Abstract
Interleukin (IL) 6 is a proinflammtory cytokine produced by antigen-presenting cells and nonhematopoietic cells in response to external stimuli. It was initially identified as a B cell growth factor and inducer of plasma cell differentiation in vitro and plays an important role in antibody production and class switching in vivo. However, it is not clear whether IL-6 directly affects B cells or acts through other mechanisms. We show that IL-6 is sufficient and necessary to induce IL-21 production by naive and memory CD4+ T cells upon T cell receptor stimulation. IL-21 production by CD4+ T cells is required for IL-6 to promote B cell antibody production in vitro. Moreover, administration of IL-6 with inactive influenza virus enhances virus-specific antibody production, and importantly, this effect is dependent on IL-21. Thus, IL-6 promotes antibody production by promoting the B cell helper capabilities of CD4+ T cells through increased IL-21 production. IL-6 could therefore be a potential coadjuvant to enhance humoral immunity.
IL-6 is produced by APCs such as macrophages, dendritic cells, and B cells, but also by nonhematopoietic cells (e.g., epithelial and endothelial cells, astrocytes, and fibroblasts) in response to external stimuli such as TNF-α, IL-1β, platelet-derived growth factor, or bacterial and fungal components. It binds to a receptor complex consisting of the specific IL-6Rα and the gp130 signal transducing unit that activates the transcription factor STAT3, among other signaling molecules. IL-6 is a pleiotropic cytokine that plays an important role in acute-phase protein synthesis, bone metabolism, central nervous system function, growth and drug response of tumors, and the immune response (for reviews see references 1, 2). Regarding its function in the immune system, IL-6 is now thought of as an important link between innate and adaptive immunity, mediating several aspects of B and T cell responses (3).
The role of IL-6 in CD4+ T cell function is multifaceted. IL-6 influences T cell effector functions by promoting Th2 cell differentiation through up-regulation of NFATc2 and c-maf (4, 5). It also blocks IFN-γ signaling through increased expression of silencer of cytokine signaling 1, thereby inhibiting Th1 cell differentiation (5). In the presence of TGF-β, IL-6 promotes Th17 cell differentiation through STAT3-mediated up-regulation of the transcription factor retinoic acid receptor-related orphan receptor γt (6–10). Although some of the molecular mechanisms used by IL-6 to mediate these different responses are known, it is still unclear how these various effects on CD4+ T cells are orchestrated.
IL-6 was initially characterized as a factor that enhances antibody production in a B cell line (11), and overexpression of IL-6 in mice causes plasmocytosis, suggesting that IL-6 can promote the differentiation of B cells into plasma cells (12). Likewise, IL-6–deficient mice show reduced antigen-specific IgG1, IgG2a, and IgG3 levels upon immunization with a T cell–dependent antigen, although IgM levels were not affected (13). Further evidence for a role of IL-6 in IgG production has come from experiments using a transgenic mouse expressing a truncated form of gp130. These mice are unable to activate STAT3 upon IL-6 exposure and show reduced levels of most antibody isotypes after immunization with a T-dependent antigen (14). However, expression of the transgene is not restricted to B cells, leaving open the possibility that other cells may require a functional gp130 receptor. Likewise, B cell–specific deletion of STAT3 results in impaired plasma cell differentiation and diminished antibody responses (15), but this transcription factor is also activated by other cytokines.
The cytokine IL-21 has been shown to play a major role in antibody production by promoting the differentiation of B cells into plasma cells both in mice and humans (16, 17). Accordingly, IL-21 promotes the production of IgG1, IgG2a, and IgG3, but it has an inhibitory effect on IgE production (18). The inhibition of IgE is mediated by IL-21–induced up-regulation of Id2 that negatively regulates class switching to IgE (19). Similar to IL-6, IL-21 activates predominantly STAT3, but through binding to its specific receptor and the common γ chain signal transducing unit that it shares with other members of the IL-2 family of cytokines (20). In B cells, IL-21–induced STAT3 activation down-regulates B cell lymphoma 6 and up-regulates expression of B lymphocyte–induced maturation protein 1, thereby promoting plasma cell differentiation (21).
In this report, we identify IL-21 as the only cytokine that is specifically induced in CD4+ T cells by IL-6 early upon antigen stimulation. IL-6 is necessary to mediate IL-21 production in naive CD4+ T cells during antigen stimulation. Furthermore, we show that IL-6 promotes antibody production in B cells indirectly by up-regulation of IL-21 expression in CD4+ T cells and that this IL-21 then acts on B cells.
RESULTS AND DISCUSSION
Regulation of early gene expression by IL-6 during the activation of CD4+ T cells
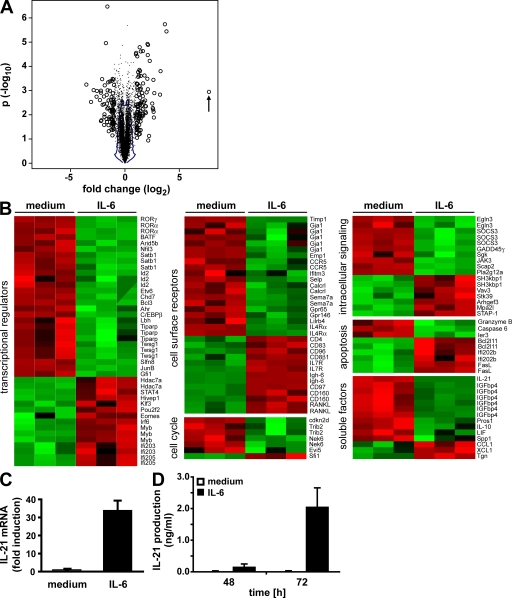
IL-6 has been associated with different aspects of T cell activation, differentiation, and survival (2). However, little is known about the changes in gene expression induced by IL-6 stimulation early during T cell activation. To further characterize early effects of IL-6 on CD4+ T cells, we performed microarray analysis. Sorted CD4+ NK1.1− T cells were activated with anti-CD3 and anti-CD28 mAbs in the absence or presence of IL-6 for 16 h. For each condition, three independent preparations of RNA were used for hybridization of gene arrays. For analysis of microarray data, we explored the use of the 1% contour on the joint distribution of fold change and p-value derived from sample label permutations (Fig. 1 A), which resulted in significance at P = 0.1, as well as identification of 343 genes, with a false discovery rate of 13%. 132 differentially regulated genes (190 probe sets) were found when using a twofold threshold on the probe set intensity ratio, IL-6 treated to untreated (Fig. 1 B; and Fig. S1 and Table S1, available at http://www.jem.org/cgi/content/full/jem.20081571/DC1). The microarray results were validated by real-time RT-PCR of several genes positively or negatively affected by the IL-6 treatment (Fig. S2).
