Abstract
The m-AAA protease is a conserved hetero-oligomeric complex in the inner membrane of mitochondria. Recent evidence suggests a compartmentalization of the contiguous mitochondrial inner membrane into an inner boundary membrane (IBM) and a cristae membrane (CM). However, little is known about the functional differences of these subdomains. We have analyzed the localizations of the m-AAA protease and its substrate cytochrome c peroxidase (Ccp1) within yeast mitochondria using live cell fluorescence microscopy and quantitative immunoelectron microscopy. We find that the m-AAA protease is preferentially localized in the IBM. Likewise, the membrane-anchored precursor form of Ccp1 accumulates in the IBM of mitochondria lacking a functional m-AAA protease. Only upon proteolytic cleavage the mature form mCcp1 moves into the cristae space. These findings suggest that protein quality control and proteolytic activation exerted by the m-AAA protease take place preferentially in the IBM pointing to significant functional differences between the IBM and the CM.
INTRODUCTION
Mitochondria feature two nested membranes, a smooth outer membrane surrounding the inner membrane, which exhibits numerous infoldings, the cristae. The contiguous inner membrane is subdivided into two morphologically and also presumably functionally distinct subcompartments, namely the cristae membrane (CM) and the inner boundary membrane (IBM), running parallel to the outer membrane. These membrane domains are connected by narrow tubular cristae junctions (Frey et al., 2002; Mannella, 2006).
The cristae are particular numerous in cells with high energy demand such as muscle or liver cells. In line with a role in energy generation, protein complexes required for oxidative phosphorylation are enriched in the CM (Gilkerson et al., 2003). Perhaps not surprisingly, the translocases of the inner membrane that mediate the import of nuclear encoded proteins have been shown to be enriched in the IBM (Vogel et al., 2006; Wurm and Jakobs, 2006). Apart from protein import, very little is known about processes that are preferentially localized in the IBM.
Many proteins of the inner membrane assemble into larger protein complexes. Such assembly processes require extensive quality control mechanisms to avoid the accumulation of deleterious malfolded or misassembled proteins (Carr and Winge, 2003; Fontanesi et al., 2006). Central components of the proteolytic control system are two conserved AAA proteases that degrade misfolded or nonassembled inner membrane proteins: the i-AAA protease exposes its catalytic site to the intermembrane space (IMS), whereas the m-AAA protease is active at the matrix (for review see Juhola et al., 2000; Koppen and Langer, 2007). The m-AAA protease is a hetero-oligomeric complex composed of the homologous subunits AFG3L2 and paraplegin in humans (Atorino et al., 2003) and Yta10 (Afg3) and Yta12 (Rca1) in the budding yeast Saccharomyces cerevisiae (Arlt et al., 1996). Mutations in the m-AAA protease cause severe defects in various organisms, including respiratory deficiency in budding yeast (Arlt et al., 1998), accumulation of aberrant mitochondria in paraplegin-deficient mice (Ferreirinha et al., 2004) and neurodegeneration in humans (Nolden et al., 2005). Next to its role in protein surveillance, the m-AAA protease processes the ribosomal subunit MrpL32 and is essential for the maturation of the heme-binding reactive oxygen scavenger protein Ccp1 in S. cerevisiae (Esser et al., 2002; Nolden et al., 2005).
A bipartite presequence targets the nuclear encoded Ccp1 into the IMS. The hydrophobic sorting signal initially triggers insertion of newly imported precursor Ccp1 (pCcp1) protein into the mitochondrial inner membrane. Subsequently, the presequence is cleaved off in a two-step process by the m-AAA protease and the rhomboid protease Pcp1 (Rbd1; Esser et al., 2002; Michaelis et al., 2005). For maturation of pCcp1, the m-AAA protease mediates the ATP-dependent vectorial dislocation of the precursor protein and positions it within the lipid bilayer for intramembrane cleavage by Pcp1 (Tatsuta et al., 2007). Finally, mature Ccp1 (mCcp1) is released as a soluble protein into the IMS.
In this study we demonstrate that the m-AAA protease and the membrane anchored pCcp1 are preferentially localized in the IBM, giving new insights into the functional differences between the IBM and the CM.
MATERIALS AND METHODS
Growth Media, Strain Construction, and Cloning
Growth media were described previously (Sherman, 2002). A detailed listing of the yeast strains used in this study is given in Supplementary Table S1. For epitope-tagging of CCP1, the tagging vector pUGFP (Andresen et al., 2004) containing a KanMX4-cassette as selection marker was used as a PCR template with the primers GCG CCC AGT CCA TTT ATT TTC AAG ACT TTA GAG GAA CAA GGT TTA GGA TCC TCT GGA TGT TGT CCT and GTA TGG CTT TCC ATT TTT CAT AGA CGT ACC GTA CAA ACG TAT TAG ACT ATA GGG AGA CCG GCA GAT. For tagging of QCR2, genomic DNA isolated from a QCR2-mRFP:kanMX4 yeast strain was used as a template for PCR with the primers CCG TGG TAT CTT CCA ACA TC and CCG TTT ATT TCA GCC TTG CG. For targeted gene disruptions, PCR fragments were generated using genomic DNA isolated from Δyta10::kanMX4 and Δyta12::kanMX4 strains, respectively, using the corresponding A and D primer pairs as designed by the Saccharomyces Genome Deletion Project (www-sequence.stanford.edu/group/yeast_deletion_project). Disruption of MDM10 was performed anew before each experiment by replacing the gene by a kanamycin or a nourseothricine-resistance cassette, as described earlier (Wurm and Jakobs, 2006).
