Abstract
Structural studies of caspase-1 reveal that the dimeric thiol protease can exist in two states: in an on-state, when the active site is occupied, or in an off-state, when the active site is empty or when the enzyme is bound by a synthetic allosteric ligand at the dimer interface ∼15 Å from the active site. A network of 21 hydrogen bonds from nine side chains connecting the active and allosteric sites change partners when going between the on-state and the off-state. Alanine-scanning mutagenesis of these nine side chains shows that only two of them—Arg286 and Glu390, which form a salt bridge—have major effects, causing 100- to 200-fold reductions in catalytic efficiency (kcat/Km). Two neighbors, Ser332 and Ser339, have minor effects, causing 4- to 7-fold reductions. A more detailed mutational analysis reveals that the enzyme is especially sensitive to substitutions of the salt bridge: even a homologous R286K substitution causes a 150-fold reduction in kcat/Km. X-ray crystal structures of these variants suggest the importance of both the salt bridge interaction and the coordination of solvent water molecules near the allosteric binding pocket. Thus, only a small subset of side chains from the larger hydrogen bonding network is critical for activity. These form a contiguous set of interactions that run from one active site through the allosteric site at the dimer interface and onto the second active site. This subset constitutes a functional allosteric circuit or “hot wire” that promotes site-to-site coupling.
Keywords: caspase-1, allostery, alanine-scanning mutagenesis, H-bonding, cooperativity
Introduction
Allostery, which refers to functional coupling between sites on proteins, is central to biological regulation. This phenomenon has been classically examined in hemoglobin and aspartate transcarbamoylase (ATCase), and has been generalized to many other regulatory enzymes involved in metabolic and signaling pathways, as well as in membrane and nuclear receptors (for a recent review, see Changeux and Edelstein1). While structural changes brought on by allostery are well-documented, the functional roles of amino acid side chains that couple conformational changes are less well understood. In this study, we begin to systematically examine the functional importance of amino acid side chains in coupling conformational changes between sites in caspase-1.
Caspases are dimeric thiol proteases that drive cellular processes such as apoptosis and inflammation. These enzymes have at least two conformational states that are observed crystallographically and represent both active and inactive zymogen conformations (for reviews, see Fuentes-Prior and Salvesen2 and Shi3). The active conformation of caspase-1 has been crystallized with substrate mimics bound to the active site, and these represent the on-state conformation of the enzyme.4,5 Recently, specific allosteric small-molecule ligands have been captured at a cysteine residue at the dimer interface of apoptotic caspases-3 and -7,6 as well as the inflammatory caspase-1.4 These ligands bind to a site ∼15 Å from the active site and trap an inactive conformation of caspases. This off-state conformation is similar to the inactive zymogen form of caspase-76 or the ligand-free form of caspase-1,5 and therefore represents a natural state of these proteases (Fig. 1).
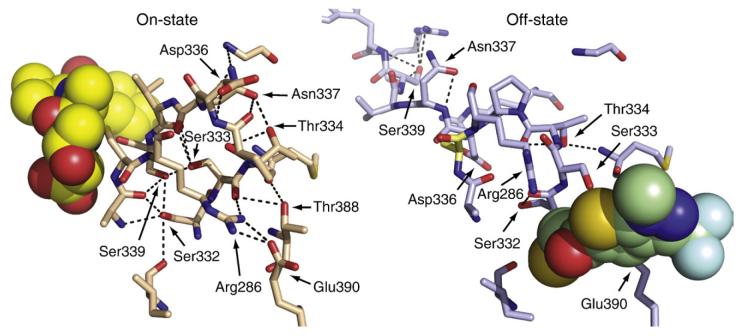
Fig. 1.
Locations of the residues involved in the H-bonding network (highlighted in green sticks) of dimeric caspase-1: containing the active-site inhibitor z-VAD-FMK (on-state, top; PDB ID code 2HBQ); ligand-free (off-state, middle; PDB ID code 1SC1); or containing an allosteric inhibitor (off-state, bottom; PDB ID code 2FQQ). The active-site Cys285 (C285A in off-state structures) is shown in orange, and the inhibitors are shown in spheres. The Glu390 residue is hidden behind the allosteric inhibitor in the bottom structure.
Caspase-1 shows positive cooperativity, indicating that binding of substrate at one active site enhances catalysis at the second site.4 Inspection of the X-ray structures for the active and allosterically inhibited enzyme suggests that coupling may be mediated by a network of 21 hydrogen bonding (H-bonding) interactions primarily involving nine side chains (Fig. 2). This network runs from one active site through the allosteric site to the second active site. Eight of the nine H-bonding residues in the network are completely conserved among inflammatory caspases-1 and -5, and a close homolog caspase-4. (Asp336 in caspase-1 is a histidine in caspases-4 and -5.) Overall, procaspase-1 has about a 45% sequence identity with procaspases-4 and -5, which have a 66% sequence identity with each other. One key interaction in caspase-1 appears to be a conserved salt bridge between Arg286 and Glu390, as the allosteric inhibitors in caspase-1 directly disrupt this network by preventing the salt bridge from forming.4
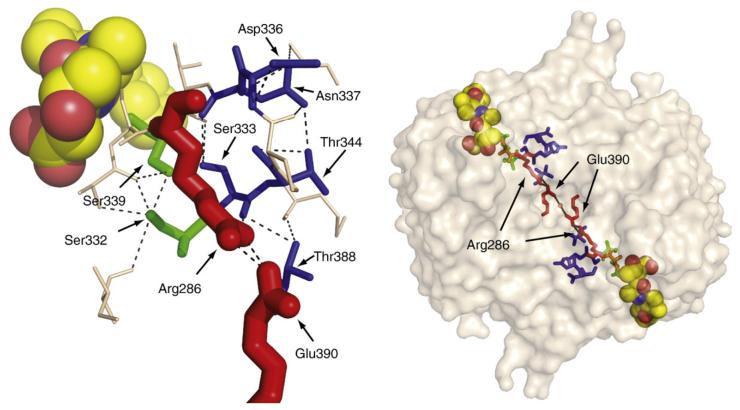
Fig. 2.
Residues that form an H-bonding network and a salt bridge connecting the active and allosteric ligand sites of caspase-1. In the on-state conformation (left), many H-bond interactions involving polar side chains that are not preserved in the off-state conformation (right) are formed. Dashed lines indicate a distance of <3.5 Å between two polar atoms. Yellow spheres represent the z-VAD-FMK active-site inhibitor in the on-state structure (PDB ID code 2HBQ);4 green spheres represent the allosteric inhibitor in the off-state structure (PDB ID code 2FQQ).4 The active-site cysteine (Cys285) is replaced with alanine (orange) in the off-state structure, and Thr388 is hidden behind the allosteric inhibitor.
