Abstract
In a gene trap screen for genes expressed in the primitive streak and tail bud during mouse embryogenesis, we isolated a mutation in Jade1, a gene encoding a PHD zinc finger protein previously shown to interact with the tumor suppressor pVHL. Expressed sequence tag analysis indicates that Jade1 is subject to posttranscriptional regulation, resulting in multiple transcripts and at least two protein isoforms. The fusion Jade1-β-galactosidase reporter produced by the gene trap allele exhibits a regulated expression during embryogenesis and localizes to the nucleus and/or cytoplasm of different cell types. In addition to the primitive streak and tail bud, β-galactosidase activity was found in other embryonic regions where pluripotent or tissue-specific progenitors are known to reside, including the early gastrulation epiblast and the ventricular zone of the cerebral cortex. Prominent reporter expression was also seen in the extraembryonic tissues as well as other differentiated cell types in the embryo, in particular the developing musculature. We show that the gene trap mutation produces a null allele. However, homozygotes for the gene trap integration are viable and fertile. Database searches identified a family of Jade proteins conserved through vertebrates. This raises the possibility that the absence of phenotype is due to a functional compensation by other family members.
Patterning of the mammalian embryo along the anteroposterior (A/P) axis involves complex morphogenetic and tissue diversification events taking place during gastrulation and organogenesis in the primitive streak and tail bud. The appearance of the streak on the prospective posterior side of the embryo at the onset of gastrulation (6.5 days postcoitum [d.p.c.]) constitutes the first morphological asymmetry indicating the polarity of the A/P axis. In the streak, cells lose contact with the epiblast (primitive ectoderm) and migrate anteriorly to give rise to the mesoderm and definitive endoderm. The axis is laid down progressively in a rostrocaudal sequence. Early during gastrulation, progenitors for the axial tissues are located throughout the epiblast (26). At later stages, however, lineage analysis studies support the existence of a resident pool of progenitors in the streak and its descendant, the tail bud, which can maintain itself and give rise to the entire postcranial axis (4, 35, 52, 63). Several genes expressed in these regions were shown to be involved in maintenance of progenitor populations, specification, and patterning of mesoderm or morphogenetic movements (reviewed in reference 53). One such gene is T (Brachyury). Heterozygotes for a loss of function mutation in T have short or absent tails (7), whereas homozygotes die at midgestation lacking structures posterior to the forelimb (3, 66). T, the founder member of the T-box family of transcription factors, is one of the earliest markers of nascent mesoderm. It is expressed in the streak during gastrulation and in the tail bud until the end of axial elongation at 13.5 d.p.c (3, 64). Thus, the primitive streak and tail bud play a pivotal role in the formation and patterning of axial tissues. However, the basic mechanisms underlying development of the axis are still not clearly understood, and the genes directing these processes remain largely unknown.
Gene trapping is an attractive method for creation of insertional mutations in embryonic stem (ES) cells. This is specifically designed to enrich for intragenic integration events, since it makes use of promoterless reporter constructs that need to integrate downstream of a gene's regulatory sequences to activate reporter expression (reviewed in reference 49). Gene trap insertions result in production of fusion transcripts consisting of the reporter and upstream endogenous sequences. Thus, mutated genes can be readily identified by methods such as 5′ rapid amplification of cDNA ends (5′ RACE) or inverse PCR (55, 57). Reporter expression can be monitored in ES cell-derived chimeric or transgenic embryos, and in the majority of cases examined, it accurately reflects the expression pattern of the trapped gene. Moreover, the availability of ES cells and the development of techniques for their differentiation allows for in vitro preselection of integrations in genes expressed in specific lineages or responding to specific cues (10, 11, 59).
We undertook a gene trap screen in differentiating ES cells to identify novel genes expressed in the primitive streak and tail bud by screening in vitro for coexpression of the gene trap reporter and T. In this report, we describe the characterization of a gene trap insertion into a mouse gene encoding a PHD zinc finger protein. A human cDNA clone, Jade-1, corresponding to a short transcript of this gene, was recently identified in a two-hybrid screen for proteins interacting with pVHL (von Hippel-Lindau syndrome protein) tumor suppressor (69). During embryogenesis, the fusion Jade1-β-galactosidase (β-Gal) reporter showed a restricted expression pattern, in particular in extraembryonic tissues and in embryonic regions known to harbor pluripotent or tissue-specific progenitors. We show that integration of the gene trap vector generates a null allele of Jade1. However, mice homozygous for this allele do not show any obvious phenotypic defects.
MATERIALS AND METHODS
Culture, electroporation, and screening of gene trap ES cell clones.
E14TG2a ES cells (17) were cultured as described in reference 48, except that they were grown in Glasgow’s minimal essential medium supplemented with 0.25% sodium bicarbonate, 0.1% nonessential amino acids, 4 mM glutamine, 2 mM sodium pyruvate, 0.1 M 2-mercaptoethanol, 10% heat-inactivated fetal calf serum, and 100 U of leukemia inhibitory factor (LIF)/ml. ES cells (108) were electroporated with a 150-μg/ml equimolar mixture of pGT1,2,3 HindIII linearized vectors as described previously (64) and selected with 100 μg of G418/ml for 10 days. Resistant clones were differentiated by removal of LIF for 72 h and screened by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) staining and in situ hybridization with a T riboprobe (61) as described previously (51).
Production of transgenic mouse line, breeding, and genotyping.
Chimeric mice were produced by injection of E148 ES cells into C57BL/6 blastocysts as described previously (41). F1 animals heterozygous for the gene trap mutation Jade1gtE148ISCR were obtained by mating male chimeras with C57BL/6 females. F2 to F5 heterozygotes (129/Ola:C57BL/6 mixed background) were intercrossed to generate homozygous individuals. All work with animals was carried out under United Kingdom Project license 60/2107.
Genotyping of animals was routinely performed by X-Gal staining of tail biopsy specimens. Homozygosity for the gene trap was initially determined by quantitative Southern blotting and analysis following identification of the trapped gene by PCR. Blots were probed with a BglII/BamHI fragment corresponding to the engrailed-2 intron by standard procedures (44), and the intensities of the endogenous engrailed-2 gene (two-copy loading control) and gene trap vector-specific bands were compared. A common E148INT4 forward primer (5′-GATGTTAAGAGTGGCATCCTGG-3′) and either E148INT5 (5′-ACATCTAGGAGTGGAACACTAG-3′) or pGT/2rev (5′-CCACAACGGGTTCTTCTGTTAG-3′) reverse primer were used in separate PCRs to detect the wild-type or mutant mJade1 alleles, respectively.
5′ RACE-PCR, Northern blotting, and reverse transcription (RT)-PCR.
Total RNA was extracted from cells or embryos by using Trizol reagent (Invitrogen) according to the manufacturer's instructions. 5′ RACE-PCR was performed according to the method of Townley et al. (55). First-strand synthesis was primed with primer R1 (5′-TAATGGGATAGGTTACGT-3′). The product was poly(A) tailed, and primer R2 [5′-GGTTGTGAGCTCTTCTAGATGG(T17)-3′] was used in second-strand synthesis. First-round PCR was performed with primer R3 (5′-GGTTGTGAGCTCTTCTAGATGG-3′) and nested primer R4 (5′-AGTATCGGCCTCAGGAAGATCG-3′). In second-round PCR, 5′ biotin-R3 and R5 (5′-ATTCAGGCTGCGCAACTGTTGG-3′) primers were used. Second-round PCR products were directly sequenced with the Amplicycle sequencing kit (Perkin Elmer) with R6 (5′-GTTTTCCCAGTCACGAC-3′).
