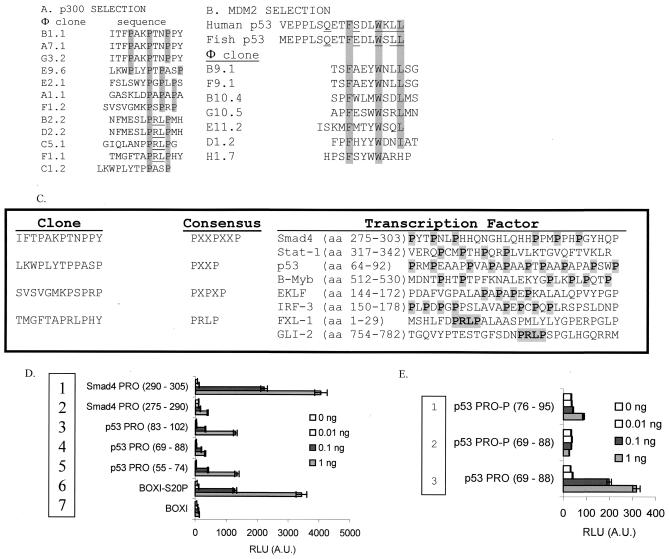
FIG. 1.
Proline repeat peptides bind p300. (A) Identification of novel p300-binding peptide motifs by phage-peptide display. The clones isolated using full-length recombinant p300 protein as a bait for phage-peptide display are highlighted, with the consensus motifs in grey, and include four representative subsets: PXXPXXP, PXXP, PRLP, and PXPXP. (B) Control reactions using MDM2. Using MDM2 as bait for phage-peptide display, clones containing the canonical MDM2-binding motif, FXXXWXXL, were isolated. (C) Sequence alignments of the four proline repeat motifs found in commonly studied transcription factors. (D and E) p300 binds directly to proline repeat motif peptides. Biotinylated peptides were used as a target for p300 in a microtiter well pull-down assay containing proline repeat peptide sequences from the aligned Smad4 (PXXPXXP, rows 1 and 2) and p53 (PXXP, rows 3 to 5) regions (D). As a control, p300 binds to the Ser20 phosphorylated LXXLL motif from the known BOX-I transactivation domain of p53 (row 6) versus unphosphorylated LXXLL motif (row 7). Phosphovariants of the PXXP domain at Thr81 (JNK site, rows 1 and 2) or the unphosphorylated PXXP domain of p53 (row 3) were used as ligands (E). The amounts of peptide captured are indicated (0, 0.01, 0.1, and 1 ng), and the amount of p300 protein bound to the indicated peptide domain was quantitated as RLU by using a peroxidase-linked secondary-antibody coupled to an anti-p300 antibody.
As a control to define peptide library integrity, purified MDM2 was used as bait, since MDM2 and p300 bind to the same N-terminal transactivation domain of the tumor suppressor protein p53. After three rounds of selection, the MDM2 consensus site FXXXWXXL was isolated (Fig. 1B). This consensus motif matches the amino acid residues involved in contacting MDM2 in the crystal structure of the MDM2-p53 complex (25). The sequence TSFAEYWNLLSG was selected three times (two are depicted in Fig. 1B), and the remaining 20 sequences (data not shown) were unique apart from the core FXXXWXXL motif. As the majority of peptides were enriched only once, this indicates that the three rounds of selection were also optimal for acquiring a consensus peptide motif.
Is it relatively surprising that we did not find LXXLL peptides enriched in this selection when using full-length p300, since many LXXLL motif-binding domains exist in p300, including C/H1, C/H3, IHD, and IBiD (Fig. 2C). The former two LXXLL-binding domains of p300 are histidine-cysteine rich and have a metal ion stabilizing the structure, while the latter two homologous LXXLL-binding domains are not necessarily stabilized by a cofactor. Since the numbers of duplicate PXXP peptides and unique PXXP peptides are relatively equivalent (Fig. 1A), the lack of LXXLL peptides enriched cannot be explained by overselection of proline-rich peptides. This appears to suggest that (i) LXXLL peptides are not stable in Escherichia coli or that the phage containing these peptides grow very slowly; (ii) all four LXXLL-binding domains of p300 are denatured under these conditions; or (iii) the PXXP peptide-binding activity of p300 is dominant in this assay. The lack of enrichment of LXXLL peptides cannot be due to the denaturing of the LXXLL-binding domains of p300, since the pure protein is active in the LXXLL peptide-binding assay and in LXXLL-dependent acetylation of p53 (see below). Further, our preliminary data indicate that, using the minimal IHD domain of p300 (12) in the phage-peptide selection procedure, we do in fact acquire peptides with a very high degree of homology to the LXXLL domain of p53 (L. Finlan and T. R. Hupp, unpublished data). This indicates that the LXXLL-containing phage-peptides are not toxic to E. coli and/or prevent phage propagation. Thus, the reason for PXXP enrichment when using phage-peptide display appears to stem from the fact that this may be a dominant activity of full-length p300 or that the p300-PXXP peptide complex is relatively more stable than LXXLL peptide complexes.
The PXXPXXP, PXXP, PXPXP, and PRLP motifs that were bound by p300 are actually present in a variety of LXXLL-containing transcription factors (Fig. 1C) and have the classic LXXLL motif flanking the PXXP repeat motif (data not shown). Two of these transcription factors stand out as relevant to this study: (i) SMAD-4 does not have a LXXLL activation domain but has an atypical activation domain that contains a proline-rich activation motif (10), and (ii) p53 has a proline repeat domain that is required for its transcription activity (40). However, the function of the polyproline domain in driving p53 activity is not defined. An in vitro peptide-binding assay was utilized to demonstrate that p300 could bind directly to these PXXP repeat domains. P300 bound both SMAD-4 PXXP repeat domains when increasing levels of the PXXP motif peptides were added to the binding assay (from 0.01 to 1 ng of peptide) (Fig. 1D, rows 1 and 2 versus the background of no peptide). All three PXXP-containing domains derived from p53 bound p300 protein (Fig. 1D, rows 3 to 5 versus background of no peptide). Phosphorylation of one PXXP repeat peptide at Thr81 (JNK site) blocked the stable binding of p300 (Fig. 1E, row 3 versus rows 1 and 2). This contrasts with the stabilizing effect of phosphorylation at Ser20 on the LXXLL motif binding to p300 (Fig. 1D, row 6 [BOX-I-S20P] versus row 7 [unphosphorylated BOX-I]) and highlights a difference in the sensitivities of the LXXLL- and PXXP-binding domains of p300 to phosphate addition.
The two phospho-LXXLL-binding domains of p300 map to an N-terminal domain named IHD (12) and a C-terminal domain named IBiD (27), and mapping of the proline repeat-binding domain of p300 was carried out by using an identical assay. An in vitro PXXP peptide pull-down assay was employed that used p300 miniprotein domains obtained from HCT116 (p53−/−) cells transfected with the indicated GAL4-p300 fusion constructs (Fig. 2A). The p300-Gal4 fusion fragments were incubated with an anti-GAL4 antibody in microtiter wells to capture the p300 fragments onto the solid phase, the corresponding biotinylated peptides (nonphosphorylated LXXLL peptide from the BOX-I domain of p53, PXXP peptide, or no peptide) were added, and p300-peptide complex stability was quantitated using streptavidin-horseradish peroxidase. As a positive control, GAL4 fused to full-length p300 protein bound specifically to the PXXP-containing peptide relative to the unphosphorylated BOX-I domain of p53 (LXXLL) or no-peptide controls (Fig. 2A, row 2 versus row 1). The minimal PXXP-binding domains in p300 were fine-mapped to two distinct regions, one in the N terminus and one in the C terminus (Fig. 2C, sites for PXXP contact [SPC-1 and -2]). The Gal4 fusion p300 miniproteins GAL4-2-337 (Fig. 2A, row 3) and Gal4-192-504 (Fig. 2A, row 4) bound specifically to the PXXP-containing peptide. This minimal PXXP-binding domain (SPC-1) in p300 flanks the N-terminal side of the peptide recognition domain in the C/H1 and IHD domains (Fig. 2C). The C-terminal site for PXXP contact was also mapped using fragments of p300. The Gal4 fusion p300 miniprotein lacking the IBiD domain, GAL4-1709-1913, bound significantly to the PXXP peptide (Fig. 2A, row 9). An immunoblot of the GAL4 fusion proteins demonstrates equivalent levels of each p300 miniprotein in each PXXP-binding reaction (Fig. 2B, lanes 1 to 7). This minimal PXXP-binding domain (SPC-2) resides between aa 1737 and 1913 (Fig. 2C). In summary, the PXXP-binding domains of p300 (SPC-1 and SPC-2) flank the phospho-LXXLL-binding domains (IBiD and IHD) and the classic C/H1 and C/H3 domains, suggesting that p300 can embrace the p53 substrate over a relatively large interface.