Figure 1.
Effects of IL-6 on the gene expression profile of CD4+ T cells early during activation. (A) FACS-sorted CD4+ T cells were activated with anti-CD3 and anti-CD28 in the absence or presence of IL-6 for 16 h. The gene expression profile from total RNA was examined by Affymetrix GeneChip analysis. In the volcano plot, open circles represent probe sets for which expression is changed more than twofold up or down after IL-6 treatment, whereas dots represent all other probe sets. The number of genes exceeding the threshold was the largest among the 10 datasets obtained by permutation of sample labels, yielding statistical significance of differential expression at P = 0.1 (the minimum value) and a false discovery rate <19% (the minimum being 10%). The blue line encloses 99% of the probe sets under the null hypothesis, obtained by permutation of sample labels. The IL-21 gene is marked with an arrow. (B) Heat maps representing the relative expression of genes among IL-6–treated and control samples. Genes were sorted within each group based on their fold change (top, highest fold induction; bottom, highest fold down-regulation). Expression statistics for each gene were centered, scaled, and mapped to a color scale. Red represents relatively low expression, whereas green represents relatively high expression. (C) Cells were purified and activated as in A, and IL-21 expression was measured by quantitative real-time RT-PCR. The means ± SEM of three experiments are shown. (D) CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs in the absence or presence of IL-6 for the indicated periods of time. Cell-culture supernatants were analyzed for IL-21 production by ELISA. The means ± SEM of four experiments are shown.
Among the 132 genes whose expression was affected by IL-6, the cytokine IL-21 was especially remarkable, with a >200-fold induction in CD4+ T cells activated in the presence of IL-6. The induction of IL-21 by IL-6 was confirmed by real-time RT-PCR using RNA from CD4+ T cells activated for 16 h (Fig. 1 C). Similar results were obtained by ELISA, where IL-21 production could only be detected in the presence of IL-6 (Fig. 1 D). Although IL-6 has been shown to induce the expression of IL-4 and IL-17 (7, 8, 10, 22), their expression was not increased by IL-6 stimulation early during activation. These results also confirm a recent study showing that induction of IL-21 by IL-6 is independent of TGF-β stimulation and Th17 cell differentiation (23). Moreover, among known cytokines IL-21 is the most potently induced one by IL-6 early during the activation of CD4+ T cells, as indicated by its high up-regulation, especially when compared with other soluble factors (Table S1).
IL-6 is required for IL-21 expression in CD4 T cells upon TCR stimulation
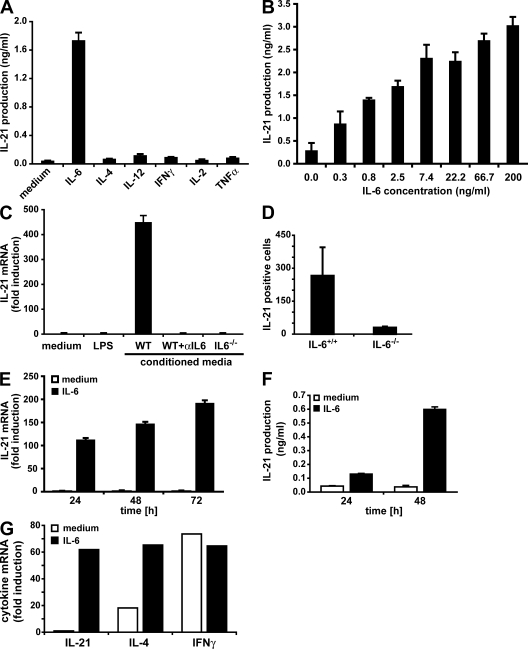
Given the high induction of IL-21 by IL-6, we examined whether other cytokines have a similar effect. CD4+ T cells were activated in the presence of IL-4, IL-12, or IFN-γ, but none of those cytokines were able to promote IL-21 production (Fig. 2 A). Neither growth and survival factors such as IL-2 and IL-7 (Fig. 2 A and not depicted) nor other proinflammatory cytokines such as TNF-α and IL-1 were able to up-regulate IL-21 production (Fig. 2 A and not depicted). IL-11, another cytokine that signals through gp130 (1), also failed to promote IL-21 production (Fig. S3, available at http://www.jem.org/cgi/content/full/jem.20081571/DC1). Moreover, IL-6 is a very efficient inducer of IL-21 expression, as a dose–response analysis revealed that IL-6 levels as low as 0.3–0.8 ng/ml were able to readily induce measurable IL-21 production (Fig. 2 B).
Figure 2.
