Abstract
Here we show a novel pathway of transcriptional regulation of a DNA-binding transcription factor by coupled interaction and modification (e.g., acetylation) through the DNA-binding domain (DBD). The oncogenic regulator SET was isolated by affinity purification of factors interacting with the DBD of the cardiovascular transcription factor KLF5. SET negatively regulated KLF5 DNA binding, transactivation, and cell-proliferative activities. Down-regulation of the negative regulator SET was seen in response to KLF5-mediated gene activation. The coactivator/acetylase p300, on the other hand, interacted with and acetylated KLF5 DBD, and activated its transcription. Interestingly, SET inhibited KLF5 acetylation, and a nonacetylated mutant of KLF5 showed reduced transcriptional activation and cell growth complementary to the actions of SET. These findings suggest a new pathway for regulation of a DNA-binding transcription factor on the DBD through interaction and coupled acetylation by two opposing regulatory factors of a coactivator/acetylase and a negative cofactor harboring activity to inhibit acetylation.
The Sp/KLF (for Sp1- and Krüppel-like factor) family of zinc finger transcription factors has received recent attention due to important roles in developmental, differentiation, and oncogenic processes, among others (2, 3, 35). It is comprised of over 15 mammalian family members which have in common three similar C2H2-type zinc fingers at the carboxyl terminus which comprises the DNA-binding domain (DBD). Sp/KLF family members include the founding ubiquitous factor Sp1 (9), the erythroid differentiation factor EKLF/KLF1 (27), and the tumor suppressor gene KLF6/GBF/Zf9/COPEB, which we and others identified as a cellular factor possibly involved in human immunodeficiency virus type 1 transcription (18, 32, 44). It was recently shown by gene knockout studies that the proto-oncogene KLF5/BTEB2/IKLF (40, 42) is important for cardiovascular remodeling in response to stress (41). Contrary to initial expectations that this family of factors would likely have redundant functions, they in fact have important individual biological functions. However, the underlying mechanisms governing their specific functions and regulation are poorly understood.
We have studied the regulatory mechanisms of action of Sp/KLF family members in the past and have shown differential regulation through interaction and acetylation on the DBD by the coactivator/acetylase p300 (45). Acetylation is an important nuclear regulatory signal which regulates transcriptional processes with biological implications, including regulation of development, differentiation, and oncogenesis (5, 10, 31), which closely resembles the roles of Sp/KLF family members. We therefore thought that the Sp/KLF factors may be differently regulated by acetylation and showed that the coactivator/acetylase p300, but not the MYST-type acetylase Tip60, specifically interacts and acetylates Sp1 but not KLF6 through the zinc finger DBD and that DNA binding inhibits this interaction and acetylation (45). While much is known of acetylation in general, its regulation and implications are still poorly understood, especially its negative regulation.
Studies on negative regulation of acetylation have been centered mainly on the role of histone deacetylases, which are categorized into three classes based on sequence characteristics, subcellular localization, and catalytic properties (17, 33). We have shown an additional pathway involving negative regulation by DNA binding (45), and others have shown that a set of molecules inhibit the acetylation of histones by masking the protein from acetylation (e.g., inhibitors of histone acetylation [INHAT]) (39). Acetylation is therefore regulated at multiple levels by both catalytic and noncatalytic processes.
Here we show a regulatory pathway of acetylation involving the oncogenic regulator SET, a subunit of a complex previously shown to inhibit histone acetylation by masking the protein (INHAT) (39), through interaction with the DNA-binding transcription factor KLF5. Our findings suggest a new transcriptional regulatory pathway through the DBD by convergence of two opposing regulatory pathways involving p300 and SET through coupled interaction and acetylation.
MATERIALS AND METHODS
Preparation of recombinant epitope-tagged protein.