Northern blot hybridization was performed according to standard procedures (44) with 10 μg of RNA. A 0.3-kb fragment complementary to the endogenous mJade1 cDNA sequence, 3′ to the gene trap integration site, was amplified by RT-PCR on RNA extracted from wild-type ES cells with primers E148/1 (5′-GACCTGAAGATCGAAAGCCTTC-3′) and mEST (5′-GATATCGACGTAGCCTAACGCT-3′), cloned into Topo-PCR2.1 vector (Invitrogen), and used to probe the blot.
RT-PCR on RNA extracted from 13.5-d.p.c. embryos was performed by using Superscript II (Invitrogen) according to the manufacturer's instructions. A poly(dT) primer was used for first-strand synthesis while fragments specific to wild-type or mutant transcripts were PCR-amplified with forward primer E148/2 (5′-GCAGCAGTGAGGATTCTGACGA-3′) and reverse primer mEST (wild type) or R5 (lacZ, mutant).
X-Gal staining of whole-mount embryos and sections.
Embryos were recovered and tested at different developmental stages. Noon of the day when a vaginal plug was detected was defined as 0.5 d.p.c. Embryos up to 9.5 d.p.c. were dissected in phosphate-buffered saline and stained with X-Gal as described in reference 2. Older embryos (10.5 to 12.5 d.p.c.) were treated and stained as described in reference 60. Cryosections (15 μm thick) were processed and stained with X-Gal as young embryos (fixation time, 10 min). Paraffin sections of stained embryos were prepared as described in reference 22.
Sequence analysis.
The full-length mJade1 mRNA (BN000281) encoding Jade1L, short mJade1 mRNA for Jade1S (BN000282), and alternative noncoding exons (BN000283 and BN000284) were predicted from contigs of cDNAs and expressed sequence tags (ESTs) from unigene cluster Mm.28483. Additional family members in both mouse and other vertebrate species were identified by Blast searches. These sequences have also been submitted to the TPA database (BN000275 to BN000280 and BN000285 to BN000289). Multiple-sequence alignments and phylogenetic analyses were carried out with Clustalx. CpG islands and candidate promoters were identified by using CpG Island Searcher (http://www.uscnorris.com/cpgislands/) (56) and PROSCAN 1.7 (http://bimas.dcrt.nih.gov/molbio/proscan/).
Nucleotide sequence accession numbers.
The following TPA database accession numbers were obtained in this study: full-length mJade1 mRNA encoding Jade1L, BN000281; short mJade1 mRNA encoding Jade1S, BN000282; alternative noncoding exons, BN000283 and BN000284; additional family members from mouse and other vertebrate species, BN000275 to BN000280 and BN000285 to BN000289.
RESULTS
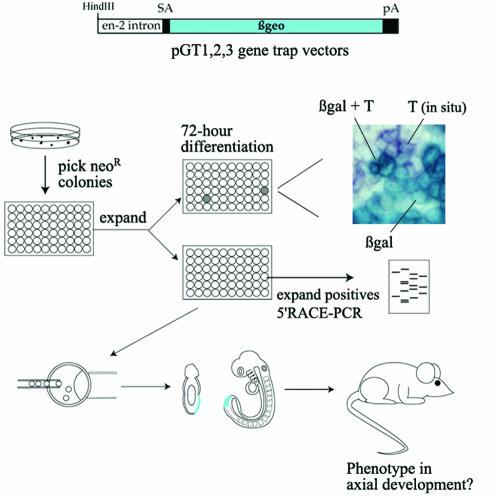
A small-scale gene trap screen was performed following the electroporation of pGT1,2,3 vectors in ES cells (Fig. 1) (64). To enrich for integrations in genes expressed in the primitive streak and tail bud during embryogenesis, 279 neomycin-resistant clones were prescreened in vitro for cells coexpressing the gene trap reporter, lacZ, and T, marker of the streak and tail bud (Fig. 1). Expression of T was shown to be upregulated in patches of cells growing in monolayers during the first 96 h of differentiation in the absence of LIF, with a maximal increase between 48 and 72 h (47). Based on this observation, ES cell clones were subjected to differentiation for 72 h and double stained for expression of lacZ, with X-Gal, and T mRNA by in situ hybridization. β-Gal activity was detected in 191 clones (68%). One hundred sixty-eight clones showed nonubiquitous reporter expression, and of these, 26 exhibited various proportions of cells expressing both β-Gal and T. E148 was one of the clones presenting a high proportion of coexpressing cells and was selected for further analysis (Fig. 2A). Chimeric embryos generated by injection of this clone into blastocysts and examined during gastrulation and organogenesis stages exhibited a restricted expression pattern including the primitive streak and tail bud (data not shown).
FIG. 1.
Gene trap screen to identify mutations in genes expressed in primitive streak and tail bud. pGT1,2,3 vectors are introduced into ES cells, and G418r colonies, representing integrations in the open reading frames, were selected. The different pGT vectors lack a translation initiation codon but allow selection of integrations in each of the three different reading frames. One set of duplicated colonies is screened for overlapping expression of β-Gal reporter activity (light blue) and T mRNA (purple) after in vitro differentiation, and positive clones are expanded from a master plate. After 5′ RACE PCR to identify endogenous mRNA fused to the vector sequence, ES cell clones corresponding to bona fide integrations in the open reading frame of genes are injected into blastocysts to create chimeric embryos, where expression of β-Gal in the primitive streak is monitored. Lines testing positive are passed through the germ line to create transgenic mice which are used to extensively characterize reporter expression and examine embryonic or postnatal homozygous phenotypes.
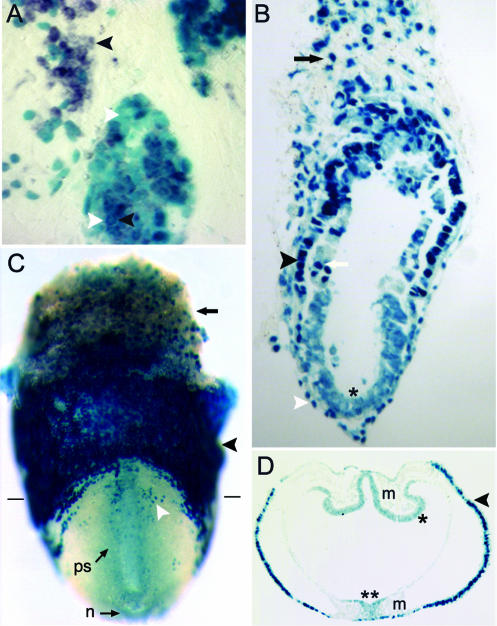
FIG. 2.
Reporter expression in differentiating E148 ES cells and early embryos. (A) Partially overlapping expression of T mRNA (black arrowheads) and β-Gal (white arrowheads) in differentiating E148 ES cells. Approximately 40% of E148 β-Gal-positive cells also express T. β-Gal is localized in the nucleus. (B) Longitudinal section of a 6.5-d.p.c. embryo heterozygous for the E148 integration. β-Gal is strongly expressed in the extraembryonic visceral (black arrowhead) and parietal (white arrowhead) endoderm as well as in the extraembryonic ectoderm (white arrow) and trophoblast (black arrow). Cells in the epiblast (asterisk) express lower levels of β-Gal protein. (C) Posterior view of a whole-mount ∼7.5-d.p.c. embryo, showing high level expression in the trophoblast (arrow) and visceral endoderm (black arrowhead) and lower expression in the primitive streak and node. Scattered cells (white arrowhead) in the embryonic endoderm express high levels of β-Gal protein. Horizontal lines indicate the approximate level and plane of the section shown in panel D. (D) Transverse section of the embryo shown in panel C, showing expression in the visceral endoderm and anterior neurectoderm (asterisk). In the primitive streak (double asterisk), cells in the ectoderm rapidly downregulate reporter expression after ingression to the adjacent mesodermal layer. ps, primitive streak; n, node; m, mesoderm.
