Abstract
Human CRM1 (hCRM1) functions in the Rex-mediated mRNA export of human T-cell leukemia virus type 1 (HTLV-1) as an export receptor and as an inducing factor for Rex multimerization on its cognate RNA. Although there are only 24 amino acid differences between hCRM1 and rat CRM1 (rCRM1), rCRM1 can hardly support Rex activity, suggesting a role for rCRM1 as a determinant restricting the host range of HTLV-1. Here, we used a series of mutants, which were generated by interchanging residues of these CRM1s, to examine the relationship of hCRM1 functions. The functions for Rex multimerization and binding to nuclear export signals are mapped to different amino acid residues, and these are separable, suggesting that CRM1 not only functions as an export receptor but also participates in the formation of the RNA export complex through higher-ordered interaction with Rex. The region for the interaction with RanBP3, comprising four residues (amino acids [aa] 411, 414, 474, and 481), and the region for Rex multimerization, including two residues (aa 411 and 414), form an overlapped domain. Our results provide the molecular basis underlying the species-specific ability of HTLV-1 to propagate in human cells.
Human CRM1 (hCRM1) belongs to the family of importin β-related nuclear transport receptors, also referred to as karyopherins, and has been shown to export some types of RNA (6, 14, 21, 31, 34) as well as a number of proteins that carry the leucine-rich type of nuclear export signal (NES) (11, 13, 32, 36). Nuclear export by hCRM1 across the nuclear pore complex (NPC) is regulated by the GDP/GTP cycle of a small GTPase, Ran. Most nuclear Ran is in GTP-bound form, but cytoplasmic Ran is GDP bound. This reflects the nuclear localization of regulator of chromosome condensation 1 (RCC1), which specifically catalyses the exchange of guanine nucleotides on Ran (4) and cytoplasmic location of a GTPase-activating protein, RanGAP (5). hCRM1 binds to export substrates only within the nucleus, since this binding requires simultaneous association of hCRM1 with RanGTP (11, 36). This ternary complex is translocated to the cytoplasm across the NPC through a possible hydrophobic interaction between hCRM1 and nucleoporins. Following this, RanGTP in the complex is converted to RanGDP by RanGAP in concert with RanBP1 (and probably RanBP2), leading to dissociation of the complex and release of the substrate into the cytoplasm (3, 23).
Recently, Ran-binding protein 3 (RanBP3) (10, 26, 29, 30) has been shown to bind to RCC1 in a Ran-dependent manner and to increase the nucleotide exchange activity of RCC1, resulting in high concentrations of RanGTP in the vicinity of RCC1. Furthermore, it has been shown that RanBP3 directly binds to hCRM1 and thereby recruits hCRM1 to RCC1. Consequently, RanBP3 acts as a scaffold protein by which components of the export complex are concentrated around the RCC1 site, thus promoting complex assembly (30). RanBP3 continues to interact with hCRM1 to stabilize the hCRM1-substrate-RanGTP complex, thereby forming a quaternary complex.
As detailed above, to efficiently export proteins hCRM1 must possess functional domains that interact with numerous proteins including NES, RanGTP, RanBP3, and nucleoporins. A previous report demonstrated that the residues assigned as Asp 716 and Lys 810, as well as other residues in the neighborhood of Lys 810 in hCRM1, are involved in binding to Rev protein of human immunodeficiency virus type 1, which carries the NES (2). Another report demonstrated that a region between residues 566 and 720 in hCRM1 might be essential for NES binding (33). The amino-terminal 150 amino acids (aa) of hCRM1 have a sequence that is homologous to that of other members of the importin-β family (12, 16). Indeed, a region between residues 61 and 160 was shown to be essential for the interaction of RanGTP with hCRM1 (33). Furthermore, the cysteine residue at 528 in hCRM1 has been demonstrated to bind directly to leptomycin B (LMB), a specific inhibitor of CRM1 function (24). Despite the progress which has been made in understanding CRM1 function, precise mapping of the functional domains of hCRM1 has not been accomplished yet, partly because the interaction of hCRM1 with one component affects its binding with other components.
RNA export is generally considered to be mediated by its surrounding RNA binding proteins, which recruit export receptors, with the exception of tRNA, which is directly transported by exportin-t (1, 25). For example, spliceosomal small nuclear RNA export is initiated by recognition of its 5′-terminal cap structure by the CBP20/80 complex, which associates with a protein termed phosphorylated adaptor for RNA export (PHAX). PHAX subsequently recruits CRM1 through its NES (31). Therefore, the processes of RNA and protein export share common mechanisms. However, since RNA usually exists as a large ribonucleoprotein (RNP) complex, its export has additional requirements, for example, the necessity for energy to facilitate movement in the nucleus (7) and remodeling of the Balbiani ring RNP complex during translocation through the NPC (28).
One of the most extensively studied examples of RNA export is that of human retroviruses, including human T-cell leukemia virus type 1 (HTLV-1) and human immunodeficiency virus; these viruses encode posttranscriptional regulator proteins Rex and Rev, respectively. The regulator proteins escort unspliced and incompletely spliced viral mRNAs to the cytoplasm and enhance the expression of enzymatic and structural proteins encoded by these mRNAs (8, 35). Rex and Rev directly bind both the viral mRNA and hCRM1 in the presence of RanGTP via their RNA binding domains and NES, respectively. Therefore, Rex and Rev act as adaptor proteins that bridge the target mRNA and the export receptor. Notably, multimer formation of Rex/Rev along the viral mRNA is required for RNA export (9, 27, 37), although export of Rex/Rev proteins themselves does not require their multimerization. Multimerization would allow a number of Rex/Rev proteins to shield RNA, thus preventing the attachment of factors that are involved in splicing and nuclear retention of RNA and consequently increasing the number of CRM1 molecules that associate with the complex. The association of multiple CRM1 molecules may be important in overcoming the factors that retain RNA in the nucleus and allow RNA to pass efficiently through NPCs.
Multimer formation requires not only intact multimerization domains, which contain the sites for Rex-Rex or Rev-Rev interaction, but also hCRM1 accompanying RanGTP (17, 18, 19). The essential role of CRM1 in Rex multimerization was confirmed by the observation that rat CRM1 (rCRM1), which exports Rex as efficiently as hCRM1 but does not support Rex multimerization, is barely able to transport viral RNA (18). Two possible mechanisms by which CRM1 induces Rex/Rev multimerization can, therefore, be envisaged. The association of CRM1 with the NES region of Rex/Rev proteins may trigger a conformational change in Rex/Rev that leads directly to their multimerization. Alternatively, binding of CRM1 to the NES may be necessary, but not sufficient, and additional interactions between Rex/Rev and CRM1 may be required for multimerization. It is important to unequivocally determine which scenario is correct, not least because the second possibility may suggest direct involvement of CRM1 in formation of the RNA export complex besides its function as an export receptor. Specifically, the second hypothesis predicts that hCRM1 possesses a domain required for its additional interaction with Rex to induce Rex-Rex interaction. Given that rCRM1 and hCRM1 are highly homologous (they differ by only 24 aa residues out of a total of 1,071), these two proteins are particularly suitable for determining the structure-function relationships of CRM1, including its induction of Rex-Rex interaction.
In this study, using a series of CRM1 mutants that was generated by interchanging hCRM1 and rCRM1 residues, we investigated the amino acid residues in hCRM1 that are critical for supporting Rex activity. Our analyses revealed a novel functional domain, in the region spanning residues 411 to 481, which is involved in binding of hCRM1 to RanBP3. Moreover, we identified two amino acids (residues 411 and 414) that are crucial for Rex multimerization, supporting the view that simple association of hCRM1 with the NES of Rex does not suffice but that a higher-ordered interaction between hCRM1 and Rex is required for RNA export.
MATERIALS AND METHODS
Cloning and plasmid construction.
