Abstract
The linker histone H1 is involved in maintaining higher-order chromatin structures and displays dynamic nuclear mobility, which may be regulated by posttranslational modifications. To analyze the effect of H1 tail phosphorylation on the modulation of the histone's nuclear dynamics, we generated a mutant histone H1, referred to as M1-5, in which the five cyclin-dependent kinase phosphorylation consensus sites were mutated from serine or threonine residues into alanines. Cyclin E/CDK2 or cyclin A/CDK2 cannot phosphorylate the mutant in vitro. Using the technique of fluorescence recovery after photobleaching, we observed that the mobility of a green fluorescent protein (GFP)-M1-5 fusion protein is decreased compared to that of a GFP-wild-type H1 fusion protein. In addition, recovery of H1 correlated with CDK2 activity, as GFP-H1 mobility was decreased in cells with low CDK2 activity. Blocking the activity of CDK2 by p21 expression decreased the mobility of GFP-H1 but not that of GFP-M1-5. Finally, the level and rate of recovery of cyan fluorescent protein (CFP)-M1-5 were lower than those of CFP-H1 specifically in heterochromatic regions. These data suggest that CDK2 phosphorylates histone H1 in vivo, resulting in a more open chromatin structure by destabilizing H1-chromatin interactions.
Consisting of a central globular domain flanked by two lysine-rich, positively charged amino (N)- and carboxy (C)-terminal tails, the mammalian linker histone H1 plays important roles in the stabilization of higher-order chromatin structure, in the inhibition of DNA replication, and in transcriptional regulation (39). Histone H1 binds to the nucleosomal core particle near the entry and exit point of DNA, although its exact location within the 165-bp chromatosome remains controversial (14, 48, 50). Mammals possess up to five somatic histone H1 variants, termed H1a to H1e (nomenclature is from reference 42). Two other H1 variants, H10 and H1t, are found in differentiated cells and testes, respectively. The variants have been suggested to have different functions in cell cycle progression and gene expression (8).
The phosphorylation of histone H1 at its N- and C-terminal tails during the cell cycle influences its function. Phosphorylation of H1 increases during the transition from G1 to S phase, reaching a limited maximum during S phase (45). Additional phosphorylation occurs at the G2-M transition, resulting in maximal phosphorylation during mitosis. It has been shown that histone H1 phosphorylation increases or decreases transcription of specific genes (3, 17, 18) and that phosphorylated H1 is localized to RNA splicing centers (13), suggesting a regulatory role for phosphorylation.
Whereas phosphorylated H1a, H1c, and H1e can contain four phosphate groups, H1b and H1d contain five, corresponding to the number of conserved cyclin-dependent kinase phosphorylation sequence motifs located at the tails (45). Consistent with these findings, cdc2 has been implicated as the major in vivo G2 kinase for H1 (30). Recent data suggest that CDK2 is another in vivo H1 kinase and is perhaps responsible for the H1 phosphorylation observed during the transition from G1 to S phase (3, 5, 12, 23). At each stage of the cell cycle, H1b is the most highly phosphorylated of any of the H1 variants.
As summarized above, histone H1 is involved in maintaining chromatin higher-order structure. Specifically, linker histones can both direct and stabilize the in vitro folding of nucleosomal arrays into compact, condensed structures (1, 9, 28). While many of the studies investigating H1 function have been performed using in vitro systems, analyses with Tetrahymena strongly suggest that H1 also regulates higher-order structure in vivo (6). The globular domain of linker histones binds to DNA in the nucleosome, while the tails are believed to stabilize the folded chromatin fibers (22). Early reconstitution experiments demonstrated that phosphorylation of the histone H1 tails diminishes H1's ability to condense chromatin (29). More recently, others have shown that while in vivo phosphorylation does not influence H1 binding to mononucleosomes, in vitro aggregation of polynucleosomes is decreased by linker histone phosphorylation (46).
The consequence of increased H1 phosphorylation appears to be the relaxation of chromatin structure (13, 23, 47). Accordingly, dephosphorylated H1 is located in the electron-dense chromatin bodies of Tetrahymena macronuclei, whereas phosphorylated H1 is present at higher levels in the surrounding euchromatin (35). Relaxed or decondensed chromatin is suggested to facilitate the activities of the replication and transcription machineries on DNA (11, 13, 21). However, consistent with the observation that the highest levels of H1 phosphorylation occur during mitosis, a model suggesting that phosphorylation drives chromosome condensation by promoting H1-H1 protein interactions via the proteins' globular domains was proposed (6). An alternative model postulates that phosphorylation of H1 weakens tail-DNA interactions and decreases H1-H1 globular domain binding, resulting in a decondensed chromatin state (41). Support for the latter model comes from experiments which have demonstrated that while unphosphorylated linker histone inhibits the activity of ATP-dependent chromatin-remodeling enzymes on nucleosomal arrays, in vitro phosphorylation of the histone before incorporation into the arrays can restore enzyme activity by relaxing the topological constraints induced by unphosphorylated histone H1 (26).
Importantly, in vivo evidence to support the latter model was obtained by analyzing the mobility of green fluorescent protein (GFP)-tagged histone H1 in living cells by using the technique of fluorescence recovery after photobleaching (FRAP) (31, 36). In these FRAP experiments, fluorescently tagged histones were expressed in cells, followed by photobleaching of specific nuclear regions. The relative level of recovery of the protein can be measured within the bleached area. GFP-H1 recovered within several minutes, whereas GFP-H2B did not show appreciable recovery over the same time period (31). Deletion of the C-terminal tail increased the rate of recovery, suggesting that the H1 tail is involved in stabilizing H1-chromatin association. Furthermore, inhibition of kinase activity decreased the level of recovery of GFP-H1. Recently, with the use of histone H1 mutants, it has been shown that phosphorylation of the histone tails and an as yet undescribed ATP-dependent process both increase H1-chromatin dissociation (16). However, these studies were performed using Tetrahymena H1, which lacks the central globular domain contained in the mammalian H1. Thus, histone H1 is in nuclear dynamic equilibrium, and phosphorylation of its tails is suggested to alter H1-chromatin binding (31, 36). Interestingly, GFP-H1 recovered to a lesser extent in heterochromatic regions than in euchromatin, suggesting a more statically bound GFP-H1 in heterochromatin (36).
We set out to determine whether direct phosphorylation of the H1 tails influences the dynamic mobility of histone H1 in vivo and to identify the responsible kinase(s) in mammalian cells. The serine or threonine residues in the five cdc/CDK phosphorylation consensus sites were mutated into alanines, followed by the fusion of GFP to the carboxy-terminal end of either wild-type histone H1b or unphosphorylated mutant histone, termed M1-5. FRAP experiments were performed to compare the mobilities of the two chimeric proteins in different cell lines, which exhibited different CDK2 activities. GFP-M1-5 recovered to a lesser extent in two immortalized cell lines studied but not in a third cell line that was shown to have much lower intrinsic CDK2 activity. Consistent with these results, cells at G0, where CDK2 activity is very low, and cells in which the CDK2 inhibitor p21 is overexpressed displayed decreased GFP-H1b mobility. Lastly, by specifically analyzing heterochromatic regions, we observed that cyan fluorescent protein (CFP)-tagged M1-5 recovers considerably slower than CFP-H1b in heterochromatin although their rates of recovery are the same in euchromatin, suggesting that the statically bound form of GFP-H1b observed previously in heterochromatin is in the unphosphorylated state.
MATERIALS AND METHODS
Plasmids.
