Abstract
The archetypal TATA-box deficient G+C-rich promoter of the murine adenosine deaminase gene (Ada) requires a 48-bp minimal self-sufficient promoter element (MSPE) for function. This MSPE was used to isolate a novel full-length cDNA clone that encodes a 66-kDa murine G+C-rich promoter binding protein (mGPBP). The mGPBP mRNAs are ubiquitously expressed as either 3.0- or 3.5-kb forms differing in 3′ polyadenylation site usage. Purified recombinant mGPBP, in the absence of any other mammalian cofactors, binds specifically to both the murine Ada gene promoter's MSPE and the nonhomologous human Topo IIα gene's G+C-rich promoter. In situ binding assays, immunoprecipitation, and Western blot analyses demonstrated that mGPBP is a nuclear factor that can form complexes with TATA-binding protein, TFIIB, TFIIF, RNA polymerase II, and P300/CBP both in vitro and in intact cells. In cotransfection assays, increased mGPBP expression transactivated the murine Ada gene's promoter. Sequestering of GPBP present in HeLa cell nuclear extract by immunoabsorption completely and reversibly suppressed extract-dependent in vitro transcription from the murine Ada gene's G+C-rich promoter. However, transcription from the human Topo IIα gene's TATA box-containing G+C-rich promoter was only partially suppressed and the adenovirus major late gene's classical TATA box-dependent promoter is totally unaffected under identical assay conditions. These results implicate GPBP as a requisite G+C-rich promoter-specific transcription factor and provide a mechanistic basis for distinguishing transcription initiated at a TATA box-deficient G+C-rich promoter from that initiated at a TATA box-dependent promoter.
Promoters that govern the transcription of mammalian genes by RNA polymerase II fall broadly into three types: the classical TATA box-dependent promoters, the initiator element (Inr)-dependent promoters, and the G+C-rich promoters, which also have been referred to as CpG islands. Transcription initiation at the TATA box-dependent promoters is dictated by the direct interaction of the TATA box with the TATA-binding protein (TBP) as a first and rate-limiting step (reviewed in reference 8). Likewise, transcription initiation at Inr-dependent promoters occurs when the Inr element interacts with sequence-specific Inr-binding proteins (35, 36). The mechanism by which transcription initiation sites within G+C-rich promoters are recognized by the RNA polymerase II transcription machinery is currently unelucidated.
Transcription initiation of a large number of mammalian genes, including most housekeeping and central nervous system genes and many highly regulated genes that control cell growth and differentiation, is under the control of G+C-rich promoters (2, 20, 23, 26, 28, 31). This class of promoter is not found in either the Drosophila melanogaster or yeast genome. Because a common characteristic of this type of promoter is the presence of a noncanonical TATA box and one or more Sp1 binding sites upstream of the major transcription initiation site, several reports have claimed that those sequences are the key functional elements in G+C-rich promoters (6, 18). However, reports that challenged these claims, in some cases even with respect to the same promoter, also have been published (2, 24, 25). The lack of obvious conserved sequence motifs shared among different G+C-rich promoters, with the exception of the Sp1 binding sites and the “noncanonical” TATA boxes, provides further impetus for investigations to elucidate how transcription can initiate nonrandomly at these precise genome locations.
The murine Ada gene has an archetypal G+C-rich promoter that contains multiple Sp1 binding sites and the noncanonical TATA-like element TAAAAAA. Neither the Sp1 binding sites nor the TAAAAAA sequence is required for basal or enhancer-activated promoter function (1, 2; C.-Y. Yeung et al., unpublished data). Instead, the minimal self-sufficient promoter activity resides within a 48-bp minimal self-sufficient promoter element (MSPE) that displays an imperfect dyad symmetry (2). Many other G+C-rich promoters contain elements with similar secondary structure-forming potential around their transcription initiation sites (2). We previously demonstrated that there are nuclear protein-binding sites within the Ada gene's MSPE (2). This observation led us to attempt the cloning and characterization of the nuclear protein responsible for directing the assembly of the RNA polymerase II transcription initiation complex at this promoter.
A multimerized MSPE probe was used to identify and clone a full-length cDNA that encodes a ubiquitously expressed 66-kDa murine protein. This 66-kDa protein, designated the murine G+C-rich promoter binding protein (mGPBP), can bind specifically to both the murine Ada gene's MSPE and the nonhomologous human Topo IIα gene's G+C-rich promoter, as revealed by competitive gel mobility shift assays. The coding sequence of mGPBP displays domain homologies with those encoding several known transcription factors. Purified recombinant mGPBP generated from Escherichia coli was used to raise polyclonal rabbit antiserum against the protein. In situ immunostaining studies showed that mGPBP is localized in the nucleus. Immunoprecipitation and Western blot analyses revealed that GPBP can form a complex with TBP, transcription fraction IIB (TFIIB), TFIIF, RNA polymerase II, and P300/CAAT binding protein (CBP) both in vitro and in vivo. In cotransfection experiments performed on various mammalian cells, elevated mGPBP expression was found to transactivate a murine Ada gene promoter-controlled reporter gene. Immunoabsorption-induced sequestering of GPBP from HeLa cell nuclear extract resulted in a complete suppression of extract-dependent in vitro transcription directed by the mouse Ada gene's G+C-rich promoter. This suppression was reversed by the replenishment of the nuclear extract with purified recombinant mGPBP. The GPBP requirement for transcription initiated at this G+C-rich promoter is promoter specific. Parallel experiments showed that similar immunoabsorption of GPBP in HeLa nuclear extract had no adverse effect on transcription initiated at the adenovirus major late gene's classical TATA box-dependent promoter and only a partial and reversible suppressive effect on the human Topo IIα gene's G+C-rich promoter, which contains a perfect TATAAA consensus element.
These results thus describe the discovery of a novel requisite promoter-specific transactivating transcription factor with demonstrable abilities to both bind a typical G+C-rich promoter and interact with multiple transcription factors that comprise the core mammalian RNA polymerase II transcription initiation complex. Since GPBP is not required for transcription directed by the adenovirus major late gene's classical TATA box-dependent promoter, the results also provide definitive proof that transcription initiated at the murine Ada gene's G+C-rich promoter is mechanistically distinct from that initiated at a classical TATA box-dependent promoter.
MATERIALS AND METHODS
mGPBP gene cloning.
A mouse brain cDNA λgt11 bacteriophage expression library of 3 × 107 clones was screened with a 32P-labeled multimerized Ada promoter fragment C (2) probe. The restriction fragment C was isolated from a plasmid, gel purified, and multimerized by ligation end to end in the presence of T4 ligase. Multimers containing four or five copies of the fragment were gel purified, end labeled with polynucleotide kinase and [γ-32P]ATP, and used to screen the expression library according to the method of Singh et al. (34). The full-length mGPBP clones were generated by using, for the 5′ rapid amplification of cDNA ends (RACE), the oligonucleotide primer 5′-CAGGCTGGAGCAAAGTCATGCTGCGCC-3′ and the AP1 primer in the first PCR and the oligonucleotide primer 5′-AGGTCCAGTCTCTCCAACTCAGTGAAAC-3′ and the AP2 primer in the nested PCR. For the 3′ RACE, the first PCR was performed with the oligonucleotide primer 5′-CTGATCTGTGACTTCAAGTTTGGACC-3′ and the AP1 primer and the oligonucleotide primer 5′-TGACGATGTGTGAAGGAGATCCTCACAGC-3′ with the AP2 primer were used for the nested PCR according to the recommendations of the manufacturer of the Marathon-ready mouse brain cDNA templates and Advantage cDNA PCR kit (Clontech), whose products were used for the PCRs.
Northern blot analyses.
Mouse multiple-tissue Northern blots (MTN; Clontech) were hybridized with mGPBP cDNA probes and washed under standard conditions (29). The probes used were gel-purified, individually cloned mGPBP cDNA restriction fragments. The blots used were stripped as previously described (C.-Y. Yeung et al., unpublished data) and reprobed with a β-actin cDNA probe as a loading control. All probes were synthesized by the random hexanucleotide-priming method (14) and purified with a Bio-Gel P-30 column (Bio-Rad).
mGPBP expression constructs.
The mGPBP cDNA's open reading frame (ORF) sequence was inserted downstream of the hexahistidine tag coding sequence of pET28a (Novagen) to yield the bacterial expression vector pETmGPBP, which can synthesize recombinant His-tagged mGPBP upon IPTG (isopropyl-β-d-thiogalactopyranside) induction. The bacterial expression vector used to synthesize the recombinant glutathione S-transferase (GST)-mGPBP fusion protein was generated by inserting the mGPBP cDNA's ORF downstream of the GST cDNA sequence in plasmid pGEX-4T-1 (Pharmacia) to yield construct pGEX-mGPBP. These bacterial expression constructs were transformed into E. coli strains BL21/DE3 and BL21, respectively, for the production of recombinant mGPBP. The eukaryotic mGPBP expression vector pCDNA-mGPBP was generated by inserting the mGPBP cDNA's ORF into the pCDNA3 expression vector (Invitrogen). The hemagglutinin antigen (HA)-tagged (15) mGPBP eukaryotic expression construct pCDNAHA-mGPBP was generated by inserting the HA tag coding sequence upstream of, and in frame with, the mGPBP cDNA's ORF in pCDNA-mGPBP.