To construct pSRαhrCRM1 and pSRαrhCRM1, the N-terminal half of the hCRM1 or rCRM1 coding region (corresponding to aa 1 to 679 of each CRM1) was amplified by PCR using the primer pair 5′-GTT CAA TCT CTG GTA ATC TAT GCC AGC-3′ and 5′-GCT GCT TGA CTG TCT CGG GGT CTT T-3′ (primer 1) or the primer pair 5′-AGG AAG GAG CAG TTG GTT CAA TCT CTG GTA A-3′ and primer 1, with pSRαhCRM1 or pSRαrCRM1, respectively, as a template. The C-terminal half of the hCRM1 or rCRM1 coding region (corresponding to aa 680 to 1071) was amplified using the primer pair 5′-TAT CCT GAA AGA CCC CGA GAC AGT CAA-3′ (primer 2) and 5′-AAC GGT ACC CGC ACT AGT CAC ACA TTT CTT CTG GAA TCT CAT GTG GAT-3′ or the primer pair primer 2 and 5′-AGG ACA AAC GCT GCA CAG GGA AA-3′. These four fragments were blunt ended by Pfu DNA polymerase treatment and digested with AvaI, and the C-terminal fragments were digested with KpnI. The digested N- and C-terminal fragments were then cloned into pSRα296 (17) in the appropriate combinations.
To exchange amino acids in the hCRM1 sequence for the corresponding residues from the rCRM1 sequence or vice versa, PCR-based mutagenesis was carried out using a QuikChange site-directed mutagenesis kit (Stratagene) in accordance with the manufacturer's instructions. The sequences of all of the constructs were confirmed by DNA sequencing.
To construct pSRαRex-HA, the Rex coding region from pSRαRex was amplified by PCR using the primer pair 5′-ATG CCC AAG ACC CGT CGG AGG C-3′ and 5′-GGA ATT CTA AGC GTA GTC TGG GAC GTC GTA TGG GTA CGT GGG GCA GGA GGG GCC AGG TGA T-3′, which encodes a hemagglutinin (HA) tag. The PCR product was blunt ended and digested with EcoRI and cloned into the PstI-digested, blunt-ended, and EcoRI-digested pSRα296. The activity of Rex-HA protein was the same as that of nontagged wild-type Rex (data not shown). The plasmids, including pSRαRex, pSRαRexM64, pSRαTAgRexM64, pCDMβ-galactosidase (pCDMβ-gal), pGAL4, pRexVP, pSRαhCRM1, pSRαrCRM1, pGAL-hCRM1, pGAL-rCRM1, pG5BCAT, and pDM128RxRE, have all been described previously (17, 18).
The recombinant protein expression plasmids pET3dRanQ69L (20) and pGEX-2TRanBP3 (38), which expresses glutathione S-transferase (GST)-RanBP3 type b, were kind gifts from Y. Yoneda and I. Macara, respectively.
Cell culture and transfection.
HeLa cells and rat REF52 cells were maintained in an atmosphere of 5% CO2 at 37°C in Dulbecco's modified Eagle's medium that was supplemented with 10% fetal bovine serum. Transfection was carried out using Lipofectamine Plus reagents (GIBCO-BRL Life Technologies) according to the manufacturer's instructions. The total amount of DNA in each experiment was kept constant by adding pSRα296. All transfection experiments were performed in duplicate and repeated at least twice.
Measurement of Rex activity.
HeLa cells were transfected with a mixture that included 0.05 μg of pSRαRex, 0.2 μg of pSRαTagRexM64, 0.3 μg of various pSRαCRM1s, and 0.5 μg of pDM128RxRE, which expresses chloramphenicol acetyltransferase (CAT) protein, depending on the degree of Rex activity. All samples were also transfected with 0.1 μg of pCDMβ-gal for the normalization of transfection efficiency. At 24 h posttransfection, the cells were lysed and the amount of CAT was quantified using a CAT ELISA kit (Boehringer Mannheim) according to the manufacturer's instructions. The β-galactosidase (β-Gal) activity was measured by assays employing standard colorimetric methods. The CAT/β-Gal ratio was calculated for each sample to represent Rex activity.
REF52 cells were transfected with 0.5 μg of pDM128RxRE-0.5 μg of pSRαRex-0.5 μg of various pSRαCRM1s-0.1 μg of pCDMβ-gal. At 24 h posttransfection, the cells were harvested and the CAT/β-Gal ratio was calculated.
Measurement of Gag expression.
REF52 cells were transfected with 0.5 μg of K30 (which is an infectious HTLV-1 molecular clone)-0.01 μg of various pSRαCRM1s-0.1 μg of pCDMβ-gal. At 48 h posttransfection, the medium of the cell culture was centrifuged at low speed to remove the cell debris. The clarified supernatant was ultracentrifuged at 40,000 × g for 60 min, the resultant sediment (comprising viral particles) was lysed in lysis buffer with a RETRO-TEK HTLV p19 Antigen ELISA kit (ZeptoMetrix), and the amount of Gag protein was quantified in accordance with the manufacturer's recommended procedure. On the other hand, the cells were harvested to estimate the β-Gal activity and the Gag/β-Gal ratio was calculated.
In vivo assay of protein-protein interaction.
Protein-protein interactions in REF52 cells were analyzed using a mammalian two-hybrid system (17). The cells were transfected with a mixture of 0.2 μg of the plasmid expressing various GAL-CRM1s in combination with 0.2 μg of pRexVP and 0.6 μg of pG5BCAT, a reporter plasmid which expresses CAT protein when GAL4-fused CRM1s interact with Rex-VP. All samples were also transfected with 0.1 μg of pCDMβ-gal. At 24 h posttransfection, the cells were harvested and the CAT/β-Gal ratio was calculated.
Detection of Rex multimerization.
HeLa cells were transfected with 0.3 μg of various pSRαCRM1s, 0.5 μg of pDM128RxRE, and 0.05 μg of pSRαRexM64, which expresses RexM64 harboring mutations in the multimerization domain (17). All samples were also transfected with 0.1 μg of pCDMβ-gal. At 24 h posttransfection, the cells were harvested and the CAT/β-Gal ratio was calculated for each sample.
Immunofluorescence microscopy.
REF52 cells were transfected with a mixture of 0.1 μg of pSRαRex-HA along with 0.3 μg of pSRα296 or various pSRαCRM1s. At 24 h posttransfection, the cells were fixed with a 2% paraformaldehyde-phosphate-buffered saline solution. After perforation with 0.1% NP-40, the cells were incubated with rat anti-HA monoclonal antibody (clone 3F10; Roche) and affinity-purified chicken anti-hCRM1 antibody for 2 h followed by incubation with Cy3- and fluorescein isothiocyanate (FITC)-conjugated secondary antibody for 1 h. The stained cells were observed using an Axiovert 135 system (Carl Zeiss).
Western blot analysis.
Proteins dissolved in sample buffer were applied to sodium dodecyl sulfate-polyacrylamide electrophoresis gels and transferred to nitrocellulose filters by using standard techniques. Mouse anti-GAL4 monoclonal antibody (Santa Cruz Biotechnology) and affinity-purified chicken anti-hCRM1 antibody (18) were used as primary antibodies to detect GAL-fused proteins and CRM1s, respectively. Horseradish peroxidase- or alkaline phosphatase-conjugated anti-immunoglobulin G antibodies (Promega) were used as secondary antibodies. Immunoreactive bands were visualized using an ECL Plus chemiluminescent substrate (Amersham Pharmacia Biotech) followed by the use of a LAS-1000 Plus system (Fujifilm) or BCIP (5-bromo-4-chloro-3-indolylphosphate)/Nitro Blue Tetrazolium solution.
Expression and purification of recombinant proteins.
The recombinant RanQ69L protein was expressed in Escherichia coli, purified, and then charged with GTP according to the method previously reported (18).