The zinc finger region/DBD of human KLF5 (KLF5 ZF/DBD) (40) (a kind gift of C. Teng) was PCR amplified and subcloned into BamHI-digested 6His-pET11d (45). Protein expression and purification were done essentially as described previously, with modification by use of HiTrap heparin and HisTrap columns (Amersham Pharmacia Biotech) (45). Glutathione S-transferase (GST)-tagged constructs KLF5 wt (wild type), KLF5-ΔDBD, KLF5-DBD, KLF5-zinc finger 1, KLF5-zinc finger 2, and KLF-zinc finger 3 were similarly PCR amplified and inserted into pGEX vectors (Amersham Pharmacia Biotech) (44, 45). Hexahistidine-tagged SET/TAF-Iβ construct (29) (a kind gift of K. Nagata) was transformed into the BL21-Gold(DE3)pLysS strain, induced, and then purified with Probond resin (Invitrogen) and buffer C containing 20 mM imidazole for washing and 200 mM imidazole for elution similar that described previously (45). SET/TAF-Iβ deletion constructs were in part a generous gift from K. Nagata, otherwise they were constructed by PCR mutagenesis. The p300 histone acetyltransferase (HAT) domain constructs have been described previously (45) (a kind gift of Y. Nakatani). All procedures were done at 4°C. Zinc finger peptides were synthesized commercially by Hokkaido System Science Co. Ltd. Product purity was more than 95%, and the molecular weights of synthesized peptides were confirmed by mass spectral analysis.
Cell culture and preparation of nuclear extract.
C2/2 rabbit vascular smooth muscle cells (VSMCs) (48) were maintained in Dulbecco's modified Eagle medium (Sigma), and HeLa S3 cells were maintained in Joklik's modified Eagle medium (Sigma) supplemented with 10% fetal bovine serum with 100 μg of streptomycin/ml and 100 U of penicillin G/ml. Nuclear extract was prepared as described previously (7). The final supernatant was dialyzed against buffer B (25 mM HEPES [pH 7.9], 10% glycerol, 150 mM KCl, 100 μM ZnSO4, 5 mM 2-mercaptoethanol, 1 mM phenylmethylsulfonyl fluoride, 0.5 μg of leupeptin/ml, and 1 μg of pepstatin/ml), centrifuged for 15 min at 18,000 × g, and used as nuclear extract.
Isolation of factors associating with KLF5 ZF/DBD.
Fifty micrograms of hexahistidine-tagged KLF5 ZF/DBD was bound to 5 μl of equilibrated Probond nickel-chelating resin (Invitrogen) by rotating for 6 h. Following five washes with buffer B, the resin bound with KLF5 ZF/DBD was then incubated for 6 h with 725 μg of C2/2 nuclear extract. After 10 washes with buffer B containing 20 mM imidazole, the bound proteins were eluted with buffer B containing 500 mM imidazole. Samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by Coomassie brilliant blue staining. All procedures were done at 4°C.
Protein identification by MALDI-TOF (MS).
Protein bands were excised from a Coomassie brilliant blue-stained SDS-PAGE gel and washed three times in 50% acetonitrile-25 mM ammonium bicarbonate (pH 8), and 100% acetonitrile was added. Following removal of acetonitrile, the gel slices were dried and the protein band was in-gel digested with 15 μg of trypsin/ml in 25 mM ammonium bicarbonate (pH 8) at 37°C overnight. Gel slices were then soaked in 50% acetonitrile-5% tetrahydrofuran, and the supernatant was collected and dried up. The dried digest was reconstituted by the addition of 50% acetonitrile-0.1% tetrahydrofuran, mixed with α-cyano-4-hydroxycinnamic acid, and analyzed by matrix-assisted laser desorption-ionization time of flight (mass spectrometry) [MALDI-TOF (MS)] (Voyager-DE STR; Applied Biosystems). Database searches were performed against the nonredundant National Center for Biotechnology Information database by using Protein Prospector programs, version 3.2.1, developed by the University of California—San Francisco MS facility. MS analysis of peptide acetylation was done essentially as described previously (16).
Protein-protein interaction assay.
GST fusion proteins were immobilized to glutathione-Sepharose 4B resin and incubated with histidine-tagged proteins in a buffer containing 20 mM HEPES (pH 7.6 at 4°C), 20% glycerol, 0.2 mM EDTA, 0.1% Triton X-100, 100 mM NaCl, and 100 μM ZnSO4. Reactions were carried out at 4°C for 3 h, and the mixtures were washed two times in the same buffer. Bound proteins were resolved on an SDS-PAGE (10% polyacrylamide) gel, transferred to a nitrocellulose membrane, immunoblotted with anti-HIS probe (G-18) antibody (Santa Cruz Biotechnology) or anti-FLAG (M2) antibody (Sigma), and detected with enhanced chemiluminescence Western blotting detection reagents essentially according to the manufacturer's instructions (Amersham Pharmacia Biotech). Histidine tag pull-down assays were done with Probond resin (Invitrogen) and blotted with anti-GST antibody (Santa Cruz) when appropriate.
