Abstract
Human leukocyte antigen (HLA) gene polymorphism plays a critical role in protective immunity, disease susceptibility, autoimmunity, and drug hypersensitivity, yet the basis of how HLA polymorphism influences T cell receptor (TCR) recognition is unclear. We examined how a natural micropolymorphism in HLA-B44, an important and large HLA allelic family, affected antigen recognition. T cell–mediated immunity to an Epstein-Barr virus determinant (EENLLDFVRF) is enhanced when HLA-B*4405 was the presenting allotype compared with HLA-B*4402 or HLA-B*4403, each of which differ by just one amino acid. The micropolymorphism in these HLA-B44 allotypes altered the mode of binding and dynamics of the bound viral epitope. The structure of the TCR–HLA-B*4405EENLLDFVRF complex revealed that peptide flexibility was a critical parameter in enabling preferential engagement with HLA-B*4405 in comparison to HLA-B*4402/03. Accordingly, major histocompatibility complex (MHC) polymorphism can alter the dynamics of the peptide-MHC landscape, resulting in fine-tuning of T cell responses between closely related allotypes.
MHC class I (MHC-I) molecules—termed HLA-A, HLA-B, and HLA-C in humans and H2-K, H2-D, and H2-L in mice—are highly polymorphic molecules that function to present antigenic peptides to the αβ TCR expressed by CTLs. Maintenance of HLA polymorphism reflects natural selection for enhanced protective immunity against the pathogens encountered by humans (1). The MHC molecules can differ from each other by either >30 amino acids or only a few amino acids (micropolymorphism). This MHC-I polymorphism is generally concentrated in the antigen-binding cleft, controlling the size and diversity of the peptide repertoire presented by each HLA molecule. For instance, HLA-B44 molecules will preferentially bind peptides that possess a P2-glutamate (2). Alleles from each of the HLA families are distributed in virtually all human populations, indicating that their maintenance reflects an adaptable strategy for protective immunity toward diverse microbial ligands. Although it is recognized that HLA polymorphism can alter the peptide-MHC (pMHC) landscape, it remains unclear at the atomic level how HLA polymorphism directly affects TCR recognition.
Despite the multitude of potential pMHC landscapes, the TCR–pMHC interaction achieves remarkable specificity, the underlying basis of which has been addressed in part by several structural and biophysical studies on the TCR–pMHC interaction (3–5). For example, a consensus-docking mode between the TCR and the pMHC is preserved in which the Vα and Vβ domains are positioned over the α2 and α1 helices, respectively. Moreover, a recurrent theme in the interactions between MHC molecules and Vβ8.2-TCRs has been observed (6, 7). More globally, it is apparent that three positions (65, 69, and 155 in MHC-I) are invariably contacted by the TCR and, thus, may represent a minimal MHC restriction element (8). However, in the mouse-based systems, the structures of TCRs ligated to H2-Kb and H2-Ld MHC molecules have only been reported, and in human-based MHC-I systems, TCR co-complex structures have been limited to HLA-A2, HLA-B8, and HLA-B35 (3, 5). Accordingly, the potential diversity as well as the possible generalities of the TCR–pMHC interaction may be under- or overestimated, respectively, by the limited number of distinct HLA allotypes that have been studied. Although the H2-Kb mutant system has to an extent unraveled the impact of micropolymorphism on MHC structure and function (9, 10), it is nevertheless important to study the impact of naturally selected microvariation in HLA allotypes found in outbred human populations.
The HLA-B44 family, which comprises 60 alleles, participates in important antiviral, antitumor, and minor antigen-specific responses. Within this large HLA supertype, HLA-B*4402 is the predominant HLA-B44 allele in most Caucasian populations, HLA-B*4403 is the more common allele in African and Asian populations, and HLA-B*4405 is a relatively rare allotype. Although these three HLA-B44 allotypes differ only at two positions, 116 and 156 (HLA-B*4405: 116-Tyr, 156-Asp; HLA-B*4402: 116-Asp, 156-Asp; HLA-B*4403: 116-Asp, 156-Leu), both of which are buried within the antigen-binding cleft, the impact of these micropolymorphisms on antigen presentation and TCR recognition is marked (11, 12). For example, mismatching of HLA-B*4402 and HLA-B*4403 is associated with transplant rejection and acute graft-versus-host disease, indicating that they represent significant barriers to clinical transplantation (13). In addition, although HLA-B*4402 and HLA-B*4403 are dependent on tapasin for the loading of their peptide cargo, HLA-B*4405 is tapasin independent, and this feature engenders this allotype with a reduced susceptibility to viral evasion of immune responses (12).
HLA-B44+ individuals recognize an immunodominant determinant (EENLLDFVRF, abbreviated to EENL) from the EBV nuclear antigen 6 (EBNA6, also called EBNA3C) (14). This viral epitope is presented by HLA-B*4402, HLA-B*4403, and HLA-B*4405, but there is a distinct hierarchy in the CTL responses to these pMHC complexes. Namely, HLA-B*4405+ individuals respond most strongly to this determinant, and CTLs preferentially lyse target cells presenting HLA-B*4405EENL in comparison to HLA-B*4402EENL and HLA-B*4403EENL targets (14). Moreover, this preferential recognition of HLA-B*4405EENL target cells is observed irrespective of the HLA-B44 subtype of the CTLs, or whether the EENL epitope is exogenously added or endogenously processed (14). Interestingly, one such prototypical HLA-B*4405EENL–restricted CTL, termed DM1 (TRAV26-1/TRBV7-9), was observed to persist in an EBV-infected individual for at least 11 yr, suggesting that such CTL clonotypes are important factors in controlling such persistent viral infections (15).
In this paper, we have examined how this DM1 TCR interacts with the EENL determinant when bound to the three HLA-B44 allotypes. These findings provide insight into the complex interactions controlling HLA-B44 restriction and reveal how micropolymorphisms can affect epitope conformation that subsequently fine tunes the responding CTL repertoire.
RESULTS
The DM1 TCR exhibits differential affinity for the HLA-B44 allotypes
Given that CTL clones isolated from HLA-B44+ individuals lyse target cells presenting HLA-B*4405EENL better than the HLA-B*4402 and HLA-B*4403 counterparts, we sought to gain an understanding of whether this reflected a difference in the affinity of the interaction between the TCR and the HLA-B44 allotypes. A previous analysis of HLA-B*4405EENL tetramer–sorted cells from the HLA-B*4405+ donor DM revealed that a single clonotype (termed DM1) dominated the EENL-specific repertoire in this donor (15). Furthermore, this clonotype remained dominant in this donor for an extended period, with “nucleotide-identical” TCRs dominating the EENL-specific repertoire in blood samples taken in 1994 and 2005. Accordingly, we chose the DM1 TCR for our analysis, which is encoded by the TRAV26-1*02 TRAJ13*02, TRBV7-9*01 TRBJ2-1*01, and TRBD1*01 genes.
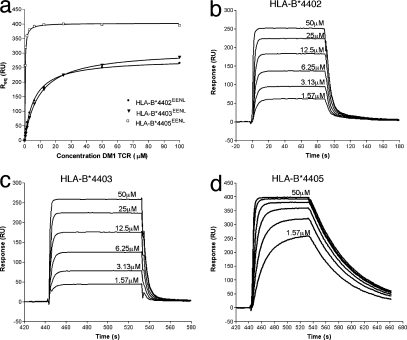
First, we expressed, refolded, and purified the DM1 TCR (see Materials and methods) (16) and showed that it interacted with anti-TCR mAbs (unpublished data). Next, we conducted surface plasmon resonance (SPR) studies between the DM1 TCR and the refolded and purified HLA-B*4405EENL, HLA-B*4403EENL, and HLA-B*4402EENL allotypes in which the pMHC was coupled to the sensor chip. The DM1 TCR bound to HLA-B*4405EENL with a relatively high affinity of 0.3 ± 0.1 μM (Fig. 1 and Table I), whereas the DM1 TCR bound to HLA-B*4402EENL and HLA-B*4403EENL with an affinity of 6.3 ± 0.3 μM and 9.4 ± 0.2 μM, respectively (Fig. 1 and Table I). Interestingly, the binding kinetics of the interaction also differed between the HLA-B44 allotypes. The disassociation rate (Koff) of the DM1 TCR for the HLA-B*4405EENL was 10-fold slower than from the HLA-B*4402/3EENL allotypes (Fig. 1 and Table I). Thus, the DM1 TCR interacted with the HLA-B*4405EENL complex with >10-fold higher affinity in comparison to HLA-B*4402EENL and HLA-B*4403EENL. These SPR studies reflect the intrinsic ability of DM1 to more readily recognize the cognate HLA-B*4405EENL complex despite the hidden nature of the polymorphic HLA-B44 residues.
Figure 1.
