SUMMARY
Detection of nucleic acids and induction of type I interferons (IFNs) are principal elements of antiviral defense, but can cause autoimmunity if misregulated. Cytosolic DNA detection activates a potent, cell-intrinsic antiviral response through a poorly defined pathway. In a screen for proteins relevant to this interferon-stimulatory DNA (ISD) response, we identify 3’ repair exonuclease 1 (Trex1). Mutations in the human trex1 gene cause Aicardi-Goutieres Syndrome (AGS) and chilblain lupus, but the molecular basis of these diseases is unknown. We define Trex1 as an essential negative regulator of the ISD response and delineate the genetic pathway linking Trex1 deficiency to lethal autoimmunity. We show that single-stranded DNA derived from endogenous retroelements accumulates in Trex1-deficient cells and that Trex1 can metabolize reverse-transcribed DNA. These findings reveal a cell-intrinsic mechanism for initiation of autoimmunity, implicate the ISD pathway as the cause of AGS, and suggest an unanticipated contribution of endogenous retroelements to autoimmunity.
INTRODUCTION
Detection of foreign nucleic acids is an ancient form of host defense. In vertebrates, nucleic acid detection activates a program of antiviral defense designed to neutralize the spread of infection. This antiviral program is coordinated by type I interferons (IFNs), which direct a multifaceted response to restrict viral replication within infected cells, alert neighboring cells to the presence of infection, and expand effector lymphocytes to provide long-term and specific protection against the virus (Stark et al., 1998; Stetson and Medzhitov, 2006b).
Two complementary systems link nucleic acid detection to the IFN-mediated antiviral response. One system consists of several Toll-like receptors (TLRs), which are transmembrane sensors expressed on sentinel immune cells that sample endosomal cargo for nucleic acids (Takeda et al., 2003). As such, TLRs comprise a non-cell autonomous mechanism whereby uninfected immune cells sense viral infection by detecting free viral particles or viral nucleic acids within phagocytosed apoptotic cells (Pichlmair and Reis e Sousa, 2007). A second, more broadly expressed system detects viral nucleic acids within the infected cell itself. This system is exemplified by the cytosolic RNA helicases RIG-I and MDA5, which signal activation of a cell-intrinsic antiviral response through the adaptor protein IPS-1 (also known as MAVS, CARDIF, or VISA; reviewed in Pichlmair and Reis e Sousa, 2007). Recently, a cytosolic antiviral pathway that detects DNA was described (Ishii et al., 2006; Martin and Elkon, 2006; Okabe et al., 2005; Stetson and Medzhitov, 2006a). This system, termed the interferon-stimulatory DNA (ISD) response, is analogous to the RIG-I and MDA5 RNA helicases in that it is cell-intrinsic. However, the ISD pathway engages a distinct signaling cascade that is IPS-1-independent (Kumar et al., 2006; Sun et al., 2006), possibly through activation of the candidate ISD sensor DAI (Takaoka et al., 2007). Although little is known about the upstream signaling events that distinguish the ISD response from RIG-I- and MDA5-mediated RNA recognition, both pathways activate potent type I IFN production through the transcription factor interferon regulatory factor 3 (IRF3) (Ishii et al., 2006; Stetson and Medzhitov, 2006a).
Together, these two types of nucleic acid detection systems – TLRs and cytosolic sensors – account for essentially all IFN-mediated antiviral immunity (Koyama et al., 2007). However, discrimination of viral from self nucleic acids is imperfect, and recent studies have shown that defective clearance of self-derived nucleic acids can cause severe, IFN-associated autoimmunity. For example, deficiency for the extracellular DNAse I causes a lupus-like syndrome in mice (Napirei et al., 2000), and DNAse I mutations in humans are associated with lupus (Yasutomo et al., 2001). One major mechanism by which these extracellular nucleic acids cause autoimmunity is through activation of TLRs on autoreactive B cells (Leadbetter et al., 2002). TLR7 and TLR9 are important for autoantibody production in a murine model of lupus (Christensen et al., 2006), and a spontaneous gene duplication of murine TLR7 predisposes to autoimmunity (Pisitkun et al., 2006; Subramanian et al., 2006). A complex of extracellular DNA and the antimicrobial peptide LL37 activates TLR9-dependent IFN production by plasmacytoid dendritic cells (pDCs) in human psoriasis (Lande et al., 2007). Notably, liver macrophages in mice lacking the lysosomal DNAse II become engorged with the ejected nuclei of erythrocyte precursors and develop a TLR-independent IFN response to this undigested DNA (Okabe et al., 2005; Yoshida et al., 2005).
Importantly, all of these examples involve non-cell autonomous mechanisms in that the source of the accumulated nucleic acids is distinct from the cells that detect them. In contrast, it is unclear whether accumulation of self-nucleic acids within cells can activate cytosolic sensors because a situation where this occurs has not yet been identified. Given the importance of these cell-intrinsic nucleic acid sensors in antiviral defense, a number of fundamental questions regarding the principles of self/non-self discrimination arise. Specifically, are there dedicated mechanisms that limit excessive activation of cytosolic nucleic acid sensors? What happens when these mechanisms break down? And, can cell-intrinsic nucleic acid detection cause autoimmunity if improperly controlled?
Here, we developed an approach to isolate proteins relevant to the ISD response and identified 3’ repair exonuclease 1 (Trex1) as an ISD-binding protein. Trex1 is the most abundant 3’->5’ DNA exonuclease in cells (Hoss et al., 1999; Lindahl et al., 1969; Mazur and Perrino, 1999). Loss of function mutations in the human Trex1 gene cause Aicardi-Goutieres Syndrome (AGS; Crow et al., 2006a). AGS presents in infancy as a severe encephalitis with intracranial calcifications, lymphocyte infiltrates and elevated type I IFN levels in cerebrospinal fluid, and demyelination of motor neurons with accompanying psychomotor retardation (Rice et al., 2007b). Many of these symptoms resemble those caused by congenitally acquired infection with cytomegalovirus or rubella virus (Crow et al., 2003; Sanchis et al., 2005), but no viral pathogen has been detected in AGS patients. Different Trex1 mutations also cause monogenic chilblain lupus and are found more frequently in people with systemic lupus erythematosus than in healthy individuals, suggesting that a common mechanism may underlie these disorders (Alarcon-Riquelme, 2006; Lee-Kirsch et al., 2007a; Lee-Kirsch et al., 2007b; Rice et al., 2007a). Trex1-deficient mice exhibit dramatically reduced postnatal survival; the principal cause of mortality in these mice is circulatory failure caused by inflammatory myocarditis (Morita et al., 2004).
Despite the emerging evidence linking Trex1 deficiency to autoimmunity, little is known about the specific mechanisms that lead to disease in the absence of Trex1. Herein, we define Trex1 as a negative regulator of the ISD response and delineate the genetic pathway linking cell-intrinsic initiation of autoimmunity to type I IFN-dependent autoantibody production and autoimmune pathology. We identify single-stranded DNA fragments derived from endogenous retroelements that accumulate in Trex1-deficient hearts and show that Trex1 can metabolize reverse-transcribed DNA of these endogenous retroelements, revealing a source of Trex1 substrates that may trigger autoimmunity.
RESULTS
Identification of Trex1 as an ISD-binding protein
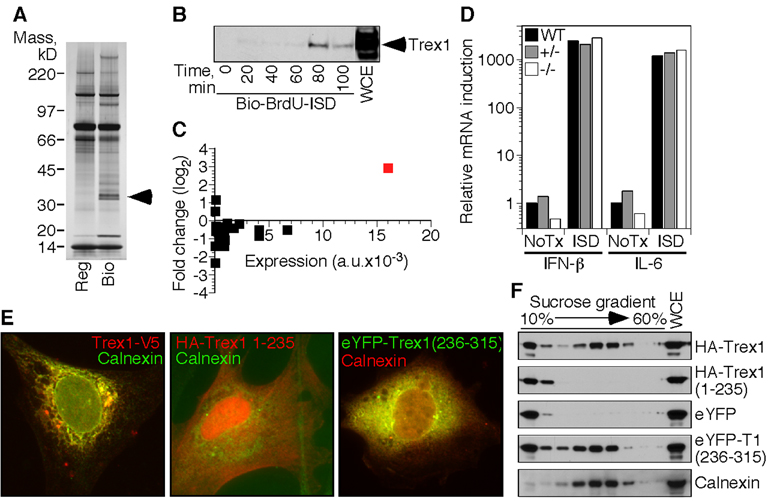
We designed an unbiased biochemical approach to identify proteins involved in ISD recognition based on direct binding to cytosolic DNA in live cells. We took advantage of the fact that the ISD response is sequence-independent and synthesized biotinylated oligonucleotides with several bromodeoxyuridine (BrdU) nucleosides in place of thymidine; BrdU more readily crosslinks to closely associated proteins upon exposure to ultraviolet (UV) light. We transfected the BrdU-modified ISD into macrophages, UV-treated the cells 90 minutes later, and recovered the biotinylated DNA/protein complexes from cell lysates. Mass spectrometry analysis identified a prominent recovered protein as Trex1 (Figure 1A), which we confirmed by western blot using antisera to Trex1 (Figure 1B).