Although the regions of p300 that bind p53 were originally reported to be within the C/H1 and C/H3 domains (15), more recent reconstitution studies have identified two new phospho-LXXLL-binding domains named IHD (12) and IBiD (27). With these added to the PXXP-binding domains in p300, there are at least six domains of p300 that can contact p53 under various conditions. Although the IHD and IBiD domains may be higher-affinity binding regions for p53, this does not exclude a contribution of the C/H1 and C/H3 domains of p300 since it is not clear how p300 embraces the tetravalent p53 substrate. For example, for p53 tetramer acetylation to occur there may be a requirement for one C/H1 domain to bind to one monomer, IHD to bind to another monomer, IBiD to bind to a third subunit, and a PXXP-binding domain to contact a fourth monomer. Further, it is possible that the in vitro assay we used to measure p300 activity unfolds the C/H1 and C/H3 domains, thus releasing IHD and/or IBiD domains to compensate. The main assay used to access p300 protein integrity is the docking-dependent acetyltransferase activity of p300. This purified fraction of p300 is active in histone acetylation, p53-phospho-LXXLL binding, PXXP-peptide binding, p53 protein binding, and LXXLL-dependent (docking-dependent) p53 acetylation. To maintain p300 protein activity and conformation from the outset, EDTA was not included in the lysis buffers nor was EDTA included in the GAL4-p300 fusion protein-binding assay. However, p53 is purified in buffers containing 0.1 mM EDTA (22). This amount of EDTA does not disrupt folding of the p53 tetramer nor does it dissociate zinc from the protein. When diluted into reaction buffers with purified p300, the final concentration of EDTA ranges from 1 to 3 μM, which is not known to disrupt the very stable Cys/His structure. Further, p300-coactivated transcription by p53 in vivo is phospho-LXXLL dependent (11) and is inhibited by transfected IBID or IHD domains (12), suggesting that the in vitro character of p300 reflects its in vivo activity. Thus, it appears that in vitro and in vivo, the C/H1 or C/H3 domain of p300 contributes to p53 binding to a lesser extent than does IBiD or IHD. We are presently developing single, double, triple, and quadruple mutants of C/H1, IHD, C/H3, and IBiD in p300 to determine which are the major docking domains on p300 for the LXXLL motif of p53.
Proline repeat peptides inhibit p53 acetylation.
The addition of consensus site DNA to reactions containing p53, p300, and acetyl-coenzyme A (CoA) stimulates p53 acetylation (12). Further, docking of p300 to the LXXLL motif of p53 is essential for DNA-dependent acetylation of p53, thus indicating two possibly interrelated conformational constraints to p53 tetramer acetylation (12). These two conformational constraints are released by consensus site DNA and presumably by a second p300-docking interface. If the proline repeat motif were in fact the second conformationally sensitive interface driving p53 acetylation, then there should be a connection between p300 binding to the proline repeat domain and p53 acetylation. We investigated whether the adjacent PXXP repeat motif of p53 was also required for mediating acetylation.
Using the p300 acetylation reaction where acetyl-CoA promotes p53 acetylation in a sequence-specific DNA-dependent manner (Fig. 3B, lane 2 versus lane 1), the PXXP repeat peptide was found to be a specific inhibitor of DNA-dependent acetylation (Fig. 3B, lanes 8 to 10 versus lanes 2 to 6). Histone acetylation was not affected by the PXXP repeat peptide (Fig. 3D, lanes 5 to 7 versus lanes 1 to 4). In the absence of both DNA and acetyl-CoA, the PXXP repeat peptide reduced the stable binding of p300 to p53 (Fig. 3E). Furthermore, the PXXP repeat peptide attenuated acetylation of p53 in the complete acetylation reaction when p53 was captured in ELISA format (Fig. 3F), similar to that seen in the immunoblot (Fig. 3B). The PXXP peptide, at concentrations ranging from 50 to 200 μM, is a more potent destabilizer of the p53-p300 protein complex than is the phospho-LXXLL peptide (Fig. 3E), and the extent of destabilization mirrors the extent of inhibition of p53 acetylation (Fig. 3F). These data suggest that the proline repeat motif is the second interaction site we were looking for and indicate that p300 contacts two contiguous peptide domains on p53 (LXXLL and PXXP).
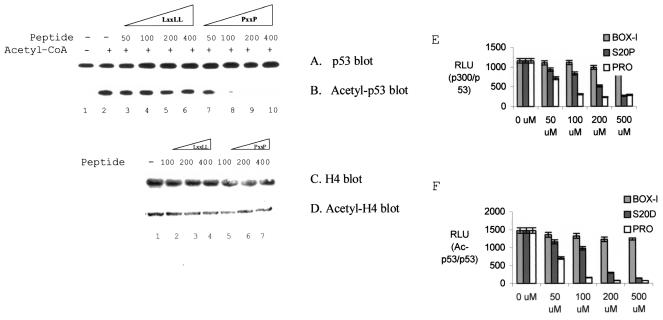
FIG. 3.
Proline repeat peptides inhibit DNA-dependent acetylation of p53. (A and B) p300-mediated acetylation of p53 is inhibited by PXXP peptides. Acetylation reactions using p300 and p53 were carried out with or without acetyl-CoA and with the addition of various concentrations of peptides (50 to 400 ng): LXXLL peptide (lanes 3 to 6) or the PXXP peptide (lanes 7 to 10; proline repeat derived from p53; aa 55 to 74; see Fig. 1). Shown are results for total p53 (A) and acetylated p53 (B). (C and D) Histone acetylation is not inhibited by PXXP. Acetylation reactions contained 1 μg of histone H4 as the substrate. Acetylation reactions were carried out with various concentrations of peptides (100 to 400 ng): LXXLL peptide or PXXP peptide. Shown are results for total histone (C) and acetylated histone (D). (E) Quantitation of p300-p53 complex stability in the presence of the phospho-LXXLL or PXXP peptide. Acetylation reactions were carried out as described for panels A and B but without acetyl-CoA and consensus site DNA and with various concentrations of peptides as indicated: BOX-I (LXXLL), Ser20 phospho-LXXLL (S20-P), or PXXP peptide (concentrations in micromolar). After capturing p53 with the monoclonal antibody ICA-9, the relative levels of p300 bound to p53 was quantified by using an anti-p300 antibody followed by a peroxidase-conjugated secondary antibody coupled to chemiluminescence. p300 binding is expressed in RLU. (F) Quantitation of p53 acetylation in the presence of the phospho-LXXLL or PXXP peptide. Acetylation reactions were carried out as described for panels A and B with acetyl-CoA, consensus site DNA, and various concentrations of peptides as indicated: BOX-I (LXXLL), Ser20 phospho-LXXLL (S20-P), or PXXP peptide. After capturing total p53 with the monoclonal antibody ICA-9, the relative levels of acetylation of p53 were normalized to total p53 protein levels by using an anti-p53 polyclonal antibody (CM-1) and the anti-p53 acetylation polyclonal antibody (RLU).
There is some apparent inconsistency between the assays we used to assess p300 binding to PXXP peptides. First, the PXXP peptides in solution were more effective inhibitors of p300-catalyzed acetylation than the phospho-LXXLL peptides (Fig. 3F). Consistent with this, PXXP peptides were enriched in phage-peptide display (Fig. 1A), suggesting that proline repeat rather than leucine repeat peptides bind better in solution to p300. Second, both PXXP and phospho-LXXLL peptides disrupt the p53 tetramer-p300 complex (Fig. 3E), indicating that disruption of one docking interaction prevents the other from compensating. However, the PXXP peptides bind more poorly to p300 in the solid-phase ELISA than the phospho-LXXLL peptide (Fig. 1D), and this assay may underestimate the avidity of p300 for PXXP peptides. It is possible that the PXXP peptides are hindered sterically by attachment to the solid phase and that this attenuates p300-PXXP peptide complex stability. For example, we found that a phosphospecific monoclonal antibody binding to a phosphopeptide can be hindered by attachment of the peptide to the solid phase but the monoclonal antibody can bind when the phosphopeptide is cross-linked to bovine serum albumin and attached indirectly to the solid phase (data not shown). Thus, according to the results of three different assays, the affinities of the PXXP and phospho-LXXLL peptides for p300 are different: phage-peptide display suggested that the PXXP peptide-p300 complex is much more stable than the LXXLL-p300 complex, the ELISA suggested that the phospho-LXXLL peptide binds better to p300 than does the PXXP peptide, and the acetylation assay showed that the PXXP peptides are more effective inhibitors.