Differential affinity and binding kinetics of the DM1 TCR for the HLA-B44 allotypes. (a) Steady-state (or R equilibrium) binding of the three immobilized allotypes (HLA-B*4402EENL, HLA-B*4403EENL, and HLA-B*4405EENL) to the DM1 TCR. (b–d) Binding of increasing concentrations of the DM1 TCR (1.57–50 μM) to HLA-B*4402EENL (b), HLA-B*4403EENL (c), and HLA-B*4405EENL (d). Data are representative of at least four experiments. Kinetic association and dissociation rate constants were evaluated using the BIAevalution software. The DM1 TCR displayed a 10-fold higher affinity and a 10-fold slower dissociation rate constant for HLA-B*4405EENL over the other allotypes.
Table I.
Binding constants for the interaction of the DM1 TCR with HLA-B*4402/3/5 (determined by SPR)
| Immobilized ligand | Analyte | Kd (μM) | Kon (×104/Ms) | Koff (1/s) | Kd calc (μM) |
|---|---|---|---|---|---|
| HLA-B*4402EENL | DM1 TCR | 6.3 ± 0.3 | 1.4 | 0.2 | 14 |
| HLA-B*4403EENL | DM1 TCR | 9.4 ± 0.2 | 1.3 | 0.25 | 16 |
| HLA-B*4405EENL | DM1 TCR | 0.3 ± 0.1 | 6.8 | 0.02 | 0.23 |
| aDM1 TCR | HLA-B*4405EENL | 1 ± 0.5 | ND | ND | ND |
ND, not determined.
For cross-validation, the affinity for the DM1 TCR–HLA-B*4405EENL interaction was determined. This was similar irrespective of whether the DM1 TCR or the pMHC was coupled to the chip (note that the levels for the HLA-B*4402EENL and HLA-B*4403EENL refolds were too low to permit these MHCs to be the analytes).
Central mobility of the peptide in the binary HLA-B44EENL complexes
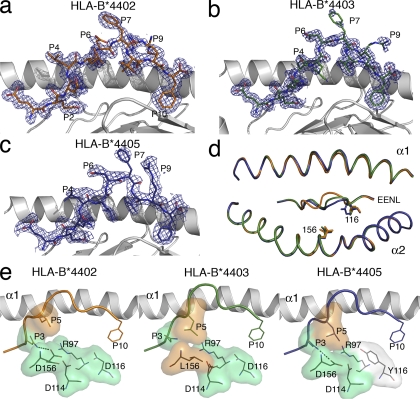
To begin to address whether the affinity differences between the DM1 TCR–HLA-B*4405EENL and the DM1 TCR–HLA-B*4402/03EENL complexes were caused by differences in the pMHC structures, we determined the structures of HLA-B*4402EENL, HLA-B*4403EENL, and HLA-B*4405EENL to 1.7, 1.6, and 2.1 Å resolution, respectively (Table S1, available at http://www.jem.org/cgi/content/full/jem.20082136/DC1). The three pMHC complexes crystallized in the same space group with isomorphous unit cells, and the bound peptide did not participate in crystal contacts. Accordingly, any conformational differences observed can be solely attributed to the buried polymorphic residues (positions 116 and 156) between the three allotypes. Position 116 is located at the F-pocket on the floor of the antigen-binding cleft, whereas position 156 is located on the α2 helix forming part of the D/E pocket on HLA-B44.
In all three HLA-B44EENL complexes, the peptide bound in an extended conformation bulged centrally around the P6-Asp and P7-Phe. A degree of mobility in the peptide around the P6–P7 position was evident, especially in the HLA-B*4405EENL complex (Fig. 2, a–c). As expected, P2-Glu and P10-Phe represented the main anchor residues for the B-pocket and F-pocket of HLA-B44, respectively (2), whereas the surface exposed residues P4-Leu, P6-Asp, P7-Phe, and P9-Arg represented potential TCR contact points.
Figure 2.
Structures of HLA-B*4402, HLA-B*4403, and HLA-B*4405 presenting EENLLDFVRF show marked differences in the centrally bulged region of the epitope. (a–c) Structure of the EENL peptide bound to (with associated 2Fo-Fc electron density contoured at 1σ) HLA-B*4402 (a), HLA-B*4403 (b), and HLA-B*4405 (c). The α2 helix has been removed for clarity. Potential TCR contact points are at P4, P6, P7, and P9. The central region (P5–P9) of the epitope showed mobility in all three allotypes. (d) An overlay of the three HLA-B44 allotypes (HLA-B*4402, orange; HLA-B*4403, green; HLA-B*4405, blue) revealing the minimal movement of the α helices and the movement of the central region (P5–P7) of the Cα backbones of the peptides. (e) The hydrophobic (orange) Leu at P5 of the epitope in HLA-B*4402 is pushed away from the unfavorable hydrophilic (green) patch composed of R97, D114, D116, and D156, resulting in movement of the peptide backbone toward the α1 helix. In HLA-B*4403 and HLA-B*4405, this patch is less hydrophilic because of either the hydrophobic Leu at position 156 or the partially hydrophobic Tyr at position 116 (white), and can accommodate the hydrophobic P5 residue.
Previous crystal structures of HLA-B*4402 and HLA-B*4403 in complex with a self-peptide revealed that the 156 polymorphism affected both the heavy chain and peptide conformation (11, 17). However, superposition of the antigen-binding clefts of the three HLA-B44EENL allotypes showed virtually no movement of the α1 and α2 helices (root mean square distance [r.m.s.d.] of <0.1 and 0.2 Å, respectively), indicating that the impact of the 156 polymorphism on the pMHC landscape is peptide dependent (Fig. 2 d). However, the positioning of the peptide within the antigen-binding cleft differed markedly between the three allotypes, and these differences were attributable to the interactions between the polymorphic residues at the bound peptide (Fig. 2 d). The r.m.s.d. of the superposed peptides ranged from 0.4 to 0.9 Å, with the greatest movement around the mobile, centrally bulged residues of P5-Leu to P7-Phe (with an r.m.s.d. of up to 1.9 Å). As a consequence, the peptide is positioned more closely to the α2 helix in the HLA-B*4405EENL structure, whereas in the HLA-B*4402EENL structure it is positioned more closely to the α1 helix, and in HLA-B*4403EENL it is more centrally located within the antigen-binding cleft (Fig. 2 d).
In HLA-B*4403, Leu156 formed favorable van der Waals (vdw) interactions with P5-Leu, whereas in HLA-B*4402 and HLA-B*4405, Asp156 was observed to “push away” the P5-Leu and instead formed an H bond to the P3-Asn, and was also able to form a network of water-mediated interactions. In HLA-B*4402, the unfavorable P5-Leu–Asp156 interaction is exacerbated by Asp114 and the polymorphic Asp116, thereby creating a marked electronegative pocket at the C-terminal end of the antigen-binding cleft that is mitigated to an extent by Arg97 (Fig. 2 e). In HLA-B*4405, however, position 116 is replaced by a Tyr, which will not only form more favorable interactions with the aromatic P10-Phe, namely pi–pi interactions, but will also serve to reduce the electronegative potential around the C-terminal region of the cleft (Fig. 2 e). Accordingly, the differing hydrophobic and electrostatic properties of the antigen-binding clefts of the HLA-B44 allotypes impart differing conformations of the centrally bulged portion of the bound epitope (P5–P9) that displays a more marked degree of mobility in HLA-B*4405.
The DM1 TCR recognizes the C-terminal end of the peptide
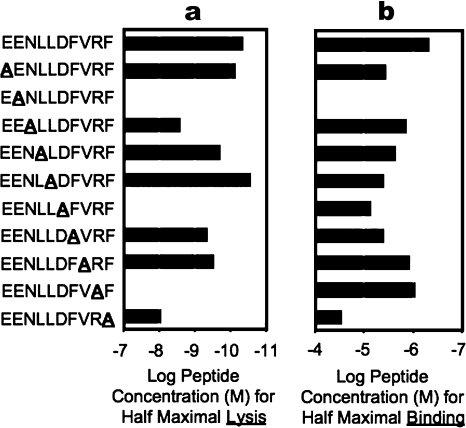
To begin to gain an understanding of how the DM1 TCR engaged the HLA-B*4405EENL complex, we undertook an alanine replacement scan of the epitope and assayed for CTL killing by the DM1 clone and for peptide–HLA-B*4405 binding efficiency. As expected, mutation of the P2-Glu and P10-Phe anchor residues markedly reduced binding to HLA-B*4405 (Fig. 3). Of the surface-exposed residues, substitutions at position 8 reduced T cell recognition by ∼10-fold, whereas substitutions at positions 6 and 9 abrogated recognition by the T cells (Fig. 3). Accordingly, it appeared that the DM1 TCR focused on the P6–P9 region of the peptide, the region that displayed the greatest variation among the three HLA-B44EENL allotypes.
