Abstract
Subtelomeric genes are either fully active or completely repressed and can switch their state about once per 20 generations. This meta-stable telomeric position effect is mediated by strong repression signals emitted by the telomere and relayed/enhanced by weaker repressor elements called proto-silencers. In addition, subtelomeric regions contain sequences with chromatin partitioning and antisilencing activities referred to as subtelomeric antisilencing regions. Using extensive mutational analysis of subtelomeric elements, we show that ARS consensus sequence (ACS)-containing proto-silencers convert to antisilencers in several replication factor mutants. We point out the significance of the B1 auxiliary sequence next to ACS in mediating these effects. In contrast, an origin-derived ACS does not convert to antisilencer in mutants and its B1 element has little bearing on silencing. These results are specific for the analyzed ACS and in addition to the effects of each mutation (relative to wild type) on global silencing. Another line of experiments shows that Mcm5p possesses antisilencing activity and is recruited to telomeres in an ACS-dependent manner. Mcm5p persists at this location at the late stages of S phase. We propose that telomeric ACS are not static proto-silencers but conduct finely tuned silencing and antisilencing activities mediated by ACS-bound factors.
INTRODUCTION
Genes positioned close to telomeres are either fully active or completely repressed. They switch their state of expression once per 20 generations on average. This meta-stable telomeric position effect (TPE) is governed by strong repression signals emitted by the telomeres (Fourel et al., 2002a; Tham and Zakian, 2002; Rusche et al., 2003). Weaker repressor elements in the core X- and Y′-subtelomeric elements, called proto-silencers, can relay and enhance repression signals away from telomeres (Fourel et al., 2002a). To date, proto-silencer activity has been demonstrated for ARS consensus sites (ACS) and for the binding sites for Rap1p and Abf1p (Boscheron et al., 1996; Fourel et al., 1999; Pryde and Louis, 1999; Lebrun et al., 2001). In addition, subtelomeric core X- and Y′-elements contain sequences, which display chromatin partitioning or antisilencing activities and are referred to as subtelomeric antisilencing regions (STARS) (Fourel et al., 1999, 2004; Pryde and Louis, 1999). The state of gene expression at individual telomeres is determined by the complex interplay between the telomere, proto-silencers, and antisilencers (Fourel et al., 1999; Pryde and Louis, 1999; Lebrun et al., 2001; Fourel et al., 2002a, 2004; Rehman et al., 2006); however, it is not clear whether and how these elements contribute to the variegated nature of TPE.
ACS is a conserved 11-base pair sequence found in all yeast origins of replication, in subtelomeric core X- and Y′-elements and in the silencers of the mating type loci. ACS binds origin recognition complex (ORC) and is essential for replication initiation at origins and for silencing at the mating type loci (Bell and Stillman, 1992; Diffley and Cocker, 1992; Rusche et al., 2003). A less conserved B1 element upstream of ACS serves as an additional docking site for ORC and contributes to, but is not essential for replication initiation (Marahrens and Stillman, 1992; Rao and Stillman, 1995; Lee and Bell, 1997). The B1 element of the HMR-E silencer is dissimilar to the B1 elements found in origins (Palacios DeBeer and Fox, 1999; Palacios DeBeer et al., 2003). It confers high affinity to ORC and contributes to robust silencing of HMR (Palacios DeBeer and Fox, 1999; Palacios DeBeer et al., 2003). The role of B1 and ACS in subtelomeric core X- and Y′-elements has not been addressed.
TPE is modulated by a significant number of genes. For example, mutations in general regulators of silencing such as the SIR genes, in the ORC genes, ABF1 and RAP1, reduce the silencing at telomeres, the mating type loci and at the ribosomal gene loci (Aparicio et al., 1991; Pryde and Louis, 1999; Roy and Runge, 2000). In contrast, genes involved in telomere maintenance and localization (HDF1, HDF2, MRE11, XRS2, and RAD50) specifically affect silencing at telomeres (Boulton and Jackson, 1998; Laroche et al., 1998; Fisher and Zakian, 2005; Mueller et al., 2006). Finally, the histone methylase COMPASS (Mueller et al., 2006), the histone acetyl transferase NuA4 (Babiarz et al., 2006; Clarke et al., 2006), the histone chaperone CAF-1 (Mueller et al., 2006; Tamburini et al., 2006), cohesin (Suter et al., 2004; Chang et al., 2005), SUM1 (Irlbacher et al., 2005), and SCP160 (Marsellach et al., 2006) were also implicated in telomeric silencing.
There is a puzzling link between DNA replication and silencing. The establishment of silent chromatin requires the passage through S-phase (Miller and Nasmyth, 1984), but does not require DNA replication per se (Dubey et al., 1991; Kirchmaier and Rine, 2001; Li et al., 2001). Even so, many genes that affect silencing encode DNA replication factors. These include ORC2, ORC5, MCM5, MCM10, CDC44, CDC45, POL30 (Ehrenhofer-Murray et al., 1995; Dillin and Rine, 1997; Fox et al., 1997; Ehrenhofer-Murray et al., 1999; Dziak et al., 2003; Suter et al., 2004; Liachko and Tye, 2005). Paradoxically, an earlier comprehensive study on telomeric silencing found that mutations in orc2 and orc5 have little effect on gene expression in the vicinity of ACS (Pryde and Louis, 1999). Similarly, the deletion of ACS in the HMR-E silencer actually increased HMR repression in orc2 mutants (Palacios DeBeer and Fox, 1999). On the other hand, we have demonstrated that mutations in orc2, orc5 and mcm5 exert their effects on subtelomeric silencing mostly via ACS (Rehman et al., 2006). In an attempt to resolve this apparent inconsistency on the role of ACS in silencing, we further explored the ACS-mediated effects in different mutants. Intriguingly, we noticed that in certain mutants the subtelomeric ACS alter their behavior and act as antisilencers.
MATERIALS AND METHODS
Yeast Strains and Reporter Constructs
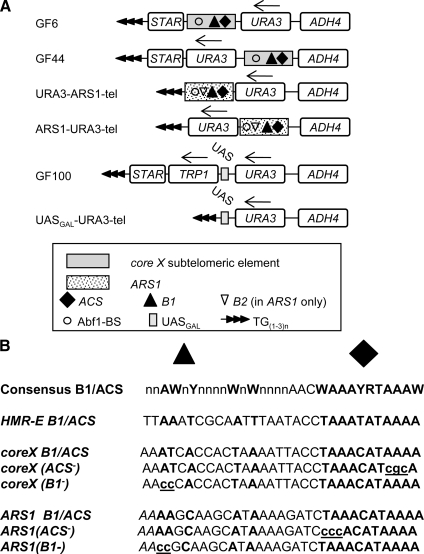
The yeast strains used are shown in Supplemental Table 1. All experiments were performed at the permissive temperature of 23°C. The constructs used are shown in Figure 1A. GF6 and GF44 were described in Fourel et al. (1999); GF100 in (Fourel et al., 2002b); URA3-tel and URA3-UASGAL4-tel in (Jacobson and Pillus, 2004), ARS1-URA3-tel, and URA3-ARS1-tel in (Rehman et al., 2006). GF6(ACS−), GF44(ACS−), GF6(B1−), GF44(B1−), ARS1(ACS−)-URA3-tel, ARS1(B1−)-URA3-tel, and URA3-ARS1(B1−)-tel were generated by site-directed mutagenesis of GF6, GF44, ARS1-URA3-tel, and URA3-ARS1-tel as depicted in Figure 1B; GF6(ACS−ΔSTAR) and GF6(B1−ΔSTAR) were produced by removing STAR elements from GF6(ACS−) and GF6(B1−). All reporter constructs include a portion of ADH4 and a telomeric TG(1−3)n repeat to direct their insertion between ADH4 and the telomere of the VII-L arm. After transformation with the constructs cells were selected on SC-ura (and SC-ura-trp in the case GF100) plates and variegated reporter expression was confirmed by restreaking positive clones on SC-ura and SC/FOA plates (see below). Three colonies from each transformation were then subjected to analysis by the reporter activity assays described below.
Figure 1.