To begin to address the significance of the PXXP repeat domain to p53-dependent transcription, we fused a 30-aa PXXP repeat peptide (aa 64 to 93) to EGFP for in vivo transfection. The cotransfection of either phosphomimetic LXXLL (S20D) or PXXP peptide-GFP fusions with the p53 gene attenuated p53 activity from the p21 or bax promoters (Fig. 4A and B). p300 binding to p53 is required for p53 stabilization after DNA damage (42), and we investigated whether the EGFP-LXXLL phosphomimetic peptide or the EGFP-PXXP peptide could neutralize p300 and destabilize p53 protein in vivo. Notably, the cotransfection of either LXXLL or PXXP-EGFP fusions alone did not affect basal p53 protein levels (Fig. 4C, lanes 6 and 8 versus lane 2). However, the cotransfection of the LXXLL and PXXP-EGFP fusions together induced a striking decrease in steady-state p53 protein levels (Fig. 4C, lane 10 versus lane 2). The levels of the EGFP fusions remain equivalent in each transfection (Fig. 4D, lanes 2, 4, 6, 8, and 10). The cotransfection of LXXLL and PXXP-EGFP fusions alone or together was sufficient to attenuate endogenous p21 protein levels (Fig. 4E, lanes 6, 8, and 10 versus lanes 2 and 4), which is consistent with the data showing that p21-Luc reporter activity is reduced by the peptides (Fig. 4A). This decrease in p53 protein steady-state levels by cotransfection of both LXXLL and PXXP-EGFP fusions is due to proteosome-dependent degradation (Fig. 4F, lane 2 versus lane 1). These data indicate that although disrupting either of the two p300-docking sites attenuates p53 activity in cells, disrupting both p300-binding motifs is required to promote p53 protein degradation. These in vivo data are consistent with in vitro data showing that the phospho-LXXLL or PXXP peptides inhibit p53 acetylation (Fig. 3F).
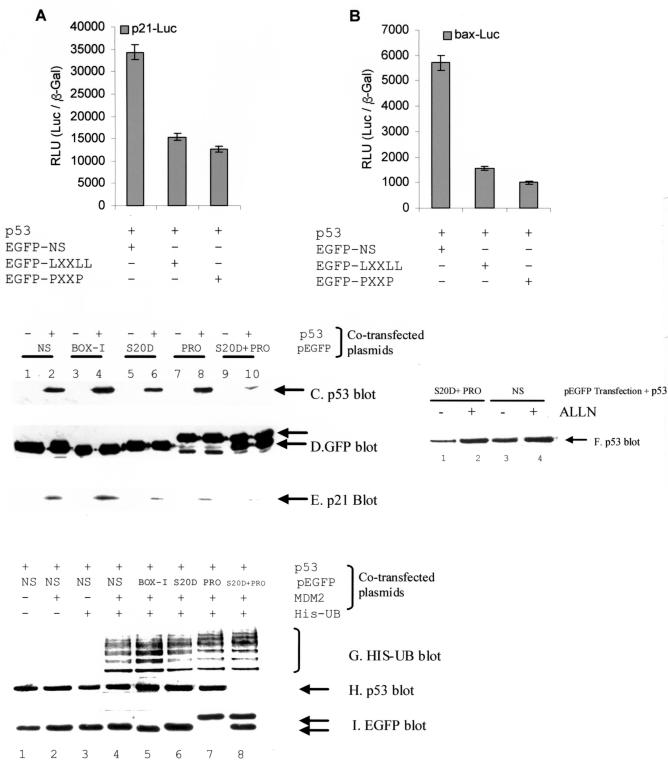
FIG. 4.
p300-binding peptides attenuate p53 activity and destabilize p53 protein steady-state levels in vivo. (A and B) Competitive inhibition of p53-dependent transcription by p300-binding peptides. pCMV-p53 (1 μg) was cotransfected with the luciferase reporters (1 μg of p21-Luc [A] or bax-Luc [B] and the transfection control [pCMVβ-Gal] and either the GFP-NS [1 μg; control], GFP-LXXLL phosphomimetic [1 μg], or GFP-PXXP [1 μg] peptide). The RLU is expressed as a ratio of Luc to the internal transfection control (pCMVβ-Gal). (C to E) p53 and p21 protein levels are destabilized by cotransfection of GFP fusion p300-binding peptides. pCMV-p53 (1 μg) and the indicated GFP fusion construct (1 μg; NS, nonspecific; BOX-I, the MDM2-binding peptide; S20D, the phosphomimetic LXXLL; PRO, proline repeat) and the levels of the indicated proteins were determined by immunoblotting. (F) Destabilization of p53 by p300 targeting is mediated by the proteosome. Acetyl-Leu-Leu-norleucine (ALLN) was added for 2 h after transfection of the indicated plasmids for 24 h, and the levels of p53 protein are as indicated. (G to I) p53 ubiquitination by MDM2 is not altered after cotransfection of GFP fusion p300-binding peptides. pCMV-p53 (1 μg), the indicated GFP fusion construct (1 μg), pCMV-MDM2, or pCMV-His-ubiquitin was transfected for 24 h, and the levels of the indicated proteins were determined by immunoblotting. MDM2-dependent ubiquitinated products were purified using a His pull-down assay, blotted with p53 antibodies, and are shown in panel G.
The effects of the LXXLL and PXXP-GFP fusion proteins on MDM2-dependent ubiquitination of p53 were also examined (Fig. 4G). The cotransfection of the indicated EGFP constructs with His-Ub, MDM2, and p53 demonstrates that MDM2-dependent ubiquitination of p53 is not affected by the p300-binding peptides (Fig. 4G, lanes 6 to 8 versus lane 4). However, there was a marginal increase in p53 ubiquitination when using the MDM2 peptide inhibitor (BOX-I EGFP fusion) (Fig. 4G, lane 5 versus lane 4). The cotransfection of the LXXLL and PXXP-EGFP fusion proteins together resulted in p53 protein destabilization (Fig. 4H, lane 8 versus lanes 6 and 7). The transfected EGFP peptides were expressed at similar levels (Fig. 4I). Thus, two independent p300-binding domains (LXXLL and PXXP) are required to stabilize p53 protein.
It is curious that total p53 protein can be destabilized by the GFP fusion peptides that bind p300 without affecting MDM2-dependent p53 ubiquitination. Since the PXXP and DLXXLL GFP fusion peptides do not bind MDM2 in vivo, we expected few changes in MDM2-dependent monoubiquitination of p53. The monoubiquitinated p53 was pulled out by using a HIS pull-down assay, and this pool represents a very small pool of the total p53 protein. Therefore, the depletion of p53 protein levels (Fig. 4H) by the p300 binding peptides is more representative of the total pool of p53 than the minor proportion of p53 that is HIS monoubiquitinated. Presumably, therefore, in this cotransfection system, p53 ubiquitination is uncoupled from protein degradation.
Proline repeat transactivation motif promotes DNA-dependent acetylation of p53 by p300 in vitro and in vivo.
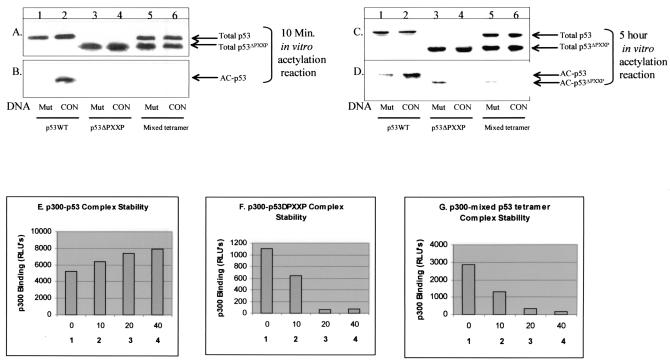
The mechanism of proline-directed acetylation of p53 was determined by examining the effects of deleting the proline repeat domain on p53 acetylation. Sequence-specific DNA-dependent acetylation was observed with wild-type p53 (Fig. 5B, lane 2 versus lane 1), while the p53ΔPXXP tetramer displayed no acetylation (Fig. 5B, lane 4 versus lane 2). The mixed wild-type p53/p53ΔPXXP tetramer was also found to be defective in acetylation (Fig. 5B, lane 6 versus lane 2). Thus, despite the fact that this mixed tetramer had four LXXLL motifs and two PXXP motifs (six of eight docking motifs), p300 cannot acetylate wild-type p53/p53ΔPXXP, suggesting a dominant negative effect of proline repeat deletion on p53 conformation. After incubation of the acetylation reaction mixture in the presence of nonspecific DNA for 5 h instead of 10 min (12), we could begin to see p53ΔPXXP tetramer and wild-type p53/p53ΔPXXP tetramer acetylation in the presence of nonspecific DNA (Fig. 5D, lanes 3 and 5 versus lane 1). The inclusion of consensus site DNA prevented acetylation of the proline-deleted p53 tetramer and the mixed wild-type p53/p53ΔPXXP tetramer (Fig. 5D, lanes 4 and 6 versus lanes 3 and 5), further indicating that the DNA-bound conformation of p53 requires the PXXP motif to mediate acetylation.