Figure 1. Biochemical identification of Trex1 as an ISD-binding protein.
A) Proteins specifically recovered with BrdU-substituted, biotinylated ISD were visualized by silver staining. Arrows indicate the bands identified as Trex1.
B) Cells transfected with BrdU-substituted, biotinylated ISD were exposed to ultraviolet light at the indicated times post transfection. Trex1 association with recovered ISD was visualized by western blot. WCE=whole cell extract.
C) We examined microarray data (Stetson and Medzhitov, 2006a) of macrophages treated for four hours with ISD and plotted the fold change in expression in log2 format versus the sum of untreated and treated expression values for all 3’->5’ DNA exonucleases in the mouse genome (Shevelev and Hubscher, 2002). Trex1 is indicated in red.
D) Bone marrow-derived macrophages of the indicated Trex1 genotype were transfected with ISD. IFN-β and IL-6 mRNA expression four hours post-transfection was measured by quantitative RT-PCR, normalized to HPRT expression within each sample, and compared to untreated controls to calculate the relative expression.
E) HeLa cells were transfected with vectors encoding tagged, full-length Trex1 (left panel), Trexl lacking the c-terminal 76 amino acids (middle panel) or YFP fused to the c-terminal 76 amino acids of Trex1 (right panel), and visualized by immunofluorescence microscopy. Images are at 400x magnification.
F) 293T cells were transfected with expression vectors for haemagluttinin (HA)-tagged Trexl, HA-Trex1 (1–235), eYFP, or eYFP fused to Trex1 (236–315). Extracts from mechanically disrupted cells were centrifuged through a 10–60% sucrose gradient, and the position of each protein in harvested fractions was determined by western blot for HA (top two panels), eYFP (third and fourth panels), or calnexin (bottom panel).
Analysis of microarray data revealed that, in addition to its steady-state abundance, transcription of the trex1 gene is robustly induced by ISD stimulation (Figure 1C). Of all 3’->5’ DNA exonucleases in the mouse genome, Trex1 is uniquely ISD-inducible, which suggested a more specialized role for Trex1 in antiviral immunity (Figure 1C). We found that Trex1-deficient cells had an intact ISD response to transfected DNA, thus ruling out the possibility that Trex1 is the ISD sensor itself (Figure 1D).
Consistent with a previous report (Chowdhury et al., 2006), we found that Trex1 was cytosolic and localized to the endoplasmic reticulum (ER; Chowdhury et al., 2006). The C-terminal region of Trex1 that follows the nuclease domain was necessary for this ER localization: a truncated form of Trex1 lacking the last 79 amino acids was located diffusely throughout the cell (Figure 1E). Notably, a missense mutation in human Trex1 resulting in a similarly truncated protein causes autosomal dominant retinal vasculopathy with cerebral leukodystrophy (RVCL; OMIM 192315), underscoring the importance of this ER localization (Richards et al., 2007). Fusion of the C-terminal 79 amino acids of Trex1 to the C-terminus of yellow fluorescent protein (YFP) conferred ER localization identical to that of full-length Trex1 (Figure 1E; Richards et al., 2007).
Trex1 contains a predicted transmembrane helix at its c-terminus, suggesting that it may be an integral membrane protein of the ER (Lee-Kirsch et al., 2007b). However, Trex1 lacks an identifiable signal sequence for ER insertion, either at its N-terminus or within the 79 C-terminal amino acids that dictate ER localization. When we examined the migration of epitope-tagged Trex1 in sucrose gradients, we unexpectedly found two distinct pools of Trex1: a buoyant fraction that migrated at the top of the gradient, and a dense fraction that co-eluted with calnexin, an ER-resident, transmembrane protein (Figure 1F). Removal of the Trex1 C-terminus eliminated the dense pool, while fusion of this C-terminus to YFP resulted in a dual distribution identical to that of full-length Trex1 (Figure 1F). The dense form of Trex1 was also eliminated by pretreatment of cell extracts with detergent (0.5% NP-40; data not shown). These data suggest that Trex1 is not a transmembrane protein, and that a fraction of Trex1 interacts through its C-terminus with ER membranes or with another ER-resident protein to determine its localization.
Trex1 is a negative regulator of the ISD pathway
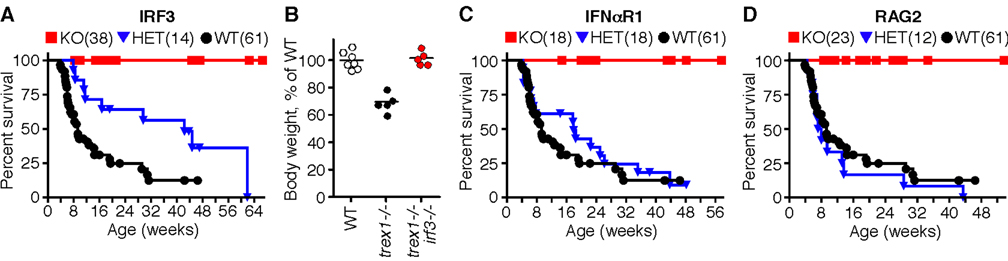
Trex1 is an ISD-inducible DNA exonuclease, but it is not the sensor of the ISD pathway. We therefore hypothesized that Trex1 may play a regulatory role in the ISD pathway by metabolizing DNA ligands of this cell-intrinsic antiviral response. Because IRF3 is essential for the antiviral ISD response (Stetson and Medzhitov, 2006a), we examined the contribution of the ISD-IRF3 pathway by breeding Trex1-deficient mice to IRF3-deficient mice (Sato et al., 2000). Importantly, IRF3 dependence distinguishes the ISD pathway from TLR9-dependent recognition of DNA, which activates type I IFNs in pDCs through the related IRF7 transcription factor (Honda et al., 2005; Kawai et al., 2004). Trex1-deficient mice in our colony succumbed to lethal autoimmunity at a median age of nine weeks (Figure 2A). In contrast, genetic ablation of IRF3 prevented disease in Trex1-deficient mice, as evidenced by complete rescue from mortality and restoration of normal body weight (Figure 2A, 2B).
Figure 2. Genetic dissection of the Trex1 knockout phenotype.
A) Survival curves for Trex1-deficient mice of the indicated irf3 genotype. Trex1−/− irf3+/+ mice include mice generated both by intercrossing trex1+/− irf3+/− mice as well as by separately intercrossing trex1+/− irf3+/+ mice. These latter mice are also included as “WT” for the crosses in figure 2C and 2D. The number of mice of each genotype is indicated in parentheses, and symbols represent individual mice.
B) Body weights of mice of the indicated genotype were measured at 4.5 weeks of age, and compared to the average weight of sex-matched, WT littermates. For these measurements trex1+/− mice were considered WT.
C) Survival curves for Trex1-deficient mice of the indicated ifnαr1 genotype.
D) Survival curves for Trex1-deficient mice of the indicated rag2 genotype.
We performed two additional crosses to further dissect the phenotype of Trex1-deficient mice. Detection of intracellular DNA by the ISD pathway activates potent type I IFN production, and elevated type I IFNs are a hallmark of AGS and lupus (Crow et al., 2003; Pascual et al., 2006; Theofilopoulos et al., 2005). Similar to Trex1/IRF3 double knockout (DKO) mice, Trex1-deficient mice lacking the type I IFN receptor (IFNαR1) were completely protected from mortality (Figure 2C). Finally, we found that lymphocytes were essential for disease because RAG2 deficiency fully rescued Trex1-deficient mice from mortality (Figure 2D).