For expression of the hybrid protein Su9(1-69)-MrpL32(72-183), the respective fragment of MRPL32 was amplified from yeast genomic DNA with the primers GCG CGC GGT ACC GCA GTT CCT AAA AAA AAA G and GCG CGC CTC GAG CTA GTC CTT TTT TAA AGT CC and cloned in the vector pVT100U-mtGFP (Westermann and Neupert, 2000) via KpnI/XhoI, replacing the coding sequence of green fluorescent protein (GFP) with that of MrpL32(72–183), as described (Nolden et al., 2005). A yeast strain expressing Ccp1-GFP and Qcr2-monomeric red fluorescent protein (mRFP) was first transformed with the resulting plasmid pVT100U-mtMrpL32(72–183) and then, in a second step, a targeted gene disruption of YTA12 was performed. For expression of s-Mgm1*, a yeast strain expressing Ccp1-GFP and Qcr2-mRFP was first transformed with pRS313 s-Mgm1* (Herlan et al., 2003) and then, in a second step, a targeted gene disruption of PCP1 was performed.
Hydrogen Peroxide Growth Assay
Cells were grown to logarithmic growth phase (OD600 = 1) in YPD medium and diluted 1:200 in fresh growth medium. Hydrogen peroxide was then added (final concentrations: 0, 1, 2, 4, 8, and 16 mM), and the cells were incubated for 1 h at 37°C with continuous agitation. Ten microliters of the cell suspensions was spotted onto YPD agar plates that were incubated for 3 d at 33°C.
Biochemical Characterization and Western Analysis
Isolation of yeast mitochondria was performed as described (Diekert et al., 2001). To separate soluble from integral membrane proteins, mitochondria (100 μg) were suspended in SEM-buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS/KOH, pH 7.2) and extracted in 100 mM Na2CO3. Subsequently proteins were separated by ultracentrifugation (45 min, 110,000 × g). To determine the membrane association of the different forms of Ccp1-GFP, mitochondria (100 μg) were disrupted by suspending them in 20 mM HEPES (pH 7.4). Then the disrupted mitochondria were extracted in 200 mM Na2CO3 before ultracentrifugation (10 min, 35,000 × g). To discriminate proteins of the outer membrane from those of the inner membrane, mitochondria (100 μg) were incubated in SEM-buffer (250 mM sucrose, 1 mM EDTA, 10 mM MOPS/KOH, pH 7.2) with or without proteinase K (final concentration 10 μg/ml) for 30 min on ice. The inhibition of protease activity was achieved by adding complete protease inhibitor cocktail (Roche, Mannheim, Germany). For Western analysis, proteins were precipitated with 10% (wt/vol) TCA. After separation by SDS-PAGE, the samples were transferred to a membrane and detected using the indicated antibodies.
Quantitative Immuno-Electron Microscopy
Cells were grown in complete liquid media containing 2% (wt/vol) galactose at 30°C and harvested during early log-phase. For the electron microscopy (EM) of the yeast cells, ultrathin cryosections were prepared as described previously (Tokuyasu, 1973; Kreykenbohm et al., 2002). In brief, the yeast cells were fixed with 2% (wt/vol) formaldehyde (in 50%, vol/vol, growth medium) for 30 min at room temperature. After centrifugation, they were postfixed with 4% (wt/vol) formaldehyde (in PBS) overnight, followed by 2 h with 4% (wt/vol) formaldehyde and 0.1% (wt/vol) glutaraldehyde (both steps on ice). After two additional washings with PBS and 0.02% (wt/vol) glycine, cells were embedded in 10% (wt/vol) gelatin, cooled on ice, and cut into small blocks. The blocks were infused with 2.1 M sucrose and 0.4% (wt/vol) formaldehyde overnight. After washing in 2.3 M sucrose and 0.02% (wt/vol) glycine, the blocks were mounted on metal pins and frozen in liquid nitrogen. Ultrathin sections (80 nm) were cut in an ultracryomicrotome (Leica Microsystems, Wetzlar, Germany) using a diamond knife (Diatome, Biel, Switzerland). For immunolabeling, sections were incubated with a polyclonal rabbit antibody against GFP (Abcam, Cambridge, United Kingdom, 1:200) for 20 min, followed by incubation with protein A-gold (10 nm) for 20 min. After several washing steps (five times for 3 min in TBS/0.5%, wt/vol, BSA; five times for 3 min in TBS followed by five times for 3 min in TBS/0.1% (wt/vol) Tween20), sections were contrasted with uranyl acetate/methyl cellulose (Liou et al., 1996) for 10 min on ice, embedded in the same solution, and examined with an electron microscope (Philips, Mahwah, NJ; CM120).
For the quantification ≥100 individual images corresponding to >100 gold particles were analyzed. To this end the distance of a gold particle to the mitochondrial boundary was determined. Gold particles closer than 20 nm were counted as being localized to the mitochondrial rim, i.e., to the IBM or to the inner boundary membrane space (IBMS). Those gold particles within the mitochondria at a distance of more than 20 nm were counted as being localized in the mitochondrial interior, i.e., the CM or the cristae space (CS).
To determine the relative surface areas of the CM and the IBM, >25 EM images of the respective yeast cells were analyzed. All scrutinized images showed good structural preservation, as judged by a clearly distinguishable IBM. The relative surfaces of the CM and the IBM were determined by fitting freeforms (open multipoint polygons) to the EM images. The analysis was performed using the AnalySIS5 software (Soft Imaging System GmbH, Münster, Germany).
Confocal Fluorescence Microscopy
For the microscopy of cells exhibiting enlarged mitochondria, strains were freshly transformed to delete MDM10. Single colonies were microscopically analyzed 5–9 d after transformation. Only colonies with more than 50% of the cells exhibiting enlarged mitochondria were analyzed. The diameter of the analyzed enlarged mitochondria was between 0.8 and 2.0 μm. Images were recorded with a beam scanning confocal microscope (TCS SP5, Leica Microsystems) equipped with an HCX PL APO CS 63× oil immersion objective. Dual color images were recorded sequentially. Each image was averaged twice. Except for contrast stretching, no image processing was applied.