The H1b phosphorylation mutant M1-5 was created by successive rounds of in vitro site-directed mutagenesis by using pcDNA3.1/HygroH1b (FLAG) as the starting template. pcDNA3.1/HygroH1b (FLAG) contains the coding sequence of the human H1b gene in front of an oligonucleotide coding for the FLAG epitope. The in vitro site-directed mutagenesis was accomplished using the Quikchange site-directed mutagenesis kit (Stratagene) with five sets of primers (25 to 28 bp in length) each centered around one of the phosphorylated serine or threonine residues within the five potential cyclin/CDK2 phosphorylation motifs (see Fig. 1). These five mutagenic primer pairs contained alterations which resulted in a serine- or threonine-to-alanine change at the following sites: residue 18 (ACT→GCT), residue 146 (ACC→GCC), residue 154 (ACC→GCC), residue 172 (AGC→GCC), and residue 187 (AGC→GCC).
FIG. 1.
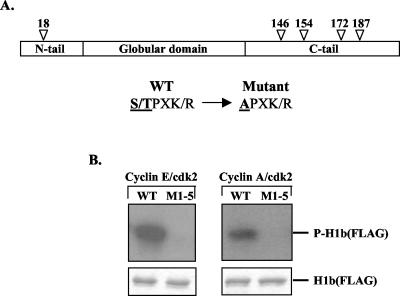
Cyclin E/CDK2 and cyclin A/CDK2 phosphorylate wild-type but not mutant H1b in vitro. (A) Schematic diagram representing the tripartite structure of the human linker histone H1b consisting of a globular domain flanked by an N- and a C-terminal tail. Below are the sequences of the cyclin-dependent kinase phosphorylation consensus motifs from wild-type and mutant H1, with the phosphorylated residues underlined. The arrows indicate the locations of these five residues within H1b. Each of these serine or threonine residues within wild-type H1b (FLAG) was mutated into an alanine residue in the H1b phosphorylation mutant M1-5 (FLAG). (B) In the top panel, results of an in vitro kinase assay demonstrate that the cyclin E/CDK2 and cyclin A/CDK2 complexes phosphorylate the wild-type (WT) H1b (FLAG) protein but not the phosphorylation mutant M1-5 (FLAG). The bottom panel shows results of a kinase assay performed in parallel. The gel was then stained with Coomassie, showing that equal amounts of FLAG-tagged H1b protein were loaded.
Both the H1b and M1-5 coding regions were excised from pcDNA3.1/Hygro (FLAG) and cloned into the pEGFP-N1 (Clontech) and pECFP-N1 (Clontech) vectors. The GFP or CFP coding sequence is located 3′ of the histone coding sequence. The cytomegalovirus promoter drives expression. For the cell cycle and p21 infection experiments, the coding sequences for GFP-H1b and GFP-M1-5 were excised from the pEGFP-N1 vector and cloned into the bicistronic vector pEFIRES-N (25), kindly provided by Steven Hobbs (Cancer Research Campaign Center for Cancer Therapeutics, Institute of Cancer Research, London, United Kingdom). The pEF-1α promoter drives the expression of the chimeric proteins in the pEFIRES-N vector. This promoter was used because it functions in serum-starved cells. The construction of the yellow fluorescent protein (YFP)-tagged lac repressor containing a nuclear localization sequence has been previously described (37).
In vitro kinase assays.
Purified FLAG-tagged Hib [H1b (FLAG)] and FLAG-tagged M1-5 [M1-5 (FLAG)] proteins used as phosphorylation substrates in the in vitro kinase assay were prepared as follows. Wild-type H1b (FLAG) and M1-5 (FLAG) coding regions were subcloned from the pcDNA3.1/Hygro vector into the bacterial expression vector pET-3d (Stratagene). The expression clones, pET-3dH1b WT (FLAG) and pET-3dH1b M1-5 (FLAG), were used to transform Escherichia coli BL21 (DE3) cells and were induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). After lysis, the cell lysates were precipitated with 5% perchloric acid to remove bacterial proteins. The bacterially expressed FLAG-tagged wild-type and M1-5 H1b proteins were then purified from the resulting supernatant by precipitation with 15% trichloric acid, washed once with acidified acetone (0.5 ml of concentrated HCl/100 ml) and twice with acetone, dried in a vacuum desiccator, dissolved in Tris-buffered saline, and subsequently passed over an anti-FLAG M2 affinity column (Sigma). Protein concentrations were determined using the Pierce Coomassie Plus kit (Pierce).
The kinase complexes were coimmunoprecipitated from Sf9 cell lysates containing baculovirus-expressed cyclin E/CDK2 and cyclin A/CDK2 (Pharmingen). Cell lysates (40 μg) were incubated with 4 μg of CDK2 (M2) rabbit polyclonal antibody (Santa Cruz) and Protein A/G PLUS agarose (Santa Cruz) in a final volume of 500 μl of ELB (150 mM NaCl, 50 mM HEPES [pH 7.5], 5 mM EDTA, 0.1% NP-40, Complete protease inhibitor [Roche]) for 18 h at 4°C with gentle agitation. The beads were then washed three times with ELB and once with 1× kinase buffer (20 mM Tris [pH 7.5], 10 mM MgCl2, 5 mM MgCl2, 2.5 mM MnCl2, 1 mM dithiothreitol buffer) before being resuspended in 1× kinase buffer, 3 μCi of [γ-32P]ATP (Perkin-Elmer), 14 μM ATP, and 2 μg of H1b (FLAG) protein to a final volume of 15 μl. After incubation at 30°C for 15 min, sodium dodecyl sulfate (SDS) loading dye was added and the reaction mixtures were heated to 99°C for 5 min before being analyzed by SDS-12% polyacrylamide gel electrophoresis. After electrophoresis, the wet gel was exposed to autoradiography. As a loading control, the above-described assay was done in duplicate and the resulting SDS-PAGE gel was stained with Coomassie dye for visualization of the proteins.
Endogenous CDK2-associated kinase activities of asynchronously growing HeLa, WI-38 VA, and WI-38 cells (Fig. 2C) were determined as described above. Equal amounts of protein were used for each immunoprecipitation. The kinase substrate was calf thymus histone H1 (Boehringer Mannheim).
FIG. 2.
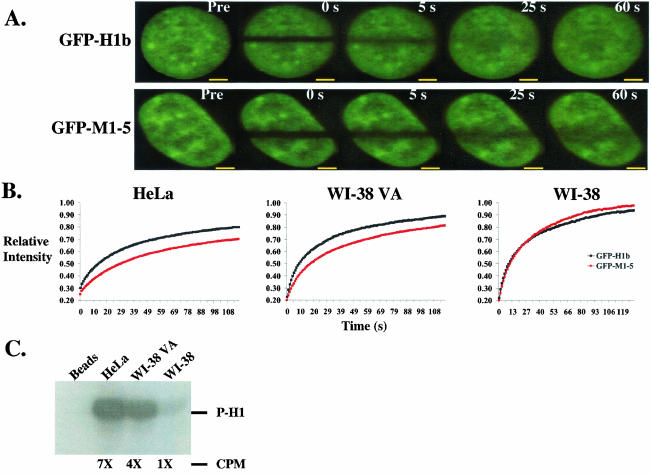
GFP-M1-5 recovers to a lesser extent than GFP-H1b in HeLa and WI-38 VA cells but not in WI-38 cells. (A) Representative immunofluorescence of HeLa cells transfected with either GFP-H1b (top panel) or GFP-M1-5 (bottom panel) and photobleached for FRAP analysis as described in Materials and Methods. An image was taken before bleaching (Pre), immediately after bleaching (0 s), and then every 0.5 s until the level of recovery within the bleached region stabilized. Comparison at 25 and 60 s shows that GFP-H1b recovered more in the bleached region than did GFP-M1-5. Bar, 3 μM. (B) Quantitative analysis comparing the relative levels of recovery of GFP-H1b and GFP-M1-5 in HeLa, WI-38 VA, and WI-38 cells. Each data point represents the mean of the levels of recovery, measured at 0.5-s intervals, of at least 10 cells from one experiment. GFP-M1-5 did not recover to the same level as did GFP-H1b in HeLa and WI-38 VA cells. Experiments were done in triplicate, and similar results were measured for the different experimental sets. Although not shown, error bars at each time point were calculated as twice the standard error of the mean and showed no significant overlap for FRAP experiments done in HeLa and WI-38 VA cells. Analysis using the standard Student t test exhibited high statistical significance (see the text). (C) Histone H1 kinase activity of anti-CDK2 immunoprecipitated from cell lysates of exponentially growing, asynchronous HeLa, WI-38 VA, and WI-38 cells. The position of 32P-labeled histone H1 (P-H1) is indicated. Sevenfold (7×) more CDK2 kinase activity is present in HeLa cells and fourfold (4×) more CDK2 kinase activity is present in WI-38 VA cells than in WI-38 cells (1×), as determined by radioactivity quantitation measured as counts per minute (CPM).
