Abstract
A defect in the Werner syndrome protein (WRN) leads to the premature aging disease Werner syndrome (WS). Hallmark features of cells derived from WS patients include genomic instability and hypersensitivity to certain DNA-damaging agents. WRN contains a highly conserved region, the RecQ conserved domain, that plays a central role in protein interactions. We searched for proteins that bound to this region, and the most prominent direct interaction was with poly(ADP-ribose) polymerase 1 (PARP-1), a nuclear enzyme that protects the genome by responding to DNA damage and facilitating DNA repair. In pursuit of a functional interaction between WRN and PARP-1, we found that WS cells are deficient in the poly(ADP-ribosyl)ation pathway after they are treated with the DNA-damaging agents H2O2 and methyl methanesulfonate. After cellular stress, PARP-1 itself becomes activated, but the poly(ADP-ribosyl)ation of other cellular proteins is severely impaired in WS cells. Overexpression of the PARP-1 binding domain of WRN strongly inhibits the poly(ADP-ribosyl)ation activity in H2O2-treated control cell lines. These results indicate that the WRN/PARP-1 complex plays a key role in the cellular response to oxidative stress and alkylating agents, suggesting a role for these proteins in the base excision DNA repair pathway.
Werner syndrome (WS) is a rare autosomal recessive disorder characterized by premature aging and the early onset of cancer (26). WS is caused by mutations in the gene (WRN) encoding the Werner syndrome protein (WRN) (51). WRN is a multifunctional protein which possesses three catalytic activities, which reside in the amino terminus (3′-5′ exonuclease) and the central part of the protein (DNA-dependent ATPase and 3′-5′ helicase) (5). The RecQ conserved (RQC) motif resides in the C-terminal part of WRN and is present in almost all of the RecQ family members (51). Recently, we have shown that a region of 144 amino acids (aa) of the RQC domain binds to and stimulates flap endonuclease 1 (FEN-1) (7), binds to the Bloom's syndrome protein (49) and telomere repeat binding factor 2 (30), and contains a nuclear localization signal-dependent nucleolar targeting sequence (48). Thus, the highly conserved RQC domain of WRN appears to play a very important role in mediating WRN protein interactions and in regulating the nuclear trafficking (nucleolar targeting) of the protein. In an effort to better understand the functional role(s) of this domain, we performed a series of pull-down experiments to identify proteins that specifically bind to the WRN RQC domain. The most prominent binder that we identified was poly(ADP-ribose) polymerase 1 (PARP-1), whose binding represents a novel protein interaction with WRN.
PARP-1 is a nuclear enzyme belonging to the DNA damage surveillance network. The protein responds to DNA damage by transferring 50 to 200 molecules of ADP-ribose to various nuclear proteins, including transcription factors, histones, and PARP-1 itself (8). This poly(ADP-ribosyl)ation activity of PARP-1 appears to be important for maintaining genomic integrity (47), and it has been associated with longevity (8, 16, 52). Furthermore, PARP-1 is activated by several DNA-damaging agents, such as hydrogen peroxide (H2O2), alkylating agents (including methyl methanesulfonate [MMS]), bleomycin, and radiation.
Our finding in the present study that PARP-1 is the most prominent protein associated with the WRN RQC domain prompted further analysis of this protein interaction and an investigation of the biological significance of this interaction. We report here that WS cells are deficient in the poly(ADP-ribosyl)ation pathway after H2O2 and MMS treatment. After DNA damage, PARP-1 becomes auto-poly(ADP-ribosyl)ated in WS cells, but the poly(ADP-ribosyl)ation of other cellular proteins in WS cells is undetectable by immunofluorescence and Western blot analysis. The WRN/PARP-1 interaction appears to depend on the poly(ADP-ribosyl)ation state of PARP-1. In further support of a biological function of the WRN/PARP-1 interaction, we show that overexpression of the PARP-1 binding domain of WRN strongly inhibits the poly(ADP-ribosyl)ation activity in H2O2-treated control cell lines. Altogether, these results indicate that the WRN/PARP-1 complex plays a key role in the cellular response to oxidative stress and DNA alkylation damage. The DNA damage caused by these reagents is generally repaired by the base excision repair (BER) machinery. A role for the WRN/PARP-1 complex in BER is discussed.
MATERIALS AND METHODS
Cell lines.
TERT-1604 (telomerase-immortalized normal fibroblasts) and TERT-3141 (telomerase-immortalized WS fibroblasts) were generously provided by Jerry W. Shay. These cell lines were grown in Dulbecco's modified Eagle's medium (GIBCO BRL, Life Technologies) supplemented with penicillin-streptomycin (GIBCO BRL) and 10% fetal bovine serum (FBS). The control cell lines (WI38, MRC-5, HFL-1, and HeLa) and WS cell lines (AG03141C, AG00780H, AG05229C, AG05233C, AG12795, and AG11395) were from Coriell Cell Repositories. Cells were cultured (37°C, 5% CO2) in minimum essential medium with 15% FBS, essential and nonessential amino acids, vitamins, 2 mM l-glutamine, and penicillin-streptomycin.
Recombinant proteins.
PARP-1 (generously provided by Gilbert de Murcia) and His-tagged WRN were purified from insect cells as described previously (15, 31).
Cloning and expression of pDsRed1-C1-PARP-1.
The PARP-1 sequence from the pARP vector (generously provided by Gilbert de Murcia) was subcloned into SalI sites of the pDsRed1-C1 vector (Clontech). The correct orientation was verified by HindIII digestion.
GST pull-down assays from HeLa NE.
The pull-down experiments using HeLa nuclear extracts (NE) were essentially as previously described (7). Glutathione S-transferase (GST)-WRN RQC (aa 949 to 1092) and GST-WRN (aa 1072 to 1236) proteins were expressed in Escherichia coli and purified as previously described (7). In these experiments, approximately 750 μg (per sample) of HeLa NE was incubated for 2 h at 4°C with glutathione-Sepharose beads (Amersham Biosciences) saturated with GST-WRN fragments. After extensive washing, a fraction of the bound proteins was analyzed either by Coomassie staining or Western blotting with an anti-PARP-1 monoclonal antibody (1:2,000; clone 2-C-10; Sigma).
MALDI-MS.
Matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS) was performed by the HHMI/Keck facility at Yale University (http://info.med.yale.edu/wmkeck).
Coimmunoprecipitation assay.
Approximately 1 mg of HeLa NE was precleared with protein A-Sepharose (Amersham Biosciences) for 1 h at 4°C and then incubated with either rabbit anti-PARP-1 antibodies (1:50; Alexis Corp.) or control rabbit antibodies for 16 h at 4°C. The subsequent steps were as previously described (49). For the Western blot analysis, the membranes were probed with anti-WRN (1:250; Transduction Laboratories) or anti-PARP-1 (1:2,000; clone 2-C-10; Sigma) antibodies for 16 h at 4°C.
Transfection and colocalization of enhanced green fluorescent protein (EGFP)-WRN and red fluorescent protein (RFP)-PARP-1 in living HeLa cells.
HeLa cells were cotransfected with pEGFPC3-WRN (49) and pDsRed1C1-PARP-1 by using the Calphos mammalian transfection kit (Clontech) as described by the manufacturer. Sixteen hours posttransfection, the cells were analyzed as previously described (48).
Characterization of the direct WRN/PARP-1 interaction by ELISA.
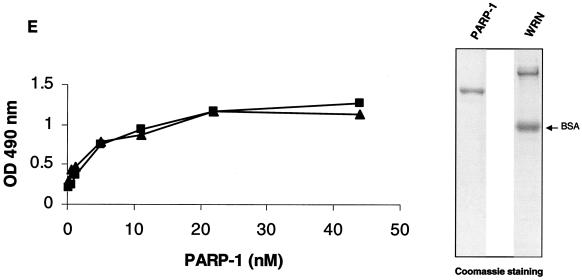
The enzyme-linked immunosorbent assay (ELISA) was performed basically as previously described (49). For the coating step, 80 ng of WRN (10 nM/well)/well diluted in carbonate buffer was added to the corresponding wells for 16 h at 4°C. For the binding step, serial dilutions of PARP-1 (ranging between 0.3 and 44 nM/well) were added to the corresponding wells for 90 min at 37°C. The binding reaction was done either in the presence or absence of ethidium bromide (30 μg/ml) as described in the Fig. 1 legend. Bound PARP-1 was detected with rabbit anti-PARP-1 antibodies.