RESULTS
Yta10-GFP and Yta12-GFP Complement the Untagged Proteins
The m-AAA protease is composed of Yta10 and Yta12 in budding yeast. To enable the light microscopy and EM analysis of the distribution of the m-AAA protease within the inner membrane of mitochondria, we genetically fused Yta10 and Yta12 with the GFP. To ensure close to normal expression levels, the respective fusion gene replaced the native gene in the genome, leaving the promoter region unchanged.
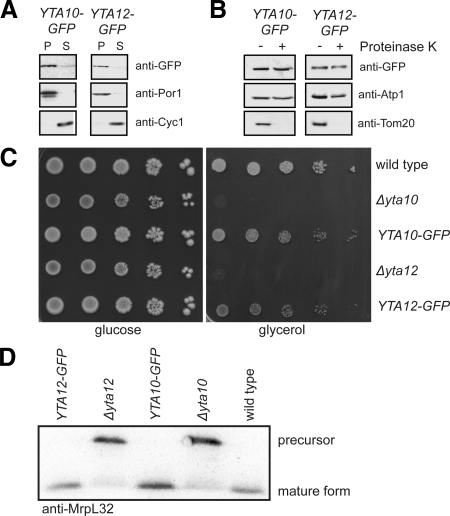
To verify the correct localization and integrity of the fusion proteins, biochemical subfractionation experiments were performed. Sodium carbonate extraction of purified mitochondria identified Yta10-GFP and Yta12-GFP as membrane proteins (Figure 1A). Incubation of intact mitochondria with proteinase K did dot affect the proteins, in full agreement with previous reports on Yta10 and Yta12 as being integral proteins of the mitochondrial inner membrane (Figure 1B; Pajic et al., 1994; Arlt et al., 1998). Furthermore, no degradation products of Yta10-GFP or Yta12-GFP were detected on Western blots (Supplementary Figure S1).
Figure 1.
Yta10-GFP and Yta12-GFP are functional. (A) Yta10-GFP and Yta12-GFP are integrated into mitochondrial membranes. Mitochondria isolated from cells expressing Yta10-GFP or Yta12-GFP were extracted with sodium carbonate. The membrane pellet (P) and the soluble supernatant (S) were analyzed by SDS-PAGE and immunoblotting using GFP-specific antiserum to detect the fusion proteins. As a control, antisera against the membrane protein porin and the soluble protein cytochrome c were used. (B) Yta10-GFP and Yta12-GFP are inner membrane proteins. Isolated mitochondria were subjected to proteinase K treatment to digest outer membrane proteins. After this treatment the samples were analyzed by SDS-PAGE and immunoblotting using GFP-specific antiserum. As a control, antisera against the outer membrane protein Tom20 and Atp1, a subunit of the F1F0-ATP-Synthase of the inner membrane, were used. (C) Restoration of respiratory growth of Δyta10 and Δyta12 cells upon expression of Yta10-GFP and Yta12-GFP, respectively. Tenfold serial dilutions of logarithmically growing cultures were spotted onto plates containing glucose or glycerol as sole carbon sources and incubated for 7 d at 30°C. (D) MrpL32 is processed in cells expressing Yta10-GFP and Yta12-GFP as in wild-type cells. Isolated mitochondria were analyzed by SDS-PAGE and immunoblotting using MrpL32-specific antiserum.
Strains expressing Yta10-GFP or Yta12-GFP grew on fermentable and nonfermentable carbon sources at wild-type rates, whereas Δyta10 and Δyta12 strains were respiration deficient (Figure 1C). Further, yeast strains defective in Yta10 or Yta12 are deficient in the proteolytic processing of the mitochondrial ribosomal protein MrpL32 (Figure 1D; Nolden et al., 2005). Cells expressing Yta10-GFP or YTA12-GFP process MrpL32 as wild-type cells (Figure 1D), demonstrating that the m-AAA protease exhibits normal proteolytic activity in these strains. We conclude that Yta10-GFP and Yta12-GFP are expressed at endogenous levels, are correctly inserted into the mitochondrial inner membrane, and are functional.
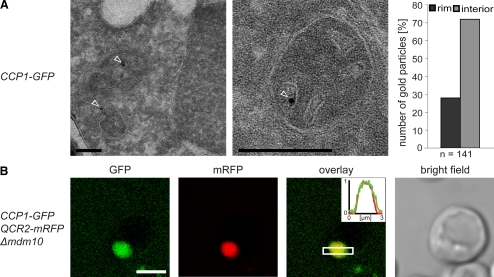
m-AAA Protease Is Enriched in the IBM
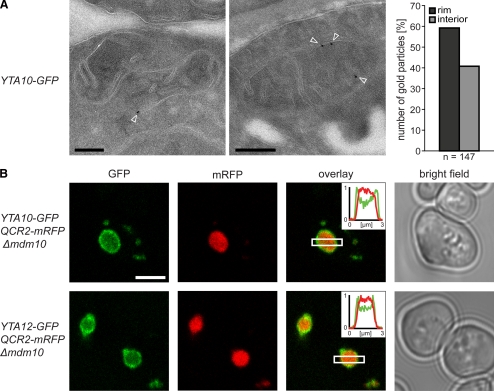
To determine the distribution of the m-AAA protease within the mitochondrial inner membrane, we performed quantitative immuno-EM on chemically fixed cryosectioned yeast cells. The used GFP antibody was highly specific and gave very little unspecific labeling in control experiments. The mitochondria of cells expressing Yta10-GFP were decorated on average with one or two gold particles. The majority (59%) of the gold particles were found at the IBM (Figure 2A). All cells analyzed by EM were grown in galactose-containing medium. We found that in galactose-grown cells the surface area of the CM was on average almost identical to the surface area of the IBM. Hence these data suggest that the m-AAA protease is preferentially, albeit not exclusively, localized in the IBM.
