Abstract
Modification by acetylation occurs at ɛ-amino lysine residues of histones and transcription factors. Unlike phosphorylation, a direct link between transcription factor acetylation and cellular growth or apoptosis has not been established. We show that the nuclear androgen receptor (AR), a DNA-binding transcriptional regulator, is acetylated in vivo. The acetylation of the AR is induced by ligand dihydrotestosterone and by histone deacetylase (HDAC) inhibitors in living cells. Direct AR acetylation augmented p300 binding in vitro. Constructs mimicking neutral polar substitution acetylation (ARK630Q, ARK630T) enhanced p300 binding and reduced N-CoR/HDAC/Smad3 corepressor binding, whereas charged residue substitution (ARK630R) reduced p300 binding and enhanced corepressor binding. The AR acetylation mimics promoted cell survival and growth of prostate cancer cells in soft agar and in nude mice and augmented transcription of a subset of growth control target gene promoters. Thus, transcription factor acetylation regulates coactivator/corepressor complex binding, altering expression of specific growth control genes to promote aberrant cellular growth in vivo.
Prostate cancer is the second leading cause of cancer death in American males. Although potentially curable by radical prostatectomy or radiation therapy, metastatic disease is common at presentation and may occur subsequently in patients treated with curative intent. The androgen receptor (AR) is a classical nuclear receptor (NR) that binds testosterone and is required for the induction of male secondary sexual characteristics. The AR conveys several dissociable functions, including transactivation, transrepression, growth regulation, basal activity, and context-dependent cell survival or apoptosis functions. Aberrant AR function plays an important role in prostate cancer (1). The wild-type AR can induce cellular differentiation or cellular apoptosis in prostate cancer cells (2, 16, 47). Both AR-dependent and AR-independent mechanisms contribute to prostate cancer cellular growth. Somatic missense AR gene mutations have been detected in prostate cancer cell lines, xenografts, and primary and metastatic forms of prostate cancer (39-41).
The NR superfamily, of which the AR is a member, are transcriptional regulators that coordinate important metabolic and differentiation functions (29, 42). Transcriptional activity of NR is regulated by ligand-dependent recruitment of coactivator/corepressor proteins. In the presence of ligand, the most carboxyl-terminal helix 12 (H12) of NR folds over the ligand-binding hydrophobic pocket, creating structural surfaces that bind the basal transcriptional apparatus and recruit coactivators required for efficient transactivation (29, 42). Corepressor function involves interaction with SIN3 and a histone deacetylase (HDAC) function (3, 15, 30). Several findings suggest a role for corepressor proteins with HDAC activity in at least one component of the AR's functions. Liganded AR transcriptional activity is induced by trichostatin A (TSA) (7), the HDAC binding protein Smad3 inhibited liganded AR activity (7, 8, 13), HDAC1 bound to the AR in vivo, and HDAC1 binding to the AR was dissociated by the ligand dihydrotestosterone (DHT) (7). In the presence of hormone, corepressors dissociate from the NR. As corepressor and coactivators show substantial overlap in their site of binding to the NR (19), coactivator recruitment may be contingent upon corepressor disengagement (29, 42).
The cointegrator proteins p300 and CBP augment NR activity through several functions. They recruit coactivators and serve as a molecular bridge between the NR and the basal transcription apparatus. The cointegrator proteins p300/CBP augment NR activity, in part related to their intrinsic histone acetyltransferase (HAT) activity (31, 37, 38). The intrinsic HAT activity acetylates both histones and nonhistone proteins to regulate their activity (37). Histone acetylation may contribute to nucleosome destabilization, facilitating transcription factor binding to specific target DNA sequences in the promoter region of a target gene (31, 38). Direct acetylation of lysine residues in either histone or transcription factors neutralizes the residue's positive charge and increases its polarity, events that are mimicked by a glutamine substitution. The AR is directly acetylated, and mutation of the residues acetylated in vitro abrogates acetylation of the full-length AR in living cells (7). Although loss-of-function mutation of transcription factor lysine residues implicate acetylation in transcription factor activity, the physiologic effect of transcription factor acetylation in regulating cellular growth remains to be determined (4, 45). The current studies show for the first time that AR transcription factor acetylation regulates resistance to therapeutic agents and determines prostate cell growth in vivo.
MATERIALS AND METHODS
Reporter genes and expression vectors.
The androgen-responsive synthetic reporter constructions MMTV-Luc and PSA-Luc, cyclin D1-Luc, cyclin E-Luc, p21Cip1-Luc, cdc25A-Luc and the expression vectors pCMVHA-p300, HA-TIP60, TIP60ΔHAT, pSG5SRC-1a, pSG5ARA70, pSG5ARA55, and hARwt (pcDNA3) were described previously (7, 8, 36). ARK630Q and ARK630T were derived by PCR-directed amplification and cloned into pcDNA3. The expression plasmids encoding pFlag-CMV-2-Ubc-9, Flag-Smad3, Smad3ΔC, pCEP4-MEKK1wt, pCMX-FlagN-CoR, pCMV-HDAC1, and pCMV-EGFP were described previously (7, 8).
Cell culture, DNA transfection, chemicals, and luciferase assays.
The reporter assays, cell culture, DNA transfection, and luciferase assays were performed as previously described (7). The prostate cancer cell line DU145 and the 293 T-cell line were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin. Cells were incubated in medium containing 10% charcoal-stripped fetal bovine serum prior to experimentation with dihydrotestosterone (DHT) (7). Statistical analyses was performed with the Mann-Whitney U test.
Apoptosis, colony formation, soft agar assays, and nude mouse tumor implantation.
Apoptosis was detected by morphological analysis of green fluorescent protein (GFP)-transfected cells (7, 8). At least 200 cells were counted with a fluorescent microscope, and cells were scored for blebbing and chromatin condensation by an investigator blinded to the experimental condition. Stable cell lines were assessed for cellular proliferation and colony formation. We seeded 2 × 105 DU145 cells into six-well plates and transfected them 24 h later with 2.5 μg of wild-type AR, AR acetylation mutants, or the control vector, pcDNA3. Forty-eight hours after transfection, the cells were seeded into a 15-cm plate, and G418 (2 mg/ml) was included in the medium for selection of the transfected cells. Single colonies were harvested on day 14 and maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 500 μg of G418 per ml.
AR expression was determined by Western blotting, and three independent colonies for wild-type AR and each of the AR mutants were selected. Anchorage-independent growth of stable cell lines was assayed by soft agar growth and in nude mice. Apoptosis was determined within tumors of similar volumes from nude mice assessed by terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) staining (ApopTag Red in situ apoptosis detection kit; Intergene, Purchase, N.Y.), evaluating five fields at 40×, counting at least 400 cells per field. The tetrazolium salt (MTT) assay was performed as previously described (8).
Western blots, immunoprecipitation, and immunoprecipitation-HAT assays.
Antibodies used for Western blot analysis and immunoprecipitation as previously described include anti-AR (N-20), anti-p300 (Santa Cruz), anti-M2-Flag (Sigma), anti-acetyl-lysine (Upstate Biotech), anti-acetyl-lysine motif p53320 (12, 17, 27, 28), anti-HDAC1 (Upstate Biotech), and guanine disassociation inhibitor (GDI) as a protein-loading control (36). Immunoprecipitation-Western blot analysis was performed with DU145 stable cell lines expressing the wild-type AR or 293T cells transfected with pcDNA3AR, pcDNA3ARK630Q, pcDNA3ARK630T, pCMV-HA-p300, pCMX-FlagN-CoR, or the expression vector control (7, 8, 36). The DU145ARwt stable cells were treated with vehicle (ethanol) or DHT (10 nM) for 12 h, TSA (1 μM) or suberoylanilide hydroxamic acid (SAHA, 5 μM; Aton Pharma Inc.) for 6 h. The cell lysates were harvested and subjected to immunoprecipitation as shown in the figures.