FIG. 5.
The PXXP transactivation motif of p53 is required for sequence-specific DNA-dependent acetylation by p300. (A and B) The PXXP motif of p53 is required for DNA-dependent acetylation by p300 in vitro. Human recombinant p53 (lanes 1 to 2), p53ΔPXXP (lanes 3 to 4), or p53/p53ΔPXXP mixed tetramers (lanes 5 to 6) were purified from insect cells and incubated in the linear range for the enzyme assay (10 min) with p300 and p53 consensus site oligonucleotide DNA (CON, lanes 2, 4, or 6) or nonspecific DNA (Mut, lanes 1, 3, and 5). Relative acetylation was quantitated by Western blotting using an anti-p53 acetylation antibody (B) and normalized to total p53 protein by using the monoclonal antibody DO-1 (A). (C and D) The PXXP motif of p53 is required for DNA-dependent acetylation by p300 in vitro. Details are as described for panels A and B, except that reactions were carried out for 5 h instead of 10 min. (E to G) The PXXP motif of p53 is required for maximal p300-p53 complex stability in the presence of consensus site DNA. Insect cell-purified human recombinant P53 (E), p53ΔPXXP (F), or p53/p53ΔPXXP mixed tetramers (G) were captured onto the solid phase of a microtiter well with the ICA-9 anti-p53 monoclonal antibody, including a titration of p53 consensus site DNA as indicated in panels 1 to 4 (0, 10, 20, or 40 ng). The captured p53 isoforms were incubated with buffer containing insect cell-expressed recombinant human p300 protein, and the amount of p300 bound to p53 was quantitated using anti-p300 antibodies. The levels of the p53 protein isoforms were determined using an anti-p53 antibody and quantitated as the ratio of p300 bound to total p53 by using enhanced chemiluminescence and expressed as RLU.
The acetylation of p53 in the presence of nonspecific DNA after 5 h of incubation reflects a reaction with a very low specific activity and may also reflect changes in p53 conformation over this time frame. The original acetylation study on p53 (18) demonstrated that bacterially expressed p53 can be acetylated, though the stoichiometry of acetylation on p53 protein was not determined nor was the percentage of p53 in the folded conformation determined. The folded and unfolded ratios of recombinant p53 protein can be quantified with monoclonal antibodies specific for the denatured or unfolded forms of the protein (20). The p53 protein used in our experiments is derived from insect cells, where the majority of the protein is folded (22). The enhanced 5-h incubation that resulted in some basal p53 acetylation in the absence of consensus site DNA may be due to the unfolding of the p53 tetramer, resulting in loss of the conformational constraint on acetylation (see Fig. 10). The native p53 tetramer is known to “breathe” in an equilibrium between unfolded and folded states (9), and this breathing presumably permits weak docking-independent and DNA-independent acetylation to occur.
FIG. 10.
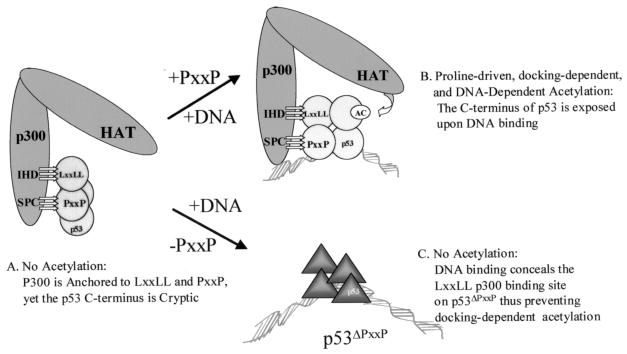
DNA induces a conformational change that mediates proline-directed p53 acetylation by p300. (A) In the absence of DNA, p300 can dock to the p53 tetramer via LXXLL and PXXP binding, but conformational constraints in the native p53 tetramer prevent acetylation. (B) Sequence-specific DNA binding changes the conformation of p53 (34), thus permitting acetylation to occur in a PXXP-dependent manner. Thus, DNA binding does not change p300 binding as much as it activates p53 acetylation, suggesting that in the DNA-free state the C terminus of p53 is cryptic with respect to acetylation. (C) When the PXXP domain is deleted, p300 cannot acetylate DNA-bound p53ΔPXXP due to the inability of p300 to form stable contacts with the p53ΔPXXP tetramer. However, the p53ΔPXXP tetramer can be acetylated by p300 in the absence of DNA, with the proline deletion essentially converting the p53ΔPXXP tetramer to a histone-like substrate which can be acetylated in a docking-independent and DNA-independent manner. These latter data also suggest that DNA binding creates a specific interface in p53 for p300 and that the PXXP domain forms an integral part of this interface. Thus, both the PXXP/LXXLL domains and the C-terminal acetylation motif act concertedly after DNA binding to permit p300-catalyzed acetylation. There is a precedent for the C-terminal acetylation domain of p53 being cryptic in the DNA-free state. The p300 acetylation sites in p53 are within the epitope for monoclonal antibody PAb421. When p53 is DNA free, the PAb421 epitope is cryptic and DNA binding exposes the epitope. This led to the postulation that long-range allosteric effects mediate DNA binding by p53 (19). The cryptic nature of the p53 acetylation motif in the DNA-free conformation of p53 builds into the tetramer an intrinsic negative regulatory mechanism to prevent acetylation until the tetramer is promoter bound.
How does consensus site DNA prevent p53ΔPXXP tetramer acetylation? Two potential mechanisms could account for this: (i) after DNA binding, the p53ΔPXXP tetramer binds p300, but the acetyltransferase activity cannot engage; or (ii) after DNA binding, the p53ΔPXXP tetramer changes conformation to preclude p300 binding. To determine whether DNA binding by the p53ΔPXXP tetramer prevented p300 binding, we quantitated p300-p53ΔPXXP complex stability in the absence and presence of consensus site DNA (from 0 to 40 ng) (Fig. 5E to G). In the absence of consensus site DNA, wild-type p53 bound significantly to p300 (Fig. 5E, column 1) and the p300-p53 complex was marginally stabilized further by addition of increasing amounts of consensus site DNA oligonucleotide (from 10 to 40 ng) (Fig. 5E, columns 2 to 4 versus column 1). Deletion of the PXXP motif did not prevent p300 from complexing with p53 (Fig. 5F, column 1), presumably due to the LXXLL activation domain in the tetramer. However, p53ΔPXXP binding to p300 was completely destabilized by the addition of consensus site DNA (Fig. 5F, columns 2 to 4 versus column 1). Further, a mixed p53/p53ΔPXXP tetramer was similarly destabilized from p300 by DNA (Fig. 5G, columns 2 to 4 versus column 1), indicating that the PXXP domain on wild-type p53 could not rescue the p300-p53/p53ΔPXXP complex. This explains why DNA-complexed p53ΔPXXP cannot be acetylated by p53 (Fig. 5B and D), as p300 cannot form an interface with a DNA-bound p53 tetramer lacking PXXP repeat motifs. Thus, it is the proline repeat motif that places conformational constraints on p53 acetylation.
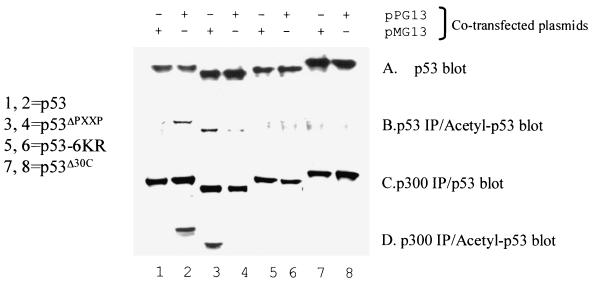
It was important to evaluate the contribution of the PXXP motif to DNA-dependent acetylation in vivo. We previously developed an in vivo consensus site DNA-dependent acetylation assay for p53 (12). After cotransfection of genes encoding p53 and p53ΔPXXP (Fig. 6A, lanes 1 to 8) along with either nonspecific or consensus site plasmid DNA, cells were lysed after 24 h and the transfected p53 protein was assayed for both in vivo acetylation and binding to endogenous p300 protein. After immunoprecipitation of transfected p53 protein and immunoblotting with acetylation-specific p53 antibodies, consensus site plasmid DNA-dependent acetylation was observed (Fig. 6B, lane 2 versus lane 1). After immunoprecipitation of endogenous p300 and immunoblotting with an anti-p53 antibody, the amounts of transfected wild-type p53 bound to p300 were similar after the cotransfection of either consensus site or nonspecific plasmid DNA (Fig. 6C, lane 2 versus lane 1). However, after immunoprecipitation of endogenous p300 and immunoblotting with an anti-acetyl-p53 antibody, only wild-type p53 cotransfected with consensus site plasmid DNA was acetylated (Fig. 6D, lane 2 versus lane 1).