Figure 2.
The m-AAA protease is preferentially localized in the IBM. (A) Localization of Yta10-GFP by quantitative immuno-EM in mitochondria of S. cerevisiae cells using GFP-specific antiserum. Left, representative images. The arrowheads point to gold particles. Right, quantification of the distribution of Yta10-GFP in the IBM (rim) and the CM (interior). (B) Live-cell fluorescence microscopy of Yta10-GFP or Yta12-GFP in cells exhibiting enlarged mitochondria due to a lack of Mdm10. Qcr2-mRFP was coexpressed to label the CM. Single confocal sections are displayed. Insets, normalized intensity profiles along the indicated area. Scale bars, (A) 200 nm; (B) 3 μm.
To verify this finding, we utilized the enlarged mitochondria of Δmdm10 cells, which have previously been used as an in vivo model system to study protein distributions within the inner membrane (Wurm and Jakobs, 2006). With a typical diameter of 1–2 μm, these mitochondria are large enough to visualize proteins being enriched in the IBM or the inner boundary membrane space (IBMS), respectively, using fluorescence microscopy. Next to Yta10-GFP or Yta12-GFP, we tagged Qcr2 (Cor2), a subunit of respiratory chain complex III, with the mRFP. We found Qcr2-mRFP in the cristae containing interior of enlarged mitochondria (Figure 2B), in full agreement with previous data showing an enrichment of Qcr2 in the CM (Gilkerson et al., 2003; Vogel et al., 2006). Unlike Qcr2-mRFP, both Yta10-GFP and Yta12-GFP were found to be enriched at the rim of the enlarged mitochondria (Figure 2B) confirming a preferential localization of the m-AAA protease in the IBM.
Because the immuno-EM (Figure 2A) and the live cell fluorescence microscopy (inset Figure 2B) revealed a substantial fraction of Yta10 and Yta12 also at the CM, we conclude that the m-AAA protease is enriched in the IBM, but is not exclusively localized in this subdomain of the inner membrane.
Ccp1-GFP Is Functional and Processed Properly
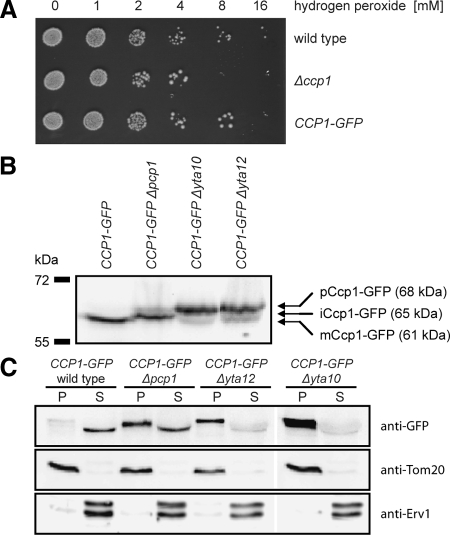
The m-AAA protease is essential for the maturation of Ccp1, a mitochondrial heme-binding reactive oxygen species (ROS) scavenger (Esser et al., 2002; Tatsuta et al., 2007). The elevated sensitivity of Δccp1 cells against increased hydrogen peroxide concentrations was restored upon expression of Ccp1-GFP from the genomic locus, demonstrating the functionality of the fusion protein (Figure 3A). After import into mitochondria, the precursor pCcp1 is initially inserted with a single membrane-spanning domain in the inner membrane with the large carboxy-terminal domain protruding in the IMS (Daum et al., 1982; Kaput et al., 1989). In wild-type cells, the precursor protein is processed by the consecutive action of m-AAA protease and Pcp1 to the soluble mature form (mCcp1), which is released into the IMS. In strains without Pcp1, an intermediate form of Ccp1 (iCcp1) accumulates (Esser et al., 2002; Michaelis et al., 2005). We found that Ccp1-GFP is processed to mCcp1-GFP and iCcp1-GFP in wild-type and Δpcp1 cells, respectively, whereas in cells without functional m-AAA protease only pCcp1-GFP was detected (Figure 3B), fully corroborating previous reports on untagged Ccp1. No unspecific degradation products of the Ccp1-GFP were found in any of these strains (Supplementary Figure S1). To determine whether pCcp-GFP, iCcp1-GFP, and mCcp-GFP are soluble or membrane bound, we performed sodium carbonate extractions of the respective purified mitochondria (Figure 3C). Although pCcp1-GFP in Δyta10 or Δyta12 cells was solely found in the membrane pellet, mCcp1-GFP of wild-type cells was in the soluble supernatant. We found iCcp1-GFP to various extents in the pellet and the soluble fraction, corroborating previous reports on a weak membrane attachment of this intermediate (Michaelis et al., 2005). We conclude that the GFP-tagged Ccp1 is functional and is processed by the m-AAA protease and Pcp1 similar to the untagged Ccp1.
Figure 3.