For immunoprecipitation with anti-acetyl-p53320, the cell lysates were precleared with anti-p53 antibody (p53 Pab240; Santa Cruz Biotech), and the supernatants were subjected to immunoprecipitation with anti-acetyl-p53320. In vitro acetylation assays were performed with baculovirus-purified p300 as a source for HAT and AR peptides as previously described (7). Glutathione S-transferase (GST) pull-down experiments with the acetylated AR and p300 were conducted as described (33). The AR fusion protein was expressed from GST-AR624-676 as it is efficiently acetylated by p300 (7), incubated with 1 mM acetyl-coenzyme A, and pull-down was conducted with p300. The Western blotting for the AR associated with p300 was conducted with the GST antibody.
In vivo acetylation.
Murine hepatocellular lysate were extracted in cell lysis buffer (20 mM HEPES [pH 7.5], 0.1 M KCl, 0.4 mM EDTA, 0.2% NP-40, 10 mM β-mercaptoethanol, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg of pepstatin per ml, 1 μg of NaVO4 per ml, 5 mM sodium butyrate, 50 μM TSA) and immunoprecipitated with normal rabbit IgG, anti-AR, or anti-acetyl-lysine antibody. The immunoprecipitate was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and blotted with either anti-AR or anti-acetyl-lysine antibodies (7).
RESULTS
AR acetylation site governs ligand sensitivity and specificity.
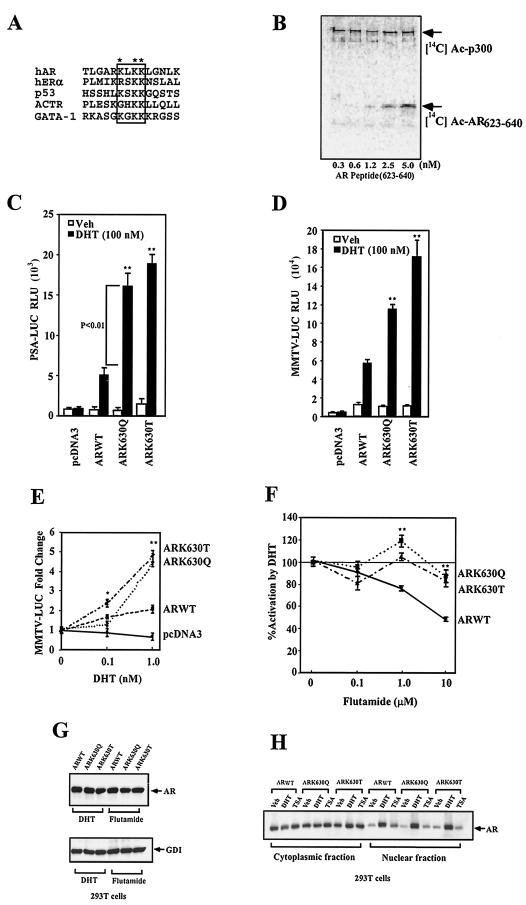
The AR is acetylated in cultured cells, requiring an acetylation motif (KxKK) which is conserved with other NR and with the tumor suppressor p53 (7, 37) (Fig. 1A ). A peptide containing the AR acetylation motif was sufficient for acetylation by p300 (Fig. 1B). Of the three lysine residues within the motif, we chose to study ARLys630 in more detail. An AR lysine (Lys630-Ala) substitution was most defective in ligand-induced transactivation and abrogated AR acetylation in cultured cells (7). The acetylated ARK630 residue was replaced with glutamine (Lys630-Glu) to create an acetylation mimic. Since acetylation of lysine neutralizes its positive charge and increases its hydrophobicity, comparison was made to an AR containing a polar uncharged threonine residue substitution of ARK630, a somatic mutation that occurs in prostate cancer patients.
FIG.1.
AR acetylation site regulates ligand sensitivity and specificity. (A) The AR acetylation motif indicating lysine 630 (*), with homology shown to the acetylation motif of human p53 and ACTR proteins. (B) In vitro histone acetyltransferase (HAT) assays used wild-type AR peptides incubated with baculovirus-purified p300 and [14C]acetyl coenzyme A. (C) Activity of the PSA-Luc and (D) MMTV-Luc reporters was assessed in DU145 cells with the wild-type and mutant ARs and is shown as relative luciferase activity (mean ± standard error of the mean for n > 6 separate transfections). (E) The expression plasmids encoding wild-type AR or AR mutants were transfected and examined for DHT dose responsiveness in DU145 cells with the MMTV-Luc reporter. Cells were treated with either vehicle or DHT (24 h). (F) The wild-type AR and AR acetylation site mutants were assessed for responsiveness to flutamide with induction shown compared to equal amounts of the empty expression vector cassette (pSG5). The data are means ± standard error of the mean for n > 9. (G) The wild-type AR and AR acetylation site mutants were transfected into 293T cells, treated with 100 nM DHT or 10 nM flutamide for 24 h. The cell lysates were then subjected to Western blotting with the antibodies to the AR or GDI as the protein-loading control. (H) The wild-type AR and AR acetylation site mutants were transfected into 293T cells, treated either with vehicle, DHT (100 nM), or TSA (100 nM) for 24 h. Western blotting of the nuclear and cytoplasmic fractions is shown.
Transcriptional activity of the ARK630Q was greater than that of wild-type AR with either the androgen-responsive prostate-specific antigen gene (PSA) promoter (wild-type AR 4.5-fold, ARK630Q 10-fold, and ARK630T 15-fold) (Fig. 1C) or androgen-responsive mouse mammary tumor virus (MMTV)-luciferase (Luc) reporter gene (wild-type AR 4.3-fold, ARK630Q10-fold, and ARK630T 14-fold; P < 0.01) (Fig. 1D). The AR acetylation site mimic was more active at lower DHT concentrations than the wild-type AR (Fig. 1E) (at 0.1 nM DHT, wild-type AR 1.5-fold, ARK630T 2.5-fold; at 1 nM DHT, wild-type AR 1.8-fold, ARK630Q 4.5-fold, and ARK630T 5-fold). The DHT antagonist flutamide antagonized DHT-induced wild-type AR activity; however, the AR acetylation mimic was relatively resistant (Fig. 1F).
In order to determine the mechanism of enhanced ligand-dependent transactivation by ARK630Q and ARK630T of the androgen-responsive reporter genes, we examined the relative abundance of the AR wild-type and acetylation mutants. In AR-deficient 293T cells, transient transfection of the AR expression vector and subsequent Western blot analysis demonstrated that the relative abundance of AR was similar for the AR wild type and the acetylation mutant in the presence of DHT or flutamide (Fig. 1G). In addition, the relative distribution of the wild type and acetylation mimic mutant AR between the nuclear and cytoplasmic fractions was similar in the presence of either DHT or TSA (Fig. 1H). Together, these studies suggest that the AR acetylation mutants are expressed similarly to wild-type AR in cultured cells and are subcellularly distributed like the AR wild type.
AR is acetylated in vivo.