Purification of recombinant mGPBP.
Recombinant mGPBP (His tagged) used to raise anti-mGPBP antiserum was purified under denaturing conditions with Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) by following the vendor's instructions and then isolated as a single excised band following sodium dodecyl sulfate-8.5% polyacrylamide gel electrophoresis (SDS-8.5% PAGE). The recovered mGPBP protein was concentrated by using ULTRAFREE-MC 10,000NMWL filter units (Millipore).
Recombinant His-tagged mGPBP used for functional transcription or DNA binding assays was purified under native conditions with Qiagen's Ni-NTA agarose by following the manufacturer's protocol with minor modifications. After incubation of bacterial lysate with a 50% Ni-NTA agarose slurry at 4°C for 2 h, the resin-bound protein was eluted from the agarose matrix and the protein fractions containing the purified protein were then pooled and dialyzed twice for 60 min each, first against 500 ml of dialysis buffer (20 mM Tris-HCl [pH 7.5], 50 mM KCl, 10 mM MgCl2, 10 μM ZnSO4, 1 mM EDTA, 20% glycerol, 0.5 mM dithiothreitol [DTT], 0.2 mM phenylmethylsulfonyl fluoride [PMSF]) containing 0.25 M NaCl and then against 500 ml of the same dialysis buffer without NaCl. The GST-mGPBP fusion protein used for the pull-down assay was purified according to a published protocol (16) with minor modifications. After IPTG induction, the bacterial cells were pelleted and resuspended in phosphate-buffered saline (PBS) containing 10 mM DTT, 0.5 mM PMSF, and 2 μg each of leupeptin, pepstain A, and antipain/ml. The cells were then sonicated and subsequently treated with 1/10 volume of 10% Triton X-100. The GST fusion protein in the supernatant was then purified by one-step affinity chromatography using glutathione-Sepharose beads (Pharmacia) according to the vendor's recommendations.
EMSAs.
The probes were isolated as restriction fragments from plasmids and radiolabeled with [α-32P]dCTP by using Klenow fragments. Electrophoretic mobility shift assays (EMSAs) were performed as previously described (10) with minor modifications. Binding-reaction mixtures containing purified bacterially expressed recombinant proteins were prepared in binding buffer (10 mM Tris-HCl [pH 7.5], 60 mM KCl, 5 mM MgCl2, 0.5 mM DTT, 0.1 mM EDTA, 10 μM ZnCl2, 0.05% NP-40, 12 to 15% glycerol) containing 0.5 μg of poly(dI/dC) (total volume, 20 μl). After the binding reaction mixtures had been incubated for 10 min at room temperature, the radiolabeled probe was added for an additional 20 min of incubation at room temperature. The mixture was then separated in 4% acrylamide-bisacrylamide (29:1) nondenaturing polyacrylamide gel containing 1× Tris-borate-EDTA buffer.
Generation of anti-mGPBP antiserum and purification of the antibodies.
Immunization was performed at the Immunological Resource Center at the University of Illinois. Purification of antibodies from the antiserum was carried out with the ImmunoPure(A/G) immunoglobulin G purification kit (Pierce), as suggested by the manufacturer. Eluted antibodies were combined, dialyzed against PBS, and concentrated with centrifugal filtration units (Millipore).
Antibodies.
The antibodies used were anti-HA monoclonal antibodies (clone 12CA5; Boehringer Mannheim), anti-RNA polymerase II C-terminal domain monoclonal antibodies (Promega), anti-HA monoclonal antibodies conjugated with agarose, rabbit anti-TFIIB polyclonal antibody C-18, rabbit anti-TFIIF RAP30 polyclonal antibody C-17, anti-TBP monoclonal antibody 58C9, rabbit anti-CBP polyclonal antibody A-22 (Santa Cruz Biotechnology), and anti-nucleoporin p62 monoclonal antibody clone 53 (Transduction Laboratories).
Indirect immunofluorescence.
HeLa cells (5 × 105) were plated in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal calf serum in 60-mm-diameter dishes containing glass coverslips and incubated at 37°C overnight. The following day the cells were transfected with 5 μg of either the pCDNAHA-mGPBP expression construct or the pCDNA-HA control vector by a calcium phosphate transfection protocol as described previously (2). Sixteen hours after transfection, the cells were rinsed extensively with PBS, treated with fresh medium, and incubated for an additional 48 h. The cells were then rinsed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. After fixation the cells were permeabilized with 0.2% Triton X-100-PBS for 5 min, rinsed, and stained with either the anti-mGPBP (αN-80) antiserum- or preimmune antiserum-derived antibodies that had been purified by binding to protein A-conjugated beads or an antibody against HA (2 μg/ml) at 37°C for 1 h. After being washed four times (5 min/wash) with PBS, the cells were stained with either rabbit or mouse fluorescein-conjugated secondary antibodies (1:200 dilution) (Amersham) at 37°C for an additional hour. The coverslips were then washed extensively, mounted in antifade solution (Vector Lab) containing 0.25 μg of DAPI (4′,6′-diamidino-2-phenylindole) to counterstain the nuclei, and photographed with a Zeiss Axiovert microscope with an attached Princeton Instruments charge-coupled device camera.
Preparation of nuclear extracts.
Nuclear extracts were prepared as previously described (4) with minor modifications. Mouse Cl-1D cells were collected and washed with 10 volumes of PBS once. The cells were then suspended in 5 volumes of cold buffer A (10 mM HEPES-KOH [pH 7.9]; 10 mM KCl; 1.5 mM MgCl2; 0.1 mM EGTA; 0.5 mM DTT; 0.5 mM PMSF; 2 μg each of antipain, leupeptin, and pepstain/ml) and incubated on ice for 15 min. Cells were lysed by 20 strokes of a Dounce homogenizer (B pestle) and a further 2 to 5 strokes in the presence of 0.3 to 0.4% Nonidet P-40 (NP-40). The lysed cells were centrifuged at 1,300 × g for 10 min. The pelleted nuclei were washed three times with buffer A without NP-40, centrifuged again as described above, resuspended in 1 to 2 volumes of cold buffer B (10 mM HEPES-KOH [pH 7.9], 0.1 mM EGTA, 0.5 mM DTT, 400 mM NaCl, 5% glycerol, 0.5 mM PMSF), and incubated on ice for 60 min. The solution was then centrifuged at 48,000 × g for 1 h. The supernatant was dialyzed twice against 500 ml of dialysis buffer (20 mM HEPES-KOH [pH 7.9], 75 mM NaCl, 0.1 mM EDTA, 0.5 mM DTT, 20% glycerol, 0.5 mM PMSF) for 1 h and cleared by centrifugation in a microcentrifuge for 15 min. HeLa cell nuclear extract was from Promega.
Western blotting.
Cells were lysed in radioimmunoprecipitation assay buffer (25 mM Tris [pH 8.2], 50 mM NaCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 0.1% sodium azide) containing 1 mM PMSF, 10 μg of aprotinin/ml, and 10 μg of leupeptin/ml. Cell lysates were incubated on ice for 15 min and centrifuged at 12,000 × g for 10 min at 4°C. The protein concentration of the cleared lysates was determined with the Bio-Rad protein assay kit. Equal amounts of protein per lane were analyzed by SDS-8.5% PAGE. Western blotting was carried out with Amersham polyvinylidene difluoride membranes according to the vendor's recommendations.
Pull-down assay.
Immobilization of bacterially expressed GST and GST-mGPBP proteins was accomplished by incubating 1 to 5 μg of the proteins with 100 μl of a 50% slurry of glutathione-Sepharose beads (Pharmacia) for 1 h at 4°C. The protein-bound beads were collected and washed three times with PBS buffer. The beads were then resuspended in binding buffer (20 mM HEPES-KOH [pH 7.9], 75 mM NaCl, 0.1 mM EDTA, 0.5 mM DTT, 20% glycerol, 0.5 mM PMSF) (17). About 1/60 of the beads were subjected to SDS-8.5% PAGE to confirm that GST proteins were bound to the beads. The rest of the beads were incubated with 100 μg of Cl-1D cell nuclear extract on a rotator overnight at 4°C. The beads were collected and washed three times with washing buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1 mM EDTA, 0.1 to 0.3% NP-40). The bound proteins were eluted with SDS sample buffer and subjected to SDS-4 to 20% PAGE. Western blotting was then carried out on the gels.
Coimmunoprecipitation analyses.
Cells were collected by centrifugation in PBS 48 h after DNA transfection. Cells from each 100-mm-diameter dish were treated with 60 μl of lysis buffer containing 20 mM HEPES, pH 7.9, 400 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM DTT, 0.5 mM PMSF, 10% glycerol, and 1.5 μl of a protease inhibitor cocktail (Sigma) (17). Cell lysates were incubated for 30 min at 4°C and centrifuged at 13,000 × g for 10 min at 4°C. The supernatant from each 100-mm dish was treated with 300 μl of lysis buffer without NaCl and incubated with the agarose bead-bound anti-HA antibody (Santa Cruz Biotechnology) overnight at 4°C. The beads were pelleted and washed three or four times with 1 ml of buffer W (20 mM Tris-HCl [pH 8.0], 100 mM NaCl, 0.1 to 0.3% NP-40, 1 mM EDTA, 1 mM DTT, 0.5 mM PMSF). Bound proteins were analyzed by Western blotting using a 4 to 20% gradient polyacrylamide precast SDS-PAGE gel (BioWittaker Molecular Applications).