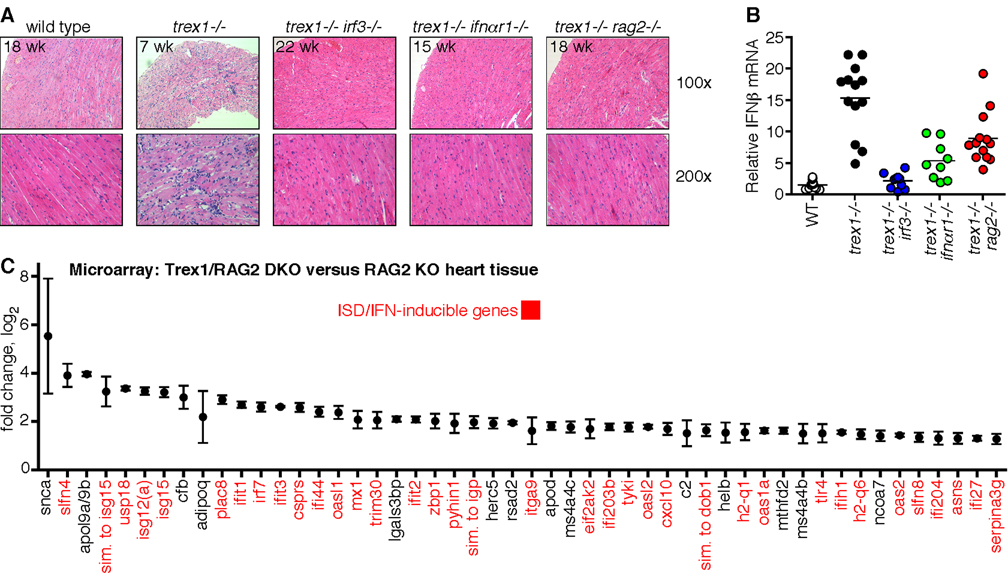
Most Trex1-deficient mice die of circulatory failure caused by severe inflammatory myocarditis (Morita et al., 2004). All hearts examined from Trex1-deficient mice in our colony had evidence of profound inflammation, with thinning of the ventricular wall, mononuclear cell infiltrates, and disruption of normal heart muscle morphology (Figure 3A). These pathological features were absent in Trex1-deficient mice lacking IRF3, IFNαR1, or RAG2 (Figure 3A). Interestingly, despite the restoration of normal heart anatomy in Trex1-deficient mice on all three rescued backgrounds, we found distinct effects of each rescuing genotype on type I IFN expression in heart tissue. IFN-β RNA expression was dramatically increased in Trex1-deficient hearts and completely restored to wild-type levels in Trex1/IRF3 DKO hearts, further emphasizing the primary role of IRF3 in this IFN-dependent pathology (Figure 3B). Trex1/IFNαR1 DKO hearts had low but detectably elevated levels of IFN-β RNA, consistent with disruption of the positive feedback loop of type I IFN expression that is amplified by IFNαR1 signaling (Figure 3B). In contrast, hearts of Trex1/RAG2 DKO mice retained significantly elevated levels of IFN-β RNA (Figure 3B). Importantly, these data indicate that expression of type I IFNs alone is not sufficient to cause disease; rather, it is the IFN-dependent mobilization of lymphocytes that precipitates autoimmunity in this setting.
Figure 3. Amelioration of autoimmunity through intervention at three phases of disease.
A) Paraffin-embedded heart tissue sections from mice of the indicated age and genotype were stained with H&E. The top panels are at 100x and the bottom panels are at 200x magnification.
B) IFNβ mRNA levels in hearts of mice of the indicated genotype were determined by quantitative RT-PCR, normalized to HPRT expression within each sample, and compared to WT littermates to calculate the relative IFNβ expression.
C) Gene expression analysis of heart tissue from three pairs of Trex1/RAG2 DKO and RAG2 KO littermates, aged 18, 19, and 28 weeks. Data are presented in log2 format and represent means and standard deviations of differential expression in Trex1-deficient hearts compared to wild-type hearts.
The Trex1/RAG2 DKO mice revealed an unexplored stage of disease, after initiation and before lymphocyte-dependent pathology. To further characterize this post-initiation phase, we performed a microarray analysis comparing heart tissue of Trex1/RAG2 DKO hearts to RAG2 KO littermate controls. We found that ISD- and IFN-inducible genes comprised over 75% of the top 50 genes with increased expression levels in Trex1-deficient hearts, further emphasizing the role of the ISD pathway in initiating disease (Figure 3C; Stetson and Medzhitov, 2006a).
Taken together, these findings indicate that Trex1 substrates are ligands of the ISD pathway. Accumulation of these substrates within Trex1-deficient cells initiates IRF3-dependent type I IFN production, which in turn recruits and activates lymphocytes to attack the affected tissues. Genetic ablation of IRF3, IFNαR1, or RAG2 ameliorates disease at three discrete phases, thus defining a genetic pathway connecting unregulated cell-intrinsic DNA detection by the ISD pathway to lethal autoimmunity.
The ISD pathway is required for autoantibody production in Trex1-deficient mice
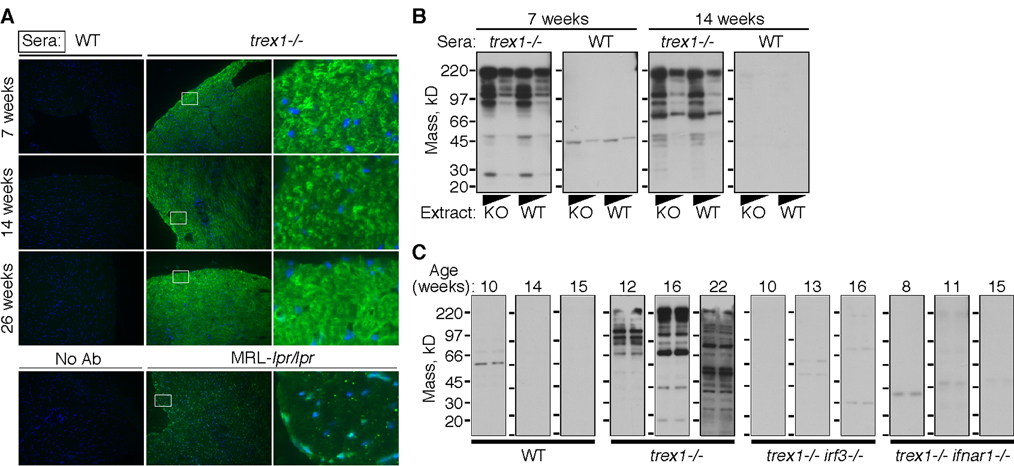
Because lymphocytes are essential for the autoimmune pathology caused by loss of Trex1 function, we tested for the presence of autoantibodies in Trex1-deficient mice. Sera from wild-type mice showed no detectable reactivity with heart tissue sections (Figure 4A, left panels). In contrast, IgG antibodies from Trex1-deficient mice strongly stained heart tissue (Figure 4A, middle and right panels). Sera from different Trex1 KO mice reproducibly detected similar cytosolic structures (Figure 4A). This cytosolic staining pattern was different from the nuclear and punctate cytosolic distribution of autoantigens detected by autoantibodies from MRL-lpr/lpr mice that develop systemic autoimmunity (Figure 4A, bottom row). Autoantibodies from Trex1-deficient mice detected knockout and wild type heart extracts equivalently, demonstrating that the autoantigens were not specific to Trex1 KO hearts (Figure 4B, 4C). Moreover, we observed a trend towards broader autoreactivity with sera from older Trex1-deficient mice, consistent with epitope spreading (Figure 4B, 4C).
Figure 4. The ISD pathway is essential for autoantibodies in Trex1-deficient mice.
A) IgG autoantibodies were detected by incubating Trex1/RAG2 DKO heart tissue sections with sera of the indicated genotype and then with fluorescently labeled secondary antibodies to mouse IgG. IgG signal is colored green, and DAPI staining of nuclei is in blue. The images in the left and middle panels are at 200x magnification, and the right panels are 1600x digital zooms of the fields demarcated by the white boxes.
B) IgG autoantibodies were detected by western blot against heart protein extracts, prepared from Trex1/RAG2 DKO (KO) or RAG2 KO (WT) mice and used neat or diluted 1:5.
C) Heart extracts were prepared as described for Figure 1B and incubated with sera harvested from individual mice of the indicated age and genotype. For each western, the left lane is KO extract and the right lane is WT extract.
We next tested whether the ISD pathway was linked to autoantibody production in Trex1-deficient mice. Sera from all singly Trex1-deficient mice tested demonstrated strong reactivity to heart extracts of both wild type and Trex1 KO mice (Figure 4C). In contrast, serum autoantibodies were undetectable in similarly aged Trex1 KO mice lacking either IRF3 or IFNαR1 (Figure 4C). These data reveal an important connection between the ISD pathway and the generation of autoantibodies, and suggest a probable explanation for the contribution of lymphocytes to disease in Trex1-deficient mice.
