Abstract
A defining feature of eukaryotic polytopic protein biogenesis involves integration, folding, and packing of hydrophobic transmembrane (TM) segments into the apolar environment of the lipid bilayer. In the endoplasmic reticulum, this process is facilitated by the Sec61 translocon. Here, we use a photocross-linking approach to examine integration intermediates derived from the ATP-binding cassette transporter cystic fibrosis transmembrane conductance regulator (CFTR) and show that the timing of translocon-mediated integration can be regulated at specific stages of synthesis. During CFTR biogenesis, the eighth TM segment exits the ribosome and enters the translocon in proximity to Sec61α. This interaction is initially weak, and TM8 spontaneously dissociates from the translocon when the nascent chain is released from the ribosome. Polypeptide extension by only a few residues, however, results in stable TM8-Sec61α photocross-links that persist after peptidyl-tRNA bond cleavage. Retention of these untethered polypeptides within the translocon requires ribosome binding and is mediated by an acidic residue, Asp924, near the center of the putative TM8 helix. Remarkably, at this stage of synthesis, nascent chain release from the translocon is also strongly inhibited by ATP depletion. These findings contrast with passive partitioning models and indicate that Sec61α can retain TMs and actively inhibit membrane integration in a sequence-specific and ATP-dependent manner.
INTRODUCTION
Eukaryotic membrane protein biogenesis is facilitated in the endoplasmic reticulum (ER) membrane by a protein-conducting channel known as the Sec61 translocon (Johnson and van Waes, 1999; Higy et al., 2004; Rapoport et al., 2004; Pitonzo and Skach, 2006). Because the efficiency with which proteins fold is dependent upon their environment, the translocon performs a critical role by ensuring that lumenal, cytosolic, and transmembrane domains are delivered into their proper cellular compartment as they emerge from the ribosome exit tunnel (Pitonzo and Skach, 2006). This is accomplished through a series of coordinated interactions between the translocon and the translating ribosome that control both an axial translocation pathway into the ER lumen and a lateral integration pathway into the ER membrane (Crowley et al., 1994; Do et al., 1996; Liao et al., 1997; Meacock et al., 2002; McCormick et al., 2003).
An important and long-standing question in protein biogenesis involves the mechanism by which hydrophobic transmembrane (TM) segments are recognized and transferred from the aqueous environment required for protein synthesis into the apolar core of the lipid bilayer. This process involves a series of distinct steps that begin as the nascent TM helix forms within the ribosome exit tunnel and enters the translocon through Sec61α, a major component of the translocation channel (Do et al., 1996; Laird and High, 1997; Mothes et al., 1997; Meacock et al., 2002; Woolhead et al., 2004; Cannon et al., 2005; Sadlish et al., 2005; Saurí et al., 2005). In contrast to lumenal protein domains which pass through the translocon into the ER, most native TMs terminate translocation and transiently reside in proximity to Sec61α, thereby redirecting the elongating polypeptide beneath the base of the ribosome and into the cytosol (Thrift et al., 1991; Martoglio et al., 1995; Do et al., 1996; Meacock et al., 2002; McCormick et al., 2003; Sadlish et al., 2005; Buck et al., 2007). The precise nature of these initial TM-Sec61 interactions remains unknown, but photocross-linking and structural studies suggest that nascent TMs enter a fixed binding site in Sec61α, possibly adjacent to TM2b and TM7, that restricts random movement of the helix (Do et al., 1996; Plath et al., 1998, 2004; McCormick et al., 2003; Cannon et al., 2005; Sadlish et al., 2005). As TMs leave this binding site, they either transiently interact with other translocon components or are released directly into the lipid bilayer. In this study, we refer to final release of the TM into the bilayer as the committed step of membrane integration to distinguish it from earlier stages where the TM has terminated translocation but remains in close contact with translocon components.
For polytopic proteins that must achieve a compact fold within the bilayer, the timing of TM integration will profoundly impact the biophysical environment in which helical packing is initiated (Pitonzo and Skach, 2006). Factors that alter integration kinetics therefore have important implications for protein folding, particularly during formation of interactions between noncontiguous TMs. In this regard, photocross-linking studies of ribosome-bound integration intermediates have implicated two potential mechanisms of integration. Nascent chain cross-linking to lipids has suggested that some TMs integrate rapidly into the ER membrane after Sec61 insertion by passive thermodynamic partitioning through a lateral opening in the Sec61αβγ heterotrimer (Martoglio et al., 1995; Heinrich et al., 2000; van den Berg et al., 2004; Cannon et al., 2005). However, integration of many and perhaps most native TMs seems to occur via a stepwise progression through distinct proteinaceous environments that is temporally and mechanistically controlled at specific stages of synthesis. For example, TMs in native and engineered proteins can exhibit different phases of cross-linking to Sec61α, TRAM, and other translocon components as the nascent polypeptide elongates (Do et al., 1996; Liao et al., 1997; Meacock et al., 2002; McCormick et al., 2003; Sadlish et al., 2005; Saurí et al., 2005). Multiple TMs in polytopic proteins can also accumulate within or adjacent to the translocon before final integration into the lipid bilayer (Ota et al., 1998; Meacock et al., 2002; McCormick et al., 2003; Sadlish et al., 2005; Wilson et al., 2005; Kida et al., 2007). Both integration models, however, predict that when the nascent chain is released from the ribosome, either physiologically (at the end of translation) or prematurely (by puromycin), nascent TM-Sec61 contacts are generally lost (Mothes et al., 1994; Do et al., 1996; Sadlish et al., 2005; Saurí et al., 2005; Ismail et al., 2006). These and additional studies have led to the prevailing view that Sec61α recognizes structurally diverse TMs via relatively weak and nonspecific hydrophobic contacts (Hessa et al., 2005, 2007) and that it provides a passive conduit for membrane integration once the peptidyl-tRNA bond has been cleaved. Despite the important implications for membrane protein folding, few studies have investigated how structural features of nascent polypeptides might influence the timing and molecular events that govern TM integration.
To investigate whether the translocon might use an active rather than passive mechanism for integrating polytopic protein TMs, we used a photocross-linking approach to examine the cotranslational proximity of a native TM to Sec61α during synthesis of TM7 and TM8 from the cystic fibrosis transmembrane conductance regulator (CFTR). CFTR is an epithelial, chloride channel that exhibits the typical domain architecture of ATP-binding cassette transporters (Riordan et al., 1989). It contains two transmembrane domains (TMDs), two cytosolic nucleotide-binding domains (NBDs), and a unique regulatory domain (R) (Figure 1A). Inherited mutations in the CFTR gene cause cystic fibrosis, most commonly by disrupting a critical, but as yet uncharacterized, folding step in the ER (Cheng et al., 1990; Du et al., 2004; Kleizen et al., 2005). Although CFTR N- and C-terminal TMDs exhibit a similar six-spanning transmembrane topology, they acquire this topology via different translocation and integration events, in part because of polar residues located within their TM segments (Lu et al., 1998; Carveth et al., 2002). Full-length CFTR has also been reported to exhibit prolonged interactions with ER biosynthetic machinery before release into the lipid bilayer (Oberdorf et al., 2005). This has led to speculation that polar interactions required for proper helical packing and TMD assembly may contribute to inefficient and relatively slow folding kinetics (Therien et al., 2001; Wigley et al., 2002; Partridge et al., 2004; Oberdorf et al., 2005).
Figure 1.
Characterization of CFTR TM8 read-through polypeptides. (A) Diagram of putative structure and topology of CFTR. Closed circles show location of two N-linked glycosylation sites in ECL4. Arrows represent approximate location of CFTR truncation sites in ECL4 (aa 935, 957, 967, 977, and 987), in TM9 (aa 997 and 1007), and in TM10 (aa 1017 and 1027). Methionine residue 837 initiates translation of TM7-8 construct. (B) TM8 sequence showing location of UAG sites (912, 913, and 914). Putative boundaries of CFTR TM8 are shown. (C) In vitro translation of truncated mRNAs in the presence and absence of Lys-tRNACUA (even- and odd-numbered lanes, respectively). Polypeptides that terminated at the amber codon (nRT) were partially glycosylated at two N-linked consensus sites (downward arrows). Read-through (RT) of the UAG codon generated larger polypeptides (asterisks) that were also singly or doubly glycosylated (g and gg, respectively). (D) As in C, except that glycosylation was suppressed by addition of the tripeptide NYT during translation.