Ccp1-GFP is functional and processed correctly. (A) The growth phenotype of Δccp1 cells on elevated hydrogen peroxide concentrations is suppressed by the expression of Ccp1-GFP. Cells were grown to midlogarithmic growth phase and diluted in growth medium with the indicated hydrogen peroxide concentration. After 1 h at 37°C, 10 μl of the cell suspensions was spotted onto agar plates. (B) Analysis of Ccp1-GFP processing. Isolated mitochondria from wild-type, Δpcp1, Δyta10, or Δyta12 cells, each expressing Ccp1-GFP from the genomic locus, were analyzed by SDS-PAGE and analyzed by immunoblotting with GFP-specific antiserum to detect the fusion proteins. (C) Analysis of the membrane association of mCcp1, iCcp1, and pCcp1. Mitochondria isolated from cells expressing Ccp1-GFP and with the indicated genotypes were extracted with 200 mM sodium carbonate. The membrane pellet (P) and the soluble supernatant (S) were analyzed by SDS-PAGE and immunoblotting using GFP-specific antiserum. As control, antibodies against an integral protein of the outer membrane, Tom20, and a soluble protein of the IMS, Erv1, were utilized.
pCcp1-GFP Is Preferentially Localized in the IBM
Next, we set out to determine the distribution of pCcp1 between the CM and the IBM. To this end we exploited the fact that in Δyta12 cells pCcp1-GFP is inserted into the inner membrane but no further processing is occurring (Figure 3, B and C).
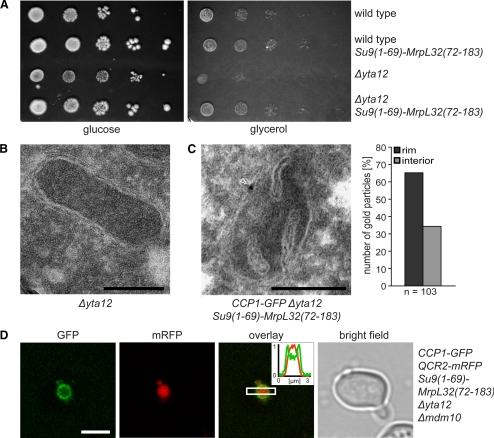
However, Δyta12 cells are respiration deficient (Figure 4A), and their mitochondria exhibit a strongly reduced number of cristae (Figure 4B), preventing a meaningful analysis of the submitochondrial distribution of pCcp1 in these cells. Because the respiration deficiency phenotype of Δyta10 cells is due to lack of mature MrpL32 (Nolden et al., 2005), we reasoned that the impaired maturation of MrpL32 might also determine the absence of cristae in the mitochondria of Δyta12 cells. To examine this possibility, we expressed mature MrpL32, targeted to the mitochondrial matrix by the residues 1–65 of subunit 9 of the F1FO-ATPase of Neurospora crassa in Δyta12 cells. We found that expressing the hybrid protein Su9(1-69)-MrpL32(72-183) suppresses the respiratory phenotype of cells lacking a functional m-AAA protease (Figure 4A), corroborating previous reports (Nolden et al., 2005). Moreover, the inner structures of these mitochondria were indistinguishable from wild-type mitochondria (Figure 4C). Analysis of the relative surface areas of CM and IBM revealed that the surface ratio of these two subdomains was on average practically identical in cells expressing Ccp1-GFP and Δyta12 cells expressing Su9(1-69)-MrpL32(72-183) and Ccp1-GFP (ratio CM/IBM = 0.98 ± 0.37 and 0.95 ± 0.33, respectively). Thus expression of matrix targeted mature MrpL32 fully restores cristae formation in Δyta12 cells, allowing the analysis of the submitochondrial distribution of pCcp1 in these cells.
Figure 4.
The precursor form of Ccp1 (pCcp1) is preferentially localized in the IBM. (A) Expression of the hybrid protein Su9(1-69)-MrpL32(72-183) suppresses the respiratory phenotype of Δyta12 cells. Tenfold serial dilutions of logarithmically growing cultures were spotted onto plates containing glucose or glycerol as sole carbon sources and incubated for 6 d at 30°C. (B) Electron micrograph showing that the mitochondria of Δyta12 cells exhibit a strongly reduced number of cristae. (C) Left, exemplary EM image of a mitochondrion of a cell lacking Yta12, but expressing the hybrid protein Su9(1-69)-MrpL32(72-183). The mitochondrion was immunogold-labeled for Ccp1-GFP; the arrowhead points to a gold particle. Right, quantification of the distribution of pCcp1-GFP in the IBM (rim) and the CM (interior). (D) Live-cell fluorescence microscopy of Ccp1-GFP coexpressed with the CM-marker Qcr2-mRFP in Δyta12 cells. To enable the light microscopic analysis, the mitochondria were enlarged by deleting MDM10. Single confocal sections are displayed. Scale bars, (B and C) 200 nm; (D) 3 μm.
Next, we performed quantitative immuno-EM on cells lacking the m-AAA subunit Yta12 but expressing Su9(1-69)-MrpL32(72-183) and Ccp1-GFP. Using a GFP-specific antibody, 66% of the gold particles were found at the IBM (Figure 4C), suggesting an enrichment of the membrane-anchored pCcp1 in this subdomain.
To verify this observation, we analyzed the localization of Ccp1-GFP in enlarged mitochondria of Δmdm10 cells lacking Yta12, but expressing Su9(1-69)-MrpL32(72-183). Additionally, the CM-marker Qcr2-mRFP was expressed in these cells to visualize differential protein sorting within the inner membrane. Using live cell fluorescence microscopy, we found that Qcr2-mRFP is localized in the cristae containing interior of the enlarged mitochondria, whereas pCcp1-GFP is strongly enriched in the IBM, confirming the EM data (Figure 4D). We conclude that Qcr2 and pCcp1 are differently distributed in the inner membrane, with the membrane-anchored precursor pCcp1 being preferentially localized in the IBM.
iCcp1-GFP Is Not Enriched at the Mitochondrial Rim
To determine whether the extended membrane anchor of pCcp1 is required for the enrichment of pCcp1 in the IBM, we analyzed the submitochondrial distribution of the intermediate form iCcp. In Δpcp1 cells, due to the action of the m-AAA protease, a substantial part of the membrane anchor of pCcp1 is removed. The resulting iCcp1-GFP is only weakly attached to the inner membrane (Figure 3C), suggesting that it is able to dissociate from the inner membrane into the IMS in vivo (Michaelis et al., 2005).