Identification of endogenous Trex1 DNA substrates
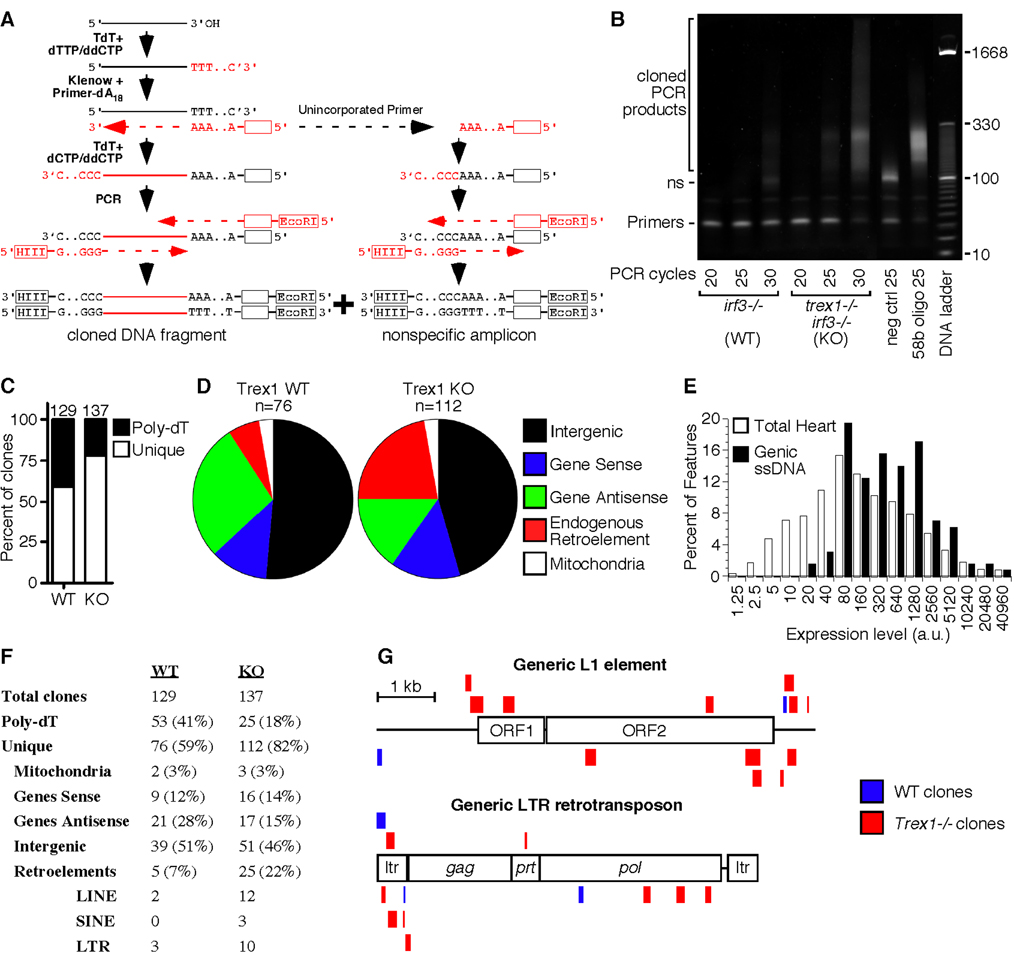
The data presented above suggest that accumulation of Trex1 DNA substrates activates the ISD pathway, raising questions about the nature and the source of this DNA. We developed a method to purify, amplify, and directionally clone these endogenous Trex1 DNA substrates directly from hearts of Trex1-deficient mice (Figure 5A). We homogenized hearts in the presence of 200µM digitonin to selectively disrupt cholesterol-rich plasma membranes and minimize extraction of other organelles. Purified DNA was tailed with dTTP using terminal deoxynucleotidyl transferase (TdT), and then copied with Klenow DNA polymerase and a primer specific for the poly-dT DNA tail. This complementary DNA strand was then tailed with dCTP to yield single-stranded DNA with two defined ends. PCR amplification yielded two types of products: correctly tailed and copied DNA fragments, and a smaller, nonspecific amplicon of unincorporated primer from the Klenow extension step (Figure 5A).
Figure 5. Identification of endogenous Trex1 substrates.
A) Schematic of the strategy used to tag and amplify DNA fragments purified from heart tissue.
B) DNA was purified from pooled hearts of three irf3−/− mice and three trex1−/− irf3−/− mice and PCR-amplified for the indicated number of cycles. The negative control revealed the nonspecific (ns) amplicon, and the positive control amplified a tailed, 58 base DNA oligonucleotide.
C) The percentage of unique clones and clones with an endogenous Poly-dT stretch immediately 3’ of the insert are indicated, with the total number of clones per genotype above each bar.
D) The relative percentage of each type of DNA fragment within the pool of unique clones.
E) Expression levels of 45,037 features from Affymetrix microarray analysis of wild-type heart tissue, compared to the expression levels of the 129 features corresponding to genes with recovered intragenic ssDNA clones, pooled from WT and KO samples. The x-axis is plotted exponentially.
F) Assignment and percentages of all recovered clones from WT and KO heart tissue.
G) ssDNA fragments that mapped to endogenous retroelements were plotted to scale against a generic, consensus sequence of L1 (top) and LTR (bottom) elements. Sense fragments are above each element, and antisense matches are below. For the fragments that mapped to LTRs, only the 5’ mapping is shown to avoid redundancy.
Using this method, we compared the abundance of the DNA recovered from wildt-ype and Trex1-deficient hearts, both of which were on an irf3−/− background to eliminate any confounding contribution of inflammation. The DNA recovered from Trex1 knockout hearts was more abundant and more complex than DNA recovered from wild-type hearts based on three criteria. First, Trex1 knockout PCR products amplified five cycles earlier than wild-type products, suggesting that the knockout heart extracts contained at least 32 times more DNA than the wild-type extracts (Figure 5B). Second, PCR of the wild-type samples amplified more of the nonspecific amplicon, whereas all of the Klenow primer was incorporated into larger products in the knockout samples (Figure 5B). Finally, we found that the wild-type samples had a significantly higher proportion of clones with an endogenous (genome-encoded) poly-dT stretch immediately downstream of the cloned sequence (Figure 5C). These clones were probably derived from contaminating genomic DNA that was primed with the poly-dA primer and copied during the Klenow extension step.
After eliminating duplicate clones and clones with endogenous 3’ poly-dT sequences from analysis, we categorized the inserts of 112 knockout and 76 wild-type clones (Figure 5D, 5F, and Table S1). We noted several features of these DNA fragments that collectively provided clues to the origin and nature of endogenous Trex1 substrates. Interestingly, over a third of the recovered DNA fragments in both wild-type and knockout samples mapped within genes, a dramatic increase over the ~1% contribution of genic sequence to the mass of the mouse genome (Figure 5D). Moreover, these DNA fragments mapped preferentially to genes with abundant expression levels in heart tissue (Figure 5E). The genic DNA fragments corresponded mostly to intronic sequence, and both sense and antisense DNA fragments were recovered. Further analysis may reveal quantitative differences in the recovery of these fragments that may contribute to the ~32-fold increase in DNA purified from knockout hearts (Figure 5B). At present, however, the significance of these genic DNA fragments remains unclear, and the frequency of their recovery was qualitatively similar between wild-type and knockout samples.
Remarkably, DNA fragments derived from endogenous retroelements were abundantly represented among the clones derived from knockout hearts, but not among wild-type clones (Figure 5D, 5F, 5G). Of the 25 retroelement sequences recovered from knockout samples, 12 were derived from L1 retrotransposons, 10 were from Long Terminal Repeat (LTR) endogenous retroviruses, and 3 were SINE elements (Figure 5F). We did not observe any complementary clones (clones derived from both strands of the same DNA fragment). Although we did not sequence to saturation in this analysis, this observation suggests that the recovered DNA fragments were single-stranded. Trex1 is a single-stranded DNA exonuclease, and single-stranded DNA was recently shown to accumulate in the cytosol of Trex1-deficient cells (Yang et al., 2007), suggesting that this technique revealed authentic Trex1 DNA substrates.
We mapped the ssDNA fragments to generic, consensus L1 and LTR retroelement sequences and noted trends that suggested clues about their biogenesis. L1-derived DNA fragments mapped preferentially to the 3’ end of the consensus L1 sequence, which may reflect the abundance of defective, 5’-deleted L1 elements in the mouse genome (Figure 5G; (Ostertag and Kazazian, 2001). We observed an antisense strand bias with the LTR retrotransposon-derived DNA fragments, but not with the L1-mapping DNA (Figure 5G). Taken together, these data suggest that single-stranded DNA fragments derived from endogenous retroelements accumulate within heart cells of Trex1-deficient mice.