To better understand the importance of residues in the H-bonding network in caspase-1 for enzyme activity, we employed alanine-scanning mutagenesis. This approach has been proven effective for the systematic identification of functional “hot spots” in protein interfaces7 (for a recent review, see Moreira et al.8). Here, alanine-scanning mutational analysis shows that only four of the nine side chains, including the salt bridge, are significantly important for enzyme activity. These form a contiguous circuit of residues, or “hot wire,” that runs from one active site to the dimer interface and onto the allosteric site. Such allosteric circuits may be revealed by mutational analysis in other cooperative enzymes and help to identify important functional determinants for allosteric coupling and activity.
Results
Mutational analysis of the H-bonding network in caspase-1
Active forms of caspase-1 have been crystallized while bound to the active-site inhibitors Ac-WEHDCHO9 and z-Val-Ala-Asp-fluoromethylketone (z-VAD-FMK)4 [Fig. 1, top; Protein Data Bank (PDB) ID code 2HQB]. They have also been crystallized as active-site ligand-free enzyme5 (Fig. 1, middle; PDB ID code 1SC1) and in the allosterically inhibited form4 (Fig. 1, bottom; PDB ID code 2FQQ). The active-site ligand-free and allosterically inhibited conformations are nearly identical, suggesting that they represent a natural off-state of the protease. The on-state and the off-state appear to be in dynamic equilibrium. These states can be trapped in caspase-14 or caspase-76 with site-specific covalent inhibitors to either the active site or the allosteric site. The labeling is mutually exclusive, since for both enzymes, the binding of an active-site inhibitor blocks the binding of an allosteric inhibitor, and vice versa.
The on-state of caspase-1 shows a complex network of 21 protein—protein H-bonds that connects the active site to the allosteric site (Table 1). In contrast, only 12 H-bonds are present in the off-state; of these, only one is preserved in the on-state. The nine side chains that dominate these H-bonding interactions form a contiguous network between the active site and the allosteric site, including a central Arg286-Glu390 salt bridge (Fig. 2). We wished to probe how important this H-bonding network is to the activity of caspase-1. Thus, we systematically replaced with alanines the nine H-bonding residues whose side chain positions change significantly (>3–5 Å) when caspase-1 switches from the on-state to the off-state (Table 1). The variants were expressed, purified, and kinetically analyzed as described in Materials and Methods.
Table 1.
Potential intramolecular hydrogen bonds formed by polar side chains
| Side chain | Active-site ligand-bound structure | Active-site ligand-free structure |
|---|---|---|
| Arg286 | Arg286 Nη1:Glu390Oε1 | No intramolecular contacts |
| Arg286 Nη1: Ser333 O | ||
| Arg286 Nη2:Glu390Oε1 | ||
| Ser332 | Ser332 Oγ: Ser339 Oγ | Ser332 Oγ: Ser332 O |
| Ser332 Oγ: Ala284 O | ||
| Ser332 Oγ: Ala284 N | ||
| Ser332 Oγ: Ile282 O | ||
| Ser333 | Ser333 Oγ: Ser339 Oγ | No intramolecular contacts |
| Ser333 Oγ: Asn337 O | ||
| Ser333 Oγ: Val388 O | ||
| Thr334 | Thr334 Oγ1:Asn337 Oδ1 | No intramolecular contacts |
| Thr334 Oγ1: Thr334 O | ||
| Asp336 | Asp336 Oδ1:Arg240Nη1 | Asp336 Oδ1: Asp336 O |
| Asp336 Oδ1: Asp336 N | Asp336 Oδ1: Arg383 Nη1 | |
| Asp336 Oδ2:Arg240Nη1 | Asp336 Oδ2: Arg383 Nη1 | |
| Asn337 | aAsn337 Oδ1: Thr334 Oγ1 | Asn337 Oδ1: Asn337 N |
| Asn337 Oδ1: Pro335 O | ||
| Asn337 Nδ2: Asn337 N | Asn337 Nδ2: Asn337 O | |
| Asn337 Nδ2: Gly291 N | Asn337 Nδ2: Asn337 N | |
| Ser339 | aSer339 Oγ: Ser333 Oγ | Ser339 Oγ: Arg341 O |
| aSer339 Oγ: Ser332 Oγ | Ser339 Oγ: Trp340 N | |
| Ser339 Oγ: Ala284 O | ||
| Thr388 | Thr388 Oγ1: Ser333 O | Thr388 Oγ1: Glu390 Oε1 |
| Thr388 Oγ1: Met386 | Thr388 Oγ1: Thr388 O | |
| Glu390 | aGlu390 Oε1: Arg286 Nη1 | aGlu390 Oε1: Thr388 Oγ1 |
| aGlu390 Oε1: Arg286 Nη2 | ||
| Glu390 Oε2: Arg391 N | ||
| Total number of unique 2H-bonds | 21 | 12 |
Potential H-bonds were based on PDB ID code 2HBQ for active-site ligand-bound structure and on PDB ID code 1SC1 for active-site ligand-free structure.
Indicates potential hydrogen bond listed earlier in this table under an interacting partner.
The effects of alanine substitution differed widely (Table 2)†. Five of the variants (S333A, T334A, D336A, N337A, and T388A) caused minor (2-fold or less) reductions in kcat/Km. In contrast, two variants caused moderate [about 4-fold (S332A) and 7-fold (S339A)] reductions in kcat/Km, and two caused much larger [230-fold (R286A) and 130-fold (E390A)] reductions. Generally, the effects showed up as a mixture of both increases in Km and decreases in kcat. For hydrolysis of amides, acylation is rate-limiting; thus, Km approximates Kd for substrate binding.10 Therefore, much of these effects reflects reduced binding affinity of the substrate, although for the S332A variant, most of the effects appeared in kcat.
Table 2.
Kinetic data for alanine scan mutants of the caspase-1 H-bonding network
| Construct |
Km (μM) |
kcat (S-1) |
kcat/Km (M-1·S-1) |
kcat/Km ratioa |
nHill | H-bonds |
|
|---|---|---|---|---|---|---|---|
| Active | Apo | ||||||
| Wild-type | 12 | 0.65 | 5.3 × 104 | 1 | 1.4 | ||
| R286A | 230 | 0.055 | 2.4 × 102 | 230 | 1.8 | 3 | 0 |
| S332A | 3.2 | 0.055 | 1.4 × 104 | 3.7 | 1.4 | 4 | 1 |
| S333A | 15 | 0.38 | 2.5 × 104 | 2.0 | 1.8 | 3 | 0 |
| T334A | 18 | 0.55 | 3.1 × 104 | 1.7 | 1.7 | 2 | 0 |
| D336A | 21 | 0.78 | 3.8 × 104 | 1.4 | 1.5 | 3 | 3 |
| N337A | 13 | 0.41 | 3.0 × 104 | 1.8 | 1.7 | 4 | 3 |
| S339A | 25 | 0.20 | 7.9 × 103 | 6.7 | 1.6 | 3 | 2 |
| T388A | 8.6 | 0.24 | 2.8 × 104 | 1.9 | 1.6 | 2 | 2 |
| E390A | 100 | 0.041 | 4.0 × 102 | 130 | 1.3 | 2 | 2 |
| R286A/ E390A |
430 | 0.014 | 3.2 × 101 | 1700 | 1.6 | ||
Standard errors are based on data collected in triplicate and are within 10% of reported values.
kcat/Km ratio relative to wild-type.