The plasmid pGEX-2TRanBP3 was used to prepare recombinant RanBP3 protein with a NH2-terminal GST tag. The expression of recombinant GST-RanBP3 protein was induced using 1 mM isopropyl-β-d-thiogalactopyranoside in the E. coli strain BL21(DE3) Gold. The E. coli cells were lysed in buffer A (50 mM Tris HCl [pH 8.0], 500 mM NaCl, 2 mM MgCL2, 1 mM dithiothreitol, 1 μg each of aprotinin, leupeptin, and pepstatin per ml) by freezing-thawing and sonication. After centrifugation (100,000 × g, 30 min), the clarified lysate was applied to glutathione-Sepharose 4B (Amersham Pharmacia) equilibrated with buffer A and incubated with rotation at 4°C for 30 min. The resin was washed three times with buffer A and once with buffer B (50 mM Tris HCl [pH 8.0], 50 mM NaCl, 1 mM MgCL2, 1 mM dithiothreitol, 1 μg each of aprotinin, leupeptin, and pepstatin per ml), and then 10 mM reduced glutathione was added to elute proteins. The eluate was applied to a Hi-Trap Q column and separated with a linear gradient of buffer C (50 mM Tris HCl [pH 8.0], 1 mM MgCL2, 1 mM dithiothreitol, 1 μg each of aprotinin, leupeptin, and pepstatin per ml) containing 50 to 500 mM NaCl. The fractions containing GST-RanBP3 protein were applied to Superdex 200 (Amersham Pharmacia Biotech) equilibrated with transport buffer (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 0.5 mM EGTA, 2 mM dithiothreitol, 1 μg each of aprotinin, leupeptin, and pepstain per ml) and then concentrated using a Centricon YM 10 centrifugal filter unit (Amicon-Millipore). For preparation of recombinant nontagged RanBP3, GST-RanBP3 (eluted by the reduced glutathione as mentioned above) was cleaved by thrombin before purification with a Hi-Trap Q column.
GST proteins were expressed by pGEX6P and purified with glutathione-Sepharose 4B.
In vitro transcription and translation system.
Coupled transcription and translation was performed using TNT T7 Quick for PCR DNA (Promega). PCR DNA products used as templates in this system were prepared as follows. Template DNA for Rex-HA protein synthesis was prepared by PCR using the primer pair 5′-CAG ATT TAA TAC GAC TCA CTA TAG GGA AAA ACC ACC ATG CCC AAG ACC CGT CGG AG-3′ and 5′-GGA ATT CTA AGC GTA GTC TGG GAC GTC GTA TGG GTA CGT GGG GCA GGA GGG GCC AGG TGA-3′ and pRex-VP as a template. The first-named primer contains the T7 promoter, the Kozak sequence, and a Rex gene-specific element. For each CRM1 synthesis, the primer pair 5′-CAG ATT TAA TAC GAC TCA CTA TAG GGA AAA ACC ACC ATG CCA GCA ATT ATG ACA ATG TTA G-3′ and 5′-AAC GGT ACC CGC ACT AGT CAC ACA TTT CTT CTG GAA TCT CAT GTG GAT-3′ and various pGAL-CRM1s as a template were used. The first-named primer contains the T7 promoter, the Kozak sequence, and an hCRM1 gene-specific element.
Interaction of CRM1s with Rex-HA in vitro.
CRM1 and Rex-HA were produced (using a transcription-translation system) in one reaction tube. These were incubated with or without 4 μM of GTP-charged RanQ69L protein and 2 mM GTP for 30 min at 30°C in 50 μl of binding buffer A (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 20 mM sodium acetate, 2 mM magnesium acetate, 2 mM dithiothreitol, 0.01% NP-40). After incubation, 2 μl of each reaction mixture was dissolved in sample buffer and used as an input fraction and 40 μl of each reaction mixture was incubated with 0.2 μg of anti-HA monoclonal antibody, which had been immobilized on 20 μl of protein G-Sepharose, for 2 h at 4°C in 200 μl of binding buffer A. The resins were collected by low-speed centrifugation and washed four times with 1 ml of binding buffer A, following which sample buffer was added to the resins. CRM1 proteins in the input and bound fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis followed by Western blot analysis using anti-hCRM1 antibody.
In vitro binding of CRM1s to NES peptide.
Various CRM1s were translated in 60 μl of transcription-translation reaction solution. Then, 2-μl aliquots were mixed with sample buffer (input) and 25-μl aliquots were incubated with NES peptide, which had been conjugated to biotinylated bovine serum albumin (BSA) and immobilized on avidin-agarose, in the absence or presence of 5 μM GTP-charged RanQ69L protein and 1 mM GTP for 2 h at 4°C in 240 μl of binding buffer B (20 mM HEPES [pH 7.3], 110 mM potassium acetate, 5 mM sodium acetate, 2 mM magnesium acetate, 0.5 mM EGTA, 2 mM dithiothreitol, 0.01% NP-40). The NES peptide used in this assay was CYELALKLAGLDINK, which is derived from the protein kinase A inhibitor (PKI). After incubation, the resins were recovered by low-speed centrifugation and washed four times with 1 ml of binding buffer B, following which sample buffer was added to the resins.
In some experiments, recombinant RanBP3 protein was added to the binding reaction mixture at a final concentration of 0.3 μM or 0.9 μM.
CRM1 binding to RanBP3.
Various CRM1s were translated in 50 μl of transcription-translation reaction solution. Aliquots (2 μl) were mixed with sample buffer (input), and 20-μl aliquots were incubated with 4 μg of GST or GST-RanBP3, which had been immobilized on 20 μl of glutathione-Sepharose 4B, for 1 h at 4°C in 200 μl of binding buffer B. The resins were recovered by low-speed centrifugation and washed four times with 1 ml of binding buffer B, following which sample buffer was added to the resins.
RESULTS
Four amino acid residues of hCRM1 are important for Rex activity.
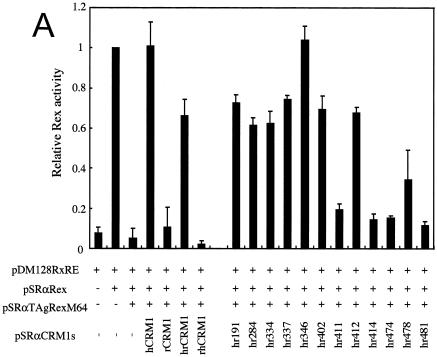
To measure the activity of various CRM1s in Rex-mediated mRNA export, we used a transient transfection assay (18).This assay includes pDM128RxRE and pSRαTAgRexM64, which expresses TAgRexM64, a dominant-negative Rex mutant that inhibits Rex activity by sequestering hCRM1 with wild-type Rex in HeLa cells. Hakata et al. previously demonstrated that overexpression of hCRM1 completely rescues Rex activity from inhibition by TAgRexM64 (17). This effect was reproduced in the present study. In contrast, rCRM1 did not restore Rex activity (Fig. 1A ). When we employed LMB, a specific inhibitor of CRM1 (24), to knock out endogenous CRM1 as an alternative method, overexpression of hCRM1, but not rCRM1, restored Rex activity again (reference 18 and data not shown). Taken together, these results reconfirm that rCRM1, in contrast to hCRM1, does not efficiently function as a cofactor of Rex.
FIG. 1.