Trex1 metabolizes DNA of endogenous retroelements
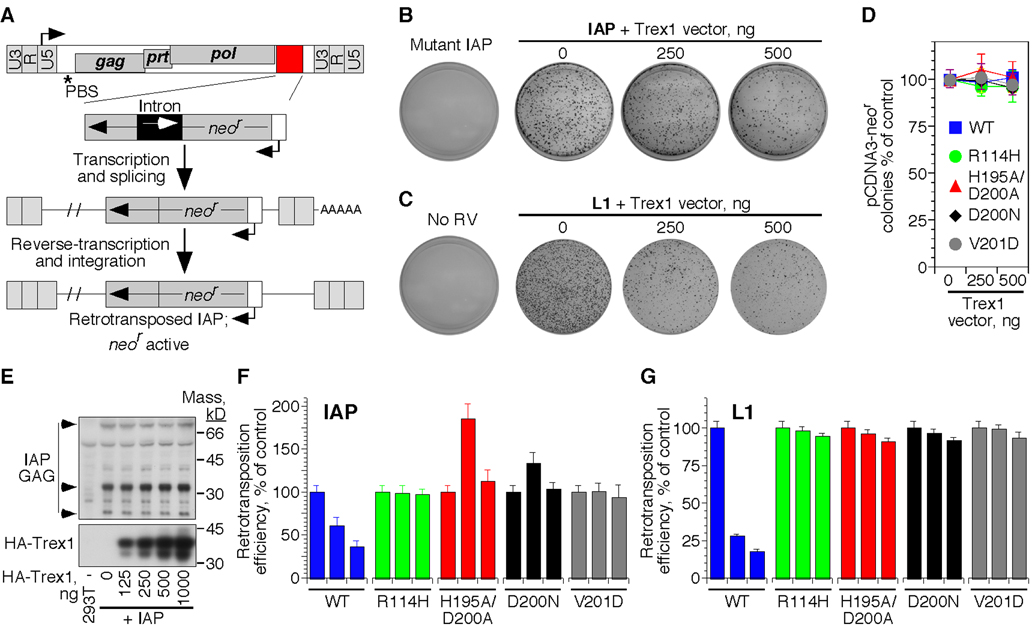
To further examine whether endogenous retroelements are relevant substrates of Trex1, we used two genetically marked retroelements corresponding to the two major classes of DNA fragments recovered from Trex1-deficient hearts: a human L1 element (Esnault et al., 2000) and a LTR-containing murine intracisternal type A particle (IAP; Dewannieux et al., 2004). The marked elements contain a neomycin resistance cassette (neor) in reverse orientation to the retroviral coding sequence; this neor gene is disrupted by a forward-facing intron that precludes its expression from the plasmid (Figure 6A). Following transcription of the marked element into mRNA, splicing out of the intron, reverse transcription, and integration into the host cell genome, the genomic neor gene becomes active (Dewannieux et al., 2004).
Figure 6. Trex1 can metabolize endogenous retroelement DNA.
A) Schematic of the marked IAP element used to measure IAP retrotransposition efficiency. The L1 element was designed with a similar selectable cassette.
B) HeLa cells were transfected with mutant or wild-type marked IAP, alone or together with the indicated amounts of Trex1 expression vector.
C) HeLa cells were transfected with the marked L1 element, alone or with the indicated amounts of Trex1 expression vector.
D) HeLa cells were transfected with pCDNA3 (which encodes plasmid-based neomycin resistance), alone or with the indicated concentrations of each Trex1 expression vector.
E) HEK 293T cells were transfected with the WT IAP, alone or with the indicated concentrations of HA-tagged Trex1. GAG expression and processing 48 hours post transfection was visualized by Western blot.
F) Relative IAP retrotransposition efficiency in the presence of the indicated Trex1 expression vectors. For each Trex1 construct, the left bar is 0 ng, the middle bar is 250 ng, and the right bar is 500 ng of expression vector. Data are mean and SEM of triplicate measurements and are representative of five independent experiments.
G) Relative L1 retrotransposition efficiency was quantitated in the presence of the indicated mutant forms of Trex1. Data are representative of three independent experiments.
HeLa cells transfected with the wild-type IAP plasmid, but not a vector with a nonsense mutation in the GAG open reading frame, formed abundant neomycin-resistant colonies, with each colony representing a single retrotransposition event (Figure 6B; Dewannieux et al., 2004). When we co-transfected an expression vector for Trex1, we observed a dramatic reduction in IAP retrotransposition efficiency to less than 40 percent of that in control cells (Figure 6B). Similarly, Trex1 reduced L1 retrotransposition efficiency by over 80% (Figure 6C). In both cases, the blockade was specific for the retroelement, as ectopic Trex1 expression had no effect on conventional, plasmid-based neomycin resistance at these concentrations (Figure 6D). Consistent with the specificity of Trex1 enzymes for retroviral DNA but not for plasmid DNA, Trex1 had no measurable effect on expression of the IAP GAG protein or protease-dependent processing of GAG (Figure 6E).
We next examined the effect of AGS-causing Trex1 mutations on IAP and L1 retrotransposition. In the IAP assays, we observed distinct effects of the different Trex1 mutations on retrotransposition that correlated with the recessive or dominant nature of each disease-causing allele. Trex1 R114H and Trex1 V201D, which cause recessive AGS, had no effect on IAP or L1 retrotransposition, consistent with the loss of function observed for these mutants in exonuclease assays (Figure 6F; Crow et al., 2006a). In contrast, co-transfection of the D200N catalytic mutant that causes autosomal dominant AGS or a similar H195A/D200A mutant resulted in a substantial increase in IAP retrotransposition efficiency over that observed in control cells (Figure 6F; Rice et al., 2007a). Trex1 functions as a dimer (Mazur and Perrino, 2001), suggesting that the increase in IAP retrotransposition may reflect inhibition of endogenous Trex1 by the catalytic mutants. We did not observe an enhancement of retratransposition efficiency by the catalytic mutant forms of Trex1 with the L1 element, perhaps reflecting distinct cellular locations of reverse transcription for LTR and L1 retrotransposons or different thresholds for inhibition (Figure 6G). Collectively, these findings suggest that Trex1 specifically targets reverse-transcribed endogenous retroviral DNA, revealing a potential connection between endogenous retroelements, the ISD pathway, and autoimmune pathology.
DISCUSSION
Nucleic acid recognition is an essential innate immune strategy for detecting viral infection, yet its very nature raises fundamental questions of self/non-self discrimination. We describe here a new mechanism of autoimmunity wherein defective metabolism of intracellular nucleic acids activates a cell-intrinsic autoimmune response. Specifically, we identify Trex1 as an ISD-inducible, negative regulator of the ISD response. We genetically define a pathway linking excessive activation of the ISD response to autoantibody production and lethal autoimmunity, thus providing mechanistic insight into the underlying cause of AGS in humans, with relevance to other diseases associated with Trex1 mutations. Finally, we identify potential Trex1 substrates by showing that fragments of endogenous retroelement DNA accumulate in the hearts of Trex1-deficient mice and that Trex1 can metabolize reverse-transcribed DNA of these elements.
Recently, another group reported cell cycle abnormalities and chronic activation of DNA damage checkpoint signaling in cell lines derived from Trex1-deficient mice and from human AGS patients, raising the possibility that these defects may be linked to the diseases caused by loss of Trex1 (Yang et al., 2007). We did not observe these abnormalities – slower growth in culture, chronic activation of p53, and upregulation of p21 – in Trex1-deficient primary fibroblasts (Figure S1A, S1B). Activation of the ISD pathway occurs independently of DNA damage sensors, including ATM, DNA-PK, p53, and ATR (Stetson and Medzhitov, 2006a), and we did not find any evidence for DNA damage-inducible gene transcription in our microarray analysis of Trex1/RAG2 DKO heart tissue (Figure 3C). Finally, it is unclear how chronic checkpoint signaling and cell cycle defects in Trex1-deficient cells would be corrected by genetic ablation of IRF3, IFNαR1, or RAG2. Together, these findings indicate that dysregulated activation of the ISD pathway, rather than chronic checkpoint activation, is the principal cause of autoimmunity in Trex1-deficient mice. It is possible, however, that the ISD pathway may be linked to the cell cycle as part of the antiviral response, perhaps through IFN-dependent induction of p53 (Takaoka et al., 2003).
More broadly, our findings establish that cytosolic nucleic acid sensors can initiate autoimmunity if improperly controlled. It seems that promiscuity is a general property of antiviral nucleic acid sensors; additional mechanisms that limit access of ligands to these sensors impose a higher threshold for activation that cannot be designed into the sensors themselves. Trex1 likely represents only one example of such threshold-setting mechanisms for cytosolic nucleic acid sensors. Extending this logic, we propose that analogous pathways might limit the activation of RIG-I and MDA5 by self-RNA, and that breakdown of these mechanisms would result in a similar, cell-intrinsic autoimmune response.