It is useful to interpret these data in the context of the structure of the on-state and the off-state of caspase-1. Four of the H-bonding side chains—Ser332, Ser333, Ser339, and Thr388—form a cluster that lies behind loop 2 (residues 285–290), which contains the catalytic Cys285. This cluster appears to stabilize this region, which forms the floor of the substrate-binding site in the on-state (Fig. 2). In the ligand-free or allosteric ligand-bound conformations (off-states), these loops undergo a large conformational change, breaking almost all of these H-bonds (Fig. 2). We determined the structure of the T388A variant in complex with the z-VAD-FMK active-site inhibitor. The Arg286 and Glu390 side chains involved in the salt bridge interaction adopted different rotamers in the T388A variant (PDB ID code 2H54), but the structure was otherwise virtually identical with the wild-type enzyme (data not shown). The S332A and S339A substitutions, which are located most centrally in this cluster directly under the active site, cause larger reductions in kcat/Km compared to substitutions at the more peripherally located Ser333 and Thr388 positions.
Another part of the H-bonding network formed in the on-state of caspase-1 involves the side chains of Thr334, Asp336, and Asn337 (Fig. 2). In the on-state conformation, these side chains make polar interactions that appear to stabilize loop 3 (residues 332–346). Loop 3 contains Arg341 and Trp340, which directly contact the P1 and P2 substrate residues, respectively. In the off-state, Asp336 forms a new salt bridge with Arg383, but Thr334 makes no intramolecular H-bonds, and Asn337 only interacts with its backbone amide and carbonyl (Fig. 2). Alanine substitutions at these three residues cause only minor (within 2-fold) reductions in kcat/Km. The dominant effect appears at the Arg286-Glu390 salt bridge, where alanine substitutions cause >100-fold reductions in kcat/Km (Table 2). These effects are comparable to alanine substitutions in subtilisin that break direct H-bonds stabilizing the oxyanion transition state.11
The magnitude of these individual functional effects is not readily rationalized based on the change in H-bond inventory between the on-state and the off-state or on the absolute number of H-bonds present in each state (Table 2). For example, some substitutions that break a net of three H-bonds when going from on-state to off-state (R286A, S332A, and S333A) cause reductions in kcat/Km ranging from 2- to 200-fold. A similar range (2- to 200-fold) is seen when the net change in H-bond inventory is 0 or 1 (D336A, N337A, S339A, T388A, and E390A). Side chains seen engaged in many or few H-bonds in either state do not predict the effects of alanine substitution either. Thus, the extent of H-bonding or change in H-bonding is not a good predictor of its functional effect on the allosteric network.
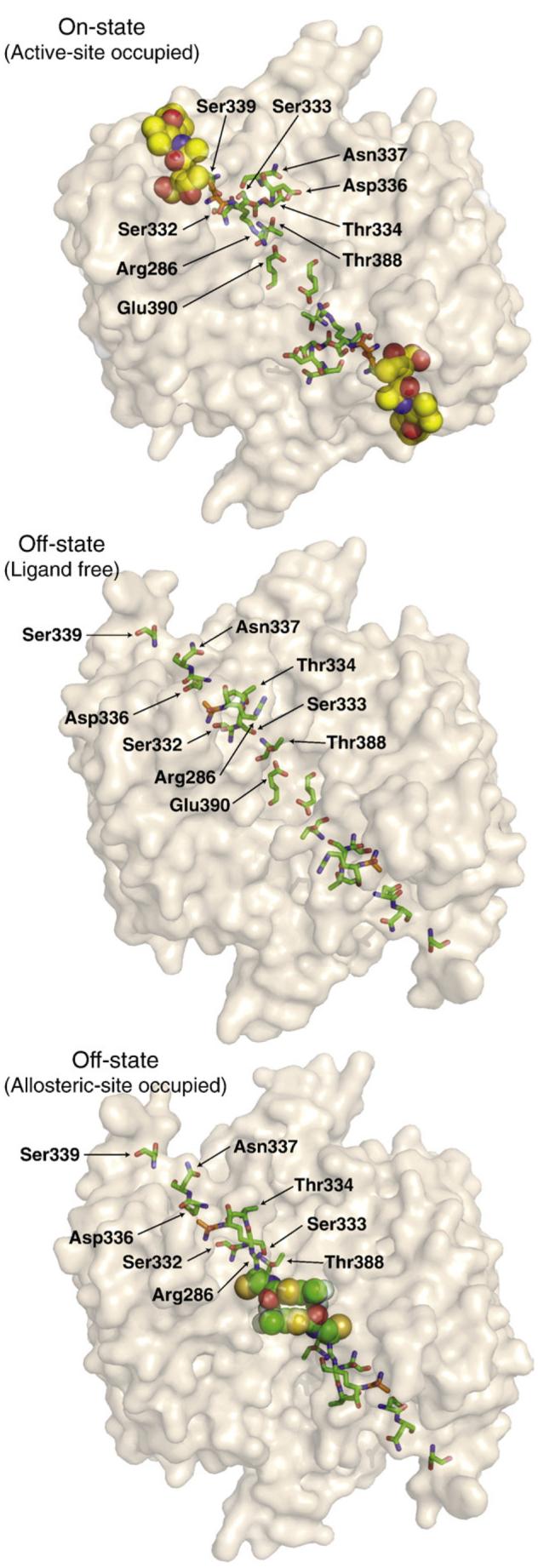
If one maps the magnitude of alanine substitution effects on the side chains as they sit in the active conformation of caspase-1, one can see that the most functionally critical side chains (Ser332, S339, Arg286, and Glu390) form a circuit that connects the active site and the allosteric site (Fig. 3, left). Viewing this circuit in the context of a dimeric protease, these residues effectively connect one active site to the central allosteric cavity and, by symmetry, to the second active site (Fig. 3, right). We have found that caspase-1 shows positive cooperativity with a Hill coefficient nHill of ∼1.4 (Table 2 and Scheer et al.4), indicating that binding at one active site enhances activity at the second active site. The values of nHill can theoretically range from 1 to 2 for a positive cooperative dimeric enzyme. The nHill value we find for caspase-1 is comparable to that of a classically positive cooperazyme such as glycogen phosphorylase (nHill =1.612,13).