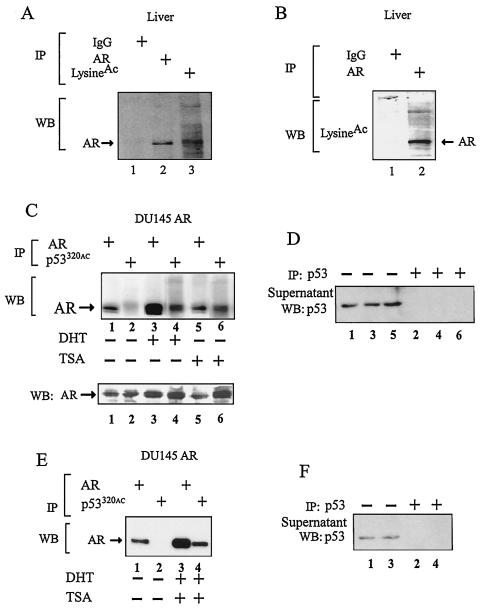
To determine whether the AR is acetylated in living cells in vivo, immunoprecipitation of murine hepatic tissue was performed with antibodies to either the AR or acetylated lysine residues, with subsequent Western blotting for the AR. The anti-acetyl-lysine antibody efficiently precipitated the AR (Fig. 2A). In the reciprocal immunoprecipitation with the AR antibody, Western blotting with the anti-acetyl-lysine antibody demonstrated AR immunoreactivity (Fig. 2B). In view of the conservation of the AR acetylated lysine motif with the acetylated p53 lysine motif (7), we used previously well characterized antibodies generated to an acetylated lysine motif peptide of p53 (27) in immunoprecipitation studies of stable DU145-AR cell lines. Subsequent to clearing of p53, the anti-acetyl-lysine-p53 antibody immunoprecipitate was subjected to Western blot for coprecipitated AR (Fig. 2C). The amount of AR precipitated by antibody to the acetylated lysine motif was increased by the addition of either DHT or TSA (Fig. 2C, lane 2 versus 4 and 6).
FIG. 2.
AR acetylation in vivo. (A) The cell lysate was extracted from wild-type male liver and subjected to immunoprecipitation (IP) with either normal rabbit IgG, anti-AR, or anti-acetyl-lysine antibody. The immunoprecipitate was separated by SDS-PAGE and blotted with anti-AR antibody (Upstate Biotech). (B) The mouse liver lysate was also immunoprecipitated with normal rabbit IgG or anti-AR antibody and blotted with anti-acetyl-lysine antibody. (C, E) The DU145ARwt stable cell lines were treated with vehicle (ethanol) or DHT (100 nM) for 12 h and then treated with TSA (30 nM) for 6 h. Immunoprecipitation was done with anti-AR or anti-acetyl-lysine peptide (p53320) as indicated. Prior to immunoprecipitation with anti-acetyl-p53320, cell lysates were first cleared with an anti-p53 antibody (anti-p53[1-393]; sc-4246; Santa Cruz Biotech). The immunoprecipitate was resolved by SDS-PAGE and blotted with anti-AR antibody. (D, F) Western blot (WB) of cell lysates precleared with anti-p53 antibody (sc-4246).
Western blotting for AR expression in the DU145-AR wild-type stable cell line demonstrated a DHT-dependent increase in the amount of AR (Fig. 2C, bottom panel, Western blot AR lane 2 versus 4). The approximately twofold increase in the abundance of AR by Western blotting suggests that the increase in the amount of acetylated AR that was immunoprecipitated in the presence of DHT may be due to an increase in both absolute and relative amounts of AR in the presence of DHT. The preclearing by immunoprecipitation with p53 was saturating (Fig. 2D, lanes 1, 3, and 5 versus 2, 4, and 6).
The addition of DHT with TSA also increased the amount of AR coprecipitated with the anti-acetyl-lysine motif antibody (Fig. 2E, lane 2 versus 4). Again, the preclearing with p53 was saturating (Fig. 2F, lanes 1 and 3 versus 2 and 4). Together, these studies demonstrate that the AR is acetylated in vivo and that, in prostate cancer cells, the addition of DHT or TSA increases the amount of acetylated AR.
AR coactivation is acetylation site dependent.
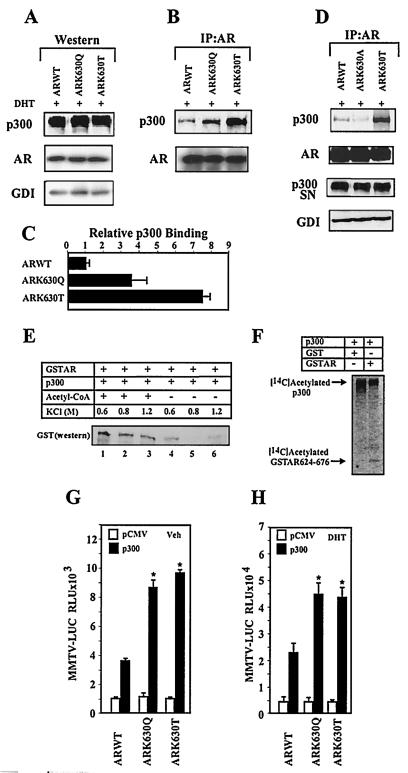
Acetylated lysine residues form a docking site recognized by bromodomain-containing proteins (21, 24, 33). The coactivator proteins p300/CBP contain a bromodomain, and an 18-amino-acid polypeptide containing the AR acetylation site was sufficient for binding and acetylation by either p300 (Fig. 1A, B) or p300/CBP-associated factor (P/CAF) (7). The binding of p300 to wild-type AR and the AR mutants was compared by immunoprecipitation-Western blot analysis in 293T cells transfected with expression vectors for p300 and either wild-type AR or the AR mutants. After 24 h of DHT treatment, the cell lysates were subjected to either Western blotting (Fig. 3A) or immunoprecipitation-Western blotting (Fig. 3B).
FIG. 3.
Direct AR acetylation enhances p300 binding. (A, B, and D) Extracts from cells transfected with the AR expression vectors were subjected to Western blot analysis (A) or AR immunoprecipitation Western blotting (B and D) to detect p300 and the AR. The AR immunoprecipitation supernatant (SN) was immunoblotted for p300 to show the proportion of p300 not bound to the AR. (C) The relative binding of the AR mutants to p300 were shown as mean ± standard error of the mean (n = 3). (E) Acetylation of the AR enhances p300 binding in vitro. GST pull-down was performed with p300 and either acetylated AR (lanes 1 to 3) or unacetylated AR (lanes 4 to 6). Western blotting of the GST-AR pull-down product is shown. (F) p300 in vitro HAT assay. Baculovirus-expressed p300 protein (100 ng) was incubated with either GST or GST-AR624-676 in the presence of [14C]acetyl coenzyme A, resolved by SDS-PAGE, and exposed to a phosphoimaging screen for 24 h. The autoradiograms of the 14C-acetylated p300 and AR fusion protein are indicated. (G and H) Increased p300-mediated trans-activation of AR acetylation mutants. The MMTV-Luc reporter was assessed with the wild-type AR and AR acetylation site mutants, with p300 or equal amounts of the empty expression vector cassettes (pCMV), and treated with vehicle or DHT (10−7 M) for 24 h. The data shown were normalized to the DHT-induced activity for the AR construct (mean ± standard error of the mean, n > 9 separate transfections).
The wild-type AR and AR mutants were expressed to similar levels, and the relative abundance of p300 was similar between samples in transfected cells (Fig. 3A). In three separate immunoprecipitation-Western blot assays increased amounts of ARK630Q or ARK630T were associated with p300 compared with wild-type AR (Fig. 3C). Conversely, a tiny nonpolar substitution, ARK630A, showed reduced binding of p300 (Fig. 3D), consistent with a role for an acetylation-induced increase in polarity and/or hydrophobicity in docking to bromodomain coactivators. We examined whether direct acetylation of recombinant AR could alter the affinity for p300 in pull-down experiments. In the presence of p300 and acetyl-coenzyme A, the AR fusion protein 624-676 is efficiently acetylated (7). Under the same conditions, a semiquantitative GST pull-down experiment was conducted in the presence and absence of acetyl-coenzyme A (Fig. 3E). p300 bound the AR fragment in the pull-down, and the association of p300 with the unacetylated AR was disrupted above 0.6 M salt. p300 binding with acetylated AR held up under more stringent salt washing conditions, suggesting that AR acetylation enhances p300 binding (Fig. 3E, lane 2 versus 5).