Reporter expression in E148 mice during embryogenesis.
lacZ expression was examined in heterozygotes at various developmental stages. At 6.5 d.p.c., β-Gal activity was observed in all tissues but at various levels and in various subcellular compartments (Fig. 2B). The strongest expression was seen in the extraembryonic region, in particular in the visceral and parietal endoderm, which showed nuclear localization of the fusion protein. Expression levels were heterogeneous in the extraembryonic ectoderm and trophoblast, whereas in a small proportion of cells in these tissues, β-Gal was seen in both the nucleus and cytoplasm. Weaker expression that was clearly not restricted to the nucleus was apparent in the epiblast. During gastrulation, extraembryonic β-Gal activity remained strong and widespread, whereas expression in the embryo proper became progressively restricted (Fig. 2C). By late gastrulation, stained cells were seen in the streak and node, while expression was sharply downregulated in nascent mesoderm (Fig. 2D). In the remaining ectodermal layer, β-Gal activity was restricted to the neurepithelium of the head folds. Cells in the neurectoderm and the streak showed both cytoplasmic and nuclear β-Gal activity.
At 9.5 d.p.c., expression remained moderately high throughout the neurectoderm (Fig. 3A) and the late primitive streak (Fig. 3B). Strong de novo activation of mesodermal expression occurred in the presomitic mesoderm, concordant with the formation of new somites (Fig. 3B and C). β-Gal activity was restricted to the anterior half of the condensing and most recently formed somites while it seemed weaker and uniformly distributed in more differentiated somites (Fig. 3A and C).
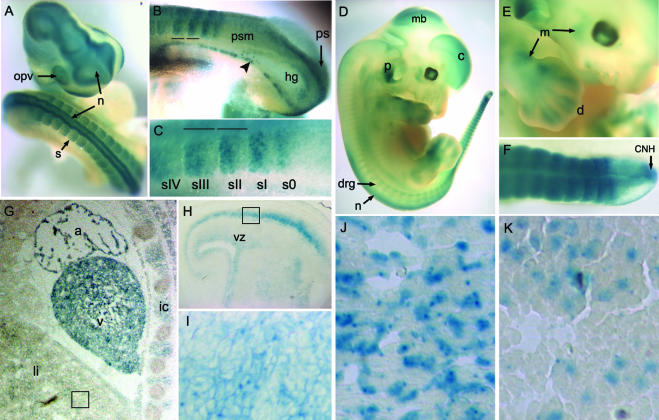
FIG. 3.
Reporter expression in E148 mice during organogenesis and later fetal development. (A) A 9.5-d.p.c. embryo showing strong β-Gal expression in the developing neural tube, optic vesicle, and somites. (B) Enlarged view of the posterior end of a 9.5-d.p.c. embryo, showing expression in the ectodermal layer of the primitive streak. De novo upregulation of β-Gal expression occurs in somites emerging from the presomitic mesoderm. Horizontal lines in panels B and C indicate the limits of somites sII and sIII. A subset of cells throughout the gut, overlapping with the presumed location of primordial germ cells at this stage (16), show intense staining (arrowhead). (C) Enlarged view of the presomitic mesoderm and posterior somites. β-Gal activity is present in the anterior compartment of the nascent somite (s0) and three most recently formed somites (sI to sIII). In more mature somites (sIV and panel A), the level of β-Gal activity is lower and more evenly distributed throughout the somite. (D) Lateral view of a 12.5-d.p.c. embryo showing expression in the cerebral cortex and other neural tissue, in the pinna of the ear, and in somites and limb buds. (E) Enlargement of the embryo shown in panel D. β-Gal expression is high in developing muscle and in cells surrounding the digits. (F) Tail of a 12.5-d.p.c. embryo showing β-Gal activity in somites and the CNH of the tail bud. (G) Detail from a sagittal section of a 15.5-d.p.c. embryo. Expression is strong in the heart and in intercostal muscles, and more moderate expression is found in liver. The box shows the approximate region enlarged in panel K. (H) Sagittal section through the head of a 15.5-d.p.c. embryo, showing β-Gal activity in the ventricular zone, containing progenitors for the cortex, and in a subset of more differentiated cells. The box indicates the approximate region enlarged in panel I. (I) Differentiating cells in the cerebral cortex show cytoplasmic localization of β-Gal protein. (J) In muscle cells, expression is strong and localized primarily to the nucleus. (K) Liver cells show weaker nuclear expression. n, neural tube; s, somite; opv, optic vesicle; psm, presomitic mesoderm; ps, primitive streak; hg, hindgut; c, cerebral cortex; mb, midbrain; drg, dorsal root ganglia; p, pinna; m, muscle; d, digit; a, atrium; v, ventricle of the heart; li, liver; ic, intercostal muscle; vz, ventricular zone.
At 12.5 d.p.c., β-Gal activity predominated in the nervous system and developing muscles (Fig. 3D and E). Interestingly, expression in the tail bud was particularly high in the chordoneural hinge (CNH), a region encompassing the most posterior part of the neural tube and shown to contain progenitor cells for elongation of the axis (4) (Fig. 3F).
Reporter expression at 15.5 d.p.c. was similar to that seen at 12.5 d.p.c., with the highest β-Gal levels in parts of the nervous system and the developing musculature (Fig. 3G and H). Weaker expression was observed in the liver. In the central nervous system, expression was seen in the neural progenitors in the ventricular zone of the cortex (Fig. 3H) and also at higher levels in more differentiated progeny (Fig. 3H and I). While β-Gal localization was mainly cytoplasmic in the neural tissue (Fig. 3I), it was nuclear in all the musculature examined (Fig. 3J) and in the subset of cells that showed expression in the liver (Fig. 3K). Strong cytoplasmic β-Gal activity was also observed in deposits of brown fat, the submandibular gland, the gut epithelium, and epithelial structures in the kidney and testis (data not shown).
Molecular characterization of the E148 integration.
Fluorescent in situ hybridization (FISH) and Southern blot analysis revealed a single vector insertion at the proximal region of chromosome 3 (Fig. 4 and data not shown). A unique endogenous sequence (154 bp), which corresponds to a single open reading frame fused in frame with βgeo, was identified by 5′ RACE-PCR. In the initial database searches, this sequence showed homology only to ESTs indicating that integration had occurred into a novel gene. The contigs made with mouse and human ESTs aligned to unique genomic sequences (Ensembl) in syntenic regions of mouse chromosome 3 and human chromosome 4. These findings indicated that these ESTs, including the E148 endogenous sequence, correspond to single orthologous genes in humans and mice. While our study was in progress, a human cDNA clone corresponding to this gene was identified in a two-hybrid screen for proteins interacting with the tumor suppressor pVHL and was named Jade-1 (69). Hereafter, we use the names mJade1 and JADE1 to define the gene interrupted in E148 mice and its human orthologue, respectively, while the term Jade1 designates both orthologues.
FIG. 4.