Importantly, the cell-intrinsic mechanism we describe here is distinct from the two other, well-established mechanisms that cause autoimmunity. First, defects in central or peripheral lymphocyte tolerance can lead to the pathological accumulation of self-reactive lymphocytes that cause disease (Goodnow, 2007); in this case, an innate immune contribution may be unnecessary (Gray et al., 2007). Second, activation of non-cell autonomous innate immune recognition through TLRs clearly contributes to numerous autoimmune syndromes (Marshak-Rothstein and Rifkin, 2007). Cell-intrinsic initiation of autoimmunity therefore defines a third class, with distinct requirements for regulation and unique mechanisms that precipitate lymphocyte-dependent autoimmunity.
We demonstrate that the ISD pathway is linked to autoantibody production in Trex1-deficient mice. Current models of autoreactive B cell activation focus on the contribution of TLR signaling to autoantibody production (Marshak-Rothstein and Rifkin, 2007). Whether the autoantibody response we document here is fully independent of TLRs will require further exploration. One interesting possibility is that cell-intrinsic initiation of autoimmunity by the ISD pathway in Trex1-deficient mice precedes a TLR-dependent contribution to autoantibody production. For example, the ISD ligands that accumulate inside a Trex1-deficient cell undergoing apoptosis could become TLR9 ligands after phagocytosis of that cell by a macrophage or dendritic cell, akin to what was recently shown for viral RNA (Schulz et al., 2005).
Another key to understanding the pathogenesis of AGS and related diseases is the nature of the Trex1 substrates that accumulate in its absence. We provide evidence that endogenous retroviral DNA is a relevant Trex1 substrate. Species- and tissue-specific expression of endogenous retroelements may explain both the highly stereotyped symptoms of Trex1 deficiency and the fact that these symptoms are different in humans and mice. A key question that remains is how Trex1 metabolizes DNA that is formed by such distinct reverse transcription mechanisms: LTR retrotransposons probably undergo reverse transcription in the cytosol, whereas reverse transcription of L1 mRNA is thought to occur in the nucleus (Ostertag and Kazazian, 2001). One possibility is that Trex1 is recruited to reverse-transcribed DNA regardless of its location. Notably, loss of function mutations in the genes encoding the human RNase H2 enzyme also cause AGS (Crow et al., 2006b), suggesting that RNase H2 may be involved in metabolism of retroviral RNA-DNA hybrids. Others have observed regulated nuclear translocation of Trex1 (Chowdhury et al., 2006; Yang et al., 2007), although we did not see this after ISD stimulation (data not shown).
The identification of Trex1 substrates as ISD ligands has interesting implications for the mechanics of the ISD pathway. Typically, double-stranded DNA is used to experimentally activate the ISD response. However, Trex1 is a single-stranded DNA exonuclease (Mazur and Perrino, 2001), and single-stranded DNA oligonucleotides efficiently activate the ISD pathway (data not shown), suggesting that ISD sensing may involve processing of double-stranded DNA.
More generally, our results may reconcile a long-standing debate over the proposed contribution of endogenous retroviruses to autoimmunity (Colmegna and Garry, 2006; Lower, 1999; Stoye, 1999). This debate focuses on three proposed mechanisms: disruption of an important gene by de novo transposition, activation of T cells by retroviral antigens, or immunomodulatory properties of the retroviral proteins themselves. We suggest that accumulation of reverse-transcribed DNA, rather than any inherent activity of the retroviral proteins, is the key factor that determines the involvement of endogenous retroviruses in autoimmunity. Importantly, neither the coding specificity of the retroviral DNA nor its competence for retrotransposition is relevant for detection by the ISD pathway, suggesting that even defective retroelements can cause autoimmunity if their reverse-transcribed DNA fails to be properly metabolized. Indeed, of the retroelement-derived ssDNA fragments that we could definitively map to a single “parent” location, all were derived from defective retrotransposons (Table S1 and data not shown).
Finally, our findings suggest that the ISD pathway comprises a hitherto unknown cell-intrinsic retroviral detection system. In the case of AGS, the ISD pathway may be inappropriately triggered by endogenous retroviral DNA, but it is likely that infectious retroviruses trigger the same response. We found that treatment of Trex1-deficient mice with the reverse transcriptase (RT) inhibitor AZT did not rescue from lethality (Supplementary Figure 2). However, the mouse genome contains hundreds of diverse RT enzymes, only a fraction of which is susceptible to inhibition by AZT, and it is unclear which of them might be responsible for generating Trex1 substrates. Detection of reverse-transcribed DNA may be relevant to other cellular abnormalities, including genotoxic stress and oncogenic transformation, two conditions known to activate expression of endogenous retroelements (Nelson et al., 2003; Rudin and Thompson, 2001). It is intriguing to note that the ISD pathway (that is, the antiviral response to natural, phosphodiester DNA) is absent from every transformed cell line we have examined (data not shown), suggesting that a functional ISD response may be incompatible with outgrowth of immortalized cells.
In conclusion, we define Trex1 as a negative regulator of the ISD response and identify endogenous Trex1 substrates. Just as commensal bacteria outnumber our own cells by four or five orders of magnitude, endogenous retroelements outnumber our genes by at least 100-fold. Both have the potential to be detected by the immune system and cause autoimmune disease. Therefore, specific mechanisms evolved to prevent this, and Trex1 represents the first example of a mechanism to prevent autoimmunity caused by endogenous retroelements. The cell-intrinsic initiation of autoimmunity described here provides a model for integrating this mechanism into other established models of autoimmunity.
EXPERIMENTAL PROCEDURES
Mice and reagents
C57Bl/6 mice mice were purchased from Jackson Laboratories (Bar Harbor, ME). Trex1-deficient mice and immortalized cell lines (Morita et al., 2004) were generously provided by Dr. Deborah Barnes and Dr. Tomas Lindahl (Cancer Research UK). Irf3−/− mice (Sato et al., 2000) were kindly provided by Dr. Tadatsugu Taniguchi (University of Tokyo). RAG2-deficient mice were purchased from Jackson Laboratories (Bar Harbor, ME), and IFNαR1-deficient mice on a C57Bl/6 background were a gift from Warren Shlomchik (Yale University). The marked IAP (DJ33-440N1) and L1 elements were generated as previously described (Dewannieux et al., 2004; Esnault et al., 2000). All murine Trex1 cDNAs were cloned, mutated, and expressed in pCMV-HA (Clontech) using conventional techniques. Antisera to Trex1 (BD Biosciences), hemagluttinin (HA) epitope (Cell Signaling Technology), V5 (Invitrogen), calnexin (kindly provided by Dr. Peter Cresswell), and the IAP GAG (generously provided by Drs. Hal Bogerd and Bryan Cullen) were used for western blots and immunofluorescence.
ISD in vivo pulldowns
~109 bone marrow-derived macrophages were pretreated with chloroquine (50µM) to neutralize lysosomal pH and then transfected with 25 µg per plate of BrdU-substituted, biotinylated 85 base pair ISD complexed with Lipofectamine 2000 (Invitrogen; ISD sequence available upon request). 90 minutes later, the plates were exposed to ultraviolet light for five minutes in a Stratalinker 1800 (Stratagene). Cells were lysed in 1% SDS, 50mM Tris pH 8.0, 10mM EDTA supplemented with Complete protease inhibitors (Roche). Lysates were diluted in 1% Triton X-100, 150mM NaCl, 50mM Tris pH8.0, and rotated overnight with streptavidin-M280 Dynabeads (Dynal Biotech). Washes were done in the above buffer with either 150mM or 500mM NaCl, followed by a wash in 1% SDS, 1.5M NaCl. Protein/DNA complexes were boiled off the beads, separated using gradient SDS-PAGE, and visualized with silver staining for resolution or coomassie blue for mass spec preparative gels. Protein bands were excised and identified at the Stanford PAN facility.
Sucrose gradients
293T cells (2.5×106 per 10 cm dish) were transfected with 2 µg of the indicated expression vectors and harvested 48 hours later into perm buffer (150mM NaCl, 50mM HEPES, Complete protease inhibitors (Roche) with 25 µg/ml digitonin). Cells were mechanically disrupted by four passages through a 27-gauge needle, and extracts were cleared three times at 800g. 1.1 ml of cleared extracts was layered onto 10.8 ml 10–60% sucrose gradients, centrifuged in a SW-41 rotor at 40,000 rpm for 2 hours at 4°C, and fractionated before separation by SDS-PAGE and western blotting.