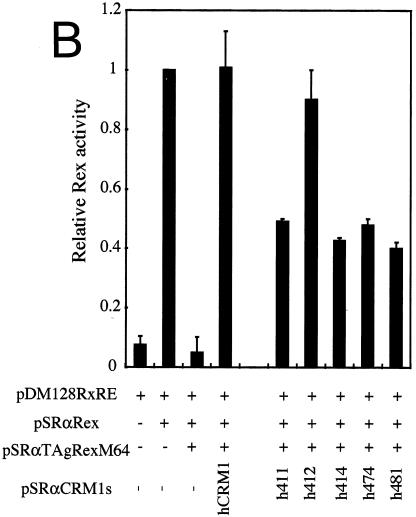
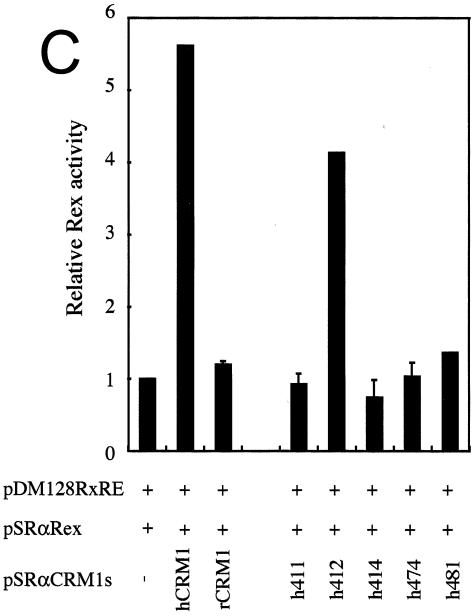
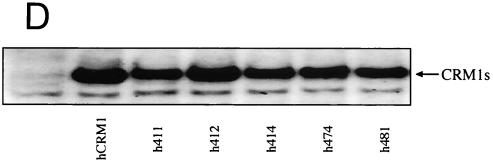
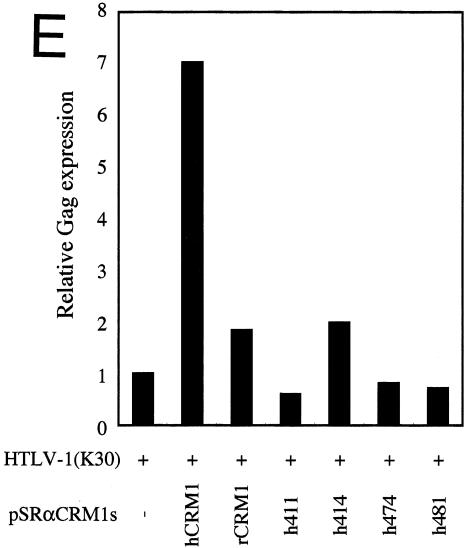
The amino acid residues of hCRM1 required for Rex activity. (A) The ability of CRM1s to support Rex activity. HeLa cells were transfected with the indicated plasmids. After cell lysis, the amount of CAT and the activity of β-Gal were measured and CAT/β-Gal ratios were calculated. The ratio for the control sample without pSRαTAgRexM64 and pSRαCRM1s was arbitrarily set at 1. The amount of CAT and the β-Gal activity in control samples were over 300 pg and 3.0 × 10−3 U, respectively. Error bars represent standard deviations. (B) Restoration of Rex activity by overexpression of CRM1s in HeLa cells. The experimental procedure was the same as that described for panel A. (C) Effect of overexpressing CRM1s on Rex activity in REF52 cells. REF52 cells were transfected with the indicated plasmids. At 24 h of posttransfection, CAT/β-Gal ratios were calculated. The ratio for the sample without pSRαCRM1s was arbitrarily set at 1. (D) Western blot analysis of various CRM1s. A fraction of each sample used in the experiments described for panel C was subjected to Western blot analysis using the anti-hCRM1 antibody to examine CRM1 protein synthesis. This antibody was raised with the peptide, which represents the carboxy-terminal region of hCRM1 and has a sequence different from that of rCRM1, so it does not recognize endogenous rCRM1. (E) Effect of CRM1s on Rex-mediated Gag expression from HTLV-1 molecular clone. At 48 h posttransfection, Gag/β-Gal ratios were calculated. The ratio for the sample without pSRαCRM1s was arbitrarily set at 1. (F) A schematic representation of hCRM1 functional domains. The amino acid sequence of hCRM1 is shown. Experiments characterizing the RanGTP binding domain (broken line) (33), LMB binding residue (arrowhead) (24), Rev-interacting amino acids (asterisks) (2), and the domain binding to NES (underline) (33) have been previously reported. The residues 411, 414, 474, and 481 in hCRM1 are indicated in bold characters, and the corresponding residues of rCRM1 are indicated under the hCRM1 sequence in single-letter amino acid code.
To identify the region of hCRM1 that is critical in supporting Rex activity, we constructed plasmids that express chimeric proteins of hCRM1 and rCRM1. One of these chimeras (designated hrCRM1) consists of the N-terminal half of hCRM1 (aa 1 to 679) and the C-terminal half of rCRM1 (aa 680 to 1072), and the other is a reverse chimera (termed rhCRM1) (Fig. 1A). Each N-terminal or C-terminal half of hCRM1 has 12 residues that differ from those of rCRM1. We compared the abilities of the two chimeric CRM1s to rescue Rex activity from inhibition by TAgRexM64. As shown in Fig. 1A, overexpression of hrCRM1 could restore Rex activity with over half the efficiency of hCRM1, whereas there was no enhancement of Rex activity by rhCRM1. These results indicated that the N-terminal region of hCRM1 is involved in mediating Rex activity.
To identify amino acids in the N-terminal half of hrCRM1 that are necessary for Rex activity, we made plasmids that express mutants in which one hCRM1-specific amino acid was replaced with the corresponding rat residue. These CRM1s were named in accordance with the specific amino acid changes; for example, in hr191, the human-type residue at position 191 of hrCRM1 was replaced with the corresponding rat residue. Western blot analysis confirmed that all the CRM1 proteins were produced at similar levels in cells (data not shown). In examining whether these mutants could restore Rex activity, we found four mutants (hr411, hr414, hr474, and hr481) that had very little capacity for reinstating activity similar to that of rCRM1 (Fig. 1A). The CRM1 mutants hr191, hr284, hr334, hr337, hr346, hr402, and hr412 restored Rex activity as efficiently as hrCRM1, while hr478 had an intermediate capacity. These data suggested that the four residues at positions 411, 414, 474, and 481 are the most important for support of Rex activity and that residue at position 478 might affect Rex activity weakly. Thus, in the following experiments we focused on the four residues at positions 411, 414, 474, and 481. The positions of these four residues are schematically represented in Fig. 1F in relation to the positions of other functional domains of CRM1.
We then constructed plasmids that express the same series of hCRM1-based mutants as to these four residues (h411, h414, h474, and h481) and a plasmid, h412, in which residue 412 of hCRM1 was replaced with the corresponding rat residue, as a control. As hrCRM1-based mutants, these corresponding mutants showed reduced abilities to restore TAgRexM64-inhibited Rex activity, whereas h412 showed the almost same activity as hCRM1 (Fig. 1B). To further confirm the importance of these four residues, we investigated whether overexpression of these CRM1s could directly enhance Rex activity in rat cells, as these experimental conditions are simpler than the TAgRexM64 system. In agreement with our previous observations, hCRM1, but not rCRM1, could promote Rex activity in rat cells. Under similar conditions, all of the mutants, with the exception of h412, had little effect (Fig. 1C). Western blot analysis showed expression levels similar to those of the CRM1s (Fig. 1D). These results clearly suggest that residues 411, 414, 474, and 481 of hCRM1 are critical for efficient Rex activity.
Next we tested whether these four residues are critical for the Rex-dependent Gag expression from the HTLV-1 molecular clone, as is the case in the results of the above-described experiments performed using a reporter plasmid, pDM128RxRE. As expected, the results indicated the significance of the four residues in Gag production (Fig. 1E). We obtained similar results even when the different amounts of CRM1 expression plasmids were transfected (data not shown). These results obviously confirm the findings obtained from the experiments using pDM128RxRE.
CRM1s with single-residue substitutions have decreased affinity for Rex in cells.