FIG. 6.
The PXXP motif of p53 is required for sequence-specific DNA-dependent acetylation by p300 in vivo. pCMV-P53 (lanes 1 and 2), pCMV-p53ΔPXXP (lanes 3 and 4), pCMV-p53-6KR (lanes 5 and 6; mutated acetylation sites), or pCMV-p53Δ30 (lanes 7 and 8; lacking acetylation sites) were cotransfected into p53−/− cells in the presence of consensus site plasmid DNA (pG13-CAT, lanes 2, 4, 6, and 8) or plasmid DNA without the consensus site (pMG13-CAT, lanes 1, 3, 5, and 7). (A) The amount of total p53 in each transfection was quantified by direct immunoblotting with the monoclonal antibody DO-1. (B) To quantitate p53 acetylation in vivo, total p53 protein was immunoprecipitated with a mixture of ICA-9 and DO-1 monoclonal antibodies and the levels of acetylated p53 were quantitated by immunoblotting with the acetylation-specific antibody. (C and D) Endogenous P300 protein was immunoprecipitated with an anti-p300 antibody, and levels of total p53 protein isoforms (C) or acetylated p53 isoforms (D) bound to p300 were quantitated by immunoblotting with either the acetylation-specific antibody or a p53 protein antibody to normalize to the total p53 protein.
Notably, the converse was observed when the proline deletion mutant, p53ΔPXXP, was used. After immunoprecipitation of endogenous p300 and immunoblotting with an anti-p53 antibody, the amounts of p53ΔPXXP-p300 complexes were similar after the cotransfection of either consensus site or nonspecific plasmid DNA (Fig. 6C, lane 4 versus lane 3). Furthermore, after cotransfection of p53ΔPXXP with consensus site plasmid DNA, the p53ΔPXXP in complex with p300 was not acetylated (Fig. 6D, lane 4 versus lane 2). However, transfection of p53ΔPXXP with the mutant consensus site supercoiled plasmid permitted significant acetylation of p53ΔPXXP (Fig. 6D, lane 3 versus lane 1). After immunoprecipitation of transfected p53ΔPXXP protein and immunoblotting with acetylation-specific p53 antibodies, acetylation was also observed only in the presence of nonspecific plasmid DNA (Fig. 6B, lane 3 versus lane 4). This p300-cataylzed acetylation of p53ΔPXXP in the absence of consensus site DNA can be seen in vitro with long incubation times (Fig. 5D, lanes 3 and 5) and further indicates that in vivo, consensus site DNA places an absolute requirement for the proline repeat motif to drive p53 acetylation.
There is an apparent contradiction in data showing the effects of consensus site DNA on the stability of the p300-p53ΔPXXP mutant. While in vitro studies using purified proteins indicated that p53ΔPXXP cannot bind to p300 in the presence of consensus site DNA, after transfection the p300 still binds the p53ΔPXXP mutant in vivo, though the mutant p53 is not acetylated. It is possible that in vivo cotransfection would force the interactions of p53ΔPXXP and p300 even in the presence of transfected consensus site plasmid DNA, since it is known that p300 can recruit cotransfected proteins into nuclear inclusions or particles (16). Further, using purified proteins in vitro, a dose-dependent titration can be better controlled under conditions where the intrinsic stability of the complex can be assessed. Alternatively, differences in complex stability and the assay conditions could account for the differences. The solid-phase ELISA used to demonstrate that DNA prevents formation of a stable complex between p300 and p53ΔPXXP involves incubations with primary and secondary antibodies with washings over a 150-min time period, while the immunoprecipitation from cells involves quick washing times of only 15 min. The longer washing times in the ELISA would ensure that weakly bound p300-p53ΔPXXP complexes were not detected. This may be the most likely explanation for the discrepancy, since basal p53 acetylation by p300 in the presence of nonspecific DNA results in destabilization of the p300-p53 complex, while p53 acetylation by p300 in the presence of consensus site DNA results in stabilization of the p300-p53 complex (12). Thus, deletion of the PXXP domain may prevent p53 acetylation when the tetramer is DNA bound, most likely by increasing the off-rate rather than decreasing the on-rate for p300.
Deletion of a single PXXP motif attenuates p53 activity in vivo.
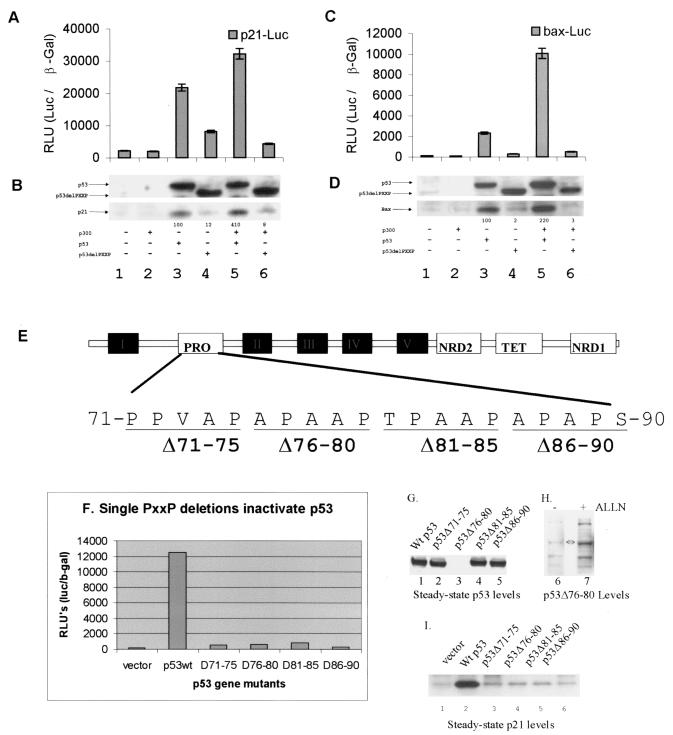
Presumably, since p53ΔPXXP cannot be acetylated by p300 when bound to consensus site DNA in vitro or in vivo, the proline deletion mutant should be inert as a transcription factor in vivo. Saos-2 (p53−/−) cells were cotransfected with p53 or p53ΔPXXP genes with or without p300 and the corresponding p53-dependent luciferase reporter constructs (p21 or bax). p53-dependent transactivation of the p21 and bax promoters was attenuated by deletion of the PXXP domain of p53 (Fig. 7A and C, lanes 4 versus lanes 3) despite the similarity in expression levels of the wild-type and mutant p53 proteins (Fig. 7B and D, lanes 4 versus lanes 3). The p53ΔPXXP mutant failed to synergize with p300 protein, and its activity was inhibited on the p21 and Bax promoters relative to that of wild-type p53 (Fig. 7A and C, lanes 6 versus lanes 5). Since the nature of this transcription assay depends on reporter constructs, which have not necessarily been processed with eukaryotic chromatin assembly proteins and subsequent chromatin remodeling enzymes, the response from the endogenous p21 and bax promoters was normalized by Western blotting of endogenous proteins. Induction of p21 and Bax proteins was reduced after transfection of the p53ΔPXXP mutant relative to what was seen with wild-type p53 (Fig. 7B and D, lanes 3 and 5 versus lanes 4 and 6).
FIG. 7.
Deletion of a single PXXP motif attenuates p53 activity in vivo. (A to D) Deletion of the entire proline repeat domain inhibits p53. The transactivation activity of p53 and p53ΔPXXP on the p21 (A) and bax (C) luciferase reporter promoters (RLU) is expressed as the ratio of p21-Luc or bax-Luc to the internal transfection control (pCMVβ-Gal). Expression levels of transfected p53 forms and endogenous target gene products are shown in panels B and D as follows: (B) transfected p53 protein (lanes 3 and 5) or p53ΔPXXP protein (lanes 4 and 6) and endogenous p21 protein and (D) transfected p53 protein (lanes 3 and 5) or p53ΔPXXP protein (lanes 4 and 6) and endogenous Bax protein were quantitated by Western blotting with an anti-p53, anti-p21, or anti-Bax protein antibody and enhanced chemiluminescence. In each transfection, 1 μg of pCMV-p53 or pCMV-p53ΔPXXP alone or with 5 μg of pCMVβ-p300 was added as indicated. (E) Map of the clustered PXXP repeat motifs in the N-terminal activation domain of p53 and site of the PXXP deletions used in this study. (F) The transactivation activity of p53 and individual p53ΔPXXP deletion mutants on the p21 luciferase reporter promoter (RLU) is expressed as a ratio of p21-Luc to the internal transfection control (pCMVβ-Gal). (G and I) Expression levels of transfected p53ΔPXXP deletion mutant p53 proteins (G) and endogenous p21 protein (I) were quantified by immunoblotting. (H) The unstable PXXP deletion mutant (76-80) was stabilized by ALLN. Twenty-four hours after transfection of p53Δ76-80, ALLN was added for 2 h prior to harvesting of the cells and p53 was blotted with DO-1 monoclonal antibody.