To consider the alternative possibility, that in the presence of AR, acetyl-coenzyme A may enhance acetylation of p300 and thereby regulate AR binding and function, we conducted in vitro acetylation experiments with the AR fusion protein and assessed the autoacetylation of p300 (Fig. 3F). The addition of AR peptide did not increase p300 autoacetylation, suggesting that augmentation of p300 acetylation did not independently contribute to increasing the binding of p300 to the AR. p300 overexpression enhanced activity of both the unliganded (Fig. 3G) and liganded wild-type AR (Fig. 3H). The AR acetylation site mutants were activated relatively more by p300 either in the absence (9- to 10-fold) (Fig. 3G) or in the presence (4.5-fold) of ligand (Fig. 3H). Since p300 may serve as a platform to recruit AR coactivators, we examined the role of the AR acetylation site mimic in regulating AR coactivation by ARA55, ARA70, SRC1a, TIP60, and Ubc-9 (34). Each of the AR coactivators augmented activity of the liganded AR mutants more than wild-type AR (Table 1).
TABLE 1.
Activation by AR coactivatorsalegend
| Coactivator | Activation (fold)
|
||
|---|---|---|---|
| Wild-type AR | ARK630Q | ARK630T | |
| ARA55 | 5.2 | 7.2 | 13.5 |
| ARA70 | 6.1 | 7.5 | 9.6 |
| SRC-1 | 3.3 | 7.7 | 10.9 |
| TIP60 | 4.2 | 6.6 | 10.2 |
| Ubc-9 | 20.1 | 29.8 | 72.0 |
aThe MMTV-Luc reporter was transfected into DU145 cells with the expression vector for hAR and the AR coactivators as detailed in Materials and Methods, and cells were treated with DHT (10−7 M) for 24 h. Data are means ± standard errors of the means compared with the control empty expression vector.
AR acetylation site regulates corepressor binding and function.
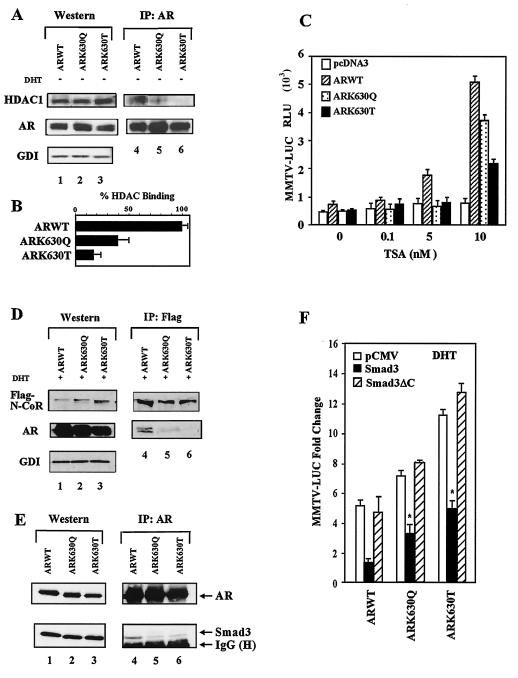
The repression function of several unliganded NR, mediated by N-CoR/SMRT (nuclear receptor corepressor/silencing mediator for retinoid and thyroid hormone receptors), involves recruitment of HDACs, TBL1, or basal transcriptional components (TFIIB, TAFII32, TAFI70) (5, 8, 11, 18). Furthermore, the AR binds to HDAC1 in the liver in vivo (5, 8, 11, 18). AR-deficient 293T cells transfected with wild-type AR and the AR mutants showed similar levels of AR and HDAC1 expression by Western blotting (Fig. 4A, lanes 1 to 3). Saturating AR immunoprecipitation with subsequent Western blotting for the AR and HDAC1 showed that HDAC1 binding to the AR mutants was reduced by 60 to 85% (Fig. 4B). The specific HDAC inhibitor TSA induced wild-type AR activity with MMTV-Luc (1.8-fold at 5 nM and 5.2-fold at 10 nM) (Fig. 4C) consistent with the binding of HDAC1 to the AR. In contrast, the AR acetylation site mutants were not activated by TSA at 5 nM and conveyed reduced TSA induction at 10 nM (Fig. 4C) (P < 0.05, n = 9).
FIG. 4.
Reduced N-CoR and HDAC1 binding of AR acetylation site mutants. (A) 293T cells transfected with the ARs were subjected to either direct Western blotting or AR immunoprecipitation and Western blotting for HDAC1 and AR. GDI served as the loading control. (B) The binding of HDAC1 to the AR is shown as percent binding for multiple separate transfections. (C) DU145 cells were cotransfected with the MMTV-Luc reporter and the expression vector for the wild-type AR, mutant AR, or empty expression vector cassette (pcDNA3) and treated with TSA for 24 h. (D) 293T cells were transfected with the ARs and Flag-N-CoR, treated with DHT (10−7M), and the cellular extracts were subjected to Western blotting or immunoprecipitation (anti-M2-Flag antibody for N-CoR) and subsequent AR Western blotting. (E) AR immunoprecipitation of cells transfected with AR acetylation site mutants and Smad3 with sequential Western blotting for Smad3. (F) DU145 Cells were transfected with the MMTV-Luc reporter and expression vectors for AR, Smad3, or Smad3ΔC. The DHT-induced change in MMTV-Luc activity is shown as the mean ± standard error of the mean.
The N-CoR N-terminal repression domain contacts NR and interacts with HDAC complexes (19). Western blot analysis of AR-deficient 293T cells transfected with AR and Flag-tagged N-CoR revealed similar levels of N-CoR and AR proteins (Fig. 4D, lanes 1, 2, and 3). Anti-Flag antibody immunoprecipitation showed similar amounts of N-CoR (Fig. 4D, lanes 4, 5, and 6) with reduced levels of AR mutants in the N-CoR immunoprecipitate (Fig. 4D, lane 4 versus 5 and 6). AR immunoprecipitation with sequential Western blotting for AR or Smad3 demonstrated similar levels of AR but reduced Smad3 binding to the AR acetylation site mutant (Fig. 4E). Smad3, a component of the N-CoR complex (14), inhibited liganded wild-type AR function, and deletion of the Smad3 carboxyl-terminal AR-binding region, abrogated repression (Fig. 4F). The AR acetylation site mutant remained three- to fivefold more active than wild-type AR. Together these studies suggest that the reduced binding of N-CoR/HDAC/Smad3 to the AR mutants correlated with reduced TSA responsiveness.
AR acetylation site mimic regulates prostate cellular growth and apoptosis in vivo.