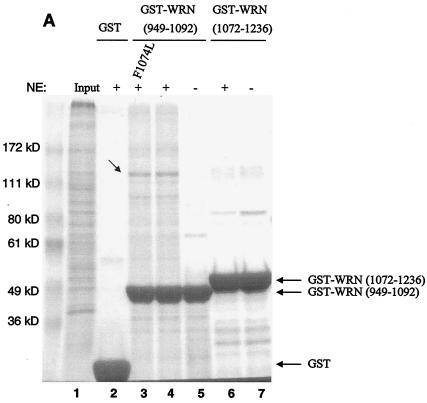
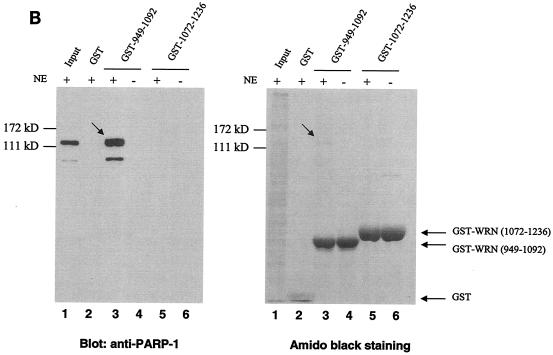
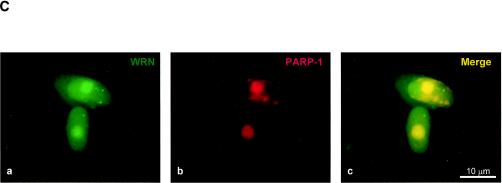
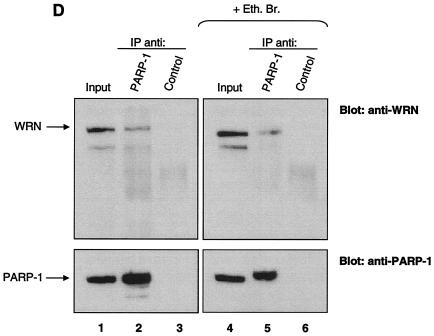
FIG. 1.
Identification of PARP-1 as a WRN RQC domain-interacting protein. (A) HeLa NE (lane 1) was incubated either with recombinant GST alone (lane 2), the GST-WRN RQC domain (aa 949 to 1092; lanes 3 to 5), or GST-WRN (aa 1072 to 1236; lanes 6 and 7). Input corresponds to 5% of the total NE used in the binding reaction. Bound proteins were eluted and analyzed by Coomassie staining. Arrow, ∼113-kDa protein that bound specifically and tightly to the GST-WRN RQC domain and that was identified as PARP-1 by mass spectrometry. F1074L corresponds to a GST-WRN RQC domain containing a point mutation that is present in some WS patients (23). (B) Bound proteins (in panel A) were also analyzed by Western blotting with anti-PARP-1 antibodies (left). Amido black staining indicates the relative amount of GST-WRN recombinant proteins loaded (right). Arrows, PARP-1. (C) WRN and PARP-1 reside in the nucleoli of living HeLa cells. WRN and PARP-1 were coexpressed in HeLa cells as EGFP (green; a) and RFP (red; b) fusion proteins, respectively. After transfection (∼20 h) the living cells were analyzed by confocal microscopy as described in Materials and Methods. (D) WRN and PARP-1 were coimmunoprecipitated (IP) from HeLa NE either in the absence (lanes 1 to 3) or the presence (lanes 4 to 6) of ethidium bromide (Eth. Br.; 30 μg/ml) with anti-PARP-1 antibodies (lanes 2 and 5) but not with the control antibodies (lanes 3 and 6), as indicated by Western blotting with anti-WRN antibodies (top) and anti-PARP-1 antibodies (bottom). Inputs are 10% (lanes 1 and 4), and the loaded fractions in lanes 2, 3, 5, and 6 correspond to 50%. (E) Recombinant WRN and PARP-1 proteins interact directly as shown by ELISA. The wells werecoated with purified recombinant WRN (10 nM/well). After blocking with BSA, the wells were incubated with serial dilutions of recombinant PARP-1 (0.3, 0.6, 1.2, 5.5, 11, 22, and 44 nM/well) either alone (triangles) or with 30 μg of ethidium bromide/ml (squares). Bound PARP-1 was detected with rabbit anti-PARP-1 antibodies, followed by colorimetric analysis. All values were corrected for the background signal (PARP-1 binding to BSA). The purity of the recombinant proteins used in this work is shown by SDS-PAGE and Coomassie staining (right).
Detection of PAR polymer formation in cells.
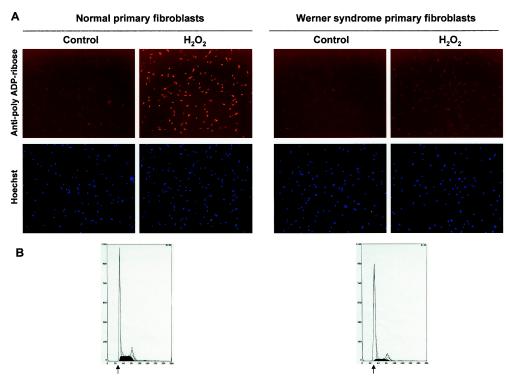
Exponentially growing cells (normal and WS cells; see Table 1) were either untreated or arrested in early S phase with 5 mM hydroxyurea (HU) for 18 h at 37°C and 5% CO2. The medium containing HU was removed, and the cells were incubated with either serum-free medium alone, H2O2 (500 μM), bleomycin (35 μM or 50 μg/ml), or MMS (1 mM) for 10 min at 37°C and 5% CO2. For the Western blot analysis to detect poly(ADP-ribose) (PAR) polymer formation, the cells were washed once with ice-chilled 1× phosphate-buffered saline (PBS), scraped, spun down, and resuspended in sodium dodecyl sulfate (SDS)-protein sample buffer. The lysates were analyzed by immunoblotting with anti-PAR antibodies (1: 2,500; Alexis Corp.). The Western blot analysis was performed with two different WS primary cell lines (AG03141C and AG00780H) and two normal primary cell lines (MRC-5 and WI-38). For the immunofluorescence assays, the cells (after H2O2 treatment) were washed as described above and fixed with 3% paraformaldehyde in 1× PBS for 15 min at 4°C. Following the permeabilization step (0.4% Triton X-100 in 1× PBS, 15 min at room temperature [RT]), the coverslips were incubated with blocking buffer (1% nonfat milk-0.1% Tween-20 in 1× PBS) for 1 h at RT. The cells were then incubated with anti-PAR antibodies (1:500) for 16 h at 4°C. After washing and incubation with the corresponding Texas Red-conjugated secondary antibodies (1:200; Vector Laboratories) and Hoechst 33342 (1:5,000; Molecular Probes), the coverslips were mounted on Vectashield (Vector Laboratories). The cells were then visualized with an Axiovert 200 M microscope (Zeiss; 10× lens) by using separate channels for the analysis of red fluorescence and blue fluorescence. The images were analyzed with the AxioVision, version 3.0, program (Zeiss). The immunofluorescence assays were performed with the cells described in Table 1.
TABLE 1.
Cell lines used in this study
| Cell line | PAR polymer formationa (positive nuclei/total nuclei [%]) | PDLb (range) |
|---|---|---|
| WS | ||
| Primary | ||
| AG03141C | 23/683 (3.3) | 10-13 |
| AG00780H | 25/591 (4.2) | 10-15 |
| AG05229C | 20/361 (5.5) | 9-12 |
| AG05233C | 14/182 (7.6) | 10-15 |
| AG12795 | 15/500 (3) | 13-16 |
| Transformed | ||
| AG11395 | 46/557 (8.2) | |
| TERT-3141 | 56/256 (21.8) | |
| Control | ||
| Primary | ||
| WI38 | 586/609 (96.2) | 20-35 |
| MRC-5 | 1,666/1,824 (91.3) | 20-35 |
| HFL-1 | 200/375 (53.3) | 8-11 |
| Transformed | ||
| HeLa | 742/762 (97.3) | |
| TERT-1604 | 792/800 (99) |
The assay of PAR polymer formation after oxidative stress in the cell lines was carried out as described for Fig. 2 (HU treatment of cells and subsequent immunofluorescence analysis using anti-PAR antibodies).
PDL, population doubling.
PARP-1 poly(ADP-ribosyl)ation assay using purified proteins.