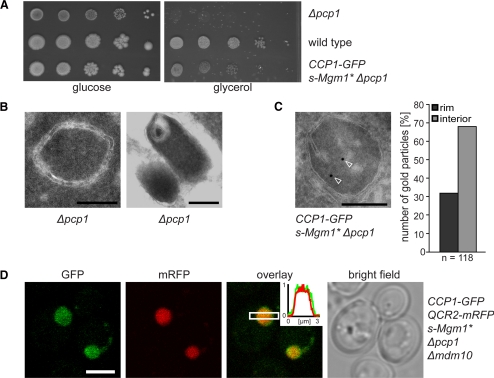
In addition to the impaired processing of iCcp1, cells lacking Pcp1 are also defective in the processing of the dynamin-related GTPase Mgm1 into the short isoform s-Mgm1, which results in fragmentation of mitochondria and in the loss of respiration competence of the cells (Herlan et al., 2003; Sesaki et al., 2003). The expression of s-Mgm1 containing an N-terminal extension of the targeting and sorting signals of cytochrome b2 (s-Mgm1*), partially suppressed the loss of respiration competence in Δpcp1 cells (Figure 5A; Herlan et al., 2003). We found that the expression of s-Mgm1* also partially restored the generation of cristae in mitochondria of Δpcp1 cells (Figure 5, B and C). Hence we utilized Δpcp1 cells expressing s-Mgm1* to study the submitochondrial distribution of iCcp1-GFP.
Figure 5.
The intermediate form of Ccp1 (iCcp1) is not enriched at the IBM of mitochondria. (A) Expression of s-Mgm1* partially suppresses the respiratory growth defect of Δpcp1 cells. Tenfold serial dilutions of logarithmically growing cultures were spotted onto agar plates containing glucose or glycerol as sole carbon source and incubated for 7 d at 30°C. (B) Δpcp1 cells exhibit mitochondria with aberrant internal structures. Frequently, mitochondria without cristae (left) or with onion-shaped membrane inclusions (right) were observed. (C) The mitochondria of Δpcp1 cells expressing s-Mgm1* frequently contain cristae. Exemplary EM image of a mitochondrion of a Δpcp1 cells expressing s-Mgm1* immunogold labeled for Ccp1-GFP; the arrowheads point to gold particles. Right, quantification of the distribution of iCcp1-GFP revealed an enrichment (68% of all gold particles) of iCcp1-GFP within the mitochondrial interior. (D) Live-cell fluorescence microscopy of both Ccp1-GFP and the CM-marker Qcr2-mRFP in Δpcp1 Δmdm10 cells. Single confocal sections are displayed. Inset, normalized intensity profiles along the indicated area. Scale bars, (B and C) 200 nm; (D) 3 μm.
Both, quantitative immuno-EM (Figure 5C) and live cell fluorescence microscopy of cells harboring enlarged mitochondria (Figure 5D) revealed that iCcp1-GFP is not enriched at the IBM. 68% of all gold particles were found in the mitochondrial interior. Because the low abundance of Pcp1 prevented a determination of the submitochondrial localization of this protein, the localization of the processing of iCcp1 cannot be deduced from this data. We conclude that the extended membrane anchor of pCcp1 is required for the enrichment of pCcp1 at the IBM.
Mature Ccp1 Is Localized in the Cristae Space
Next, we determined the submitochondrial localization of mature soluble Ccp1-GFP. In mitochondria of cells exhibiting functional m-AAA and Pcp1 proteases, the membrane anchor of Ccp1-GFP is fully cleaved off, and the mature protein is released into the IMS (Figure 3, B and C). Immuno-EM demonstrated a large fraction of mature Ccp1-GFP (71% of all gold particles) to be localized to the CS (Figure 6A). The shift in the submitochondrial distribution of Ccp1-GFP from the rim to the interior of mitochondria upon proteolytic processing from the precursor to the mature form was further confirmed by the analysis of the localization of Ccp1-GFP in enlarged mitochondria of living Δmdm10 cells. In these mitochondria, we found the localization of mature Ccp1-GFP indistinguishable from that of Qcr2-mRFP (Figure 6B).
Figure 6.
Mature Ccp1 (mCcp1) is distributed within the IMS. (A) Quantitative immuno-EM on wild-type cells expressing Ccp1-GFP. Left, representative immuno-EM images decorated with GFP-specific antiserum. The arrowheads point to gold particles. Right, quantification of the distribution of mCcp1-GFP in the IBMS (rim) and the CS (interior). (B) Live-cell fluorescence microscopy of Ccp1-GFP and the CM-marker Qcr2-mRFP in enlarged mitochondria of Δmdm10 cells. Single confocal sections are displayed. Inset, normalized intensity profiles along the indicated area. Scale bars, (A) 200 nm; (B) 3 μm.
Taken together, we conclude that the m-AAA protease is enriched in the IBM and that the membrane-anchored pCcp1 is preferentially localized in this subdomain of the inner membrane. On processing by the m-AAA protease and Pcp1, the mature Ccp1 is released and moves into the CS. These results demonstrate that with the m-AAA protease, central components of the mitochondrial protein surveillance machinery are preferentially localized in the inner boundary membrane.
DISCUSSION
The contiguous inner membrane of mitochondria consists of two morphologically distinct subdomains, namely the IBM that parallels the outer membrane and the infoldings of the inner membrane, the CM. A major conclusion from this study is that the m-AAA protease is preferentially localized in the IBM. Likewise, pCcp1, which is a substrate of the m-AAA protease, is enriched in the IBM of mitochondria lacking a functional m-AAA protease. This strongly indicates that the m-AAA protease processes pCcp1 preferentially in the inner boundary membrane.