Figure 3.
Alanine scanning mutagenesis of the peptide reveals a C-terminal focus. The impact of single amino-acid substitutions within the EENLLDFVRF peptide upon CTL recognition and HLA-B*4405 binding revealed P6 and P9 to be critical for TCR recognition. (a) EENL-specific CTL clones were tested for recognition of a panel of altered peptide ligands in which a single alanine substitution was introduced at each peptide position. A range of peptide concentrations was used in the chromium release assays, and the concentration required for half-maximum lysis was determined from the dose–response data. (b) MHC peptide-binding assays were also conducted at a range of concentrations for their ability to stabilize HLA-B*4405 expression. The concentration of peptide required for half-maximum HLA-B*4405 stabilization was calculated. Experiments were conducted once, with each sample tested in duplicate.
Overview of the DM1 TCR–HLA-B*4405EENL complex
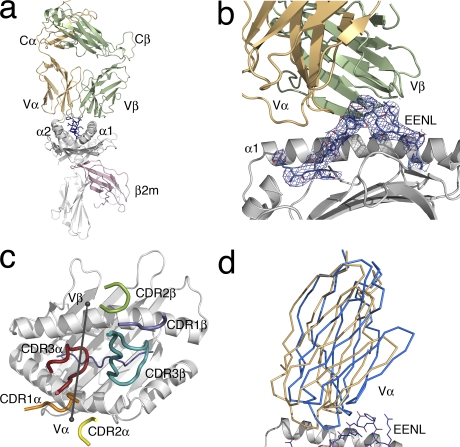
To gain a further understanding of why the DM1 TCR recognizes HLA-B*4405EENL more effectively than HLA-B*4402/3EENL, we determined the structure of the DM1 TCR in its nonliganded state and ligated to HLA-B*4405EENL to a 2.75 and 3.5 Å resolution, respectively (Fig. 4 a and Table S1). The data collection and structure determination of the ternary complex was challenging, as the crystal form exhibited a cell edge close to 700 Å with three complexes in the asymmetric unit. Given that we had solved the structures of the nonliganded counterparts of the ternary complex, the structure nevertheless refined readily at this resolution, and moreover, the electron density at the DM1 TCR–HLA-B*4405EENL interface was unambiguous (Fig. 4 b). The three complexes in the asymmetric unit were structurally identical. An overlay of the three complexes showed an overall r.m.s.d. of <1.4 Å, where major differences were in the differing juxtapositioning of the β2-microglobulin (β2m) domain (the r.m.s.d. without the β2m domain is 0.7 Å). Thus, unless explicitly stated, structural analysis is restricted to one complex in the asymmetric unit.
Figure 4.
Overview of the DM1 TCR in complex with HLA-B*4405EENL. (a) The DM1–HLA-B*4405EENL complex (DM1 TCR Vα, orange; DM1 TCR Vβ, green; HLA-B*4405, gray; EENL peptide, blue; β2m, pink). (b) A closer view of the DM1 TCR binding to the HLA-B*4405EENL, showing the clear electron density for the peptide (2Fo-Fc map contoured at 1σ). (c) Relative docking angle of the DM1 TCR on the pMHC (80°). For docking orientation, an axis was drawn through the points at the center of mass of the Vα and Vβ domains. The positioning of the CDR loops of the DM1 TCR on HLA-B*4405 is also shown (CDR1α, orange; CDR2α, yellow; CDR3α, red; CDR1β, slate; CDR2β green; CDR3β, teal). (d) Scissoring action of the Vα domain compared with the Vβ domain upon ligation. Superposition of the Vβ domains of the nonliganded and liganded DM1 TCR revealed movement of the Vα domain by 6°. The nonliganded Vα domain of the TCR is shown in blue, and the liganded Vα domain of the TCR is shown in orange.
The DM1 TCR docked at an angle of 80° across the long axis of the HLA-B*4405EENL binding cleft, and accordingly, the DM1 TCR docked within the range of previously determined TCR–pMHC structures (Fig. 4 c) (5). The DM1 TCR was essentially centrally located above HLA-B*4405EENL and formed five H bonds, eight salt bridges, and a large number of vdw interactions (Table II). The total buried surface area (BSA) was ∼2,200 Å2, which lies within the range of previously determined TCR–pMHC-I structures (3).
Table II.
DM1–HLA-B*4405EENL interactions
| TCR residue | MHC or peptide residues | Bond type | Gene segment | |
|---|---|---|---|---|
| CDR1α | Gly 30 | Ala 158 | vdw | Vα |
| CDR1α | Asn 31 | Gln 155, Tyr 159, Leu 163 | vdw | Vα |
| CDR1α | Tyr 33 | Arg 151, Gln 155 | vdw | Vα |
| CDR2α | Leu 57 | Arg 151, Glu 154 | vdw | Vα |
| FW | Glu 67Oe1, Oe2 | Arg 151NH1, NH2 | salt bridge | Vα |
| FW | Glu 67 | Arg 151 | vdw | Vα |
| FW | Glu 84 | Thr 41 | vdw | Vα |
| CDR3α | Tyr 110 | Arg 62, Gln 65, Ile 66, | vdw | Jα |
| CDR3α | Gln 111 | Gln 65 | vdw | Jα |
| CDR1β | Asn 30Nδ2 | Glu 76Oε1 | H bond | Vβ |
| CDR1β | Asn 30 | Glu 76 | vdw | Vβ |
| CDR2β | Gln 57Nε2 | Glu 76Oε2 | H bond | Vβ |
| CDR2β | Gln 57 | Gln 72, Thr 73, Glu 76 | vdw | Vβ |
| CDR2β | Asn 58Oδ1 | Gln 72Nε2 | H bond | Vβ |
| CDR2β | Asn 58 | Gln 72, Glu 76 | vdw | Vβ |
| CDR2β | Glu 59 | Gln 72 | vdw | Vβ |
| FW | Leu 66 | Thr 69 | vdw | Vβ |
| FW | Glu 67 | Glu 65 | vdw | Vβ |
| CDR3β | Asp 110Oδ1 | Lys 146NZ | salt bridge | Dβ-N |
| CDR3β | Asp 110 | Lys 146 | vdw | Dβ-N |
| CDR3β | Ser 112 | Ala 150 | vdw | Jβ |
| CDR3α | Tyr 110 | Leu 4, Asp 6 | vdw | Jα |
| CDR3α | Trp 107 | Asp 6, Phe 7 | vdw | N region |
| CDR3α | Gly 109 | Leu 5 | vdw | Jα |
| CDR1β | Asn 30 | Arg 9 | vdw | Vβ |
| CDR1β | Asn 30Nδ2 | Arg 9NH2 | H bond | Vβ |
| CDR1β | Arg 31NH1, NH2 | Asp 6Oδ1, Oδ2 | salt bridge | Vβ |
| CDR1β | Arg 31Nε | Arg 9NH1 | H bond | Vβ |
| CDR1β | Arg 31 | Asp 6, Phe 7, Arg 9 | vdw | Vβ |
| CDR3β | Arg 107NH1, NH2 | Asp 6Oδ1, Oδ2 | salt bridge | Vβ-N |
| CDR3β | Arg 107 | Asp 6 | vdw | Vβ-N |
| CDR3β | Arg 109 | Arg 9 | vdw | Dβ-N |
| CDR3β | Asp 110° | Arg 9 Nε | salt bridge | Dβ-N |
| CDR3β | Asp 110 | Phe 7, Arg 9 | vdw | Dβ-N |
| CDR3β | Asp 111 | Arg 9 | vdw | N-Jβ |
| CDR3β | Ser 112 | Phe 7 | vdw | Jβ |
| CDR3β | Tyr 113 | Phe 7 | vdw | Jβ |
Superscripted designations indicate the atom involved in hydrogen bonding. The vdw cutoff distance was ≤4 Å. D, diversity; FW, framework residue; J, joining; N, nongermline encoded; V, variable.
The Vα and Vβ domains contributed roughly equally (54.6 and 45.4%, respectively) to the BSA at the HLA-B*4405EENL interface. Moreover, all six complementarity determining region (CDR) loops of the DM1 TCR contributed to the HLA-B*4405EENL interaction, albeit to varying degrees, which is consistent with the variable usage of CDR loops observed in TCR–pMHC-I interactions (3, 18–20). The CDR1α and CDR2α loops of DM1 TCR contributed 18 and 7.8%, respectively, and the CDR1β and CDR2β loops contributed 7 and 11.8%, respectively, to the BSA. In contrast, the CDR3 loops played a more prominent role, with the CDR3α and CDR3β loops contributing 19.6 and 17%, respectively, of the BSA at the DM1 TCR–HLA-B*4405EENL interface.