We now show that during cotranslational integration, the eighth TM segment of CFTR can be actively retained within the translocon adjacent to Sec61α even after cleavage of the peptidyl-tRNA bond. This interaction is dependent upon both the presence and orientation of an acidic residue, Asp924, within the putative TM8 hydrophobic core, and it is partially maintained by glutamate but not arginine substitution. Moreover, TM8 release from Sec61α after peptidyl-tRNA cleavage is ATP dependent and stimulated by maneuvers that remove ribosomes from the ER membrane. These findings are in striking contrast to passive integration models and indicate that sequence-specific and ATP-dependent interactions with Sec61α confer a previously unappreciated level of regulatory control to the timing of TM integration.
MATERIALS AND METHODS
Plasmid Construction
CFTR expression plasmids encoding wild-type and D924V mutant have been described previously (Carveth et al., 2002) and encode a methionine ATG translation start codon followed by CFTR residues Glu838 to Tryp1204. Truncated cDNA templates were generated by polymerase chain reaction (PCR) amplification using a sense oligonucleotide (TAGAGGATCTGGCTAGCGAT) that hybridizes to the vector SP64 plasmid (approximate base pairs 2726) and the following antisense oligonucleotides: CACACCTCTGAAGAATCCCAT(935), CACAACAGAATGTAACATTTTGTG(957), CACCGTGTTGAGGGTTGACAT(967), CACACTAGTTTTCAACGTGTTGAG(971), CACGAATCTATTAAGAATCCC(977), CACAAGGTCATCCAAAATTGC(987), CACCTGGATGAAGTCAAATATGG(997), AACTGCTATAGCTCCAATCACAAT(1007), AACAAAGATGTAGGGTTGTAAAACTGC(1017), and CACAAAAGCCACTATCACTGGCACTGT(1027). For all constructs, the last codon was converted to valine, which forms a stable peptidyl-tRNA bond and minimizes variation in spontaneous hydrolysis during the experiment. Mutants encoding D924E, D924R, and A923D/D924V as well as glycosylation mutants were generated by standard techniques using PCR overlap extension as described previously (Carveth et al., 2002) using the following sense (and corresponding antisense) oligonucleotides: GGGGCTAGCACTCATAGTAGAAATAACAG(N894A), CATTCTAGAGCGAACAGCTATGCAGTGATTAT(N900A), and GACAAAGGGGCTAGCACTCATTCTAGAGCGAAC(N894A/N900A). All PCR-amplified cDNA was verified by direct sequencing.
In Vitro Transcription, Translation, and Photocross-Linking
In vitro transcription and translation conditions were carried out as described previously (Oberdorf and Skach, 2002). Briefly, transcription was performed using SP6 RNA polymerase under standard conditions at 40°C for 1 h. mRNA was isolated by phenol/chloroform extraction and stored at −80°C. In vitro translations were carried out in the dark at 24°C for 30–45 min in rabbit reticulocyte lysate (RRL) supplemented with [35S]methionine (Foster et al., 2000) with the following modifications. Dithiothreitol was replaced with 2 mM reduced glutathione to reduce Nε-(5-azido-2-nitrobenzoyl (ANB) quenching. Before use, canine rough microsomes were treated with 25 mM EDTA, 250 mM sucrose, 50 mM triethanolamine, pH 7.5, and 1 mM dithiothreitol on ice for 15 min to remove endogenous ribosomes; isolated; and added at a final concentration OD280 = 2–4. To block N-linked glycosylation, the tripeptide NYT (200 μM) was added at the start of translation. Where indicated, nascent chains were released from ribosome by incubation in 1 mM puromycin for 10 min at 24°C unless otherwise stated. Synthetic aminoacyl-tRNAs, Lys-tRNACUA and εANB-lys-tRNACUA were synthesized as described previously (McCormick et al., 2003; Sadlish et al., 2005) and added to translation reactions at a final concentration of 1 μM. Translation products were irradiated on ice with collimated beam (300–350 nM) of UV light from an Oriel 500W mercury arc lamp. Membranes were collected by pelleting at 180,000g × 10 min through buffer containing 0.5 M sucrose, 20 mM HEPES, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, and 100 mM dithiothreitol. The membrane pellet containing targeted ribosome-nascent chain complexes was solubilized in 0.5% (wt/vol) SDS and 10 mM Tris-HCl, pH 8.0. Except where indicated, all samples were digested in 0.05 mg/ml RNaseA for 15 min at room temperature immediately before SDS-polyacrylamide gel electrophoresis (PAGE).
Ribosome Stripping EDTA/High Salt Treatment
After translation, EDTA (20 mM) or NaCl (0.3 or 1 M) was added to samples and incubated for 10 min at 24°C. Samples were then either photolyzed directly by UV or pelleted and resuspended in fresh translation mixture (minus radioactive methionine) in the presence or absence of 1 mM puromycin as indicated. Membranes were then collected by centrifugation, solubilized in 0.5%SDS, digested with 0.05 mg/ml RNaseA, and subjected to SDS-PAGE.
Carbonate Extraction
Translation products were diluted 100-fold in either sodium carbonate (0.1 M Na2CO3, pH 11.5) or Tris (0.25 M sucrose and 0.1 M Tris, pH 7.5). Samples were incubated on ice for 30 min and then centrifuged at 187,000 × g for 30 min. Pellets were solubilized in 1% SDS and 0.1 M Tris, pH 8.0, and equivalent amounts of supernatant or solubilized membranes were analyzed by SDS-PAGE. Bovine prolactin and a chimeric transmembrane protein containing the immunoglobulin G TM segment S.gG.ST.P (described in Skach and Lingappa, 1994) were used as a secretory and transmembrane control, respectively, and were mixed before extraction and SDS-PAGE.
Autoradiography, Quantitation, and Immunoprecipitation
Samples were analyzed by SDS-PAGE on 12–17% gels, destained, and treated with EN3HANCE (PerkinElmer Life and Analytical Sciences, Boston, MA) and subjected to autoradiography or phosphorimaging. Representative gels were scanned with an UMax III scanner and Adobe Photoshop (Adobe Systems, Mountain View, CA). Gels for quantitation were placed onto Kodak phosphor screens and analyzed with a Personal FX PhosphorImager and QuantityOne software (Bio-Rad, Hercules, CA). For immunoprecipitation, equivalent amounts of read-through translation products, determined by phosphorimaging as described previously (Sadlish et al., 2005) were denatured in 1% SDS and 100 mM Tris-Cl, pH 8.0, diluted in 1 ml of buffer B [1% (vol/vol), Triton X-100, 100 mM NaCl, 2 mM EDTA, and 100 mM Tris-Cl, pH 8.0), and rotated overnight with 5 μl of protein A Affigel (Bio-Rad) and 1 μl of peptide-specific rabbit antisera raised against Sec61α (CEIFVKEQSEVGSMGALLF), TRAM (CH3CON-CADSPRNRKEKSS) or TRAPα (CH3CON-CLPRKRAQKRSVGSDE). Beads were washed three times with 0.5 ml of ice-cold buffer B and twice with 0.5 ml of ice-cold 100 mM NaCl/100 mM Tris-Cl, pH 8.0. Samples were then analyzed by autoradiography as described above.
RESULTS
Experimental Strategy
To understand how the translocon facilitates integration of polytopic protein TMs, we used a photocross-linking approach to characterize cotranslational interactions between Sec61 and arrested integration intermediates derived from CFTR TMD2. CFTR biogenesis involves integration and folding of two six-spanning TM domains that are separated by 508 cytosolic residues (Figure 1A). During TMD2 topogenesis, TM7 and TM8 reinitiate and terminate translocation, respectively, thereby establishing topology of the fourth intra- and extracellular loops (ICL4 and ECL4, respectively) (Carveth et al., 2002). Both TMs cotranslationally achieve a membrane spanning topology. However, TM7 is required for proper orientation of TM8 due to an acidic residue, Asp924, near the center of the predicted TM8 hydrophobic core (Carveth et al., 2002).