A tempting explanation of the different protein compositions of the IBM and the CM is based on topological reasoning as the IBM is facing the outer membrane, whereas the CMs are facing each other. This appears to be a likely explanation for the enrichment of the TIM23 complex in the IBM (Wurm and Jakobs, 2006), because the TIM23 complex interacts at least during protein translocation with the TOM complex in the outer membrane. The results of this study suggest that additional mechanisms determining the predominant localization of proteins in the IBM exist.
On import, the nuclear encoded Ccp1 is inserted into the IBM. Because the precursor form of Ccp1 is loaded with heme in wild-type cells, it could be speculated that the machinery mediating this process might retain the precursor Ccp1 in the IBM. However, the attachment of heme is not a prerequisite for proteolytic processing of Ccp1 (Esser et al., 2002), and we found that the addition of the heme synthesis inhibitor succinyl acetone to enlarged mitochondria does not influence the localization of pCcp1-GFP (not shown). Furthermore, in wild-type cells, pCcp1 exists only in minute quantities, whereas it accumulates to substantial amounts in mitochondria lacking a functional m-AAA protease. Hence the stoichiometric occurrence of a binding partner of pCcp1 in Δyta12 cells appears rather unlikely, suggesting that the localization of precursor Ccp1 is determined differently.
One suggestion is that the cristae junctions might restrict the movement of pCcp1 from the IBM to the CM. Also, the preferential localization of the m-AAA protease in the IBM may be explained by such a barrier function of the highly bended inner membrane at the cristae junctions because the m-AAA protease has no known direct or indirect binding partner in the outer membrane. Because mCcp1 is soluble in the IMS and iCcp is only weakly attached to the inner membrane, with a tendency to dissociate into the IMS, it is tempting to assume that mCcp1 and iCcp1 may diffuse unrestrictedly through the openings of the cristae junctions. Because the relative sizes of the CS and the IBMS are not known, a conclusion on a subcompartmentalization of the IMS based on the data presented in this study is not possible; the data are, however, in full agreement with the hypothesis that mCcp1 is uniformly distributed in the IMS of wild-type cells.
The protein distributions between CM and IBM observed in this study are not binary. Rather, we demonstrated enrichment of the m-AAA protease and of the precursor form of Ccp1 in the IBM, but we also found these proteins in the CM, albeit at lower levels. Thus, if the cristae junctions act as gate-keepers, they appear not to be strict barriers, but may allow the passage of some proteins, possibly in a regulated manner.
Most nuclear encoded integral inner membrane proteins are released laterally into the lipid phase of the membrane during the import process. For proteins that are exported from the matrix into the inner membrane, another insertion machinery exists (for reviews see Rehling et al., 2004; Neupert and Herrmann, 2007). The major substrates of this machinery are mitochondrial encoded hydrophobic proteins that assemble with nuclear encoded proteins into supercomplexes involved in oxidative phosphorylation. The m-AAA protease is likely to have a safeguard function in these assembly processes. Thus the preferential localization of the m-AAA protease in the IBM points to the intriguing idea that the insertion, assembly, and quality control of the proteins of the mitochondrial inner membrane may predominantly take place in the IBM, suggesting that the functional differences of the IBM and the CM are more significant than previously anticipated.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Kai Hell, University of Munich, Thomas Langer, University of Cologne, and Doron Rapaport, University of Tübingen, for providing antisera. We thank Andreas Reichert, University of Frankfurt, for the plasmid pRS313-s-Mgm1*. We thank Stefan W. Hell for continuous support. We acknowledge Rita Schmitz-Salue for excellent technical assistance, Jaydev Jethwa for carefully reading the manuscript, and Ralf Reski for helpful discussions. This work was supported by a Grant JA 1129/3 from the Deutsche Forschungsgemeinschaft to S.J.
Abbreviations used:
- CM
cristae membrane
- CS
cristae space
- GFP
green fluorescent protein
- IBM
inner boundary membrane
- IMS
intermembrane space
- mRFP
monomeric red fluorescent protein
- IBMS
inner boundary membrane space
- EM
electron microscopy.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E07-11-1112) on November 19, 2008.