Plasticity and the recognition of HLA-B44
Our findings represent the first report of a TCR ligated to HLA-B44 and thereby provided us with an opportunity to gain an insight into the structural requirements underscoring HLA-B44 restriction. As we determined the structure of the DM1 TCR and HLA-B*4405EENL in the nonliganded state, we were able to address the extent of plasticity that facilitates HLA-B44 restriction. Indeed, upon ligation, the Vα domain of the DM1 TCR changed orientation by 6° relative to the Vβ domain (Fig. 4 d), in a manner analogous to that observed recently (20), and the CDR3β loop of the DM1 TCR changed conformation appreciably upon pMHC ligation.
The DM1 TCR contacted HLA-B*4405 at positions 65, 69, and 155, three positions that have been invariably contacted in all TCR–pMHC-I structures determined to date (5, 8), despite positions 65 and 69 representing polymorphic sites among the HLA alleles. To enable recognition, HLA-B*4405 moved minimally upon ligation (the overall r.m.s.d. was 0.45 Å), although rearrangement of four MHC residues (Glu65, Glu76, Arg151, and Gln155) was observed.
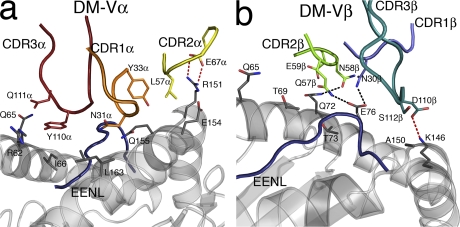
The CDR1α and CDR2α loops exclusively contacted residues spanning 155–163 from the α2 helix (Table II and Fig. 5 a). Asn 31α nestled in a shallow pocket that was flanked by Leu163 and Gln155, the latter of which also interacted with Tyr 33α (Fig. 5 a). Interestingly, Gln155, previously described as a gatekeeper residue, shifted conformation upon TCR ligation to form interactions with CDR1α as well as the peptide. The CDR2α played a limited role in interacting with HLA-B44 (Table II and Fig. 5 a), with Leu 57α forming vdw interactions with Glu154 and Arg151, the latter of which also moved to salt bridge with framework residue Glu 67α (Table II and Fig. 5 a). The CDR1β and CDR2β contacted residues spanning 69–76 of the α1 helix (Table II), with the role of CDR1β being restricted to interacting with Thr73 and Glu76 (Fig. 5 b), a residue that moved ∼2.5 Å to contact the CDR1β and CDR2β loops. The CDR2β loop comprised several polar residues (Gln 57β, Asn 58β, and Glu 59β) and, accordingly, was observed to form several H bonds to the α1 helix of HLA-B*4405 (Fig. 5 b).
Figure 5.
DM1 interactions with HLA-B*4405EENL. (a) Interactions with the HLA-B*4405 heavy chain mediated by the Vα domain of the DM1 TCR (CDR1α, orange; CDR2α, yellow; CDR3α, red; HLA-B*4405 heavy chain, gray; EENL epitope, blue). (b) Interactions with the HLA-B*4405 heavy chain mediated by the Vβ domain of the DM1 TCR (CDR1β, slate; CDR2β, green; CDR3β, teal; HLA-B*4405 heavy chain, gray; EENL epitope, blue).
In addition to participating in peptide-mediated interactions, the CDR3 loops also contacted the HLA-B44 heavy chain. CDR3α made several vdw contacts with the α1 helix (residues 62–66) that were centered on the aromatic residue Tyr 110α. Upon engagement, the CDR3β loop swung ∼90° and moved up to 5.8 Å to interact with the C terminus of the peptide (discussed in Epitope recognition by the DM1 TCR), and also formed a salt bridge between Asp 110β and Lys146 of the α2 helix. Accordingly, the structure of the nonliganded and ligated DM1 TCR provided insight into the large degree of plasticity that was required to enable HLA-B44 restriction.
Similar TCR usage against different HLA alleles
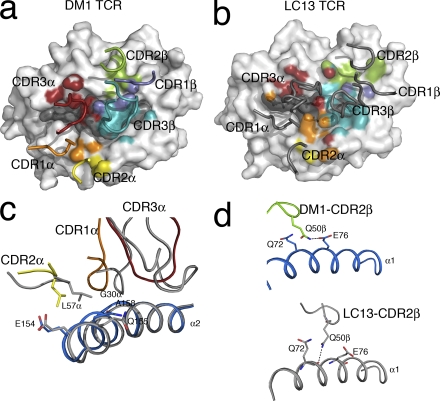
The DM1 TCR (TRAV26-1*02;TRAJ13*02;TRBV7-9*01;TRBJ2-1*01;TRBD1*01) uses Vα and Vβ gene segments similar to those of the LC13 TCR (TRAV26-2*01;TRAJ52*01/TRBV7-8*03;TRBJ2-7*01;TRBD1/2), which interacts with HLA-B*0801 bound to the FLRGRAYGL antigen (21), as well as HLA-B44 when bound to an alloantigen. The DM1 and LC13 TCRs share similarities in the CDR1α, CDR2α, CDR1β, and CDR2β loops (for sequence conservation see Table S2, available at http://www.jem.org/cgi/content/full/jem.20082136/DC1) (22, 23). In comparison to the LC13 footprint on HLA-B8FLRGRAYGL, the CDR1α and CDR2α loops are in roughly equivalent locations, although the positioning of the corresponding loops in the Vβ domain is more divergent (Fig. 6, a and b). From examination of all known TCR–pMHC co-crystal structures, it has been noted that TCRs often use a Tyr/Phe in the CDR1α to interact with position 155 (24). Indeed, both DM1 and LC13 use an aromatic residue to contact Gln155, as well as a Gly 30α to contact Ala158 (Fig. 6 c). Moreover, although CDR2α played a minor role in interacting with HLA-B44, both the DM1 TCR and the LC13 TCR use a Leu at the same position in CDR2α to conserve the interaction with Glu154 (Fig. 6 c). Although it has been suggested that a Ser in the CDR2α loop will contact the MHC at position 154–155 (24), this has not been observed in the DM1 TCR complex or other TCR–pMHC complexes. Although the positioning of the CDR2β loops is quite different, the sandwiching of Gln 50β between Gln72 and Glu76 is maintained (Fig. 6 d). Accordingly, there are some similarities, as well as notable differences, in the manner in which very similar Vα and Vβ gene segments interact with differing pMHC landscapes.
Figure 6.
DM1–pMHC interactions compared with the LC13–pMHC structure, revealing similar positioning of the CDRα loops. (a) DM1 TCR “footprint” on HLA-B*4405EENL. HLA-B*4405EENL (gray) is shown in surface representation. The pMHC residues are colored according to the relevant contacting CDR loop. The positions of the CDR loops are also shown (CDR1α, orange; CDR2α, yellow; CDR3α, red; CDR1β, slate; CDR2β, green; CDR3β, teal). (b) LC13 footprint on HLA-B*0801FLR. HLA-B*0801FLR (gray) is shown in surface representation. The pMHC residues are colored according to the relevant contacting CDR loop (CDR1α, orange; CDR2α, yellow; CDR3α, red; CDR1β, slate; CDR2β, green; CDR3β, teal). The LC13 CDR loops are shown in gray. (c) Similar positioning of the CDR1α loops (CDR1α, orange; CDR2α, yellow; CDR3α, red) of DM1 compared with LC13 (gray) allowed conservation of interactions with residues E154, Q155, and A158 on the α2 helix. (d) Although the positioning of the Vβ loops of the two TCRs was divergent, the packing of Q50β of the CDR2β loop of DM1 (green) and LC13 (gray) between Q72 and E76 was maintained.
Epitope recognition by the DM1 TCR
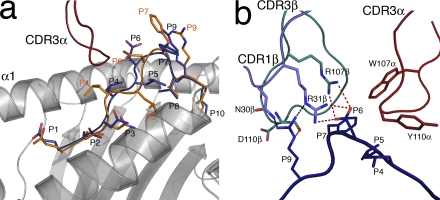
Although the HLA-B*4405 heavy chain did not change conformation upon DM1 TCR ligation, we observed an alteration in the conformation of the peptide (P4–P7) upon ligation (Fig. 7 a) analogous to the peptide “bulldozing” observed in the ELS4 TCR–HLA-B*3501EPLP structure (25). Upon ligation, the CDR3α loop reorientated the P4-Leu side chain, which consequently appeared to push the peptide toward the α1 helix, resulting in the complete shift of the backbone conformation of P5-Leu and P6-Asp (Fig. 7 a). In addition, the side chain of P7-Phe, which was mobile in the nonliganded form (Fig. 4 c), became completely ordered via interactions with the CDR3β loop.
Figure 7.