IL-6 is sufficient and necessary to induce IL-21 expression during the activation of naive and memory CD4+ T cells. (A and B) FACS-sorted CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs in the absence or presence of the indicated cytokines (A) or the indicated IL-6 concentrations (B). After 3 d, cell-culture supernatants were analyzed for IL-21 production by ELISA. The means ± SEM of three (A) or four (B) experiments are shown. (C) FACS-purified CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs in the presence of medium alone, LPS, conditioned media (CM) from LPS-stimulated mouse splenocytes, CM plus 10 μg/ml of a neutralizing anti–IL-6 (αIL-6) mAb, or CM from LPS-stimulated IL-6−/− splenocytes. IL-21 expression was measured by quantitative real-time RT-PCR. One representative experiment out of two is shown (error bars represent SD). (D) C57BL/6 and IL-6−/− mice were immunized s.c. with 200 μg OVA in CFA. CD4+ T cells prepared from spleens and lymph nodes were analyzed 6 d later for the frequency of IL-21–producing cells by ELISPOT. The means ± SD of three immunizations are shown (P < 0.05). (E) FACS-purified CD4+ CD25− NK1.1− CD44low naive cells were activated with anti-CD3 and anti-CD28 mAbs in the absence or presence of IL-6 for the indicated periods of time. IL-21 expression was measured by quantitative real-time RT-PCR. One representative experiment out of two is shown (error bars represent SD). (F) CD4+ T cells were isolated from AND TCR transgenic mice, and activated with 5 μM cyt c peptide and APCs in the absence or presence of IL-6 for the indicated periods of time. IL-21 production was examined by ELISA. One representative experiment out of two is shown (error bars represent SD of a triplicate determination). (G) FACS-purified CD4+ CD25− NK1.1− CD44high memory cells were activated with anti-CD3 and anti-CD28 mAbs in the absence or presence of IL-6 for 24 h. Cytokine expression was measured by quantitative real-time RT-PCR. Values are presented as fold induction over naive CD4+ T cells activated for 24 h. One representative experiment out of two is shown.
To determine whether IL-6 was required for IL-21 production by CD4+ T cells, spleen cells from wild-type or IL-6−/− mice were treated with LPS, and the conditioned media were added to CD4+ T cells activated with anti-CD3 and anti-CD28 mAbs. Only CD4+ T cells activated in the presence of the LPS-conditioned media from wild-type splenocytes expressed IL-21, whereas this expression was abrogated in the presence of a neutralizing anti–IL-6 mAb (Fig. 2 C). In addition, conditioned media from IL-6−/− splenocytes stimulated with LPS did not promote IL-21 expression (Fig. 2 C). As a control, LPS alone added to activated CD4+ T cells did not induce IL-21 expression (Fig. 2 C). Thus, APC-derived IL-6 is required for IL-21 production by CD4+ T cells upon activation in vitro. To show that IL-6 is required for IL-21 expression in vivo, IL-6−/− and wild-type mice were immunized with OVA, and CD4+ T cells were analyzed for IL-21 production by ELISPOT analysis. The number of IL-21–producing CD4+ T cells was severely impaired in IL-6−/− compared with wild-type mice (Fig. 2 D). Collectively, these results demonstrate that IL-6 is required for the production of IL-21 by CD4+ T cells upon antigen stimulation in vitro and in vivo. Although a recent paper reported that IL-21 can induce its own expression through STAT3 (24), our results indicate that the initial IL-21 expression in CD4+ T cells is mediated by IL-6.
IL-21 has been shown to be produced by differentiated Th2 and Th17 effector CD4+ T cells upon restimulation as well as by NKT cells (25–29). To further demonstrate the direct effect of IL-6 on IL-21 production by naive CD4+ T cells, we activated sorted CD4+ CD44low NK1.1− T cells in the presence or absence of IL-6. IL-21 expression was essentially undetectable in purified naive CD4+ T cells activated in the absence of IL-6 even after 3 d of stimulation (Fig. 2 E). In contrast, high IL-21 expression could be detected in activated naive CD4+ T cells treated with IL-6 as soon as 24 h after initial stimulation (Fig. 2 E). Similarly, naive CD4+ T cells from AND TCR transgenic mice (recognizing pigeon cytochrome c [cyt c]) produce IL-21 only in the presence of IL-6 upon stimulation with cyt c peptide and APCs (Fig. 2 F).
Memory CD4+ T cells are known for their ability to produce high amounts of effector cytokines such as IFN-γ and IL-4 rapidly after antigen stimulation (30). To test whether IL-21 expression was similar to other effector cytokines up-regulated early in memory CD4+ T cells, sorted CD4+ CD44high T cells were stimulated with anti-CD3 and anti-CD28 mAbs in the absence or presence of IL-6. As expected, memory CD4+ T cells expressed relatively high levels of IFN-γ and IL-4 regardless of the presence of IL-6 (Fig. 2 G). However, similar to naive CD4+ T cells, memory cells did not produce IL-21 in the absence of IL-6 but did have high levels of expression when IL-6 was present (Fig. 2 G). Thus, production of IL-21 by naive and memory CD4+ T cells upon antigen stimulation requires the presence IL-6.