Recombinant WRN and PARP-1 proteins (0.5 μM final concentration) were preincubated in 1× ribosylation buffer (10 mM Tris-HCl [pH 8], 1 mM MgCl2, 1 mM dithiothreitol) for 15 min at RT. The poly(ADP-ribosyl)ation reaction was initiated by adding NAD+ (1 mM) and activated DNA (0.1 mg/ml) either in the absence or presence of 3-aminobenzamide (3-AB; 10 mM). The reactions were terminated after 5 or 10 min by adding 2× SDS-protein sample buffer and heating. The samples were analyzed by Western blotting with anti-PAR antibodies, an anti-PARP-1 antibody, and anti-WRN antibodies consecutively.
Combined ribosylation and ELISA method to detect poly(ADP-ribosyl)ation of H1 histones.
H1 histones (200 μg/ml in carbonate buffer) were used to coat ELISA wells for 2 h at 37°C. After the blocking step, either 500, 50, or 5 nM recombinant PARP-1 was added in the absence or presence of WRN (80 nM). The poly(ADP-ribosyl)ation reaction was initiated by adding NAD+ (100 μM final concentration) and activated DNA (100 μg) for 30 min at RT. The specific PARP inhibitor 3-AB was added (12 nM final concentration) to the ribosylation reaction as a negative control. The PAR polymer was detected with anti-PAR antibodies and horseradish peroxidase-conjugated secondary antibodies. The bound antibodies were detected by colorimetric analysis as described for the ELISA method.
PARP-1 activity blotting.
The auto-poly(ADP-ribosyl)ation reaction of PARP-1 on a nitrocellulose membrane was performed exactly as indicated on the website http://parplink.u-strasbg.fr. Following the reaction, the membrane was processed for Western blotting with anti-lamin B antibodies (1:500; Santa Cruz Biotechnology).
PARP-1 poly(ADP-ribosyl)ation assay and binding with GST-WRN fragments.
The PARP-1 auto-poly(ADP-ribosyl)ation assay was performed as indicated on the website http://parplink.u-strasbg.fr. Briefly, 10 μg of recombinant PARP-1 was incubated either with 1× ribosylation buffer (10 mM Tris-HCl [pH 8], 1 mM MgCl2, 1 mM dithiothreitol) alone or with 1× ribosylation buffer plus NAD+ (1 mM) and activated DNA (0.1 mg/ml; final volume per sample, 100 μl) for 30 min at RT. Next, 100 μl (per sample) of binding buffer (3% bovine serum albumin [BSA]-0.1% Tween 20 in 1× PBS) was added. The samples were then incubated with glutathione-Sepharose beads saturated either with GST-WRN RQC (aa 949 to 1092) or GST-WRN (aa 1072 to 1236) for 2 h at RT. After extensive washing, the bound proteins were analyzed by Western blotting with anti-PARP-1 and anti-PAR monoclonal antibodies (Sigma and Alexis Corp., respectively) as the primary antibodies. Subsequent steps were as previously described (49).
Inhibition of PAR polymer formation by overexpressing EGFP-WRN RQC (aa 949 to 1092).
The cloning and expression of EGFP-WRN RQC (aa 949 to 1092) were as previously described (48). TERT-1604 cells were transfected as described above (colocalization analysis) with either pEGFP-WRN RQC (aa 949 to 1092) or pEGFP alone. Thirty-six hours posttransfection, the cells were arrested in early S phase and treated with H2O2 as described previously for PAR polymer formation.
RESULTS
To search for proteins that interact with the important RQC domain of WRN, we first purified a GST fragment containing the WRN RQC domain (aa 949 to 1092). We then performed pull-down experiments using HeLa NE and this GST-RQC fragment of WRN. Using mass spectrometry, we identified PARP-1, a 113-kDa protein, as the most prominent binder to the GST-WRN RQC domain (aa 949 to 1092) fusion protein (Fig. 1A). This suggests that the WRN/PARP-1 interaction is likely to be of significant biological importance. We confirmed this protein interaction by Western blotting using anti-PARP-1 antibodies (Fig. 1B, lane 3). GST-WRN (aa 1072 to 1236) did not bind PARP-1 (Fig. 1B, lane 5), demonstrating the specificity of the binding to the WRN RQC domain. Based on the amount of loaded sample, the GST-WRN RQC precipitated ∼30% of nuclear PARP-1. To investigate whether this interaction also occurs in vivo, we next performed colocalization and coimmunoprecipitation studies. As shown in Fig. 1C, EGFP-WRN and RFP-PARP-1 colocalized in the nucleoli of living HeLa cells. This result is consistent with previous reports that each of these proteins localizes to the nucleoli (1, 11, 23, 48). In addition, WRN and PARP-1 specifically coimmunoprecipitated from HeLa NE with anti-PARP-1 antibodies (Fig. 1D, lane 2). The coimmunoprecipitation was not affected by the presence of the DNA intercalator ethidium bromide (Fig. 1D, lanes 5 and 6), indicating that WRN/PARP-1 complex formation was not mediated by DNA. Based on the amount of loaded extract (Fig. 1D legend), the anti-PARP-1 antibodies coimmunoprecipitated ∼40% of endogenous PARP-1 and ∼10% of WRN. Thus, ∼25% of PARP-1 is in a protein complex with WRN in HeLa NE. This result is consistent with the pull-down experiment using GST-WRN RQC (see above). WRN and PARP-1 proteins also interact directly in vitro, as demonstrated by ELISA using purified recombinant proteins (Fig. 1E). This interaction was not disrupted by the presence of ethidium bromide, demonstrating a direct protein-protein interaction, and ruling out the possibility that DNA, which could potentially be present in the purified recombinant proteins, mediated the binding (Fig. 1E). Thus, WRN and PARP-1 reside in the nucleoli and interact directly through the WRN RQC domain.
We next explored whether the physical interaction between PARP-1 and WRN was associated with a functional consequence. A major activity of PARP-1 involves the poly(ADP-ribosyl)ation of a large number of proteins after cellular stress. H2O2 has been used at 0.5 to 1 mM for 10 to 15 min as an oxidative agent to study the activation of cellular PARP-1 (27). To examine whether the lack of WRN alters this cellular activity of PARP-1, we exposed WS and normal primary fibroblasts to H2O2. Evidence indicates that WRN and other RecQ helicases play important roles at sites of replication forks (29). Thus, we induced replication fork arrest (in early S phase) by treatment of the cells with HU prior to the exposure to oxidative stress (Fig. 2B). After H2O2 treatment (500 μM, 10 min), the cells were fixed and analyzed by immunofluorescence for PAR polymer formation with specific anti-PAR antibodies. As shown in Fig. 2A, normal cells demonstrated strong PAR synthesis after H2O2 treatment. In contrast, WS cells were severely deficient in PAR synthesis. Under these experimental conditions (500 μM H2O2, 10 min), more than 95% of the cells remained viable (data not shown), ruling out the possibility that we were detecting a phenomenon caused by toxicity. We obtained the same results with a lower dose of H2O2 (50 μM, 10 min) (data not shown). We confirmed and extended these results using five control cell lines and seven WS cell lines (Table 1). An average of 4.7% of the primary WS cells were PAR polymer formation positive, whereas the primary control cells averaged 80% positive. The transformed WS cell lines displayed somewhat more PAR polymer formation than the primary WS cells, but still much less than the transformed control cell lines. Thus, all WS cell lines tested showed a dramatic defect in PAR polymer formation after oxidative stress.
FIG. 2.
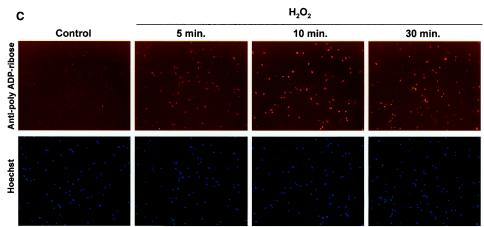
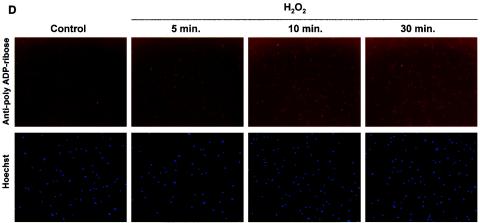
WS cells are deficient in the poly(ADP-ribosyl)ation of cellular proteins after H2O2 treatment. (A) Immunofluorescence. Normal and WS primary fibroblasts (arrested in early S phase [B]) were incubated with H2O2 (500 μM) for 10 min at 37°C. Fixed cells were probed with anti-PAR antibodies. Control panels correspond to untreated cells. Nucleus staining by Hoechst 33342 is shown. (B) Synchronization in early S phase (see Materials and Methods) prior to H2O2 treatment is confirmed by propidium iodide staining and flow cytometry analysis. Arrows, G1-S phase boundary. (C and D) Time course of PAR polymer formation. After H2O2 treatment, normal (C) and WS (D) primary cells were fixed at the indicated times and processed as for panel A. Similar results were obtained with two different WS primary cell lines (AG03141C and AG00780H) and two normal primary cell lines (MRC-5 and WI-38).