DM1-mediated peptide interactions. (a) Conformational movement of the EENL peptide upon ligation by the DM1 TCR revealing plasticity of the pMHC surface. The nonligand-bound conformation is shown in orange, and the ligand-bound conformation of the EENL epitope is shown in blue. (b) DM1 interactions with the EENL peptide. A cluster of charged interactions between the DM1 TCR and the peptide provides specificity. This specificity is driven by CDR3α (red), CDR1β (slate), and CDR3β (teal). Salt bridges are shown as red dashed lines, and H bonds are shown as black dashed lines.
The repositioning of the peptide enabled the CDR1β, CDR3α, and CDR3β loops to interact with positions 3–7 and 9 of the determinant. CDR1β played a major role in interacting with the peptide courtesy of some large side chains bridging the gap between the Cα backbones of the CDR1β loop and peptide (specifically, P6-Asp salt bridges to Arg 31β and the P9-Arg H bonds to Asn 30β and Arg 31β; Fig. 7 b). The CDR3α loop interacted with the middle region of the peptide (Fig. 7 b) in which Tyr 110α and Trp 107α played a prominent role in this interaction. Tyr 110α packed against P4-Leu and Trp 107α interacted with P6-Asp and P7-Phe. Notably an analysis of the α chain repertoire of HLA-B*4405EENL–specific TCRs from other HLA-B*4405+, EBV-exposed individuals (Table S3, available at http://www.jem.org/cgi/content/full/jem.20082136/DC1) revealed that this Trp is highly conserved despite this residue being encoded entirely from the N region. Furthermore, Trp is encoded by a single codon (TGG), and there is therefore a theoretically low (1 out of 64) probability of this codon appearing by coincidence (26). This bulky Trp residue also helped to orientate the CDR3β loop, which abutted the C-terminal end of the peptide in an antiparallel manner. The main chain of Ser 112β and Tyr 113β “sat” on the P7-Phe, whereas the P9-Arg protruded into a niche formed between the CDR1β and CDR3β loops, forming an H bond to Asp 110βO (Fig. 7 b). The interactions include a salt bridge between P6-Asp and Arg 107β as well as P9-Arg and Asp 110β (Fig. 7 b). The cluster of charged interactions between the DM1 TCR (Arg 31β and Arg 107β) and the peptide (P6-Asp and P9-Arg) was reminiscent of the highly specific interaction between the KK50.4–HLA-ELIL interaction (27), and is likely to be a key driver in determining the specificity of the DM1 TCR–HLA-B*4405EENL interaction, which is consistent with the peptide substitution data. Accordingly, the DM1 TCR made a number of critical interactions with the centrally bulged region of the epitope, which is the region that displayed the greatest degree of variability between the HLA-B*4405EENL, HLA-B*4402EENL, and HLA-B*4403EENL binary complexes.
DISCUSSION
There are ∼3,000 different HLA alleles (28) clustered into nine HLA supertypes (29), the maintenance of which within the human population is critical for protective immunity, disease susceptibility, autoimmunity, and drug hypersensitivity. Despite the importance of micropolymorphism in protective immunity, data are limited on how this directly relates to TCR recognition. This study provides simultaneous insight into HLA-B44 restriction, and how micropolymorphism between the HLA-B*4405, HLA-B*4403, and HLA-B*4402 allotypes finely tunes the CTL response toward an EBV determinant. The micropolymorphisms between these allotypes are not accessible to the TCR, and they did not appreciably affect the conformation of the MHC. Nevertheless, the DM1 TCR recognized the HLA-B*4405 allotype, presenting an EBV epitope, with a 10-fold increase in affinity when compared with HLA-B*4403 and HLA-B*4402. This is attributable to the altered conformation and dynamics of the peptide within the respective antigen-binding clefts. Specifically, residues 4–7 of the EBV epitope varied between the allotypes, with the epitope displaying a greater degree of mobility in HLA-B*4405. The significance of this variability and the dynamics of the epitope was apparent in the DM1 TCR–HLA-B*4405EENL complex, whereby the DM1 TCR not only contacted this mobile region but also caused a radical reshaping of the peptide upon ligation. In addition, the slow association rates (Kon) for the DM1 TCR interaction with all three pMHC complexes are consistent with marked conformational rearrangement occurring at the TCR–pMHC interface, which further reflects the underlying flexibility of the TCR–pMHC interface (30). Interestingly, the on rate for the DM1 TCR–HLA-B*4405EENL interaction was approximately sixfold higher when compared with the interactions with HLA-B*4403EENL and HLA-B*4402EENL. This is consistent with the increased flexibility of the EENL epitope in HLA-B*4405 facilitating the DM1 TCR docking, whereas the more “rigid” HLA-B*4403EENL and HLA-B*4402EENL complexes would be less favorable for DM1 TCR engagement.
Although structural studies have been enormously informative in addressing how the TCR can engage the pMHC, they are somewhat limited in their scope, as the subset of different MHC alleles that have been solved in complex with the TCR is relatively small. For example, of the 16 unique TCR–pMHC-Ia structures that have been determined to date, seven are with H2-Kb, five are with HLA-A2, two are with HLA-B35, and one each is with H2-Ld and HLA-B8 (3). Accordingly, any generalities proposed could potentially be skewed toward a particular MHC allele. For example, although common pairwise interactions are observed in defined Vβ8.2–MHC interactions (6, 7), this generality does not extend to Vβ8.2–CD1d interactions (31, 32). The HLA-B*4405–restricted DM1 TCR exhibits similar Vα and Vβ usage to the LC13 TCR, whose cognate antigen is HLA-B8, but also alloreacts on HLA-B44 (33). Although the respective Vβ domains showed a large degree of variation in docking, there was some similarity in the interaction mode within the CDR1 and CDR2 loops. However, mutational mapping has shown that for the LC13–HLA B8FLR interaction, the energetic driving force for the interaction rested not in the CDR1 and CDR2 loops but in the CDR3 loops (18). Perhaps the different docking modes of the LC13 and DM1 TCRs may relate to a given V gene “selecting” an orientation that is specific for a given MHC allotype. The distinct docking modes observed may represent two examples of several potential germline-encoded modes of interaction, with each being “selected” to optimize interactions with the distinct peptides presented by the specific MHC allotype.
Studies on the experimental H2-Kb/Kbm3 system have indicated how a buried polymorphism can affect peptide presentation that leads to an increase in Vβ-mediated interactions (10). Similarly, studies in the H2-Kb/Kbm8 system provided some evidence to suggest that very subtle differences between the alleles, which may include differing pMHC dynamics, can affect TCR recognition (9). The effect of buried HLA polymorphism on peptide conformation/dynamics has implications in graft-versus-host disease. Namely, donor T cells that have escaped thymic deletion caused by being weakly autoreactive toward a given pMHC may recognize very similar recipient pMHC complexes with much greater affinity because of the changes in the peptide conformation/dynamics. For example, the effect of MHC micropolymorphism on TCR recognition has been observed in comparative studies between HLA-B*3501 and HLA-B*3508. (34, 35). Similarly, mismatching of HLA-B*4402 and HLA-B*4403, two allomorphs that differ by just one amino acid, is a known barrier in transplantation and has been termed a “taboo mismatch” (13). In this case, the polymorphism was shown to subtly affect the peptide repertoire, the conformation of the peptide, and the MHC itself (11). Moreover, within the HLA-B*3501/B*3508 setting, it has also been established that the immunogenicity of a viral epitope is controlled by the conformation of the peptide within the antigen-binding cleft (36), and our data resonate with these findings. Such effects on TCR recognition of viral determinants may be an important consideration for future vaccination strategies against viruses, as markedly different CTL responses to the same viral epitope may be elicited in individuals expressing closely related HLA types.
TCR degeneracy is considered to be an important factor enabling TCRs to bind a wide range of potential pMHC combinations with the limited number of T cells present in the periphery. Conversely, we illustrate how a single HLA polymorphism alters the dynamics of the pMHC landscape and affects TCR recognition, thereby providing direct insight into the heightened specificity of a TCR and the subtleties of TCR recognition.
MATERIALS AND METHODS
Protein expression, purification, and crystallization.
The DM1 TCR was expressed and purified essentially as previously described (27) using an engineered disulfide linkage in the constant domains between TRAC48 and TRBC57. In brief, the α and β chains of the DM1 TCR were expressed separately in Escherichia coli and purified from inclusion bodies before being refolded before purification. HLA-B*44021-276, HLA-B*44031-276, HLA-B*44051-276, and β2m were also expressed separately in E. coli and purified from inclusion bodies before being refolded before purification, as previously described (17). An excess of peptide was required to drive refolding (e.g., 45 mg of HLA-B44 heavy chain, 15 mg of β2m, and 50 mg of peptide were mixed, resulting in ∼2–3mg of pure refolded protein). Purified HLA-B*4405EENL was mixed with an excess of purified DM1 TCR, and the DM1 TCR–HLA-B*4405EENL complex was purified using a gel filtration column (Sephadex-200; GE Healthcare). Crystals were grown by the hanging-drop, vapor-diffusion method at 20°C with a protein/reservoir drop ratio of 1:1.