Induction of antibody production by IL-6 requires up-regulation of IL-21 expression in CD4+ T cells
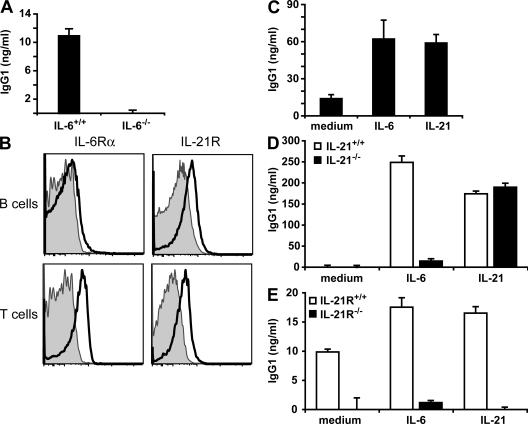
Both IL-6 and IL-21 have been shown to promote antibody production by B cells. Several studies demonstrated a direct effect of IL-21 on plasma cell differentiation through up-regulation of the transcription factor B lymphocyte–induced maturation protein 1 (16, 17, 21). Although in vitro experiments using T–B cell co-culture and in vivo studies indicate that IL-6 is required for antibody production (13), the molecular mechanism remains unknown, and it is unclear whether this is a direct effect on B cells. Because IL-6 promotes IL-21 production by CD4+ T cells, it was possible that these two pathways are closely linked to provide T cell help to B cells. The analysis of antibody production in vitro with wild-type or IL-6−/− total spleen cells activated with anti-CD3 mAb confirmed the contribution of IL-6 to IgG1 production (Fig. 3 A). However, analysis of IL-6Rα cell-surface expression showed that resting splenic B cells had almost undetectable levels, whereas it was present on a substantial fraction of CD4+ T cells (Fig. 3 B). In contrast, IL-21R expression could be detected on B cells as well as CD4+ T cells, in accordance with a previous paper (20). In addition, IL-21 promoted IgG1 production in total splenocytes from IL-6−/− mice activated with anti-CD3 mAb, similar to IL-6 (Fig. 3 C). Collectively, these results suggest that IL-6 mediates antibody production by inducing IL-21 expression in CD4+ T cells. To test for this possibility, we established a co-culture of CD4+ T cells activated with anti-CD3 mAb and B cells in the presence of exogenous IL-6. CD4+ T cells were isolated from wild-type and IL-21−/− mice, whereas B cells were obtained from IL-6−/− mice, because IL-6 is known to be also produced by B cells. In the absence of IL-6, IgG1 levels were practically undetectable (Fig. 3 D). IL-6 strongly increased IgG1 production when B cells were cultured together with CD4+ T cells from wild-type mice but failed to induce IgG1 in the presence of IL-21−/− CD4+ T cells (Fig. 3 D). Thus, the effect of IL-6 on antibody production appears to be dependent on IL-21 produced by CD4+ T cells. Addition of exogenous IL-21 could compensate for IL-21 deficiency, indicating that IL-21−/− CD4+ T cells are functional despite their lack of IL-21 production (Fig. 3 D).
Figure 3.
IL-6 requires IL-21 expression in CD4+ T cells to induce IgG1 production. (A) Total splenocytes from wild-type and IL-6−/− mice were stimulated with anti-CD3 mAb for 7 d. IgG1 levels in the cell supernatant were measured by ELISA. Results are representative of three experiments performed in triplicate. (B) B220+CD19+ B cells and CD4+ T cells from the spleens of C57BL/6 mice were analyzed for IL-6Rα and IL-21R expression (bold-line histograms) by flow cytometry. Shaded histograms are control-stained cells. Representative profiles of one mouse out of five analyzed are shown. (C) Total splenocytes of IL-6−/− mice were activated with anti-CD3 mAb and 100 ng/ml IL-6 or 100 ng/ml IL-21 where indicated. After 7 d, IgG1 levels in the supernatant were determined by ELISA. (D) CD4+ T cells from IL-21−/− mice or wild-type littermates were activated with anti-CD3 mAb in the presence of B cells from IL-6−/− mice and the indicated cytokines. After 7 d, IgG1 levels in the supernatant were determined by ELISA. (E) CD4+ T cells from wild-type mice were activated with anti-CD3 mAb in the presence of B cells from IL-21R−/− mice or wild-type littermates and the indicated cytokines. After 7 d, IgG1 levels in the supernatant were determined by ELISA. The means ± SEM of a triplicate determination from one representative experiment out of three performed are shown in A and C–E.
In addition to its effects on B cells, IL-21 has been shown to promote Th2 cell differentiation by inhibiting Th1 cell differentiation (25). It can also induce Th17 cell differentiation when TGF-β is present (27–29). To exclude that IL-6–induced IL-21 promoted antibody production through its effect on the differentiation of CD4+ T cells rather than acting directly on B cells, we activated wild-type CD4+ T cells with anti-CD3 mAb and co-cultured them in the presence of B cells from wild-type and IL-21R−/− mice. IL-6 augmented IgG1 production in wild-type B cells compared with the medium control (Fig. 3 E). In contrast, B cells that were deficient for IL-21R did not produce IgG1 when stimulated with IL-6 despite co-cultured CD4+ T cells that were capable of receiving IL-21 signals (Fig. 3 E). Accordingly, IL-21 promoted IgG1 production only when IL-21R was present on B cells but not in IL-21R−/− B cells, suggesting that IL-21 induced by IL-6 acted directly on B cells to increase IgG1 levels (Fig. 3 E). These results show that IL-6 can promote antibody production by inducing IL-21 expression in CD4+ T cells, and IL-21 subsequently acts on B cells.
Administration of IL-6 during influenza immunization promotes specific IgG production through IL-21
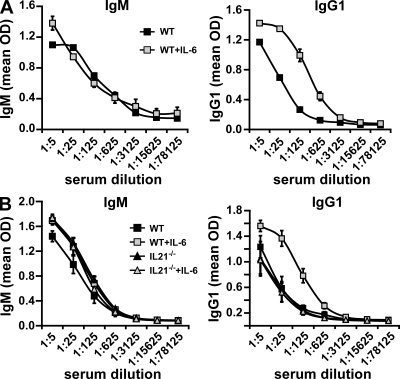
The indirect effect of IL-6 on antibody production through IL-21 generated by CD4+ T cells suggested that IL-6 could be a potent enhancer of the humoral immune response, because small amounts of IL-6 could result in an increased and prolonged production of IL-21, which would increase antibody production by B cells. We therefore investigated whether administration of IL-6 during viral immunization could boost the antiviral specific antibody response. We immunized wild-type mice with inactive PR8 influenza virus (in the absence of adjuvant) in combination with IL-6 or PBS, and analyzed virus-specific antibody production. Influenza-specific IgM serum levels were not affected by the administration of IL-6 (Fig. 4 A), indicating that IL-6 did not provide overall survival signals to B cells. In contrast, the presence of IL-6 during immunization substantially enhanced the levels of influenza-specific IgG1 (Fig. 4 A). Thus, IL-6 enhances the in vivo antibody response.
Figure 4.
IL-6 enhances antibody responses to inactive influenza virus immunization through IL-21. (A) Wild-type C57BL/6 mice were injected s.c. with 108 egg infectious units of inactive PR8 virus alone or together with IL-6. After 30 d, serum was harvested and influenza-specific IgM and IgG1 levels were determined by ELISA. (B) IL-21−/− mice and wild-type littermates were treated as in A, and influenza-specific IgM and IgG1 levels were determined by ELISA. Experiments were performed twice with five mice per group. Error bars indicate SD.