Our strategy was to use a synthetic suppressor tRNA Nε-(5-azido-2-nitrobenzoyl)-lysine-tRNACUA (εANB-Lys-tRNACUA) to introduce a photoactive cross-linking probe at engineered amber (UAG) stop codons near the N terminus of TM8 (Figure 1, B and C). mRNAs were then truncated and translated in vitro in the presence of ER microsomes to recapitulate events involved in the integration process. When the ribosome encounters the UAG codon in this system, it either terminates translation and releases the nascent chain, or it incorporates the εANB-Lys probe and continues translating to the end of the mRNA. Because these mRNAs lack a terminal stop codon the nascent chain remains attached to the ribosome via the covalent peptidyl-tRNA bond, thus generating an arrested intermediate that mimics the conformational state of the nascent chain at the specific stage of synthesis determined by the truncation site. When activated by UV light, the ANB moiety forms a reactive nitrene that covalently cross-links adjacent alkyl and nucleophilic groups within reach of the 12-Å linker. By comparing photoadducts obtained for nascent chains that are systematically truncated at different lengths, it is therefore possible to reconstruct the molecular environment of the TM at sequential steps along the integration pathway. To simplify this analysis, CFTR N-terminal domains (TMD1, NBD1, and R) were removed, and Met837 was used as the translation initiation codon (Figure 1A). Importantly, the topogenic behaviors of TM7 and TM8 are similar when TMD2 is expressed alone or together with TMD1 (Carveth et al., 2002). Cotranslational interactions with the translocon should therefore reflect the initial biogenesis events experienced by the full-length polypeptide.
Incorporation of an εANB-Lys Probe into CFTR-TM8
We first characterized CFTR-derived polypeptides containing a UAG codon at residue 913 that were truncated between residues 935 and 987 (Figure 1C). In the absence of εANB-Lys-tRNACUA, translation terminates before TM8 and generates an ∼10-kDa polypeptide that undergoes partial glycosylation at two N-linked consensus sites, Asn894 and Asn900 (Figure 1C, odd-numbered lanes). ECL4 is therefore translocated into the ER lumen by TM7 when translocation is terminated at residue 913 (Figure 1C and Supplemental Figure 1). Glycosylation was verified by inhibition with a tripeptide NYT (Figure 1, compare C and D). Addition of εANB-Lys-tRNACUA to the translation reaction resulted in tRNA-dependent read-through of the UAG codon and a progressive increase in polypeptide size (Figure 1, C and D). These “read-through” polypeptides were also glycosylated but only for truncations beyond residue 967 (≥130 amino acid residues [aa]) when the ribosome-tethered ECL4 had extended sufficiently into the ER lumen to be modified by oligosaccharyltransferase (OST) (Figure 1C, lanes 6, 8, 10, and 12). Although glycosylation efficiency is <100% due to the relatively short length of the connecting loop, these data demonstrate that nascent ECL4 progressively passes through the translocon coincident with nascent chain elongation until residues 957–977 are synthesized.
TM8 Is Retained by Sec61α after Nascent Chain Release from the Ribosome
To determine when TM8 enters and leaves the translocon during TMD2 synthesis, the nascent chain was sequentially truncated between residues 935 and 997, and an εANB-Lys probe was incorporated at one of three adjacent residues (912, 913, and 914) near the N terminus of TM8 (Figures 1B and 2). At truncation 935, when the probe is 21–23 residues from the peptidyltransferase center, two UV-dependent photoadducts were observed (Figure 2) that migrated at ∼40 and 60 kDa and resembled those described previously for ribosomal proteins L4 and L17 (Liao et al., 1997; McCormick et al., 2003; Woolhead et al., 2004). Further extension of the nascent chain (truncation ≥957 aa) resulted in a single major photoadduct that shifted migration by ∼38–40 kDa. This photoadduct was observed at varying intensity for all three probe insertion sites, although the strongest photocross-linking was to residue 913 at truncations 971–987. For truncations beyond residue 997, photoadducts became difficult to distinguish from background when total translation products were evaluated but were still visible up to truncation 1027 (after synthesis of TM9) when glycosylation was inhibited (Supplemental Figure 2). Most subsequent experiments were therefore performed in the presence of NYT to improve photoadduct visualization. Immunoprecipitation of equal amounts of read-through translation product revealed that residue 913 cross-linked to Sec61α at all truncations between 957 and 1027 (Figure 3). Together, these data indicate that TM8 enters the translocon shortly after exiting the ribosome at a chain length between 935 and 957 aa and remains adjacent to translocon proteins for synthesis of at least 70 additional residues before emergence of TM9 from the ribosome.
Figure 2.
Photocross-linking to CFTR integration intermediates. CFTR mRNAs, codons 837 through the stated truncation site, containing UAG codons at the sites indicated were transcribed and translated in the presence of εANB-lys-tRNACUA as described in Materials and Methods. Puromycin (Puro) was added for 10 min where indicated, and samples were irradiated with UV light (UV) to activate the ANB moiety. Single and double asterisks indicate migration of nonglycosylated nonread-through and read-though polypeptides, respectively. Photoadducts are indicated by upward arrowheads. Diagrams show predicted location of CFTR nascent polypeptide and TM7-8 within the ribosome translocon complex. Location of εANB-Lys probe is shown by circle (aa 913). The number of residues separating the εANB-Lys probe site from the ribosome peptidyl-transferase center is indicated by d.
Figure 3.
Identification of TM8-Sec61α photoadducts. CFTR mRNAs either lacking a UAG codon (−UAG) (lanes 1–10) or with a UAG at codon 913 (lanes 11–20) were truncated at indicated residues and expressed in the presence of NYT peptide and ε-ANB-lys-tRNACUA. Equal amounts of read-through polypeptides, determined by phosphorimaging, were immunoprecipitated with Sec61α antisera and analyzed by SDS-PAGE as described in Materials and Methods. Bands migrating between 14 and 30 kDa represent nonspecific adsorption of nascent polypeptides to protein A beads.
A surprising result of the TM8 cross-linking profile was that the sensitivity of photoadducts to puromycin treatment changed markedly during synthesis of residues 957–977. On initial entry of TM8 into the translocon, the Sec61 photoadduct was largely abolished by puromycin (Figure 2, truncations 957–967). Thus, at this stage of synthesis, TM8 leaves the proximity of Sec61α upon cleavage of the peptidyl-tRNA bond. As the nascent chain was extended, however, TM8 entered a different environment in which photoadducts to residue 913 and to a lesser extent 912 persisted even after puromycin addition (Figure 2, truncations 971, 977, 987; and Supplemental Figure 2). This suggested either that puromycin was unable to release the nascent polypeptide from the ribosome or that TM8 remained bound to the translocon after peptidyl-tRNA bond cleavage.
Because TM retention by the translocon would have significant implications for membrane integration, we examined the nature of TM8 cross-links in greater detail. We first ruled out the trivial possibility that puromycin failed to cleave the peptidyl-tRNA bond by showing that addition of puromycin to the translation mixture efficiently removed the tRNA moiety from the nascent chain (Figure 4). Persistent TM8 photoadducts must therefore be due to retention of the nascent chain within the translocon independent of its attachment to the ribosome.
Figure 4.
TM8 remains proximal to Sec61α after peptidyl-tRNA cleavage. (A) CFTR residues 837–977 were expressed in the presence of NYT peptide. Downward arrow shows peptidyl tRNA band that persists after SDS-PAGE and is effectively removed by treatment with puromycin (Puro) for 10 min, RNase, or both. (B) Autoradiograph of immunoprecipitated translation products containing the ε-ANB-Lys moiety at residue 913. Photoadducts were recovered with Sec61α antisera both before and after puromycin treatment. No visible photoadducts were detected with antisera against TRAP α (lanes 5–8), TRAM (lanes 9–12), or nonimmune sera (NIS). Equal amounts of translation products used for each immunoprecipitation reaction.
To identify TM8 photoadducts after ribosome release, equal amounts of translation products (truncated at residue 977) were immunoprecipitated with antisera raised against three major translocon components—Sec61α, TRAM, or TRAPα—that exhibit similar electrophoretic mobility on SDS-PAGE (Görlich and Rapoport, 1993; Fons et al., 2003). As shown in Figure 4B, the photoadduct to residue 913 was recovered predominantly with Sec61α antisera both before and after puromycin treatment. Thus, TM8 remains proximal to Sec61α after peptidyl-tRNA bond cleavage. Faint photoadducts to TRAM were also observed upon longer exposure (D.P and W.S., unpublished observations). These findings represent a striking contrast with previous photocross-linking studies of aquaporin and other membrane proteins (Mothes et al., 1994; Do et al., 1996; McCormick et al., 2003; Sadlish et al., 2005; Daniel et al., 2008) and demonstrate that the nascent chain is not necessarily released spontaneously from Sec61α upon release from the ribosome.