Cytogenetic mapping of E148 integration. FISH in metaphase chromosome spreads of E148 ES cells with a probe specific to the gene trap vector detects a single chromosomal integration site on the proximal region of chromosome 3 (red). This location was confirmed by using a chromosome 3-specific paint (green).
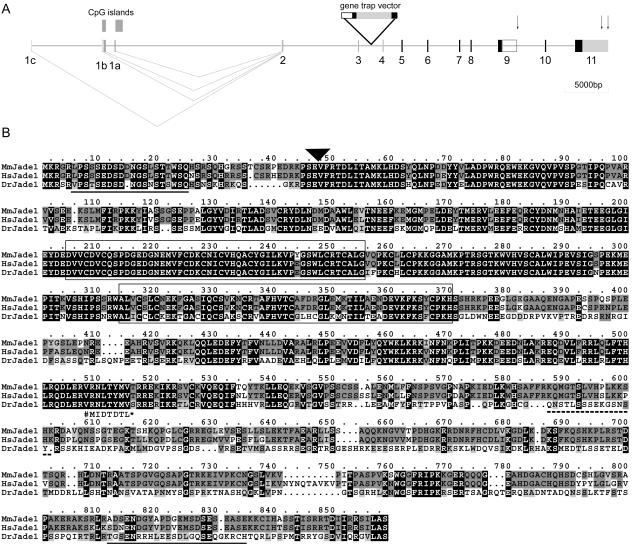
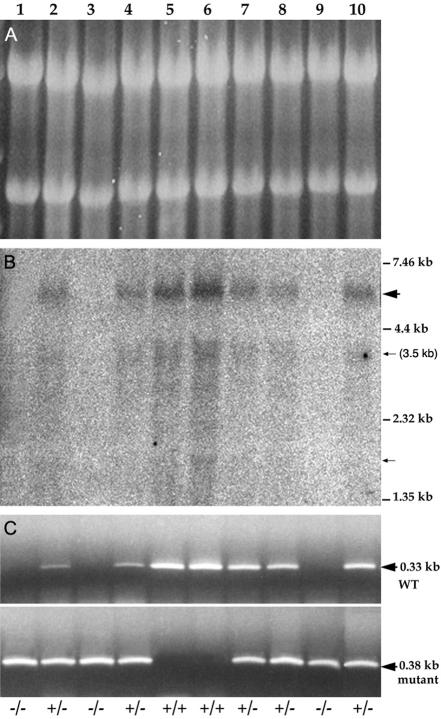
The alignment of human and mouse ESTs and cDNAs to the genomic sequences generated a map of exons (Fig. 5A) and indicated the presence of alternative exons at both ends of Jade1. The production of alternative transcripts was further supported by the detection of multiple bands in Northern blots (see Fig. 7B and reference 69 for JADE1). A major transcript of approximately 6 kb was detected, which corresponds well with the size of the transcript estimated by alignment of all ESTs. Some faint bands suggesting the presence of minor, lower-molecular-weight products were also seen. One of these smaller transcripts (∼3.5 kb) coincides with the size of the JADE1 cDNA clone (accession no. AF520952) isolated by Zhou et al. (69).
FIG. 5.
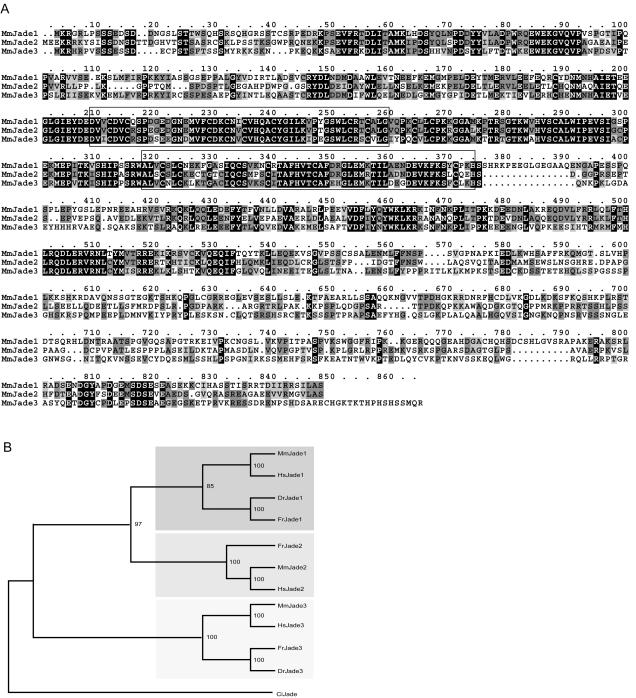
Jade1 gene structure and multiple-protein alignment of mouse, human, and zebra fish Jade1. (A) Mouse Jade1 is encoded by 10 exons (black boxes 2 to 11). The gene trap insertion occurred between exons 3 and 4. Noncoding exons are shown in numbered grey boxes. Potential polyadenylation signals are indicated by arrows. The box outlined at the end of exon 9 indicates an alternative 3′ untranslated region sequence that would give rise to a ∼3.5-kb transcript and the short Jade1 isoform (Jade1S), equivalent to the published human Jade1 protein (69). EST and cDNA alignments indicate that at least three alternative 5′ noncoding exons (1a to 1c) are used. A promoter scan identified two putative promoter regions in proximity to exons 1a and 1b that coincide with CpG islands. (B) Multiple alignment of mouse Jade1 (Mm) with the putative Jade1 orthologues in human (Hs) and zebra fish (Dr) reveals extensive homology. Residues identical in all three homologues are filled black while those identical only in two of these and conserved amino acid substitutions are filled dark and light grey, respectively. The PHD finger domains are boxed. A potential bipartite NLS is indicated by a dashed underline. Although this NLS is not conserved in the zebra fish, two possible overlapping bipartite NLS are present at positions 635 to 651 and 648 to 664 of DrJade1. Two putative PEST sequences (PESTFIND score > 10) found in both mouse and human Jade1 are underlined in solid black. The position of the gene trap insertion is indicated by the black triangle. The published human Jade1 protein sequence (GenPept accession no. AAM95612) is shorter than the sequence shown here; AAM95612 is identical up to the position marked # below the aligned sequences (corresponding to the end of exon 9) but is truncated at a stop codon (*) after 7 variant amino acids encoded by intron 9.
FIG. 7.
Wild-type Jade1 transcripts are absent in embryos homozygous for the E148 integration. Panels A and B show Northern blot analysis of total RNA prepared from 13.5-d.p.c. embryos (lanes 1 to 10) of a heterozygous intercross. The probe detects an mJade1 sequence immediately 3′ to the insertion site. (A) An ethidium bromide-stained gel loading control shows the presence of intact RNA. (B) In wild-type and heterozygous embryos a major ∼6-kb transcript is present (wide arrow). Lower-molecular-size faint bands (indicated by small arrows) are also detected, indicating the presence of smaller minor transcripts, including one at 3.5 kb. No transcripts are detected in homozygous embryos. (C) RT-PCR analysis performed on the same RNA samples with primers from the endogenous Jade1 sequence lying 5′ and 3′ to the E148 insertion (top panel). A wild-type band is detected in all heterozygous and wild-type embryos. A second RT-PCR with the same 5′ primer and a 3′ primer specific to the gene trap vector sequence amplifies a band in all heterozygous and homozygous embryos (bottom panel). No wild-type product is detectable in homozygous mutant embryos (lanes 1, 3, and 9).