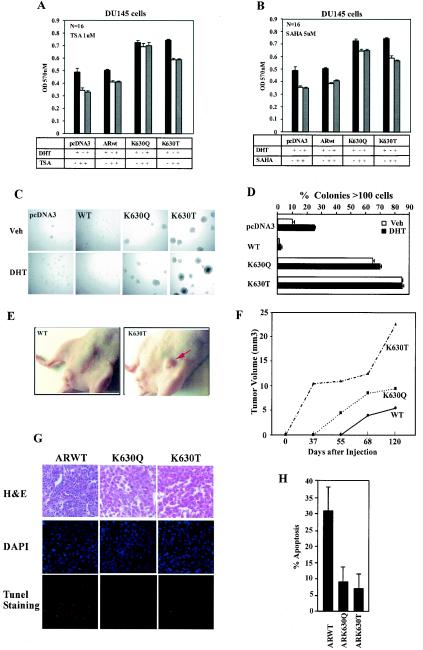
To determine the biological properties regulated by AR acetylation, stable prostate cancer cell lines (DU145) expressing either wild-type AR or the AR acetylation site mimic or control vector pcDNA3 were examined for effects on cellular growth and apoptosis. In view of the reduced binding of HDAC1 to the AR acetylation mutants and reduced N-CoR binding in the presence of DHT, we examined DU145 stable cell lines expressing either wild-type AR, ARK630Q or ARK630T for cellular growth and effects of HDAC inhibition. The indirect measure of cellular proliferation, the MTT assay, was used first to assess the effects of the HDAC inhibitor TSA (Fig. 5A ) and SAHA (Fig. 5B). The cellular proliferation rate of the AR acetylation mutant stable cell lines was increased in the presence of DHT compared to the AR wild type. The addition of TSA reduced MTT activity by 20% in the wild-type AR cells. The AR acetylation mutants demonstrated increased MTT activity in the presence of TSA compared to the AR wild type. Cellular proliferation was also increased in the AR acetylation mutant cell lines compared to the wild-type AR line. The cellular proliferation was relatively resistant to inhibition by the HDAC inhibitor SAHA.
FIG.5.
AR acetylation mutants convey contact-independent growth. (A and B) MTT assay of DU145 stable cell lines expressing either pcDNA3, wild-type AR, or AR acetylation site mutants. Equal numbers of cells were seeded into 96-well plates, treated with either DHT, TSA, or SAHA for 24 h, and the MTT assay was conducted, measuring absorbance at 570 nm. (C and D) DU145 cells stably expressing wild-type AR or AR acetylation site mutants were seeded in soft agar. Phase contrast image of the colonies from a representative experiment is shown (magnification, ×100). Colony numbers and size (percentage of colonies with >100 cells) are shown at day 14. (E and F) Nude mice were implanted with 106 cells of stable lines expressing either wild-type AR or AR acetylation site mutants. The mean volume of DU145 tumors grown in nude mice were shown at each time point. (G and H) Apoptosis in implanted tumors was assessed by TUNEL staining for wild-type AR, ARK630Q, and ARK630T (n = 4).
Colony formation was next assessed in DU145 cells expressing either the empty vector, wild-type AR, or the AR acetylation mutants. The size and number of soft agar colonies of ARK630T and ARK630Q were substantially increased compared with wild-type AR in either the presence or absence of ligand (Fig. 5C and D and data not shown). The major growth advantage in colonies containing greater than a hundred cells was observed in the absence of ligand, suggesting that a basal-level function of the AR acetylation mutants may contribute in an important manner to the size of colonies. The mechanism by which DHT enhances colony formation of DU145 in the absence of the AR may involve receptor-independent effects of DHT through other receptors or intracellular kinases and remains to be determined (22, 25). The AR acetylation site mutations conferred a growth advantage on stable prostate cancer cell lines compared with wild-type AR in vivo when implanted in nude mice, forming tumors approximately 20 days earlier and achieving a volume twice that of wild-type AR. The ARK630T clones formed tumors within 37 days (Fig. 5E and F), and at 120 days (mean = 22.5 mm3) and were some 4.5-fold larger than the wild-type AR (mean = 5.2 mm3) (Fig. 5F, n = 5). Apoptosis by TUNEL staining was reduced three- to fourfold in tumors harboring the AR acetylation mimic mutants compared with the wild-type AR-expressing clones (Fig. 5G and H, n = 4).
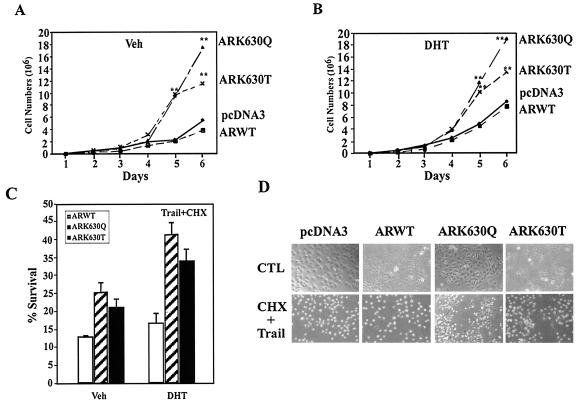
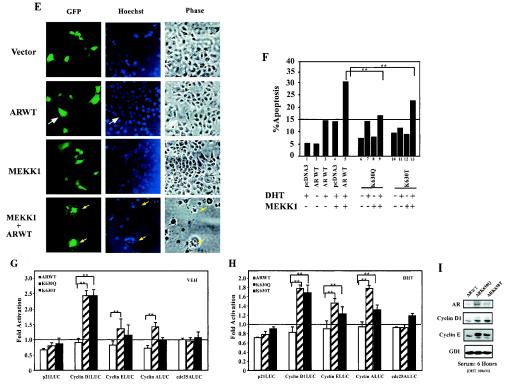
The cellular proliferation rate of the ARK630Q and ARK630T stable lines, assessed by cell counting, was also greater than that of wild-type AR (Fig. 6A ). A significant difference in cell numbers was observed at 5 days, where a trend was observed at day 4 in the presence of DHT (Fig. 6B). These results are consistent with the doubling time of DU145 cells (36 h) (44). Apoptosis induced by TRAIL and cycloheximide was reduced twofold in the ARK630Q and ARK630T lines compared with wild-type AR (Fig. 6C). Experiments were conducted on at least three stable lines on three separate experiments. Mitogen-activated protein kinase kinase kinase 1 (MEKK1) induces prostate cancer cell apoptosis in an AR-dependent manner (2) (Fig. 6E). In the presence of DHT and wild-type AR expression, with morphological analysis of GFP-transfected cells as previously described (20), cellular apoptosis was 15%. MEKK1 increased liganded wild-type AR apoptosis rates from 15% to 30 to 35% (Fig. 6F, lane 3 versus 5), again consistent with prior studies. MEKK1-induced apoptosis of ARK630T was, however, reduced 50% compared with wild-type AR and was abolished in cells expressing ARK630Q (Fig. 6F, lane 5 versus 9 and 13).
FIG. 6.
AR acetylation site governs cellular proliferation and MEKK1-dependent apoptosis. (A and B) Enhanced cellular proliferation rate of AR acetylation site mutants. A total of 2 × 104 DU145 cells stably transfected with the expression vector for either wild-type AR, AR mutants, or control vector pcDNA3 were seeded and treated either with vehicle (A) or DHT (10−7M) (B). The representative results of three independent experiments are shown. (C) AR acetylation site prostate cancer cell lines are resistant to TRAIL-induced apoptosis. Cell survival rates of DU145 stable cell lines exposed to TRAIL (10 ng/ml) and cycloheximide (CHX; 10 μg/ml) are shown compared with untreated cells (100%). (D) Phase contrast of cell lines is shown. (E, F). AR acetylation site mutants evade MEKK-1-mediated apoptosis. DU145 cells were transfected with MEKK1, AR, and pCMV-GFP as indicated and treated with either vehicle or DHT (10 nM) for 24 h. The morphology of the transfected DU145 cells is shown in phase contrast. (E) White arrows indicate GFP-positive cells, and yellow arrows indicate GFP-positive cells showing chromatin condensation. (F) The graph represents independent experiments in which 200 green fluorescent cells were counted and scored for cytoplasmic blebbing and chromatin condensation; *, P < 0.01 for the effect of MEKK1 on liganded AR-induced apoptosis. (G and H) AR acetylation site mutants alter regulation of cell cycle control genes. Reporter assays showing regulation of cell cycle control promoters by wild-type AR or AR acetylation site mutants in DU145 cells treated with vehicle (G) or DHT (H). (I) Western blot analysis of DU145 stable cell lines stably expressing wild-type AR and AR acetylation mutants. Cells were starved for 24 h and harvested 6 h after treatment (10% charcoal-stripped fetal bovine serum plus 100 nM DHT). GDI served as a protein-loading control.