Cell culture and transfections.
Monolayer cultures of murine Cl-1D LM (thymidine kinase−) fibroblast cells derived from bone marrow stromal cells of a (C57BL/6J × C3H/HeJ)F1 mouse and human embryonic kidney (HEK) 293 cells were maintained in 10% fetal calf serum in DMEM. Transfections into 293 and Cl-1D cells for transactivation assays were carried out with Lipofectamine Plus (for 3 h), as suggested by the manufacturer (Gibco BRL). The transfected cells were maintained in medium for about 24 to 36 h before being collected for luciferase assays. All transfection assays were repeated at least three times with different DNA preparations. All coimmunoprecipitation assays were performed using Cl-1D cells that had been transfected by the calcium phosphate coprecipitation technique as previously described (2). The coimmunoprecipitation assays were performed on the cells 45 to 48 h after transfection.
Luciferase assay.
For each 60-mm plate of Cl-1D or 293 cells, 0.2 μg of the luciferase reporter gene under the control of the murine Ada gene promoter was cotransfected with various amounts of the mGPBP expression plasmid (pCDNA-mGPBP) and 0.1 μg of Rous sarcoma virus-β-galactosidase (β-Gal) plasmid. The total amount of plasmid used per plate was brought up to 2 μg with the empty pCDNA3 vector. The transfected cells were washed with PBS and then cultured in 10% fetal calf serum in DMEM for 24 to 36 h prior to being harvested for reporter gene expression analyses. The luciferase activities of the transfected-cell lysates were measured with a luciferase assay system (Promega) and the VICTOR2 multilabel counter (Wallac). The obtained values were then normalized according to the β-Gal activity as previously published (29).
In vitro transcription assays.
In vitro transcription reactions were carried out as previously described (12) with minor modifications. Supercoiled template DNA was purified by banding twice in centrifuged CsCl gradients (29). The templates used consist of G-less cassette reporters either without a promoter (construct pC2AT- [30]), with the adenovirus major late promoter (construct PMLC2AT [30]), with the murine Ada gene promoter (pmADAPC2AT; generated by PCR), or with the human Topo IIα gene promoter (see Fig. 8A). Transcription reaction mixtures (total volume, 25 μl) containing 8 mM HEPES (pH 7.9), 40 mM KCl, 6 mM MgCl2, 0.08 mM EDTA, 0.2 mM DTT, 8% glycerol, 30 U of RNase T1, 100 ng of template DNA, 9 to 10 μg of HeLa nuclear extract (Promega), and different amounts of antibodies and purified recombinant protein were incubated at 30°C for 10 min prior to the addition of 0.2 mM ATP, CTP, and GTP to 0.2 mM each, of UTP to 8 nM, of 3′-O-methyl-GTP (Pharmacia) to 0.05 mM, and of [α-32P]UTP (800 Ci/mmol) to 1 μM. The reaction mixtures were then incubated for 1 h at 30°C. The reactions were stopped by the addition of 175 μl of stop solution (0.3 M Tris-HCl [pH 7.4], 0.3 M sodium acetate, 0.5% SDS, 2 mM EDTA, 25 μg of tRNA/ml) and an extraction control 200-bp end-labeled DNA fragment. The reaction solutions were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol). The nucleic acids in the reaction were ethanol precipitated, air dried, and dissolved in 4 μl of nuclease-free water. An equal volume of loading dye (98% formamide, 10 mM EDTA, 0.1% xylene cyanol, 0.1% bromophenol blue) was added to the solution of in vitro-transcribed RNA. These RNA solutions were then heated at 90°C for 10 min and electrophoresed in a 6% denaturing (7 M urea) polyacrylamide gel containing 0.5× Tris-borate-EDTA buffer. The amounts of radiolabeled transcript and extraction control bands were then quantified with a PhosphorImager (Molecular Dynamics).
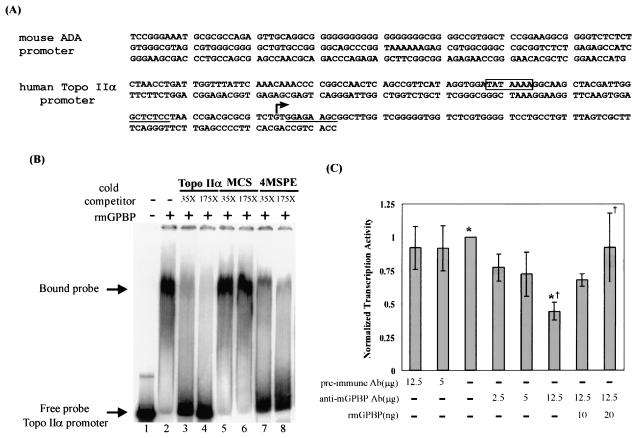
FIG. 8.
The TATA box containing the Topo IIα gene's G+C-rich promoter binds specifically to mGPBP but is only partially dependent on its presence for promoter function. (A) The human Topo IIα gene promoter shows no obvious sequence homology to the Ada gene's MSPE. The sequences displaying an imperfect dyad symmetry flanking the major transcription initiation site (arrow) are underlined. The consensus TATA element in the Topo IIα gene promoter is boxed. (B) Electrophoretic mobility of the 32P-labeled Topo IIα gene promoter probe (lane 1, arrow) was retarded (bound probe) by the presence of purified recombinant mGPBP (lane 2). This retardation of the probe can be reversed by competition with excess unlabeled probe (lanes 3 and 4) or linked quadruple copies of the MSPE derived from the Ada gene promoter (lanes 7 and 8), but not by excess copies of a 200-bp plasmid sequence that shows no secondary conformational changes under negative superhelicity (lanes 5 and 6). (C) In vitro transcription of supercoiled reporter genes driven by the Topo IIα gene promoter with HeLa nuclear extract was partially suppressed by the presence of anti-GPBP antibodies but was unaffected by the presence of preimmune antibodies. The suppressive effect of the anti-GPBP antibodies can be fully reversed by the addition of 20 ng of purified recombinant mGPBP to the reaction mixture. The statistical significance (P) for the difference of transcription levels in the absence and presence of anti-mGPBP antibodies (*) was found by a paired Student t test to be <0.000003 (n = 5), and P for the difference of transcription levels in the presence of the anti-mGPBP antibodies with and without additional recombinant mGPBP (†) was found by a paired Student t test to be <0.028 (n = 5).
Nucleotide sequence accession number.
The complete cDNA sequence corresponding to the 3.0-kb mGPBP mRNA has been assigned GenBank accession no. AY382529.
RESULTS
Molecular cloning of mGPBP.
The 48-bp MSPE fragment C within the murine Ada gene promoter (Fig. 1 of reference 2) was multimerized and used as a labeled probe to screen a λgt11 bacteriophage mouse brain cDNA expression library for cloned proteins that bind to this MSPE. Screening with the negative control Sp1 binding consensus motif probe (TGGGCGGGGC) eliminated clones expressing proteins that bind nonspecifically to random G+C-rich sequences. Out of 3 × 107 clones, 2 clones were identified after tertiary screens and were further examined by subcloning, restriction digestion, and sequence analysis.
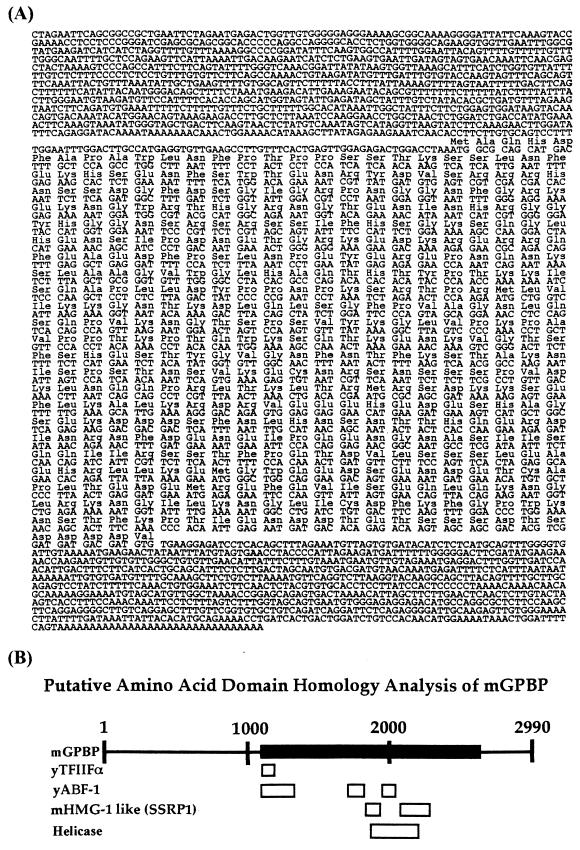
FIG. 1.
mGPBP sequence and sequence homology. (A) Complete cDNA sequence corresponding to the 3.0-kb murine GPBP mRNA, including the deduced amino acid sequence encoded by the ORF (GenBank accession number AY382529). Sequence homology searches revealed small regional sequence homology on the amino acid sequence level with several known proteins as shown in panel B. Solid box, ORF; open boxes, regions homologous to the yeast TFIIFα subunit (yTFIIFα), yABF-1, murine SSRP1 (mHMG-1 like), and helicases.