There are three PXXP repeats and one PXPXP motif in the p53 activation domain from amino acids 71 to 90 (Fig. 7E), and we evaluated whether deletion of any proline repeat motif alters p53 activity and steady-state levels. Deletion mutants of each PXXP motif produced the recombinant proteins p53Δ71-75, p53Δ76-80, p53Δ81-85, and p53Δ86-90, and any one of these PXXP motif mutants attenuated p53-dependent transcription from the p21 reporter (Fig. 7F) and failed to induce endogenous p21 protein (Fig. 7I). Further, although the p53Δ72-75, p53Δ81-85, and p53Δ86-90 proteins were expressed at levels similar to that of wild-type p53 protein, p53Δ76-80 was expressed at very low levels (Fig. 7G). p53Δ76-80 protein was degraded by a proteosome-dependent pathway (Fig. 7H, lane 7 versus lane 6). A titration of lower amounts of p53 DNA in the transfection also revealed that p53Δ81-85 was expressed at very low levels (data not shown), indicating that the core proline repeat region, 76-APAAPTPAAP-85 containing the JNK phosphorylation site, is a major determinant of p53 protein steady-state levels. However, sites outside this core (aa 76 to 85) proline repeat region are likely to be important under differing conditions, since the Pro72-Arg72 polymorphism also has reduced specific activity under the conditions described above (data not shown).
p53 acetylation in vivo is a post-DNA-binding event.
The role of the C-terminal domain of p53 is twofold. It functions as a negative regulatory domain whose phosphorylation stimulates in vitro and in vivo the specific DNA-binding function of p53 (7, 23). The molecular basis for p53 protein latency requires the C-terminal domain to destabilize and unfold the core DNA-binding domain by allosteric effects (5), suggesting that p53 is part of a growing group of regulatory proteins that are native and unfolded in the absence of posttranslational modification (13). Phosphorylation of p53 in the C-terminal domain neutralizes the destabilizing effect of the C terminus on the core domain of p53 and maintains p53 conformation and activity (30).
The C-terminal domain of p53 also functions as a positive regulatory domain that is acetylated and that recruits coactivator complexes (4, 14). A molecular explanation for why acetylation recruits coactivators may be found in the ability of acetyl-CoA to stabilize the p300-p53AC complex in the presence of consensus site DNA (12). Thus, although the role for acetylation of p53 remains relatively unclear (33), two complementary models are emerging. First, when “latent” fractions of p53 are used, acetylation can stimulate the DNA-binding activity of p53 by neutralizing basic residues implicated in nonspecific DNA binding (18). Second, when “kinase-activated” p53 protein is used, acetylation is a post-DNA-binding event and its role is to stabilize the p300-p53 AC complex (12). The proline repeat deletion mutant of p53 can be used to investigate which of these two models is favored in vivo. This relies on our observation that p53ΔPXXP is acetylated in vivo in the absence of consensus site DNA, but p53ΔPXXP cannot be acetylated in vivo in the presence of consensus site DNA (Fig. 6). If p53ΔPXXP was acetylated in vivo before p21 promoter binding, then acetylated p53ΔPXXP should be detected at the p21 promoter by ChIP. However, if p53ΔPXXP is acetylated only after DNA binding, then because the p53ΔPXXP-DNA complex cannot bind stably to p300 (Fig. 5), there should be no acetylated p53ΔPXXP cross-linked to the p21 promoter.
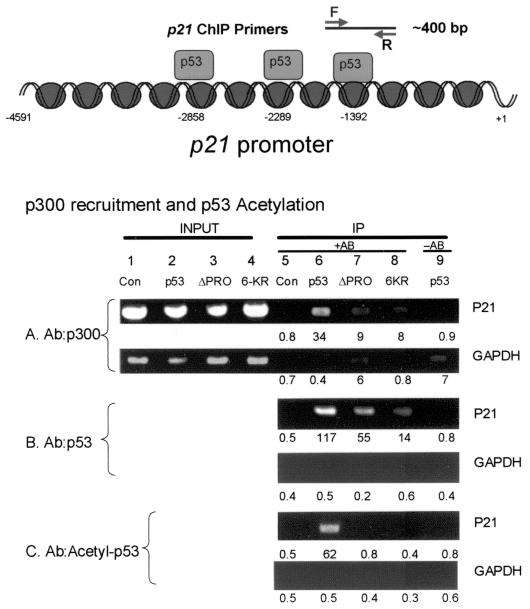
Transfection of p53 and two p53 mutants (PXXP deletion and the acetylation-defective mutant 6KR) into cells followed by formaldehyde cross-linking of protein-DNA complexes, immunoprecipitation of sonicated protein-DNA complexes, and PCR amplification with p21- (Fig. 8, bottom panel) and GAPDH-specific primers after reversing the chemical cross-link demonstrated a unique requirement for PXXP in promoting p53 acetylation at the p21 promoter. Using cell lysates transfected with the wild-type p53 gene (Fig. 8, lanes 6) or vector-only control (Fig. 8, lanes 5), immunoprecipitation of p300 (Fig. 8A) resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 8A, lane 6 [34 relative luciferase units {RLU}] versus lane 5 [0.8 RLU]). Further, immunoprecipitation of p53 resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 8B, lane 6 [117 RLU] versus lane 5 [0.5 RLU]). The p53 bound was acetylated, since immunoprecipitation with acetyl-p53 antibodies resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 8C, lane 6 [62 RLU] versus lane 5 [0.5 RLU]). As a PCR control for each amplification, GAPDH showed equivalent background levels ranging from 0.5 to 0.7 RLU (Fig. 8A to C, GAPDH panels).
FIG. 8.
The PXXP motif is required to acetylate p53 at the p21 promoter in vivo. HCT116 p53−/− cells were transfected with various p53 pCMV DNAs encoding p53 (lanes 2 and 6), p53ΔPXXP (lanes 3 and 7), and the nonacetylatable mutant, p53-6KR (lanes 4 and 8). Following transfection of the indicated construct, cross-linking of protein-DNA complexes with formaldehyde, immunoprecipitation, and processing of the samples as described in Materials and Methods, the released DNA was PCR amplified using primers to the p21 promoter (as indicated in the bottom panel) or GAPDH promoter. The immunoprecipitation was carried out with antibodies specific for p300 (A), p53 (B), and acetylated p53 (C). The data are plotted as input DNA or as immunoprecipitated DNA. Quantitation of the bioluminescence is depicted below each lane. Controls include DNA amplified in reactions processed from cells without antibody in the immunoprecipitation reaction (lane 9) or with vector control only (lanes 1 and 5). The diagram at top depicts the region of the p21 promoter that was focused onto isolate p53-bound transcription complexes.
The key question addressed next was whether deletion of p53's PXXP motif prevents it from being acetylated at promoter sites in vivo. Using cell lysates transfected with the p53ΔPXXP gene (Fig. 8, lanes 7) or vector-only control (Fig. 8, lanes 5), immunoprecipitation of p53ΔPXXP resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 8B, lane 7 [55 RLU] versus lane 5 [0.5 RLU]), indicating that a relatively large amount of p53ΔPXXP protein is chromatin bound. However, the p53ΔPXXP bound was not acetylated, since immunoprecipitation with acetyl-p53 antibodies did not result in the coprecipitation and PCR amplification of the p21 promoter (Fig. 8C, lane 7 [0.8 RLU] versus lane 5 [0.5 RLU]). This absence of acetylation can be explained by the inability of p53ΔPXXP in complex with DNA to bind to p300 (Fig. 5). Similar data were obtained using ChIP from the bax promoter (data not shown). Thus, p53ΔPXXP is not acetylated before DNA binding in vivo, confining acetylation of p53 in vivo to a proline-driven post-DNA-binding event. These data indicate that the proline deletion mutant behaves like an acetylation mutant, consistent with the sequence-specific DNA dependence in proline-directed acetylation.