Schematic representation of the constructs used in this study. (A) Telomeric repeats are represented as solid arrowheads; the core X as gray rectangle; ARS1 as dotted rectangle; STAR, URA3, TRP1, and the portion of ADH4 used for targeted integration in the VII-L telomere as open rectangles. Arrows above TRP1 and URA3 indicate the orientation of the gene. ACS (solid diamond), B1 (solid triangle), B2 (inverted open triangle), and Abf1 binding site (open circle) are depicted within ARS1 and core X. The Gal4 binding site in GF100 and URA-UASGAL-tel is shown as an open rectangle. (B) The consensus B1/ACS sequence (solid rectangle and solid triangle indicate the positions of B1 and ACS) is shown on top. The HMR-E, core X and ARS1 sequences encompassing B1/ACS are shown in capital letters with the consensus bases shown in bold. The mutated sequences in the ACS− and B1− constructs are shown in small letters and are underlined.
GAL4DBD-Fusion Protein Expression Cassettes
The coding sequence of ORC1, ORC2, and MCM3 were produced by polymerase chain reaction (PCR) of genomic DNA from W303. MCM5 was produced by PCR from pIP122CDC46 (Gauthier et al., 2002). The mcm5-461 and mcm5-1 alleles were produced by PCR with genomic DNA from the corresponding strains in Supplemental Table 1. All fragments were cloned into pGBKT7 to produce GAL4DBD fusion cassettes and then subcloned in pRS315 (CEN4, ARS1, LEU2). pGAL4DBD-GCN5, pGAL4DBD-SIR1 and pGAL4DBD-MCM10 have been described in Jacobson and Pillus (2004) and Liachko and Tye (2005), respectively. The expression plasmids were transformed into cells with previously integrated URA3-tel, URA3-UASGAL4-tel, or GF100 constructs.
Reporter Activity Assays
URA3 expression renders the cells sensitive to fluoroorotic acid (FOA). Hence, the proportion of cells with repressed URA3 was assessed as percentage of FOA resistant cells (%FOAR). The proportion of cells expressing TRP1 in the GF100 construct was assessed by growth on SC-trp plates. For the selection of the GAL4DBD-fusion protein expression plasmids, additional nutrients (leucine or histidine) were omitted as appropriate.
For each measurement, three colonies were grown in nonselective (YPD) medium for 20–30 generations. Serial 1:10 dilutions from each individual culture were spotted on SC, SC/FOA, or SC-trp plates as appropriate. The %FOAR or %TRP+ was calculated as the average number of colonies on SC/FOA or SC-trp plates divided by the average number of colonies on SC plates. The average values ± SE (calculated in Excel; Microsoft, Redmond, WA) of n triplicate measurements were then calculated and are shown in Supplemental Tables 2–6.
Chromatin Immunoprecipitation (ChIP)
Cells were arrested in mitosis by 12 μg/ml nocodazole and released from the block as described in Rehman et al. (2006). Samples of 50 ml were cross-linked at the time points indicated in text. Cross-linking was as in Kurdistani and Grunstein (2003) with modifications. Briefly, 50-ml cultures at OD600 = 1.2 were suspended in 20 ml of PBS/10 mM dimethyl adipimidate for 15 min at 25°C, and then 30 ml of PBS/1.6% formaldehyde was added for 25 min. After quenching with glycine cells were suspended in 2.5 ml of LIP (Lysis/IP) buffer (50 mM Tris-HCl, 140 mM NaCl, 3 mM EDTA, 1× protease inhibitor cocktail (P8215; Sigma-Aldrich, St. Louis, MO) and broken with glass beads. Chromatin and cell debris were collected by spinning, suspended in LIP and sonicated for 12 min in Bioruptor (Diagenode, Liege, Belgium) at maximum output. Debris was removed by spinning, and an aliquot was uncross-linked to confirm an average size of DNA of 500 base pairs. The samples received Triton X-100 and Na-deoxycholic acid to 0.5 and 0.05%, and then they were immunoprecipitated with 10 μg of anti-MCM5 (Sc-6680; Santa Cruz Biotechnology, Santa Cruz, CA) antibody for 3 h. Load, wash and eluate samples were uncross-linked and analyzed by PCR with primers specific for ARS1, ARS305, ARS501, and core X. Core X signals were also confirmed by slot-blot. The primers for the core X element in GF44 and GF44(ACS−) anneal to the plasmid backbone of the integrating constructs and do not amplify genomic core X. Primer sequences are available upon request.
RESULTS
Experimental Strategy
Recently, we correlated the telomeric antisilencing defects of several mutants (mcm5-461, orc2-1, orc5-1, and cdc45-1) to the presence of ACS (Rehman et al., 2006). These mutations significantly weaken telomeric silencing (see Supplemental Tables 2 and 3), whereas the deletion of ACS diminished these effects (Rehman et al., 2006). A separate study has shown that deletion of sas2 also requires intact ACS for its effect on silencing (Ehrenhofer-Murray et al., 1997). cdc7-1 displays both ACS-dependent and ACS-independent effects (Rehman et al., 2006). cdc6-1 and Δsir2 reduced silencing independently of ACS (Ramachandran et al., 2006; Rehman et al., 2006).
In the current study, we present extensive mutational analyses of ACS and the auxiliary B1 element in these mutant strains. Two well characterized constructs, GF6 and GF44 (Fourel et al., 1999; Rehman et al., 2006), contain a conserved subtelomeric core X elements plus a STAR element and a URA3 reporter (Figure 1A). Two other constructs, ARS1-URA3-tel and URA3-ARS1-tel (Rehman et al., 2006) contain the 150-base pair genomic sequence encompassing the ARS1 origin of DNA replication (Figure 1A).
The ACS- and B1-mutations in core X and in ARS1 are depicted in Figure 1B. ACS is highly conserved and plays key roles in ORC binding and in origin and silencer function. It has been shown that single A→C mutations in it abolishes both ORC binding and the stability of ARS1-harboring minichromosomes (Marahrens and Stillman, 1992; Rao and Stillman, 1995). In our constructs, we have substituted AAA with CCC or CGC so we expect these substitutions to preclude the binding to ORC and any effects associated with ACS. The neighboring B1 element is loosely conserved (Palacios DeBeer et al., 2003) with AWnY forming its core (Figure 1B). It acts as a secondary docking site for ORC (Rao and Stillman, 1995; Lee and Bell, 1997). In ARS1, single mutations in AW (A839A840) (Rao and Stillman, 1995) reduced but did not abolish binding to ORC and also decreased the stability of ARS1-harboring minichromosomes. Total replacement of B1 by a linker did not produce stronger effects. In HMR-E, substitutions in B1 reduced its affinity to ORC and converted it to a poor silencer but a good origin of replication (Palacios DeBeer et al., 2003). We have mutated the AW doublets in ARS1 and core X to CC. We expect these mutations to decrease, but not abolish, the binding to ORC. In terms of function, we expect the mutations to preclude any effects mediated by B1.
All mutant and wild-type constructs were integrated at the same position in VII-L telomere of mcm5-461, orc2-1, orc5-1, cdc45-1, Δsas2 cdc6-1, cdc7-1, and Δsir2 or the corresponding wild-type strains MCM5 and W303. Three colonies (per experiment) were streaked on SC-ura and SC/FOA plates to confirm variegated expression and then grown for 30 generations in YPD medium to allow for telomeric expression equilibrium to be reached under nonselective conditions. The %FOAR for the three colonies was assessed by the FOAR reporter activity assay and then average %FOAR and errors for (n) independent triplicate experiments were calculated (Supplemental Tables 2 and 4).
Finally, we divided the average %FOAR value in the (ACS−) and (B1−) constructs where the effects of ACS are abolished, by the average %FOAR value in the constructs with intact B1/ACS elements (Figures 2–4). Hence, the calculated values specifically assess the net silencing contribution of ACS and B1 at these loci independently and above the effects on global silencing of the gene mutations. Because mcm-461, orc2-1, orc5-1, cdc46-1, cdc6-1, and cdc7-1 affect essential genes, we assume that these mutations are still capable of mediating effects through the essential ACS element. In these graphs, values below 1 signify antisilencing effect of the destroyed element, whereas values above 1 signify silencing effects of the destroyed element.