TM8 Release from Sec61α Is Dependent upon ATP
Based on the above-mentioned findings, we next investigated how TM8 membrane integration might be regulated. As shown in Figure 5A, puromycin cleaved the peptidyl-tRNA bond within 30 s after addition to the translation reaction (Figure 5A, compare lanes 1 and 2). Yet, TM8 photoadducts showed relatively little change during the following 10 min (Figure 5A, lanes 7–11), and only a gradual decrease within 1 h (Figure 5C, lanes 7–10, and D). In the absence of puromycin, there was no detectable hydrolysis of the peptidyl-tRNA bond or loss of TM8 photocross-linking (Figure 5, B and C, lanes 3–6). Remarkably, when ATP was depleted from rabbit reticulocyte lysate (RRL) before photolysis, TM8 release from Sec61α was blocked as demonstrated by persistent photocross-linking (Figure 5C, lanes 11–14, and D). This was not due to loss of puromycin effectiveness, because the peptidyl-tRNA bond was efficiently cleaved even after prolonged apyrase treatment (Supplemental Figure 3). We also observed no ATP-dependent change in nascent chain stability during these experiments (Figure 5C, RT band), indicating that gradual loss of cross-linking observed in the presence of ATP was not caused by degradation of the nascent chain.
Figure 5.
TM8 release from Sec61α is ATP dependent. CFTR integration intermediates (residues 837–977) containing ε-ANB-Lys at residue 913 were translated in the presence of NYT for 30 min, incubated in puromycin for indicated time and analyzed by SDS-PAGE. (A) In the absence of puromycin (Puro) most of the read-through polypeptide remains bound to tRNA (downward arrow, lane 1). Peptidyl-tRNA release was essentially complete 30 s after addition of 1 mM puromycin (lanes 2–6), but peptidyl tRNA-cleaved chains continue to cross-link Sec61α upon UV exposure (upward arrowheads, lanes 7–11). (B) Translation was carried out as in A, but cycloheximide (20 μg/ml) was added at the end of translation. In the absence of puromycin, peptidyl-tRNA bond remains intact in RRL at 24°C for at least 2 h. (C) Translation products (generated as described in A) were incubated in puromycin for times indicated and then exposed to UV irradiation. TM8-Sec61α cross-linking was stable for 2 h in the absence of puromycin (lanes 3–6) but gradually decreased within 60 min after puromycin addition (lanes 7–10). ATP depletion (apyrase) stabilized TM8-Sec61α cross-linking (lanes 11–14). (D) Results from similar experiments shown in C were quantitated, and intensity of the photoadduct was plotted at each time point up to 60 min (n = 4 ± SEM). (E) Translation reactions were incubated as indicated before photocross-linking as in C (lanes 1–6). For lanes 7–10, microsomes were pelleted after translation, resuspended in a mock translation reaction containing RRL and UV irradiated after 10- or 30-min incubation with puromycin. (F) Photocross-linking was carried out as in C under conditions indicated (lanes 1–5). TM8 integration intermediates treated with puromycin were released from the translocon after 45 min (lanes 4–5). When these microsomes were isolated before UV and photocross-linking was subsequently performed in fresh RRL ± ATP, no TM8 photocross-links were observed (lanes 6 and 7). All translations were performed in the presence of the NYT tripeptide.
To determine whether ATP depletion resulted in irreversible retention of TM8 within the translocon, photocross-linking was either performed directly (Figure 5E, lanes 1–6), or reactions were treated with apyrase for 30 min and microsomes were isolated and resuspended in fresh RRL before photolysis (Figure 5E, lanes 7–10). ATP depletion yielded puromycin resistant TM8 photocross-links as expected (Figure 5E, compare lanes 5–6). When ATP-depleted microsomes were resuspended in the presence of fresh RRL (+ATP), TM8-Sec61 photocross-links were again present after 10 min but decreased within 30 min of puromycin treatment (Figure 5E, lanes 7–10). Using a similar strategy, TM8 was first released from Sec61α by puromycin treatment followed by incubation for 45 min in the presence of ATP (Figure 5F, lanes 1–5). Microsomes were then isolated, and incubated in RRL in the presence and absence of ATP (Figure 5F, lanes 6 and 7). Under both conditions, TM8-Sec61α photocross-links were not restored. Thus, effects of ATP depletion on TM8 retention are reversible, but once released from the translocon, TM8 does not reestablish interactions with Sec61α in this system. Together, these findings strongly argue that as TM8 exits the ribosome, it is actively retained within the translocon at a site closely adjacent to Sec61α and slowly released into the ER membrane in an ATP-dependent and regulated manner.
Peptidyl–tRNA-independent TM8–Sec61α Interaction Requires an Intact Ribosome Translocon Complex
Puromycin behaves as a peptidyl-tRNA analogue to effect transfer of the nascent polypeptide from the P-site to the A-site of the ribosome. Although the nascent chain is released from the peptidyltransferase center after attachment to puromycin, this does not necessarily remove ribosomes from the ER membrane (Neuhof et al., 1998; Potter and Nicchitta, 2002; Schaletzky and Rapoport, 2006). Because ribosome binding to the ER stabilizes Sec61α oligomerization and may also facilitate recruitment of other translocon components (Snapp et al., 2004), we examined whether the ribosome was needed for TM8 retention within the translocon. Consistent with this hypothesis, EDTA treatment abolished TM8 photocross-linking independently of puromycin effects (Figure 6A, lanes 3 and 8, respectively). Photocross-linking was also decreased by 1 M but not 300 mM NaCl (Figure 6A, lanes 5 and 10 and 4 and 9, respectively). Thus, the presence of an intact ribosome–translocon complex seems to be required for both tethered and peptidyl–tRNA-independent interactions of the nascent chain with Sec61α.
Figure 6.
TM8-Sec61α cross-linking requires an intact ribosome translocon complex. (A) Integration intermediates containing CFTR residues 837–977 were translated in the presence of ε-ANB-lys-tRNACUA, and reactions were treated with EDTA, NaCl and/or puromycin for 10 min as indicated before UV irradiation, SDS-PAGE, and autoradiography. Read-through bands containing ε-ANB-Lys (asterisk) and photoadducts (upward arrowheads) are indicated. Results show that both EDTA and 1 M NaCl, but not 300 mM NaCl, disrupted TM8 photocross-linking. (B) Samples were analyzed directly as described in A (lanes 1–6) or preincubated for 10 min with no addition (lanes 7–10), 20 mM EDTA (lanes 11–14), or 1.0 M NaCl (lanes 15–18). Membranes were then pelleted and resuspended in a mock translation reaction and subjected to UV irradiation in the presence and absence of puromycin. Ribosome removal with either treatment eliminated Sec61 cross-links that were not restored upon resuspension in normal translation conditions. (C) Translation reactions were treated with nothing (lanes 1–4) EDTA (lanes 5–8), or NaCl (lanes 9–12) as described in B and then incubated at pH 7.5 or 11.5 as indicated. Membranes were pelleted by centrifugation and equivalent amounts of supernatant (S) and pellet (P) were analyzed directly by SDS-PAGE. Lanes 13–16 show results for a control secretory (Sec) protein (bovine prolactin) and transmembrane protein (TM) (S.gG.ST.P; described in Skach and Lingappa, 1994) treated under identical conditions. Asterisk shows peptidyl-tRNA bond.
To ensure that TM8 photocross-linking was not affected by differences in experimental conditions (e.g., high ionic strength), microsomes were treated with EDTA or high salt, pelleted, and resuspended under the same conditions as translation. Results show that treatment with either EDTA or NaCl resulted in a loss of Sec61 photoadducts (Figure 6B, lanes 1–6) that were not restored when microsomes were isolated and resuspended in fresh RRL (Figure 6B, lanes 7–18). Thus, once the nascent chain is released from the translocon by ribosome removal, it is not recruited back to Sec61α. This would be expected if nascent chains were integrated into the lipid bilayer upon ribosome removal. Consistent with this, nascent chains treated with either EDTA or NaCl were also resistant to alkaline extraction in 0.1 M sodium carbonate (Figure 6C, lanes 5–12). Unfortunately, we were unable to demonstrate specific integration of TM8 because TM7 is required for nascent chain targeting, and integration of either TM would potentially render the nascent polypeptide resistant to extraction. Indeed, nascent chains were also carbonate resistant even in the absence of EDTA and NaCl treatment (Figure 6C, lanes 1–4).