Coimmunoprecipitation assay.
One microgram of anti-KLF5 rat monoclonal antibody (KM1785) (41) or 1 μg of control rat immunoglobulin G (IgG) (sc-2026; Santa Cruz Biotechnology) was bound to 10 μl of protein G-Sepharose (Amersham Pharmacia Biotech) by rotating for 6 h in buffer B at 4°C. Following three washes with buffer B, protein G-Sepharose with antibody was then rotated with 1 mg of cell extract protein for 6 h. After 10 washes with radioimmunoprecipitation assay buffer, washed immunoprecipitates were subjected to SDS-PAGE and immunoblotting with SET/TAF-Iβ-specific antibody (a generous gift of K. Nagata) (30).
Immunoconfocal fluorescence microscopy.
Murine VSMCs were prepared from the thoracic aortas of 8-week-old mice according to the enzyme digestion method (4). Cells were plated on glass coverslips, fixed with 4% formaldehyde, blocked with 3% bovine serum albumin in phosphate-buffered saline-Tween 20, incubated with anti-SET/TAF-Iβ mouse monoclonal antibody (a kind gift of K. Nagata) (30) and anti-KLF5 rat monoclonal antibody (KM1785), exposed to anti-mouse IgG antibody conjugated with fluorescein isothiocyanate and anti-rat IgG antibody conjugated with rhodamine isothiocyanate as the secondary antibodies, and then examined by confocal microscopy with a Leica TCS 4D equipped with an argon-krypton laser.
Gel shift DNA-binding assay.
The gel shift DNA-binding assay was done essentially as described previously (45). A DNA oligomer containing the KLF5 binding sequence, 5′-ATGGGCATGAGGGCCAGCCTATGAGA-3′ (SE1), was used to analyze the DNA binding of KLF5 ZF/DBD (48). For mutant analysis, the underlined nucleotides GGGCC were replaced by TTTAA. For control NF-κB gel shifts, commercially available NF-κB probe (Promega) and recombinant NF-κB p50 protein (Promega) were used. The details (e.g., protein combinations) of individual experiments are given in the figure legends.
Cotransfection reporter assay.
Plasmid constructs for the reporters, SMemb/NMHC-B and PDGF-A chain, and the effector pCAG-KLF5 have been described previously (41, 48). pCHA-SET/TAF-Iβ was a kind gift of K. Nagata (30). pCI-p300 and pCI-p300ΔHAT constructs were kind gifts of V. Ogryzko. Transient transfection assays were done by seeding cells (50,000 cells/24-well plate) 24 h prior to transfection and then transfected with 100 ng of the reporter plasmid and a total of 1 μg of either vector, pCAG-KLF5, or pCHA-SET/TAF-Iβ in combination, as described in the figure legends, by liposome-mediated transfer (Tfx-20; Promega) according to the manufacturer's instructions. Cells were harvested after 48 h and then subjected to an assay of luciferase activity (luciferase assay system; Promega) (Lumat LB9501; Berthold). The luciferase activity was normalized to the protein concentration of the cell lysates measured according to the Bradford method (Bio-Rad). For NF-κB control experiments, commercially available NF-κβ reporter was used (Stratagene), with coexpression of NF-κβ p50 and p65 expression plasmids (K. Aizawa, T. Suzuki, N. Kada, A. Ishihara, K. Kowase, T. Matsumura, K. Sasaki, Y. Munemasa, I. Manabe, M. Kurabayashi, T. Collins, and R. Nagai, submitted for publication).
Construction of point mutant expression construct.
Site-directed mutagenesis was used to construct the mutant pCAG/KLF5-K369R, which involves a lysine-to-arginine mutation at amino acid residue position 369 of KLF5. PCR was done with the primers 5′-GACGACCATCCACTACTGCGATT-3′ and 5′-TCTCCAAATCGGGGTTACTCCTT-3′, with pCAG/KLF5 as a template and KOD Plus (Toyobo) polymerase. The construct was sequenced for verification.