Cell culture, transfection, and adenoviral infection.
HeLa, WI-38 VA (13 subline 2RA), and WI-38 cells were purchased from the American Type Culture Collection. These cell lines were maintained at 37°C and 5% CO2 in Dulbecco's modification of Eagle's medium (DMEM; Cellgro Inc.) supplemented with 10% fetal bovine serum (FBS). AO3_1 cells were cultured as described previously (32).
From a confluent T-75 flask, HeLa, WI-38 VA, or WI-38 cells were split 1:20, 1:10, or 1:6, respectively, into 60-mm-diameter dishes containing acid-etched coverslips. After 18 h, cells were transfected using the FuGENE 6 reagent (Roche Molecular Biochemicals). For each transfection, the total amount of DNA was kept at 2 μg. The amounts of expression plasmids used were 0.25, 0.5, and 2.0 μg for HeLa, WI-38 VA, and WI-38 cells, respectively. The ratio of the amount of DNA (in micrograms) to the volume of FuGENE 6 reagent (in microliters) was kept at 1:3 for each transfection. Transfection was allowed to proceed for 48 h. For cell synchronization experiments using WI-38 cells, transfection was allowed to proceed for 24 h, followed by the addition of DMEM with 0.1% FBS for 48 h. To analyze cells at late G1, 10% FBS was added back for 12 h. For adenovirus infection, the 24-h transfection of WI-38 VA cells was followed by the addition of DMEM with 0.1% FBS for 48 h. Cells were then infected for 36 h with adenovirus expressing either E. coli β-galactosidase (β-Gal) or p21 (kindly provided by M. Rijnkels and J. W. Harper, respectively, Baylor College of Medicine, Houston, Tex.) at a multiplicity of infection of 100, as previously described (19). For experiments done with the AO3_1 cell line, cells were cotransfected with 0.4 μg of YFP-Lac and 0.4 μg of either CFP-H1b or CFP-M1-5
FRAP.
FRAP experiments were performed on a Zeiss LSM 510 confocal microscope, as previously described (44). Briefly, cells were transferred to a live-cell closed chamber (Bioptechs, Inc.) with the appropriate medium recirculated by a peristaltic pump. One prebleach image was acquired, followed by bleaching at the appropriate wavelength for 50 iterations over the selected region. Images were taken every 0.5 s after bleaching. Fluorescence intensities were obtained using the LSM software, and data were analyzed using Microsoft Excel, as described previously (40). The times required for recovery of 50% of fluorescence intensity were calculated for each individual cell and averaged.
RESULTS
Cyclin E/CDK2 and cyclin A/CDK2 phosphorylate wild-type histone H1b but not mutant histone M1-5 in vitro.
Previous studies have suggested that one in vivo kinase for the linker histone H1 is CDK2 and that phosphorylation of histone H1 may influence its binding affinity for chromatin (see above). To establish directly the importance of specific H1b phosphorylation sites in controlling mobility, we generated a mutant H1b, M1-5, in which the five potential CDK/cyclin phosphorylation consensus sites located within the tail domains were mutated from serine or threonine residues to alanines (Fig. 1A).
To compare the extents of phosphorylation of histone H1b and M1-5 by CDK2/cyclins, we carried out in vitro kinase assays using purified FLAG-tagged wild-type H1b or M1-5 as the phosphorylation substrate and baculovirus-produced cyclin E/CDK2 and cyclin A/CDK2 as potential kinases. These complexes have been shown to phosphorylate H1 in vitro in a manner indistinguishable from intracellular generation during late G1 (23). Whereas histone H1b is phosphorylated by both cyclin E/CDK2 and cyclin A/CDK2 in vitro, M1-5 is not phosphorylated by either kinase complex (Fig. 1B). Similarly, as determined by Western analysis using an antibody specific for phosphorylated H1 (Upstate), stably expressed FLAG-tagged H1 is phosphorylated in vivo whereas FLAG-tagged M1-5 is not (T. K. Hale and R. E. Herrera, unpublished data).
GFP-M1-5 recovers more slowly and to a lesser extent than GFP-H1b in HeLa and WI-38 VA cells but not in WI-38 cells.
In order to study the role of phosphorylation on the nuclear dynamics of histone H1b, we first fused GFP to the carboxy termini of both H1b and M1-5. GFP tagging of the linker histone or mutant histone does not significantly influence the proper assembly or function of the fusion proteins in chromatin (16, 31, 36). Furthermore, the salt-dissociation rate of the chimeric proteins is similar to that of endogenous H1 (A. Contreras and R. E. Herrera, unpublished data). HeLa cells were transiently transfected with the chimeric proteins, and the dynamic properties of the two GFP-tagged proteins in the nuclei of living cells were compared using FRAP. Based upon visual inspection of fluorescence intensities, the transfected cells expressed varying levels of the chimeric proteins. Due to the alterations of chromatin structure observed with H1 overexpression (20), only lower-expressing cells were chosen for analysis. These lower-expressing cells did not exceed a calculated average fluorescence intensity of 150 arbitrary confocal units.
After photobleaching of a strip of approximately 1 μm in width across the nucleus, the extent of recovery of fluorescence signal was recorded in the bleached region every 0.5 s until the intensity signal stabilized. These data were used to analyze protein mobility by calculating the time required for 50% of the fluorescence signal to recover over the bleached region (rate of recovery, represented by t1/2). In addition, the extents of recovery of the GFP-tagged histones were measured by analyzing the percentage of fluorescence signal that recovered within the photobleached region. This percentage measures the fraction of molecules that is mobile (27). Differences between the extents of recovery among proteins measured in the same cell type reflect differences in immobile fractions of the chimeric proteins.
In HeLa cells, GFP-H1b recovered to 47% during the first 5 s after bleaching, in contrast to GFP-M1-5, which recovered to only 37% over the same time period (Fig. 2A and B). This difference observed in the extents of recovery at 5 s postbleaching was maintained throughout the course of the experiment. The level of recovery of the intensity signal for both chimeric proteins over the bleached region stabilized after approximately 100 s. After this time period, GFP-H1b recovered to 82%, whereas GFP-M1-5 recovered to 72%. This difference is highly statistically significant as calculated by the standard Student t test (P = 0.001). In addition, GFP-H1 recovered twice as fast as GFP-M1-5, as the t1/2 values for the proteins were 6.0 and 15.2 s, respectively (P = 0.003) (Table 1). Though not included in this analysis, cells with high fluorescence intensities and normal morphology exhibited the same differences in the rates and extents of recovery of the wild-type and mutant proteins (data not shown).