To investigate whether the PAR synthesis deficiency of WS cells after cellular stress was due to a difference in the kinetics of the PAR polymer formation, we performed time course experiments. The primary fibroblasts were fixed and analyzed for PAR formation at 5, 10, and 30 min after H2O2 treatment (Fig. 2C and D). Normal cells presented strong PAR polymer formation at all the tested times (with the strongest signal at 10 min; Fig. 2C), whereas WS cells were similar to untreated controls at all time points (Fig. 2D). Following longer incubation times (4 h after damage) no detection of signal corresponding to PAR synthesis was observed in WS cells (data not shown). Thus, the lack of PAR polymer formation in WS cells after damage was not due to a difference in kinetics.
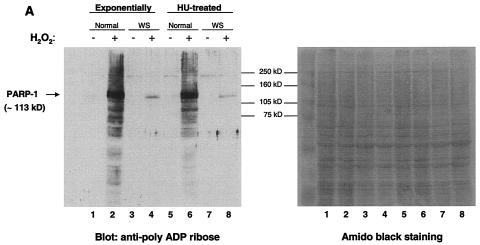
To extend and confirm the results obtained by immunofluorescence, we investigated the pattern of poly(ADP-ribosyl)ated cellular proteins after damage. Cell lysates from normal and WS primary cells (untreated and treated with H2O2) were analyzed by Western blotting using anti-PAR antibodies (Fig. 3A). The use of the Western blotting technique allows for the visualization of the various poly(ADP-ribosyl)ated cellular proteins. After damage, normal cells displayed strong anti-PAR reactivity by numerous proteins and especially by an ∼113-kDa protein (the same molecular mass as PARP-1) (Fig. 3A, lanes 2 and 6). Interestingly, the WS cell lysates showed only one damage-induced ribosylated protein (Fig. 3A, lanes 4 and 8), which was ∼113 kDa and which most likely represents auto-poly(ADP-ribosyl)ated PARP-1. We obtained the same results using cell lysates from either exponentially growing cells (Fig. 3A, lanes 1 to 4) or HU-treated cells (Fig. 3A, lanes 5 to 8), indicating that the lack of poly(ADP-ribosyl)ation of cellular proteins in WS cells is not limited to S phase. Thus, the Western blotting results support and extend our immunofluorescence findings. In sharp contrast to normal cells, WS cells are almost completely devoid of poly(ADP-ribosyl)ated cellular proteins following treatment with H2O2. Indeed, in WS cells, no protein other than PARP-1 appears to be poly(ADP-ribosyl)ated under these conditions. The Western blotting results were confirmed by using two different WS primary cell lines (AG03141C and AG00780H) and two normal primary cell lines (MRC-5 and WI-38).
FIG. 3.
(A) Western blot analysis of the pattern of poly(ADP-ribosyl)ated proteins after H2O2 treatment. Exponentially growing normal and WS primary cells (lanes 1 to 4) and HU-treated cells (lanes 5 to 8) were either untreated (lanes 1, 3, 5, and 7) or treated (lanes 2, 4, 6, and 8) with H2O2 (500 μM) for 10 min at 37°C. Cellular proteins were separated by SDS-PAGE, and the poly(ADP-ribosyl)ation of cellular proteins was detected by immunoblotting with anti-PAR antibodies. The amount of protein loaded is indicated by amido black staining (right). (B) WS cells express an active PARP-1 protein. Equivalent amounts of cell extracts from the same number of normal (lanes 1, 2, 5, and 6) and WS (lanes 3, 4, 7, and 8) cells, treated as for panel A, were analyzed either for PARP-1 expression levels (blot anti-PARP-1, lanes 1 to 4) or for PARP-1 activity (auto-poly[ADP-ribosyl]ation reaction; lanes 5 to 8). The Western blots with anti-lamin B indicate the amount of protein loaded. (C) WS cells are deficient in the poly(ADP-ribosyl)ation pathway after H2O2 and MMS treatment. Normal (lanes 1, 2, 5, 6, 9, and 10) and WS (lanes 3, 4, 7, 8, 11, and 12) cells were either untreated (lanes 1, 3, 5, 7, 9, and 11) or treated with bleomycin (35 μM; lanes 2 and 4), H2O2 (500 μM; lanes 6 and 8), or MMS (1 mM; lanes 10 and 12) for 10 min at 37°C. Proteins from the same number of cells were separated by SDS-PAGE, and the poly(ADP-ribosyl)ation of cellular proteins was detected as for panel A. Samples from the same experiment were analyzed with anti-lamin B antibodies as a loading control. Similar results were obtained with two different WS primary cell lines (AG03141C and AG00780H) and two normal primary cell lines (MRC-5 and WI-38).
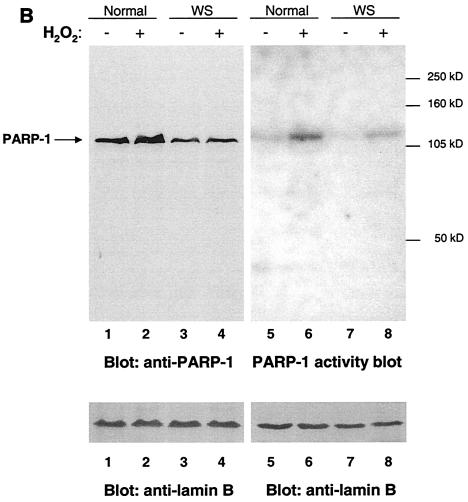
To determine whether the poly(ADP-ribosyl)ation defect in WS cells was due to a lack of PARP-1 protein or expression of an inactive PARP-1, we tested normal (MRC-5 and WI-38) and WS (AG03141C and AG00780H) cell lysates for the presence of PARP-1 (Western blotting) and for PARP-1 activity (PARP-1 activity blotting) (Fig. 3B). The result of the Western blotting confirms that PARP-1 is expressed in WS cells. The expression level of PARP-1 in WS cells was consistently about one-half of that in normal cell lines (Fig. 3B, compare lanes 1 and 2 with 3 and 4) over several experiments. In the PARP-1 activity blotting, we detected the expected auto-poly(ADP-ribosyl)ation activity of the PARP-1 protein. When PARP-1 was activated (i.e., after DNA damage), this automodification activity increased. This increase is visualized in Fig. 3B (compare lane 5 with 6 and 7 with 8). Importantly, the PARP-1 activity blot showed only one ∼113-kDa H2O2-induced auto-poly(ADP-ribosyl)ated protein (PARP-1) in both normal and WS cell lysates. The differences in the automodification intensities (Fig. 3B, compare lanes 6 and 8) appear to correlate with the expression levels of PARP-1 (Fig. 3B, compare lanes 2 and 4). We obtained the same results using cell lysates from both S-phase-arrested cells (Fig. 3B) and exponentially growing cells (data not shown). Thus, WS cells express an active PARP-1 protein. It may be slightly less active than it is in normal cells, but the lack of ribosylation in WS cells cannot be explained solely on the basis of the reduced PARP-1 activity. Altogether these results suggest that, in WS cells, PARP-1 detects the DNA damage produced by oxidative stress, but the modification of other cellular proteins is impaired.