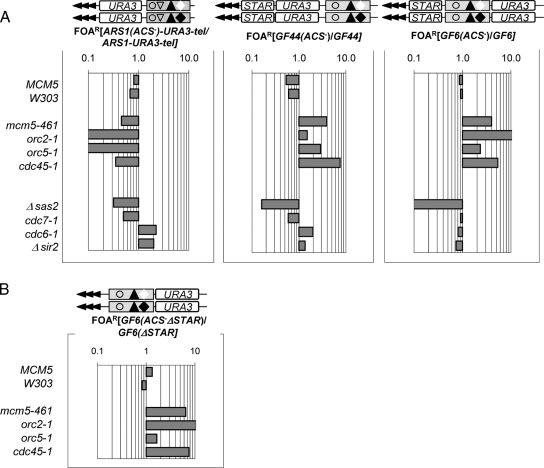
Figure 2.
ACS-mediated silencing effects in ARS1 and core X. ARS1-URA-tel, GF6, GF44, ARS1(ACS−)-URA3-tel, GF6(ACS−), GF44(ACS−) (diagrams shown on top) were inserted in the VII-L telomere of wild-type (MCM5, W303) and mutant (mcm5-461, orc2-1, orc5-1, cdc45-1, Δsas2, cdc7-1, cdc6-1, and Δsir2) strains, and %FOAR was determined for each construct/strain combination. Raw data are shown in Supplemental Table 2. (A) The ratio %FOAR(ACS−)/%FOARACS as indicated above each graph represents the effect of destruction of ACS in the construct shown on top in the strains shown on the left. In these exponential graphs, values below 1 depict derepression and values above 1 depict increased repression. (B) The ratio %FOARGF6(ACS-ΔSTAR)/%FOARGF6(ΔSTAR) shows the effect of destruction of ACS in the absence of STAR element.
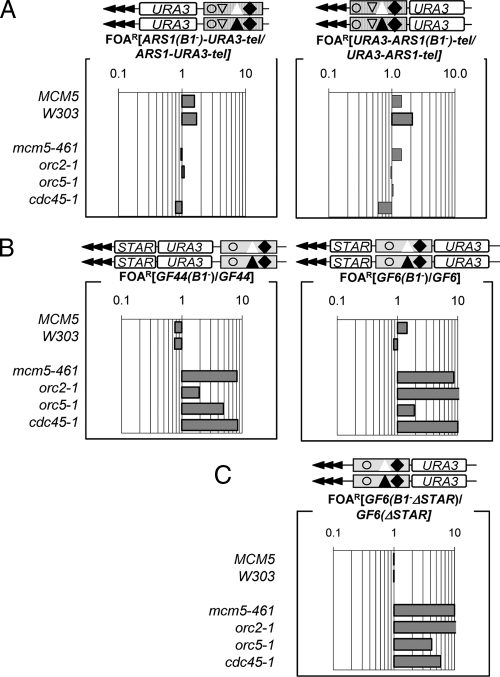
Figure 3.
B1-mediated silencing effects in ARS1 and core X. ARS1-URA-tel, URA-ARS1-tel, GF6, GF44, ARS1(B1−)-URA-tel, URA-ARS1(B1−)-tel, GF6(B1−), and GF44(B1−) were inserted in the VII-L telomere of wild type (MCM5, W303) and mutant (mcm5-461, orc2-1, orc5-1, and cdc45-1) strains and %FOAR was determined for each construct/strain combination. Raw data are shown in Supplemental Table 3. (A) The ratio %FOAR(B1−)/%FOARB1 for ARS1-URA-tel and URA-ARS1-tel is plotted to show the effect of destruction of B1 in the strains shown on the left. (B) The ratio %FOAR(B1−)/%FOARB1 for GF44 and GF6 is plotted to show the effect of destruction of B1 in the strains shown on the left. (C) The ratio %FOARGF6(B1−ΔSTAR)/%FOARGF6 shows the effect of destruction of B1 in the absence of STAR element.
Figure 4.
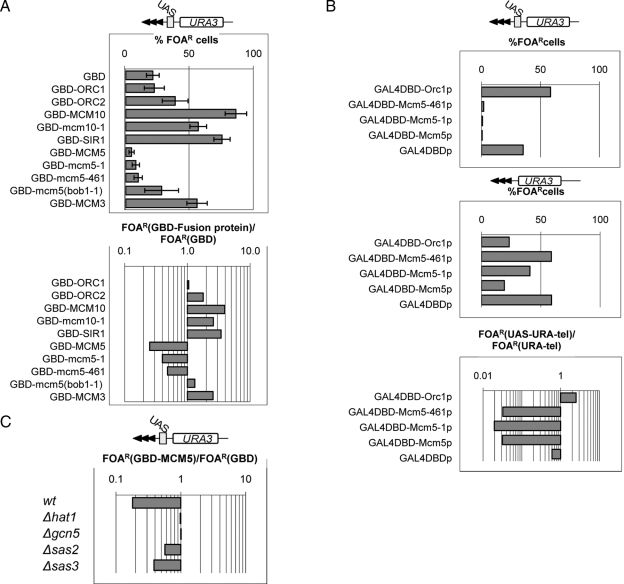
Mcm5p has antisilencing activity. (A) Wild-type cells (W303) harboring URA3-UASGAL-tel (shown on the top) were transformed with plasmids carrying expression cassettes for the GBD-fusion proteins shown on the left. Percentage of FOAR for each recombinant protein is plotted in the top graph. Raw data are shown in Supplemental Table 5. The ratio %FOARGAL4-fusion/%FOARGAL4DBD is shown in the bottom graph and indicates the effect of each recombinant protein. Values below 1 depict antirepression activity, whereas values above 1 depict repression activity. (B) LPY1030 cells harboring URA-UASGAL-tel (top) and LPY1029 cells harboring URA3-tel (middle) were transformed with plasmids expressing the proteins shown on the left. Percentage of FOAR was measured for each recombinant protein in each strain and was plotted. Raw data are shown in Supplemental Table 5. The ratio %FOARURA3-UAS-tel/%FOARURA3-tel (bottom) shows how the effect of these proteins depends on the UASGAL site. (C) Wild-type cells (BY4742) and Δhat1, Δgcn5, Δsas2, and Δsas3 isogenic deletion mutants harboring URA3-UASGAL-tel (shown on the top) were transformed with plasmids expressing GBD-Mcm5p or GAL4DBD, respectively. Percentage of FOAR for each recombinant protein was measured. Raw data are shown in Supplemental Table 5. The ratio %FOARGAL4-Mcm5p/%FOARGAL4DBD indicates the effect of each gene deletion on the antisilencing activity of Mcm5p.
Silencing Effects of ACS
We used the ACS−/ACS ratios for the ARS1-URA-tel, GF44, and GF6 constructs to specifically assess the silencing effects of ACS in each individual mutant. As expected, in all three constructs the ablation of ACS decreased the silencing of URA3 in the MCM5 and W303 strains by 2- to 4-fold, thus reiterating that in wild-type context ACS acts as a proto-silencer (Figure 2A). Unexpectedly, however, the destruction of ACS had entirely different effect in the mcm5-461, orc2-1, orc5-1, and cdc45-1 mutants. In ARS1-URA-tel the elimination of ACS in ARS1 caused reduction of repression, but to a significantly larger extent as compared with wild-type cells (Figure 2A). In contrast, the elimination of ACS in the core X elements in GF44 and GF6 consistently increased repression. We also noticed a great position variation in the magnitude of increase in URA3 repression in orc2-1 (Figure 2A, compare GF6 and GF44) but not in other mutants (Figure 3A). Although we do not completely understand these effects of orc2-1, they suggest that this particular allele may alter the directionality of silencing driven by ACS as reported earlier for the mating type loci (Zou et al., 2006a,b). The other three mutants had similar effects on GF44 and GF6. We concluded that in mcm5-461, orc2-1, orc5-1, and cdc45-1 the core X-ACS displayed properties typical for an antisilencer (Figure 2A).