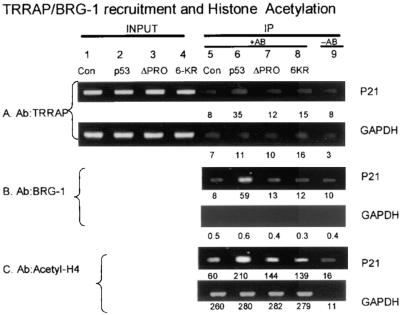
We evaluated whether the proline repeat domain can influence the recruitment of additional transcription factors implicated in chromatin remodeling and activation of gene expression (Fig. 9). Using cell lysates transfected with the wild-type p53 gene (Fig. 9, lanes 6) or vector-only control (Fig. 9, lanes 5), immunoprecipitation of TRRAP (Fig. 9A) resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 9A, lane 6 [35 RLU] versus lane 5 [8 RLU]). Furthermore, immunoprecipitation of BRG-1 resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 9B, lane 6 [59 RLU] versus lane 5 [8 RLU]). Also, enhancement of histone acetylation was p53 dependent, since immunoprecipitation with acetylhistone antibodies resulted in the coprecipitation and PCR amplification of the p21 promoter (Fig. 9C, lane 6 [210 RLU] versus lane 5 [60 RLU]). In contrast to TRRAP and BRG-1 recruitment when wild-type p53 was used, p53ΔPXXP was unable to recruit a significant amount of BRG-1 or TRRAP. Using cell lysates transfected with the p53ΔPXXP gene (Fig. 9, lanes 7), the use of antibodies to TRRAP, BRG-1, or acetylhistone failed to immunoprecipitate the p21 promoter (Fig. 9A to C, lanes 7 versus lanes 5 and 6).
FIG. 9.
The PXXP motif is required to recruit TRRAP and BRG-1 to the p21 promoter in vivo. HCT116 p53−/− cells were transfected with various p53 pCMV DNAs encoding p53 (lanes 2 and 6), p53ΔPXXP (lanes 3 and 7), and the nonacetylatable mutant, p53-6KR (lanes 4 and 8). Following transfection of the indicated construct, cross-linking of protein-DNA complexes with formaldehyde, immunoprecipitation, and processing of the samples as described in Materials and Methods, the released DNA was PCR amplified using primers to the p21 promoter (as indicated in the bottom panel) or GAPDH promoter. The immunoprecipitation was carried out with antibodies specific for TRRAP (A), BRG-1 (B), and acetylated histone H4 (C). The data are plotted as input DNA or as immunoprecipitated DNA. Quantitation of the bioluminescence is depicted below each lane. Controls include DNA amplified in reactions processed from cells without antibody in the immunoprecipitation reaction (lane 9) or with vector control only (lanes 1 and 5).
DISCUSSION
Regulation of eukaryotic gene expression is a dynamic process regulated by signal transduction networks that assemble and disassemble a variety of protein-protein machines. Characterizing these fundamental protein-protein interactions and their regulation will provide a molecular basis for understanding how the control of gene expression is linked to biological pathways such as differentiation, development, and cell cycle control. The coactivator p300 plays a central role in signal integration with transcriptional components, allowing for gene expression changes in response to a variety of stimuli (8). Understanding the mechanism whereby p300 and p53 cooperate as tumor suppressors will shed light on mechanisms that modulate cancer progression.
One key molecular stage in the cooperation between these two proteins links sequence-specific DNA binding by p53 and acetyltransferase activity of p300 (12). Conformational restraints on p53 acetylation by p300 are overcome by p53 binding to its consensus site DNA (Fig. 10). DNA-dependent acetylation of p53 requires p300 docking to the LXXLL domain of p53 through at least two subdomains in p300, named IHD and IBiD (12). Importantly, however, the ability of consensus site DNA to override conformational restraints on p53 acetylation by p300 suggested to us that p300 contacts an undefined conformationally sensitive interaction site on p53. Such a model is supported by nuclear magnetic resonance studies showing that the surface of p53 changes conformation when in complex with consensus site DNA (34). This has led to our search for a second, conformationally sensitive domain on p53 that binds p300 and led to the identification of the PXXP repeat domain as the flexible motif driving sequence-specific DNA-dependent acetylation of p53. The direct interaction of the proline repeat motif of p53 with p300 is consistent with the importance of the proline repeat domain for p53 activity (3, 40). However, we show an absolute requirement for the proline repeat motif on p53 activity from transfected p21 or bax reporter templates, while other studies have shown that p53ΔPXXP can be active from transfected p21 reporter templates (3).
There have been other molecular studies on proteins that interact with the proline repeat domain of p53, but none of these proteins explains the direct contribution of the PXXP domain to stress-activated transcription. Deletion of the proline repeat domain of p53 sensitizes p53 to MDM2-dependent degradation (6), presumably because this deletion releases p53 from a “stable” partner protein. The key PXXP-binding protein that stabilizes p53 appears to be p300 based on four criteria. First, p300 is essential for stabilizing p53 in response to DNA damage (42). Second, p300 has an intrinsic proline repeat-binding activity (Fig. 1). Third, proline repeat peptides inhibit sequence-specific DNA-dependent p53 acetylation (Fig. 3). Fourth, the LXXLL and PXXP-GFP fusion peptides can together destabilize p53 in vivo (Fig. 4). These latter data are inconsistent with a role for p300 in degrading p53 (17), unless the sequestration of the PXXP- and LXXLL-binding domains of p300 by the LXXLL and PXXP-GFP fusion peptides frees up alternate domains in p300 to degrade p53 protein. Other proteins can bind to the PXXP domain, including histone deacetylases and proline isomerases. Histone deacetylase binding to the PXXP domain switches on the transrepression function of p53 (43), which is activated in response to hypoxia, thus indicating a fundamental difference between classic DNA damage and oxygen deprivation. It may be that the energy limitations in cells orchestrating a p53 response in cells undergoing anaerobic metabolism induce a more passive switch-off of gene expression, rather than the more energetically demanding DNA damage response. The ability of proline isomerase to stimulate p53 identifies another proline-related modifier of p53 (41). It will be interesting in the future to examine whether proline isomerases or hydroxylases antagonize or synergize with p300 and affect DNA-dependent acetylation of p53.
In summary, a direct function for the proline domain in mediating the transactivation function of p53 was identified: the PXXP motif binds p300. There are therefore two distinct motifs (LXXLL and PXXP) in p53 that bind directly to p300 and that are required for p300-catalyzed acetylation. The ubiquity of the PXXP domain in many transcription factors (Fig. 1) flanking the classic LXXLL motif expands on the core p300-activation motifs. Our preliminary data indicate that proteins known to bind p300 and having both the LXXLL and PXXP motifs, including IRF-3 and E-KLF, do in fact require both motifs for maximal transcription activity (data not shown). The intrinsic conformational constraints on native, folded, p53 tetramer acetylation by p300 can be overcome after p53 DNA binding via p300 contacting two transactivation domains in p53 (Fig. 10). This built-in restraint presumably ensures that acetylation of p53 only occurs at promoters in vivo and reveals an allosteric role for DNA in controlling protein-protein interactions at a promoter (26). The fact that protein kinases and proline isomerases modify the core LXXLL and PXXP transactivation domains of p53 identifies signal transduction pathways, like CHK2 and proline isomerase, that stimulate p300 coactivated p53-dependent transcription. Furthermore, the realization that many transcription factors have LXXLL motifs contiguous to PXXP motifs expands our understanding of the basal p300 activation domain interactions at a promoter that may scaffold, bridge, or mediate the recruitment of multiprotein complexes at promoters in vivo.
Acknowledgments
D.D. was supported by a PhD studentship from the BBSRC. T.R.H. is supported by a program grant from Cancer Research UK, an MRC Career Establishment grant, and the Association for International Cancer Research.