TABLE 1.
Time required for 50% recovery of fluorescence (t1/2) and percent recovery
| Cell line | Histone and/or relevant experimental conditions | t1/2 (s) ± 2 × SEM | P value (t test) | % Recovery ± 2 × SEM | P value (t test) |
|---|---|---|---|---|---|
| HeLa | GFP-H1 | 6.0 ± 1.8 | 0.003 | 82 ± 3.6 | 0.001 |
| GFP-M1-5 | 15.2 ± 5.0 | 72 ± 4.2 | |||
| WI-38 | GFP-H1 | 3.7 ± 1.4 | 0.9 | 89 ± 6.0 | 0.32 |
| GFP-M1-5 | 3.6 ± 1.2 | 93 ± 4.0 | |||
| Cells in G0 | 6.0 ± 1.0 | <0.001 | 83 ± 2.0 | 0.04 | |
| Asynchronous cells | 3.1 ± 0.6 | 89 ± 4.2 | |||
| Cells in G0 | 6.0 ± 1.0 | 0.04 | 83 ± 2.0 | 0.04 | |
| Cells in mid to late G1 | 4.4 ± 0.8 | 87 ± 2.5 | |||
| WI-38 VA | GFP-H1 | 4.1 ± 1.0 | 0.004 | 89 ± 4.0 | 0.01 |
| GFP-M1-5 | 7.5 ± 1.6 | 82 ± 2.8 | |||
| GFP-H1, p21-expressing cells | 16.1 ± 3.4 | 0.001 | 71 ± 3.4 | 0.005 | |
| GFP-H1, β-Gal-expressing cells | 8.3 ± 1.4 | 81 ± 4.6 | |||
| GFP-M1-5, p21-expressing cells | 10.1 ± 1.8 | 0.70 | 78 ± 3.2 | 0.73 | |
| GFP-M1-5, β-Gal-expressing cells | 10.9 ± 3.4 | 79 ± 6.4 | |||
| AO3_1 | CFP-M1-5, lac array | 25.5 ± 7.0 | 69 ± 9.1 | ||
| CFP-M1-5, nucleus | 19.7 ± 5.6 | 0.22a | 86 ± 9.8 | 0.04a | |
| CFP-H1, lac array | 12.4 ± 3.0 | 0.004a | 87 ± 8.2 | 0.008a | |
| CFP-H1, nucleus | 14.0 ± 3.0 | 0.009a | 89 ± 6.0 | 0.005a |
P values are for comparisons with GFP-M1-5 in the lac arrays.
To determine whether the differences observed in HeLa cells in the rates and extents of recovery of wild-type histone H1b and the mutant histone are observed in different cell types, we carried out FRAP experiments with WI-38 human normal diploid lung fibroblasts and with the counterpart WI-38 VA cell line. WI-38 cells have a finite passage number, whereas WI-38 VA cells are immortalized due to stable expression of simian virus 40 large T antigen. Similar to what was observed in HeLa cells, GFP-M1-5 recovered to a lesser extent than GFP-H1b in WI-38 VA cells. After 100 s, GFP-M1-5 recovered to 82%, whereas GFP-H1b recovered to 89% (P = 0.01) (Fig. 2B). The exchange rate for GFP-H1b (t1/2 = 4.1 s) in WI-38 VA cells was also significantly faster than that for GFP-M1-5 (t1/2 = 7.5 s) (P = 0.004) (Table 1). Interestingly, no difference in mobilities of GFP-H1b and GFP-M1-5 in WI-38 cells was observed in the recovery (Fig. 2B; Table 1).
Because CDK2 has been suggested as an in vivo kinase for histone H1 (3, 5, 23), we compared the CDK2 activity levels of these three different cell lines. Histone H1 purified from calf thymus was used as the phosphorylation substrate in measuring the CDK2-associated kinase activity from lysates of asynchronously growing cells. As shown in Fig. 2C, WI-38 cells have the lowest level of CDK2 activity. Compared to WI-38 cells, WI-38 VA cells and HeLa cells have four- and sevenfold greater levels of CDK2 activity, respectively (Fig. 2C). Thus, the levels of recovery of the fusion proteins directly correlate with CDK2 activity. When CDK2 activity is low, moderate, or high, the difference in levels of recovery between the two chimeras is low, moderate, or high, respectively.
GFP-H1b recovers to a lesser extent in WI-38 cells at G0 than in cells at mid to late G1.
A correlation between G1 progression, CDK2 activity, and H1 phosphorylation in WI-38 cells has been reported previously (23). Briefly, at G0, when cells have low CDK2 activity, low levels of phosphorylated histone H1 are observed. However, as cells progress through mid to late G1, increasing levels of histone H1 phosphorylation correlate with increasing CDK2 activity. The FRAP experiments depicted in Fig. 2 indicated that a correlation exists between CDK2 activity and the levels of recovery of the proteins. We therefore wanted to investigate whether this correlation is also apparent during changes in CDK2 activity during the cell cycle.
To test whether the difference in CDK2 activity observed during G1 progression correlates with a change in the level of recovery of GFP-H1b in WI-38 cells, we performed FRAP experiments with transfected cells that were either serum starved for 72 h or serum starved before the readdition of serum for 12 h (at which point the cells are in late G1 [23]). For comparison, FRAP was also performed with an asynchronous population of transfected WI-38 cells. As shown in Fig. 3 and presented in Table 1, GFP-H1b recovered to a higher level in G1 and asynchronous cells than in those arrested in G0. After 45 s, when the intensity within the photobleached region of the nucleus stabilized, GFP-H1b recovered to 83, 87, and 89% in cells at G0 and mid to late G1 and in asynchronous cells, respectively. P values for G0 versus G1 or asynchronous cells are both 0.04. The t1/2 values for GFP-H1 in these cells are 6.0, 4.4, and 3.1 s, respectively. The exchange rate observed in the G0 cells is significantly different from those observed in the asynchronous cells and in cells in mid to late G1 (Table 1). Thus, the recovery rate of GFP-H1 is lower in cells that are in phases of the cell cycle that have low CDK2 activity. The dynamics of GFP-M1-5 are independent of cell cycle status, as no change in GFP-M1-5 mobility is observed when cells in G0 are compared to those in late G1 (A. Contreras and R. E. Herrera, unpublished data).
FIG. 3.
GFP-H1b recovers least in WI-38 cells at G0. A quantitative analysis of results from FRAP experiments was carried out to compare the relative levels of recovery of GFP-H1b in asynchronous cells, cells at late G1, and serum-starved cells (G0). For analysis of WI-38 cells at G0 and late G1, cells were transfected with GFP-H1b for 24 h, followed by serum starvation in growth medium with 0.1% FBS. For cells at late G1, growth medium supplemented with 10% FBS was added back for 12 h. The level of recovery of GFP-H1b is lowest in cells at G0. Experiments were done as described above but in duplicate. Error bars did not significantly overlap between results for asynchronous cells and G0 cells or between results for cells at late G1 and those for cells at G0.
p21 expression decreases the recovery and exchange rates of GFP-H1b, but not those of GFP-M1-5, in WI-38 VA cells.