TM8-Sec61 Photocross-Linking Is Independent of N-linked Glycosylation
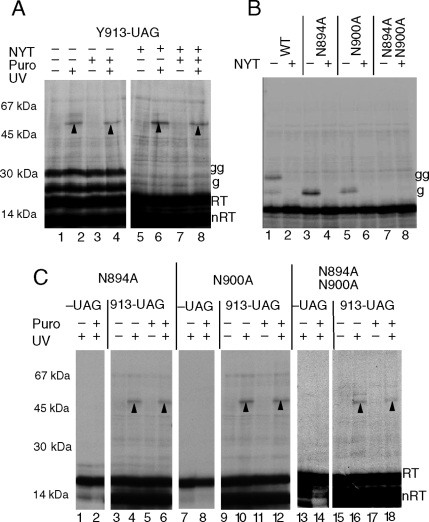
As nascent glycoproteins are translocated into the ER lumen, they are recognized by OST via a tripeptide consensus sequence NXS/T. This recognition event is mediated at least in part by the STT3 subunit of OST, and the interaction is strengthened for mutant consensus sites (QXS/T) that are unable to accept the core oligosaccharide moiety (Nilsson et al., 2003). We therefore tested whether OST interaction with the poorly used CFTR consensus sites at Asn 894 and Asn900 might contribute to persistent TM8 association with Sec61α. Photocross-linking in the presence and absence of an acceptor peptide (NYT) demonstrated that blocking glycosylation had no effect on TM8 cross-linking (Figure 7A). Similarly, removal of one or both N-linked consensus sites (Asn to Ala substitution) eliminated nascent chain glycosylation but did not change the cross-linking pattern (Figure 7, B and C). The two N-linked glycosylation sites in ECL4 therefore do not contribute to peptidyl–tRNA-independent TM8–Sec61α interactions.
Figure 7.
N-linked glycosylation sites do not alter TM8-Sec61α photocross-linking. (A) Translation of CFTR residues 837–977 in the absence (lanes 1–4) and presence (lanes 5–8) of the glycosylation inhibitor, NYT revealed no difference in TM8 photoadducts (upward arrowheads). (B) Replacement of Asn consensus sites with Ala eliminates glycosylation at residue 894 (lane 3), residue 900 (lane 5), or both (lane 7). Migration of singly and doubly glycosylated polypeptides are indicated as g and gg, respectively. (C) Photocross-linking to constructs lacking N-linked glycosylation sites at residues 894 and 900 revealed no detectable effect on TM8 photoadducts (upward arrowheads) in the presence or absence of puromycin. Control constructs (−UAG) lack the ε-ANB-Lys probe. Translations in C were performed with acceptor peptide.
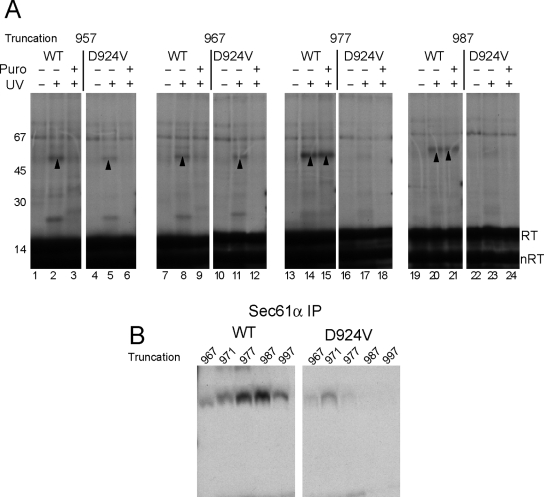
Aspartate 924 Is Responsible for TM8 Retention in the Translocon
An interesting feature of TM8 is the presence of an aspartate, Asp924, residue located near the center of its predicted hydrophobic core. Because aspartate residues stabilize helix–helix interactions in apolar environments (Zhou et al., 2000; Adamian, 2001; Hermansson and von Heijne, 2003; Buck et al., 2007), we tested whether Asp924 might contribute to TM8 retention within the translocon. In both wild-type (WT) and D924V mutants, TM8 photocross-links were observed during initial stages of translocon insertion at truncations 957 and 967 (Figure 8). However, photoadducts to D924V decreased abruptly at truncation sites beyond residue 967 (Figure 8B), and no peptidyl–tRNA-independent photocross-links to residue 913 were observed (Figure 8A). Thus, the D924V mutant leaves the proximity of Sec61 at an earlier stage of synthesis than its wild-type counterpart and fails to exhibit peptidyl–tRNA-independent TM8-Sec61α interactions. When Asp924 was converted to glutamate, D924E, nascent chains truncated at residue 977 also continued to cross-link Sec61α after puromycin treatment, albeit to a lesser extent than wild type (Figure 9, A and B). This was not simply due to the presence of a charged residue, because arginine substitution (D924R) reduced photocross-linking in tethered nascent chains and also eliminated Sec61α photocross-linking after peptidyl-tRNA cleavage (Figure 9B).
Figure 8.
Asp924 causes persistent TM8 photocross-linking to Sec61α after puromycin release. (A) Photocross-linking to integration intermediates (residues 837–977) containing WT and mutant (D924V) TM8 revealed similar UV-dependent photoadducts (upward arrowheads) for truncations 957 and 967 that disappeared after puromycin treatment. Bands migrating at 67 kDa and higher were present in the absence of UV and represent nonspecific background. At truncations 977 and 987, photoadducts to WT TM8 increased in intensity and were peptidyl-tRNA independent (lanes 14 and 15 and 20 and 21), whereas photoadducts were absent for the D924V mutant (lanes 17, 18, 23, and 24). (B) Immunoprecipitation of photoadducts with Sec 61α antisera confirmed that the D924V mutation decreases Sec61α cross-linking at the same stage of synthesis when photocross-linking to WT TM8 becomes independent of the peptidyl-tRNA bond.
Figure 9.
Sequence-specific control of TM8-Sec61α photocross-linking. (A) Photocross-linking to CFTR integration intermediates (residues 837–977) containing the ε-ANB-Lys probe at residue 913 in which Asp924 was mutated to glutamine (E), arginine (R), or valine (V). Samples were treated with puromycin and UV irradiation as indicated. (B) Immunoprecipitation of WT and mutant TM8 photoadducts with Sec61α antiserum revealed that both photocross-linking efficiency and persistence of cross-linking after puromycin treatment were highly favored by acidic residues, with aspartate greater than glutamate. Equivalent amounts of read-through translation products were used for immunoprecipitation. (C) Integration intermediates containing the ε-ANB-Lys probe at residues 912, 913, or 914 were truncated at residue 977, photolyzed, and immunoprecipitated with Sec61α antisera. A923D, D924V mutations decreased Sec61α photocross-linking (to residue 913) after puromycin treatment (compare lane 4 with lane 10) but significantly increased the peptidyl-tRNA–independent photocross-linking to residue 912 (lanes 2 and 8). Thus, TM8 release from Sec61α is highly sensitive to the presence as well as orientation of acidic residues. Immunoprecipitations used equivalent amounts of read-through products.
The relative intensity of photoadducts to residues 912, 913, and 914 (Figures 2 and 9C) are consistent with the notion that as TM8 inserts into Sec61α, it enters a binding site that restricts random motion of the helix. If this were the case, and if Asp924 interacted specifically with an adjacent polar residue, then such an interaction should be dependent not only on the presence but also the relative orientation of the acidic side chain. We therefore moved the aspartate group 1 residue toward the N terminus to position 923 (A923D and D924V), which would be located at approximately the same distance from the peptidyl-transferase center but would face a different side of the TM8 helix. Before puromycin treatment, this change did not affect photocross-linking to residues 912 and 914 but modestly decreased photoadducts to residue 913 (Figure 9C). However, repositioning the polar group eliminated puromycin resistant cross-links to residue 913 and markedly stabilized photocross-links to residue 912 after cleavage of the peptidyl-tRNA bond (Figure 9C compare lanes 2 and 4 with lanes 8 and 10). Because the ANB probes are located 9–11 residues from the Asp residue, the orientation of the entire TM segment with respect to Sec61α therefore seems to be influenced by the specific location of the aspartate side chain. Together, these data suggest that TM8 can make sequence-dependent contacts while it resides within the translocon through a specific interaction with Asp924.