To determine whether the effect of IL-6 on in vivo antiviral antibody response is also dependent on IL-21, we immunized wild-type and IL-21−/− mice with inactive influenza virus in the presence or absence of IL-6. Influenza-specific IgM levels were comparable in IL-21−/− and wild-type mice (Fig. 4 B), in agreement with previous studies showing independence of IgM from IL-21 (18). In contrast, IL-6 promoted the production of IgG1 in wild-type mice, but it had no effect in IL-21−/− mice (Fig. 4 B). Collectively, these results show that administration of IL-6 enhances antibody response, but this effect is dependent on endogenous IL-21 production.
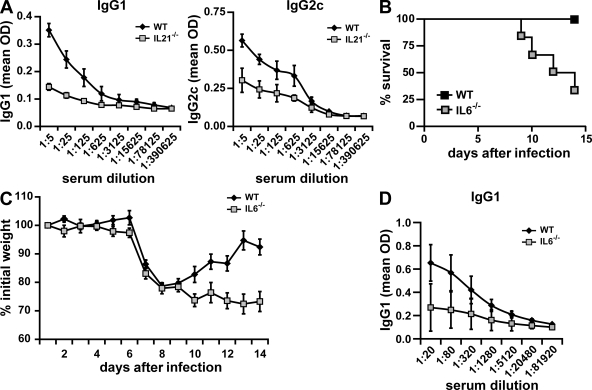
Role of IL-21 and IL-6 in antibody response to influenza infection
Despite the established role of IL-21 in generation of plasma cells and antibody production, it remains unknown whether IL-21 plays a role in antibody response during influenza infection or any other viral infection. We therefore examined the contribution of endogenous IL-21 to the production of virus-specific antibodies in response to a sublethal dose (0.1 LD50) of the live PR8 influenza virus using wild-type and IL-21–deficient mice. 14 d after infection, influenza-specific antibody titers in serum were determined. Influenza-specific IgG1 and IgG2c levels were severely reduced in infected IL-21−/− mice compared with wild-type mice (Fig. 5 A). Thus, IL-21 plays a role in antibody response to influenza. We also examined the contribution of endogenous IL-6 to the antibody production against infection with PR8 infection using IL-6−/− mice. Interestingly, we found in repeated experiments that IL-6–deficient mice were highly susceptible to influenza virus infection, because a sublethal dose of live PR8 was sufficient to cause a relatively high rate of mortality in these mice but not in wild-type mice (Fig. 5 B), indicating that IL-6 is essential for protection against influenza virus. Unlike infected wild-type mice, infected IL-6−/− mice could not recover the weight loss caused by the acute viral infection (Fig. 5 C), suggesting that these mice could not clear or control the virus. Analysis of PR8-specific antibody production in the surviving IL-6−/− mice showed a reduction in virus antibody titers (Fig. 5 D). IL-6 is therefore a key cytokine for resolution of influenza virus infection.
Figure 5.
Role of IL-21 and IL-6 in influenza virus infection. (A) Wild-type and IL-21−/− mice were infected i.n. with a sublethal dose (0.1 LD50) of active PR8 virus, and after 2 wk, influenza-specific IgG1 and IgG2c levels in serum were determined by ELISA. (B–D) Wild-type and IL-6−/− mice were infected i.n. with a sublethal dose (0.05 LD50) of active PR8 virus. Animals were monitored daily, and the time to death was recorded as the day after infection. Kaplan-Meier survival curves were generated using Prism 4.03 software (GraphPad Software, Inc.), and the log-rank test was used to determine the significance of differences between the strains (B). Weight was also measured daily in the surviving mice. The percentage of the initial weight is shown (C). Serum levels of virus-specific IgG1 were determined in the surviving mice (six out of six for wild-type mice and two out of six for IL-6−/− mice) 14 d after infection (D). Data represent groups of five (A) or six (B–D) mice from one experiment out of two performed. Error bars indicate SD.
In summary, we show that IL-6 is a very specific and effective inducer of IL-21 production by CD4+ T cells, and that it is sufficient and required for IL-21 production by naive (and probably memory) CD4+ T cells upon antigen stimulation in vitro and in vivo. Although it was proposed that IL-21 is preferentially expressed by effector Th17 cells (24, 27–29), a recently published study (23), as well as our results in this report, show that IL-6, in the absence of TGF-β, can induce the production of IL-21 in CD4+ T cells. IL-21 is also produced by the T follicular helper (Tfh) subset of CD4+ T cells that localizes in B cell follicles and provides help to B cells (31). Because follicular dendritic cells produce significant amounts of IL-6 (13), it is possible that the presence of IL-6 in follicles can contribute to the production of IL-21 by these Tfh cells. A study published at the time of submission of this manuscript supports the need of IL-6 for the generation of Tfh cells (32).
Although IL-6 has been long considered as a B cell growth factor and mediator of plasma cell differentiation, our data in this report show the requirement of IL-21 for IL-6 to induce antibody production in vitro and in vivo. We propose that IL-6 enhances antibody production indirectly by acting on naive (and likely memory) antigen-specific CD4+ T cells to promote their differentiation into Tfh cells that produce high levels of IL-21. Accordingly, we also show that both IL-6 and IL-21 play a role in the immune response during influenza infection, and administration of IL-6 during viral immunization substantially increased virus-specific antibody response without the need of additional coadjuvants, but in an IL-21–dependent manner. The use of IL-6 as coadjuvant during immunization may therefore be a promising tool to achieve successful vaccination against specific pathogens whose clearance is highly dependent on humoral immunity.