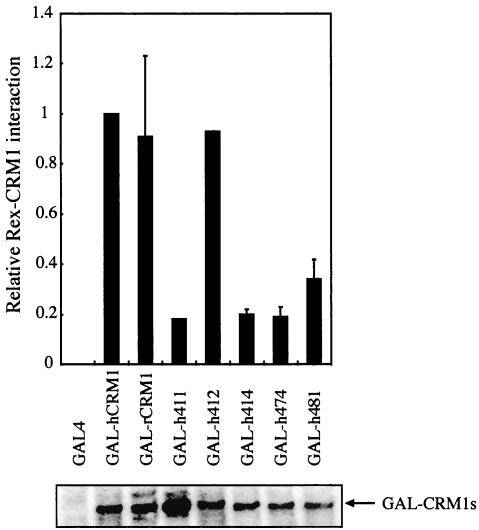
One possible explanation for the above-described results is that CRM1s with substitutions at position 411, 414, 474, or 481 might not be able to efficiently interact with Rex in the cell. To assess this possibility, we fused these CRM1s with the GAL4 domain for measurement (using two-hybrid assays) of their binding affinities for Rex-VP. The results revealed that the mutated CRM1s had a distinctly lower affinity for Rex-VP than hCRM1 and h412 (Fig. 2). Western blot analysis confirmed that all of the GAL-CRM1s were expressed at similar levels. These results indicated that the four amino acids at positions 411,414, 474, and 481 are involved in binding to Rex in cells.
FIG. 2.
In vivo interaction of Rex with CRM1 mutants in which one amino acid is replaced. REF52 cells were transfected with the plasmid expressing GAL-CRM1s in combination with pRexVP, pG5BCAT, and pCDMβ-gal. The cells were harvested and subjected to CAT and β-Gal assays, and CAT/β-Gal ratios were calculated. The ratio for the control sample, which detected the interaction between GAL-hCRM1 and Rex-VP, was arbitrarily set at 1. The amount of CAT and β-Gal activity in control samples were over 400 pg and 3.0 × 10−3 U, respectively. GAL4 nonfusion protein, expressing only a GAL4 region, was used as a negative control. A fraction of each sample was subjected to Western blot analysis using the anti-GAL4 monoclonal antibody to examine GAL-CRM1 expression.
Ability of CRM1s with single-residue substitutions to export Rex.
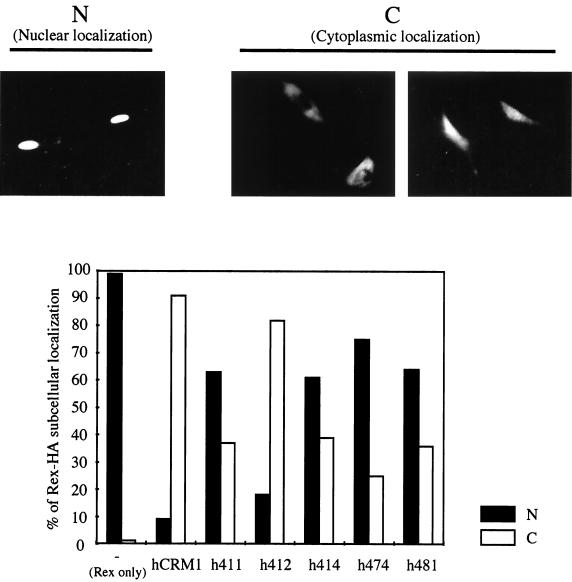
Next, to investigate the ability of the CRM1s to export Rex protein, the expression plasmids of CRM1s and Rex with an HA tag were cotransfected into REF52 cells and an indirect immunofluorescent assay was performed. Under microscopic observation, double-positive cells, which express both CRM1 and Rex-HA, were divided into the following two classes in accordance with localization of the Rex-HA protein. One class included cells in which Rex-HA is mainly localized in the nucleus, and the other included cells in which Rex-HA is mainly localized in the cytoplasm and throughout the cell (Fig. 3). The percentage of classified cells relative to total cells counted was calculated. In agreement with our previous observations, without overexpression of CRM1s, Rex-HA was observed in the nucleus (Fig. 3), whereas hCRM1 or h412 coexpression resulted in dominant cytoplasmic localization of Rex-HA, reflecting enhanced export. In contrast, cytoplasmic migration of Rex-HA was significantly reduced when the four mutant CRM1s were overexpressed, indicating that the levels of ability of these CRM1s to function as export receptors are lower than that of hCRM1, which is consistent with the in vivo binding results (Fig. 2).
FIG. 3.
The subcellular localization of Rex in the presence of overexpressed CRM1s. REF52 cells were transfected with 0.1 μg of pSRαRex-HA along with 0.3 μg of pSRαCRM1s or pSRα296. The subcellular localizations of Rex-HA and CRM1s were visualized with Cy3- and FITC-conjugated antibodies. Subcellular localization pattern of Rex-HA in the Cy3 and FITC double-positive cells were divided into two classes. The cells in which Rex-HA was mainly localized to the nucleus are designated N. The cells which contained Rex-HA throughout the cell or mainly in the cytoplasm are designated C. More than 100 cells were counted, and the percentages of cells in each class relative to the total cell count (double positive) were calculated. Three independent experiments were performed, and similar results were obtained.
The four CRM1s form the complex Rex-CRM1-RanQ69L in vitro.
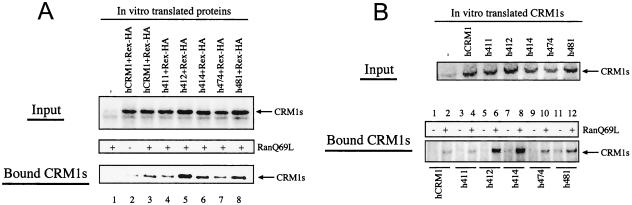
To examine the binding of CRM1s to Rex-HA in vitro, the CRM1s and Rex-HA were synthesized with an in vitro transcription-translation system. Equivalent amounts of all CRM1s were translated (Fig. 4A; input CRM1s). After the binding reaction, CRM1 proteins coprecipitated with Rex-HA in both the absence and presence of GTP-loaded RanQ69L—a Ran mutant that is resistant to GTP hydrolysis—were examined by immunoblot analysis. The amount of hCRM1 in the bound fraction increased when RanQ69L was added, suggesting that this binding was specific (Fig. 4A, lane 2 versus 3; bound CRM1s). Unexpectedly, in contrast to the in vivo results, all the CRM1s investigated had affinity for Rex-HA in the presence of RanQ69L similar to (h411, h414, h474, and h481) or apparently higher than (h412) that of hCRM1 (Fig. 2 and 4A, lanes 3 to 8). These results suggest that the four CRM1 mutants possess the ability to efficiently, and specifically, bind to Rex.
FIG. 4.
In vitro binding of CRM1s to Rex or NES peptide in the presence of RanQ69L. (A) Binding to Rex. CRM1 and Rex-HA were incubated in the absence or presence of GTP-charged RanQ69L protein for 30 min at 30°C. A portion of these samples was added to sample buffer and used as an input fraction (Input), and the remaining portion was further incubated with anti-HA monoclonal antibody bound to protein G-Sepharose The precipitated proteins were dissolved in sample buffer and subjected to Western blot analysis using anti-hCRM1 antibody (Bound CRM1s). (B) Binding to NES peptide. In vitro-translated CRM1s, a portion of which was used as an input fraction (Input), were incubated with NES peptide (which had been chemically conjugated to biotinylated BSA and immobilized on avidin-agarose) in the absence or presence of GTP-charged RanQ69L. After incubation, the precipitated proteins were dissolved in sample buffer (Bound CRM1s). The amount of translated CRM1s subjected to binding assays was 12.5 times that of the input fraction.
The four CRM1s can form the NES peptide-CRM1-RanQ69L complex.
To determine whether the in vitro binding capacity of the four mutant CRM1s is a general feature of the leucine-rich type NES, we used the NES peptide derived from PKI in place of translated Rex-HA protein in the in vitro binding assay (Fig. 4B). As seen with the experiment performed using Rex-HA, none of the mutant CRM1s obliterated the binding to NES peptide immobilized on biotinylated-BSA avidin-agarose but instead showed affinities similar to or stronger than those of hCRM1 (Fig. 4B, lanes 2, 4, 6, 8, 10, and 12). In the absence of RanQ69L, precipitated CRM1s were not detectable or, when any were present (lane 11), were at considerably lower levels than those seen in the sample containing RanQ69L (lane 12). Collectively, these two in vitro binding assays indicated that the four residues, which are located in positions 411, 414, 474, and 481 in hCRM1, are not directly involved in the formation of the NES-CRM1-RanQ69L ternary complex.