DISCUSSION
In this study, we used site-specific photocross-linking to identify several novel features that control lateral transfer of a native polytopic protein TM from the Sec61 translocon into the hydrophobic environment of the ER membrane. First, we found that nascent polypeptides can form stable interactions within the translocon that inhibit TM release from Sec61α even after cleavage of the peptidyl-tRNA bond. Second, these interactions are dependent on nascent chain structure and involve not only the presence but also the precise orientation of an acidic side chain group in the hydrophobic TM segment. Third, TM retention by Sec61 requires an intact ribosome translocon complex. And fourth, the kinetics of TM release from Sec61α can be regulated in an ATP-dependent manner. These findings are in stark contrast to passive partitioning models of integration and indicate that the ribosome and translocon cooperatively play an active role in the recognition and integration of TMs into the ER membrane.
Photocross-linking to truncated CFTR integration intermediates revealed that during TMD2 synthesis, TM8 contacts Sec61α as it exits the ribosome, and remains in proximity as the nascent chain is extended for at least 70 additional residues. The asymmetry of photocross-links to adjacent sites on different faces of the putative helix (residues 912, 913, and 914) further indicates that TM8 rapidly enters a translocon-binding site that restricts free rotational movement (McCormick et al., 2003; Plath et al., 2004; Sadlish et al., 2005). At relatively short nascent chain lengths (e.g., truncation 957), Sec61α photocross-linking abruptly decreased when the polypeptide was released from the ribosome, indicating that the peptidyl-tRNA bond is initially responsible for retaining the nascent chain within the Sec61-binding site complex. Unexpectedly, synthesis of only a few additional residues markedly changed the character of TM8 photoadducts, whereby TM8 continued to form asymmetric Sec61α cross-links even after the nascent chain was released from the ribosome. TM8 interactions with the translocon therefore change in a fundamental way during a brief period of synthesis. The predominant photocross-links to Sec61α versus TRAM or TRAP, lack of involvement of N-linked glycosylation, and the role of Asp924 strongly suggest that Sec61α can recognize and stably interact with TMs in a sequence specific manner before membrane integration.
One of the most unexpected findings of this study was that TM8 release from Sec61α was blocked by ATP depletion. Integration of hydrophobic TMs into the lipid bilayer is generally considered to be a thermodynamically favorable process, and this is also predicted for the putative TM8 sequence shown in Figure 2 (transfer free energy from water to octanol, ΔG = −6.48 kcal/mol, Wimley et al., 1996). Thus, under certain conditions, the translocon seems to actively inhibit integration of ribosome-released nascent chains. Because no core translocon components are known to bind or hydrolyze ATP, this finding suggests a novel means by which the final stage of integration might be regulated. One possibility is that as residues 968–971 are synthesized, TM8 enters a binding site within Sec61α with a lower free energy than it would have in the lipid bilayer and remains in this site until favorable interactions with Asp924 are disrupted. Alternatively, TM8 could be physically shielded from the bulk lipid of the ER membrane by a conformational rearrangement of the translocon components. Regardless of the mechanisms, our results extend previous findings and demonstrate that integration of certain TMs can be regulated in an energy dependent manner by protein–protein interactions within the translocon (Do et al., 1996; Liao et al., 1997). Of course, TMs with different energetic requirements might also integrate via other mechanisms as reported previously (Martoglio et al., 1995; Heinrich et al., 2000). The ER chaperone BiP uses ATP to open and close the lumenal face of the translocon pore (Haigh and Johnson, 2002; Alder et al., 2005). BiP is not required for TM8 retention, however, because photocross-linking was unaffected by depletion of ER lumenal contents (Supplemental Figure 4). Integration might also be controlled by the phosphorylation status of translocon components such as TRAPα, Sec61β, or TRAM (Prehn et al., 1990; Gruss et al., 1999). Although further studies are needed to define the precise role of ATP, our results indicate that the timing and mechanism of polytopic protein TM integration is much more complex than appreciated previously.
Although this study is focused primarily on defining events of CFTR biogenesis, we realize that the substrates examined here contain only a small portion of CFTR and are therefore unlikely to reach a native folded state. Persistent TM8 cross-linking to Sec61α may therefore reflect nascent chains that are destined for retrotranslocation and/or degradation (Nakatsukasa and Brodsky, 2008). Interestingly, persistent TM8-Sec61 photocross-linking does not seem to involve substrate recruitment back to the translocon, because once nascent chains are released (e.g., by EDTA or high salt), they do not efficiently reassociate with Sec61. This raises the possibility that the translocon could provide a potential means to distinguish proteins that are not yet able to achieve a stable conformation in the ER membrane. If so, then the final stage of integration could provide a regulatory mechanism to abort nascent chain release into the bilayer in a manner analogous to preemptive quality control of translocation that was recently been described for secretory proteins (Kang et al., 2006). We should note, however, that no retrotranslocation or degradation of CFTR fragments was observed under the photocross-linking conditions used in this study. Moreover, TM8 retention was dependent on ribosome binding, which would not be expected for degradation substrates. The current data are therefore most consistent with a role of the translocon in facilitating TMD folding, and additional work is needed to determine how early events of membrane integration, and degradation might be linked.
Our results also demonstrate an important mechanistic distinction between membrane integration and translocation termination. During CFTR biogenesis, TM8 functions as a stop transfer sequence to terminate translocation of the fourth extracellular loop (ECL4) and prevent the fourth intracellular loop from entering the ER lumen (Carveth et al., 2002; Sadlish and Skach, 2004). Because glycosylation efficiency of ECL4 increases only until truncation 971, translocation of the nascent chain is likely terminated shortly after TM8 engages the Sec61α-binding site. However, TM8 continues to cross-link Sec61α long after translocation has ceased. Moreover, translocation termination is accomplished by both WT TM8 and the D924V mutant (Carveth et al., 2002), but integration is delayed only when an acidic residue is present. Thus, although recent studies have defined the biological code for translation termination (Hessa et al., 2005; Hessa et al., 2007), our findings indicate that different structural properties of the nascent chain, and likely different interactions with translocon components, govern the final stage of integration.
How then does Asp924 prevent TM8 integration? Aspartate residues exhibit the strongest propensity to form hydrogen bonds (e.g., with Asp, Glu, Asn, and Gln) and stabilize helix–helix interactions in detergent micelles and lipid bilayers (Choma et al., 2000; Zhou et al., 2000; Adamian 2001; Gratkowski et al., 2001; Walters and DeGrado, 2006). If TM8 resided between TMs 2 and 7 of Sec61α as has been suggested by the SecY crystal structure (van den Berg et al., 2004; Tian and Andricioaei, 2006), then Asp924 could potentially form polar contacts with Gln92, Gln294, Asn300, or Gln306. However, it is also possible that Asp924 might interact with other regions of Sec61α or even other components in the fully assembled translocon (Beckmann et al., 1997; Hegde and Lingappa, 1997; Menetret et al., 2005; Kida et al., 2007; Skach, 2007). Aspartate residues have also been implicated in intramolecular interactions between nascent TMs before membrane integration (Meindl-Beinker et al., 2006) and in the folding of polytopic protein domains in the bilayer (Adamian, 2001; Buck et al., 2007). It is interesting in this respect that CFTR-TM7 also contains an acidic residue (Glu873) and that CFTR-TM7 is required for TM8 stop transfer activity (Carveth et al., 2002). In addition, removal of TM7 and insertion of TM8 into a chimeric protein where it fails to terminate translocation (Carveth et al., 2002), eliminated tRNA-independent photocross-linking (Pitonzo and Skach, unpublished observations). Thus, the mechanism of TM8 integration may reflect a competition between intermolecular polar interactions with Sec61α and intramolecular interactions that are established during early stages of helical packing.