We considered that this surprising behavior of ACS in the mutants could be a nonspecific effect of overall reduction in silencing. For this reason, we performed similar assays in four other mutants. As mentioned, Δsas2 requires intact ACS for its effect (Ehrenhofer-Murray et al., 1997), cdc7-1 has both ACS-dependent and ACS-independent effects (Rehman et al., 2006), whereas cdc6-1 and Δsir2 act independently of ACS (Ramachandran et al., 2006; Rehman et al., 2006). As expected, in Δsas2 cells the destruction of ACS exacerbated the loss of silencing in all three constructs albeit to a different level compared with wild-type cells (Figure 2A). The cdc7-1 mutation showed little difference compared with the wild-type cells. The cdc6-1 and the Δsir2 mutations showed mild increase in repression in ARS1-URA-tel and GF44 and derepression in GF6. In these two mutants, we see weak correlation to the position of URA3 relative to the telomere and the proto-silencer rather than to the nature of the proto-silencer. In summary, these four mutants did not show any unexpected effects. Importantly, they indicated that the conversion of ACS to antisilencer in core X (GF44 and GF6) is specific for mcm5-461, orc2-1, orc5-1, and cdc45-1 and not general consequence of deregulation of silencing.
Finally, we tested whether the observed effects of the core X-ACS element could somehow be driven by the neighboring STAR element. In Figure 2B we show that the deletion of STAR in GF6(ACS−) does not alter the pattern of repression in mcm5-461, orc2-1, orc5-1, and cdc45-1.
Silencing Effects of B1
At least two studies have reported functional dissimilarities between different B1 elements that relate to the replicator or silencer efficiency (Palacios DeBeer and Fox, 1999; Palacios DeBeer et al., 2003; Chang et al., 2008). We therefore asked whether the apparent differences of ACS in ARS1 and core X could be extended to their B1 elements. We mutated the B1 elements in the ARS1-URA-tel, URA-ARS1-tel, GF44, and GF6 and used the B1−/B1 ratios to evaluate the contribution of B1 to telomeric silencing. The destruction of B1 in ARS1 only marginally affected silencing in the mutants and actually slightly increased repression in the wild-type strains (Figure 3A). Switching of the position of ARS1 and URA3 relative to the telomere did not make difference in the lack of effect of the destruction of B1 (Figure 3A, compare ARS1-URA-tel and URA-ARS1-tel). We concluded that, independently of position or orientation, the B1 element of ARS1 has no bearing on silencing at telomeres. In contrast, the destruction of the coreX-B1 in GF6 and GF44 produced very similar gain of repression in mcm5-461, orc2-1, orc5-1, and cdc45-1 compared with the elimination of core X-ACS (compare Figure 2A and Figure 3B). Again, the level of repression in orc2-1 was dependent on the position of the ACS/B1 element (Figure 3B). Finally, we tested whether the observed effects of mutations in the B1 element could somehow be driven by the neighboring STAR element. In Figure 3C, we show that the deletion of STAR in GF6(B1−) does not alter the pattern of repression in any of the mutants (compare Figure 3, A and B, with C).
The significance of the data presented in Figures 2 and 3 is twofold. First, we show that, unlike ARS1-derived B1/ACS, subtelomeric core X-derived B1/ACS convert from proto-silencers in wild-type cells to antisilencers in four replication/silencing factor mutants. Second, we demonstrate that, unlike ARS1-B1, the core X-B1 element has a profound effect on silencing.
Mcm5p Has Antisilencing Activity
The conversion of coreX-B1/ACS from proto-silencer to antisilencer suggests that a hypothetic ACS-binding antisilencing factor could be more active in the analyzed mutants. This idea prompted us to look for possible antisilencing activities in transfactors, which are known to associate with ACS. We exploited an assay, which has been extensively used to identify general antisilencing activities out of the context of ACS or other silencers (Fourel et al., 1999, 2002b, 2004; Jacobson and Pillus, 2004; Liachko and Tye, 2005). It measures the expression of reporters inserted in the VII-L telomere upon the tethering of recombinant GAL4-fusion proteins via a nearby UASGAL1-10 site (Figure 4). Using this assay, we tested the silencing activities in an array of proteins, which are known to associate with ACS. The open reading frames of the ORC1, ORC2, MCM3, MCM5, mcm5-461, mcm5-1 (Dziak et al., 2003) (see Supplemental Figure 1 for details on these mutations), mcm5(bob1-1)(Hardy et al., 1997), MCM10, mcm10-1 (Liachko and Tye, 2005), and SIR1 alleles, respectively, were fused to a GAL4DBD expression cassette and subcloned in low copy plasmids (pRS315). The produced plasmids were then transformed into a wild-type (W303) strain containing the URA3-UASGAL-tel reporter inserted in the VII-L telomere (Jacobson and Pillus, 2004). The expression of the recombinant proteins was confirmed by Western blot with anti-GAL4 antibodies (data not shown). Next, using the FOAR assay, we evaluated how these proteins influenced the repression of URA3 (Figure 4 and Supplemental Table 5). The specific effect of each protein was calculated as the ratio of FOAR cells expressing GAL4DBD-fusion proteins divided by FOAR cells expressing GAL4DBD alone (Figure 4A). Again, values below 1 signify derepression and values above 1 signify repression.
In tune with several previous reports (Rusche et al., 2003; Liachko and Tye, 2005), the tethering of Orc1p, Orc2p, Sir1p, Mcm10p, and the mutant Mcm10-1p increased the repression (albeit at different levels) at the VII-L telomere relative to GAL4DBD alone. However, Mcm5p or the mutant Mcm5-461p and Mcm5-1p proteins displayed distinct antisilencing activity. The mutations in Mcm5-461p and Mcm5-1p only moderately reduced the antisilencing effect of the wild-type protein (Figure 4A); however, the Mcm5(bob1-1)p protein showed no antisilencing activity. Because the mcm5(bob1-1) mutation by-passes the requirement for CDC7 (Hardy et al., 1997), we speculate that the antisilencing activity of Mcm5p is at least partially regulated by CDC7. Another member of the MCM family, Mcm3p, showed silencing rather than antisilencing properties (Figure 4A). This important control indicates that antisilencing is specific for Mcm5p and not a general feature of the MCM proteins.
We tested whether the antisilencing activity of the Mcm5p proteins was due to side effects of overexpression not mediated by the UASGAL4 site. In Figure 4B, we show that the expression of wild-type GAL4-Mcm5p causes threefold derepression on a UASGAL4-less cassette (LPY1029; (Jacobson and Pillus, 2004) relative to GAL4DBD alone, whereas the mutant proteins had weaker effects. GAL4-Orc1p also reduced repression by approximately threefold. However, the ratio %FOAR(URA3-UASGAL-tel)/FOAR(URA-tel) indicated that the antisilencing effect of the Mcm5 proteins is 80–90 times stronger on the URA3-UASGAL-tel cassette (LPY1030; Jacobson and Pillus, 2004), whereas Orc1p increased silencing by approximately twofold (Figure 4B). We conclude that it is the tethering of the Mcm5 proteins that is mostly responsible for the derepression of URA3-UASGAL-tel. Consequently, we concluded that most of the tested proteins, which build up prereplicative complexes and presumably also associate with subtelomeric ACS, possess silencing activities. Among them, Mcm5p clearly displayed antisilencing activity.
Next, we asked what could be the mechanism, which is engaged in this antisilencing activity of Mcm5p. We performed similar tethering assays in strains with deletions in four nonessential histone-acetyl-transferase genes: Δhat1, Δgcn5, Δsas2, and Δsas3 (Figure 4C). It is noteworthy that Δsas2 and Δsas3 dramatically derepress truncated telomeres, but it had significantly weaker effect on the more “complex” constructs including GF6, GF44, and URA-UAS-tel (Supplemental Figure 3). Whereas the deletion of SAS2 and SAS3 only slightly reduced the antisilencing effect of GAL4DBD-Mcm5p, the deletion of HAT1 and GCN5 essentially abolished it (Figure 4C). We concluded that the antisilencing effect of Mcm5p is mediated by the histone-acetyl-transferases HAT1 and GCN5 and thus conforms with a general expectation of an antisilencing factor.