Predicted translation starts at the first exon common to all transcripts (exon 2). The major 6-kb transcript is predicted to encode a protein of 834 amino acids and approximately 94 kDa while the 3.5-kb transcript results in a truncated form of the protein (510 amino acids, 58 kDa). Here, we designate the short and long protein isoforms Jade1S and Jade1L, respectively. An ∼61-kDa band corresponding to Jade1S was seen in Western blots with an antiserum raised against a 20-amino-acid peptide in the C-terminal sequence of both human and mouse Jade1S (69). Although the upper part of this blot was not shown, Zhou et al. mentioned the presence of an additional band at 95 kDa (see discussion in reference 69), supporting the existence of the long isoform.
Insertion of the vector occurred in the third intron leading to the production of a 47-amino-acid truncated protein (Fig. 5). PCR amplification of the integration site with primers hybridizing to the 5′ intronic sequence flanking this site and lacZ provided further confirmation that the gene trap vector had inserted into mJade1 (data not shown).
Jade1 belongs to a subfamily of PHD zinc finger proteins.
Jade1L contains two PHD zinc finger domains, two strong candidate PEST motifs, and a bipartite nuclear localization signal (NLS) (Fig. 5B). Interestingly, this NLS is not included in either Jade1S or the truncated mJade1-β-Gal protein produced by the gene trap allele. Conservation is high throughout the amino acid sequence of human and mouse Jade1. Strong conservation, albeit at lower levels in the C-terminal third of the sequence, also exists between human and mouse Jade1 and a zebra fish Jade1 orthologue (Fig. 5B).
Significant similarity to Jade1 was also detected in ESTs and cDNAs corresponding to genomic sequences mapping to mouse chromosomes 11 and X and syntenic regions of human chromosomes 5 and X, respectively. We designate these genes Jade2 (mouse chromosome 11 and human 5) and Jade3 (mouse and human chromosome X). The overall exon-intron organization of these genes is similar to that of Jade1. Extensive homology, extending well beyond the PHD zinc finger domains, was observed between the deduced amino acid sequences of Jade1, 2, and 3. (Fig. 6A and data not shown). The strongest conservation can be seen in the N-terminal part and, in particular, the interfinger domain. Shorter amino acid stretches are identical in the region C-terminal to the PHD fingers. It is therefore probable that these highly conserved domains and C-terminal motifs characterize and define a novel subfamily of PHD zinc finger proteins. All three Jade members were also identified in Fugu rubripes while a single Jade homologue was found in the complete Ciona intestinalis genomic sequence. Finally, only two Jade genes were found to date among zebra fish ESTs. Figure 6B shows a phylogenetic tree constructed by comparison of members of this subfamily in different chordates.
FIG. 6.
The Jade protein family. (A) Multiple alignment of mouse Jade1 and the closest related mouse sequences (Jade2 and Jade3) identified in blast searches. Shading is the same as described in the legend to Fig. 5. The PHD fingers are conserved in all three family members (boxed), but there is also extensive conservation outside these domains. (B) A neighbor-joining tree of the Jade protein family. The tree was generated from an alignment of mouse (Mm), human (Hs), zebra fish (Dr), Fugu rubripes (Fr), and Ciona intestinalis (Ci) Jade sequences and rooted with the C. intestinalis sequence. Groups of Jade proteins are indicated by grey rectangles.
Phenotypic analysis.
To investigate any phenotypic defects resulting from homozygosity for the E148 gene trap insertion, transgenic F1 offspring were backcrossed to C57BL/6 animals for several generations and the resulting heterozygotes were intercrossed. The results of this analysis revealed that homozygous animals are viable and fertile, showing no obvious morphological abnormalities compared to their wild-type and heterozygote littermates. However, the number of homozygotes obtained at weaning age was significantly less than expected according to the Mendelian ratio (28 of 184, or 15.2%; 0.05 > P > 0.01). Further examination is therefore required to exclude a more subtle or genetic background-specific effect of gene trap integration on the viability of E148 homozygous mice.
Endogenous transcripts are disrupted by the gene trap integration.
One possible explanation for the absence of overt phenotypic defects in E148 homozygous mice is that the gene trap integration has failed to generate a null allele of mJade1. Inefficient use of the polyadenylation signal and splice acceptor of the gene trap construct may result in splicing around the vector, such that sufficient quantities of wild-type transcript are generated to sustain normal function (9, 33, 43, 58). To test this possibility, a Northern blot was performed on total RNA prepared from 13.5-d.p.c. embryos of an intercross litter. As can be seen in Fig. 7B, a probe complementary to the mJade1 sequence, 3′ of the gene trap integration, fails to detect the major 6 kb and shorter mJade1 transcripts in homozygous embryos. The absence of trace quantities of wild-type transcript in homozygotes was further confirmed by RT-PCR with two sets of primers, each amplifying a sequence specific to the wild-type or trapped allele (Fig. 7C). These findings indicate that endogenous transcripts are efficiently disrupted by the gene trap insertion.
DISCUSSION
We have characterized a gene trap insertion isolated in an in vitro screen designed to enrich for mutations in genes expressed in the primitive streak and tail bud during mouse embryogenesis. Reporter expression is detected in these structures as well as in a subset of other tissues during development. We have shown that the E148 mouse line carries a mutation in mJade1, encoding a PHD zinc finger protein. A short Jade1 isoform, encoded by a human cDNA clone previously identified, was shown to interact with the tumor suppressor pVHL (69). We demonstrated that integration of the vector efficiently disrupts transcription of wild-type mJade1. Since the fusion protein produced by the trapped allele contains only a 47-amino-acid fragment of mJade1, it seems extremely unlikely that this protein retains any endogenous function. Therefore, we conclude that mJade1gtE148ISCR is a null allele. However, mice homozygous for the gene trap allele do not present any overt phenotypic defects. Database searches led to identification of a family of Jade genes conserved throughout vertebrates.
Posttranscriptional regulation of Jade1 expression.
Comparative analysis of the genomic and cDNA sequences suggested that primary Jade1 transcripts in both mice and humans are subject to alternative splicing and polyadenylation. These posttranscriptional regulatory events result in production of several alternative mRNAs and at least two protein isoforms. In most of these transcripts, variation is limited to the sequence and/or size of untranslated sequences. Although these sequences do not contribute to the composition of the protein, they are known to regulate the stability and translation of transcripts (37, 39, 45, 56). The stability of gene products can also be regulated at the protein level by PEST sequences, such as these found in Jade1, that contribute to protein degradation (5, 40). The study reported by Zhou et al. (69) shows that Jade1 is a short-lived protein, unless stabilized by pVHL. Since two strong candidate PEST sequences are present in Jade1L, while only one is found in Jade1S, these isoforms may show different degradation rates.
The subcellular localization may also vary between Jade1 isoforms. Immunohistochemistry with an antibody that potentially recognizes both isoforms revealed a differential nuclear or cytoplasmic localization of this protein in a number of cell types (69). The mJade1-β-Gal reporter was also localized to the nuclei of certain cell types, although this fusion protein does not include any sequence known to function as an NLS. It seems, therefore, that the short amino acid sequence (47 amino acids) at the N terminus of endogenous mJade1, present in the mJade1-β-Gal protein, is sufficient for its transport to the nucleus, at least in these cell types. However, the bipartite NLS, included only in the predicted Jade1L, may direct a more exclusively nuclear localization of this isoform.
Molecular characteristics of the Jade family.