Fig. 3.
The left panel shows a size- and color-coded representation of residues important for wild-type caspase-1 activity. Larger residues had a larger impact when replaced with alanine; red, green, and blue indicate a >100-fold, >2-fold, and <2-fold decrease in kcat/Km relative to wild-type. The allosteric site is located to the right of Glu390. The right panel shows an expanded view of the caspase-1 dimer. A circuit of residues connects the two active sites to the central allosteric site via the Arg286-Glu390 salt bridges. The yellow spheres represent the z-VAD-FMK active-site inhibitor. This figure is derived from the active-site bound on-state structure (PDB ID code 2HBQ).4
We determined the impact of alanine substitutions in the H-bonding network on the Hill coefficient. Interestingly, most mutations preserve or even enhance the nHill value, indicating that binding of the substrate at one active site can still couple through to the other active site. Our data suggest that cooperativity is a robust property in that it is not significantly impacted by single amino acid substitutions. The kinetic parameters reported here are nearly the same as those previously reported for wild-type caspase-1 and the salt bridge mutants. However, the Hill coefficient for the E390A mutant was 1.0 in our previous work and 1.3 in this study.4 We believe that this modest difference reflects the newer data being collected at a higher enzyme concentration and with more replicates.
Conservative substitutions of the Arg286-Glu390 salt bridge
To further dissect the importance of the central salt bridge for caspase-1 activity, we introduced less dramatic changes to the side chains by mutagenesis and determined their X-ray structures. The Arg286 in the large subunit was replaced with a lysine (PDB ID code 2H4Y), and the Glu390 in the small subunit was replaced with an aspartate (PDB ID code 2H4W). We tested the effects of these substitutions, individually and together, on caspase-1 activity (Table 3). The conservative replacement of Arg286 with Lys caused the largest reduction—a 150-fold decrease in catalytic efficiency (kcat/Km). This decrease resulted from both decreasing kcat and increasing Km. In contrast, the conservative replacement of Glu390 with Asp caused only a 2-fold decrease in kcat/Km. Interestingly, the effect of the R286K/E390D variant (PDB ID code 2H51) was intermediate between the two single amino acid substitutions; the 37-fold decrease in activity of this variant relative to that of wild-type suggests that the E390D substitution is able to partly restore the catalytic function lost in the R286K variant. The effects on the nHill value were small, as seen with the single-site alanine substitutions.
Table 3.
Kinetic data for caspase-1 salt bridge variants
| Construct |
Km (μM) |
kcat (S-1) |
kcat/Km (M-1·S-1) |
kcat/Km ratioa |
nHill |
|---|---|---|---|---|---|
| R286K | 320 | 0.12 | 3.6 × 102 | 150 | 1.5 |
| E390D | 8.6 | 0.30 | 3.5 × 104 | 1.5 | 1.4 |
| R286K/E390D | 60 | 0.086 | 1.4 × 103 | 37 | 1.3 |
Standard errors are based on data collected in triplicate and are within 10% of reported values.
kcat/Km ratio relative to wild-type (Table 2).
To better understand the effects of these substitutions on the active form of caspase-1, we determined X-ray structures of the three enzyme variants labeled with the active-site inhibitor z-VAD-FMK to trap the active form of the enzyme and compared them with active site labeled wild-type caspase-1 (Table 3). The enzyme structures were determined at 1.8–2.1 Å resolution and refined with the covalent active-site inhibitor bound (Table 4). These showed that the overall structures of the variant enzymes were virtually identical with the wild-type protein in the active conformation, except for subtle changes in the key salt bridge interaction (Fig. 4). Interestingly, the R286K variant that showed the most drastic effect on catalytic activity was the only variant to maintain the direct salt bridge Lys286-Glu390 (Fig. 4a and b). In both the E390D variant and the R286K/E390D variant, a water-mediated salt bridge is formed (Fig. 4c and d).
Table 4.
Crystallographic data and refinement statistics
| Mutation | E390D | R286K | R286K E390D | T388A |
|---|---|---|---|---|
| PDB ID | 2H4W | 2H4Y | 2H51 | 2H54 |
| Space group | P43212 | P43212 | P43212 | P43212 |
| Cell constants a, b, c (Å) | 63.3, 63.3, 161.3 | 63.3, 63.3, 141.6 | 63.2, 63.2, 141.5 | 63.2, 63.2, 162.1 |
| X-ray source | R-Axis IV | R-Axis IV | SSRL BL 1.5 | R-Axis IV |
| Wavelength (Å) | 1.54 | 1.54 | 1.16 | 1.54 |
| Resolution (Å) | 20–2.0 | 20–1.9 | 20–2.1 | 20–1.8 |
| Number of observations | 72,514 | 65,566 | 59,667 | 215,595 |
| Number of reflections | 22,946 | 23,520 | 17,438 | 31,401 |
| Completeness (%)a | 92.0 (98.9) | 96.8 (90.6) | 95.6 (82.7) | 96.0 (89.3) |
| Mean I/(σI) | 6.6 (2.5) | 6.1 (2.6) | 8.8 (3.4) | 8.8 (2.0) |
| Rmerge on Ib | 0.057 (0.268) | 0.065 (0.237) | 0.049 (0.148) | 0.042 (0.364) |
| Cutoff criteria | I ← 3σ(I) | I ← 3σ(I) | I ← 3σ(I) | I ← 3σ(I) |
| Model and refinement statistics | ||||
| Resolution range (Å) | 20–2.0 | 20–1.9 | 20–2.1 | 20–1.8 |
| Number of reflectionsc | 19,985 (1079) | 19,735 (1070) | 15,528 (834) | 28,552 (1522) |
| Completeness (%) | 91.8 | 88.4 | 93.8 | 95.7 |
| Cutoff criterion | |F|>0.0 | |F|>0.0 | |F|>0.0 | |F|>0.0 |
| Number of residues | 256 | 250 | 254 | 258 |
| Number of water molecules | 179 | 189 | 215 | 271 |
| r.m.s.d. bond lengths (Å) | 0.006 | 0.006 | 0.008 | 0.006 |
| r.m.s.d. bond angles (°) | 0.888 | 0.879 | 1.001 | 1.287 |
| Luzzatti error (Å) | 0.263 | 0.243 | 0.246 | 0.246 |
| Correlation factord | 0.927 | 0.925 | 0.921 | 0.935 |
| Rcryste | 21.03 | 20.31 | 17.96 | 20.31 |
| Rfree | 24.66 | 23.55 | 23.63 | 23.71 |
| Ramachandran plot statisticsf | ||||
| Most favored | 200 (89.3%) | 197 (89.5%) | 204 (91.5%) | 202 (89.4%) |
| Additionally allowed | 23 (10.3%) | 22 (10.0%) | 18 (8.1%) | 23 (10.2%) |
| Generously allowed | 1 (0.4%) | 1 (0.5%) | 1 (0.4%) | 1 (0.4%) |
| Disallowed | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) |
| Overall G-factorg | 0.2 | 0.1 | 0.1 | 0.2 |
Numbers in parenthesis indicate high-resolution shells: E390D, 2.07–2.0 Å; R286K, 1.97–1.9 Å; R286K/E390D, 2.17–2.1 Å; T388A, 1.86–1.8 Å.