The role of p21 in inhibiting G1 progression by blocking the activity of CDK/cyclins has been well characterized (43). Infection of cells with an adenovirus expressing p21 has been shown to block the activity of CDK2 (19). Recent data suggest that p21 expression leads to a decrease in phosphorylation of histone H1 and CDK2 kinase activity in serum-starved retinoblastoma (Rb) null mouse embryo fibroblasts (A. Morrison and R. E. Herrera, unpublished data). These cells maintain high levels of CDK2, due to the derepression of the cyclin E gene, even in G0. It has been shown that the only active CDK in these G0-arrested Rb−/− cells is CDK2 (24). Therefore, p21 expression in G0 Rb null mouse embryo fibroblasts most likely only blocks the activity of CDK2.
As shown in Fig. 2 and 3, CDK2 may play a role in influencing the recovery of GFP-H1b in living cells. To test this observation more directly, we wanted to analyze the effect of p21 expression on the mobility of GFP-H1b in WI-38 VA cells at G0. WI-38 VA cells have high CDK2 activity at G0 due to the inactivation of the Rb protein by large T antigen, which leads to derepression of the cyclin E promoter and cyclin E/CDK2 activation. Therefore, when cells are arrested in G0, CDK2 is the only active CDK in these cells. WI-38 VA cells were transfected with GFP-H1b for 16 h, followed by serum starvation. After 48 h of serum starvation, cells were infected for 30 h with either β-Gal-expressing adenovirus (adeno-β-Gal) or p21-expressing adenovirus (adeno-p21). FRAP experiments were then carried out following adenovirus infection as described before.
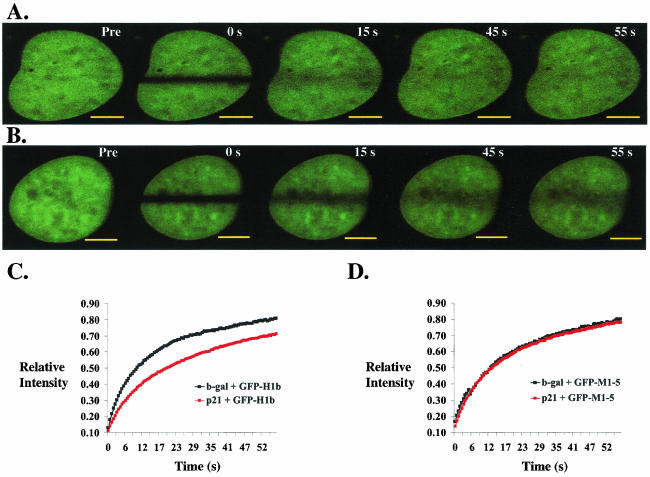
Infection with adeno-p21 (Fig. 4B) decreased the recovery of GFP-H1b compared with that in cells infected with adeno-β-Gal (Fig. 4A). Fifteen seconds after bleaching, GFP-H1b recovered to 58% in cells infected with adeno-β-Gal but only to 45% in adeno-p21-infected cells (Fig. 4C). After the level of intensity recovery had stabilized, GFP-H1b recovered to 81% in adeno-β-Gal-infected cells but, in adeno-p21-infected cells, the extent of recovery was 71% (P = 0.005). Furthermore, the exchange rate of GFP-H1b decreased nearly twofold, as the t1/2 value for p21-infected cells was 16.2 s, compared to 8.3 s for the β-Gal-expressing cells (P = 0.001) (Table 1).
FIG. 4.
p21 overexpression decreases the recovery of GFP-H1b, but not that of GFP-M1-5, in WI-38 VA cells at G0. (A and B) Representative immunofluorescence of WI-38 VA cells transfected with GFP-H1b, serum starved for 48 h, infected with adenovirus expressing either β-Gal (A) or p21 (B), and photobleached for FRAP analysis as described in Materials and Methods. An image was taken before bleaching (Pre), immediately after bleaching (0 s), and then every 0.5 s until the level of recovery within the bleached region stabilized. Comparison of the immunofluorescent images 15 s postbleaching shows that the extent of recovery of GFP-H1b is lower in cells infected with adenovirus expressing p21. Bar, 3 μM. (C and D) Quantitative analyses comparing the relative recoveries of GFP-H1b (C) and GFP-M1-5 (D) in WI-38 VA cells infected with adenovirus expressing either β-Gal or p21. Each data point represents the mean of the levels of recovery, measured at 0.5-s intervals, of at least 10 cells from one experiment. Infection with adeno-p21 decreases the extent of recovery of GFP-H1b (p21 + GFP-H1b) compared to that observed in the case of infection with adeno-β-Gal (b-gal + GFP-H1b). No change in recovery of GFP-M1-5 is measured in cells infected with either adeno-p21 (p21 + GFP-M1-5) or adeno-β-Gal (b-gal + GFP-M1-5). Experiments were done as described above. Error bars did not significantly overlap between the curves of p21 plus GFP-H1b and β-Gal plus GFP-H1b.
To test whether these differences in rates and extents of recovery are due to a change in the phosphorylation status of GFP-H1b and to ensure that the recovery difference seen with the transfection of GFP-H1b is not due to overexpression of β-Gal, FRAP experiments were carried out in WI-38 VA cells transfected with GFP-M1-5. As shown in Fig. 4D and Table 1, the recovery of GFP-M1-5 is not affected in cells overexpressing p21 compared to that in cells infected with adeno-β-Gal. Taken together, these results demonstrate that the mobility of GFP-histone H1 that is unable to be phosphorylated is decreased compared to that of GFP-histone H1 that can be phosphorylated and that CDK2 activity directly or indirectly determines the phosphorylation of histone H1 in vivo.
The mobility of CFP-M1-5 is reduced compared to that of CFP-H1b in heterochromatin.
Previous studies have compared wild-type GFP-H1 dynamics in heterochromatic versus euchromatic regions, defining these regions according to morphological criteria (36). We wanted to analyze the mobility of our GFP-tagged chimeric proteins more directly in both heterochromatic and more euchromatic regions of the nucleus to test whether phosphorylation status may influence histone H1 dynamics in these regions.
In order to target our FRAP analysis to heterochromatic regions, we took advantage of a lac repressor-based system that allowed for the direct visualization of a heterochromatic chromosome arm generated by gene amplification (2, 49). AO3_1 CHO cells, in which lac operator repeats and coamplified genomic DNA form an ∼90-Mbp highly condensed heterochromatic chromosomal array (32), were cotransfected with YFP-tagged Lac and either CFP-tagged H1b or CFP-M1-5. YFP-Lac binds to the lac operator repeats within the array, and thus the heterochromatic region can be visualized as a 0.5- to 1.0-μM mass (Fig. 5A) by fluorescence microscopy. For FRAP analysis, photobleaching was performed across a region of the nucleus that contains the heterochromatic array (Fig. 5A). The extent and rate of recovery of CFP-H1b or CFP-M1-5 were measured over the heterochromatic array and over the “nonarray” portion of the nucleus (Fig. 5B, inset).
FIG. 5.
CFP-M1-5 recovers to a lesser extent than CFP-H1b in heterochromatin. (A) Representative immunofluorescence of AO3_1 CHO cells cotransfected with CFP-H1b (green) and YFP-Lac (red) as determined by FRAP analysis. YFP-Lac is targeted to a heterochromatic array containing lac repeats. An image was taken before bleaching (Pre), immediately after bleaching (0 s), and then every 0.5 s until the level of recovery within the bleached region stabilized. Bleached regions contained the entire heterochromatic array, as shown in panel A. (B) Quantitative analysis comparing the relative recoveries of CFP-H1b and CFP-M1-5 in either heterochromatin (CFP-H1b : Lac and CFP-M1-5 : Lac, respectively) or the portion of the nucleus not containing the array (CFP-H1b : Nuc and CFP-M1-5 : Nuc) (see inset). Each data point represents the mean of the levels of recovery, measured at 0.5-s intervals, of at least 10 cells from one experiment. CFP-M1-5 recovered to a lesser extent than CFP-H1b in the heterochromatic array (Lac) but not in general nuclear regions (Nuc). Experiments were done as described above. Error bars did not significantly overlap between results for CFP-H1b : Lac and those for CFP-M1-5 : Lac.