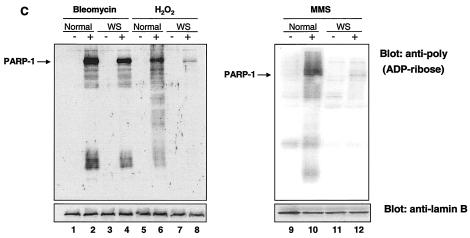
To further investigate the nature of the PARP-1 activation pathway in WS cells, we exposed the cells to other DNA-damaging agents, such as bleomycin and MMS. Bleomycin is a known gamma-irradiation-mimicking agent and DNA break inducer, and we used experimental conditions for PAR polymer formation reported previously (40) (see Materials and Methods). MMS is an alkylating agent which produces DNA lesions mainly repaired by BER (13). The pattern of poly(ADP-ribosyl)ated proteins in the cells exposed to these DNA-damaging agents was analyzed by Western blotting with anti-PAR antibodies. As shown in Fig. 3C, WS cells were proficient in the poly(ADP-ribosyl)ation pathway after bleomycin treatment (Fig. 3C, compare lanes 2 and 4), whereas they were deficient after MMS treatment (Fig. 3C, compare lanes 10 and 12). As demonstrated previously, WS cells were deficient in the poly(ADP-ribosyl)ation of cellular proteins after H2O2 treatment (Fig. 3C, compare lanes 6 and 8). In conclusion, we observed that the PARP-1 response in WS cells after treatment with bleomycin appears to be normal, while it is severely impaired after treatment with H2O2 and MMS. Interestingly, treatment of normal cells with different DNA-damaging agents resulted in different patterns of poly(ADP-ribosyl)ated cellular proteins (Fig. 3C, compare lanes 2, 6, and 10), suggesting the activation of different repair pathways.
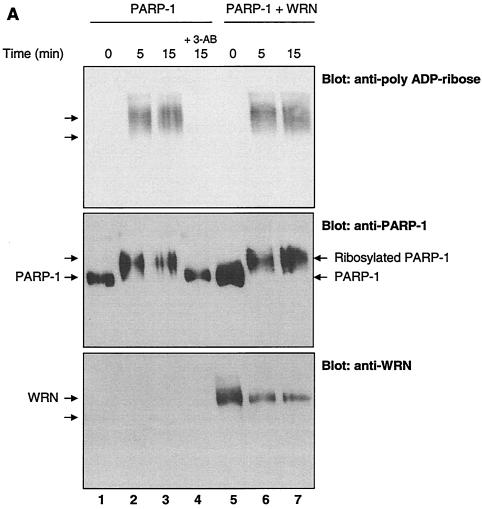
We next tested whether WRN affected the PARP-1 ribosylation function biochemically using purified proteins. Recombinant PARP-1 was incubated with or without recombinant WRN in the presence of NAD+. The proteins were resolved by SDS-10% polyacrylamide gel electrophoresis (PAGE) and analyzed by Western blotting with anti-PAR, anti-PARP-1, and anti-WRN antibodies consecutively (Fig. 4A). As shown in the top section (blot anti-PAR), the poly(ADP-ribosyl)ation reaction (PAR polymer formation) was detected as a typical smear for this type of posttranslational modification, and it was not affected by the presence of WRN (Fig. 4A, compare intensity of signals in lanes 2 and 3 with that in lanes 6 and 7). In the middle section (blot anti PARP-1), we detected a shift in the PARP-1 mobility after incubation with NAD+ (lanes 2, 3, 6, and 7), indicative of an extensive automodification of PARP-1. Again, the presence of WRN (lanes 6 and 7) did not affect the PARP-1 shift. As expected, the presence of the poly(ADP-ribosyl)ation inhibitor 3-AB (lane 4) completely abolished PAR polymer formation and subsequently the PARP-1 shift. Finally, in the bottom section (blot anti-WRN), no change in the WRN mobility in SDS-PAGE gel was observed (Fig. 4A, compare lane 5 with 6 and 7), suggesting that recombinant WRN is not significantly poly(ADP-ribosyl)ated by PARP-1. Thus, WRN does not affect the autocatalytic activity of PARP-1 in vitro. To address the question of whether WRN affects the PARP-1 ribosylation of other substrates, we used an experiment combining in vitro ribosylation and ELISA (Fig. 4B). Here, we detected PARP-1-dependent PAR polymer formation on H1 histones used to coat ELISA wells, both in the presence (Fig. 4B, bars 4 to 7) and absence (Fig. 4B, bars 1 to 3) of WRN. As shown in Fig. 4B, WRN (at different molar ratios relative to PARP-1) did not affect the polymer formation, as detected by anti-PAR antibodies. The presence of the PARP-1 inhibitor 3-AB completely inhibited the reaction (Fig. 4B, bar 7). Thus, WRN does not affect the poly(ADP-ribosyl)ation of H1 histones by PARP-1.
FIG. 4.
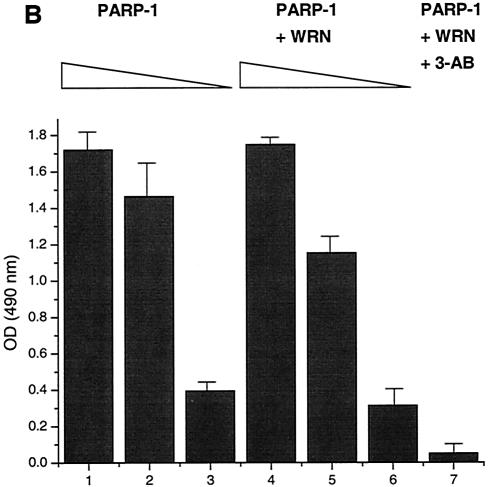
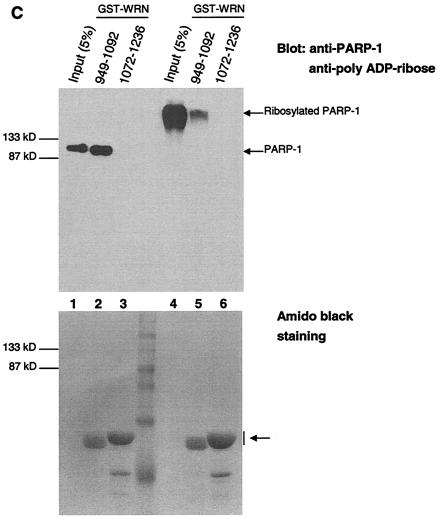
The autocatalytic activity of PARP-1 is unaffected by WRN in vitro. (A) Poly(ADP-ribosyl)ation activity of recombinant PARP-1 either alone (lanes 1 to 3) or with the inhibitor 3-AB (lane 4) or recombinant WRN (lanes 5 to 7). The reactions were terminated at the indicated times, and the samples were analyzed by SDS-10% PAGE and Western blotting with anti-PAR antibodies (top), anti-PARP-1 antibodies (middle), and anti-WRN antibodies (bottom). Arrows, SDS-PAGE mobilities of unmodified WRN and PARP-1. The increased amount of WRN detected in lane 5 is not due to the lack of reactivity of the anti-WRN antibodies but rather reflects larger amounts of WRN and PARP-1 loaded (compare lane 5 with 6 and 7 [middle and bottom]). (B) H1 histones were used to coat ELISA wells as described in Materials and Methods. After the blocking step, either 500 (bars 1, 4, and 7), 50 (bars 2 and 5), or 5 nM (bars 3 and 6) recombinant PARP-1 was added in the absence (bars 1 to 3) or presence (bars 4 to 7) of WRN (80 nM). The poly(ADP-ribosyl)ation reaction was initiated by adding NAD+ and activated DNA for 30 min at RT. The specific PARP inhibitor 3-AB was added to the ribosylation reaction (bar 7) as a negative control. The PAR polymer was detected with anti-PAR antibodies and horseradish peroxidase-conjugated secondary antibodies. The bound antibodies were detected by colorimetric analysis. OD, optical density. (C) Poly(ADP-ribosyl)ation of PARP-1 strongly decreases, but does not abolish, binding to the WRN RQC domain. GST-WRN RQC (aa 949 to 1092) (lanes 2 and 5) and GST-WRN (aa 1072 to 1236) (lanes 3 and 6) were incubated for 2 h at RT with either recombinant PARP-1 (lanes 1 to 3) or auto-poly(ADP-ribosyl)ated PARP-1 (lanes 4 to 6). The bound proteins were eluted and analyzed by SDS-7.5% PAGE and Western blotting with a mixture of anti-PARP-1 and anti-PAR antibodies. Amido black staining (bottom) shows the amount of GST-WRN fusion proteins (arrow) loaded.