The timing of TM integration likely has important physiological implications for polytopic protein folding. In mature CFTR, Asp924 functionally interacts with Arg347 in TM6 of the first CFTR transmembrane domain (Cotten and Welsh, 1999). This raises the question as to where noncontiguous yet interacting polar TM residues might reside during the synthesis of large intervening cytosolic domains. We recently showed that full-length CFTR expressed in Xenopus oocytes exhibits prolonged association with ER biosynthetic machinery after translation has been completed and that CFTR release into the bilayer is also ATP dependent (Oberdorf et al., 2005). The delayed integration behavior demonstrated here for isolated CFTR TMs therefore parallels the behavior of native CFTR maturation in cells and supports the prediction that formation of native polar interactions may contribute to the slow release of CFTR into the membrane (Oberdorf et al., 2005). Consistent with this notion, recent studies of the Aquaporin 1 water channel have demonstrated that hydrogen bond formation between polar residues in TM2 and TM5 play a critical role in early biogenesis (Buck et al., 2007). When this bond is disrupted (in a TM2 mutant), an unpaired aspartate residue in TM5 prevents monomer folding and results in retention of immature protein in a large protein complex or aggregate (Buck et al., 2007). An intriguing possibility arising from these observations is that the translocon may facilitate polytopic protein biogenesis by transiently stabilizing noncontiguous polar residues and thereby maintain a folding-competent state that allows efficient formation of TM–TM interactions in the native structure. If this were the case, then the Sec61 translocon may play a critical role in enabling TMs to acquire polar residues that would otherwise constrain folding efficiency. Although further studies are clearly needed to unravel the details of this hypothesis, it is likely that a deeper understanding into the mechanics of translocon function will shed new insight into these poorly understood aspects of membrane protein folding.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Kent Matlack for generously providing TRAM and TRAPα antisera and to K. Rusterholtz for technical assistance. This work was supported by National Institutes of Health grants DK-51818 and GM-53457 (to W.R.S.) and GM-26494 (to A.E.J.), American Cystic Fibrosis Foundation (to W.R.S.), American Heart Association Grant-in-Aid 0755832Z (to W.R.S.), the Robert A. Welch Foundation Chair grant BE-0017 (to A.E.J.), and the Manpei Suzuki Diabetes Foundation (to Y.M.).
Abbreviations used:
- aa
amino acid residue
- ANB
Nε-(5-azido-2-nitrobenzoyl)
- RRL
rabbit reticulocyte lysate
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-09-0902) on November 19, 2008.
REFERENCES
- Adamian L.L.J. Helix-helix packing and interfacial pairwise interactions of residues in membrane proteins. J. Mol. Biol. 2001;311:891–907. doi: 10.1006/jmbi.2001.4908. [DOI] [PubMed] [Google Scholar]
- Alder N., Shen Y., Brodsky J., Hendershot L., Johnson A. The molecular mechanisms underlying BiP-mediated gating of the Sec61 translocon of the endoplasmic reticulum. J. Cell Biol. 2005;168:389–399. doi: 10.1083/jcb.200409174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R., Bubeck D., Grassuicci R., Panczek P., Verschoor A., Blobel G., Frank J. Alignment of conduits for the nascent polypeptide chain in the ribosome-Sec61 complex. Science. 1997;278:2123–2126. doi: 10.1126/science.278.5346.2123. [DOI] [PubMed] [Google Scholar]
- Buck T., Wagner J., Grund S., Skach W. A novel tripartite structural motif involved in aquaporin topogenesis, monomer folding and tetramerization. Nat. Struct. Mol. Biol. 2007;14:762–769. doi: 10.1038/nsmb1275. [DOI] [PubMed] [Google Scholar]
- Cannon K. S., Or E., Clemons W., Shibata Y., Rapoport T. Disulfide bridge formation between SecY and a translocating polypeptide localizes the translocation pore to the center of SecY. J. Cell Biol. 2005;169:219–225. doi: 10.1083/jcb.200412019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carveth K., Buck T., Anthony V., Skach W. Cooperativity and flexibility of cytsic fibrosis transmembrane conductance regulator transmembrane segments participate in membrane localization of a charged residue. J. Biol. Chem. 2002;277:39507–39514. doi: 10.1074/jbc.M205759200. [DOI] [PubMed] [Google Scholar]
- Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- Choma C., Gratowski H., Lear J., DeGrado W. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Mol. Biol. 2000;7:161–166. doi: 10.1038/72440. [DOI] [PubMed] [Google Scholar]
- Cotten J., Welsh M. Cystic fibrosis-associated mutations at arginine 347 alter the pore architecture of CFTR: evidence for disruption of a salt bridge. J. Biol. Chem. 1999;274:5429–5435. doi: 10.1074/jbc.274.9.5429. [DOI] [PubMed] [Google Scholar]
- Crowley K., Liao S., Worrell V., Reinhart G., Johnson A. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Daniel C. J., Conti B., Johnson A. E., Skach W. R. Control of translocation through the Sec61 translocon by nascent polypeptide structure within the ribosome. J. Biol. Chem. 2008;283:20864–20873. doi: 10.1074/jbc.M803517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H., Falcone D., Lin J., Andrews D., Johnson A. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Du K., Sharma M., Lukacs G. The ΔF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat. Struct. Mol. Biol. 2004;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- Fons R., Bogert B., Hegde R. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster W., Helm A., Turnbull I., Gulati H., Yang B., Verkman A., Skach W. Identification of sequence determinants that direct different intracellular folding pathways for AQP1 and AQP4. J. Biol. Chem. 2000;275:34157–34165. doi: 10.1074/jbc.M000165200. [DOI] [PubMed] [Google Scholar]
- Görlich D., Rapoport T. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Gratkowski H., Lear J., DeGrado W. Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. USA. 2001;98:880–885. doi: 10.1073/pnas.98.3.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss O., Feik P., Frank R., Dobberstein B. Phosphorylation of components of the ER translocation site. Eur. J. Biochem. 1999;260:785–793. doi: 10.1046/j.1432-1327.1999.00215.x. [DOI] [PubMed] [Google Scholar]
- Haigh N., Johnson A. A new role for BiP: closing the aqueous translocon pore during protein integration into the ER membrane. J. Cell Biol. 2002;156:261–270. doi: 10.1083/jcb.200110074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde R., Lingappa V. Membrane protein biogenesis: regulated complexity at the endoplasmic reticulum. Cell. 1997;91:575–582. doi: 10.1016/s0092-8674(00)80445-6. [DOI] [PubMed] [Google Scholar]
- Heinrich U., Mothes W., Brunner J., Rapoport T. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hermansson M., von Heijne G. Inter-helical hydrogen bond formation during membrane protein integration into the ER membrane. J. Mol. Biol. 2003;334:803–809. doi: 10.1016/j.jmb.2003.10.019. [DOI] [PubMed] [Google Scholar]
- Hessa T., Bihlmaler K., Lundin C., Boekel J., Andersson H., Nilsson I., White S., von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Higy M., Junne T., Spiess M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry. 2004;43:12716–12722. doi: 10.1021/bi048368m. [DOI] [PubMed] [Google Scholar]
- Ismail N., Crawshaw S., High S. Active and passive displacement of transmembrane domains during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 2006;119:2826–2836. doi: 10.1242/jcs.03018. [DOI] [PubMed] [Google Scholar]
- Johnson A., van Waes M. The translocon: a dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999;15:799–842. doi: 10.1146/annurev.cellbio.15.1.799. [DOI] [PubMed] [Google Scholar]
- Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Sakaguchi M. Two translocating hydrophilic segments of a nascent chain span the ER membrane during multispanning protein topogenesis. J. Cell Biol. 2007;179:1441–1452. doi: 10.1083/jcb.200707050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleizen B., Vlijmen T., de Jonge H., Braakman I. Folding of CFTR is predominantly cotranslational. Mol. Cell. 2005;20:277–287. doi: 10.1016/j.molcel.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Laird V., High S. Discrete cross-linking products identified during membrane protein biosynthesis. J. Biol. Chem. 1997;272:1983–1989. doi: 10.1074/jbc.272.3.1983. [DOI] [PubMed] [Google Scholar]
- Liao S., Lin J., Do H., Johnson A. Both lumenal and cytosolic gating of the aqueous translocon pore are regulated from inside the ribosome during membrane protein integration. Cell. 1997;90:31–42. doi: 10.1016/s0092-8674(00)80311-6. [DOI] [PubMed] [Google Scholar]