RanBP3 binding to CRM1s.
The above-described results, indicating that the four mutant CRM1s interact with Rex with lower efficiency than hCRM1 in a two-hybrid assay system (Fig. 2 and 3) although the same CRM1s have a full capacity to bind to Rex or NES peptide in vitro (Fig. 4), led us to hypothesize that another cellular factor supports export complex formation in cells. RanBP3 is a possible candidate for mediating this putative function, which was evidently perturbed by mutation of the four critical CRM1 residues. This protein has been reported to function as a scaffold protein for promotion of the efficient assembly of the protein export complex (30), and it stabilizes the interaction between hCRM1 and an export substrate via direct interaction with hCRM1, thereby forming the RanBP3-RanGTP-hCRM1-NES quaternary complex (10, 26). Thus, we investigated whether the mutant CRM1s, which exhibited reduced affinity for Rex in vivo, are able to bind to RanBP3. GST-RanBP3 immobilized on the affinity resin was incubated with in vitro-translated CRM1s, and the recovered CRM1s were visualized (Fig. 5A). HCRM1 and h412 were detected in the bound fraction, but the other CRM1s were not. These results were completely consistent with the data from the two-hybrid assay (Fig. 2), implying that positions 411, 414, 474, and 481 of hCRM1 are required for the binding to RanBP3 and that the reduced cellular affinities of the four mutant CRM1s for Rex result from poor binding between CRM1 proteins and RanBP3.
FIG. 5.
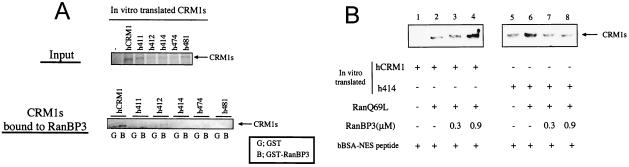
Affinity of CRM1s for RanBP3. (A) In vitro binding of CRM1s to RanBP3. In vitro-translated CRM1s, a portion of which was used as an input fraction (Input), were incubated with GST or GST-RanBP3 immobilized on glutathione-Sepharose 4B. After incubation, the precipitated proteins were subjected to Western blot analysis. The amount of translated CRM1s subjected to the binding assay was 10 times that of the input fraction (CRM1s bound to RanBP3). (B) Effect of RanBP3 on NES-RanQ69L-CRM1 ternary complex formation. In vitro-translated hCRM1 (lanes 1 to 4) or h414 (lanes 5 to 8), RanQ69L, and RanBP3 were added to biotinylated BSA (bBSA)-NES peptide immobilized on avidin-agarose as indicated. After incubation, the precipitated proteins were dissolved in sample buffer.
RanBP3 has been demonstrated to bind hCRM1 and to enhance the interaction of hCRM1 with NES peptide and RanGTP at optimal concentrations (10). Thus, we added various amounts of RanBP3 to the in vitro binding reaction to examine the effect of the presence of RanBP3 on the affinity of CRM1s for the NES peptide derived from PKI, with the reaction being performed in the presence of RanQ69L (Fig. 5B). Addition of RanBP3 at 0.9 μM promoted the interaction between NES and hCRM1 in the presence of RanQ69L (lane 4). However, RanBP3 did not enhance the interaction of h414-NES-RanQ69L at any concentration (lane 6 to 8), confirming that this mutant CRM1 could not bind to RanBP3. Similar results were obtained using other mutants (h411, h474, and h481) (data not shown).
A mutant, h411/414, can bind to both RanBP3 and Rex.
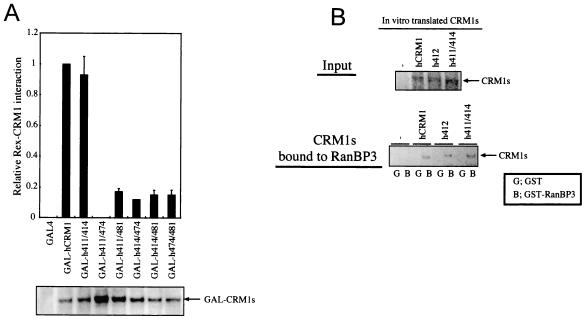
To further analyze the importance of residues 411, 414, 474, and 481 of hCRM1, we expressed mutant CRM1s with two rat-type amino acids among these four residues in hCRM1 as GAL4-fused proteins and examined (using two-hybrid assays) their ability to bind to Rex-VP (Fig. 6A). One mutant, h411/414, in which aa 411 and 414 of hCRM1 were substituted for rat residues, exhibited binding similar to that of hCRM1. Western blot analysis confirmed that all of these CRM1s were expressed at a high level.
FIG. 6.
Binding characterization of two amino acid-substituted CRM1s. (A) In vivo interaction of CRM1s with Rex. REF52 cells were treated as described for Fig. 2 except for the plasmids that expressed two amino acid-substituted CRM1s as a GAL4 fusion protein. A portion of each sample was subjected to Western blot analysis to confirm GAL-CRM1 expression. (B) In vitro binding of h411/414 to RanBP3. As shown in Fig. 5A, in vitro-translated CRM1s were incubated with GST or GST-RanBP3 immobilized on glutathione-Sepharose 4B.
Next, we investigated whether h411/414 could bind to RanBP3 in vitro in a manner similar to that of hCRM1. As shown in Fig. 6B, this mutant had the ability to bind to RanBP3. None of CRM1s were precipitated by a GST tag alone. Taken together, these results suggest that efficient binding of h411/414 to RanBP3 results in high affinity of this mutant for Rex in cells, as was observed for hCRM1 and h412.
Residues at 411 and 414 of hCRM1 are required for Rex multimerization.
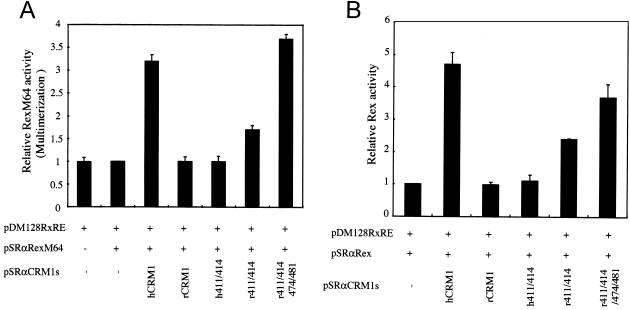
Since a mutant, h411/414, could bind efficiently to Rex in vivo, we next examined the ability of this mutant to support Rex multimerization. For this purpose we employed RexM64, which is not functional in the Rex-mediated mRNA export pathway, because it cannot multimerize. We have previously reported that when hCRM1 is overexpressed, the ability of RexM64 to multimerize is restored and, therefore, that it can function like a wild-type Rex (17). This provided evidence that hCRM1 can support Rex multimerization (Fig. 7A). In contrast, rCRM1 did not restore the activity of RexM64, implying that rCRM1 cannot support multimerization (Fig. 7A). Thus, by measuring the restoration of RexM64 activity when a CRM1 of interest was overexpressed, we were able to determine whether its CRM1 can support multimerization. When h411/414 was overexpressed RexM64 activity was not restored, suggesting that in similarity to an rCRM1 mutant, h411/414 cannot support the multimerization event (Fig. 7A). This was in agreement with the observation that h411/414 cannot enhance wild-type Rex activity in rat cells (Fig. 7B). These results indicate that residues 411 and 414 of hCRM1 are important for Rex multimerization. To confirm this hypothesis, we expressed r411/414 (which has human-type residues only at positions 411 and 414 in rCRM1) along with RexM64 (Fig. 7A). As expected, r411/414 restored RexM64 activity, suggesting that it can support Rex multimerization and promote Rex activity in rat cells (Fig. 7B). These results clearly demonstrated that these two amino acids in hCRM1 are critical for Rex multimerization. We also observed that r411/414/474/481, in which residues 411, 414, 474, and 481 of rCRM1 are changed to human-type residues, can fully support Rex multimerization and activity (Fig. 7). We summarized the results obtained from this study in Table 1.