The poly(ADP-ribosyl)ation of PARP-1 affects its ability to interact with DNA and specific proteins. For example, XRCC1 binds only poly(ADP-ribosyl)ated PARP-1 (27, 34). The binding results presented in Fig. 1E (ELISA) indicate that WRN binds efficiently to unmodified PARP-1. To determine whether the ribosylation state of PARP-1 influences WRN binding, we performed an in vitro binding assay (Fig. 4C). Unmodified PARP-1 and auto-poly(ADP-ribosyl)ated recombinant PARP-1were incubated with either GST-WRN RQC (aa 949 to 1092) or GST-WRN (aa 1072 to 1236). Following the incubation, the bound proteins were analyzed by Western blotting with anti-PARP-1 and anti-PAR antibodies. GST-WRN RQC bound unmodified PARP-1 with an efficiency of ∼70 to 75% (Fig. 4C, lanes 1 and 2). However, when PARP-1 was extensively auto-poly(ADP-ribosyl)ated, the binding efficiency was reduced to about 7 to 10% (compare input lanes, 1 and 4, with binding lanes, 2 and 5). GST-WRN (aa 1072 to 1236) did not bind PARP-1 (Fig. 4B, lanes 3 and 6). Thus, the poly(ADP-ribosyl)ation of PARP-1 strongly inhibits its in vitro binding to the WRN RQC domain.
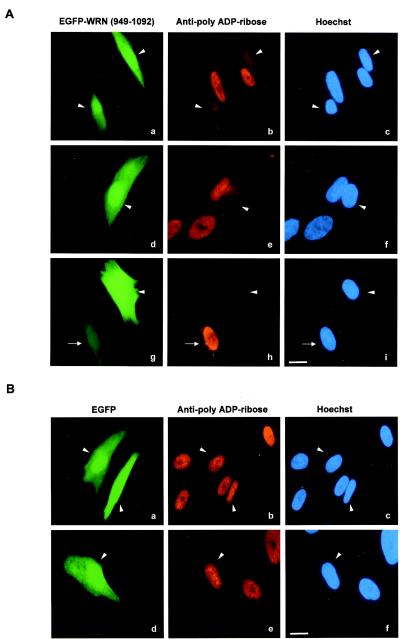
To determine the role of the WRN/PARP-1 physical interaction in the poly(ADP-ribosyl)ation of cellular proteins after damage, the PARP-1 binding domain of WRN (RQC domain) was overexpressed as an EGFP fusion protein in a control cell line. The cells were then treated with H2O2 (Fig. 5). The overexpression of the PARP-1 binding domain of WRN strongly reduced the poly(ADP-ribosyl)ation reaction, as detected with anti-PAR antibodies (Fig. 5A, compare PAR polymer synthesis signals for transfected and untransfected cells). In contrast, overexpression of EGFP alone did not affect the damage response reaction (Fig. 5B). The extent of EGFP-WRN RQC-expressing cells showing inhibition of ribosylation correlated with the level of overexpression. As shown in Fig. 5A (g and h), when the expression of EGFP-WRN RQC was low, there was no inhibitory effect. Approximately 80% of the cells (33 of 40) that expressed high levels of EGFP-WRN RQC were defective in the poly(ADP-ribosyl)ation reaction after exposure to H2O2.
FIG.5.
Overexpression of the PARP-1 binding domain of WRN strongly reduces PAR polymer formation after H2O2 treatment. TERT-1604 cells were transfected with either pEGFP-WRN (aa 949 to 1092) (A) or pEGFP alone (B). Thirty-six hours posttransfection, the cells were synchronized (early S phase) with HU for an additional 18 h. The cells were then incubated with H2O2 (500 μM) for 10 min at 37°C. After fixation, the cells were probed with anti-PAR antibodies. Arrowheads, EGFP-WRN- (aa 949 to 1092) and EGFP-transfected cells; arrow (A, g), one cell with low EGFP-WRN (aa 949 to 1092) expression. Bars (A, i, and B, f), 10 μm. Pictures correspond to representative transfected cells showing high-level of expression either of EGFP-WRN (aa 949 to 1092) (A) or EGFP alone (B). Approximately 80% (33 of 40) of the EGFP-WRN RQC-overexpressing cells showed less PAR polymer formation than nontransfected cells.
DISCUSSION
Here we identify PARP-1 as one of the strongest binders to the WRN RQC domain using HeLa nuclear extracts. Our results indicate that the WRN/PARP-1 complex plays a key role in the poly(ADP-ribosyl)ation pathway after treatment with agents that invoke DNA base damage. We demonstrate that the poly(ADP-ribosyl)ation of cellular proteins other than PARP-1 is severely defective after H2O2 and MMS treatment in primary cells derived from WS patients. To our knowledge, this represents the first human disease displaying a defect in the cellular-stress-dependent poly(ADP-ribosyl)ation pathway. We have shown that PARP-1 is active in WS cells but that its ability to ribosylate proteins other than itself after DNA damage is severely hampered. We demonstrate this PAR polymer formation defect in WS cells both by immunofluorescence and Western blot analysis. The role of WRN in this process is also demonstrated by the marked inhibition of PAR polymer formation following overexpression of the PARP-1 binding region of WRN (Fig. 5). Overexpression of this region of WRN may disrupt WRN/PARP-1 complex formation via competition for binding to endogenous WRN. Thus, the normal process of poly(ADP-ribosyl)ation of nuclear proteins after DNA damage may be impaired. This result suggests a direct and important role for the WRN/PARP-1 complex in the early transmission of signals after H2O2 and MMS treatment.
After DNA damage, PARP-1 uses NAD+ as a substrate in the poly(ADP-ribosyl)ation reaction. The deficient PARP-1 response that we observe in WS cells is not likely to be due to lower levels of NAD+ because WS cells exhibit much higher levels of NAD+ than age-matched controls (9). Alternatively, the lack of ribosylation in WS cells after oxidative stress could be due to a differential protein expression and/or activity levels of oxygen radical scavengers, such as catalase. However, we did not detect any differences in catalase protein levels between normal and WS cells (data not shown). This result is in agreement with a previous report showing that the enzymatic activities of Cu,Zn superoxide dismutase, Mn superoxide dismutase, catalase, and glutathione peroxidase 1 in WS cells were normal (24).
The deficiency in PAR polymer formation in WS cells after oxidative stress was initially observed when the cells were treated with HU before exposure to H2O2 (Fig. 2). This pretreatment arrests the cells in S phase, where the RecQ helicases are thought to have important roles (29). WS cells are known to be particularly sensitive to certain compounds in S phase (32, 33, 35, 36) and are known to have a longer S phase than normal cells (37, 46). DNA damage introduced in S phase presents obvious problems to the cells, but more so to WS cells, and thus there is a higher risk of genomic instability during this stage of the cell cycle in WS cells than in normal cells. In addition, a cellular arrest step minimizes the possibility that the differences in the DNA damage response are due to variations in the percentage of cells in different phases of the cell cycle. However, we observed that exponentially growing WS cells presented the same deficiency in the poly(ADP-ribosyl)ation of cellular proteins after H2O2 treatment by Western blotting (Fig. 3A) as did the HU-treated cells. Thus, the deficiency in the poly(ADP-ribosyl)ation pathway occurs both in S-phase-arrested and exponentially growing WS cells.
A previous study reported that HU treatment induced apoptosis in WS lymphoblasts (33). Our study was conducted mainly with primary WS fibroblasts (Table 1), and we observed that, 2 days after HU release, the WS fibroblasts remained viable and without any signs of apoptosis (data not shown).
Exposure of cells to H2O2 generates various types of macromolecular damage, including some DNA breaks, but the most frequent DNA modifications are oxidized DNA bases (3, 25). These DNA lesions are generally repaired by the BER pathway (4). MMS treatment produces alkylated DNA bases, and it is well known that these types of DNA lesions are mainly repaired by BER (13). In contrast, the compound bleomycin is a radiation-mimicking agent and mainly introduces a large frequency of single-strand and double-strand DNA breaks (3, 38). The PARP-1 response to bleomycin operates properly in WS cells. In contrast, specific DNA base modifications induced by H2O2 and MMS are not properly sensed (via the ribosylation pathway) in WS cells. According to several reports, PARP-1 is an active participant in BER (10, 39, 41). Our present results suggest that the WRN/PARP-1 complex is also involved in this repair process. WRN interacts with other proteins that play a role in BER (polymerase δ [Pol δ], PCNA, FEN-1, replication protein A [RPA], and p53), suggesting a function for WRN in this process (6, 7, 18, 20, 43, 45). WRN and PARP-1 bind some common BER components (e.g., p53) (14, 22, 43), further supporting the hypothesis that they might share intracellular pathways. Also, recent in vitro data demonstrate that WRN is an active player in the BER process and physically interacts with Pol β (17). Altogether, these results suggest that the WRN/PARP-1 complex is directly involved in the repair and/or sensing of specific DNA lesions produced by oxidative stress and alkylating agents. Interestingly, a very recent study reported that PARP-1-null WrnΔhel/Δhel (helicase-defective) mice show genetic cooperation between WRN and PARP-1, suggesting that both proteins work together in the processing of DNA damage (19).