.
Numbers in parentheses indicate the number of reflections used to calculate the Rfree factor.
Correlation factor between the structure factors and the model, as calculated by SFCHECK.14
Rcryst = Σhkl|Fo(hkl) − Fc(hkl)|/Σhkl|Fo(hkl)|, where Fo and Fc are observed and calculated structure factors, respectively.
Computed with PROCHECK.15
Overall G factor is a measure of the overall normality of the structure and is obtained from an average of all the different G factors for each residue in the structure. It is essentially a log-odds score based on the observed distributions of these stereochemical parameters.15
Fig. 4.
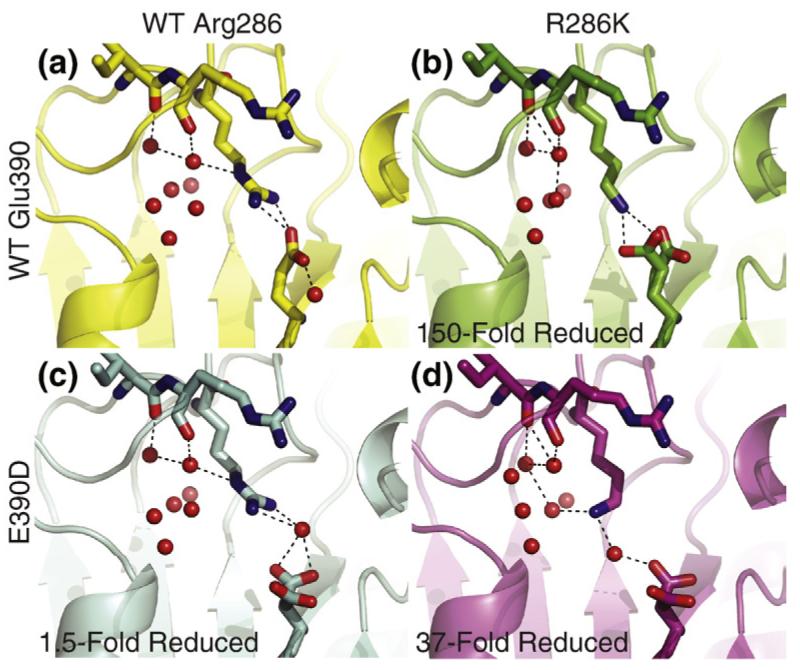
Comparison of the salt bridge interaction in wild-type caspase-1 and variants in the active conformation. (a) The top left panel shows the configuration of the Arg286-Glu390 salt bridge in wild-type caspase-1. The γ-amine of Arg286 coordinates a water molecule in the solvent pocket. (b) The top right panel shows the direct salt bridge formed between lysine at position 286 and Glu390; the ε-amine of the lysine does not coordinate solvent water molecules. (c) The bottom left panel shows the indirect salt bridge formed between Arg286 and the shortened acidic side chain of aspartate at position 390. (d) The bottom right panel shows the configuration of the indirect salt bridge formed in the R286K/E390D variant, which is able to restore activity lost in the R286K variant. The ε-amine of lysine bends back towards the solvent pocket to a position where it both coordinates a solvent water molecule and maintains an indirect salt bridge with the aspartate.
What could explain the lower activity of the R286K variant even when the salt bridge is maintained? In the wild-type structure, solvent water molecules sit in the pocket behind Arg286, one of which is coordinated by the carbonyl of Arg240 and the γ-amine of Arg286 (Fig. 4a). In the R286K variant, this water is only coordinated by the carbonyl of Arg240; the ε-amine of the lysine is held away from this pocket to form a salt bridge with Glu390 and is unable to coordinate solvent water molecules (Fig. 4b). In contrast, the γ-amine of Arg286 in the E390D variant maintains coordination of water, while the terminal amine forms an indirect salt bridge with the aspartate in the 390 position (Fig. 4c). The indirect salt bridge is completed by a water molecule that sits between the γ-amine of Arg286 and the aspartate. In the R286K/E390D variant, the lysine and aspartate are far too separated to form a direct salt bridge. Instead, the ε-amine of lysine bends back toward the solvent pocket into a position where it is able to both coordinate a solvent water molecule and maintain an indirect salt bridge with the aspartate via solvent water (Fig. 4d). Thus, perhaps stabilizing interactions with the solvent pocket and maintaining a salt bridge interaction between the allosteric site and the active site are both important for preserving catalytic activity in caspase-1. It is also possible that these static structures of the on-state do not reveal the true stability of the on-form relative to the off-form. Although we see that the salt bridge can form in the R286K/E390D variant when trapped by binding the covalent active-site inhibitor, the enzyme is clearly challenged to reach the active conformation.
Discussion
These mutational and structural studies begin to reveal the critical features of an extensive and conserved H-bonding network that couples the functional sites in caspase-1. The alanine-scanning experiments indicate that only four of the nine H-bonding side chains have a significant effect on activity, especially the Arg286-Glu390 interaction. These form a contiguous chain of interactions that connects the active and allosteric sites in the protease.
It is remarkable that so many of the H-bonding pairs seen in the on-state are not preserved in the off-state of caspase-1 (Table 1, Fig. 2). From structural inspection alone, it is difficult to predict which of the residues involved in this extensive H-bonding network are most critical for enzyme activity. Neither the extent of H-bonding in one state nor the changes in H-bonding between on-state and off-state predict which residues should be most important (Table 2). Including bound water molecules in the H-bond inventory still does not improve predictive power (see Supplemental Table S1). We conclude that changes in H-bonding patterns are not clear predictors of the effects on the activity we observe. In general, the substitutions had fairly small effects on the Hill coefficient. Even the 130- and 230-fold reductions in activity seen for the E390A and R286A caused only slight changes in the Hill coefficient, suggesting that cooperativity is a fairly robust property of the enzyme.
The effects of conservative substitutions in the critical Arg286-Glu390 salt bridge on both the structure and the function of the enzyme were somewhat surprising. One conservative shortening substitution (R286K) caused a 150-fold reduction in activity, whereas another one (E390D) had only a small 2-fold reduction.12 This substitution, combined with the R286K, caused partial restoration from the single R286K variant. The structures of the active forms of the enzyme, driven by reaction with the active-site titrant z-VAD-FMK, did not provide an obvious structural interpretation for the functional effects. For example, we see that the direct salt bridge is preserved in the dramatically reduced R286K variant; however, for the E390D variant, a water-mediated salt bridge is seen between Asp390 and Arg286. The most active variants are those that preserve both the salt bridge between positions 286 and 390 and the interaction with the water cluster. These latter interactions are lost in the R286K variant and preserved in the more active R286K/E390D variant.