MATERIALS AND METHODS
Mice
B10.BR and C57BL/6J mice (The Jackson Laboratory) were housed under sterile conditions at the animal care facility at the University of Vermont and used for experiments between 8 and 14 wk of age. IL-21 gene–deficient mice were obtained from the Mutant Mouse Regional Resource Center at the University of California, Davis, and were backcrossed to C57BL/6J. TCR transgenic AND mice specific for pigeon cyt c, as well as IL-6 and IL-21R gene–deficient mice, as previously described (18, 33, 34). Immunizations were performed with UV-inactivated A/PR/8/34 (PR8) influenza A virus (H1N1) injected s.c. together with 500 ng recombinant IL-6 (R&D Systems), as indicated in the figures. For live infections, mice were inoculated intranasally (i.n.) during light isoflurane anesthesia with 0.05 or 0.1 LD50 (2.5 or 5 × 103 egg infectious units/ml) of PR8 virus in 100 μl PBS. Procedures that involved mice were approved by the Institutional Animal Care and Use Committees from the University of Vermont and the Trudeau Institute.
Cell preparation and activation
CD4+ T cells from lymph nodes and spleens were isolated by negative selection, as previously described (35), depleting cells expressing CD8, CD11b, MHC class II, and NK1.1. For FACS isolation, negatively selected CD4+ T cells were stained with anti-CD4–PE–Cy5.5, anti-NK1.1–PE, and anti-CD25–FITC plus anti-CD44–allophycocyanin (BD) and sorted on a FACSAria (BD) by gating in the low forward scatter and low side scatter lymphocyte population. Antibodies against B220, CD19, IL-6Rα (BD), and IL-21R (eBioscience) were also used for flow cytometry.
CD4+ T cells were activated with plate-bound 5 μg/ml anti-CD3 (2C11) and 1 μg/ml of soluble anti-CD28 (BD) mAbs in Bruff's medium in the presence or absence of different cytokines: 10 ng/ml TNF-α, 20 ng/ml IL-1α, 50 U/ml IL-2, 103 U/ml IL-4, 100 ng/ml IL-6, 3.5 ng/ml IL-12, 10 ng/ml IFN-γ (all obtained from R&D Systems), 100 ng/ml IL-11 (Genetics Institute), 100 ng/ml IL-21 (PeproTech), or 10 ng/ml LPS (Sigma-Aldrich).
B cells were purified by positive selection using anti-CD19 MACS beads (Miltenyi Biotec) according to the manufacturer's instructions and were generally >95% CD19+B220+. For co-culture experiments, CD4+ T cells were stimulated with 5 μg/ml of plate-bound anti-CD3 (2C11) and mixed with B cells in a 1:3 T/B cell ratio.
ELISAs
For IL-21 detection, the mouse-specific IL-21 DuoSet kit was used according to the manufacturer's protocol (R&D Systems). IgG1 levels were determined with anti–mouse IgG1 as capture antibody and horseradish peroxidase (HRP)–conjugated anti–mouse IgG1 as detection antibody, with mouse IgG1 as standard (all obtained from SouthernBiotech). Influenza-specific antibody isotypes were determined using UV-inactivated influenza PR8 as antigen. Antibody levels in serially diluted serum samples were analyzed with HRP-conjugated antibodies specific for mouse IgM, IgG1, and IgG2c (SouthernBiotech).
ELISPOT assay
IL-21–producing CD4+ T cells were detected according to a protocol adapted from a previously published paper (36). In brief, CD4+ T cells were activated with anti-CD3 and anti-CD28 mAbs in the presence or absence of IL-6 or an anti–IL-6 mAb. After 45 h, cells were harvested and counted, and 104 cells per well were plated in anti–IL-21–coated ELISPOT plates. Cells were incubated for 4 h in medium alone or medium containing 5 ng/ml PMA and 250 ng/ml ionomycin. Plates were developed as previously described (36) using a biotinylated anti–IL-21 mAb, and the total number of IL-21–producing cells was determined in an ELISPOT reader (CTL-ImmunoSpot S5 ELISPOT reader; Cellular Technology Ltd.).
Analysis of RNA
Total RNA was extracted from cells by using the RNAeasy kit (QIAGEN) as recommended by the manufacturer. RNA was reverse transcribed into cDNA, as previously described (37). mRNA levels were quantified with a TaqMan system with Assay-on-Demand sets (Applied Biosystems) and normalized to the levels of β2-microglobulin present in the same sample.
Microarray.
Fragmented biotinylated antisense cRNA was prepared according to the Affymetrix standard labeling protocol and hybridized to Mouse 430a 2.0 GeneChips (Affymetrix) at the Microarray Core Facility of the University of Vermont. Microarray data have been deposited in the Gene Expression Omnibus under accession no. GSE7459.
Data analysis.
The signal intensity for each probe on each chip was calculated from scanned images using GeneChip Operating Software (Affymetrix). Signal intensities were analyzed using R (38) and BioConductor (39). Probe intensities were normalized using the Qspline method (40), a cubic spline normalization using quantiles. An expression statistic was calculated for each probe set on each chip using the robust multichip average method (41, 42). The difference of population means, IL-6 treated minus control, referred to as log2 (IL-6/control), and a p-value (using the two-sided t test) were calculated for each probe set after each permutation of sample labels. Heat maps were drawn using the Bioconductor library Heatplus, written by Alexander Plona (available at http://www.bioconductor.org/packages/2.3/bioc/vignettes/Heatplus/inst/doc/Heatplus.pdf).
Online supplemental material
Fig. S1 shows IL-6–induced changes in gene expression for genes with a miscellaneous or unknown function. Fig. S2 shows a validation of several IL-6 target genes found in the microarray by real-time RT-PCR. Fig. S3 shows the lack of IL-21 production by CD4+ T cells after IL-11 treatment. Table S1 lists all genes with an at least twofold change in gene expression level after IL-6 treatment during the activation of CD4+ T cells. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20081571/DC1.
Supplementary Material
Acknowledgments
We thank T. Hunter, S. Tighe, and the DNA Sequencing Facility for assistance with real-time RT-PCR analysis and the microarray analysis.
This work was supported by National Institutes of Health (NIH) grant P01AI045666 (to M. Rincon) and the Vermont Genetics Network through grant P20 RR16462 from the IDeA Network of Biomedical Research Excellence program of the National Center for Research Resources, a component of the NIH (to J.P. Bond).