Based on the observations reported here, we speculate that WRN and PARP-1 function in vivo in the early cellular response to DNA damage. The WRN/PARP-1 complex may be recruited to the damaged site as part of the DNA damage-sensing step. In support of this notion, we have previously reported that WRN exonuclease activity is blocked in vitro at sites of certain oxidative DNA base lesions (21). The formation of this complex may then allow PARP-1 to efficiently poly(ADP-ribosyl)ate nuclear target proteins and to subsequently transmit signals to DNA repair proteins. XRCC1 specifically binds ribosylated PARP-1 (27), and, as mentioned above, WRN binds to RPA, p53, FEN-1, Pol δ, and PCNA. Altogether, these interactions indicate that the WRN/PARP-1 complex may function at the site of DNA damage and recruit factors for the early assembly of DNA repair. The poly(ADP-ribosyl)ated proteins are then released from the site of DNA damage, allowing access for components of the DNA repair machinery (52). Our observation (Fig. 4C) that WRN binds less efficiently to PARP-1 when it is auto-poly(ADP-ribosyl)ated is in accordance with this model because, after the PARP-1 automodification, WRN can perform other functions.
In this scenario, it is likely that, after DNA damage, these protein-protein and DNA-protein interactions are more stable when all the involved proteins are present as a complex. In the absence of WRN (i.e., WS cells), although PARP-1 is still able to sense the DNA damage, as detected by the auto-poly(ADP-ribosyl)ation modification, the subsequent modification of target proteins is strongly impaired. A logical consequence of this lack of signal transmission may be the accumulation of DNA damage or a delay in the repair process, leading to a premature senescence caused by a diminished DNA repair capacity, as was previously proposed for primary cells from WS and Cockayne syndrome patients (50). Interestingly, there is evidence that longevity is associated with a high poly(ADP-ribosyl)ation capacity (16, 28), and our data with WS cells support this hypothesis.
It is well accepted that one of the hallmarks of aging is the accumulation of damage caused by reactive oxygen species (ROS; endogenous and exogenous) (2, 4). ROS are produced during normal aerobic metabolism and inflammatory reactions, and normal cells have evolved a mechanism to detect “oxidative signals” that are potentially deleterious for macromolecules such as DNA (44). At the cellular and organismal level, high-dose exposure to oxidants may cause cytotoxicity and neoplastic transformation. Some characteristic features in WS patients involve the presence of chronic inflammatory processes (ulcers) on the legs and soft-tissue tumors (12). Oxidative stress is also implicated in the initiation of the age-related onset of cataracts. Evidence suggests that a high concentration of H2O2 in the lens (up to 660 μM) is likely to be the major cause of cataract formation in humans (42). Interestingly, the premature onset of cataracts is perhaps the most typical feature in WS patients (26). Thus, in some tissues, WS cells are continuously exposed to presumably elevated rates of ROS generation.
Our results showing a defect in the cellular response after H2O2 and MMS exposure in WS cells may explain some of the phenotypical characteristics of WS, as a consequence of persistent unrepaired or unsensed DNA damage. WS cells may have an accelerated damage accumulation process, which leads to the characteristic mutator, potentially oncogenic, and genomically unstable phenotype.
.
Acknowledgments
We thank Gilbert de Murcia and Valerie Schreiber for the kind gift of recombinant PARP-1 protein and pARP vector (containing the PARP-1 sequence) and helpful discussions. We are grateful to Fred E. Indig, David M. Wilson III, and Kazunori Hashiguchi for critical reading of the manuscript.
REFERENCES
- 1.Alvarez-Gonzalez, R., H. Spring, M. Muller, and A. Burkle. 1999. Selective loss of poly(ADP-ribose) and the 85-kDa fragment of poly(ADP-ribose) polymerase in nucleoli during alkylation-induced apoptosis of HeLa cells. J. Biol. Chem. 274:32122-32126. [DOI] [PubMed] [Google Scholar]
- 2.Barja, G. 2002. Endogenous oxidative stress: relationship to aging, longevity and caloric restriction. Ageing Res. Rev. 1:397-411. [DOI] [PubMed] [Google Scholar]
- 3.Benitez-Bribiesca, L., and P. Sanchez-Suarez. 1999. Oxidative damage, bleomycin, and gamma radiation induce different types of DNA strand breaks in normal lymphocytes and thymocytes. A comet assay study. Ann. N. Y. Acad. Sci. 887:133-149. [DOI] [PubMed] [Google Scholar]
- 4.Bohr, V. A. 2002. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic. Biol. Med. 32:804-812. [DOI] [PubMed] [Google Scholar]
- 5.Bohr, V. A., M. Cooper, D. Orren, A. Machwe, J. Piotrowski, J. Sommers, P. Karmakar, and R. Brosh. 2000. Werner syndrome protein: biochemical properties and functional interactions. Exp. Gerontol. 35:695-702. [DOI] [PubMed] [Google Scholar]
- 6.Brosh, R. M., Jr., D. K. Orren, J. O. Nehlin, P. H. Ravn, M. K. Kenny, A. Machwe, and V. A. Bohr. 1999. Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem. 274:18341-18350. [DOI] [PubMed] [Google Scholar]
- 7.Brosh, R. M., Jr., C. von Kobbe, J. A. Sommers, P. Karmakar, P. L. Opresko, J. Piotrowski, I. Dianova, G. L. Dianov, and V. A. Bohr. 2001. Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO J. 20:5791-5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burkle, A. 2001. PARP-1: a regulator of genomic stability linked with mammalian longevity. Chembiochem. Eur. J. Chem. Biol. 2:725-728. [DOI] [PubMed] [Google Scholar]
- 9.Chapman, M. L., M. R. Zaun, and R. W. Gracy. 1983. Changes in NAD levels in human lymphocytes and fibroblasts during aging and in premature aging syndromes. Mech. Ageing Dev. 21:157-167. [DOI] [PubMed] [Google Scholar]
- 10.Dantzer, F., R. G. de La, J. Menissier-de Murcia, Z. Hostomsky, G. de Murcia, and V. Schreiber. 2000. Base excision repair is impaired in mammalian cells lacking poly(ADP-ribose) polymerase-1. Biochemistry 39:7559-7569. [DOI] [PubMed] [Google Scholar]
- 11.Desnoyers, S., S. H. Kaufmann, and G. G. Poirier. 1996. Alteration of the nucleolar localization of poly(ADP-ribose) polymerase upon treatment with transcription inhibitors. Exp. Cell Res. 227:146-153. [DOI] [PubMed] [Google Scholar]
- 12.Duvic, M., and N. A. Lemak. 1995. Werner's syndrome. Dermatol. Clin. 13:163-168. [PubMed] [Google Scholar]
- 13.Friedberg, E. C., G. C. Walker, and W. Siede. 1995. DNA repair and mutagenesis, p. 135-190. American Society for Microbiology, Washington, D.C.