The nuclear localization of the mJade1-β-Gal reporter and the presence of PHD zinc fingers in all Jade family members indicate that these proteins may regulate gene expression. The function of the PHD zinc finger domain, characterized by a Cys4-His-Cys3 pattern, is still not known. However, PHD fingers were found in several proteins implicated in chromatin-mediated transcriptional regulation. These include members of the Drosophila melanogaster Trithorax and Polycomb group genes known to regulate transcription of homeotic genes (14, 32), HRX, a human Trithorax homologue implicated in acute leukemia (15, 54), and the imprinting regulator DNMT3L (1). Another characteristic shared by many PHD finger-containing proteins is that they function as part of multicomponent complexes, raising the possibility that like the closely related LIM domain (46), PHD fingers are involved in protein-protein interaction. However, interaction of Jade1 with pVHL was shown to occur independently of the PHD finger domains and is likely to be mediated by the interfinger region or the amino terminus of Jade1 (69). Interestingly, the longest contiguous stretches of amino acid identity outside the PHD finger domains were found in these two regions of Jade family members. Thus, pVHL may interact with all three Jade proteins.
Interaction with pVHL and potential role of Jade1 in placental vasculogenesis.
Endogenous Jade1S levels were shown to increase in the presence of pVHL in 293T17 human embryonic kidney cells due to the prolonged half-life of Jade1S (69). pVHL functions as a tumor suppressor, and germ line mutations of the VHL gene are the cause of von Hippel-Lindau disease, a hereditary cancer syndrome in humans (reviewed in references 19 and 31). At the molecular level, pVHL seems to be involved in the control of diverse processes. While stabilization of Jade1 was a new function attributed to pVHL (69), other better studied functions include targeting of specific proteins for polyubiquitination and subsequent degradation (29, 36) and, at least indirectly, regulation of gene expression (6, 8, 25) and mRNA stability (13, 18, 28). This functional heterogeneity is supported by the observation that pVHL forms multimeric complexes that may contain different partners. Furthermore, like Jade1, pVHL was shown to be differentially localized to the cytoplasm and nuclei of cells (27, 67). Targeted inactivation of VHL in mice results in embryonic lethality due to defective placental vasculogenesis (12). Consistent with this finding, expression of the angiogenic vascular endothelial growth factor is greatly reduced in pVHL-deficient placentas. The strong expression of the mJade1-β-Gal reporter in the progenitors of placental components (extraembryonic ectoderm and trophoblast) suggests that Jade1 may play a role in the induction of placental vasculogenesis through its interaction with pVHL.
Embryonic expression of Jade1.
The fusion mJade1-β-Gal protein was also expressed in many regions where multipotent or tissue-specific progenitors reside. During early embryogenesis, these sequentially include the pre- and early streak epiblast, the primitive streak and node, and finally, their descendant, the CNH of the tail bud. A balance between maintenance of these progenitor populations and their controlled differentiation and migration seems to be essential for the correct specification of the different embryonic lineages and elongation of the A/P axis (62, 64-66). The expression of mJade1 in these regions indicates that this gene may be involved in these processes. The finding that pVHL is important for epithelial differentiation and seems to inhibit cell migration (20, 30) further supports this possibility. pVHL also plays an important role in neuronal differentiation of central nervous system progenitor cells (21, 34). Interestingly, mJade1-β-Gal expression was seen in neural progenitors in the ventricular zone of the brain. Expression in a variety of stem cell types is also indicated by the isolation of Jade1 cDNAs from hematopoietic, mesenchymal, and trophoblast stem cell libraries.
During organogenesis stages, expression of mJade1-β-Gal is strongly activated in stripes localized to the anterior compartment of the condensing and newly formed somites. This pattern is reminiscent of genes involved in segmentation of the paraxial mesoderm and determination of A/P identity, such as Mesp2 and genes of the HoxD complex (38, 42, 68). Interestingly, global changes in chromatin structure of Hox complexes in both the primitive streak and tail bud and the nascent somites have been suggested to account for the sequential activation of progressively more 3′ genes in the clusters as axial elongation proceeds, thus conferring successively more posterior identity upon somites (23, 24). Therefore, Jade1 may function in the activation and/or repression of Hox complex genes via modulation of chromatin structure.
The high degree of conservation through vertebrate species argues for an essential function of Jade1. The absence of an overt phenotype could therefore indicate a functional compensation by other Jade family members. Examination of the libraries from which ESTs corresponding to mJade2 and mJade3 were isolated shows that these homologues are coexpressed with mJade1 in a subset of tissues. In particular, mJade3 is expressed in the early organogenesis stage embryo and an mJade3 cDNA clone was isolated in a screen for genes involved in breast cancer (50). Thus, compound mutants lacking more than one Jade member may be required to reveal the function of these genes.
Acknowledgments
We thank Ronald Wilkie for technical assistance and Muriel Lee for FISH analysis. We also thank Jean-François Nicolas, Ruth Arkell, Josh Brickman, Brian Hendrich, and Marios Stavridis for helpful comments on the manuscript.
This work was funded by an MRC Career Development Award (G120/215) to V.W. and a Ph.D. studentship from Faculty of Science and Engineering, University of Edinburgh, to E.T.
REFERENCES
- 1.Aapola, U., I. Liiv, and P. Peterson. 2002. Imprinting regulator DNMT3L is a transcriptional repressor associated with histone deacetylase activity. Nucleic Acids Res. 30:3602-3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beddington, R. S., J. Morgernstern, H. Land, and A. Hogan. 1989. An in situ transgenic enzyme marker for the midgestation mouse embryo and the visualization of inner cell mass clones during early organogenesis. Development 106:37-46. [DOI] [PubMed] [Google Scholar]
- 3.Beddington, R. S., P. Rashbass, and V. Wilson. 1992. Brachyury-a gene affecting mouse gastrulation and early organogenesis. Dev. Suppl. 157-165. [PubMed]
- 4.Cambray, N., and V. Wilson. 2002. A population of axial progenitors with extensive potency localised to the mouse chordoneural hinge. Development 129:4855-4866. [DOI] [PubMed] [Google Scholar]
- 5.Chevaillier, P. 1993. Pest sequences in nuclear proteins. Int. J. Biochem. 25:479-482. [DOI] [PubMed] [Google Scholar]
- 6.Cohen, H. T., M. Zhou, A. M. Welsh, S. Zarghamee, H. Scholz, D. Mukhopadhyay, T. Kishida, B. Zbar, B. Knebelmann, and V. P. Sukhatme. 1999. An important von Hippel-Lindau tumor suppressor domain mediates Sp1-binding and self-association. Biochem. Biophys. Res. Commun. 266:43-50. [DOI] [PubMed] [Google Scholar]
- 7.Dobrovolskaïa-Zavadskaïa, N. 1927. Sur la mortification spontanée de la queue chez la souris nouveau-née et sur l'exustence d'un caractčre hereditaire “non-viable.” C. R. Soc. Biol. 97:114-116. [Google Scholar]