Mcm5 Is a Poor Chromatin Insulator
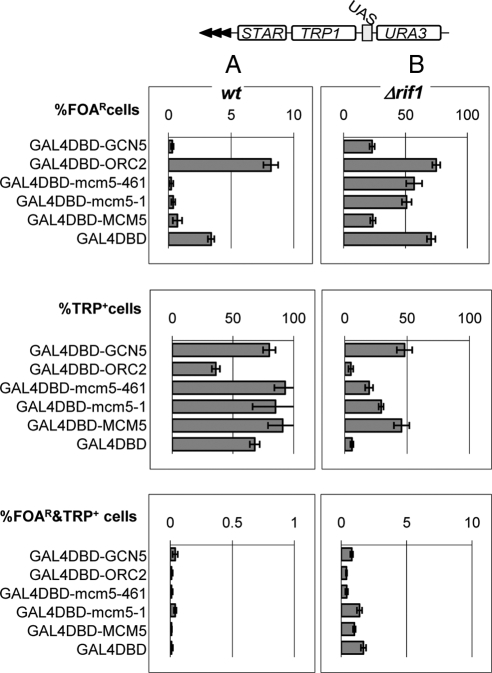
Certain proteins or DNA elements impede the spreading of chromatin modifications into adjacent genomic domains thus acting as chromatin insulators or “partitioners” (Fourel et al., 2002b, 2004; Ishii et al., 2002). We considered the possibility that the observed effects of the Mcm5 proteins are a consequence of chromatin partitioning rather than of genuine antisilencing. We addressed this issue in the GF100 strain (Figure 5; Fourel et al., 2002b), which harbors a double reporter cassette (URA3 and TRP1) and a STAR element inserted in the VII-L telomere. These two reporters are separated by UASGAL so that TRP1 is proximal to the telomeric repeats. If a protein tethered to UASGAL4 equally increases or decreases the levels of expression of TRP1 and URA3, then this protein has antisilencing or silencing activities only. However, if the tethered protein increases the levels of expression of TRP1 and URA3 remains repressed, it acts as a chromatin insulator (Fourel et al., 2002b, 2004). In this assay, the level of repression of URA3 is assessed as %FOAR cells, the level of repression of TRP1 is assessed as %TRP+ cells, whereas partitioning activity is assessed as %FOAR&TRP+ cells.
Figure 5.
MCM5 has poor insulating activity. W303 cells (A) and W303Δrif1 (B) cells harboring the GF100 reporter (shown on the top) in the VII-L telomere were transformed with plasmids carrying expression cassettes for the GDB-fusion proteins shown on the left. %FOAR, %TRP+, and %FOAR&TRP+ cells for each recombinant protein were measured and plotted in the top, middle, and bottom graphs, respectively. Raw data are shown in Supplemental Table 6.
Using the GF100 strain, the three GAL4-Mcm5 proteins were compared with GAL4DBD and to well characterized silencing (GAL4-Orc2p) (Rusche et al., 2003) or antisilencing (GAL4-Gcn5p) (Jacobson and Pillus, 2004) proteins. As expected, GAL4-Orc2p increased the %FOAR cells and decreased the %TRP+ cells, whereas GAL4-Gcn5p had the opposite effect relative to GAL4DBD alone (Figure 5A and Supplemental Table 6). The Mcm5 proteins behaved as antisilencers similarly to GAL4-Gcn5p (Figure 5A). Importantly, GAL4DBD and all fusion proteins produced <0.01% FOAR&TRP+ cells thus indicating poor partitioning activity (Figure 5A).
Because Mcm5p and Gcn5p induce significant levels of derepression at the VII-L telomere and as a result low % FOAR cells, it may be difficult to detect partitioning activity under these experimental conditions. We addressed this concern by deleting RIF1 in the GF100 strain (Fourel et al., 2002b). RIF1 counteracts the repression signals emitted by the telomere via Rap1p (Fourel et al., 2002b). Indeed, the deletion of RIF1 increased ∼10 times the repression of both URA3 and TRP1 (Figure 5B). Correspondingly, the %FOAR&TRP+ cells increased from ∼0.01 to ∼1% for all proteins tested (Figure 5B), but again no substantial difference between GAL4DBD and the fusion proteins was observed. These experiments argue that Mcm5p does not have partitioning properties under the conditions of either high or low levels of local repression.
Mcm5p Remains Associated with Core X in Late S Phase
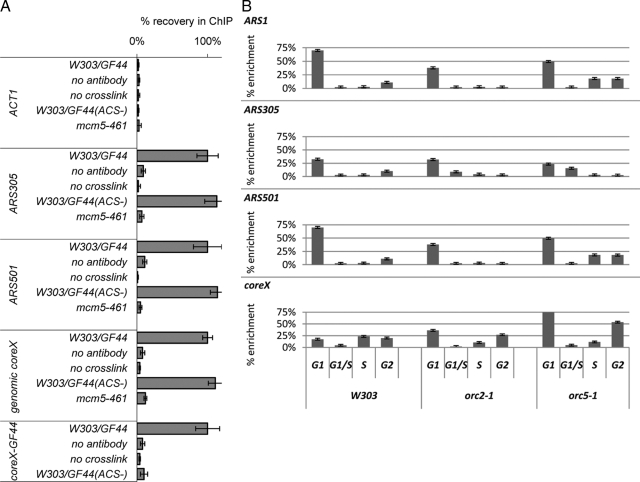
Prior studies have shown abundant association of MCM proteins with subtelomeric regions (Wyrick et al., 2001), but it is not known whether this abundance is dependent on subtelomeric ACS. We tested whether this is the case. We conducted ChIP with anti-Mcm5p antibodies in W303 strains harboring the GF44 or GF44(ACS−) constructs in the VII-L telomere. After determining the linear range for PCR in input samples (data not shown), the input and immuno-enriched DNA was quantified by PCR with primers for ACT1, ARS305, ARS501, total genomic core X, and the core X element in the VII-L telomere in GF44. The signals from the eluates relative to input in the W303 and W303/GF44 strains are plotted in Figure 6A. No specific association of MCM proteins with the ACT1 gene was detected. In contrast, comparable strong signals in both strains were observed for two origins of replication, ARS305 and ARS501, and for total genomic core X DNA. Importantly, the elimination of ACS from the core X-element in GF44 precluded the binding of Mcm5p at this particular position (Figure 6A). Finally, the anti-Mcm5p antibody did not work in the mcm5-461 strain, supporting the notion that all signals are specific for Mcm5p. We concluded that Mcm5p associates with subtelomeric core X elements in an ACS-dependent manner.
Figure 6.
(A) Mcm5p association with telomeres depends on ACS. ChIP with anti-Mcm5p antibodies (Santa Cruz Biotechnology) was performed in the W303/GF44, W303/GF44(ACS−), and mcm5-461 strains. Each experiment was performed with parallel controls where cross-linking or primary antibody incubation was omitted. The abundance of different DNAs (shown on the left) was quantified by PCR after confirming linear PCR range for the input samples. The primers for the core X element in GF44 and GF44(ACS−) anneal to the plasmid backbone of the integrating constructs and do not amplify genomic core X. core X signals were also confirmed by slot-blot. The signals in the eluates were subtracted by the signals in the final washes and then divided by the signals in the input fractions to produce “net signals.” Finally, all net signals were normalized so that the corresponding values in GF44 are 100% and then compared with the values in W303/GF44(ACS−) and mcm5-461. The figure is representative of two independent ChIP experiments. (B) Wild-type (W303), orc2-1 and orc5-1 cells (shown at the bottom top) were released from mitotic arrest and samples were collected after 10, 35, 60, and 75 min (early G1, late G1/S, S, and G2 phases of the cell cycle, respectively) as shown at the bottom. ChIP with anti-Mcm5p antibodies was conducted as described above. The signals in the eluates were subtracted by the signals in the final washes and then divided by the signals in the input fractions and plotted as % enrichment relative to input. The analyzed genomic loci are indicated in the top left corner of each graph. The figure is representative of two independent ChIP experiments.