W.J. Leonard is an inventor on patents and patent applications related to IL-21. The authors have no other conflicting financial interests.
References
- 1.Kamimura, D., K. Ishihara, and T. Hirano. 2003. IL-6 signal transduction and its physiological roles: the signal orchestration model. Rev. Physiol. Biochem. Pharmacol. 149:1–38. [DOI] [PubMed] [Google Scholar]
- 2.Kishimoto, T. 2005. Interleukin-6: from basic science to medicine–40 years in immunology. Annu. Rev. Immunol. 23:1–21. [DOI] [PubMed] [Google Scholar]
- 3.Jones, S.A. 2005. Directing transition from innate to acquired immunity: defining a role for IL-6. J. Immunol. 175:3463–3468. [DOI] [PubMed] [Google Scholar]
- 4.Yang, Y., J. Ochando, A. Yopp, J.S. Bromberg, and Y. Ding. 2005. IL-6 plays a unique role in initiating c-Maf expression during early stage of CD4 T cell activation. J. Immunol. 174:2720–2729. [DOI] [PubMed] [Google Scholar]
- 5.Diehl, S., and M. Rincon. 2002. The two faces of IL-6 on Th1/Th2 differentiation. Mol. Immunol. 39:531–536. [DOI] [PubMed] [Google Scholar]
- 6.Yang, X.O., A.D. Panopoulos, R. Nurieva, S.H. Chang, D. Wang, S.S. Watowich, and C. Dong. 2007. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 282:9358–9363. [DOI] [PubMed] [Google Scholar]
- 7.Veldhoen, M., R.J. Hocking, C.J. Atkins, R.M. Locksley, and B. Stockinger. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. [DOI] [PubMed] [Google Scholar]
- 8.Mangan, P.R., L.E. Harrington, D.B. O'Quinn, W.S. Helms, D.C. Bullard, C.O. Elson, R.D. Hatton, S.M. Wahl, T.R. Schoeb, and C.T. Weaver. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. [DOI] [PubMed] [Google Scholar]
- 9.Ivanov, I.I., B.S. McKenzie, L. Zhou, C.E. Tadokoro, A. Lepelley, J.J. Lafaille, D.J. Cua, and D.R. Littman. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 126:1121–1133. [DOI] [PubMed] [Google Scholar]
- 10.Bettelli, E., Y. Carrier, W. Gao, T. Korn, T.B. Strom, M. Oukka, H.L. Weiner, and V.K. Kuchroo. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 441:235–238. [DOI] [PubMed] [Google Scholar]
- 11.Hirano, T., T. Taga, N. Nakano, K. Yasukawa, S. Kashiwamura, K. Shimizu, K. Nakajima, K.H. Pyun, and T. Kishimoto. 1985. Purification to homogeneity and characterization of human B-cell differentiation factor (BCDF or BSFp-2). Proc. Natl. Acad. Sci. USA. 82:5490–5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suematsu, S., T. Matsuda, K. Aozasa, S. Akira, N. Nakano, S. Ohno, J. Miyazaki, K. Yamamura, T. Hirano, and T. Kishimoto. 1989. IgG1 plasmacytosis in interleukin 6 transgenic mice. Proc. Natl. Acad. Sci. USA. 86:7547–7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopf, M., S. Herren, M.V. Wiles, M.B. Pepys, and M.H. Kosco-Vilbois. 1998. Interleukin 6 influences germinal center development and antibody production via a contribution of C3 complement component. J. Exp. Med. 188:1895–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumanogoh, A., S. Marukawa, T. Kumanogoh, H. Hirota, K. Yoshida, I.S. Lee, T. Yasui, K. Yoshida, T. Taga, and T. Kishimoto. 1997. Impairment of antigen-specific antibody production in transgenic mice expressing a dominant-negative form of gp130. Proc. Natl. Acad. Sci. USA. 94:2478–2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fornek, J.L., L.T. Tygrett, T.J. Waldschmidt, V. Poli, R.C. Rickert, and G.S. Kansas. 2006. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood. 107:1085–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ettinger, R., G.P. Sims, A.M. Fairhurst, R. Robbins, Y.S. da Silva, R. Spolski, W.J. Leonard, and P.E. Lipsky. 2005. IL-21 induces differentiation of human naive and memory B cells into antibody-secreting plasma cells. J. Immunol. 175:7867–7879. [DOI] [PubMed] [Google Scholar]
- 17.Ozaki, K., R. Spolski, R. Ettinger, H.P. Kim, G. Wang, C.F. Qi, P. Hwu, D.J. Shaffer, S. Akilesh, D.C. Roopenian, et al. 2004. Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J. Immunol. 173:5361–5371. [DOI] [PubMed] [Google Scholar]
- 18.Ozaki, K., R. Spolski, C.G. Feng, C.F. Qi, J. Cheng, A. Sher, H.C. Morse III, C. Liu, P.L. Schwartzberg, and W.J. Leonard. 2002. A critical role for IL-21 in regulating immunoglobulin production. Science. 298:1630–1634. [DOI] [PubMed] [Google Scholar]
- 19.Kishida, T., Y. Hiromura, M. Shin-Ya, H. Asada, H. Kuriyama, M. Sugai, A. Shimizu, Y. Yokota, T. Hama, J. Imanishi, et al. 2007. IL-21 induces inhibitor of differentiation 2 and leads to complete abrogation of anaphylaxis in mice. J. Immunol. 179:8554–8561. [DOI] [PubMed] [Google Scholar]
- 20.Leonard, W.J., and R. Spolski. 2005. Interleukin-21: a modulator of lymphoid proliferation, apoptosis and differentiation. Nat. Rev. Immunol. 5:688–698. [DOI] [PubMed] [Google Scholar]