Construction of recombinant adenovirus vectors.
KLF5, the KLF5 K369R mutant, and SET cDNA were subcloned into the adenovirus cosmid vector pAxCAwt (Takara) at the SwaI site. 293 cells were cotransfected by the cosmid vector and restriction enzyme-treated DNA-terminal protein complex, with subsequent selection of plaques as a result of homologous recombination. The protein expression was confirmed by Western blot analysis. The titer was determined by the plaque method.
Production of stable transformant cell lines.
KLF5 and KLF5 K369R cDNA inserts were subcloned into 3× FLAG expression vectors (Sigma). Constructs were transfected into cells (3T3-3) and then selected on the basis of G418 resistance for 2 weeks.
BrdU incorporation assay.
Cells were plated on 96-well plates. Following adenovirus-mediated transfection in given experiments, bromo-2′-deoxyuridine (BrdU) incorporation was examined after 24 h over a span of 2 h by using the Biotrak cell proliferation enzyme-linked immunosorbent assay system, version 2 (Amersham Pharmacia Biotech), essentially according to the manufacturer's instructions. Experiments were done in triplicate.
Phorbol ester-induced expression of KLF5 and SET.
Cell lysates from C2/2 VSMCs stimulated with phorbol 12-myristate 13-acetate (PMA) (100 ng/ml) following 24 h of starvation (0% fetal bovine serum) were resolved on an SDS-12% PAGE gel, transferred to a nitrocellulose membrane, and then immunoblotted with anti-KLF5 monoclonal antibody (KM1785) or anti-SET/TAF-Iβ antibody (30). The relative intensity of KLF5 or SET protein in reference to Coomassie brilliant blue stain was calculated by using National Institutes of Health Image software. Total RNA was obtained by the RNeasy preparation kit (Qiagen) and reverse transcribed, and then quantitative PCR was performed with a platelet-derived growth factor A (PDGF-A) chain gene-specific primer set (5′-CAGCATCCGGGACCTCCAGCGACTC-3′ and 5′-TCGTAAATGACCGTCCTGGTCTTGC-3′) and a QuantumRNA 18S internal standard primer set (Ambion) as previously described (22). The relative intensity of the PDGF-A chain in reference to internal 18S rRNA was calculated by National Institutes of Health Image software.
Rat arterial injury model and immunohistochemistry.
After 4 weeks on their respective diets, rats weighing 400 to 450 g were anesthetized with chloral hydrate (370 mg/kg of body weight, intraperitoneally). Balloon denudation of the left common carotid artery was performed. The right common carotid artery served as a control. Fourteen days after operation, rats were euthanized with a lethal dose of anesthetic, after which the carotid arteries were perfused with 4% paraformaldehyde and phosphate-buffered saline. Each injured left carotid artery was excised from the proximal edge of the omohyoid muscle to the carotid bifurcation. The middle third of the segment was then fixed in 4% paraformaldehyde for 12 h and embedded in paraffin. Serial cross sections (6 μm thick) were cut from each sample and stained with hematoxylin-eosin or prepared for immunohistochemistry. For immunohistochemical analysis, tissue sections were preincubated with 2% bovine serum albumin and then serially treated with SET-specific antibody (KM1720) (30) or KLF5-specific antibody (KM1785). Specimens were then treated with biotinylated goat anti-mouse IgG antibody (Vector Laboratories) or biotinylated goat anti-rat IgG antibody (Chemicon) followed by avidin-biotinylated horseradish peroxidase (Vectastain ABC kit; Vector Laboratories) and developed with 0.004% H2O2 and 0.02% diaminobenzidine tetrahydrochloride.
RESULTS
Isolation of interactors of KLF5.