- 8.Duan, D. R., A. Pause, W. H. Burgess, T. Aso, D. Y. Chen, K. P. Garrett, R. C. Conaway, J. W. Conaway, W. M. Linehan, and R. D. Klausner. 1995. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 269:1402-1406. [DOI] [PubMed] [Google Scholar]
- 9.Faisst, A. M., and P. Gruss. 1998. Bodenin: a novel murine gene expressed in restricted areas of the brain. Dev. Dyn. 212:293-303. [DOI] [PubMed] [Google Scholar]
- 10.Forrester, L. M., A. Nagy, M. Sam, A. Watt, L. Stevenson, A. Bernstein, A. L. Joyner, and W. Wurst. 1996. An induction gene trap screen in embryonic stem cells: identification of genes that respond to retinoic acid in vitro. Proc. Natl. Acad. Sci. USA 93:1677-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gajovic, S., K. Chowdhury, and P. Gruss. 1998. Genes expressed after retinoic acid-mediated differentiation of embryoid bodies are likely to be expressed during embryo development. Exp. Cell Res. 242:138-143. [DOI] [PubMed] [Google Scholar]
- 12.Gnarra, J. R., J. M. Ward, F. D. Porter, J. R. Wagner, D. E. Devor, A. Grinberg, M. R. Emmert-Buck, H. Westphal, R. D. Klausner, and W. M. Linehan. 1997. Defective placental vasculogenesis causes embryonic lethality in VHL-deficient mice. Proc. Natl. Acad. Sci. USA 94:9102-9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gnarra, J. R., S. Zhou, M. J. Merrill, J. R. Wagner, A. Krumm, E. Papavassiliou, E. H. Oldfield, R. D. Klausner, and W. M. Linehan. 1996. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc. Natl. Acad. Sci. USA 93:10589-10594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gould, A. 1997. Functions of mammalian Polycomb group and trithorax group related genes. Curr. Opin. Genet. Dev. 7:488-494. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Y., T. Nakamura, H. Alder, R. Prasad, O. Canaani, G. Cimino, C. M. Croce, and E. Canaani. 1992. The t(4;11) chromosome translocation of human acute leukemias fuses the ALL-1 gene, related to Drosophila trithorax, to the AF-4 gene. Cell 71:701-708. [DOI] [PubMed] [Google Scholar]
- 16.Hogan, B., R. Beddington, F. Constantini, and E. Lacy. 1994. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Hooper, M., K. Hardy, A. Handyside, S. Hunter, and M. Monk. 1987. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonisation by cultured cells. Nature 326:292-295. [DOI] [PubMed] [Google Scholar]
- 18.Iliopoulos, O., A. P. Levy, C. Jiang, W. G. Kaelin, Jr., and M. A. Goldberg. 1996. Negative regulation of hypoxia-inducible genes by the von Hippel-Lindau protein. Proc. Natl. Acad. Sci. USA 93:10595-10599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaelin, W. G., Jr. 2002. Molecular basis of the VHL hereditary cancer syndrome. Nat. Rev. Cancer 2:673-682. [DOI] [PubMed] [Google Scholar]
- 20.Kamada, M., K. Suzuki, Y. Kato, H. Okuda, and T. Shuin. 2001. von Hippel-Lindau protein promotes the assembly of actin and vinculin and inhibits cell motility. Cancer Res. 61:4184-4189. [PubMed] [Google Scholar]
- 21.Kanno, H., F. Saljooque, I. Yamamoto, S. Hattori, M. Yao, T. Shuin, and U. Hoi-Sang. 2000. Role of the von Hippel-Lindau tumor suppressor protein during neuronal differentiation. Cancer Res. 60:2820-2824. [PubMed] [Google Scholar]
- 22.Kaufman, M. H. 1995. The atlas of mouse development. Academic Press Limited, London, United Kingdom.
- 23.Kmita, M., F. van Der Hoeven, J. Zakany, R. Krumlauf, and D. Duboule. 2000. Mechanisms of Hox gene colinearity: transposition of the anterior Hoxb1 gene into the posterior HoxD complex. Genes Dev. 14:198-211. [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo, T., J. Zakany, and D. Duboule. 1998. Control of colinearity in AbdB genes of the mouse HoxD complex. Mol. Cell 1:289-300. [DOI] [PubMed] [Google Scholar]
- 25.Kroll, S. L., W. R. Paulding, P. O. Schnell, M. C. Barton, J. W. Conaway, R. C. Conaway, and M. F. Czyzyk-Krzeska. 1999. von Hippel-Lindau protein induces hypoxia-regulated arrest of tyrosine hydroxylase transcript elongation in pheochromocytoma cells. J. Biol. Chem. 274:30109-30114. [DOI] [PubMed] [Google Scholar]
- 26.Lawson, K. A., J. J. Meneses, and R. A. Pedersen. 1991. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113:891-911. [DOI] [PubMed] [Google Scholar]
- 27.Lee, S., D. Y. T. Chen, J. S. Humphrey, J. R. Gnarra, W. M. Linehan, and R. D. Klausner. 1996. Nuclear/cytoplasmic localisation of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc. Natl. Acad. Sci. USA 93:1770-1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levy, A. P., N. S. Levy, and M. A. Goldberg. 1996. Hypoxia-inducible protein binding to vascular endothelial growth factor mRNA and its modulation by the von Hippel-Lindau protein. J. Biol. Chem. 271:25492-25497. [DOI] [PubMed] [Google Scholar]
- 29.Li, Z., X. Na, D. Wang, S. R. Schoen, E. M. Messing, and G. Wu. 2002. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J. Biol. Chem. 277:4656-4662. [DOI] [PubMed] [Google Scholar]
- 30.Lieubeau-Teillet, B., J. Rak, S. Jothy, O. Iliopoulos, W. Kaelin, and R. Kerbel. 1998. von Hippel-Lindau gene-mediated growth suppression and induction of differentiation in renal carcinoma cells grown as multicellular tumor spheroids. Cancer Res. 58:4957-4962. [PubMed] [Google Scholar]
- 31.Linehan, W. M., M. I. Lerman, and B. Zbar. 1995. Identification of the von Hippel-Lindau (VHL) gene. Its role in renal cancer. JAMA 273:564-570. [PubMed] [Google Scholar]
- 32.Mahmoudi, T., and C. P. Verrijzer. 2001. Chromatin silencing and activation by Polycomb and trithorax group proteins. Oncogene 20:3055-3066. [DOI] [PubMed] [Google Scholar]
- 33.McClive, P., G. Pall, K. Newton, M. Lee, J. Mullins, and L. Forrester. 1998. Gene trap integrations expressed in the developing heart: insertion site affects splicing of the PT1-ATG vector. Dev. Dyn. 212:267-276. [DOI] [PubMed] [Google Scholar]
- 34.Murata, H., N. Tajima, Y. Nagashima, M. Yao, M. Baba, M. Goto, S. Kawamoto, I. Yamamoto, K. Okuda, and H. Kanno. 2002. von Hippel-Lindau tumor suppressor protein transforms human neuroblastoma cells into functional neuron-like cells. Cancer Res. 62:7004-7011. [PubMed] [Google Scholar]
- 35.Nicolas, J. F., L. Mathis, C. Bonnerot, and W. Saurin. 1996. Evidence in the mouse for self-renewing stem cells in the formation of a segmented longitudinal structure, the myotome. Development 122:2933-2946. [DOI] [PubMed] [Google Scholar]
- 36.Ohh, M., C. W. Park, M. Ivan, M. A. Hoffman, T. Y. Kim, L. E. Huang, N. Pavletich, V. Chau, and W. G. Kaelin. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat. Cell Biol. 2:423-427. [DOI] [PubMed] [Google Scholar]
- 37.Pesole, G., F. Mignone, C. Gissi, G. Grillo, F. Licciulli, and S. Liuni. 2001. Structural and functional features of eukaryotic mRNA untranslated regions. Gene 276:73-81. [DOI] [PubMed] [Google Scholar]
- 38.Pourquié, O. 2000. Vertebrate segmentation: is cycling a rule? Curr. Opin. Cell Biol. 12:747-751. [DOI] [PubMed] [Google Scholar]
- 39.Qu, X., Y. Qi, and B. Qi. 2002. Generation of multiple mRNA transcripts from the novel human apoptosis-inducing gene hap by alternative polyadenylation utilization and the translational activation function of 3′ untranslated region. Arch. Biochem. Biophys. 400:233-244. [DOI] [PubMed] [Google Scholar]
- 40.Rechsteiner, M., and S. W. Rogers. 1996. PEST sequences and regulation by proteolysis. Trends Biochem. Sci. 21:267-271. [PubMed] [Google Scholar]
- 41.Robertson, E. J. 1987. Teratocarcinomas and embryonic stem cells. IRL Press, Oxford, United Kingdom.