- 21.Diehl, S.A., H. Schmidlin, M. Nagasawa, S.D. van Haren, M.J. Kwakkenbos, E. Yasuda, T. Beaumont, F.A. Scheeren, and H. Spits. 2008. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J. Immunol. 180:4805–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rincon, M., J. Anguita, T. Nakamura, E. Fikrig, and R.A. Flavell. 1997. Interleukin (IL)-6 directs the differentiation of IL-4–producing CD4+ T cells. J. Exp. Med. 185:461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suto, A., D. Kashiwakuma, S. Kagami, K. Hirose, N. Watanabe, K. Yokote, Y. Saito, T. Nakayama, M.J. Grusby, I. Iwamoto, and H. Nakajima. 2008. Development and characterization of IL-21–producing CD4+ T cells. J. Exp. Med. 205:1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wei, L., A. Laurence, K.M. Elias, and J.J. O'Shea. 2007. IL-21 is produced by Th17 cells and drives IL-17 production in a STAT3-dependent manner. J. Biol. Chem. 282:34605–34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wurster, A.L., V.L. Rodgers, A.R. Satoskar, M.J. Whitters, D.A. Young, M. Collins, and M.J. Grusby. 2002. Interleukin 21 is a T helper (Th) cell 2 cytokine that specifically inhibits the differentiation of naive Th cells into interferon γ–producing Th1 cells. J. Exp. Med. 196:969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coquet, J.M., K. Kyparissoudis, D.G. Pellicci, G. Besra, S.P. Berzins, M.J. Smyth, and D.I. Godfrey. 2007. IL-21 is produced by NKT cells and modulates NKT cell activation and cytokine production. J. Immunol. 178:2827–2834. [DOI] [PubMed] [Google Scholar]
- 27.Zhou, L., I.I. Ivanov, R. Spolski, R. Min, K. Shenderov, T. Egawa, D.E. Levy, W.J. Leonard, and D.R. Littman. 2007. IL-6 programs T(H)-17 cell differentiation by promoting sequential engagement of the IL-21 and IL-23 pathways. Nat. Immunol. 8:967–974. [DOI] [PubMed] [Google Scholar]
- 28.Nurieva, R., X.O. Yang, G. Martinez, Y. Zhang, A.D. Panopoulos, L. Ma, K. Schluns, Q. Tian, S.S. Watowich, A.M. Jetten, and C. Dong. 2007. Essential autocrine regulation by IL-21 in the generation of inflammatory T cells. Nature. 448:480–483. [DOI] [PubMed] [Google Scholar]
- 29.Korn, T., E. Bettelli, W. Gao, A. Awasthi, A. Jager, T.B. Strom, M. Oukka, and V.K. Kuchroo. 2007. IL-21 initiates an alternative pathway to induce proinflammatory T(H)17 cells. Nature. 448:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rogers, P.R., C. Dubey, and S.L. Swain. 2000. Qualitative changes accompany memory T cell generation: faster, more effective responses at lower doses of antigen. J. Immunol. 164:2338–2346. [DOI] [PubMed] [Google Scholar]
- 31.Chtanova, T., S.G. Tangye, R. Newton, N. Frank, M.R. Hodge, M.S. Rolph, and C.R. Mackay. 2004. T follicular helper cells express a distinctive transcriptional profile, reflecting their role as non-Th1/Th2 effector cells that provide help for B cells. J. Immunol. 173:68–78. [DOI] [PubMed] [Google Scholar]
- 32.Nurieva, R.I., Y. Chung, D. Hwang, X.O. Yang, H.S. Kang, L. Ma, Y.H. Wang, S.S. Watowich, A.M. Jetten, Q. Tian, and C. Dong. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaye, J., M.L. Hsu, M.E. Sauron, S.C. Jameson, N.R. Gascoigne, and S.M. Hedrick. 1989. Selective development of CD4+ T cells in transgenic mice expressing a class II MHC-restricted antigen receptor. Nature. 341:746–749. [DOI] [PubMed] [Google Scholar]
- 34.Poli, V., R. Balena, E. Fattori, A. Markatos, M. Yamamoto, H. Tanaka, G. Ciliberto, G.A. Rodan, and F. Costantini. 1994. Interleukin-6 deficient mice are protected from bone loss caused by estrogen depletion. EMBO J. 13:1189–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diehl, S., C.W. Chow, L. Weiss, A. Palmetshofer, T. Twardzik, L. Rounds, E. Serfling, R.J. Davis, J. Anguita, and M. Rincon. 2002. Induction of NFATc2 expression by interleukin 6 promotes T helper type 2 differentiation. J. Exp. Med. 196:39–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khader, S.A., G.K. Bell, J.E. Pearl, J.J. Fountain, J. Rangel-Moreno, G.E. Cilley, F. Shen, S.M. Eaton, S.L. Gaffen, S.L. Swain, et al. 2007. IL-23 and IL-17 in the establishment of protective pulmonary CD4(+) T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 8:369–377. [DOI] [PubMed] [Google Scholar]
- 37.Rincon, M., and R.A. Flavell. 1997. Transcription mediated by NFAT is highly inducible in effector CD4+ T helper 2 (Th2) cells but not in Th1 cells. Mol. Cell. Biol. 17:1522–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.The R Development Core Team. R: A Language and Environment for Statistical Computing. Version 2.8.0 (2008-10-20). Available at http://cran.r-project.org/doc/manuals/refman.pdf (accessed June 2008).
- 39.Gentleman, R.C., V.J. Carey, D.M. Bates, B. Bolstad, M. Dettling, S. Dudoit, B. Ellis, L. Gautier, Y. Ge, J. Gentry, et al. 2004. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Workman, C., L.J. Jensen, H. Jarmer, R. Berka, L. Gautier, H.B. Nielser, H.H. Saxild, C. Nielsen, S. Brunak, and S. Knudsen. 2002. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 3:R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolstad, B.M., R.A. Irizarry, M. Astrand, and T.P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 19:185–193. [DOI] [PubMed] [Google Scholar]
- 42.Irizarry, R.A., B.M. Bolstad, F. Collin, L.M. Cope, B. Hobbs, and T.P. Speed. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.