- 14.Galande, S., and T. Kohwi-Shigematsu. 1999. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 274:20521-20528. [DOI] [PubMed] [Google Scholar]
- 15.Giner, H., F. Simonin, G. de Murcia, and J. Menissier-de Murcia. 1992. Overproduction and large-scale purification of the human poly(ADP-ribose) polymerase using a baculovirus expression system. Gene 114:279-283. [DOI] [PubMed] [Google Scholar]
- 16.Grube, K., and A. Burkle. 1992. Poly(ADP-ribose) polymerase activity in mononuclear leukocytes of 13 mammalian species correlates with species-specific life span. Proc. Natl. Acad. Sci. USA 89:11759-11763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrigan, J. A., P. L. Opresko, C. von Kobbe, P. S. Kedar, R. Prasad, S. H. Wilson, and V. A. Bohr. 2003. The Werner syndrome protein stimulates DNA polymerase beta strand displacement synthesis via its helicase activity. J. Biol. Chem. 278:22686-22695. [DOI] [PubMed] [Google Scholar]
- 18.Kamath-Loeb, A. S., E. Johansson, P. M. Burgers, and L. A. Loeb. 2000. Functional interaction between the Werner Syndrome protein and DNA polymerase delta. Proc. Natl. Acad. Sci. USA 97:4603-4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebel, M., J. Lavoie, I. Gaudreault, M. Bronsard, and R. Drouin. 2003. Genetic cooperation between the Werner syndrome protein and Poly(ADP-ribose) polymerase-1 in preventing chromatid breaks, complex chromosomal rearrangements, and cancer in mice. Am. J. Pathol. 162:1559-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lebel, M., E. A. Spillare, C. C. Harris, and P. Leder. 1999. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 274:37795-37799. [DOI] [PubMed] [Google Scholar]
- 21.Machwe, A., R. Ganunis, V. A. Bohr, and D. K. Orren. 2000. Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res. 28:2762-2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malanga, M., J. M. Pleschke, H. E. Kleczkowska, and F. R. Althaus. 1998. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J. Biol. Chem. 273:11839-11843. [DOI] [PubMed] [Google Scholar]
- 23.Marciniak, R. A., D. B. Lombard, F. B. Johnson, and L. Guarente. 1998. Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl. Acad. Sci. USA 95:6686-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marklund, S., I. Nordensson, and O. Back. 1981. Normal CuZn superoxide dismutase, Mn superoxide dismutase, catalase and glutathione peroxidase in Werner's syndrome. J. Gerontol. 36:405-409. [DOI] [PubMed] [Google Scholar]
- 25.Marnett, L. J. 2000. Oxyradicals and DNA damage. Carcinogenesis 21:361-370. [DOI] [PubMed] [Google Scholar]
- 26.Martin, G. M. 1978. Genetic syndromes in man with potential relevance to the pathobiology of aging. Birth Defects Orig. Artic. Ser. 14:5-39. [PubMed] [Google Scholar]
- 27.Masson, M., C. Niedergang, V. Schreiber, S. Muller, J. Menissier-de Murcia, and G. de Murcia. 1998. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol. Cell. Biol. 18:3563-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muiras, M. L., M. Muller, F. Schachter, and A. Burkle. 1998. Increased poly(ADP-ribose) polymerase activity in lymphoblastoid cell lines from centenarians. J. Mol. Med. 76:346-354. [DOI] [PubMed] [Google Scholar]
- 29.Oakley, T. J., and I. D. Hickson. 2002. Defending genome integrity during S-phase: putative roles for RecQ helicases and topoisomerase III. DNA Repair 1:175-207. [DOI] [PubMed] [Google Scholar]
- 30.Opresko, P. L., C. von Kobbe, J. P. Laine, J. Harrigan, I. D. Hickson, and V. A. Bohr. 2002. Telomere-binding protein TRF2 binds to and stimulates the Werner and Bloom syndrome helicases. J. Biol. Chem. 277:41110-41119. [DOI] [PubMed] [Google Scholar]
- 31.Orren, D. K., R. M. Brosh, Jr., J. O. Nehlin, A. Machwe, M. D. Gray, and V. A. Bohr. 1999. Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 27:3557-3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pichierri, P., A. Franchitto, P. Mosesso, and F. Palitti. 2000. Werner's syndrome cell lines are hypersensitive to camptothecin-induced chromosomal damage. Mutat. Res. 456:45-57. [DOI] [PubMed] [Google Scholar]
- 33.Pichierri, P., A. Franchitto, P. Mosesso, and F. Palitti. 2001. Werner's syndrome protein is required for correct recovery after replication arrest and DNA damage induced in S-phase of cell cycle. Mol. Biol. Cell 12:2412-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pleschke, J. M., H. E. Kleczkowska, M. Strohm, and F. R. Althaus. 2000. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J. Biol. Chem. 275:40974-40980. [DOI] [PubMed] [Google Scholar]
- 35.Poot, M., K. A. Gollahon, M. J. Emond, J. R. Silber, and P. S. Rabinovitch. 2002. Werner syndrome diploid fibroblasts are sensitive to 4-nitroquinoline-N-oxide and 8-methoxypsoralen: implications for the disease phenotype. FASEB J. 16:757-758. [DOI] [PubMed] [Google Scholar]
- 36.Poot, M., K. A. Gollahon, and P. S. Rabinovitch. 1999. Werner syndrome lymphoblastoid cells are sensitive to camptothecin-induced apoptosis in S-phase. Hum. Genet. 104:10-14. [DOI] [PubMed] [Google Scholar]
- 37.Poot, M., H. Hoehn, T. M. Runger, and G. M. Martin. 1992. Impaired S-phase transit of Werner syndrome cells expressed in lymphoblastoid cells. Exp. Cell Res. 202:267-273. [DOI] [PubMed] [Google Scholar]
- 38.Povirk, L. F. 1996. DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Fundam. Mol. Mech. Mutagen. 355:71-89. [DOI] [PubMed] [Google Scholar]
- 39.Prasad, R., O. I. Lavrik, S. J. Kim, P. Kedar, X. P. Yang, B. J. Vande Berg, and S. H. Wilson. 2001. DNA polymerase beta-mediated long patch base excision repair. Poly(ADP-ribose)polymerase-1 stimulates strand displacement DNA synthesis. J. Biol. Chem. 276:32411-32414. [DOI] [PubMed] [Google Scholar]
- 40.Rajaee-Behbahani, N., P. Schmezer, A. Burkle, and H. Bartsch. 2000. Quantitative assessment of bleomycin-induced poly(ADP-ribosyl)ation in human lymphocytes by immunofluorescence and image analysis. J. Immunol. Methods 244:145-151. [DOI] [PubMed] [Google Scholar]
- 41.Schreiber, V., J. C. Ame, P. Dolle, I. Schultz, B. Rinaldi, V. Fraulob, J. Menissier-de Murcia, and G. de Murcia. 2002. Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem. 277:23028-23036. [DOI] [PubMed] [Google Scholar]
- 42.Spector, A., and W. H. Garner. 1981. Hydrogen peroxide and human cataract. Exp. Eye Res. 33:673-681. [DOI] [PubMed] [Google Scholar]
- 43.Spillare, E. A., A. I. Robles, X. W. Wang, J. C. Shen, C. E. Yu, G. D. Schellenberg, and C. C. Harris. 1999. p53-mediated apoptosis is attenuated in Werner syndrome cells. Genes Dev. 13:1355-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stadtman, E. R. 1992. Protein oxidation and aging. Science 257:1220-1225. [DOI] [PubMed] [Google Scholar]
- 45.Szekely, A. M., Y. H. Chen, C. Zhang, J. Oshima, and S. M. Weissman. 2000. Werner protein recruits DNA polymerase delta to the nucleolus. Proc. Natl. Acad. Sci. USA 97:11365-11370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeuchi, F., F. Hanaoka, M. Goto, M. Yamada, and T. Miyamoto. 1982. Prolongation of S phase and whole cell cycle in Werner's syndrome fibroblasts. Exp. Gerontol. 17:473-480. [DOI] [PubMed] [Google Scholar]
- 47.Tong, W. M., U. Cortes, and Z. Q. Wang. 2001. Poly(ADP-ribose) polymerase: a guardian angel protecting the genome and suppressing tumorigenesis. Biochim. Biophys. Acta 1552:27-37. [DOI] [PubMed] [Google Scholar]
- 48.von Kobbe, C., and V. A. Bohr. 2002. A nucleolar targeting sequence in the Werner syndrome protein resides within residues 949-1092. J. Cell Sci. 115:3901-3907. [DOI] [PubMed] [Google Scholar]
- 49.von Kobbe, C., P. Karmakar, L. Dawut, P. Opresko, X. Zeng, R. M. Brosh, Jr., I. D. Hickson, and V. A. Bohr. 2002. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 277:22035-22044. [DOI] [PubMed] [Google Scholar]
- 50.Weirich-Schwaiger, H., H. G. Weirich, B. Gruber, M. Schweiger, and M. Hirsch-Kauffmann. 1994. Correlation between senescence and DNA repair in cells from young and old individuals and in premature aging syndromes. Mutat. Res. 316:37-48. [DOI] [PubMed] [Google Scholar]
- 51.Yu, C. E., J. Oshima, Y. H. Fu, E. M. Wijsman, F. Hisama, R. Alisch, S. Matthews, J. Nakura, T. Miki, S. Ouais, G. M. Martin, J. Mulligan, and G. D. Schellenberg. 1996. Positional cloning of the Werner's syndrome gene. Science 272:258-262. [DOI] [PubMed] [Google Scholar]
- 52.Ziegler, M., and S. L. Oei. 2001. A cellular survival switch: poly(ADP-ribosyl)ation stimulates DNA repair and silences transcription. Bioessays 23:543-548. [DOI] [PubMed] [Google Scholar]