Next, we analyzed the subtelomeric occupancy by Mcm5p in two mutants, which display strong ACS-dependent antisilencing activity, orc2-1 and orc5-1. Wild-type (W303) and mutant cells were arrested in mitosis by nocodazole for 3 h and then washed and suspended in YPD. Samples were collected at 10, 35, 60, and 75 min after release from the point of mitotic arrest. These time points correspond to early G1, late G1/S, S, and late S/G2 phases of the cell cycle, respectively (Rehman et al., 2006). The cells were cross-linked, lysed, and chromatin was immunoprecipitated with anti-Mcm5p antibodies. The association of Mcm5p with a very early (ARS305), an early (ARS1), and a late (ARS501) origins as well as with core X DNA was measured by PCR and is plotted in Figure 6B. At all four genomic loci a high levels of association were observed in early G1 phase, which corresponds to the time of loading of MCM proteins on chromatin (Lei and Tye, 2001). The Mcm5p association markedly diminishes in the subsequent time points at the two early origins (Figure 6B, top two rows) and seems to persist longer for the late ARS501 origin (Figure 6B, third row). This pattern of occupancy is in good agreement with the expected time of firing and MCM displacement from origins (Lei and Tye, 2001). Remarkably, ChIP signals with core X-DNA were quite dissimilar compared with the pattern displayed by the origins (Figure 6B, bottom row). At core X, we observed the typical peak of ChIP signal in early G1 but also a subsequent gradual increase in mid- and late S phase in both wild-type and mutant cells. It is possible that this increase is produced by enhanced exposure of epitope on already loaded MCM proteins, for example as a result of phosphorylation. This conclusion would be in tune with most current concepts on the turnover of MCM proteins throughout the cell cycle (Lei and Tye, 2001). Although we do not favor the idea, our data do not exclude a continuous loading of MCMs at telomeres through S-phase or enrichment of MCMs via arriving replication forks. In any case we can make the minimal conclusion that considerable amounts of Mcm5p are present at telomeres through the late stages of S phase. Regardless of the mechanism, this pattern is in stark contrast to the loading and displacement of MCM proteins at origins (Figure 6A). It is notable that the two mutants have extended S phase (Rehman et al., 2006), providing opportunity for longer action of the antisilencing activity of Mcm5p.
DISCUSSION
Subtelomeric ACS Convert to Antisilencers in Replication Factors Mutants
ACS represents the essential core of all origins of DNA replication (Breier et al., 2004) and of the silencers at the mating type loci (Rusche et al., 2003). At telomeres, ACS operate as weaker proto-silencers, which relay/enhance the repression signals emitted by the telomere, whereas their destruction causes modest decrease in silencing (Fourel et al., 2002a). At telomeres, we also find other regulatory units such as antisilencers and chromatin insulators (Fourel et al., 2004). A peculiar feature of gene expression at telomeres is its metastable character, meaning that subtelomeric genes switch at low frequency from complete repression to full expression (Rusche et al., 2003). It is not clear how this meta-stability persists and why the combination of a strong silencer (the telomere) and proto-silencers (ACS, Abf1p-, and Rap1p-binding sites) can successfully establish, but then fail to maintain the repressed state. One can speculate that some of these repressor elements temporarily reduce their strength or alternatively, antisilencer elements temporary increase their activity to allow for the reversal.
In this article, we analyze the role of ACS in telomeric silencing and show that their destruction increases repression in orc2-1, orc5-1, mcm5-461, and cdc45-1 mutants (Figure 2), whereas in the corresponding wild-type strains it leads to modest derepression (Figure 2). Similar increase in repression was observed when the adjacent B1 element was eliminated (Figure 3). A straightforward interpretation of these data are that within these mutants, the subtelomeric B1/ACS elements have antisilencing activity. These effects are concurrent with the overall decrease in silencing in the mutants.
Are the reported effects indeed mediated by ACS? First, we point out that the observed conversion cannot be a mere consequence of global derepression in the mutants. For example, the cdc6-1, cdc7-1, and Δsir2 mutations caused significant reduction in telomeric silencing (Ramachandran et al., 2006; Rehman et al., 2006) but did not alter the nature of ACS (Figure 2). Second, we can reasonably assume that the orc2-1 and orc5-1 mutations as well as the other mutations do not preclude association of ORC with ACS, otherwise they would be lethal. Therefore, we favor the interpretation that in each specific genetic context our data represents the roles of ACS and B1 as occupied by ORC. Third, although it is conceivable that some of the observed effects in Figures 2 and 3 are influenced by the different genomic environment in ARS1 and in core X, our calculations (ACS−/ACS; B1−/B1) are heavily geared toward detecting the effects mediated by ACS and B1. Finally, in orc2-1 and orc5-1, we (Figure 2) and others (Palacios DeBeer and Fox, 1999; Pryde and Louis, 1999; Lebrun et al., 2001; Fourel et al., 2002b) have observed very strong de-repression uncharacteristic for other mutants. Hence, we do not exclude the possibility that the strong effects in Figure 2 are consequence of the combination of the gene mutations and the ablation of ACS and do not represent ACS-driven effects only.
A very recent article (Chang et al., 2008) mapped the essential replication activity of HMR-E to two overlapping 9/11 ACS matches, but not to the 11/11 ACS match of the silencer (ARS317). Is it then possible that the antisilencing activity at telomeres is mediated by near-ACS matches? We see no such overlapping near-ACS matches at the telomere whereas the individual near-ACS matches are eight of 11 at best. At the same time, mutation of the B1 site next to the 11/11 ACS match did not have strong effect on the replication activity of HMR-E (ARS317) and the subtelomeric ARS319 (Chang et al., 2008) but have profound effect on silencing at HMR-E (Palacios DeBeer et al., 2003) and at the telomere (our findings). These observations provide circumstantial support to the notion that the B1/ACS element rather than near-ACS matches are responsible for the silencing effects at these positions.
The conclusion that subtelomeric ACS act as an antisilencers in certain replication factor mutants challenges many earlier studies, which consistently portray ACS as silencers or proto-silencers (Fourel et al., 2002a; Rusche et al., 2003). Yet, our observations do not stand alone. A systematic study on the silencing of a reporter along the VII-L subtelomere has paradoxically shown that orc2-1 and orc5-1 cause significant derepression at most subtelomeric positions but have almost no effect in the vicinity of ACS (Pryde and Louis, 1999). In addition, similarly to the data presented here, the destruction of ACS in the HMR-E silencer in orc2-1 actually increased repression (Palacios DeBeer and Fox, 1999). We extend these observations and propose that subtelomeric ACS have weak and/or short-lived antisilencing activity, which is somehow revealed in replication factor mutants.
Functional Differences between Subtelomeric and Origin B1/ACS
In Figures 2 and 3, we show that core X-derived B1/ACS, but not origin-derived B1/ACS convert to antisilencers in the tested mutants. At the same time, the destruction of B1 does not affect the proto-silencing properties of ARS1, whereas its counterpart in core X is as important as ACS (Figures 2 and 3). It should be noted that the B1 element in ARS1 has a significant albeit not essential contribution to the origin activity of ARS1 (Marahrens and Stillman, 1992; Rao and Stillman, 1995). Incontrast, mutations in B1of HMR-E make this element a poor silencer but a good origin (Palacios DeBeer and Fox, 1999; Palacios DeBeer et al., 2003). Furthermore, the silencing, but not the replication strength of several B1/ACS combinations was correlated to their high affinity to ORC (Palacios DeBeer et al., 2003).
It is important that the B1 elements of core X (Figure 3) and B1 in HMR-E (Palacios DeBeer et al., 2003) but not B1 in ARS1 (Figure 2) were identified as contributors to silencing. Even more, the sequence of the core X-B1 (Figure 1) is related to B1 of HMR-E and synthetic silencers rather than to that of several origins of replication (Figure 1 in Palacios DeBeer et al., 2003). We anticipate the coreX-B1/ACS element to have high-affinity to ORC. Notwithstanding the affinity to ORC, it seems that B1 elements can distinguish between replicator and silencer types of ACS. There are ∼10,000 ACS matches or near matches in the yeast genome (Breier et al., 2004), but only a few hundred of them have been functionally assigned as origins and only several as silencers/proto-silencers. We propose that the adjacent B1 elements can predict the functions of ACS, which are not experimentally accessible at the threshold of genome-wide studies.