The goal of our search was to identify a G+C-rich promoter binding transcription factor which can participate in the assembly of the RNA polymerase II transcription initiation complex. Sequence analysis of the subcloned cDNA fragments derived from the two clones revealed that one cDNA insert fragment contained an ORF which had regions of sequence homology with genes encoding several known transcription factors. These factors include yeast TFIIFα, which interacts with RNA polymerase II in transcription initiation complex assembly (3, 19); yeast ARS binding factor 1 (yABF-1), which is a known transcription factor involved in DNA replication initiation in yeast (5, 22); mouse high mobility group 1-like structure-specific recognition protein 1 (SSRP1), which apparently binds to DNA through structural recognition (7, 33) and which participates in both transcription activation (13, 38) and elongation (27); and helicases, which can unwind double-stranded DNA for transcription and replication (reviewed in reference 40). Based on our expectations of the properties of the gene sought, this fragment was deemed to be a good candidate clone and was used to generate two full-length cDNA clones, 3.0 and 3.5 kb in size, by 3′ and 5′ RACE. The entire cDNA sequence of the 3,451-bp clone, including the deduced 493-amino-acid (aa) sequence encoded by the 1,479-bp ORF and the 815-bp 3′ untranslated region (UTR), is shown in Fig. 1A. This cloned gene was named the mGPBP gene.
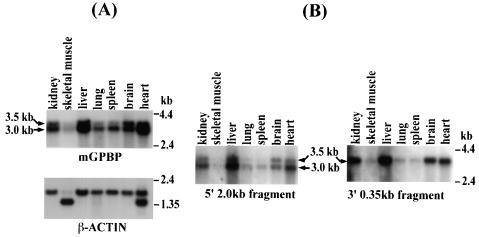
The mGPBP transcript is ubiquitously expressed as two mRNA species differing in 3′ polyadenylation site utilization.
Because G+C-rich promoters control practically all housekeeping genes, which are essential for cell survival, we predicted that, if mGPBP is critical for transcription of these genes, it would be ubiquitously expressed. The full-length mGPBP cDNA sequence was labeled and used as a probe to determine the distribution in tissue and mRNA sizes of the gene in a Northern blot analysis. The results (Fig. 2A) showed that the gene is highly expressed in all mouse tissues analyzed as two mRNA species of 3.0 and 3.5 kb. Because we had two 3′ RACE products that differed in size by ∼500 bp and because each product contained consensus AATAAA polyadenylation signaling motifs located appropriately upstream of the poly(A) tracts, we surmised that the 3.0- and 3.5-kb mRNA species may represent the same RNA with different polyadenylation site utilization. This prediction was confirmed when we cloned out a 2.0-kb fragment from the 5′ end and a 350-bp fragment from the 3′ end of the longer cDNA clone and used them as probes on the same Northern blot, which was stripped between each hybridization experiment. The results (Fig. 2B) revealed that the 5′ probe hybridized with both the 3.0- and 3.5-kb mRNA species, whereas the 3′ probe hybridized only with the 3.5-kb mRNA species. These observations indicate that both these cross-hybridizing mRNAs contain the entire ORF of the mGPBP gene and differ only in their choice of 3′ polyadenylation site usage.
FIG. 2.
Northern blot analyses to examine the mGPBP mRNA species and tissue distribution. Approximately 3 μg of poly(A+) RNA derived from the tissues indicated was loaded per lane. (A) The blot was first probed with an mGPBP-specific probe (top) and then stripped and reprobed with a β-actin probe (bottom). (B) The blot was probed with a 2.0-kb probe derived from the 5′ terminus of the mGPBP cDNA (left), stripped, and reprobed with a 350-bp probe derived from the 3′ terminus of the mGPBP cDNA corresponding to the 3.5-kb mGPBP mRNA (right). The 5′ probe hybridized with both mRNA species, whereas the 3′ probe hybridized with only the 3.5-kb mRNA.
The mGPBP ORF encodes a ubiquitously expressed 66-kDa protein.
Expression of the ORF in both bacteria (Fig. 3) and in reticulocyte lysates (data not shown) resulted in the synthesis of a novel 66-kDa protein. The presence of all or part of the 1.15-kb 5′ UTR in the mGPBP mRNA had an inhibitory effect on the translation of this mRNA in both systems (data not shown). However, only a single 66-kDa translation product was produced irrespective of the size of the 5′ UTR present in the mRNA (data not shown). The 3′ UTR of the GPBP mRNA contains multiple TAAAT repeats (Fig. 1A) that are reportedly involved in mRNA turnover control (32). The presence of either the long or short 3′ UTR also had no effect on the size of the translation product generated from the mRNA with either prokaryotic or mammalian in vitro translation systems (data not shown). To generate purified mGPBP and anti-mGPBP antibodies for functional assays, the 1,479-bp ORF of the mGPBP cDNA clone was inserted downstream of the IPTG-inducible promoter as an in-frame histidine-tagged fusion protein-encoding sequence in the PET28a bacterial expression plasmid. The resultant pETmGPBP plasmid was introduced into the E. coli BL21/DE3 strain host for recombinant mGPBP synthesis. Cell lysates from IPTG-induced and IPTG-uninduced transformed BL21 cells were prepared and analyzed by SDS-PAGE and Coomassie blue staining. Cell lysates from IPTG-induced cells displayed a prominent 66-kDa protein band that was absent from IPTG-uninduced cells (Fig. 3A, lanes 3 and 2, respectively). The His-mGPBP fusion protein purified by Ni2+ resin affinity column chromatography and SDS-PAGE was found to be >95% pure, as revealed by SDS-PAGE and Coomassie blue staining analysis (Fig. 3A, lane 4). This purified His-mGPBP was used to raise rabbit polyclonal antiserum against mGPBP. The resulting anti-mGPBP antiserum was shown by Western blot analyses to be immunoreactive against mGPBP in both BL21 cell lysate carrying IPTG-induced pETmGPBP and purified-mGPBP preparations (Fig. 3A, lanes 7 and 8, respectively) and showed no cross-reactivity with BL21 bacterial proteins (Fig. 3A, lane 6). This anti-mGPBP antiserum was used to examine the presence of GPBP in several human and mouse cell lines. All of the cell lysates analyzed by Western blotting using the anti-mGPBP antiserum as the probe revealed a prominent 66-kDa band (Fig. 3B). Since the cells used were of both mouse (Fig. 3B, lanes 2 and 5) and human (Fig. 3B, lanes 1, 3, 4, and 6) origin, the results indicate that our anti-mGPBP antiserum is immunoreactive with both mouse and human GPBP. This conclusion was confirmed by subsequent Western blot analyses performed on purified and recombinant human GPBP, which is encoded by a similarly sized ORF in our full-length human GPBP cDNA clone (C.-Y. Yeung et al., unpublished data). Thus the ORF within our full-length mGPBP cDNA clone encodes a 66-kDa protein and the translation product of that ORF in either bacterial or mammalian cells was immunoreactive to our anti-mGPBP antiserum. Tissue distribution analysis confirmed that GPBP is present in all mammalian tissues and cells examined to date, as predicted for a candidate critical regulator of housekeeping gene transcription.
FIG. 3.
Mammalian GPBP is approximately 66 kDa in size and is ubiquitously expressed. (A) Electrophoretically separated proteins in SDS-polyacrylamide gels stained with Coomassie blue. Bacterial lysate derived from a BL21 strain carrying the PET-mGPBP expression plasmid was analyzed either without (−) or with (+) IPTG induction. Lane 4, purified recombinant mGPBP; lanes 1 and 5, molecular mass markers; lanes 7 and 8, Western blot analysis of the samples used in lanes 3 and 4, respectively, using antiserum against mGPBP as the probe; lane 6, analysis of control BL21 cell (without the PET-mGPBP expression plasmid) lysate. (B) Western blot analyses of cell lysates derived from human HeLa cells (lane 1), mouse Cl-1D cells (lane 2), human JEG-3 cells (lane 3), human VA-2 cells (lane 4), mouse M2-10B4 cells (lane 5), and human 293 cells (lane 6) using the anti-mGPBP antiserum as the probe.
Recombinant mGPBP can bind specifically to the MSPE of the mouse Ada gene.