To investigate the mechanism by which AR acetylation mutant may convey enhanced cellular proliferation, we assessed the effects of the AR mutants on the activity of cell cycle control gene promoters. Compared with wild-type AR, the AR acetylation mimic mutants increased activity of the cell cycle control genes cyclin D1 two- to fourfold and the cyclin E promoter 1.5-fold (Fig. 6G and H). Activity of the p21Cip1 promoter was regulated equally by expression of either wild-type AR or an AR mutant, suggesting that a subset of growth control genes are regulated by the AR acetylation mimic mutants. Western blot analysis of the DU145 stable cell line was consistent with the reporter data. Cyclin D1 and cyclin E protein levels were both increased in the AR acetylation mutant cell lines compared to the AR wild-type cell lines when normalized to the loading control GDI (Fig. 6I). Together, these studies demonstrate that the AR acetylation mutant stable cell lines exhibit enhanced basal and DHT-induced cellular proliferation and reduced cellular apoptosis in response to TRAIL and cycloheximide. The enhanced growth advantage correlated with increased activation of the promoters for the cell-cycle control genes cyclin D1 and cyclin E.
DISCUSSION
Phosphorylation of receptors or transcription factors has been linked to altered contact-independent cellular growth (20, 23). Herein, transcription factor acetylation is shown to govern cellular growth in vivo. Prostate cancer stable cells overexpressing AR acetylation mimic mutants grew faster, formed larger numbers of colonies and colonies of greater size, and, when implanted in nude mice, grew faster than the wild-type AR lines. The AR was acetylated in vivo, and the acetylation was regulated by DHT and TSA. The acetylation site mimic mutants regulated a subset of AR functions.
The AR conveys both ligand-dependent and -independent (aporeceptor) functions. Previous studies demonstrated that the AR acetylation site did not affect trans-repression (NF-κB, Sp1), sumoylation, or global structure assessed by protease digestion (7). In the current studies, the AR acetylation site affected aporeceptor function (TRAIL-mediated apoptosis, HDAC1 binding, cellular proliferation, contact-independent growth, induction of cell cycle gene promoters), and ligand-induced activities (ARE trans-activation, N-CoR, p300 binding). As the AR acetylation mimic mutants conveyed a cellular proliferate advantage and evaded MEKK1- and TRAIL-mediated apoptosis, it is plausible that both characteristics contributed to the growth advantage identified in vivo.
Previous studies have shown that the AR is acetylated in vitro by the HATs p300, P/CAF, and TIP60 (7-10). Several lines of evidence in the current study suggest that the AR is acetylated in vivo. Reciprocal immunoprecipitation-Western blotting was conducted on mouse liver tissues. The immunoprecipitate with the AR antibody, which contained immunoreactive AR, was cross-reactive with the anti-acetyl-lysine antibody at the same mobility as the AR by Western blotting. Conversely, the anti-acetyl-lysine antibody-immunoprecipitated protein reacted specifically with the AR antibody. Second, antibodies generated against an acetylated peptide resembling the acetylated AR motif immunoprecipitated the AR. Third, the HDAC inhibitor TSA and the AR ligand DHT induced acetylation of the AR in a prostate cancer cell line.
In the current studies, the reduced binding of N-CoR/HDAC1 and increased binding of p300 to ARK630Q and ARK630T suggests that Lys630 contributes to the molecular surface that interacts with both coactivators and corepressors. Acetylation of lysine residues through neutralizing the residue's positive charge and increasing its polarity may alter intra- and intermolecular interactions. The enhanced ligand-dependent transcriptional activity of the ARK630Q and ARK630T mutants, together with enhanced p300 binding and coactivator induction, suggest that neutralization of the ARK630 charge, or changes in size or shape at the side chain, plays a key role in coactivator recruitment and trans-activation. The finding that the coactivator and corepressor surfaces of NR overlap substantially (19, 32) is compatible with a dynamic model in which enzymatic modifications of the nuclear receptors by acetylation coordinates sequential disengagement of corepressors followed by coactivator binding (29). Our observations contribute to a more general model of transcription factor regulation in which acetylation contributes to corepressor disengagement with sequential coactivator recruitment (24).
Several lines of evidence suggest that the alterations in coactivator binding of the AR acetylation mimic are specific. First, we show that acetylation of the AR physically enhances association with p300. Second, the gain-of-function acetylation mimic mutants showed enhanced binding to p300, whereas substitution of a residue that does not mimic acetylation (alanine) did not enhance p300 binding. Third, the gain-of-function acetylation mimic mutants showed reduced binding to HDAC1 and N-CoR, whereas the alanine substitution mutant showed enhanced corepressor (N-CoR) binding. Fourth, the two acetylation mimic point mutants (ARK630Q and ARK630T) had gain of function in transcriptional reporter assays, whereas the acetylation “dead” mutants, alanine (ARK630A) and arginine (ARK630R) substitutions, showed loss of ligand-induced trans-activation. Fifth, our previous studies showed that point mutation of the acetylated lysine residues abrogated immunoreactivity with the anti-acetyl-lysine antibody and that the gross conformation of the AR mutated at the lysine residue is unaltered in protease sensitivity assays. Sixth, acetylation mutations do not affect sumoylation of the AR or AR trans-repression of NF-κB and Sp1 signaling (7, 8), demonstrating that the AR mutants maintain several normal functions of the receptor and that the acetylation site affects only a subset of AR activities. Finally, the acetylation mimic AR mutants showed selective enhancement of trans-activation of a subset of growth control gene promoters (cyclin D1, cyclin E, and cyclin A). Thus, each of these selective gains of function of the AR acetylation mimic mutants are the opposite to the function of the acetylation-dead AR mutants.
Our studies demonstrate that select subsets of functions of the AR are regulated by the acetylation site of the AR, both basally and ligand induced, that together contribute to aberrant growth control. The AR conveys several distinct functions in the absence of ligand (aporeceptor) and in the presence of ligand. The unliganded functions of the AR include trans-activation of synthetic AREs, trans-activation of native promoters, trans-repression (NF-κB, Sp1), and regulation of apoptosis and growth. The current studies show a role for the AR acetylation site in the absence of ligand in regulation of cell cycle control genes, reduced apoptosis in the presence of TRAIL/cycloheximide, and altered binding of HDAC1. In the presence of ligand, the AR acetylation site mutants showed increased trans-activation of synthetic and natural AREs, either alone or in the presence of coactivators. The acetylation sites do not appear to affect the trans-repression function of synthetic reporters or sumoylation of the AR (7, 8).
In the current studies, mutation of the AR acetylation site reduced apoptosis in response to TRAIL and cycloheximide or MEKK1. The role of the AR in basal and ligand-induced apoptosis is controversial, some investigators considering the AR an inducer of apoptosis (16, 26) and others as a prosurvival factor (9, 43, 46). Here we considered that the AR acetylation mutants conveyed a “gain of function” of enhanced survival. The acetylation mimic mutant showed a growth advantage in the absence of ligand coinciding with the enhanced trans-activation of the native promoters for cell-cycle and growth control genes (cyclin D1, cyclin E and cyclin A) in the absence of ligand, suggesting that the altered regulation of these genes may contribute to the increased cellular growth.