How Can ACS Be Proto-Silencer and Antisilencer?
Many of the proteins, which work in both replication and silencing, bind ACS or regulate the action of ACS-bound proteins (Rusche et al., 2003). In DNA replication, ACS is a focal point of a rigorously regulated protein complex, which controls origin licensing and firing (Rusche et al., 2003). It would be surprising if the same ACS-bound proteins in core X are spared from similar control and do not undergo modifications during G1 and S phases. At this point, it is not clear how B1 elements (and affinity to ORC) affect these modifications. Still, it is possible that such modifications temporarily unleash antirepression activity to balance the repression signals emitted by other ACS-bound factors. In tune with this idea, we have observed that mutations in CDC7 (a protein kinase) modulate the silencing properties of proteins tethered to the telomere (Supplemental Figure 2). If alterations in ACS-bound proteins can temporarily convert these complexes to antisilencers, then mutations in them can unduly unleash antisilencing or, being also mutations in replication factor, can simply extend S phase thus prolonging the time of the ACS-driven antirepression. The net effect will lead to the observations in Figure 3.
Is Mcm5p an Antisilencing Module on ACS-bound Complexes?
We show that the Mcm5p is as strong antisilencer as the histone acetyl transferase Gcn5p, whereas other ACS associating proteins (Orc1p, Orc2p, Mcm3p, and Mcm10p) are silencers (Figures 4 and 5). Although we consider the artificial nature of our tethering assay, we stress that it recapitulated well documented effects of Gcn5p, Orc1p, Orc2p, and Mcm10p detected by similar or other assays (Rusche et al., 2003; Jacobson and Pillus, 2004; Liachko and Tye, 2005). It is noteworthy that the antisilencing activity of Mcm5p is dependent on GCN5 and HAT1 (Figure 4C). At this point, it is not clear how these two histone-acetyl-transferases communicate with Mcm5p and whether their communication is modulated by modification of the MCM complex for example by phosphorylation. Regardless, the current observations support the idea that Mcm5p could indeed be involved in antisilencing.
Equally importantly, we show that unlike origins of replication, core X associates with Mcm5p later in S phase (Figure 6B). Although we do not observe any dramatic differences in this occupation in two of the analyzed mutants, we still believe that this peculiar dissimilarity to origins provides clues on how Mcm5p may exhibit stronger antisilencing in replication factor mutants. In addition, the Mcm5p antisilencing activity is not affected by the amino acid substitutions in mcm5-461 and mcm5-1, whereas these two mutations lead to telomeric derepression (Dziak et al., 2003). We postulate that the mcm5-461, mcm5-1, and other mutations probably boost antisilencing by prolonging S phase (Rehman et al., 2006) rather than by affecting the Mcm5p antisilencing activity itself.
In conclusion, we propose that there is a dynamic interplay of weak antirepression (Mcm5p) and stronger repression (Orc1p, Orc2p, Mcm3p, and Mcm10p) signals on telomeric ACS. Mutations, which prolong the time of manifestation or boost these antisilencing activities by other means can in turn convert ACS from proto-silencers to antisilencers.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Rine, B. Stillman, S. Jacobson and I. Liachko for the generous donation of strains and plasmids. This study was supported by a grant from Natural Ssciences Engineering and Research Council-Canada (to K. Y.) and a grant from the Ligue Nationale Contree le Cancer-France (to E. G.).
Abbreviations used:
- ACS
ARS consensus sequence
- FOA
fluoroorotic acid
- MCM
minichromosome maintenance
- ORC
origin recognition complex
- STAR
subtelomeric antisilencing region
- TPE
telomeric position effect.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-01-0099) on November 12, 2008.
REFERENCES
- Aparicio O. M., Billington B. L., Gottschling D. E. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Babiarz J. E., Halley J. E., Rine J. Telomeric heterochromatin boundaries require NuA4-dependent acetylation of histone variant H2A.Z in Saccharomyces cerevisiae. Genes Dev. 2006;20:700–710. doi: 10.1101/gad.1386306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell S. P., Stillman B. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature. 1992;357:128–134. doi: 10.1038/357128a0. [DOI] [PubMed] [Google Scholar]
- Boscheron C., Maillet L., Marcand S., Tsai-Pflugfelder M., Gasser S. M., Gilson E. Cooperation at a distance between silencers and proto-silencers at the yeast HML locus. EMBO J. 1996;15:2184–2195. [PMC free article] [PubMed] [Google Scholar]
- Boulton S. J., Jackson S. P. Components of the Ku-dependent non-homologous end-joining pathway are involved in telomeric length maintenance and telomeric silencing. EMBO J. 1998;17:1819–1828. doi: 10.1093/emboj/17.6.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breier A. M., Chatterji S., Cozzarelli N. R. Prediction of Saccharomyces cerevisiae replication origins. Genome Biol. 2004;5:R22. doi: 10.1186/gb-2004-5-4-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. R., Wu C. S., Hom Y., Gartenberg M. R. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 2005;19:3031–3042. doi: 10.1101/gad.1356305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Theis J. F., Miller J., Nieduszynski C. A., Newlon C. S., Weinreich M. Analysis of chromosome III replicators reveals an unusual structure for the ARS318 silencer origin and a conserved WTW sequence within the origin recognition complex binding site. Mol. Cell Biol. 2008;28:5071–5081. doi: 10.1128/MCB.00206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke A. S., Samal E., Pillus L. Distinct roles for the essential MYST family HAT Esa1p in transcriptional silencing. Mol. Biol. Cell. 2006;17:1744–1757. doi: 10.1091/mbc.E05-07-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley J. F., Cocker J. H. Protein-DNA interactions at a yeast replication origin. Nature. 1992;357:169–172. doi: 10.1038/357169a0. [DOI] [PubMed] [Google Scholar]
- Dillin A., Rine J. Separable functions of ORC5 in replication initiation and silencing in Saccharomyces cerevisiae. Genetics. 1997;147:1053–1062. doi: 10.1093/genetics/147.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey D. D., Davis L. R., Greenfeder S. A., Ong L. Y., Zhu J. G., Broach J. R., Newlon C. S., Huberman J. A. Evidence suggesting that the ARS elements associated with silencers of the yeast mating-type locus HML do not function as chromosomal DNA replication origins. Mol. Cell Biol. 1991;11:5346–5355. doi: 10.1128/mcb.11.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dziak R., Leishman D., Radovic M., Tye B. K., Yankulov K. Evidence for a role of MCM (mini-chromosome maintenance)5 in transcriptional repression of sub-telomeric and Ty-proximal genes in Saccharomyces cerevisiae. J. Biol. Chem. 2003;278:27372–27381. doi: 10.1074/jbc.M301110200. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Gossen M., Pak D. T., Botchan M. R., Rine J. Separation of origin recognition complex functions by cross-species complementation. Science. 1995;270:1671–1674. doi: 10.1126/science.270.5242.1671. [DOI] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Kamakaka R. T., Rine J. A role for the replication proteins PCNA, RF-C, polymerase epsilon and Cdc45 in transcriptional silencing in Saccharomyces cerevisiae. Genetics. 1999;153:1171–1182. doi: 10.1093/genetics/153.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenhofer-Murray A. E., Rivier D. H., Rine J. The role of Sas2, an acetyltransferase homologue of Saccharomyces cerevisiae, in silencing and ORC function. Genetics. 1997;145:923–934. doi: 10.1093/genetics/145.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher T. S., Zakian V. A. Ku: a multifunctional protein involved in telomere maintenance. DNA Repair. 2005;4:1215–1226. doi: 10.1016/j.dnarep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Fourel G., Lebrun E., Gilson E. Protosilencers as building blocks for heterochromatin. Bioessays. 2002a;24:828–835. doi: 10.1002/bies.10139. [DOI] [PubMed] [Google Scholar]