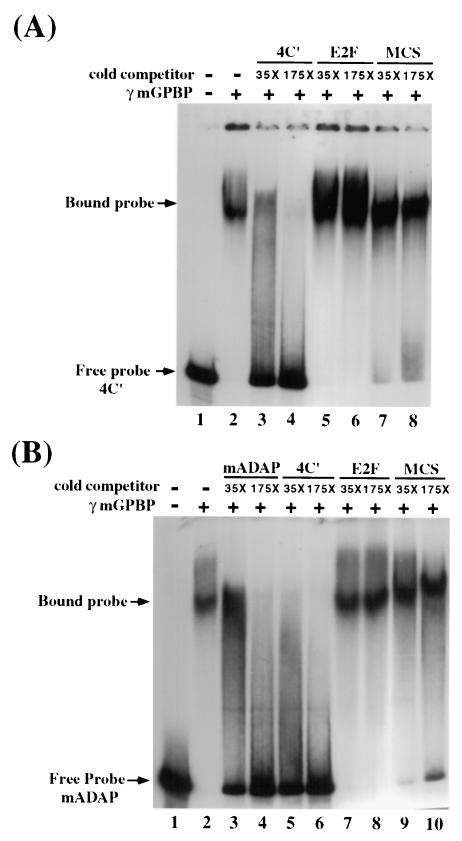
The DNA binding capability of mGPBP was examined directly in EMSAs using either the multimerized fragment C′ with imperfect dyad symmetry (see Fig. 5 of reference 2) or the 236-bp murine Ada gene promoter as the probe. The assay results are shown in Fig. 4. In Fig. 4A, the labeled DNA probe used consisted of four tandem-repeat copies of fragment C′ (4C′) that had been end ligated. Purified recombinant mGPBP bound specifically to this probe and retarded probe electrophoretic mobility (Fig. 4A, lanes 1 and 2). This binding was specifically competed out by adding excess unlabeled probes (lanes 3 and 4), but not by adding similar amounts of unlabeled E2F binding motif (lanes 5 and 6) or a 200-bp plasmid sequence (lanes 7 and 8). As indicated in Fig. 4B, a single copy of fragment C′ in the context of the labeled 236-bp mouse Ada gene promoter also bound to purified recombinant mGPBP (lanes 1 and 2). This binding was competed out by excess unlabeled probe (lanes 3 and 4) or by excess amounts of the unlabeled tandem repeated C′ fragment (4C′) probe used in Fig. 4A (lanes 5 and 6). Again, this binding was not competed out by similar excess amounts of unlabeled E2F binding sequence (lanes 7 and 8) or the 200-bp plasmid sequences (lanes 9 and 10). In similar gel mobility shift assays using the 200-bp plasmid sequences as the probe, mGPBP was unable to bind to and retard the mobility of this nonspecific DNA probe (data not shown). These results demonstrate that recombinant mGPBP can bind specifically to the 48-bp MSPE located within the 236-bp murine Ada gene promoter.
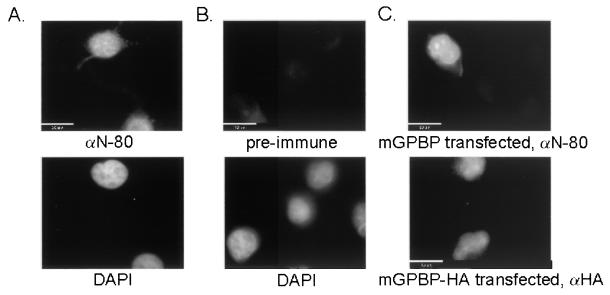
FIG. 5.
Both human and transfected mGPBP are localized in the nucleus. Human HeLa cells were stained with an antibody against mGPBP (N-80) (A, top) or an antibody from preimmune serum (B, top). The location of the nucleus was determined by counterstaining the cells with DAPI (A and B, bottom). The transfected HA-tagged mGPBP was localized with the αN-80 (C, top) and an anti-HA (C, bottom) antibodies.
FIG. 4.
The purified recombinant mGPBP can bind specifically to the mouse Ada gene's G+C-rich promoter in EMSA. (A) The DNA probe 4C′ is four copies of the MSPE C′ that were end ligated. Purified recombinant mGPBP (rmGPBP) bound specifically to the probe and caused a shift in probe electrophoretic mobility from the free-probe location (lane 1) to the bound-probe location (lane 2). This binding can be specifically competed out by adding 35-fold (lane 3) and 175-fold (lane 4) excess unlabeled probes but cannot be competed out by adding similar amounts of unlabeled E2F binding motifs (lanes 5 and 6) or a 200-bp plasmid sequence (lanes 7 and 8). (B) A single copy of the fragment C′ in the context of the labeled 236-bp mouse Ada gene promoter can also bind to and be electrophoretically retarded by the purified rmGPBP (lanes 1 and 2). This binding can be competed out by excess unlabeled probe (lanes 3 and 4) or the 4C′ probe used in panel A (lanes 5 and 6). This binding is again not competed out by unlabeled E2F binding sequences (lanes 7 and 8) or the 200-bp plasmid sequences (lanes 9 and 10).
GPBP is a nuclear protein.
Since cellular transcription factors all function within the cell nucleus, we used indirect immunofluorescence to examine the cellular localization of the GPBP. This was accomplished by using protein A affinity-purified antibodies derived from our anti-mGPBP antiserum αN-80 to immunostain fixed and permeabilized human HeLa cells, which were then counterstained with DAPI to locate the nuclei. The anti-mGPBP signal was localized to the nucleus (Fig. 5A). In control experiments using protein A affinity-purified antibodies derived from preimmune antiserum as a probe, immunoreactive signals were negligible even upon prolonged photographic exposure (Fig. 5B). The nuclear localization of GPBP was confirmed by directly transfecting the mGPBP expression vector into human HeLa cells and immunostaining for mGPBP with our anti-mGPBP antibodies (Fig. 5C, top). Definitive proof that our recombinant mGPBP was localized to the nucleus was provided by transfecting an mGPBP-HA tag fusion protein expression vector into human HeLa cells and immunostaining for the tagged protein with anti-HA antibodies (Fig. 5C, bottom). These in situ immunohistochemical analyses demonstrated that both the endogenous human GPBP and the exogenous cDNA-encoded recombinant mGPBP could translocate into the cell nucleus.
GPBP interacts with multiple key factors that participate in mammalian RNA polymerase II transcription initiation complex assembly.
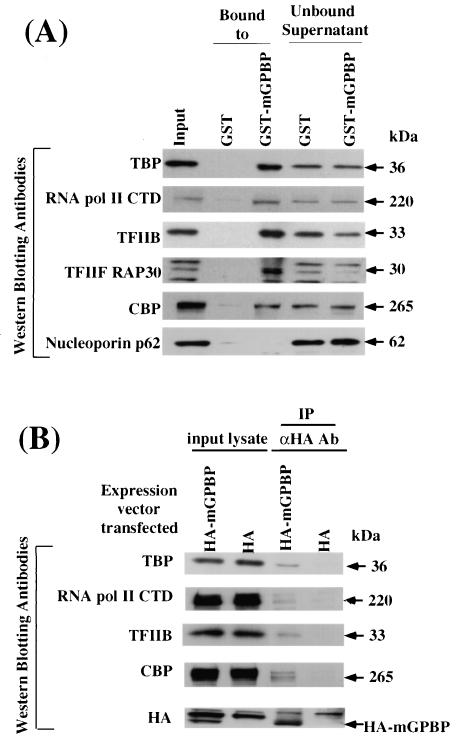
If GPBP binding to the G+C-rich promoter's MSPE can lead to the assembly of the transcription initiation complex at that location, GPBP should be able to interact with one or more transcription factors that normally participate in transcription initiation complex formation. This expectation was tested by assaying whether various nuclear transcription factors that comprise the RNA polymerase II transcription initiation complex could form complexes with immobilized recombinant mGPBP. Recombinant mGPBP fused in-frame to the GST tag was immobilized by binding the GST moiety to glutathione covalently linked to beads. Mouse Cl-1D nuclear extract proteins were then allowed to bind to the bead-immobilized mGPBP. Both the unbound proteins in the supernatant and the proteins that remained bound to the immobilized mGPBP after extensive washing were then analyzed by Western blotting using antibodies against various transcription factors as probes. The results showed that known transcription initiation complex factors, such as TBP, TFIIB, TFIIF RAP30, and RNA polymerase II, as well as the transcription factor P300/CBP, all formed complexes with the immobilized recombinant mGPBP (Fig. 6A). None of these proteins formed complexes with only the GST tag immobilized onto the same glutathione beads, and the unbound supernatant recovered from both binding assays showed that the same proteins were similarly present in both binding assays. The nuclear membrane protein nucleoporin p62, which does not participate in transcription initiation complex formation, showed no affinity to either immobilized mGPBP or GST (Fig. 6A). These in vitro results demonstrate that mGPBP can form complexes specifically with several key transcription initiation factors.
FIG. 6.