Crystals of the DM1 TCR, the HLA-B44 peptide complexes, and the DM1 TCR–HLA-B*4405EENL complex were all grown by the hanging-drop, vapor-diffusion method at 20°C with a protein/reservoir drop ratio of 1:1. The DM1 TCR was at a concentration of 10 mg/ml in TBS (10 mM Tris, 150 mM NaCl [pH 8]). Crystals appeared in 2–5 d in 0.1 M Hepes (pH 7), 0.1 M magnesium formate, 18% (weight/volume) PEG 3350. The HLA-B44 peptide complexes were at a concentration of 3 mg/ml in TBS. Crystals appeared in 1–3 d in 0.1 M citrate (pH 5.6), 0.2 M ammonium acetate, 20–30% (weight/volume) PEG 4K. The DM1 TCR–HLA-B*4405EENL complex was at a concentration of 5 mg/ml in TBS. Crystals appeared after 7 d in 0.1 M bicine (pH 9), 0.2 M lithium sulfate, 16–18% PEG 3350.
Data collection, structure determination, and refinement.
The DM1 TCR crystals were frozen in liquid nitrogen using 5% glycerol as a cryoprotectant. The HLA-B*4402/03/05EENL complexes were all frozen using 10% glycerol as the cryoprotectant. The DM1 TCR–HLA-B4405EENL crystals were dehydrated in 25% PEG 3350 for 2–5 min and flash frozen in liquid nitrogen. All data were collected on the Industrial Macromolecular Crystallography Association Collaborative Access Team beamline at the Argonne Photon Source Synchrotron using the Quantum 210 CCD detector at 100 K (ADSC). Data for the unliganded DM1 TCR and the HLA-B4402/3/5EENL binary structures were processed using either MOSFLM (37) or HKL2000 (38) software and were scaled using SCALA software (39) from the CCP4 suite (40) or scalepack (38).
The DM1 crystal belonged to the space group P21 with unit cell dimensions (Table S1) consistent with two molecules in the asymmetrical unit. The structure was determined by molecular replacement using the MOLREP program with the 1G4 TCR as the search model (available from the Protein Data Bank under accession no. 2BNU) (41). Subsequent model building was conducted using the COOT software (42), followed by maximum-likelihood refinement with the REFMAC 5 program (CCP4 suite) (40, 43). Translation, liberation, and screw-rotation (TLS) displacement refinement was used to model anisotropic displacements of defined domains. The optimal TLS domains were determined using the TLS motion determination server (44, 45) and were read into the REFMAC 5 program. Refinement was monitored by Rfree, and water molecules were included in the model if they were within hydrogen-bonding distance to chemically “reasonable” groups, and appeared in the Fo-Fc maps (where Fo and Fc are the observed and calculated structure factors, respectively) contoured at 3 sigma and had a B factor <80 Å2. Tight noncrystallographic symmetry (NCS) restraints were also used during the refinement process. The TCR was numbered according to the international ImMunoGeneTic (IMGT) unique numbering system (46), whereby the CDR1α and β loops start at residue number 27, the CDR2α and β loops start at residue number 56, and the CDR3α and β loops start at residue number 105.
The HLA-B4402/3/5EENL crystals belonged to the space group P212121 with unit cell dimensions (Table S1), each consistent with one molecule in the asymmetrical unit. The structure was determined by molecular replacement using the MOLREP program with the HLA-B*4405 as the search model (available from the Protein Data Bank under accession no. 1SYV0) (11). Subsequent model building was conducted using the COOT software (42), followed by maximum-likelihood refinement with the REFMAC 5 program (40, 43). Refinement was monitored by Rfree, and water molecules were included in the model if they were within hydrogen-bonding distance to chemically reasonable groups, and appeared in the Fo-Fc maps contoured at 3 sigma and had a B factor <80 Å2.
For the DM1 TCR–HLA-B*4405EENL complex, the data were processed and scaled using the HKL2000 software (38). The crystals belonged to the hexagonal space group P6522 with unit cell dimensions (Table S1), each consistent with three complexes in the asymmetrical unit. The structure was determined by molecular replacement using the PHASER program using the fully refined unliganded DM1 TCR and the HLA-B*4405EENL structures (minus peptide) as search models. Subsequent minimal model building was conducted using the COOT software (42), followed by maximum-likelihood refinement with the REFMAC 5 program (40, 43). TLS displacement refinement was used to model anisotropic displacements of defined domains. Again, the optimal TLS domains were determined using the TLS motion determination server (44, 45) and were read into the REFMAC 5 program. Tight NCS restraints were also used during the refinement process.
All structures were validated using the PROCHECK (47) and MOLBROBITY (48) programs. Ramachandran plots were obtained from the MOLPROBITY program (48). The final Ramachandran statistics were as follows: DM1 TCR, 92.5% (most favored region) and 98.6% (allowed region); HLA-B*4402EENL, 97.6% (most favored region) and 100% (allowed region); HLA-B*4403EENL, 97.9% (most favored region) and 99.5% (allowed region); HLA-B*4405EENL, 97.1% (most favored region) and 99.2% (allowed region); and DM1 TCR–HLA-B*4405EENL, 86.3% (most favored region) and 97.5% (allowed region). All molecular graphics representations were created using the PyMOL system (49).
SPR measurement and analysis.
All SPR experiments were conducted at 25°C on the BIAcore 3000 instrument (GE Healthcare) with HBS buffer (10 mM Hepes [pH 7.4], 150 mM NaCl, and 0.005% surfactant P20). The HBS buffer was supplemented with 1% bovine serum albumin to prevent nonspecific binding. The human HLA-specific mAb, W6/32 (50) was coupled to research grade CM5 chips with standard amine coupling. For each experiment, HLA-B*4402/3/5 was passed over two flow cells and ∼200–500 response units (RU) was captured by the antibody. The other two flow cells were used as control cells for each experiment. The DM1 TCR was injected over all four flow cells at a rate of 20 μl/min with a concentration range of 0.39–400 μM. The final response was calculated by subtraction of the response of the antibody alone from that of the antibody HLA-B44 complex. The antibody surface was regenerated between each analyte injection with Actisep (Sterogene). All experiments were conducted in duplicate.
For cross-validation of results, the reverse experiment was performed. The DM1 TCR was coupled to the CM5 chip using the 12H8 antibody (18), which is conformation specific for a region bridging the α and β constant domains of the TCR. For this experiment, 400–600 RU of the DM1 TCR was captured on the antibody, and HLA-B*4405EENL (0.6–150 μM) was injected over the flow cells at 20 μl/min. The total protein levels for the HLA-B*4402EENL and HLA-B*4403EENL refolds were too low to permit these pMHCs to be the analytes.
BIAevaluation software (version 3.1; GE Healthcare) was used for data analysis; the 1:1 Langmuir binding model was used to calculate the kinetic constants. To allow for the capture system, the model was modified to include an additional parameter for the drifting baseline for the TCR capture by 12H8. The quality of the fit between experimental data and the binding models was evaluated by monitoring the χ2 values (i.e., <10), as well as the residual plots.
Cytotoxicity assays: alanine substitution analysis.
The DM1 CTL clone was tested in duplicate for cytotoxicity in a standard 5-h chromium release assay. In brief, CTLs were assayed against 51Cr-labeled autologous PHA blast targets that were pretreated with synthetic peptide or left untreated. The percentage of specific lysis was calculated, and the peptide concentration required for half-maximum lysis was determined from dose–response curves. Peptides were synthesized by Mimotopes Ltd. Toxicity testing of all peptides was performed before use by adding peptide to 51Cr-labeled PHA blasts in the absence of CTL effectors. A β scintillation counter (Topcount Microplate; PerkinElmer) was used to measure 51Cr levels in assay supernatant samples. The mean spontaneous lysis for target cells in the culture medium was always <20%, and the variation from the mean specific lysis was <10%.
HLA peptide-binding assays.
To assess peptide binding to HLA-B*4405, HLA stabilization studies were conducted using the mutant HLA-A/B null lymphoblastoid cell line C1R (51) that had been transfected to expressed HLA-B*4405 and the herpes virus–derived protein ICP47, which inhibits the transporter associated with antigen processing. These cells were incubated in serum-free medium (AIM V; Invitrogen) with various concentrations (0.1, 1, 10, or 100 μM) of peptides at 26°C for 14–16 h, followed by incubation at 37°C for 2–3 h. HLA-B*4405 surface expression was measured by flow cytometry on a FACSCanto (BD) using an mAb to HLA-Bw4. Mean fluorescence intensity was determined, and the peptide concentration required for half-maximum mean fluorescence intensity was calculated.