Microarray analysis
Heart tissue from three pairs of male Trex1/RAG2 DKO mice and RAG2 KO littermates, aged 18, 19, and 28 weeks, was homogenized in RNA-Bee (TelTest) using a PowerGen 700 (Fisher). RNA preparation was done according to manufacturer’s instructions, and microarray hybridizations to Affymetrix Mouse Gene Chip 430 2.0 were performed at the W.M. Keck facility at Yale. Features with >2-fold increased expression in all three pairwise comparisons were presented in Figure 3C. All microarray data were deposited in the GEO database (accession #GSE11698).
Detection of autoantibodies
Trex1/RAG2 DKO hearts were fixed in 4% paraformaldehyde, cryoprotected with 30% sucrose in PBS, embedded in OCT medium and frozen. 7 µM sections were cut on a cryostat, rehydrated, blocked with 5% BSA, 0.1% Tween-20 in PBS, and incubated overnight at 4°C with sera from mice of the indicated age and genotype, diluted 1:40 in blocking buffer. Bound IgG antibodies were detected with Alexa 488-conjugated anti-mouse IgG (Molecular Probes), and sections were counterstained for 5 minutes with DAPI (10µg/ml) before mounting in Vectashield media (Molecular Probes). Hearts were homogenized using a PowerGen 700 in 1.5 ml buffer (150 mM NaCl, 50 mM HEPES, 10% glycerol, 1 mM DTT, 2 mM EDTA, Complete protease inhibitors). Triton X-100 was then added to 1%, and extracts were left on ice for 15 minutes with occasional vortexing. Extracts cleared at 16,000g were separated by SDS-PAGE, transferred to PVDF membranes, and incubated overnight at 4°C with 1:500 dilutions of sera. Autoantibodies were detected with horseradish peroxidase-conjugated anti-mouse IgG (Jackson Immunoresearch).
Trex1 substrate isolation and identification
Pooled hearts from Trex1/IRF3 DKO mice or IRF3 KO controls were homogenized in gentle extraction buffer containing 50mM HEPES, 150mM NaCl, 10% glycerol, 200µM digitonin, 1mM DTT, 2mM EDTA, and Complete protease inhibitors. Cleared extracts were treated with 1 mg/ml proteinase K (Ambion) for one hour at 55°C, extracted with phenol:chloroform, treated with RNase A (50 µg/ml) and RNase T1 (10 U/µl; Ambion) for 1.5 hours at 37°C, extracted sequentially with phenol:chloroform and then chloroform, and then precipitated, washed, and resuspended in water. Recovered material was tailed with TdT (NEB) and 92µM dTTP/8µM ddCTP in TdT buffer with 0.75mM µCoCl2 for 25 minutes at 37°C according to a published method (Liu et al., 2003). The tailed DNA was melted, primed with a poly-dA primer containing a unique 5’ sequence tag for subsequent amplification, and copied with Klenow polymerase (NEB) and dNTPs for 90 minutes at 37°C. The copied DNA was then tailed with TdT and 96µM dCTP/4µM ddCTP for 30 minutes at 37°C. PCR amplification of the tailed DNA was performed with restriction site-containing primers complementary to the poly-dC tail and to the unique tag upstream of the poly-dA stretch, and products were directionally cloned into pCDNA3. Clones were sequenced at the WM Keck facility and at Agencourt Biosciences. Insert sequences were mapped to the mouse genome using BLAST in NCBI and Ensembl, and retroelement sequences were aligned to consensus elements using Repbase and RepeatMasker.
Retrotransposition assays
5×105 HeLa cells in 60 mm dishes were transfected with 1.5 µg of the neor-marked retroviral plasmid or pCNDA3, together with the indicated expression vectors, using FuGene 6 (Roche). Three days later, cells were harvested, counted, and plated at a density of 5×105 cells per 10 cm dish. The following day, cells were put under G418 selection (0.5 mg/ml, Invitrogen). Colonies were visualized, typically a week after selection, by crystal violet staining as described (Dewannieux et al., 2004).
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Dick Winant and the Stanford PAN facility for expert mass spectrometry analysis; to Deborah Barnes and Tomas Lindahl for Trex1-deficient mice and cell lines; to Tadatsugu Taniguchi for IRF3-deficient mice; to Hal Bogerd and Bryan Cullen for IAP GAG antisera; to the WM Keck facility for microarray analysis; to Sophie Holley and Chuck Annicelli for animal care; to Arun Unni, Dominik Schenten, Noah Palm, Igor Brodsky, and Elizabeth Prescott for critical review of the manuscript; and to all members of the Medzhitov lab for insightful discussions. D.B.S. was supported by a Cancer Research Institute postdoctoral fellowship and a NIH Pathway to Independence award (1K99AI072945-01A1). R.M. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alarcon-Riquelme ME. Nucleic acid by-products and chronic inflammation. Nat Genet. 2006;38:866–867. doi: 10.1038/ng0806-866. [DOI] [PubMed] [Google Scholar]
- Chowdhury D, Beresford PJ, Zhu P, Zhang D, Sung JS, Demple B, Perrino FW, Lieberman J. The exonuclease TREX1 is in the SET complex and acts in concert with NM23-H1 to degrade DNA during granzyme A-mediated cell death. Mol Cell. 2006;23:133–142. doi: 10.1016/j.molcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Christensen SR, Shupe J, Nickerson K, Kashgarian M, Flavell RA, Shlomchik MJ. Toll-like receptor 7 and TLR9 dictate autoantibody specificity and have opposing inflammatory and regulatory roles in a murine model of lupus. Immunity. 2006;25:417–428. doi: 10.1016/j.immuni.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Colmegna I, Garry RF. Role of endogenous retroviruses in autoimmune diseases. Infect Dis Clin North Am. 2006;20:913–929. doi: 10.1016/j.idc.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Black DN, Ali M, Bond J, Jackson AP, Lefson M, Michaud J, Roberts E, Stephenson JB, Woods CG, Lebon P. Cree encephalitis is allelic with Aicardi-Goutieres syndrome: implications for the pathogenesis of disorders of interferon alpha metabolism. J Med Genet. 2003;40:183–187. doi: 10.1136/jmg.40.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow YJ, Hayward BE, Parmar R, Robins P, Leitch A, Ali M, Black DN, van Bokhoven H, Brunner HG, Hamel BC, et al. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nat Genet. 2006a;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- Crow YJ, Leitch A, Hayward BE, Garner A, Parmar R, Griffith E, Ali M, Semple C, Aicardi J, Babul-Hirji R, et al. Mutations in genes encoding ribonuclease H2 subunits cause Aicardi-Goutieres syndrome and mimic congenital viral brain infection. Nat Genet. 2006b;38:910–916. doi: 10.1038/ng1842. [DOI] [PubMed] [Google Scholar]
- Dewannieux M, Dupressoir A, Harper F, Pierron G, Heidmann T. Identification of autonomous IAP LTR retrotransposons mobile in mammalian cells. Nat Genet. 2004;36:534–539. doi: 10.1038/ng1353. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Goodnow CC. Multistep pathogenesis of autoimmune disease. Cell. 2007;130:25–35. doi: 10.1016/j.cell.2007.06.033. [DOI] [PubMed] [Google Scholar]
- Gray DH, Gavanescu I, Benoist C, Mathis D. Danger-free autoimmune disease in Aire-deficient mice. Proc Natl Acad Sci U S A. 2007;104:18193–18198. doi: 10.1073/pnas.0709160104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434:772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- Hoss M, Robins P, Naven TJ, Pappin DJ, Sgouros J, Lindahl T. A human DNA editing enzyme homologous to the Escherichia coli DnaQ/MutD protein. Embo J. 1999;18:3868–3875. doi: 10.1093/emboj/18.13.3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii KJ, Coban C, Kato H, Takahashi K, Torii Y, Takeshita F, Ludwig H, Sutter G, Suzuki K, Hemmi H, et al. A Toll-like receptor-independent antiviral response induced by double-stranded B-form DNA. Nat Immunol. 2006;7:40–48. doi: 10.1038/ni1282. [DOI] [PubMed] [Google Scholar]
- Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue J, Uematsu S, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- Koyama S, Ishii KJ, Kumar H, Tanimoto T, Coban C, Uematsu S, Kawai T, Akira S. Differential role of TLR- and RLR-signaling in the immune responses to influenza A virus infection and vaccination. J Immunol. 2007;179:4711–4720. doi: 10.4049/jimmunol.179.7.4711. [DOI] [PubMed] [Google Scholar]