The mGPBP can form complexes with multiple transcription initiation complex assembly specific factors. (A) Mouse Cl-1D cell nuclear extract proteins were allowed to bind to bead-immobilized GST-mGPBP fusion proteins. The various proteins, including the starting nuclear extract (input), proteins that bind to bead-immobilized GST or GST-mGPBP fusion proteins (lanes “bound to”), and proteins that did not bind to bead immobilized GST or GST-mGPBP fusion proteins (lanes “unbound supernatant”) were analyzed by Western blotting. The antibodies used to probe these blots were raised against TBP, the C-terminal domain of RNA polymerase II (RNA pol II CTD), TFIIB, TFIIF RAP30 subunit, P300/CBP, and the negative control, the nuclear envelope protein nucleoporin p62. The estimated molecular masses of proteins in each band based on electrophoretic mobility in comparison to protein size markers are on the right. (B) In vivo binding of mGPBP was determined by coimmunoprecipitation analyses of cell lysates. Western blot analyses showed Cl-1D cell lysate proteins after the cells were transfected with expression vectors that express either the HA tag (HA) alone or the HA tag fused to mGPBP (HA-mGPBP), without immunoprecipitation (input lysate) or after immunoprecipitation with antibodies against the HA tag (IP αHA Ab). The antibodies raised against TBP, RNA pol II CTD, TFIIB, CBP, and the HA tag were used as probes as indicated in each blot. The estimated molecular masses of proteins in each band based on electrophoretic mobility in comparison to protein size markers are shown. The migration location of the HA-mGPBP fusion protein is also shown on the right.
To examine whether the in vitro complexing of GPBP with these transcription factors also occurs in vivo, we performed a coimmunoprecipitation experiment using an HA-tagged mGPBP expression construct in mouse Cl-1D cells. Anti-HA antibodies were used to immunoprecipitate nuclear extract proteins derived from the transfected cells. As a control, nuclear extract from cells transfected with the HA expression vector containing no GPBP-encoding sequences underwent a similar immunoprecipitation procedure. The proteins that coimmunoprecipitated with anti-HA antibodies were then analyzed by Western blotting using antibodies against TBP, TFIIB, CBP, and RNA polymerase II as probes. A portion of the respective nuclear extracts prior to treatment with anti-HA antibodies (Fig. 6B, input lysate) were shown by Western blotting to contain all the requisite proteins. None of the nuclear transcription factors of interest coimmunoprecipitated with the HA tag alone. In contrast, TBP, TFIIB, CBP, RNA polymerase II (Fig. 6B), and TFIIF RAP30 (data not shown) all coimmunoprecipitated with the HA-tagged mGPBP These results demonstrate that GPBP forms complexes with these transcription factors both in vitro and in vivo.
Transcription from the mouse Ada gene's G+C-rich promoter requires GPBP.
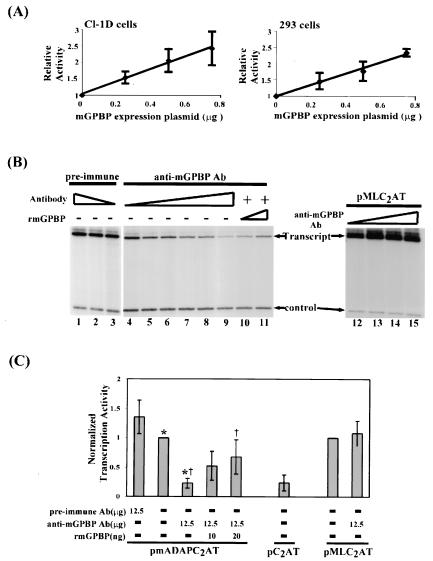
To examine whether our cloned mGPBP could transactivate a luciferase reporter gene controlled by the mouse Ada gene's G+C-rich promoter, we cotransfected both mouse Cl-1D cells and human 293 cells with a constant amount of the reporter construct and increasing amounts of the mGPBP expression vector. For both cell lines, reporter gene expression increased linearly with the amount of the mGPBP expression vector added in a dose-dependent manner (Fig. 7A). These results showed that increasing the intracellular GPBP level transactivated the G+C-rich promoter-driven reporter transcription in both cell types.
FIG. 7.
mGPBP is specifically required for transcription initiated at the mouse Ada gene's G+C-rich promoter. Cotransfection of a mouse Ada gene promoter-controlled luciferase reporter gene construct together with increasing amounts of a mGPBP expression vector into either mouse Cl-1D cells or human 293 cells resulted in a linear increase in reporter gene activity (A). In vitro transcription assays performed by coincubation of a supercoiled mouse Ada gene promoter-controlled G-less cassette reporter gene with human HeLa cell nuclear extracts showed that reporter transcript production was unaltered by the addition of increasing amounts of preimmune serum-derived antibodies (B, lanes 1 to 3). In contrast, increasing amounts of anti-mGPBP antibodies caused a gradual decrease in reporter transcript production (B, lanes 4 to 9). This immunosuppression effect can be reversed by the addition of purified recombinant mGPBP (rmGPBP; B, lanes 10 and 11). Transcription of the G-less cassette reporter gene under the control of the TATA box-dependent adenovirus major late promoter was not affected by the addition of anti-mGPBP antibodies (B, lanes 12 to 15). The assay results were repeated at least three times. (C) Normalized quantitation of the transcription assay results (using a phosphorimager), together with standard deviations of multiple repetitions. The statistical significance (P) in the difference of transcription levels in the absence and presence of anti-mGPBP antibodies (*) was found by a paired Student t test to be <0.00042 (n = 8), and P for the difference of transcription levels in the presence of the anti-mGPBP antibodies with and without additional recombinant mGPBP (†) was found by a paired Student t test to be <0.024 (n = 4).
To address the question of whether GPBP is specifically required for transcription directed by the murine Ada gene's G+C-rich promoter, we performed in vitro transcription assays using a G-less cassette reporter with either no promoter (PC2AT) or the murine Ada gene's G+C-rich promoter (pmADAPC2AT) upstream. Transcription assays were performed in the presence of HeLa cell nuclear extracts from which GPBP had been incrementally sequestered by the addition of increasing amounts of anti-mGPBP antibodies that can cross-react with human GPBP. Recovery of the reporter transcripts from the transcription reaction mixture was monitored with a labeled carrier control DNA. Figure 7B shows a representative result from experiments that were repeated three or more times. In the presence of increasing amounts of anti-mGPBP antibodies, there was a corresponding decrease in transcript production (Fig. 7B, lanes 4 to 9). When increasing amounts of purified recombinant mGPBP were added back to the antibody-treated nuclear extract, the observed antibody-induced suppression of reporter gene transcription was correspondingly reversed (Fig. 7B, lanes 10 and 11). Control experiments revealed that preimmune serum antibodies had no suppressive effect on reporter gene transcription (Fig. 7B, lanes 1 to 3). The suppressive effect of anti-mGPBP antibodies on transcription was observed only when the reporter gene was under the control of the G+C-rich promoter. Transcription of the same reporter gene under the control of the adenovirus major late gene's TATA-box dependent promoter was not diminished by the immunosequestering of GPBP in the nuclear extract (Fig. 7B, lanes 12 to 15). Quantifying all the labeled bands in several experiments with a phosphorimager allowed us to demonstrate the reproducibility of the results, as summarized in Fig. 7C. These results indicate that, although GPBP is required for transcription directed by the Ada gene's G+C-rich promoter, it is not required for transcription directed by the adenovirus major late gene's classical TATA-box dependent promoter.
The G+C-rich Topo IIα gene promoter, which contains a canonical TATA box, can also bind specifically to mGPBP.
To examine whether mGPBP is a general requisite G+C-rich promoter-dependent transcription factor, we also examined whether this protein can bind to other G+C-rich promoters, especially one that contains a consensus TATA box and G+C-rich regions that display no obvious homology to the murine Ada gene promoter, as in the case of the human topoisomerase IIα gene promoter (Fig. 8A) (41). EMSAs using the labeled Topo IIα promoter as the probe showed that the Topo IIα gene promoter can also bind specifically to purified recombinant mGPBP in the absence of any other mammalian transcription factors. This binding can be competed out by either excess unlabeled probe or excess unlabeled fragments containing four copies of the murine Ada gene MSPE but not by excess copies of a 200-bp fragment derived from the pUC2H vector, which does not adopt non-B form DNA structures under negative supercoiling conditions.
Transcription initiation at the Topo IIα gene promoter can be only partially suppressed by immunosequestration of mGPBP.
In HeLa cell nuclear extract-dependent in vitro transcription assays, we demonstrated that transcription initiated at the murine Ada gene promoter required the presence of GPBP whereas transcription initiated at a consensus TATA box-dependent adenovirus major late gene promoter did not. Since this Topo IIα gene promoter binds specifically to mGPBP but contains a canonical TATA box, we also examined how this promoter functioned when the GPBP in the nuclear extract was sequestered by immunoabsorption. Sequestering of GPBP in the HeLa nuclear extract with anti-GPBP antibodies under conditions that totally suppressed transcription initiated by the murine Ada gene promoter showed only a partial suppressive effect on transcription initiated by the Topo IIα gene promoter (Fig. 8C). This suppression can also be fully reversed by the addition of purified recombinant mGPBP to the nuclear extract. The results in Fig. 8 thus demonstrated that mGPBP can indeed bind to other G+C-rich promoters and that the presence of a canonical TATA box rendered the Topo IIα gene promoter only partially dependent on GPBP for transcription initiation, in contrast to the total dependence of the murine Ada gene promoter on GPBP for transcription in the absence of such a canonical element.