- 42.Saga, Y., and H. Takeda. 2001. The making of the somite: molecular events in vertebrate segmentation. Nat. Rev. Genet. 2:835-845. [DOI] [PubMed] [Google Scholar]
- 43.Sam, M., W. Wurst, M. Klüppel, O. Jin, H. Heng, and A. Berstein. 1998. Aquarius, a novel gene isolated by gene trapping with an RNA-dependent RNA polymerase motif. Dev. Dyn. 212:304-317. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 45.Schaaf, M. J., and J. A. Cidlowski. 2002. AUUUA motifs in the 3′UTR of human glucocorticoid receptor alpha and beta mRNA destabilize mRNA and decrease receptor protein expression. Steroids 67:627-636. [DOI] [PubMed] [Google Scholar]
- 46.Schmeichel, K. L., and M. C. Beckerle. 1998. LIM domains of cysteine-rich protein 1 (CRP1) are essential for its zyxin-binding function. Biochem. J. 331(Pt. 3):885-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmidt, C., V. Wilson, D. Stott, and R. S. P. Beddington. 1997. T promoter activity in the absence of T protein during axis formation and elongation in the mouse. Dev. Biol. 189:161-173. [DOI] [PubMed] [Google Scholar]
- 48.Smith, A. G. 1991. Culture and differentiation of embryonic stem cells. J. Tissue Cult. Methods 13:89-94. [Google Scholar]
- 49.Stanford, W. L., J. C. Cohn, and S. P. Cordes. 2001. Gene-trap mutagenesis: past, present and beyond. Nat. Rev. Genet. 2:756-768. [DOI] [PubMed] [Google Scholar]
- 50.Szelei, J., A. M. Soto, P. Geck, M. Desronvil, N. V. Prechtl, B. C. Weill, and C. Sonnenschein. 2000. Identification of human estrogen-inducible transcripts that potentially mediate the apoptotic response in breast cancer. J. Steroid Biochem. Mol. Biol. 72:89-102. [DOI] [PubMed] [Google Scholar]
- 51.Tajbakhsh, S., and D. Houzelstein. 1995. In situ hybridization and beta-galactosidase: a powerful combination for analysing transgenic mice. Trends Genet. 11:42. [DOI] [PubMed] [Google Scholar]
- 52.Tam, P. P., and R. S. Beddington. 1987. The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development 99:109-126. [DOI] [PubMed] [Google Scholar]
- 53.Tam, P. P., and R. R. Behringer. 1997. Mouse gastrulation: the formation of a mammalian body plan. Mech. Dev. 68:3-25. [DOI] [PubMed] [Google Scholar]
- 54.Tkachuk, D. C., S. Kohler, and M. L. Cleary. 1992. Involvement of a homolog of Drosophila trithorax by 11q23 chromosomal translocations in acute leukemias. Cell 71:691-700. [DOI] [PubMed] [Google Scholar]
- 55.Townley, D. J., B. J. Avery, B. Rosen, and W. C. Skarnes. 1997. Rapid sequence analysis of gene trap integrations to generate a resource of insertional mutations in mice. Genome Res. 7:293-298. [DOI] [PubMed] [Google Scholar]
- 56.van Hoof, A., and R. Parker. 2002. Messenger RNA degradation: beginning at the end. Curr. Biol. 12:R285-R287. [DOI] [PubMed] [Google Scholar]
- 57.von Melchner, H., S. Reddy, and H. E. Ruley. 1990. Isolation of cellular promoters by using a retrovirus promoter trap. Proc. Natl. Acad. Sci. USA 87:3733-3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voss, A., T. Thomas, and P. Gruss. 1998. Compensation for a gene trap mutation in the murine microtubule-associated protein 4 locus by alternative polyadenylation and alternative splicing. Dev. Dyn. 212:258-266. [DOI] [PubMed] [Google Scholar]
- 59.Voss, A. K., T. Thomas, and P. Gruss. 1998. Efficiency assessment of the gene trap approach. Dev. Dyn. 212:171-180. [DOI] [PubMed] [Google Scholar]
- 60.Whiting, J., H. Marshall, M. Cook, R. Krumlauf, P. W. Rigby, D. Stott, and R. K. Allemann. 1991. Multiple spatially specific enhancers are required to reconstruct the pattern of Hox-2.6 gene expression. Genes Dev. 5:2048-2059. [DOI] [PubMed] [Google Scholar]
- 61.Wilkinson, D. G., S. Bhatt, and B. G. Herrmann. 1990. Expression pattern of the mouse T gene and its role in mesoderm formation. Nature 343:657-659. [DOI] [PubMed] [Google Scholar]
- 62.Wilson, V., and R. Beddington. 1997. Expression of T protein in the primitive streak is necessary and sufficient for posterior mesoderm movement and somite differentiation. Dev. Biol. 192:45-58. [DOI] [PubMed] [Google Scholar]
- 63.Wilson, V., and R. S. Beddington. 1996. Cell fate and morphogenetic movement in the late mouse primitive streak. Mech. Dev. 55:79-89. [DOI] [PubMed] [Google Scholar]
- 64.Wilson, V., L. Manson, W. C. Skarnes, and R. S. Beddington. 1995. The T gene is necessary for normal mesodermal morphogenetic cell movements during gastrulation. Development 121:877-886. [DOI] [PubMed] [Google Scholar]
- 65.Yamaguchi, T. P., A. Bradley, A. P. McMahon, and S. Jones. 1999. A Wnt5a pathway underlies outgrowth of multiple structures in the vertebrate embryo. Development 126:1211-1223. [DOI] [PubMed] [Google Scholar]
- 66.Yamaguchi, T. P., S. Takada, Y. Yoshikawa, N. Wu, and A. P. McMahon. 1999. T (Brachyury) is a direct target of Wnt3a during paraxial mesoderm specification. Genes Dev. 13:3185-3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ye, Y., S. Vasavada, I. Kuzmin, T. Stackhouse, B. Zbar, and B. R. G. Williams. 1998. Subcellular localisation of the von Hippel-Lindau disease gene product is cell cycle-dependent. Int. J. Cancer 78:62-69. [DOI] [PubMed] [Google Scholar]
- 68.Zákány, J., M. Kmita, P. Alarcon, J. L. de la Pompa, and D. Duboule. 2001. Localized and transient transcription of Hox genes suggests a link between patterning and the segmentation clock. Cell 106:207-217. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, M. I., H. Wang, J. J. Ross, I. Kuzmin, C. Xu, and H. T. Cohen. 2002. The von Hippel-Lindau tumor suppressor stabilises novel plant homeodomain protein Jade-1. J. Biol. Chem. 277:39887-39898. [DOI] [PubMed] [Google Scholar]