The tumor suppressor p53 and the related p73 protein are acetylated (35), and substitution of the acetylated residues of p73 with charged arginine residues also blocked both apoptotic and p300-dependent trans-activation functions (6). Our studies demonstrate that charge changes at the AR acetylated lysine residue alter the type of coactivator/corepressor complexes recruited to the AR. Neutral polar substitutions (ARK630Q, ARK630T) enhanced p300 binding and reduced N-CoR/HDAC/Smad3 corepressor binding, whereas unacetylatable inactivating mutants (ARK630R, ARK630A) reduced p300 binding and enhanced corepressor binding (8). As all of the ARK630 mutations and the p73 mutants were resistant to the induction of apoptosis and by design are incapable of receiving an acetyl group from acetyl-coenzyme A at the native lysine residue, it is possible that the acetyl group, once transferred to the lysine acceptor, forms a platform involved in subsequent apoptosis.
The effectiveness of androgen ablation therapy in reducing prostate cancer cell growth suggests a key role for the liganded AR in aberrant prostate cell growth (1). Therapeutic resistance to the androgen antagonist hydroxyflutamide contributes to morbidity and mortality in human prostate cancer. The identification of acetylation in the current studies as a key posttranslational modification regulating AR growth control suggests that ARK630 may form an ideal target for novel tumor therapies.
.
Acknowledgments
We thank Paul A. Marks, D. Chadee, R. Evans, E. Kalkhoven, Victoria M. Richon, J. M. Kyriakis, Y. Nakatani, B. O'Malley, and N. Schreiber-Agus for plasmids and helpful discussions.
This work was supported by grants from NIH (R01CA70896, R01CA75503, R01CA86072), the Pfeiffer Foundation, the Susan Komen Breast Cancer Foundation (to R.G.P.), NIH 1 R21DK065220-01 (NIDDK) (to M.F.), R03 AG2033 (to C.A.), R01-CA65647 (to S.P.B.), and R01 CA83979 (to M.L.A.). Work conducted at the Lombardi Cancer Center was supported by Comprehensive Cancer Center Core National Institute of Health grant P30 CA51008-13.
REFERENCES
- 1.Abate-Shen, C., and M. M. Shen. 2000. Molecular genetics of prostate cancer. Genes Dev. 14:2410-2434. [DOI] [PubMed] [Google Scholar]
- 2.Abreu-Martin, M. T., A. Chari, A. A. Palladino, N. A. Craft, and C. L. Sawyers. 1999. Mitogen-activated protein kinase kinase kinase 1 activates androgen receptor-dependent transcription and apoptosis in prostate cancer. Mol. Cell. Biol. 19:5143-5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alland, L., R. Muhle, H. J. Hou, J. Potes, L. Chin, N. Schreiber-Agus, and R. A. DePinho. 1997. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature 387:49-55. [DOI] [PubMed] [Google Scholar]
- 4.Chen, D., H. Ma, H. Hong, S. S. Koh, S. M. Huang, B. T. Schurter, D. W. Aswad, and M. R. Stallcup. 1999. Regulation of transcription by a protein methyltransferase. Science 284:2174-2177. [DOI] [PubMed] [Google Scholar]
- 5.Chen, J. D., and R. M. Evans. 1995. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature 377:454-457. [DOI] [PubMed] [Google Scholar]
- 6.Costanzo, A., P. Merlo, N. Pediconi, M. Fulco, V. Sartorelli, P. A. Cole, G. Fontemaggi, M. Fanciulli, L. Schiltz, G. Blandino, C. Balsano, and M. Levrero. 2002. DNA damage-dependent acetylation of p73 dictates the selective activation of apoptotic target genes. Mol. Cell. 9:175-186. [DOI] [PubMed] [Google Scholar]
- 7.Fu, M., C. Wang, A. T. Reutens, J. Wang, R. H. Angeletti, L. Siconolfi-Baez, V. Ogryzko, M. L. Avantaggiati, and R. G. Pestell. 2000. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 275:20853-20860. [DOI] [PubMed] [Google Scholar]
- 8.Fu, M., C. Wang, J. Wang, X. Zhang, T. Sakamaki, Y. G. Yeung, C. Chang, T. Hopp, S. A. Fuqua, E. Jaffray, R. T. Hay, J. J. Palvimo, O. A. Janne, and R. G. Pestell. 2002. Androgen receptor acetylation governs trans activation and MEKK1-induced apoptosis without affecting in vitro sumoylation and trans-repression function. Mol. Cell. Biol. 22:3373-3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao, J., and J. T. Isaacs. 1998. Development of an androgen receptor-null model for identifying the initiation site for androgen stimulation of proliferation and suppression of programmed (apoptotic) death of PC-82 human prostate cancer cells. Cancer Res. 58:3299-3306. [PubMed] [Google Scholar]
- 10.Gaughan, L., I. R. Logan, S. Cook, D. E. Neal, and C. N. Robson. 2002. Tip60 and Histone Deacetylase 1 Regulate Androgen Receptor Activity through Changes to the Acetylation Status of the Receptor. J. Biol. Chem. 277:25904-25913. [DOI] [PubMed] [Google Scholar]
- 11.Glass, C. K., and M. G. Rosenfeld. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121-141. [PubMed] [Google Scholar]
- 12.Harrod, R., J. Nacsa, C. Van Lint, J. Hansen, T. Karpova, J. McNally, and G. Franchini. 2002. Human immunodeficiency virus type-1 Tat/Co-activator acetyltransferase interactions inhibit p53K320-acetylation and p53-responsive transcription. J. Biol. Chem. 278:12310-12318. [DOI] [PubMed] [Google Scholar]
- 13.Hayes, S., M. Zarnegar, M. Sharma, F. Yang, D. M. Peehl, P. ten Dijke, and Z. Sun. 2001. SMAD3 represses androgen receptor-mediated transcription. Cancer Res. 61:2112-2118. [PubMed] [Google Scholar]
- 14.Hayes, S. A., M. Zarnegar, M. Sharma, F. Yang, D. M. Peehl, P. ten Dijke, and Z. Sun. 2001. SMAD3 represses androgen receptor-mediated transcription. Cancer Res. 61:2112-2118. [PubMed] [Google Scholar]
- 15.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 387:43-48. [DOI] [PubMed] [Google Scholar]
- 16.Heisler, L. E., A. Evangelou, A. M. Lew, J. Trachtenberg, H. P. Elsholtz, and T. J. Brown. 1997. Androgen-dependent cell cycle arrest and apoptotic death in PC-3 prostatic cell cultures expressing a full-length human androgen receptor. Mol. Cell. Endocrinol. 126:59-73. [DOI] [PubMed] [Google Scholar]
- 17.Higashimoto, Y., S. Saito, X. H. Tong, A. Hong, K. Sakaguchi, E. Appella, and C. W. Anderson. 2000. Human p53 is phosphorylated on serines 6 and 9 in response to DNA damage-inducing agents. J. Biol. Chem. 275:23199-23203. [DOI] [PubMed] [Google Scholar]
- 18.Horlein, A. J., A. M. Naar, T. Heinzel, J. Torchia, B. Gloss, R. Kurokawa, A. Ryan, Y. Kamei, M. Soderstrom, C. K. Glass, et al. 1995. Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature 377:397-404. [DOI] [PubMed] [Google Scholar]