When the recovery plateau was reached at 110 s after bleaching, the relative extent of recovery of CFP-H1b over the heterochromatic array was 87% (Fig. 5B, graph). However, the extent of recovery of the CFP-M1-5 mutant was only 69% (P = 0.008). In addition, the recovery rate of CFP-H1 (12.4 ± 3.0 s) was faster than that of CFP-M1-5 (25.5 ± 7.0 s) in heterochromatin (Table 1). Interestingly, over the nonarray portion of the bleached nucleus, CFP-H1b and CFP-M1-5 recovered to 89 and 85%, respectively, and at the same rate (Table 1). The CDK2 activity in these CHO cells is sixfold less than that observed in HeLa cells (A. Contreras and R. E. Herrera, unpublished data). Thus, the major decrease in rates and extents of recovery of CFP-M1-5 compared with those of CFP-H1b is specific for heterochromatic regions.
DISCUSSION
The FRAP technique has become an essential tool for studying protein dynamics in living cells (27, 33). Intensity measurements taken after the photobleaching of a cell expressing a fluorescently tagged protein can be used to describe and compare the mobilities of different proteins. Using this technique, others have previously analyzed the nuclear dynamics of linker histone variants (16, 31, 36). A role for phosphorylation of H1 was proposed, as both treatment of cells with kinase inhibitors and mutation of the histone tails at phosphorylation consensus motifs decreased the recovery rate of H1 (16, 31). In the present study, we show that a GFP-tagged mutant histone H1 that cannot be phosphorylated (Fig. 1) recovers more slowly and to a lesser extent than the wild-type linker histone GFP-H1b in vivo (Fig. 2). Furthermore, the recovery of GFP-H1b is dependent on the activity of the cyclin-dependent kinase, CDK2 (Fig. 3 and 4). Lastly, by targeting our analysis to specific heterochromatic regions, we show that the extent and rate of recovery of CFP-M1-5 are lower than those of CFP-H1b, primarily in heterochromatin (Fig. 5).
The recovery rate of GFP-H1b, but not that of GFP-M1-5, correlates with intracellular CDK2 activity.
Similar to what is observed in Tetrahymena (16), our mutant histone chimera GFP-M1-5 is less mobile than GFP-H1b in HeLa and WI-38 VA cells. However, no difference in mobilities between the two fusion proteins is observed in the WI-38 cell line, which has significantly lower CDK2 activity than the other two cell lines studied (Fig. 2). These data are consistent with the hypothesis that unphosphorylated tails stabilize H1-chromatin interactions. The decrease in the level of recovery observed for GFP-M1-5 in the bleached area may be due to a population of GFP-M1-5 that is more immobile than that of GFP-H1b in the nonbleached areas of the nucleus (1), and/or within the bleached area, potential binding sites for H1 on chromatin may be taken up more by GFP-M1-5 than by GFP-H1b (2). As shown previously, core histone posttranslational modifications (36) and dynamic competition for chromatin binding between H1 and other DNA binding proteins (10) may also influence H1-chromatin interactions. The involvement of CDK2 in affecting recovery correlates with the idea that histone H1 may be a downstream target of CDK2 activity.
The mobility of GFP-H1b is influenced by CDK2 activity.
Although recent work demonstrated a role for an ATP-dependent process in increasing the mobility of H1 in Tetrahymena, the data did not address what specific ATP-dependent remodeling complex influences H1 mobility (16). Our FRAP data analyzing the mobility of GFP-M1-5 in the different cell lines suggest a relationship between GFP-H1b mobility and CDK2 activity. To further test whether CDK2 activity may affect H1 mobility, we set out to analyze GFP-H1 mobility in synchronized cells. The activity of cyclin E/CDK2 is low in G0 and early G1 and peaks during the transition from late G1 to S phase. As shown in Fig. 3 and Table 1, the rate of recovery of GFP-H1b is lower in cells arrested in G0 than in those in G1, when CDK2 activity is highest. Thus, at G0, WI-38 cells contain GFP-H1b that is more immobile. We hypothesize that the decrease in the rate of recovery at G0 is due to a low level of CDK2 activity, which results in a relatively less phosphorylated GFP-H1b. This in turn stabilizes the interaction between H1b and chromatin. These results do not exclude the involvement of other G1 kinases that may either directly or indirectly influence H1 mobility.
To directly test the effect of CDK2 activity on the mobility of the linker histone, we specifically blocked the activity of CDK2 by adenovirus expression of the CDK2 inhibitor protein p21 in serum-starved WI-38 VA cells expressing GFP-H1b or GFP-M1-5. Whereas the mobility of GFP-M1-5 is not affected by p21 expression, GFP-H1b mobility is decreased in cells expressing p21 (Fig. 4). Because GFP-M1-5 cannot be phosphorylated, the decrease in kinase activity due to p21 infection does not influence the mobility of the mutant chimera. These data suggest that histone H1b is a direct target for CDK2 in vivo and that in vivo phosphorylation of the linker histone variant does affect its nuclear mobility.
CFP-M1-5 recovers most slowly in heterochromatin.
In vitro experiments have shown that one function of histone H1 is to direct and stabilize higher-order chromatin structures (1, 9, 28) and that phosphorylation of the tails decreases chromatin condensation (29). Furthermore, it has been shown that the mobilities of linker histones over heterochromatin are lower than those over euchromatin, suggesting that heterochromatin contains a more immobile population of linker histones (36). We wanted to compare the mobilities of our chimeric proteins over heterochromatin to those over more euchromatic regions to test the effect of the unphosphorylated tails on the mobility of H1b in these two nuclear compartments. As shown in Fig. 5, CFP-M1-5 recovers slower and to a lesser extent than CFP-H1b over heterochromatin, which is visualized by cotransfection with YFP-Lac. A difference is not observed in the recovery rates of the tagged histones over euchromatin. Thus, the effect on mobilities of the unphosphorylated tails is largely specific for heterochromatic regions in these CHO cells and suggests that there is more immobile CFP-M1-5 than immobile CFP-H1b in this region. The lack of a mobility difference between the two proteins in the nonarray portion of the nucleus may be due to decreased CDK2 activity, which is consistent with the findings in WI-38 cells. Although the rates of recovery of M1-5 are the same in heterochromatin (lac array) versus in other areas of the nucleus (Table 1), the percent recovery of M1-5 is smaller over the array. This observation indicates that the recovery rate of M1-5 is the same throughout the nucleus and is independent of chromatin structure, yet there exists in heterochromatin a more immobile fraction of M1-5, which may prevent other M1-5 proteins from binding to the arrays. The observation that no significant change in percent recovery is seen for wild-type H1 may be due to the ability of H1, unlike M1-5, to become phosphorylated, thus facilitating the dynamic exchange of wild-type H1 in chromatin throughout the nucleus. Furthermore, these observations suggest that the factor(s) controlling H1 phosphorylation, namely CDK2, is not sequestered in any particular region of the nucleus. These data are consistent with the hypothesis that the linker histone present in heterochromatin is unphosphorylated. CDK2 activity may function in heterochromatic regions to relieve higher-order structures and may be necessary in euchromatic regions to maintain a pool of phosphorylated H1 during times in the cell cycle when a more open chromatin conformation is favored. Although CDK2 activity influences the mobility of histone H1 over more euchromatic regions, the binding of unphosphorylated H1 is likely to be more stable in condensed regions of chromatin.