REFERENCES
- 1.Ashraf, S. S., E. Anderson, K. Duke, P. T. Hamilton, and Z. Fredericks. 2003. Identification and characterization of peptide probes directed against PKCα conformations. J. Pept. Res. 61:263-273. [DOI] [PubMed] [Google Scholar]
- 2.Avantaggiati, M. L., V. Ogryzko, K. Gardner, A. Giordano, A. S. Levine, and K. Kelly. 1997. Recruitment of p300/CBP in p53-dependent signal pathways. Cell 89:1175-1184. [DOI] [PubMed] [Google Scholar]
- 3.Baptiste, N., P. Friedlander, X. Chen, and C. Prives. 2002. The proline-rich domain of p53 is required for cooperation with anti- neoplastic agents to promote apoptosis of tumor cells. Oncogene 21:9-21. [DOI] [PubMed] [Google Scholar]
- 4.Barlev, N. A., L. Liu, N. H. Chehab, K. Mansfield, K. G. Harris, T. D. Halazonetis, and S. L. Berger. 2001. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 8:1243-1254. [DOI] [PubMed] [Google Scholar]
- 5.Bell, S., C. Klein, L. Muller, S. Hansen, and J. Buchner. 2002. p53 contains large unstructured regions in its native state. J. Mol. Biol. 322:917-927. [DOI] [PubMed] [Google Scholar]
- 6.Berger, M., R. Vogt Sionov, A. J. Levine, and Y. Haupt. 2001. A role for the polyproline domain of p53 in its regulation by Mdm2. J. Biol. Chem. 276:3785-3790. [DOI] [PubMed] [Google Scholar]
- 7.Blaydes, J. P., M. G. Luciani, S. Pospisilova, H. M. Ball, B. Vojtesek, and T. R. Hupp. 2001. Stoichiometric phosphorylation of human p53 at Ser315 stimulates p53-dependent transcription. J. Biol. Chem. 276:4699-4708. [DOI] [PubMed] [Google Scholar]
- 8.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 9.Cohen, P. A., T. R. Hupp, D. P. Lane, and D. A. Daniels. 1999. Biochemical characterization of different conformational states of the Sf9 cell-purified p53His175 mutant protein. FEBS Lett. 463:179-184. [DOI] [PubMed] [Google Scholar]
- 10.de Caestecker, M. P., T. Yahata, D. Wang, W. T. Parks, S. Huang, C. S. Hill, T. Shioda, A. B. Roberts, and R. J. Lechleider. 2000. The Smad4 activation domain (SAD) is a proline-rich, p300-dependent transcriptional activation domain. J. Biol. Chem. 275:2115-2122. [DOI] [PubMed] [Google Scholar]
- 11.Dornan, D., and T. R. Hupp. 2001. Inhibition of p53-dependent transcription by BOX-I phospho-peptide mimetics that bind to p300. EMBO Rep. 2:139-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dornan, D., H. Shimizu, N. D. Perkins, and T. R. Hupp. 2003. DNA-dependent acetylation of p53 by the transcription coactivator p300. J. Biol. Chem. 278:13431-13441. [DOI] [PubMed] [Google Scholar]
- 13.Dyson, H. J., and P. E. Wright. 2002. Insights into the structure and dynamics of unfolded proteins from nuclear magnetic resonance. Adv. Protein Chem. 62:311-340. [DOI] [PubMed] [Google Scholar]
- 14.Espinosa, J. M., and B. M. Emerson. 2001. Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell 8:57-69. [DOI] [PubMed] [Google Scholar]
- 15.Goodman, R. H., and S. Smolik. 2000. CBP/p300 in cell growth, transformation, and development. Genes Dev. 14:1553-1577. [PubMed] [Google Scholar]
- 16.Grossman, S. R. 2001. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 268:2773-2778. [DOI] [PubMed] [Google Scholar]
- 17.Grossman, S. R., M. E. Deato, C. Brignone, H. M. Chan, A. L. Kung, H. Tagami, Y. Nakatani, and D. M. Livingston. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science 300:342-344. [DOI] [PubMed] [Google Scholar]
- 18.Gu, W., and R. G. Roeder. 1997. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90:595-606. [DOI] [PubMed] [Google Scholar]
- 19.Halazonetis, T. D., L. J. Davis, and A. N. Kandil. 1993. Wild-type p53 adopts a “mutant”-like conformation when bound to DNA. EMBO J. 12:1021-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hansen, S., T. R. Hupp, D. P. Lane, et al. 1996. Allosteric regulation of the thermostability and DNA binding activity of human p53 by specific interacting proteins. J. Biol. Chem. 271:3917-3924. [DOI] [PubMed] [Google Scholar]
- 21.Hupp, T. R., and D. P. Lane. 1994. Allosteric activation of latent p53 tetramers. Curr. Biol. 4:865-875. [DOI] [PubMed] [Google Scholar]
- 22.Hupp, T. R., and D. P. Lane. 1995. Two distinct signaling pathways activate the latent DNA binding function of p53 in a casein kinase II-independent manner. J. Biol. Chem. 270:18165-18174. [DOI] [PubMed] [Google Scholar]
- 23.Keller, D. M., X. Zeng, Y. Wang, Q. H. Zhang, M. Kapoor, H. Shu, R. Goodman, G. Lozano, Y. Zhao, and H. Lu. 2001. A DNA damage-induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7:283-292. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kussie, P. H., S. Gorina, V. Marechal, B. Elenbaas, J. Moreau, A. J. Levine, and N. P. Pavletich. 1996. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science 274:948-953. [DOI] [PubMed] [Google Scholar]
- 26.Lefstin, J. A., and K. R. Yamamoto. 1998. Allosteric effects of DNA on transcriptional regulators. Nature 392:885-888. [DOI] [PubMed] [Google Scholar]
- 27.Lin, C. H., B. J. Hare, G. Wagner, S. C. Harrison, T. Maniatis, and E. Fraenkel. 2001. A small domain of CBP/p300 binds diverse proteins: solution structure and functional studies. Mol. Cell 8:581-590. [DOI] [PubMed] [Google Scholar]
- 28.Manning, E. T., T. Ikehara, T. Ito, J. T. Kadonaga, and W. L. Kraus. 2001. p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol. 21:3876-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martinez-Balbas, M. A., U. M. Bauer, S. J. Nielsen, A. Brehm, and T. Kouzarides. 2000. Regulation of E2F1 activity by acetylation. EMBO J. 19:662-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nichols, N. M., and K. S. Matthews. 2002. Human p53 phosphorylation mimic, S392E, increases nonspecific DNA affinity and thermal stability. Biochemistry 41:170-178. [DOI] [PubMed] [Google Scholar]
- 31.Olivier, M., R. Eeles, M. Hollstein, M. A. Khan, C. C. Harris, and P. Hainaut. 2002. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum. Mutat. 19:607-614. [DOI] [PubMed] [Google Scholar]
- 32.Polesskaya, A., I. Naguibneva, A. Duquet, E. Bengal, P. Robin, and A. Harel-Bellan. 2001. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 21:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prives, C., and J. L. Manley. 2001. Why is p53 acetylated? Cell 107:815-818. [DOI] [PubMed] [Google Scholar]
- 34.Rippin, T. M., S. M. Freund, D. B. Veprintsev, and A. R. Fersht. 2002. Recognition of DNA by p53 core domain and location of intermolecular contacts of cooperative binding. J. Mol. Biol. 319:351-358. [DOI] [PubMed] [Google Scholar]
- 35.Sartorelli, V., P. L. Puri, Y. Hamamori, V. Ogryzko, G. Chung, Y. Nakatani, J. Y. Wang, and L. Kedes. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell 4:725-734. [DOI] [PubMed] [Google Scholar]
- 36.Shieh, S. Y., J. Ahn, K. Tamai, Y. Taya, and C. Prives. 2000. The human homologs of checkpoint kinases Chk1 and Cds1 (Chk2) phosphorylate p53 at multiple DNA damage-inducible sites. Genes Dev. 14:289-300. [PMC free article] [PubMed] [Google Scholar]
- 37.Shieh, S. Y., M. Ikeda, Y. Taya, and C. Prives. 1997. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 91:325-334. [DOI] [PubMed] [Google Scholar]
- 38.Shimizu, H., L. R. Burch, A. J. Smith, D. Dornan, M. Wallace, K. L. Ball, and T. R. Hupp. 2002. The conformationally flexible S9-S10 linker region in the core domain of p53 contains a novel MDM2 binding site whose mutation increases ubiquitination of p53 in vivo. J. Biol. Chem. 277:28446-28458. [DOI] [PubMed] [Google Scholar]
- 39.Vogelstein, B., D. Lane, and A. J. Levine. 2000. Surfing the p53 network. Nature 408:307-310. [DOI] [PubMed] [Google Scholar]
- 40.Walker, K. K., and A. J. Levine. 1996. Identification of a novel p53 functional domain that is necessary for efficient growth suppression. Proc. Natl. Acad. Sci. USA 93:15335-15340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wulf, G. M., Y. C. Liou, A. Ryo, S. W. Lee, and K. P. Lu. 2002. Role of Pin1 in the regulation of p53 stability and p21 transactivation, and cell cycle checkpoints in response to DNA damage. J. Biol. Chem. 277:47976-47979. [DOI] [PubMed] [Google Scholar]
- 42.Yuan, Z. M., Y. Huang, T. Ishiko, S. Nakada, T. Utsugisawa, H. Shioya, Y. Utsugisawa, Y. Shi, R. Weichselbaum, and D. Kufe. 1999. Function for p300 and not CBP in the apoptotic response to DNA damage. Oncogene 18:5714-5717. [DOI] [PubMed] [Google Scholar]
- 43.Zilfou, J. T., W. H. Hoffman, M. Sank, D. L. George, and M. Murphy. 2001. The corepressor mSin3a interacts with the proline-rich domain of p53 and protects p53 from proteasome-mediated degradation. Mol. Cell. Biol. 21:3974-3985. [DOI] [PMC free article] [PubMed] [Google Scholar]