Solvent accessibility calculations (Supplemental Table S2) show that Arg286 in the wild-type structure is more solvent-exposed than Lys286 in the mutant variant. Thus, it is possible that the less solvent-accessible salt bridge in the mutant is energetically less favorable due to the internal environment of the protein not fully compensating for the desolvation of the salt bridge. We calculated the solvent-exposed polar and hydrophobic surface areas for the two salt bridge residues in each of the variants. Interestingly, we find that as the exposed hydrophobic surface area increases and the exposed polar surface area decreases, the activity of the variants also decreases. We are reluctant to make strong conclusions about the importance of specific H-bonding interactions with the water cluster based on this small data set. Nonetheless, we are impressed by the functional variation in these conservative substitutions.
Nonpreserving effects for conservative substitutions of electrostatic interactions in other systems have also been reported. Studies of Ras have demonstrated that the Gln61 position is sensitive to mutations that can reduce the kcat of the RasGAP complex by at least 1000-fold. Interestingly, computational studies found that despite the direct electrostatic interactions this residue makes with other residues and substrates in the transition state, it is not directly involved chemically in GTP hydrolysis. Rather, this residue is crucial in helping to form the polar framework of the active site, and mutations of Gln61 disrupt the preorganized environment necessary for catalysis.16 The importance of electrostatic interactions has also been studied in the context of protein—protein interfaces. The structure of the p160 coactivator ACTR, in complex with the ACTR-binding domain of the CREB-binding protein CBP, identified a buried Arg-Asp salt bridge in the midst of a binding interface largely dominated by hydrophobic residues.17 Mutational studies of this salt bridge found that this interaction is especially important for specificity when discriminating between binding partners. Modification of this interaction by swapping the positions of the Arg and Asp residues to maintain the salt bridge abrogated binding.18 This observation is perhaps not surprising, as ion pairs reversal in stable local protein environments is generally destabilizing.19 These environments tend to form prepolarized sites organized to stabilize an ion pair of a given polarity such that swapping the residues in a salt bridge, as in the case of the ACTR—CBP binding interaction, will be destabilizing even when an electrostatic interaction is preserved. We have made perturbations to the electrostatic interaction in caspase-1 that we expected to be less disruptive than those in these other systems, yet we were surprised to find a significant impact on the catalytic activity in the R286K variants. It is possible that the reduction in kcat/Km seen with a R286K substitution is caused by disruption of a preorganized network of polar interactions essential for protease activity that involves not only the Arg286-Glu390 salt bridge but also surrounding interactions involving solvent water molecules.
The classic model of allosteric transitions predicts that proteins can adopt alternate conformations that are present in a preexisting equilibrium even in the absence of regulatory ligands.20 Recent studies in various systems have begun to provide direct experimental evidence for this theory and have extended it to allosteric regulation involving covalent modifications, in addition to ligand-binding events. NMR relaxation experiments on the nitrogen-regulatory protein C demonstrated that the activation of this protein by phosphorylation shifts a preexisting equilibrium between inactive and active conformations.21 In a similar vein, studies of the enzyme cyclophilin A found that the enzyme undergoes presampling of conformational states in the absence of substrate that mimics the dynamics seen during catalysis. Specific mutations cause shifts in the relative populations between conformations. In addition, cyclosporin A, which binds and inhibits cyclophilin A, shifts the enzyme to a single state. Thus, this drug works by locking the isomerase in one conformation.22 It is known that disruption of a salt bridge between the catalytic (Asp236) and regulatory (Lys143) subunits in Escherichia coli ATCase disrupts cooperative binding of the aspartate substrate.23,24 More recent work using small-angle X-ray scattering has found that disruption of this salt bridge by substitution with alanine results in an enzyme variant that exists in a reversible equilibrium between at least two states in the absence of ligands.25 This is in contrast to wild-type ATCase, which exists predominantly in a low-activity, low-affinity state. Therefore, disruption of a salt bridge in ATCase destabilizes one state, allowing the protein to exist in an equilibrium between two states in solution. In the case of caspase-1, the apo form of the enzyme crystallizes in the same conformation as the allosterically inhibited form. We believe that caspase-1, in solution, exists predominantly in an inactive conformation. The results presented here suggest that the active conformation, which is favored in the presence of an active-site ligand, is stabilized by the formation of the Arg286-Glu390 salt bridge. Disruption of this interaction could make it harder for the enzyme to visit the active conformation, leading to the decreased activity of the salt-bridge variants.
Overall, our studies point to a small set of side chains that form a contiguous circuit connecting the active and allosteric sites as being the most important for activity in caspase-1. Bioinformatic approaches analyzing coevolution of residues in large protein superfamilies show patterns of conservation and covariation suggestive of allosteric circuits. In these families, the majority of residues seem to evolve independently, while a small subset forms a linked network that is positioned for long-range communication through the structure.26 Mutational analysis at protein—protein interfaces reveals only a small subset of contact residues near the center of the binding interface drives the affinity of the interaction.8 Our alanine-scanning data on caspase-1 support a view that allostery is transmitted predominantly by a small subset of connected residues.
Materials and Methods
Caspase-1 expression and purification
Recombinant caspase-1 was prepared by expression in E. coli as insoluble inclusion body, followed by re-folding.5,27 The p20 (residues 120–297) and p10 (residues 317–404) subunits of wild-type human caspase-1 were cloned into NdeI and EcoRI restriction endonuclease sites of the pRSET plasmid (Invitrogen, Carlsbad, CA). Site-directed mutagenesis was performed using the Quik-Change Site-Directed Mutagenesis Kit from Stratagene (La Jolla, CA).
Caspase-1 subunits were expressed separately in E. coli BL21(DE3) Star cells (Invitrogen). Cells were harvested following induction of a log-phase culture with 1 mM IPTG for 4 h at 37 °C and then disrupted with a microfluidizer. The inclusion body pellets were isolated by centrifugation of lysate for 20 min at 4 °C. Pellets were washed twice with 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Hepes; pH 8.0), 300 mM NaCl, 1 M guanidine—HCl, 5mM DL-dithiothreitol (DTT), and 1% Triton X-100, and washed two more times with the same buffer without the detergent. The washed inclusion body pellets were solubilized in 6 M guanidine—HCl and 20 mM DTT, and stored frozen at −80 °C.