Model for the role of H1 phosphorylation during cell cycle progression and chromatin maintenance.
Our data are consistent with a model whereby phosphorylation of the linker histone H1 tails promotes chromatin decondensation (41). In this model, the positively charged, nonphosphorylated tails shield the negative charge of the DNA backbone to allow for chromatin fiber interactions within condensed structures. Upon phosphorylation, the negative phosphate groups weaken the interaction of the tails with DNA to allow for local decondensation of chromatin. Indeed, increased H1 phosphorylation results in a more relaxed chromatin structure (13, 23, 47) and increased accessibility to chromatin-modifying enzymes (26) and facilitates the activity of the transcriptional and replicative machineries (11, 13, 21). These results, however, differ from those of studies that suggest that the phosphorylation of the histone tails increases H1-H1 interactions and directs chromosome condensation, thus explaining the observation that peak levels of H1 phosphorylation are observed during mitosis (6, 7). Interestingly, indirect immunofluorescent analysis with antibodies against phosphorylated histone H1 shows both association (4) and dissociation (34) with mitotic chromosomes. More importantly, mitotic chromosomes and functional nuclei assemble properly in the absence of linker histones, indicating that phosphorylated linker histones may not be essential for chromatin compaction (15, 38, 51).
Our data support the model that as cells progress through the transition from late G1 to S phase, histone H1 becomes phosphorylated by cyclin E/CDK2, resulting in a more open chromatin conformation. This global, genome-wide relaxation facilitates the replication of DNA during S phase. Since cyclin E levels decrease during S phase, the phosphorylation of H1 is perhaps maintained by the increasing activity of the cyclin A/CDK2 kinase complex. The H1 phosphorylation observed during G1 or G2 may also be necessary to allow for the transcription of particular genes. Thus, the unphosphorylated form of histone H1 may stabilize the higher-order structure necessary for local facultative chromatin condensation and for general heterochromatin maintenance during interphase. Finally, as high levels of phosphorylation have been observed in cells transformed with oncogenes (13), an interesting point to consider is the effect of aberrant H1 phosphorylation on genomic instability.
Acknowledgments
We thank D. Randy Garza and M. G. Mancini for technical assistance and J. W. Harper and M. Rijnkels for generously providing the adenovirus expressing p21 and β-Gal, respectively.
This work was supported by supplemental grant RO1-CA16303-27 from the Comprehensive Minority Biomedical Branch of the National Institutes of Health (J.M.R.) and by the American Cancer Society (R.E.H.).
REFERENCES
- 1.Bednar, J., R. A. Horowitz, S. A. Grigoryev, L. M. Carruthers, J. C. Hansen, A. J. Koster, and C. L. Woodcock. 1998. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc. Natl. Acad. Sci. USA 95:14173-14178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmont, A. S. 2001. Visualizing chromosome dynamics with GFP. Trends Cell Biol. 11:250-257. [DOI] [PubMed] [Google Scholar]
- 3.Bhattacharjee, R. N., G. C. Banks, K. W. Trotter, H. L. Lee, and T. K. Archer. 2001. Histone H1 phosphorylation by Cdk2 selectively modulates mouse mammary tumor virus transcription through chromatin remodeling. Mol. Cell. Biol. 21:5417-5425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boggs, B. A., C. D. Allis, and A. C. Chinault. 2000. Immunofluorescent studies of human chromosomes with antibodies against phosphorylated H1 histone. Chromosoma 108:485-490. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury, E. M. 2002. Chromatin structure and dynamics: state-of-the-art. Mol. Cell 10:13-19. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury, E. M. 1992. Reversible histone modifications and the chromosome cell cycle. Bioessays 14:9-16. [DOI] [PubMed] [Google Scholar]
- 7.Bradbury, E. M., R. J. Inglis, and H. R. Matthews. 1974. Control of cell division by very lysine rich histone (F1) phosphorylation. Nature 247:257-261. [DOI] [PubMed] [Google Scholar]
- 8.Brown, D. T., B. T. Alexander, and D. B. Sittman. 1996. Differential effect of H1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res. 24:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carruthers, L. M., J. Bednar, C. L. Woodcock, and J. C. Hansen. 1998. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry 37:14776-14787. [DOI] [PubMed] [Google Scholar]
- 10.Catez, F., D. T. Brown, T. Misteli, and M. Bustin. 2002. Competition between histone H1 and HMGN proteins for chromatin binding sites. EMBO Rep. 3:760-766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chadee, D. N., C. D. Allis, J. A. Wright, and J. R. Davie. 1997. Histone H1b phosphorylation is dependent upon ongoing transcription and replication in normal and ras-transformed mouse fibroblasts. J. Biol. Chem. 272:8113-8116. [DOI] [PubMed] [Google Scholar]
- 12.Chadee, D. N., C. P. Peltier, and J. R. Davie. 2002. Histone H1(S)-3 phosphorylation in Ha-ras oncogene-transformed mouse fibroblasts. Oncogene 21:8397-8403. [DOI] [PubMed] [Google Scholar]
- 13.Chadee, D. N., W. R. Taylor, R. A. Hurta, C. D. Allis, J. A. Wright, and J. R. Davie. 1995. Increased phosphorylation of histone H1 in mouse fibroblasts transformed with oncogenes or constitutively active mitogen-activated protein kinase kinase. J. Biol. Chem. 270:20098-20105. [DOI] [PubMed] [Google Scholar]
- 14.Crane-Robinson, C. 1997. Where is the globular domain of linker histone located on the nucleosome? Trends Biochem. Sci. 22:75-77. [DOI] [PubMed] [Google Scholar]
- 15.Dasso, M., T. Seki, Y. Azuma, T. Ohba, and T. Nishimoto. 1994. A mutant form of the Ran/TC4 protein disrupts nuclear function in Xenopus laevis egg extracts by inhibiting the RCC1 protein, a regulator of chromosome condensation. EMBO J. 13:5732-5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dou, Y., J. Bowen, Y. Liu, and M. A. Gorovsky. 2002. Phosphorylation and an ATP-dependent process increase the dynamic exchange of H1 in chromatin. J. Cell Biol. 158:1161-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dou, Y., and M. A. Gorovsky. 2000. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Mol. Cell 6:225-231. [DOI] [PubMed] [Google Scholar]
- 18.Dou, Y., C. A. Mizzen, M. Abrams, C. D. Allis, and M. A. Gorovsky. 1999. Phosphorylation of linker histone H1 regulates gene expression in vivo by mimicking H1 removal. Mol. Cell 4:641-647. [DOI] [PubMed] [Google Scholar]
- 19.Eastham, J. A., S. J. Hall, I. Sehgal, J. Wang, T. L. Timme, G. Yang, L. Connell-Crowley, S. J. Elledge, W. W. Zhang, J. W. Harper, et al. 1995. In vivo gene therapy with p53 or p21 adenovirus for prostate cancer. Cancer Res. 55:5151-5155. [PubMed] [Google Scholar]