- Lu Y., Xiong X., Helm A., Kimani K., Bragin A., Skach W. Co- and Posttranslational mechanisms direct CFTR N-terminus transmembrane assembly. J. Biol. Chem. 1998;273:568–576. doi: 10.1074/jbc.273.1.568. [DOI] [PubMed] [Google Scholar]
- Martoglio B., Hofmann M., Brunner J., Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:207–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- McCormick P., Miao Y., Shao Y., Lin J., Johnson A. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- Meacock S., Lecomte F., Crawshaw S., High S. Different transmembrane domains associate with distinct endoplasmic reticulum components during membrane integration of a polytopic protein. Mol. Biol. Cell. 2002;13:4114–4129. doi: 10.1091/mbc.E02-04-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl-Beinker N., Lundin C., Nilsson I., White S., von Heijne G. Asn- and Asp-mediated interactions between transmembrane helices during translocon-mediated membrane protein assembly. EMBO Rep. 2006;7:1111–1116. doi: 10.1038/sj.embor.7400818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menetret J.-F., Hegde R., Heinrich S., Chandramouli P., Ludtke S., Rapoport T., Akey C. Architecture of the ribosome-channel complex derived from native membranes. J. Mol. Biol. 2005;348:445–457. doi: 10.1016/j.jmb.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Mothes W., Heinrich S., Graf R., Nilsson I., von Heijne G., Brunner J., Rapoport T. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell. 1997;89:523–533. doi: 10.1016/s0092-8674(00)80234-2. [DOI] [PubMed] [Google Scholar]
- Mothes W., Prehn S., Rapoport T. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsukasa K., Brodsky J. L. The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic. 2008;9:861–870. doi: 10.1111/j.1600-0854.2008.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhof A., Rolls M., Jungnickel B., Kalies K., Rapoport T. Binding of signal recognition particle gives ribosome/nascent chain complexes a competitive advantage in endoplasmic reticulum membrane interaction. Mol. Biol. Cell. 1998;9:103–115. doi: 10.1091/mbc.9.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I., Kelleher D., Miao Y., Shao T., Kreibich G., Gilmore R., von Heijne G., Johnson A. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J. Cell Biol. 2003;161:715–725. doi: 10.1083/jcb.200301043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdorf J., Pitonzo D., Skach W. An energy-dependent maturation step is required for release of the cystic fibrosis transmembrane conductance regulator from early endoplasmic reticulum biosynthetic machinery. J. Biol. Chem. 2005;280:38193–38202. doi: 10.1074/jbc.M504200200. [DOI] [PubMed] [Google Scholar]
- Oberdorf J., Skach W. In vitro reconstitution of CFTR biogenesis and degradation. Totowa, NJ: Humana Press, Inc; 2002. [DOI] [PubMed] [Google Scholar]
- Ota K., Sakaguchi M., von Heijne G., Hamasaki N., Mihara K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol. Cell. 1998;2:495–503. doi: 10.1016/s1097-2765(00)80149-5. [DOI] [PubMed] [Google Scholar]
- Partridge A. W., Therien A. G., Deber C. M. Missense mutations in transmembrane domains of proteins: phenotypic propensity of polar residues for human disease. Proteins. 2004;54:648–656. doi: 10.1002/prot.10611. [DOI] [PubMed] [Google Scholar]
- Pitonzo D., Skach W. Molecular mechanisms of aquaporin biogenesis by the endoplasmic reticulum Sec61 translocon. Bioch. Biophys. Acta. 2006;1758:976–988. doi: 10.1016/j.bbamem.2006.04.021. [DOI] [PubMed] [Google Scholar]
- Plath K., Mothes W., Wilkinson B., Stirling C., Rapoport T. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell. 1998;94:795–807. doi: 10.1016/s0092-8674(00)81738-9. [DOI] [PubMed] [Google Scholar]
- Plath K., Wilkinson B., Stirling C., Rapoport T. Interactions between Sec complex and prepro-alpha-factor during posttranslational protein transport into the endoplasmic reticulum. Mol. Biol. Cell. 2004;15:1–10. doi: 10.1091/mbc.E03-06-0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potter M., Nicchitta C. Endoplasmic reticulum-bound ribosomes reside in stable association with the translocon following termination of protein synthesis. J. Biol. Chem. 2002;277:23314–23320. doi: 10.1074/jbc.M202559200. [DOI] [PubMed] [Google Scholar]
- Prehn S., Herz J., Hartmann E., Kurzchalia T. V., Frank R., Roemisch K., Dobberstein B., Rapoport T. A. Structure and biosynthesis of the signal-sequence receptor. Eur. J. Biochem. 1990;188:439–445. doi: 10.1111/j.1432-1033.1990.tb15421.x. [DOI] [PubMed] [Google Scholar]
- Rapoport T., Goder V., Heinrich S., Matlack K. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 2004;14:568–575. doi: 10.1016/j.tcb.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Riordan J. R., Rommens J. M., Kerem B.-S., Alon N., Rozmahel R., Grzelczak Z., Zielenski J., Lok S., Collins F. S., Tsui L.-C. Identification of the cystic fibrosis gene: cloning and characterization of complementary DNA. Science. 1989;245:1066–1072. doi: 10.1126/science.2475911. [DOI] [PubMed] [Google Scholar]
- Sadlish H., Pitonzo D., Johnson A. E., Skach W. R. Sequential triage of transmembrane segments by Sec61α during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005;12:870–878. doi: 10.1038/nsmb994. [DOI] [PubMed] [Google Scholar]
- Sadlish H., Skach W. Biogenesis of CFTR and other polytopic membrane proteins; new roles for the ribosome-translocon complex. J. Membr. Biol. 2004;202:115–126. doi: 10.1007/s00232-004-0715-6. [DOI] [PubMed] [Google Scholar]
- Saurí A., Saksena S., Salgado J., Johnson A., Mingarro I. Double-spanning plant viral movement protein integration into the endoplasmic reticulum membrane is signal recognition particle-dependent, translocon-mediated, and concerted. J. Biol. Chem. 2005;280:25907–25912. doi: 10.1074/jbc.M412476200. [DOI] [PubMed] [Google Scholar]
- Schaletzky J., Rapoport T. Ribosome binding to and dissociation from translocation sites of the endoplasmic reticulum membrane. Mol. Biol. Cell. 2006;9:3860–3869. doi: 10.1091/mbc.E06-05-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W. The expanding role of the ER translocon in membrane protein folding. J. Cell Biol. 2007;179:1333–1335. doi: 10.1083/jcb.200711107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W., Lingappa V. Transmembrane orientation and topogenesis of the 3rd and 4th membrane-spanning regions of human P-glycoprotein (MDR1) Cancer Res. 1994;54:3202–3209. [PubMed] [Google Scholar]
- Snapp E., Reinhart G., Bogert B., Lippincott-Schwartz J., Hegde R. The organization of engaged and quiescent translocons in the endoplasmic reticulum of mammalian cells. J. Cell Biol. 2004;164:997–1007. doi: 10.1083/jcb.200312079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therien A., Grant F., Deber C. Interhelical hydrogen bonds in the CFTR membrane domain. Nat. Struct. Biol. 2001;8:597–601. doi: 10.1038/89631. [DOI] [PubMed] [Google Scholar]
- Thrift R. N., Andrews D. W., Walter P., Johnson A. E. A nascent membrane protein is located adjacent to ER membrane proteins throughout its integration and translocation. J. Cell Biol. 1991;112:809–821. doi: 10.1083/jcb.112.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P., Andricioaei I. Size, motion, and function of the SecY translocon revealed by molecular dynamics simulations with virtual probes. Biophys. J. 2006;90:2718–2730. doi: 10.1529/biophysj.105.073304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B., Clemons W., Collinson I., Modis Y., Hartmann E., Harrison S., Rapoport T. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Walters R., DeGrado W. Helix-packing motifs in membrane proteins. Proc Natl. Acad. Sci. USA. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigley W., Corboy M., Cutler T., Thibodeau, p, Oldan J., Lee M., Rizo J., Hunt J., Thomas P. A protein sequence that can encode native structure by disfavoring alternate conformations. Nat. Struct. Biol. 2002;9:381–388. doi: 10.1038/nsb784. [DOI] [PubMed] [Google Scholar]
- Wilson C., Kraft C., Duggan C., Ismail N., Crawshaw S., High S. Ribophorin I associates with a subset of membrane proteins after their integration at the Sec61 translocon. J. Biol. Chem. 2005;280:4195–4206. doi: 10.1074/jbc.M410329200. [DOI] [PubMed] [Google Scholar]
- Wimley W. C., Creamer T. P., White S. H. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry. 1996;35:5109–5124. doi: 10.1021/bi9600153. [DOI] [PubMed] [Google Scholar]
- Woolhead C., McCormick P., Johnson A. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell. 2004;116:725–736. doi: 10.1016/s0092-8674(04)00169-2. [DOI] [PubMed] [Google Scholar]
- Zhou F., Cocco M., Russ W., Brunger A., Engelman D. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 2000;7:154–160. doi: 10.1038/72430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.