DISCUSSION
The search for a DNA binding factor that can recognize a small DNA binding motif within a G+C-rich promoter and initiate the assembly of the transcription initiation complex has been the subject of intensive but, as yet, unfruitful investigation (21, 25, 37). Our work on the murine Ada gene's G+C-rich promoter has established that this promoter has neither a functional TATA box nor a potential initiator element that is larger than 4 bp (2; Hsu et al., submitted). Instead, the minimal self-sufficient promoter activity resides within a 48-bp MSPE whose sequence displays an imperfect dyad symmetry with theoretical secondary-structure-forming potential. DNA sequences with similar potential structures have also been found in many other G+C-rich promoters (2). This 48-bp MSPE DNA element was used to isolate a cloned nuclear protein that can bind both to the promoter and to key proteins that participate in the assembly of the RNA polymerase II transcription initiation complex. Our success in this endeavor may owe much to our choice of using this rather large MSPE as the probe. This MSPE does not depend on other proximal activator motifs, such as Sp1 binding sites, to exhibit promoter function (2). Smaller elements within the MSPE do not suffice as self-sufficient promoters (2; S. L. Ackerman and C.-Y. Yeung, unpublished data). By using a multimerized MSPE probe, we successfully identified a λgt11 phage cDNA expression clone that encodes a GPBP.
Because G+C-rich promoters are present upstream of many essential mammalian housekeeping genes but are not found in either the yeast or Drosophila genome, we predicted that, if GPBP expression is critical to transcription from G+C-rich promoters, it would be ubiquitously expressed in mammalian cells and would be absent from both yeast and Drosophila genomes. Northern (Fig. 2) and Western (Fig. 3B) blot analyses of various mouse tissues and mammalian cell lines confirmed the former prediction. Although there is some variability in GPBP mRNA levels in different tissues, with the highest levels being found in kidney, heart, liver, and brain (Fig. 2), the variability shows no correspondence with Ada gene expression in those tissues. This observation is consistent with our prediction that GPBP is a general transcription factor and would therefore function similarly to other general transcription factors such as TBP, which also does not show tissue-specific expression patterns that necessarily match the expression levels of all its many target promoters. This is because the attenuation of expression control usually does not reside within the minimal basal promoter element, as has been observed for the ada gene (2; R. J. Gum et al., unpublished data; Hsu et al., submitted). A sequence homology search of the DNA sequence database (which includes the entire yeast and Drosophila genome sequences) revealed no obvious mGPBP orthologs in any of the nonmammalian genomes, although small domain-specific homologies to a number of transcription factors were found (Fig. 1B). The domain-specific homology to the SSRP1 gene (Fig. 1B) is particularly intriguing. Since SSRP1 is known to bind DNA in a DNA structure-dependent manner (7, 33), it is possible that GPBP may also share that unusual property and bind to MSPE via structural recognition.
Surprisingly, although no obvious mGPBP orthologs were uncovered by the sequence search, the murine Ada gene's G+C-rich promoter functions when it is introduced into yeast cells (data not shown). We note with interest that yABF-1, which has some domain-specific homology with mGPBP (Fig. 1B), can reportedly transactivate the yeast TRP3 gene promoter, which possesses a “suboptimal” TATA box (22). Because yeast origins of replication do not function as such in mammalian cells and since no one has been able to identify a mammalian yABF-1 ortholog by low-stringency hybridization screening, yABF-1's ability to transactivate a yeast gene promoter with a noncanonical TATA box and its sequence homology with certain mGPBP domains raise the possibility that yABF-1 and mGPBP may have similar cellular functions but recognize and bind to highly divergent DNA sequences. It is certainly formally possible that yABF-1, which has no known mammalian orthologs, may also promote transcription at the mouse Ada gene's G+C-rich promoter in mGPBP-deficient yeast cells.
The predicted size of mGPBP based on the cDNA ORF sequence was confirmed by Western blot analysis of lysates of various mammalian cell lines derived from diverse tissues of origin (Fig. 3). The other immune cross-reacting proteins seen in all cell lysates (Fig. 3B) may be either degradation products of mammalian GPBP or other members of a GPBP gene family.
Purified recombinant mGPBP was shown to bind specifically to the murine Ada gene's MSPE and the sequence-divergent G+C-rich human Topo IIα gene promoter in competitive EMSA, in the absence of any other mammalian proteins. This observation is consistent with GPBP potentially being responsible for mediating the initial assembly of the transcription initiation complex.
In situ binding assays using anti-mGPBP antiserum as a probe demonstrated that endogenous human GPBP is localized to the nucleus in HeLa cells (Fig. 5A). These results are consistent with GPBP's proposed role as a mediator of transcription initiation complex assembly at the promoter, a role which would require nuclear localization.
Western blot analysis of mouse Cl-1D cell nuclear extract proteins that either form a complex with immobilized mGPBP or are coimmunoprecipitated with HA-tagged mGPBP expressed in intact cells revealed that mGPBP can form complexes specifically with TBP, TFIIB, TFIIF, RNA polymerase II, and P300/CBP both in vitro and in vivo. Since all these mGPBP-associated transcription initiation factors are known to participate in the formation of the RNA polymerase II transcription initiation complex (8, 9), these observations are consistent with the proposed role of GPBP as a mediator of transcription initiation complex assembly, following its binding to target G+C-rich promoters.
In cotransfection assays, mGPBP was found to transactivate the murine Ada gene's G+C-rich promoter (Fig. 7A) in the two different cell lines (Cl-1D and 293) used. Because all mammalian cells tested contain significant levels of endogenous GPBP (Fig. 3B), it is not surprising that the transactivating effect of the exogenous recombinant mGPBP in the transfected cells is only in the 2.5-fold range. More significantly, in the presence of high copy numbers of the cotransfected reporter gene in both Cl-1D and 293 cells, the transactivating effect of the mGPBP expression vector remained linear over the more-than-eightfold range of levels of the GPBP expression vector used (Fig. 7A). Thus, in the presence of an excess of G+C-rich promoter sequences in the cell, GPBP appeared to be rate limiting for reporter gene transcription. Moreover, GPBP activity was shown to be essential for transcription directed by the mouse Ada gene's G+C-rich promoter. In in vitro transcription assays, sequestering of GPBP in human HeLa cell nuclear extract by immunoabsorption caused a proportional and ultimately complete suppression of transcription from the Ada gene's G+C-rich promoter (with the level of transcription declining to levels indistinguishable from that for a promoterless reporter construct; Fig. 7B and C). This suppression was reversed by replenishing the GPBP-depleted nuclear extract with purified recombinant mGPBP (Fig. 7B and C). In striking contrast to the G+C-rich promoter's absolute requirement for GPBP in the in vitro transcription assay, transcription activity from the adenovirus major late gene's classical TATA-box-dependent promoter was totally unaffected by the immunoabsorption-mediated sequestering of GPBP in the HeLa cell nuclear extract (Fig. 7B and C). The G+C-rich human Topo IIα gene promoter that contains a canonical TATAAA element, on the other hand, showed only partial (and totally reversible) dependence on the presence of GPBP in similar experiments, which probably reflects this promoter's dual-promoter characteristics (i.e., both TATA box dependent and G+C rich). These results established that GPBP is required only for transcription initiated at the G+C-rich promoter but is totally dispensable for transcription initiated at a classical TATA box-dependent promoter. The observed linearity of GPBP's transactivation effect (Fig. 7A) and its absolute requirement in TATA box-deficient G+C-rich-promoter-directed transcription (Fig. 7C) are both consistent with the predicted role of GPBP as a rate-limiting factor for transcription initiated at a TATA box-deficient G+C-rich promoter.
Recently (39), the same in vitro transcription assays (with the same reporter constructs described above as templates) were used to examine transcription elongation suppression caused by immunoabsorption of a factor that associates with members of each of the three various classes (11) of transcription elongation complexes. In contrast to the results described for the immunoabsorption of GPBP, transcript production from both the G+C-rich and TATA box-dependent promoters was similarly inhibited by the addition of antibodies directed against that proposed transcription elongation factor (39). This observation suggests that elongation of transcripts initiated at both G+C-rich and TATA box-dependent promoters may utilize common elongation complexes and implicates the differential transcription-inhibitory effects displayed by GPBP sequestration as interference with a different transcription-associated process. Since GPBP can bind to a G+C-rich promoter's MSPE, can associate with all the known transcription initiation assembly factors tested, and is functionally required for transcription initiated at that promoter, the anti-GPBP antibodies probably interfered with a TATA box-independent transcription initiation complex assembly step. Whether these anti-mGPBP antibodies would interfere with transcription at non-G+C-rich, TATA box-deficient initiator element-dependent promoters and whether GPBP is similarly required for transcription initiated at all mammalian TATA box-deficient G+C-rich promoters remain unresolved questions that have become tractable with the new reagents we now have on hand.
Acknowledgments
We thank Shao-Xia Lin, Chefin Bobonis, Teresa Szal, and Sarah Bothner for excellent technical assistance. We also thank Kelly McNagny for the cDNA library and Pradip Raychaudhuri for antibody reagents and advice and discussions.