- 20.Gunjan, A., B. T. Alexander, D. B. Sittman, and D. T. Brown. 1999. Effects of H1 histone variant overexpression on chromatin structure. J. Biol. Chem. 274:37950-37956. [DOI] [PubMed] [Google Scholar]
- 21.Halmer, L., and C. Gruss. 1996. Effects of cell cycle dependent histone H1 phosphorylation on chromatin structure and chromatin replication. Nucleic Acids Res. 24:1420-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hansen, J. C. 2002. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu. Rev. Biophys. Biomol. Struct. 31:361-392. [DOI] [PubMed] [Google Scholar]
- 23.Herrera, R. E., F. Chen, and R. A. Weinberg. 1996. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts. Proc. Natl. Acad. Sci. USA 93:11510-11515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Herrera, R. E., V. P. Sah, B. O. Williams, T. P. Makela, R. A. Weinberg, and T. Jacks. 1996. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell. Biol. 16:2402-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hobbs, S., S. Jitrapakdee, and J. C. Wallace. 1998. Development of a bicistronic vector driven by the human polypeptide chain elongation factor 1alpha promoter for creation of stable mammalian cell lines that express very high levels of recombinant proteins. Biochem. Biophys. Res. Commun. 252:368-372. [DOI] [PubMed] [Google Scholar]
- 26.Horn, P. J., L. M. Carruthers, C. Logie, D. A. Hill, M. J. Solomon, P. A. Wade, A. N. Imbalzano, J. C. Hansen, and C. L. Peterson. 2002. Phosphorylation of linker histones regulates ATP-dependent chromatin remodeling enzymes. Nat. Struct. Biol. 9:263-267. [DOI] [PubMed] [Google Scholar]
- 27.Houtsmuller, A. B., and W. Vermeulen. 2001. Macromolecular dynamics in living cell nuclei revealed by fluorescence redistribution after photobleaching. Histochem. Cell Biol. 115:13-21. [DOI] [PubMed] [Google Scholar]
- 28.Howe, L., M. Iskandar, and J. Ausio. 1998. Folding of chromatin in the presence of heterogeneous histone H1 binding to nucleosomes. J. Biol. Chem. 273:11625-11629. [DOI] [PubMed] [Google Scholar]
- 29.Langan, T. A., and T. C. Chambers. 1987. H1 histone phosphorylation, cell cycle progression and chromatin structure. Prog. Clin. Biol. Res. 249:215-223. [PubMed] [Google Scholar]
- 30.Langan, T. A., J. Gautier, M. Lohka, R. Hollingsworth, S. Moreno, P. Nurse, J. Maller, and R. A. Sclafani. 1989. Mammalian growth-associated H1 histone kinase: a homolog of cdc2+/CDC28 protein kinases controlling mitotic entry in yeast and frog cells. Mol. Cell. Biol. 9:3860-3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lever, M. A., J. P. Th'ng, X. Sun, and M. J. Hendzel. 2000. Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408:873-876. [DOI] [PubMed] [Google Scholar]
- 32.Li, G., G. Sudlow, and A. S. Belmont. 1998. Interphase cell cycle dynamics of a late replicating, heterochromatic homogenously staining region: precise choreography of condensation/decondensation and intranuclear positioning. J. Cell Biol. 140:975-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lippincott-Schwartz, J., E. Snapp, and A. Kenworthy. 2001. Studying protein dynamics in living cells. Nat. Rev. Mol. Cell Biol. 2:444-456. [DOI] [PubMed] [Google Scholar]
- 34.Lu, M. J., C. A. Dadd, C. A. Mizzen, C. A. Perry, D. R. McLachlan, A. T. Annunziato, and C. D. Allis. 1994. Generation and characterization of novel antibodies highly selective for phosphorylated linker histone H1 in Tetrahymena and HeLa cells. Chromosoma 103:111-121. [DOI] [PubMed] [Google Scholar]
- 35.Lu, M. J., S. S. Mpoke, C. A. Dadd, and C. D. Allis. 1995. Phosphorylated and dephosphorylated linker histone H1 reside in distinct chromatin domains in Tetrahymena macronuclei. Mol. Biol. Cell 6:1077-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Misteli, T., A. Gunjan, R. Hock, M. Bustin, and D. T. Brown. 2000. Dynamic binding of histone H1 to chromatin in living cells. Nature 408:877-881. [DOI] [PubMed] [Google Scholar]
- 37.Nye, A. C., R. R. Rajendran, D. L. Stenoien, M. A. Mancini, B. S. Katzenellenbogen, and A. S. Belmont. 2002. Alteration of large-scale chromatin structure by estrogen receptor. Mol. Cell. Biol. 22:3437-3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohsumi, K., C. Katagiri, and T. Kishimoto. 1993. Chromosome condensation in Xenopus mitotic extracts without histone H1. Science 262:2033-2035. [DOI] [PubMed] [Google Scholar]
- 39.Parseghian, M. H., and B. A. Hamkalo. 2001. A compendium of the histone H1 family of somatic subtypes: an elusive cast of characters and their characteristics. Biochem. Cell Biol. 79:289-304. [PubMed] [Google Scholar]
- 40.Phair, R. D., and T. Misteli. 2000. High mobility of proteins in the mammalian cell nucleus. Nature 404:604-609. [DOI] [PubMed] [Google Scholar]
- 41.Roth, S. Y., and C. D. Allis. 1992. Chromatin condensation: does histone H1 dephosphorylation play a role? Trends Biochem. Sci. 17:93-98. [DOI] [PubMed] [Google Scholar]
- 42.Seyedin, S. M., and W. S. Kistler. 1979. H1 histone subfractions of mammalian testes. 1. Organ specificity in the rat. Biochemistry 18:1371-1375. [DOI] [PubMed] [Google Scholar]
- 43.Sherr, C. J., and J. M. Roberts. 1995. Inhibitors of mammalian G1 cyclin-dependent kinases. Genes Dev. 9:1149-1163. [DOI] [PubMed] [Google Scholar]
- 44.Stenoien, D. L., A. C. Nye, M. G. Mancini, K. Patel, M. Dutertre, B. W. O'Malley, C. L. Smith, A. S. Belmont, and M. A. Mancini. 2001. Ligand-mediated assembly and real-time cellular dynamics of estrogen receptor alpha-coactivator complexes in living cells. Mol. Cell. Biol. 21:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Talasz, H., W. Helliger, B. Puschendorf, and H. Lindner. 1996. In vivo phosphorylation of histone H1 variants during the cell cycle. Biochemistry 35:1761-1767. [DOI] [PubMed] [Google Scholar]
- 46.Talasz, H., N. Sapojnikova, W. Helliger, H. Lindner, and B. Puschendorf. 1998. In vitro binding of H1 histone subtypes to nucleosomal organized mouse mammary tumor virus long terminal repeat promotor. J. Biol. Chem. 273:32236-32243. [DOI] [PubMed] [Google Scholar]
- 47.Taylor, W. R., D. N. Chadee, C. D. Allis, J. A. Wright, and J. R. Davie. 1995. Fibroblasts transformed by combinations of ras, myc and mutant p53 exhibit increased phosphorylation of histone H1 that is independent of metastatic potential. FEBS Lett. 377:51-53. [DOI] [PubMed] [Google Scholar]
- 48.Travers, A. 1999. The location of the linker histone on the nucleosome. Trends Biochem. Sci. 24:4-7. [DOI] [PubMed] [Google Scholar]
- 49.Tumbar, T., G. Sudlow, and A. S. Belmont. 1999. Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145:1341-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vignali, M., and J. L. Workman. 1998. Location and function of linker histones. Nat. Struct. Biol. 5:1025-1028. [DOI] [PubMed] [Google Scholar]
- 51.Wolffe, A. 1998. Chromatin: structure and function, 3rd ed. Academic Press, San Diego, Calif.