EENL-specific T cell repertoire analysis.
T cells specific for the EENL peptide were first isolated from bulk EENL-specific CTL cultures raised from two HLA-B*4405+ family members of donor DM (from whom the DM1 CTL clone was raised) by stimulation of PBMCs with the EENL peptide, followed by expansion in rIL-2–containing medium for 10 d. Using flow cytometric cell sorting, CTLs that bound an HLA-B*4405EENL tetramer and costained with anti-CD3 and anti-CD8 mAbs (BD) were isolated. The sorting was performed on a MoFlo sorter (Dako), and purities of >96% were consistently achieved. Total RNA was extracted from the sorted CTLs using TRIzol reagent (Invitrogen). RT-PCR and PCR was performed using a One-Step PCR (Invitrogen) according to the manufacturer's guidelines. The forward primer was 1 out of 34 previously published TCR TRAV family-specific primers (52); the reverse TRAJ13-specific primer was designed in house (5′-GGATGACTTGGAGCTTTGTT-3′). PCR products were purified and cloned into the pGEM-T vector system (Promega) and sequenced using the ABI PRISM Big Dye termination reaction kit (Applied Biosystems). The IMGT database TCR gene nomenclature was used throughout (46).
Protein data bank accession codes.
The atomic coordinates and structure factors are available from the Protein Data Bank under the following accession nos: DM1 TCR–HLA-B4405EENL complex, 3DXA; DM1 TCR, 3DX9; HLA-B*4402 EENL, 3DX6; HLA-B*4403 EENL, 3DX7; and HLA-B*4405 EENL, 3DX8.
Online supplemental material.
Table S1 summarizes data collection and refinement statistics. Table S2 represents a sequence comparison between LC13 and DM1 TCR CDR loops. Table S3 shows a Vα sequence comparison between B44EENL-restricted TCRs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20082136/DC1.
Supplementary Material
Acknowledgments
We thank the staff at the Industrial Macromolecular Crystallography Association, Advanced Photon Source, Chicago for assistance with data collection.
This work was supported by the National Health and Medical Research Council (NHMRC) of Australia and the Australian Research Council (ARC). S.R. Burrows, A.W. Purcell, and M.C.J. Wilce are supported by NHMRC Senior Research Fellowships; W.A. Macdonald is supported by an NHMRC Peter Doherty Fellowship; T. Beddoe is supported by an NHMRC Career Development Award Fellowship; and J.K. Archbold is supported by a Dora Lush NHMRC Scholarship. J. Rossjohn is supported by an ARC Federation Fellowship.
The authors have no conflicting financial interests.
Abbreviations used: β2m, β2-microglobulin; BSA, buried surface area; CDR, complementarity determining region; MHC-I, MHC class I; pMHC, peptide-MHC; r.m.s.d., root mean square distance; SPR, surface plasmon resonance; vdw, van der Waals.
L.K. Ely's present address is Dept. of Molecular and Cellular Physiology, Stanford School of Medicine, Stanford, CA 94305.
References
- 1.Messaoudi, I., J.A. Patino, R. Dyall, J. LeMaoult, and J. Nikolich-Zugich. 2002. Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science. 298:1797–1800. [DOI] [PubMed] [Google Scholar]
- 2.DiBrino, M., K.C. Parker, D.H. Margulies, J. Shiloach, R.V. Turner, W.E. Biddison, and J.E. Coligan. 1995. Identification of the peptide binding motif for HLA-B44, one of the most common HLA-B alleles in the Caucasian population. Biochemistry. 34:10130–10138. [DOI] [PubMed] [Google Scholar]
- 3.Godfrey, D.I., J. Rossjohn, and J. McCluskey. 2008. The fidelity, occasional promiscuity, and versatility of T cell receptor recognition. Immunity. 28:304–314. [DOI] [PubMed] [Google Scholar]
- 4.Clements, C.S., M.A. Dunstone, W.A. Macdonald, J. McCluskey, and J. Rossjohn. 2006. Specificity on a knife-edge: the alphabeta T cell receptor. Curr. Opin. Struct. Biol. 16:787–795. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph, M.G., R.L. Stanfield, and I.A. Wilson. 2006. How TCRs bind MHCs, peptides, and coreceptors. Annu. Rev. Immunol. 24:419–466. [DOI] [PubMed] [Google Scholar]
- 6.Dai, S., E.S. Huseby, K. Rubtsova, J. Scott-Browne, F. Crawford, W.A. Macdonald, P. Marrack, and J.W. Kappler. 2008. Crossreactive T cells spotlight the germline rules for [alpha][beta] T cell-receptor interactions with MHC molecules. Immunity. 28:324–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng, D., C.J. Bond, L.K. Ely, J. Maynard, and K.C. Garcia. 2007. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon.’ Nat. Immunol. 8:975–983. [DOI] [PubMed] [Google Scholar]
- 8.Tynan, F.E., S.R. Burrows, A.M. Buckle, C.S. Clements, N.A. Borg, J.J. Miles, T. Beddoe, J.C. Whisstock, M.C. Wilce, S.L. Silins, et al. 2005. T cell receptor recognition of a ‘super-bulged’ major histocompatability complex class I-bound peptide. Nat. Immunol. 6:1114–1122. [DOI] [PubMed] [Google Scholar]
- 9.Miley, M.J., I. Messaoudi, B.M. Metzner, Y. Wu, J. Nikolich-Zugich, and D.H. Fremont. 2004. Structural basis for the restoration of TCR recognition of an MHC allelic variant by peptide secondary anchor substitution. J. Exp. Med. 200:1445–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luz, J.G., M. Huang, K.C. Garcia, M.G. Rudolph, V. Apostolopoulos, L. Teyton, and I.A. Wilson. 2002. Structural comparison of allogeneic and syngeneic T cell receptor–peptide–major histocompatibility complex complexes: a buried alloreactive mutation subtly alters peptide presentation substantially increasing Vβ interactions. J. Exp. Med. 195:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macdonald, W.A., A.W. Purcell, N.A. Mifsud, L.K. Ely, D.S. Williams, L. Chang, J.J. Gorman, C.S. Clements, L. Kjer-Nielsen, D.M. Koelle, et al. 2003. A naturally selected dimorphism within the HLA-B44 supertype alters class I structure, peptide repertoire, and T cell recognition. J. Exp. Med. 198:679–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zernich, D., A.W. Purcell, W.A. Macdonald, L. Kjer-Nielsen, L.K. Ely, N. Laham, T. Crockford, N.A. Mifsud, M. Bharadwaj, L. Chang, et al. 2004. Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J. Exp. Med. 200:13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keever, C.A., N. Leong, I. Cunningham, E.A. Copelan, B.R. Avalos, J. Klein, N. Kapoor, P.W. Adams, C.G. Orosz, P.J. Tutschka, et al. 1994. HLA-B44-directed cytotoxic T cells associated with acute graft-versus-host disease following unrelated bone marrow transplantation. Bone Marrow Transplant. 14:137–145. [PubMed] [Google Scholar]
- 14.Khanna, R., S.R. Burrows, A. Neisig, J. Neefjes, D.J. Moss, and S.L. Silins. 1997. Hierarchy of Epstein-Barr virus-specific cytotoxic T-cell responses in individuals carrying different subtypes of an HLA allele: implications for epitope-based antiviral vaccines. J. Virol. 71:7429–7435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miles, J.J., S.L. Silins, A.G. Brooks, J.E. Davis, I. Misko, and S.R. Burrows. 2005. T-cell grit: large clonal expansions of virus-specific CD8+ T cells can dominate in the peripheral circulation for at least 18 years. Blood. 106:4412–4413. [DOI] [PubMed] [Google Scholar]
- 16.Clements, C.S., L. Kjer-Nielsen, W.A. MacDonald, A.G. Brooks, A.W. Purcell, J. McCluskey, and J. Rossjohn. 2002. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr. D Biol. Crystallogr. 58:2131–2134. [DOI] [PubMed] [Google Scholar]
- 17.Macdonald, W., D.S. Williams, C.S. Clements, J.J. Gorman, L. Kjer-Nielsen, A.G. Brooks, J. McCluskey, J. Rossjohn, and A.W. Purcell. 2002. Identification of a dominant self-ligand bound to three HLA B44 alleles and the preliminary crystallographic analysis of recombinant forms of each complex. FEBS Lett. 527:27–32. [DOI] [PubMed] [Google Scholar]
- 18.Borg, N.A., L.K. Ely, T. Beddoe, W.A. Macdonald, H.H. Reid, C.S. Clements, A.W. Purcell, L. Kjer-Nielsen, J.J. Miles, S.R. Burrows, et al. 2005. The CDR3 regions of an immunodominant T cell receptor dictate the ‘energetic landscape’ of peptide-MHC recognition. Nat. Immunol. 6:171–180. [DOI] [PubMed] [Google Scholar]