Refolding of caspase-1 was performed by combining guanidine—HCl-solubilized large and small subunits (10 mg of large subunit and 20 mg of small subunit) in a 250-mL beaker, followed by rapid dilution with 100 mL of 50 mM Hepes (pH 8.0), 100 mM NaCl, 10% sucrose, 1 M non-detergent sulfobetaine 201 (NDSB-201), and 10 mM DTT. Renaturation proceeded at room temperature for 6 h. Samples were centrifuged at 16,000g for 10 min to remove precipitates, and then dialyzed overnight at 4 °C against 50 mM sodium acetate (pH 5.9), 25 mM NaCl, 5% glycerol, and 4 mM DTT. Dialyzed protein was purified by cation-exchange chromatography using a prepacked 5-mL HiTrap SP HP column (GE Healthcare Biosciences Corp., Piscataway, NJ). Protein was eluted using a linear gradient of 0–1.0 M NaCl over 20 min in a buffer containing 50 mM sodium acetate (pH 5.9) and 5% glycerol. Peak fractions were pooled, and 2-mercaptoethanol was added to a concentration of 1 mM before samples were stored frozen at −80 °C.
Enzyme kinetic analysis
For kinetic analysis of caspase-1, protein was buffer-exchanged using a NAP-5 column (Amersham) into an assay buffer containing 50 mM Hepes (pH 8.0), 50 mM KCl, 200 mM NaCl, 10 mM DTT, and 0.1% 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate. Protein concentration was determined by active-site titration;28 caspase-1 was incubated in assay buffer with titration from 0 to 2-fold stoichiometric ratio of active-site inhibitor z-VAD-FMK for 2 h at room temperature. The protein was diluted to an enzyme concentration of 50 nM, and activity was determined using the fluorogenic substrate Ac-Trp-Glu-His-Asp-7-amino-4-trifluoromethylcoumarin at 25 μM.9,29,30
Steady-state kinetic analysis was performed by titrating the enzyme with the Ac-Trp-Glu-His-Asp-7-amino-4-trifluoromethylcoumarin substrate (typically from 200 μM to 0.25 μM, but up to 2 mM for inactive variants). The enzyme concentration was set to 45 nM for all variants, except those that showed large decreases in activity: 100 nM for the R286K, R286A, and E390A variants, and 500 nM for the R286A/E390A variant. Kinetic analysis performed at 15 nM gave the same nHill, suggesting that the cooperativity we observe is not dependent on the enzyme concentration (data not shown). Data were collected for 10 min using a Spectramax M5 microplate reader (Molecular Devices, Sunnyvale, CA), with excitation, emission, and cutoff filters set to 365 nm, 495 nm, and 435 nm, respectively.
Kinetic constants Vmax, Km, and Hill coefficient nHill were calculated using GraphPad PRISM. The initial velocity (v), measured in relative fluorescence units per unit time, was plotted versus the logarithm of substrate concentration. The model used to fit the data is a sigmoidal does—response curve with variable slope; from this model, all three kinetic constants were derived. The general equation of this model is: Y = Bottom + (Top—Bottom)/(1 + 10((logEC50-X)Hill slope)), where Y is the initial velocity, X is the logarithm of the substrate concentration, and Top, Bottom, EC50 (Km), and Hill slope are free parameters fitted to the data. A standard curve using pure AFC product was used to convert relative fluorescence units into units of concentration (μM). In determining kinetic constants for caspase-1, we observed that at saturating substrate concentrations, the enzyme exhibited decreasing activity as substrate concentration increased, most likely due to product inhibition. In order to correctly fit our data using nonlinear regression, data points exhibiting product inhibition were excluded.
Crystallization, data collection, and structure determination
Crystals of caspase-1 variants in complex with inhibitors were obtained by hanging-drop vapor diffusion at 4 °C against a reservoir of 0.1 M 1,4-piperazinediethanesulfonic acid (Pipes; pH 6.0), 200 mM Li2SO4, 25% polyethylene glycol 2000 monomethyl ether (PEG 2000 MME), 10 mM DTT, 3 mM NaN3, and 2 mM MgCl2 (Arg286Lys); 0.1 M Pipes (pH 6.0), 350 mM (NH4)2SO4, 20% PEG 2000 MME, 10 mM DTT, 3 mM NaN3, and 2 mM MgCl2 (Glu390Asp); and 0.1 M Pipes (pH 6.0), 175 mM (NH4)2SO4, 20% PEG 2000 MME, 10 mM DTT, 3 mM NaN3, and 2 mM MgCl2 (R286K/E390D). All crystals for data collection were cryoprotected in mother liquors supplemented with 20% (vol/vol) glycerol for 1–2 min and immersed in liquid nitrogen.
Diffraction data were collected under standard cryogenic conditions on beamlines 1–5 at the Stanford Synchrotron Research Laboratory using a mar345 image plate detector (the Arg286Lys/Glu390Asp variant) or a Rigaku RU-3R rotating anode generator and an RAXIS-IV detector (remaining variants), processed, and scaled with Crystal-Clear (Rigaku/Molecular Structure Corporation).31 The structures were determined from single-wavelength native diffraction experiments by molecular replacement with AMoRe32 using a search model from a previously determined structure (PDB ID 1SC1). The refinement of the initial solutions with REFMAC33–35 yielded experimental electron density maps suitable for model building with the O program.36 The following residues were not visible in the electron density maps for the indicated protein—inhibitor complexes and were omitted from the refinement of the final atomic models: residues 120–131, 146–148, and 317 (the R286K variant); residues 120–124 and 145–149 (the E390D variant); and residues 120–127, 146–148, and 317 (the R286K/E390D variant). PROCHECK37 revealed no disallowed (φ, ψ) combinations and excellent stereochemistry (see Table 4 for a summary of X-ray data and refinement statistics).
Accession numbers
The atomic coordinates and structure factors have been deposited in the PDB‡. The PDB ID codes of the structures presented in this work are 2H4W (E390D mutant), 2H4Y (R286K mutant), 2H51 (R286K/E390D mutant), and 2H54 (T388A mutant).
Supplementary Material
Acknowledgements
We wish to thank our colleagues in the laboratory for useful interactions, and Sunesis Pharmaceuticals for providing support for crystallography. This work was supported by the National Institutes of Health (grant ROI-AI070292 to J.A.W., grant F32AR052602 to J.M.S., and grant GMO7618 to D.D.).
Abbreviations used
- ATCase
aspartate transcarbamoylase
- H-bonding
hydrogen bonding
- z-VAD-FMK
z-Val-Ala-Asp-fluoromethylketone
- PDB
Protein Data Bank
- Hepes
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- Pipes
1,4-piperazinediethanesulfonic acid
- PEG 2000 MME
polyethylene glycol 2000 monomethyl ether
Footnotes
Mutant nomenclature lists the wild-type amino acid and its position, followed by the mutant residue in a single-letter code.