FIG. 7.
Identification of two amino acids in hCRM1 that support Rex multimerization. (A) Restoration of the activity of RexM64 by CRM1 overexpression. HeLa cells were transfected with the indicated plasmids (0.5 μg of pDM128RxRE, 0.05 μg of pSRαRexM64, and 0.3 μg of pSRαCRM1s). All samples were also transfected with 0.1 μg of pCDMβ-gal. At 24 h posttransfection, the CAT/β-Gal ratios were calculated. The ratio for the sample without pSRαCRM1s was arbitrarily set at 1. (B) Restoration of Rex activity by overexpression of r411/414. This was done using the methods described for Fig. 1C.
TABLE 1.
The characteristics of CRM1s used in this study
| CRM1 | Support of Rex activity | Affinity for Rex in cells | Affinity for RanBP3 | Affinity for Rex and NES peptide in vitro | Support of Rex multimerization |
|---|---|---|---|---|---|
| hCRM1 | + | + | + | + | + |
| rCRM1 | − | + | +a | +b | |
| h411 | − | − | − | + | |
| h412 | + | + | + | + | |
| h414 | − | − | − | + | |
| h474 | − | − | − | + | |
| h481 | − | − | − | + | |
| h411/414 | − | + | + | − | |
| r411/414 | + | +a | + | ||
| r411/414/474/481 | + | +a | + |
Unpublished data.
Our previously published data (18).
DISCUSSION
One notable difference between the export of viral RNA and the export of protein is the requirement for multimerization of adaptor proteins, which bridge target RNAs and CRM1. hCRM1 has been shown to function dually as a promoter of multimerization and as an export receptor in Rex-mediated viral RNA export processes (17). Using a series of CRM1 mutants whose amino acids were interchanged between hCRM1 and rCRM1, we here identified two amino acids (residues 411 and 414 of hCRM1) that are crucial for Rex multimerization. These results strongly suggest that the function of hCRM1 in multimerization is independent of its function as an export receptor, since these two amino acids are located in a different region from the domain for direct binding to NES (2, 33), and that h411/414, which has a full capacity for binding to Rex, does not support Rex multimerization. Thus, to export viral RNA, binding of CRM1 to the NES of the adaptor proteins is necessary, but not sufficient, and higher-ordered interactions between Rex and CRM1 may be required. It is unlikely that the conformation of CRM1 is grossly disturbed by exchanging these two amino acids, since h411/414 has the capacity for binding RanBP3 and Rex to mediate Rex export. These observations suggest that CRM1 may directly participate in the process whereby the export complex is formed on nascent RNA. This notion is consistent with a previous report that Rev can export only nascent viral RNA in nature (22). Now, a couple of papers have reported the involvement of CRM1 in cellular mRNA export. For example, c-fos mRNA, which is devoid of introns, harbors the AUUUA sequence, which recruits HuR as an adaptor that can bind to pp32 and APRIL, which contain the NES recognized by CRM1 (15). NXF3, which is related to TAP, the mRNA export factor, and is strongly predicted to act in a certain poly(A)+ RNA export process, has also been reported to interact with CRM1 (39). Since multimerization may, therefore, be a general feature of RNA-binding proteins (including, for example, several hnRNPs) that are involved in RNA metabolism, it was of interest to investigate whether CRM1 participates in the formation of the export complex of cellular RNAs.
Four CRM1 mutants (in which residue 411, 414, 474, or 481 in the hCRM1 backbone was replaced with the corresponding rat residue) had considerably lower affinity for Rex than hCRM1, as shown by two-hybrid assay (Fig. 2) and immunofluorescence analysis (Fig. 3). Initially, these results appeared to be inconsistent with the observation that the same mutants can bind to Rex or NES peptide as efficiently as wild-type hCRM1 in the presence of RanQ69L in vitro (Fig. 4). However, this might be explained by the fact that these mutants had lost their ability to bind to RanBP3 (Fig. 5A) and therefore were unable to utilize it as a scaffold protein. Thus, the mutant CRM1s may not be actively recruited to the sites where exchange of guanine nucleotides in Ran occurs, resulting in inefficient formation of a trimeric complex comprising CRM1, Rex, and RanGTP in vivo. Moreover, the presence of RanBP3, which stabilizes the hCRM1-Rex-RanGTP complex, may also contribute to the enhanced interaction of hCRM1 with Rex in the nucleus. In contrast, concentration of the components for ternary complex formation, without supplementation of recombinant RanBP3, may be sufficiently prominent in an in vitro reaction to hide any difference in the levels of efficiency of complex formation with various CRM1s. This hypothesis was supported by the observation that supplementation of recombinant RanBP3 enhanced the complex formation with wild-type hCRM1 but not with the mutant CRM1s. Taken together, these results suggest that the four residues at positions 411, 414, 474, and 481 in hCRM1 are not involved in direct binding of hCRM1 to either RanGTP or NES. Instead, the four residues are crucial for the binding of hCRM1 to RanBP3. The fact that these CRM1 mutants with defects in RanBP3 binding (which eliminated their ability to efficiently bind to NES in cells) consequently do not support Rex function provides in vivo evidence for a critical role of RanBP3 in efficient export complex formation.
Point mutants h411 and h414, which have a single rat-type amino acid in the hCRM1 backbone at the indicated residues, could not bind to RanBP3, whereas h411/414, in which both the 411 and 414 residues were replaced with rat-type amino acids, could bind. Thus, it is conceivable that the amino acids at positions 411 and 414 in CRM1 are functionally involved in forming a binding site for RanBP3, although the direct binding of these residues to RanBP3 remains to be proven biochemically. This hypothesis is also supported by the contrasting observations that r411/414 (in which both of the indicated residues in rCRM1 backbone are human type) could support Rex activity, suggesting its binding to RanBP3, but that r411 and r414 could not support Rex activity (Fig. 7B and data not shown). Although species-specific functional linking of residues 411 and 414 is necessary for binding to RanBP3, this combination is not enough, since h474 and h481 cannot bind to RanBP3 although both of their position 411 and 414 residues are human type. Taken together, these results indicate that both the aa 411 and 414 pair and the aa 474 and 481 pair are likely to be important for RanBP3 binding. Moreover, the significance of the aa 474 and 481 pair in CRM1 function is further supported by the finding that overexpression of r411/414/474/481 restored RexM64 multimerization and Rex activity more prominently than overexpression of r411/414 (Fig. 7).
In this paper, we identified a novel functional domain which is involved in binding of hCRM1 to RanBP3 and is crucial for Rex multimerization. Although the residues that promote the dual functions of this domain overlap, these functions are distinct and separable. Moreover, while we have identified amino acids that are critical for these functions, we do not exclude the possibility that other regions of CRM1 are also implicated in RanBP3 binding and/or Rex multimerization through interaction with these amino acids. Our findings should be useful in concert with X-ray crystallography for modeling the ternary structure of CRM1. Moreover, our results clearly indicate that the process of viral RNA export from the nucleus is (at least) one determinant that restricts HTLV-1 replication in rats.
.
Acknowledgments
We thank K. Nakajima and J. Hioki for excellent technical assistance. The infectious molecular HTLV-1 clone K30 was obtained through the AIDS Research and Reference Reagent program.
This investigation was supported by grants from the Ministry of Sports and Culture (Japan) and the Ministry of Health and Welfare (Japan).