- 19.Manning, T.C., E.A. Parke, L. Teyton, and D.M. Kranz. 1999. Effects of complementarity determining region mutations on the affinity of an α/β T cell receptor: measuring the energy associated with CD4/CD8 repertoire skewing. J. Exp. Med. 189:461–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishizuka, J., G.B.E. Stewart-Jones, A. van der Merwe, J.I. Bell, A.J. McMichael, and E.Y. Jones. 2008. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its V[beta] domain. Immunity. 28:171–182. [DOI] [PubMed] [Google Scholar]
- 21.Kjer-Nielsen, L., C.S. Clements, A.G. Brooks, A.W. Purcell, M.R. Fontes, J. McCluskey, and J. Rossjohn. 2002. The structure of HLA-B8 complexed to an immunodominant viral determinant: peptide-induced conformational changes and a mode of MHC class I dimerization. J. Immunol. 169:5153–5160. [DOI] [PubMed] [Google Scholar]
- 22.Kjer-Nielsen, L., C.S. Clements, A.G. Brooks, A.W. Purcell, J. McCluskey, and J. Rossjohn. 2002. The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance. Structure. 10:1521–1532. [DOI] [PubMed] [Google Scholar]
- 23.Kjer-Nielsen, L., C.S. Clements, A.W. Purcell, A.G. Brooks, J.C. Whisstock, S.R. Burrows, J. McCluskey, and J. Rossjohn. 2003. A structural basis for the selection of dominant alphabeta T cell receptors in antiviral immunity. Immunity. 18:53–64. [DOI] [PubMed] [Google Scholar]
- 24.Marrack, P., J.P. Scott-Browne, S. Dai, L. Gapin, and J.W. Kappler. 2008. Evolutionarily conserved amino acids that control TCR-MHC interaction. Annu. Rev. Immunol. 26:171–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tynan, F.E., H.H. Reid, L. Kjer-Nielsen, J.J. Miles, M.C.J. Wilce, L. Kostenko, N.A. Borg, N.A. Williamson, T. Beddoe, A.W. Purcell, et al. 2007. A T cell receptor flattens a bulged antigenic peptide presented by a major histocompatibility complex class I molecule. Nat. Immunol. 8:268–276. [DOI] [PubMed] [Google Scholar]
- 26.Venturi, V., K. Kedzierska, D.A. Price, P.C. Doherty, D.C. Douek, S.J. Turner, and M.P. Davenport. 2006. Sharing of T cell receptors in antigen-specific responses is driven by convergent recombination. Proc. Natl. Acad. Sci. USA. 103:18691–18696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoare, H.L., L.C. Sullivan, G. Pietra, C.S. Clements, E.J. Lee, L.K. Ely, T. Beddoe, M. Falco, L. Kjer-Nielsen, H.H. Reid, et al. 2006. Structural basis for a major histocompatibility complex class Ib-restricted T cell response. Nat. Immunol. 7:256–264. [DOI] [PubMed] [Google Scholar]
- 28.Robinson, J., M.J. Waller, P. Parham, N. de Groot, R. Bontrop, L.J. Kennedy, P. Stoehr, and S.G. Marsh. 2003. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 31:311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sidney, J., B. Peters, N. Frahm, C. Brander, and A. Sette. 2008. HLA class I supertypes: a revised and updated classification. BMC Immunol. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willcox, B.E., G.F. Gao, J.R. Wyer, J.E. Ladbury, J.I. Bell, B.K. Jakobsen, and P.A. van der Merwe. 1999. TCR binding to peptide-MHC stabilizes a flexible recognition interface. Immunity. 10:357–365. [DOI] [PubMed] [Google Scholar]
- 31.Wun, K.S., N.A. Borg, L. Kjer-Nielsen, T. Beddoe, R. Koh, S.K. Richardson, M. Thakur, A.R. Howell, J.P. Scott-Browne, L. Gapin, et al. 2008. A minimal binding footprint on CD1d-glycolipid is a basis for selection of the unique human NKT TCR. J. Exp. Med. 205:939–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borg, N.A., K.S. Wun, L. Kjer-Nielsen, M.C. Wilce, D.G. Pellicci, R. Koh, G.S. Besra, M. Bharadwaj, D.I. Godfrey, J. McCluskey, and J. Rossjohn. 2007. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 448:44–49. [DOI] [PubMed] [Google Scholar]
- 33.Archbold, J.K., W.A. Macdonald, S.R. Burrows, J. Rossjohn, and J. McCluskey. 2008. T-cell allorecognition: a case of mistaken identity or déjà vu? Trends Immunol. 29:220–226. [DOI] [PubMed] [Google Scholar]
- 34.Tynan, F.E., N.A. Borg, J.J. Miles, T. Beddoe, D. El-Hassen, S.L. Silins, W.J.M. van Zuylen, A.W. Purcell, L. Kjer-Nielsen, J. McCluskey, et al. 2005. High resolution structures of highly bulged viral epitopes bound to major histocompatibility complex class I. Implications for T-cell receptor engagement and T-cell immunodominance. J. Biol. Chem. 280:23900–23909. [DOI] [PubMed] [Google Scholar]
- 35.Burrows, J.M., K.K. Wynn, F.E. Tynan, J. Archbold, J.J. Miles, M.J. Bell, R.M. Brennan, S. Walker, J. McCluskey, J. Rossjohn, R. Khanna, and S.R. Burrows. 2007. The impact of HLA-B micropolymorphism outside primary peptide anchor pockets on the CTL response to CMV. Eur. J. Immunol. 37:946–953. [DOI] [PubMed] [Google Scholar]
- 36.Tynan, F.E., D. Elhassen, A.W. Purcell, J.M. Burrows, N.A. Borg, J.J. Miles, N.A. Williamson, K.J. Green, J. Tellam, L. Kjer-Nielsen, et al. 2005. The immunogenicity of a viral cytotoxic T cell epitope is controlled by its MHC-bound conformation. J. Exp. Med. 202:1249–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leslie, A.G.W. 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 and ESF-EAMCB Newsletters on Protein Crystallography. 26.
- 38.Otwinowski, Z., and W. Minor. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276:307–326. [DOI] [PubMed] [Google Scholar]
- 39.Evans, P. 2005. Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62:72–82. [DOI] [PubMed] [Google Scholar]
- 40.Collaborative Computational Project, Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760–763. [DOI] [PubMed] [Google Scholar]
- 41.Chen, J.-L., G. Stewart-Jones, G. Bossi, N.M. Lissin, L. Wooldridge, E.M.L. Choi, G. Held, P.R. Dunbar, R.M. Esnouf, M. Sami, et al. 2005. Structural and kinetic basis for heightened immunogenicity of T cell vaccines. J. Exp. Med. 201:1243–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 43.Murshudov, G.N., A.A. Vagin, and E.J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. [DOI] [PubMed] [Google Scholar]
- 44.Painter, J., and E.A. Merritt. 2006. TLSMD web server for the generation of multi-group TLS models. J. Appl. Cryst. 39:109–111. [Google Scholar]
- 45.Painter, J., and E.A. Merritt. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450. [DOI] [PubMed] [Google Scholar]
- 46.Lefranc, M.-P., V. Giudicelli, Q. Kaas, E. Duprat, J. Jabado-Michaloud, D. Scaviner, C. Ginestoux, O. Clement, D. Chaume, and G. Lefranc. 2005. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 33:D593–D597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laskowski, R.A., M.W. MacArthur, D.S. Moss, and J.M. Thornton. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Cryst. 26:283–291. [Google Scholar]
- 48.Lovell, S.C., I.W. Davis, W.B. Arendall, P.I.W. deBakker, J.M. Word, M.G. Prisant, J.S. Richardson, and D.C. Richardson. 2003. Structure validation by C-alpha geometry: phi, psi and C-beta deviation. Proteins Struct. Funct. Genet. 50:437–450. [DOI] [PubMed] [Google Scholar]
- 49.DeLano, W.L. 2002. The PyMOL molecular graphics system. Available at http://www.pymol.org (accessed March 4, 2006).
- 50.Parham, P., C.J. Barnstable, and W.F. Bodmer. 1979. Use of a monoclonal antibody (W6/32) in structural studies of HLA-A,B,C antigens. J. Immunol. 123:342–349. [PubMed] [Google Scholar]
- 51.Storkus, W.J., J. Alexander, J.A. Payne, J.R. Dawson, and P. Cresswell. 1989. Reversal of natural killing susceptibility in target cells expressing transfected class I HLA genes. Proc. Natl. Acad. Sci. USA. 86:2361–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Panzara, M.A., E. Gussoni, L. Steinman, and J.R. Oksenberg. 1992. Analysis of the T cell repertoire using the PCR and specific oligonucleotide primers. Biotechniques. 12:728–735. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.