REFERENCES
- Andresen M., Schmitz-Salue R., Jakobs S. Short tetracysteine tags to beta-tubulin demonstrate the significance of small labels for live cell imaging. Mol. Biol. Cell. 2004;15:5616–5622. doi: 10.1091/mbc.E04-06-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H., Steglich G., Perryman R., Guiard B., Neupert W., Langer T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998;17:4837–4847. doi: 10.1093/emboj/17.16.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlt H., Tauer R., Feldmann H., Neupert W., Langer T. The YTA10–12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell. 1996;85:875–885. doi: 10.1016/s0092-8674(00)81271-4. [DOI] [PubMed] [Google Scholar]
- Atorino L., Silvestri L., Koppen M., Cassina L., Ballabio A., Marconi R., Langer T., Casari G. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 2003;163:777–787. doi: 10.1083/jcb.200304112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr H. S., Winge D. R. Assembly of cytochrome c oxidase within the mitochondrion. Acc. Chem. Res. 2003;36:309–316. doi: 10.1021/ar0200807. [DOI] [PubMed] [Google Scholar]
- Daum G., Bohni P. C., Schatz G. Import of proteins into mitochondria. Cytochrome b2 and cytochrome c peroxidase are located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 1982;257:13028–13033. [PubMed] [Google Scholar]
- Diekert K., de Kroon A. I., Kispal G., Lill R. Isolation and subfractionation of mitochondria from the yeast Saccharomyces cerevisiae. Methods Cell Biol. 2001;65:37–51. doi: 10.1016/s0091-679x(01)65003-9. [DOI] [PubMed] [Google Scholar]
- Esser K., Tursun B., Ingenhoven M., Michaelis G., Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J. Mol. Biol. 2002;323:835–843. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- Ferreirinha F., et al. Axonal degeneration in paraplegin-deficient mice is associated with abnormal mitochondria and impairment of axonal transport. J. Clin. Invest. 2004;113:231–242. doi: 10.1172/JCI20138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanesi F., Soto I. C., Horn D., Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am J. Physiol. Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- Frey T. G., Renken C. W., Perkins G. A. Insight into mitochondrial structure and function from electron tomography. Biochim. Biophys. Acta. 2002;1555:196–203. doi: 10.1016/s0005-2728(02)00278-5. [DOI] [PubMed] [Google Scholar]
- Gilkerson R. W., Selker J.M.L., Capaldi R. A. The cristal membrane of mitochondria is the principal site of oxidative phosphorylation. FEBS Lett. 2003;546:355–358. doi: 10.1016/s0014-5793(03)00633-1. [DOI] [PubMed] [Google Scholar]
- Herlan M., Vogel F., Bornhovd C., Neupert W., Reichert A. S. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J. Biol. Chem. 2003;278:27781–27788. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- Juhola M. K., Shah Z. H., Grivell L. A., Jacobs H. T. The mitochondrial inner membrane AAA metalloprotease family in metazoans. FEBS Lett. 2000;481:91–95. doi: 10.1016/s0014-5793(00)01989-x. [DOI] [PubMed] [Google Scholar]
- Kaput J., Brandriss M. C., Prussak-Wieckowska T. In vitro import of cytochrome c peroxidase into the intermembrane space: release of the processed form by intact mitochondria. J. Cell Biol. 1989;109:101–112. doi: 10.1083/jcb.109.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppen M., Langer T. Protein degradation within mitochondria: versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 2007;42:221–242. doi: 10.1080/10409230701380452. [DOI] [PubMed] [Google Scholar]
- Kreykenbohm V., Wenzel D., Antonin W., Atlachkine V., von Mollard G. F. The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur J. Cell Biol. 2002;81:273–280. doi: 10.1078/0171-9335-00247. [DOI] [PubMed] [Google Scholar]
- Liou W., Geuze H. J., Slot J. W. Improving structural integrity of cryosections for immunogold labeling. Histochem. Cell Biol. 1996;106:41–58. doi: 10.1007/BF02473201. [DOI] [PubMed] [Google Scholar]
- Mannella C. A. The relevance of mitochondrial membrane topology to mitochondrial function. Biochim. Biophys. Acta. 2006;1762:140–147. doi: 10.1016/j.bbadis.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Michaelis G., Esser K., Tursun B., Stohn J. P., Hanson S., Pratje E. Mitochondrial signal peptidases of yeast: the rhomboid peptidase Pcp1 and its substrate cytochrome C peroxidase. Gene. 2005;354:58–63. doi: 10.1016/j.gene.2005.04.020. [DOI] [PubMed] [Google Scholar]
- Neupert W., Herrmann J. M. Translocation of proteins into mitochondria. Annu. Rev. Biochem. 2007;76:723–749. doi: 10.1146/annurev.biochem.76.052705.163409. [DOI] [PubMed] [Google Scholar]
- Nolden M., Ehses S., Koppen M., Bernacchia A., Rugarli E. I., Langer T. The m-AAA protease defective in hereditary spastic paraplegia controls ribosome assembly in mitochondria. Cell. 2005;123:277–289. doi: 10.1016/j.cell.2005.08.003. [DOI] [PubMed] [Google Scholar]
- Pajic A., Tauer R., Feldmann H., Neupert W., Langer T. Yta10p is required for the ATP-dependent degradation of polypeptides in the inner membrane of mitochondria. FEBS Lett. 1994;353:201–206. doi: 10.1016/0014-5793(94)01046-3. [DOI] [PubMed] [Google Scholar]
- Rehling P., Brandner K., Pfanner N. Mitochondrial import and the twin-pore translocase. Nat. Rev. Mol. Cell Biol. 2004;5:519–530. doi: 10.1038/nrm1426. [DOI] [PubMed] [Google Scholar]
- Sesaki H., Southard S. M., Yaffe M. P., Jensen R. E. Mgm1p, a dynamin-related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol. Biol. Cell. 2003;14:2342–2356. doi: 10.1091/mbc.E02-12-0788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. In: Guthrie C., Fink G. R., editors. Methods in Enzymology. Guide to yeast genetics and molecular and cell biology. Vol. 350. London: Academic Press; 2002. pp. 3–41. [DOI] [PubMed] [Google Scholar]
- Tatsuta T., Augustin S., Nolden M., Friedrichs B., Langer T. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 2007;26:325–335. doi: 10.1038/sj.emboj.7601514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyasu K. T. A technique for ultracryotomy of cell suspensions and tissues. J. Cell Biol. 1973;57:551–565. doi: 10.1083/jcb.57.2.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel F., Bornhovd C., Neupert W., Reichert A. S. Dynamic subcompartmentalization of the mitochondrial inner membrane. J. Cell Biol. 2006;175:237–247. doi: 10.1083/jcb.200605138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann B., Neupert W. Mitochondria-targeted green fluorescent proteins: convenient tools for the study of organelle biogenesis in Saccharomyces cerevisiae. Yeast. 2000;16:1421–1427. doi: 10.1002/1097-0061(200011)16:15<1421::AID-YEA624>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Wurm C. A., Jakobs S. Differential protein distributions define two subcompartments of the mitochondrial inner membrane in yeast. FEBS Lett. 2006;580:5628–5634. doi: 10.1016/j.febslet.2006.09.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.