REFERENCES
- 1.Arts, G. J., M. Fornerod, and I. W. Mattaj. 1998. Identification of a nuclear export receptor for tRNA. Curr. Biol. 8:305-314. [DOI] [PubMed] [Google Scholar]
- 2.Askjaer, P., T. H. Jensen, J. Nilsson, L. Englmeier, and J. Kjems. 1998. The specificity of the CRM1-Rev nuclear export signal interaction is mediated by RanGTP. J. Biol. Chem. 273:33414-33422. [DOI] [PubMed] [Google Scholar]
- 3.Askjaer, P., A. Bachi, M. Wilm, F. R. Bischoff, D. L. Weeks, V. Ogniewski, M. Ohno, C. Niehrs, J. Kjems, I. W. Mattaj, and M. Fornerod. 1999. RanGTP-regulated interactions of CRM1 with nucleoporins and a shuttling DEAD-box helicase. Mol. Cell. Biol. 19:6276-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischoff, F. R., and H. Ponstingl. 1991. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature 354:80-82. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff, F. R., C. Klebe, J. Kretschmer, A. Wittinghofer, and H. Ponstingl. 1994. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc. Natl. Acad. Sci. USA 91:2587-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan, C. M., I. E. Gallouzi, and J. A. Steitz. 2000. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J. Cell Biol. 151:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calapez, A., H. M. Pereira, A. Calado, J. Braga, J. Rino, C. Carvalho, J. P. Tavanez, E. Wahle, A. C. Rosa, and M. Carmo-Fonseca. 2002. The intranuclear mobility of messenger RNA binding proteins is ATP dependent and temperature sensitive. J. Cell Biol. 159:795-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cullen, B. R. 1991. Regulation of human immunodeficiency virus replication. Annu. Rev. Microbiol. 45:219-250. [DOI] [PubMed] [Google Scholar]
- 9.Daly, T. J., R. C. Doten, P. Rennert, M. Auer, H. Jaksche, A. Donner, G. Fisk, and J. R. Rusche. 1993. Biochemical characterization of binding of multiple HIV-1 Rev monomeric proteins to the Rev responsive element. Biochemistry 32:10497-10505. [DOI] [PubMed] [Google Scholar]
- 10.Englmeier, L., M. Fornerod, F. R. Bischoff, C. Petosa, I. W. Mattaj, and U. Kutay. 2001. RanBP3 influences interactions between CRM1 and its nuclear protein export substrates. EMBO Rep. 2:926-932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 90:1051-1060. [DOI] [PubMed] [Google Scholar]
- 12.Fornerod, M., J. van Deursen, S. van Baal, A. Reynolds, D. Davis, K. G. Murti, J. Fransen, and G. Grosveld. 1997. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 16:807-816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukuda, M., S. Asano, T. Nakamura, M. Adachi, M. Yoshida, M. Yanagida, and E. Nishida. 1997. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 390:308-311. [DOI] [PubMed] [Google Scholar]
- 14.Gallouzi, I. E., C. M. Brennan, and J. A. Steitz. 2001. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA 7:1348-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gallouzi, I. E., and J. A. Steitz. 2001. Delineation of mRNA export pathways by the use of cell-permeable peptides. Science 294:1895-1901. [DOI] [PubMed] [Google Scholar]
- 16.Gorlich, D., M. Dabrowski, F. R. Bischoff, U. Kutay, P. Bork, E. Hartmann, S. Prehn, and E. Izaurralde. 1997. A novel class of RanGTP binding proteins. J. Cell Biol. 138:65-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hakata, Y., T. Umemoto, S. Matsushita, and H. Shida. 1998. Involvement of human CRM1 (exportin 1) in the export and multimerization of the Rex protein of human T-cell leukemia virus type 1. J. Virol. 72:6602-6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakata, Y., M. Yamada, and H. Shida. 2001. Rat CRM1 is responsible for the poor activity of human T-cell leukemia virus type 1 Rex protein in rat cells. J. Virol. 75:11515-11525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hakata, Y., M. Yamada, N. Mabuchi, and H. Shida. 2002. The carboxy-terminal region of the human immunodeficiency virus type 1 protein Rev has multiple roles in mediating CRM1-related Rev functions. J. Virol. 76:8079-8089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hieda, M., T. Tachibana, F. Yokoya, S. Kose, N. Imamoto, and Y. Yoneda. 1999. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J. Cell Biol. 144:645-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho, J. H., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 151:1057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iacampo, S., and A. Cochrane. 1996. Human immunodeficiency virus type 1 Rev function requires continued synthesis of its target mRNA. J. Virol. 70:8332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kehlenbach, R. H., A. Dickmanns, A. Kehlenbach, T. Guan, and L. Gerace. 1999. A role for RanBP1 in the release of CRM1 from the nuclear pore complex in a terminal step of nuclear export. J. Cell Biol. 145:645-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kudo, N., N. Matsumori, H. Taoka, D. Fujiwara, E. P. Schreiner, B. Wolff, M. Yoshida, and S. Horinouchi. 1999. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc. Natl. Acad. Sci. USA 96:9112-9117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kutay, U., G. Lipowsky, E. Izaurralde, F. R. Bischoff, P. Schwarzmaier, E. Hartmann, and D. Gorlich. 1998. Identification of a tRNA-specific nuclear export receptor. Mol. Cell 1:359-369. [DOI] [PubMed] [Google Scholar]
- 26.Lindsay, M. E., J. M. Holaska, K. Welch, B. M. Paschal, and I. G. Macara. 2001. Ran-binding protein 3 is a cofactor for Crm1-mediated nuclear protein export. J. Cell Biol. 153:1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madore, S. J., L. S. Tiley, M. H. Malim, and B. R. Cullen. 1994. Sequence requirements for Rev multimerization in vivo. Virology 202:186-194. [DOI] [PubMed] [Google Scholar]
- 28.Mehlin, H., B. Daneholt, and U. Skoglund. 1992. Translocation of a specific premessenger ribonucleoprotein particle through the nuclear pore studied with electron microscope tomography. Cell 69:605-613. [DOI] [PubMed] [Google Scholar]
- 29.Mueller, L., V. C. Cordes, F. R. Bischoff, and H. Ponstingl. 1998. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett. 427:330-336. [DOI] [PubMed] [Google Scholar]
- 30.Nemergut, M. E., M. E. Lindsay, A. M. Brownawell, and I. G. Macara. 2002. Ran-binding protein 3 links Crm1 to the Ran guanine nucleotide exchange factor. J. Biol. Chem. 277:17385-17388. [DOI] [PubMed] [Google Scholar]
- 31.Ohno, M., A. Segref, A. Bachi, M. Wilm, and I. W. Mattaj. 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101:187-198. [DOI] [PubMed] [Google Scholar]
- 32.Ossareh-Nazari, B., F. Bachelerie, and C. Dargemont. 1997. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science 278:141-144. [DOI] [PubMed] [Google Scholar]
- 33.Ossareh-Nazari, B., and C. Dargemont. 1999. Domains of Crm1 involved in the formation of the Crm1, RanGTP, and leucine-rich nuclear export sequences trimeric complex. Exp. Cell Res. 252:236-241. [DOI] [PubMed] [Google Scholar]
- 34.Pollard, V. W., and M. H. Malim. 1998. The HIV-1 Rev protein. Annu. Rev. Microbiol. 52:491-532. [DOI] [PubMed] [Google Scholar]
- 35.Rimsky, L., J. Hauber, M. Dukovich, M. H. Malim, A. Langlois, B. R. Cullen, and W. C. Greene. 1988. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature 335:738-740. [DOI] [PubMed] [Google Scholar]
- 36.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 90:1041-1050. [DOI] [PubMed] [Google Scholar]
- 37.Weichselbraun, I., J. Berger, M. Dobrovnik, H. Bogerd, R. Grassmann, W. C. Greene, J. Hauber, and E. Böhnlein. 1992. Dominant-negative mutants are clustered in a domain of the human T-cell leukemia virus type I Rex protein: implications for trans dominance. J. Virol. 66:4540-4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch, K., J. Franke, M. Köhler, and I. G. Macara. 1999. RanBP3 contains an unusual nuclear localization signal that is imported preferentially by importin-α3. Mol. Cell. Biol. 19:8400-8411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang, J., H. P. Bogerd, P. J. Wang, D. C. Page, and B. R. Cullen. 2001. Two closely related human nuclear export factors utilize entirely distinct export pathways. Mol. Cell 8:397-406. [DOI] [PubMed] [Google Scholar]