References
- 1.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 2.Fuentes-Prior P, Salvesen GS. The protein structures that shape caspase activity, specificity, activation and inhibition. Biochem. J. 2004;384:201–232. doi: 10.1042/BJ20041142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shi Y. Caspase activation: revisiting the induced proximity model. Cell. 2004;117:855–858. doi: 10.1016/j.cell.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Scheer JM, Romanowski MJ, Wells JA. A common allosteric site and mechanism in caspases. Proc. Natl Acad. Sci. USA. 2006;103:7595–7600. doi: 10.1073/pnas.0602571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romanowski MJ, Scheer JM, O’Brien T, McDowell RS. Crystal structures of a ligand-free and malonate-bound human caspase-1: implications for the mechanism of substrate binding. Structure. 2004;12:1361–1371. doi: 10.1016/j.str.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 6.Hardy JA, Lam J, Nguyen JT, O’Brien T, Wells JA. Discovery of an allosteric site in the caspases. Proc. Natl Acad. Sci. USA. 2004;101:12461–12466. doi: 10.1073/pnas.0404781101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clackson T, Wells JA. A hot spot of binding energy in a hormone—receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 8.Moreira IS, Fernandes PA, Ramos MJ. Hot spots—a review of the protein—protein interface determinant amino-acid residues. Proteins. 2007;68:803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- 9.Rano TA, Timkey T, Peterson EP, Rotonda J, Nicholson DW, Becker JW, et al. A combinatorial approach for determining protease specifi-cities: application to interleukin-1beta converting enzyme (ICE) Chem. Biol. 1997;4:149–155. doi: 10.1016/s1074-5521(97)90258-1. [DOI] [PubMed] [Google Scholar]
- 10.Gutfreund H, Sturtevant JM. The mechanism of chymotrypsin-catalyzed reactions. Proc. Natl Acad. Sci. USA. 1956;42:719–728. doi: 10.1073/pnas.42.10.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wells JA, Cunningham BC, Graycar TP, Estell DA. Importance of hydrogen-bond formation in stabilizing the transition-state of subtilisin. Philos. Trans. R. Soc. London Ser. A. 1986;317:415–423. [Google Scholar]
- 12.Madsen NB, Shechosk S. Allosteric properties of phosphorylase B: 2. Comparison with a kinetic model. J. Biol. Chem. 1967;242:3301. [PubMed] [Google Scholar]
- 13.Buchbinder JL, Guinovart JJ, Fletterick RJ. Mutations in paired alpha-helices at the subunit interface of glycogen—phosphorylase alter homo-tropic and heterotropic cooperativity. Biochemistry. 1995;34:6423–6432. doi: 10.1021/bi00019a023. [DOI] [PubMed] [Google Scholar]
- 14.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure—factor data and their agreement with the atomic model. Acta Crystallogr. Sect. D. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 15.Laskowski RA, MacArthur MW, Moss DS, Thronton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. D. 1993;26:283–291. [Google Scholar]
- 16.Shurki A, Warshel A. Why does the Ras switch “break” by oncogenic mutations? Proteins. 2004;55:1–10. doi: 10.1002/prot.20004. [DOI] [PubMed] [Google Scholar]
- 17.Demarest SJ, Martinez-Yamout M, Chung J, Chen H, Xu W, Dyson HJ, et al. Mutual synergistic folding in recruitment of CBP/p300 by p160 nuclear receptor coactivators. Nature. 2002;415:549–553. doi: 10.1038/415549a. [DOI] [PubMed] [Google Scholar]
- 18.Demarest SJ, Deechongkit S, Dyson HJ, Evans RM, Wright PE. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci. 2004;13:203–210. doi: 10.1110/ps.03366504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hwang JK, Warshel A. Why ion pair reversal by protein engineering is unlikely to succeed. Nature. 1988;334:270–272. doi: 10.1038/334270a0. [DOI] [PubMed] [Google Scholar]
- 20.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 21.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 22.Eisenmesser EZ, Millet O, Labeikovsky W, Korzhnev DM, Wolf-Watz M, Bosco DA, et al. Intrinsic dynamics of an enzyme underlies catalysis. Nature. 2005;438:17–121. doi: 10.1038/nature04105. [DOI] [PubMed] [Google Scholar]
- 23.Newton CJ, Kantrowitz ER. The regulatory subunit of Escherichia coli aspartate carbamoyltransferase may influence homotropic cooperativity and heterotropic interactions by a direct interaction with the loop containing residues 230–245 of the catalytic chain. Proc. Natl Acad. Sci. USA. 1990;87:2309–2313. doi: 10.1073/pnas.87.6.2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenstein E, Markby DW, Schachman HK. Heterotropic effectors promote a global conformational change in aspartate transcarbamoylase. Biochemistry. 1990;29:3724–3731. doi: 10.1021/bi00467a019. [DOI] [PubMed] [Google Scholar]
- 25.Fetler L, Kantrowitz ER, Vachette P. Direct observation in solution of a preexisting structural equilibrium for a mutant of the allosteric aspartate transcarbamoylase. Proc. Natl Acad. Sci. USA. 2007;104:495–500. doi: 10.1073/pnas.0607641104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat. Struct. Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 27.Scheer JM, Wells JA, Romanowski MJ. Malonate-assisted purification of human caspases. Protein Expression Purif. 2005;41:148–153. doi: 10.1016/j.pep.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Stennicke HR, Salvesen GS. Caspases: preparation and characterization. Methods. 1999;17:13–319. doi: 10.1006/meth.1999.0745. [DOI] [PubMed] [Google Scholar]
- 29.Talanian RV, Quinlan C, Trautz S, Hackett MC, Mankovich JA, Banach D, et al. Substrate specificities of caspase family proteases. J. Biol. Chem. 1997;272:9677–9682. doi: 10.1074/jbc.272.15.9677. [DOI] [PubMed] [Google Scholar]
- 30.Thornberry NA, Rano TA, Peterson EP, Rasper DM, Timkey T, Garcia-Calvo M, et al. A combinatorial approach defines specificities of members of the caspase family and granzyme B. Functional relationships established for key mediators of apoptosis. J. Biol. Chem. 1997;272:17907–17911. doi: 10.1074/jbc.272.29.17907. [DOI] [PubMed] [Google Scholar]
- 31.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr. Sect. D. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 32.Navaza J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. Sect. D. 2001;57:1367–1372. doi: 10.1107/s0907444901012422. [DOI] [PubMed] [Google Scholar]
- 33.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. Sect. D. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 35.Pannu NS, Murshudov GN, Dodson EJ, Read RJ. Incorporation of prior phase information strengthens maximum-likelihood structure refinement. Acta Crystallogr. Sect. D. 1998;54:1285–1294. doi: 10.1107/s0907444998004119. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. Sect. A. 1991;47:10–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, MacArthur MW, Moss DS, Thronton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. D. 1993;26:283–291. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.