- Kumar H, Kawai T, Kato H, Sato S, Takahashi K, Coban C, Yamamoto M, Uematsu S, Ishii KJ, Takeuchi O, Akira S. Essential role of IPS-1 in innate immune responses against RNA viruses. J Exp Med. 2006;203(7):1795–1803. doi: 10.1084/jem.20060792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, et al. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature. 2007;449:564–569. doi: 10.1038/nature06116. [DOI] [PubMed] [Google Scholar]
- Leadbetter EA, Rifkin IR, Hohlbaum AM, Beaudette BC, Shlomchik MJ, Marshak-Rothstein A. Chromatin-IgG complexes activate B cells by dual engagement of IgM and Toll-like receptors. Nature. 2002;416:603–607. doi: 10.1038/416603a. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Chowdhury D, Harvey S, Gong M, Senenko L, Engel K, Pfeiffer C, Hollis T, Gahr M, Perrino FW, et al. A mutation in TREX1 that impairs susceptibility to granzyme A-mediated cell death underlies familial chilblain lupus. J Mol Med. 2007a;85(5):531–537. doi: 10.1007/s00109-007-0199-9. [DOI] [PubMed] [Google Scholar]
- Lee-Kirsch MA, Gong M, Chowdhury D, Senenko L, Engel K, Lee YA, de Silva U, Bailey SL, Witte T, Vyse TJ, et al. Mutations in the gene encoding the 3'-5' DNA exonuclease TREX1 are associated with systemic lupus erythematosus. Nat Genet. 2007b;39:1065–1067. doi: 10.1038/ng2091. [DOI] [PubMed] [Google Scholar]
- Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J Biol Chem. 1969;244:5014–5019. [PubMed] [Google Scholar]
- Liu CL, Schreiber SL, Bernstein BE. Development and validation of a T7 based linear amplification for genomic DNA. BMC Genomics. 2003;4:19. doi: 10.1186/1471-2164-4-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lower R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. 1999;7:350–356. doi: 10.1016/s0966-842x(99)01565-6. [DOI] [PubMed] [Google Scholar]
- Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu Rev Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- Martin DA, Elkon KB. Intracellular mammalian DNA stimulates myeloid dendritic cells to produce type I interferons predominantly through a toll-like receptor 9-independent pathway. Arthritis Rheum. 2006;54:951–962. doi: 10.1002/art.21677. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3'-->5' exonucleases. J Biol Chem. 1999;274:19655–19660. doi: 10.1074/jbc.274.28.19655. [DOI] [PubMed] [Google Scholar]
- Mazur DJ, Perrino FW. Excision of 3' termini by the Trex1 and TREX2 3'-->5' exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276:17022–17029. doi: 10.1074/jbc.M100623200. [DOI] [PubMed] [Google Scholar]
- Morita M, Stamp G, Robins P, Dulic A, Rosewell I, Hrivnak G, Daly G, Lindahl T, Barnes DE. Gene-targeted mice lacking the Trex1 (DNase III) 3'-->5' DNA exonuclease develop inflammatory myocarditis. Mol Cell Biol. 2004;24:6719–6727. doi: 10.1128/MCB.24.15.6719-6727.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napirei M, Karsunky H, Zevnik B, Stephan H, Mannherz HG, Moroy T. Features of systemic lupus erythematosus in Dnase1-deficient mice. Nat Genet. 2000;25:177–181. doi: 10.1038/76032. [DOI] [PubMed] [Google Scholar]
- Nelson PN, Carnegie PR, Martin J, Davari Ejtehadi H, Hooley P, Roden D, Rowland-Jones S, Warren P, Astley J, Murray PG. Demystified. Human endogenous retroviruses. Mol Pathol. 2003;56:11–18. doi: 10.1136/mp.56.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe Y, Kawane K, Akira S, Taniguchi T, Nagata S. Toll-like receptor-independent gene induction program activated by mammalian DNA escaped from apoptotic DNA degradation. J Exp Med. 2005;202:1333–1339. doi: 10.1084/jem.20051654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH., Jr Biology of mammalian L1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol. 2006;18:676–682. doi: 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C. Innate recognition of viruses. Immunity. 2007;27:370–383. doi: 10.1016/j.immuni.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Pisitkun P, Deane JA, Difilippantonio MJ, Tarasenko T, Satterthwaite AB, Bolland S. Autoreactive B cell responses to RNA-related antigens due to TLR7 gene duplication. Science. 2006;312:1669–1672. doi: 10.1126/science.1124978. [DOI] [PubMed] [Google Scholar]
- Rice G, Newman WG, Dean J, Patrick T, Parmar R, Flintoff K, Robins P, Harvey S, Hollis T, O'Hara A, et al. Heterozygous mutations in TREX1 cause familial chilblain lupus and dominant Aicardi-Goutieres syndrome. Am J Hum Genet. 2007a;80:811–815. doi: 10.1086/513443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice G, Patrick T, Parmar R, Taylor CF, Aeby A, Aicardi J, Artuch R, Montalto SA, Bacino CA, Barroso B, et al. Clinical and molecular phenotype of Aicardi-Goutieres syndrome. Am J Hum Genet. 2007b;81:713–725. doi: 10.1086/521373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards A, van den Maagdenberg AM, Jen JC, Kavanagh D, Bertram P, Spitzer D, Liszewski MK, Barilla-Labarca ML, Terwindt GM, Kasai Y, et al. C-terminal truncations in human 3'-5' DNA exonuclease TREX1 cause autosomal dominant retinal vasculopathy with cerebral leukodystrophy. Nat Genet. 2007;39:1068–1070. doi: 10.1038/ng2082. [DOI] [PubMed] [Google Scholar]
- Rudin CM, Thompson CB. Transcriptional activation of short interspersed elements by DNA-damaging agents. Genes Chromosomes Cancer. 2001;30:64–71. [PubMed] [Google Scholar]
- Sanchis A, Cervero L, Bataller A, Tortajada JL, Huguet J, Crow YJ, Ali M, Higuet LJ, Martinez-Frias ML. Genetic syndromes mimic congenital infections. J Pediatr. 2005;146:701–705. doi: 10.1016/j.jpeds.2005.01.033. [DOI] [PubMed] [Google Scholar]
- Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- Schulz O, Diebold SS, Chen M, Naslund TI, Nolte MA, Alexopoulou L, Azuma YT, Flavell RA, Liljestrom P, Reis e Sousa C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- Shevelev IV, Hubscher U. The 3' 5' exonucleases. Nat Rev Mol Cell Biol. 2002;3:364–376. doi: 10.1038/nrm804. [DOI] [PubMed] [Google Scholar]
- Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Recognition of cytosolic DNA activates an IRF3-dependent innate immune response. Immunity. 2006a;24:93–103. doi: 10.1016/j.immuni.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006b;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Stoye JP. The pathogenic potential of endogenous retroviruses: a skeptical view. Trends Microbiol. 1999;7:430. doi: 10.1016/s0966-842x(99)01616-9. author reply 431–432. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Tus K, Li QZ, Wang A, Tian XH, Zhou J, Liang C, Bartov G, McDaniel LD, Zhou XJ, et al. A Tlr7 translocation accelerates systemic autoimmunity in murine lupus. Proc Natl Acad Sci U S A. 2006;103:9970–9975. doi: 10.1073/pnas.0603912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Sun L, Liu HH, Chen X, Seth RB, Forman J, Chen ZJ. The specific and essential role of MAVS in antiviral innate immune responses. Immunity. 2006;24:633–642. doi: 10.1016/j.immuni.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Hayakawa S, Yanai H, Stoiber D, Negishi H, Kikuchi H, Sasaki S, Imai K, Shibue T, Honda K, Taniguchi T. Integration of interferon-alpha/beta signalling to p53 responses in tumour suppression and antiviral defence. Nature. 2003;424:516–523. doi: 10.1038/nature01850. [DOI] [PubMed] [Google Scholar]
- Takaoka A, Wang Z, Choi MK, Yanai H, Negishi H, Ban T, Lu Y, Miyagishi M, Kodama T, Honda K, et al. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- Theofilopoulos AN, Baccala R, Beutler B, Kono DH. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- Yang YG, Lindahl T, Barnes DE. Trex1 Exonuclease Degrades ssDNA to Prevent Chronic Checkpoint Activation and Autoimmune Disease. Cell. 2007;131:873–886. doi: 10.1016/j.cell.2007.10.017. [DOI] [PubMed] [Google Scholar]
- Yasutomo K, Horiuchi T, Kagami S, Tsukamoto H, Hashimura C, Urushihara M, Kuroda Y. Mutation of DNASE1 in people with systemic lupus erythematosus. Nat Genet. 2001;28:313–314. doi: 10.1038/91070. [DOI] [PubMed] [Google Scholar]
- Yoshida H, Okabe Y, Kawane K, Fukuyama H, Nagata S. Lethal anemia caused by interferon-beta produced in mouse embryos carrying undigested DNA. Nat Immunol. 2005;6:49–56. doi: 10.1038/ni1146. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.