- Fourel G., Magdinier F., Gilson E. Insulator dynamics and the setting of chromatin domains. Bioessays. 2004;26:523–532. doi: 10.1002/bies.20028. [DOI] [PubMed] [Google Scholar]
- Fourel G., Miyake T., Defossez P. A., Li R., Gilson E. General regulatory factors (GRFs) as genome partitioners. J. Biol. Chem. 2002b;277:41736–41743. doi: 10.1074/jbc.M202578200. [DOI] [PubMed] [Google Scholar]
- Fourel G., Revardel E., Koering C. E., Gilson E. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 1999;18:2522–2537. doi: 10.1093/emboj/18.9.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox C. A., Ehrenhofer-Murray A. E., Loo S., Rine J. The origin recognition complex, SIR1, and the S phase requirement for silencing. Science. 1997;276:1547–1551. doi: 10.1126/science.276.5318.1547. [DOI] [PubMed] [Google Scholar]
- Gauthier L., Dziak R., Kramer D. J., Leishman D., Song X., Ho J., Radovic M., Bentley D., Yankulov K. The role of the carboxyterminal domain of RNA polymerase II in regulating origins of DNA replication in Saccharomyces cerevisiae. Genetics. 2002;162:1117–1129. doi: 10.1093/genetics/162.3.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy C. F., Dryga O., Seematter S., Pahl P. M., Sclafani R. A. mcm5/cdc46-bob1 bypasses the requirement for the S phase activator Cdc7p. Proc. Natl. Acad. Sci. USA. 1997;94:3151–3155. doi: 10.1073/pnas.94.7.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irlbacher H., Franke J., Manke T., Vingron M., Ehrenhofer-Murray A. E. Control of replication initiation and heterochromatin formation in Saccharomyces cerevisiae by a regulator of meiotic gene expression. Genes Dev. 2005;19:1811–1822. doi: 10.1101/gad.334805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii K., Arib G., Lin C., Van Houwe G., Laemmli U. K. Chromatin boundaries in budding yeast: the nuclear pore connection. Cell. 2002;109:551–562. doi: 10.1016/s0092-8674(02)00756-0. [DOI] [PubMed] [Google Scholar]
- Jacobson S., Pillus L. Molecular requirements for gene expression mediated by targeted histone acetyltransferases. Mol. Cell Biol. 2004;24:6029–6039. doi: 10.1128/MCB.24.13.6029-6039.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchmaier A. L., Rine J. DNA replication-independent silencing in. S. cerevisiae. Science. 2001;291:646–650. doi: 10.1126/science.291.5504.646. [DOI] [PubMed] [Google Scholar]
- Kurdistani S. K., Grunstein M. In vivo protein-protein and protein-DNA crosslinking for genomewide binding microarray. Methods. 2003;31:90–95. doi: 10.1016/s1046-2023(03)00092-6. [DOI] [PubMed] [Google Scholar]
- Laroche T., Martin S. G., Gotta M., Gorham H. C., Pryde F. E., Louis E. J., Gasser S. M. Mutation of yeast Ku genes disrupts the subnuclear organization of telomeres. Curr. Biol. 1998;8:653–656. doi: 10.1016/s0960-9822(98)70252-0. [DOI] [PubMed] [Google Scholar]
- Lebrun E., Revardel E., Boscheron C., Li R., Gilson E., Fourel G. Protosilencers in Saccharomyces cerevisiae subtelomeric regions. Genetics. 2001;158:167–176. doi: 10.1093/genetics/158.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D. G., Bell S. P. Architecture of the yeast origin recognition complex bound to origins of DNA replication. Mol. Cell Biol. 1997;17:7159–7168. doi: 10.1128/mcb.17.12.7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M., Tye B. K. Initiating DNA synthesis: from recruiting to activating the MCM complex. J. Cell Sci. 2001;114:1447–1454. doi: 10.1242/jcs.114.8.1447. [DOI] [PubMed] [Google Scholar]
- Li Y. C., Cheng T. H., Gartenberg M. R. Establishment of transcriptional silencing in the absence of DNA replication. Science. 2001;291:650–653. doi: 10.1126/science.291.5504.650. [DOI] [PubMed] [Google Scholar]
- Liachko I., Tye B. K. Mcm10 is required for the maintenance of transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2005;171:503–515. doi: 10.1534/genetics.105.042333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marahrens Y., Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- Marsellach F. X., Huertas D., Azorin F. The multi-KH-domain protein of Saccharomyces cerevisiae Scp160p contributes to the regulation of telomeric silencing. J. Biol. Chem. 2006 doi: 10.1074/jbc.M601671200. [DOI] [PubMed] [Google Scholar]
- Miller A. M., Nasmyth K. A. Role of DNA replication in the repression of silent mating type loci in yeast. Nature. 1984;312:247–251. doi: 10.1038/312247a0. [DOI] [PubMed] [Google Scholar]
- Mueller J. E., Canze M., Bryk M. The requirement for COMPASS and Paf1 in transcriptional silencing and methylation of histone H3 in Saccharomyces cerevisiae. Genetics. 2006 doi: 10.1534/genetics.106.055400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios DeBeer M. A., Fox C. A. A role for a replicator dominance mechanism in silencing. EMBO J. 1999;18:3808–3819. doi: 10.1093/emboj/18.13.3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios DeBeer M. A., Muller U., Fox C. A. Differential DNA affinity specifies roles for the origin recognition complex in budding yeast heterochromatin. Genes Dev. 2003;17:1817–1822. doi: 10.1101/gad.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryde F. E., Louis E. J. Limitations of silencing at native yeast telomeres. EMBO J. 1999;18:2538–2550. doi: 10.1093/emboj/18.9.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran L., et al. Evidence for ORC-dependent repression of budding yeast genes induced by starvation and other stresses. FEMS Yeast Res. 2006;6:763–776. doi: 10.1111/j.1567-1364.2006.00077.x. [DOI] [PubMed] [Google Scholar]
- Rao H., Stillman B. The origin recognition complex interacts with a bipartite DNA binding site within yeast replicators. Proc. Natl. Acad. Sci. USA. 1995;92:2224–2228. doi: 10.1073/pnas.92.6.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehman M. A., Fourel G., Mathews A., Ramdin D., Espinosa M., Gilson E., Yankulov K. Differential requirement of DNA replication factors for subtelomeric ARS consensus sequence protosilencers in Saccharomyces cerevisiae. Genetics. 2006;174:1801–1810. doi: 10.1534/genetics.106.063446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N., Runge K. W. Two paralogs involved in transcriptional silencing that antagonistically control yeast life span. Curr. Biol. 2000;10:111–114. doi: 10.1016/s0960-9822(00)00298-0. [DOI] [PubMed] [Google Scholar]
- Rusche L. N., Kirchmaier A. L., Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Suter B., Tong A., Chang M., Yu L., Brown G. W., Boone C., Rine J. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics. 2004;167:579–591. doi: 10.1534/genetics.103.024851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamburini B. A., Carson J. J., Linger J. G., Tyler J. K. Dominant mutants of the Saccharomyces cerevisiae ASF1 histone chaperone bypass the need for CAF-1 in transcriptional silencing by altering histone and sir protein recruitment. Genetics. 2006;173:599–610. doi: 10.1534/genetics.105.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham W. H., Zakian V. A. Transcriptional silencing at Saccharomyces telomeres: implications for other organisms. Oncogene. 2002;21:512–521. doi: 10.1038/sj.onc.1205078. [DOI] [PubMed] [Google Scholar]
- Wyrick J. J., Aparicio J. G., Chen T., Barnett J. D., Jennings E. G., Young R. A., Bell S. P., Aparicio O. M. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- Zou Y., Yu Q., Bi X. Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell Biol. 2006a;26:7806–7819. doi: 10.1128/MCB.01197-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Yu Q., Chiu Y. H., Bi X. Position effect on the directionality of silencer function in Saccharomyces cerevisiae. Genetics. 2006b;174:203–213. doi: 10.1534/genetics.106.055525. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.