- 19.Hu, X., Y. Li, and M. A. Lazar. 2001. Determinants of CoRNR-dependent repression complex assembly on nuclear hormone receptors. Mol. Cell. Biol. 21:1747-1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter, T. 1997. Oncoprotein networks. Cell. 88:333-346. [DOI] [PubMed] [Google Scholar]
- 21.Jacobson, R. H., A. G. Ladurner, D. S. King, and R. Tjian. 2000. Structure and function of a human TAFII250 double bromodomain module. Science 288:1422-1425. [DOI] [PubMed] [Google Scholar]
- 22.Janssen, T., R. Kiss, R. Dedecker, M. Petein, J. L. Pasteels, and C. Schulman. 1995. Influence of dihydrotestosterone, epidermal growth factor, and basic fibroblast growth factor on the cell kinetics of the PC3, DU145, and LNCaP prostatic cancer cell lines: relationship with DNA ploidy level. Prostate 27:277-286. [DOI] [PubMed] [Google Scholar]
- 23.Karin, M., and T. Hunter. 1995. Transcriptional control by protein phosphorylation: signal transmission from the cell surface to the nucleus. Curr. Biol. 5:747-757. [DOI] [PubMed] [Google Scholar]
- 24.Kouzarides, T. 2000. Acetylation: a regulatory modification to rival phosphorylation. EMBO J. 19:1176-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee, Y. F., W. J. Lin, J. Huang, E. M. Messing, F. L. Chan, G. Wilding, and C. Chang. 2002. Activation of mitogen-activated protein kinase pathway by the antiandrogen hydroxyflutamide in androgen receptor-negative prostate cancer cells. Cancer Res. 62:6039-6044. [PubMed] [Google Scholar]
- 26.Lin, H. K., S. Yeh, H. Y. Kang, and C. Chang. 2001. Akt suppresses androgen-induced apoptosis by phosphorylating and inhibiting androgen receptor. Proc. Natl. Acad. Sci. 98:7200-7205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, L., D. M. Scolnick, R. C. Trievel, H. B. Zhang, R. Marmorstein, T. D. Halazonetis, and S. L. Berger. 1999. p53 sites acetylated in vitro by PCAF and p300 are acetylated in vivo in response to DNA damage. Mol. Cell. Biol. 19:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauser, A., S. Saito, E. Appella, C. W. Anderson, W. T. Seaman, and S. Kenney. 2002. The Epstein-Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J. Virol. 76:12503-12512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna, N. J., R. B. Lanz, and B. W. O'Malley. 1999. Nuclear receptor coregulators: cellular and molecular biology. Endocrinol. Rev. 20:321-344. [DOI] [PubMed] [Google Scholar]
- 30.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A and histone deacetylase. Cell. 89:373-380. [DOI] [PubMed] [Google Scholar]
- 31.Ogryzko, V. V., R. L. Schiltz, V. Russanova, B. H. Howard, and Y. Nakatani. 1996. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 87:953-959. [DOI] [PubMed] [Google Scholar]
- 32.Perissi, V., L. M. Staszewski, E. M. McInerney, R. Kurokawa, A. Krones, D. W. Rose, M. H. Lambert, M. V. Milburn, C. K. Glass, and M. G. Rosenfeld. 1999. Molecular determinants of nuclear receptor-corepressor interaction. Genes Dev. 13:3198-3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polesskaya, A., I. Naguibneva, A. Duquet, E. Bengal, P. Robin, and A. Harel-Bellan. 2001. Interaction between acetylated MyoD and the bromodomain of CBP and/or p300. Mol. Cell. Biol. 21:5312-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Poukka, H., P. Aarnisalo, U. Karvonen, J. J. Palvimo, and O. A. Janne. 1999. Ubc9 interacts with the androgen receptor and activates receptor-dependent transcription. J. Biol. Chem. 274:19441-19446. [DOI] [PubMed] [Google Scholar]
- 35.Prives, C., and J. L. Manley. 2001. Why is p53 acetylated? Cell. 107:815-818. [DOI] [PubMed] [Google Scholar]
- 36.Reutens, A. T., M. Fu, C. Wang, C. Albanese, M. J. McPhaul, Z. Sun, S. P. Balk, O. A. Janne, J. J. Palvimo, and R. G. Pestell. 2001. Cyclin D1 binds the androgen receptor and regulates hormone-dependent signaling in a p300/CBP-associated factor (P/CAF)-dependent manner. Mol. Endocrinol. 15:797-811. [DOI] [PubMed] [Google Scholar]
- 37.Sterner, D. E., and S. L. Berger. 2000. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 64:435-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Struhl, K. 1998. Histone acetylation and transcriptional regulatory mechanisms. Genes Dev. 12:599-606. [DOI] [PubMed] [Google Scholar]
- 39.Tan, J., Y. Sharief, K. G. Hamil, C. W. Gregory, D. Y. Zang, M. Sar, P. H. Gumerlock, R. W. DeVere White, T. G. Pretlow, S. E. Harris, E. M. Wilson, J. L. Mohler, and F. S. French. 1997. Dehydroepiandrosterone activates mutant androgen receptors expressed in the androgen-dependent human prostate cancer xenograft CWR22 and LNCaP cells. Mol. Endocrinol. 11:450-459. [DOI] [PubMed] [Google Scholar]
- 40.Taplin, M. E., G. J. Bubley, Y. J. Ko, E. J. Small, M. Upton, B. Rajeshkumar, and S. P. Balk. 1999. Selection for androgen receptor mutations in prostate cancers treated with androgen antagonist. Cancer Res. 59:2511-2515. [PubMed] [Google Scholar]
- 41.Taplin, M. E., G. J. Bubley, T. D. Shuster, M. E. Frantz, A. E. Spooner, G. K. Ogata, H. N. Keer, and S. P. Balk. 1995. Mutation of the androgen-receptor gene in metastatic androgen-independent prostate cancer. N. Engl. J. Med. 332:1393-1398. [DOI] [PubMed] [Google Scholar]
- 42.Torchia, J., C. Glass, and M. G. Rosenfeld. 1998. Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol. 10:373-383. [DOI] [PubMed] [Google Scholar]
- 43.Vendola, K. A., J. Zhou, O. O. Adesanya, S. J. Weil, and C. A. Bondy. 1998. Androgens stimulate early stages of follicular growth in the primate ovary. J. Clin. Investig. 101:2622-2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vilenchik, M., A. J. Raffo, L. Benimetskay, D. Shames, and C. A. Stein. 2002. Antisense RNA down-regulation of bcl-xL expression in prostate cancer cells leads to diminished rates of cellular proliferation and resistance to cytotoxic chemotherapeutic agents. Cancer Res. 62:2175-2183. [PubMed] [Google Scholar]
- 45.Wang, C., M. Fu, S. Mani, S. Wadler, A. M. Senderowicz, and R. G. Pestell. 2001. Histone acetylation and the cell-cycle in cancer. Front. Biosci. 6:D610-D629. [DOI] [PubMed] [Google Scholar]
- 46.Ye, D., J. Mendelsohn, and Z. Fan. 1999. Androgen and epidermal growth factor down-regulate cyclin-dependent kinase inhibitor p27Kip1 and costimulate proliferation of MDA PCa 2a and MDA PCa 2b prostate cancer cells. Clin. Cancer Res. 5:2171-2177. [PubMed] [Google Scholar]
- 47.Yuan, S., J. Trachtenberg, G. B. Mills, T. J. Brown, F. Xu, and A. Keating. 1993. Androgen-induced inhibition of cell proliferation in an androgen-insensitive prostate cancer cell line (PC-3) transfected with a human androgen receptor complementary DNA. Cancer Res. 53:1304-1311. [PubMed] [Google Scholar]