This work was supported in part by NIH grant AG11623 and MUNIN Corp. award MC99-2 to C.-Y.Y., NIH grant CA72572 to J.M.L., RO1 GM44088 to V.J.K., and NIH SBIR grants CA78044, CA91376, and CA89777 to J.A.G. L.-C.H. was supported by a UIC University fellowship. Generous support for these studies was also provided by the American Lebanese Syrian Associated Charities and NIH (Cancer Center core grant 5 PO1 CA21765) to SJCRH for J.M.L. and V.J.K.
REFERENCES
- 1.Ackerman, S. L., A. G. Minden, G. T. Williams, C. Bobonis, and C.-Y. Yeung. 1991. Functional significance of an overlapping consensus binding motif for Sp1 and Zif268 in the murine adenosine deaminase gene promoter. Proc. Natl. Acad. Sci. USA 88:7523-7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackerman, S. L., A. G. Minden, and C.-Y. Yeung. 1993. The minimal self-sufficient element in a murine G+C-rich promoter is a large element with imperfect dyad symmetry. Proc. Natl. Acad. Sci. USA 90:11865-11869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aso, T., H. A. Vasavada, T. Kawaguchi, F. J. Germino, S. Ganguly, S. Kitajima, S. M. Weissman, and Y. Yasukochi. 1992. Characterization of cDNA for the large subunit of the transcription initiation factor TFIIF. Nature 355:461-464. [DOI] [PubMed] [Google Scholar]
- 4.Berger, S. L., and A. R. Kimmel (ed.). 1987. Methods in enzymology: guide to molecular cloning techniques, vol 152. Academic Press, Inc., San Diego, CA. [PubMed]
- 5.Biswas, E. E., M. J. Stefanec, and S. B. Biswas. 1990. Molecular cloning of a gene encoding an ARS binding factor from the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 87:6689-6692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blake, M. C., R. C. Jambou, A. G. Swick, J. W. Kahn, and J. C. Azizkhan. 1990. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol. Cell. Biol. 10:6632-6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bruhn, S. L., P. M. Pil, J. M. Essigmann, D. E. Housman, and S. J. Lippard. 1992. Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl. Acad. Sci. USA 89:2307-2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buratowski, S., S. Hahn, L. Guarente, and P. A. Sharp. 1989. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell 56:549-561. [DOI] [PubMed] [Google Scholar]
- 9.Chan, H. M., and N. B. La Thangue. 2001. p300/CBP proteins: HATs for transcriptional bridges and scaffolds. J. Cell Sci. 114:2363-2373. [DOI] [PubMed] [Google Scholar]
- 10.Christy, B., and D. Nathans. 1989. DNA binding site of the growth factor-inducible protein Zif268. Proc. Natl. Acad. Sci. USA 86:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conaway, J. W., A. Shilatifard, A. Dvir, and R. C. Conaway. 2000. Control of elongation by RNA polymerase II. Trends Biochem. Sci. 25:375-380. [DOI] [PubMed] [Google Scholar]
- 12.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dyer, M. A., P. J. Hayes, and M. H. Baron. 1998. The HMG domain protein SSRP1/PREIIBF is involved in activation of the human embryonic beta-like globin gene. Mol. Cell. Biol. 18:2617-2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feinberg, A. P., and B. Vogelstein. 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal. Biochem. 137:266-267. [DOI] [PubMed] [Google Scholar]
- 15.Field, J., J. Nikawa, D. Broek, B. MacDonald, L. Rodgers, I. A. Wilson, R. A. Lerner, and M. Wigler. 1988. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8:2159-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 17.Hayes, S., P. Shiyanov, X. Chen, and P. Raychaudhuri. 1998. DDB, a putative DNA repair protein, can function as a transcriptional partner of E2F1. Mol. Cell. Biol. 18:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Innis, J. W., D. J. Moore, S. F. Kash, V. Ramamurthy, M. Sawadogo, and R. E. Kellems. 1991. The murine adenosine deaminase promoter requires an atypical TATA box which binds transcription factor IID and transcriptional activity is stimulated by multiple upstream Sp1 binding sites. J. Biol. Chem. 266:21765-21772. [PubMed] [Google Scholar]
- 19.Killeen, M. T., and J. F. Greenblatt. 1992. The general transcription factor RAP30 binds to RNA polymerase II and prevents it from binding nonspecifically to DNA. Mol. Cell. Biol. 12:30-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kreidberg, J. A., and T. J. Kelly. 1986. Genetic analysis of the human thymidine kinase gene promoter. Mol. Cell. Biol. 6:2903-2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lichtsteiner, S., J. Wuarin, and U. Schibler. 1987. The interplay of DNA-binding proteins on the promoter of the mouse albumin gene. Cell 51:963-973. [DOI] [PubMed] [Google Scholar]
- 22.Martens, J. A., and C. J. Brandl. 1994. GCN4p activation of the yeast TRP3 gene is enhanced by ABF1p and uses a suboptimal TATA element. J. Biol. Chem. 269:15661-15667. [PubMed] [Google Scholar]
- 23.McGrogan, M., C. C. Simonsen, D. T. Smouse, P. J. Farnham, and R. T. Schimke. 1985. Heterogeneity at the 5′ termini of mouse dihydrofolate reductase mRNAs. Evidence for multiple promoter regions. J. Biol. Chem. 260:2307-2314. [PubMed] [Google Scholar]
- 24.Means, A. L., and P. J. Farnham. 1990. Sequences downstream of the transcription initiation site modulate the activity of the murine dihydrofolate reductase promoter. Mol. Cell. Biol. 10:1390-1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Means, A. L., and P. J. Farnham. 1990. Transcription initiation from the dihydrofolate reductase promoter is positioned by HIP1 binding at the initiation site. Mol. Cell. Biol. 10:653-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melton, D. W., D. S. Konecki, J. Brennand, and C. T. Caskey. 1984. Structure, expression, and mutation of the hypoxanthine phosphoribosyltransferase gene. Proc. Natl. Acad. Sci. USA 81:2147-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Orphanides, G., W. H. Wu, W. S. Lane, M. Hampsey, and D. Reinberg. 1999. The chromatin-specific transcription elongation factor FACT comprises human SPT16 and SSRP1 proteins. Nature 400:284-288. [DOI] [PubMed] [Google Scholar]
- 28.Rauth, S., K. G. Yang, A. M. Seibold, D. E. Ingolia, S. R. Ross, and C.-Y. Yeung. 1990. GC-rich murine adenosine deaminase gene promoter supports diverse tissue-specific gene expression. Somat. Cell Mol. Genet. 16:129-141. [DOI] [PubMed] [Google Scholar]
- 29.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 30.Sawadogo, M., and R. G. Roeder. 1985. Factors involved in specific transcription by human RNA polymerase II: analysis by a rapid and quantitative in vitro assay. Proc. Natl. Acad. Sci. USA 82:4394-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sehgal, A., N. Patil, and M. Chao. 1988. A constitutive promoter directs expression of the nerve growth factor receptor gene. Mol. Cell. Biol. 8:3160-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shaw, G., and R. Kamen. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659-667. [DOI] [PubMed] [Google Scholar]
- 33.Shirakata, M., K. Huppi, S. Usuda, K. Okazaki, K. Yoshida, and H. Sakano. 1991. HMG1-related DNA-binding protein isolated with V-(D)-J recombination signal probes. Mol. Cell. Biol. 11:4528-4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singh, H., J. H. LeBowitz, A. S. Baldwin, Jr., and P. A. Sharp. 1988. Molecular cloning of an enhancer binding protein: isolation by screening of an expression library with a recognition site DNA. Cell 52:415-423. [DOI] [PubMed] [Google Scholar]
- 35.Smale, S. T., and D. Baltimore. 1989. The “initiator” as a transcription control element. Cell 57:103-113. [DOI] [PubMed] [Google Scholar]
- 36.Smale, S. T., M. C. Schmidt, A. J. Berk, and D. Baltimore. 1990. Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc. Natl. Acad. Sci. USA 87:4509-4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sopta, M., Z. F. Burton, and J. Greenblatt. 1989. Structure and associated DNA-helicase activity of a general transcription initiation factor that binds to RNA polymerase II. Nature 341:410-414. [DOI] [PubMed] [Google Scholar]
- 38.Spencer, J. A., M. H. Baron, and E. N. Olson. 1999. Cooperative transcriptional activation by serum response factor and the high mobility group protein SSRP1. J. Biol. Chem. 274:15686-15693. [DOI] [PubMed] [Google Scholar]
- 39.Trembley, J. H., D. Hu, L. C. Hsu, C.-Y. Yeung, C. Slaughter, J. M. Lahti, and V. J. Kidd. 2002. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem. 277:2589-2596. [DOI] [PubMed] [Google Scholar]
- 40.van Brabant, A. J., R. Stan, and N. A. Ellis. 2000. DNA helicases, genomic instability, and human genetic disease. Annu. Rev. Genomics Hum. Genet. 1:409-459. [DOI] [PubMed] [Google Scholar]
- 41.Wang, Q., G. P. Zambetti, and D. P. Suttle. 1997. Inhibition of DNA topoisomerase IIα gene expression by the p53 tumor suppressor. Mol. Cell. Biol. 17:389-397. [DOI] [PMC free article] [PubMed] [Google Scholar]