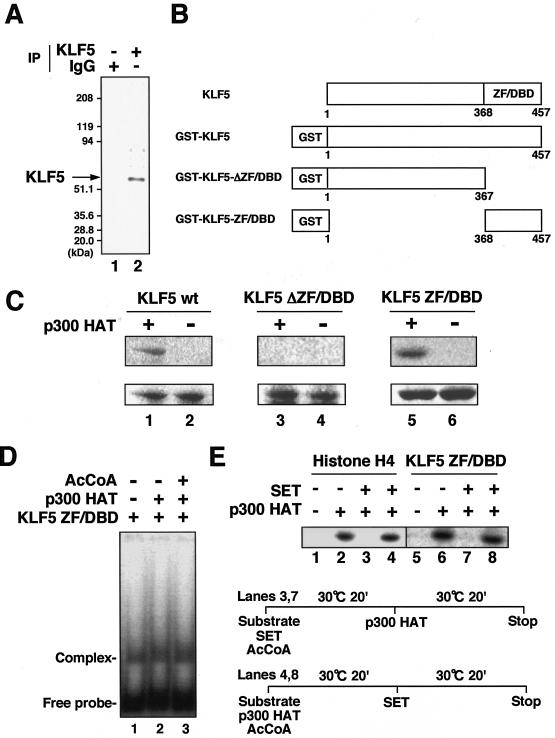
To isolate factors which regulate KLF5 by protein-protein interaction, we affinity purified interacting factors by using the ZF/DBD region, which is a potent protein-protein interface (21) that has been previously shown to mediate differential interaction with acetylase (45). Nuclear extract obtained from C2/2 VSMCs which express KLF5 were applied to hexahistidine-tagged recombinant KLF5 ZF/DBD immobilized on Ni2+-chelating resin (Fig. 1A). Bound proteins were released by imidazole and then analyzed by SDS-PAGE with Coomassie brilliant blue staining (Fig. 1B). Approximately 20 bands ranging from 30 to 200 kDa were seen when the binding reaction between the KLF5 ZF/DBD and nuclear extract was done (Fig. 1B, lane 2) and not when either nuclear extract or KLF5 ZF/DBD alone (Fig. 1B, lanes 1 and 3) was used.
FIG. 1.
Isolation of interactors of KLF5. (A) Silver-stained gel of the histidine-tagged KLF5 ZF/DBD recombinant used for interaction studies. Molecular mass markers are shown on left. (B) Isolation of factors associating with KLF5 ZF/DBD. Histidine-tagged KLF5 ZF/DBD was bound to nickel-chelating resin and subjected to VSMC C2/2 nuclear extract (NE) (lane 2). Lane 1 is nuclear extract alone, and lane 3 is recombinant protein immobilized on resin alone. Eluate was resolved by SDS-PAGE (12% polyacrylamide) and stained by Coomassie brilliant blue. The stained band, which was excised and subjected to further analysis, is indicated p39. (C) MALDI-TOF mass spectra obtained from tryptic peptides of p39. Fragment peaks assigned to SET are marked (asterisks). Peaks indicated A and B were subjected to postsource decay sequencing. (D) Partial peptide sequences of human SET. Numbering is from the initiation methionine of SET. The peptide sequences of peaks A and B obtained by postsource decay are boxed. (E) In vitro binding of KLF5 ZF/DBD and SET. Immobilized GST-KLF5 ZF/DBD fusion protein was reacted with histidine-tagged SET protein, separated by SDS-PAGE, and analyzed by immunoblotting with anti-HIS probe antibody (lane 3). Lane 1 is the input. GST protein was used as the control (lane 2). (F) In vitro binding of KLF5 full-length protein and SET. Immobilized GST-KLF5 full-length fusion protein was reacted with histidine-tagged SET protein, separated by SDS-PAGE, and analyzed by immunoblotting with anti-HIS probe antibody (lane 3). Lane 1 is the input. GST protein was used as a control (lane 2). (G) Coimmunoprecipitation of SET with KLF5. Cell lysate was immunoprecipitated with anti-KLF5 rat monoclonal antibody (lane 2) or normal rat IgG (lane 1) as a control. Bound materials were separated by SDS-PAGE and analyzed by immunoblotting with anti-SET antibody. (H) Intracellular localization of KLF5 and SET. Endogenous SET (a) (green) and KLF5 (b) (red) were detected mainly in nuclei. Confocal microscopy double-staining analysis indicates colocalization of SET and KLF5 (c) (yellow). All experiments were done at least twice with consistent findings. +, present; −, absent.
One band with an apparent molecular mass of 39 kDa was found in abundance and easily discernible from other nearby bands. MALDI-TOF (MS)-peptide mass fingerprinting, with a computer search of the National Center for Biotechnology Information mammalian database with further confirmation of the amino acid sequence by postsource decay peptide sequencing, revealed this protein to be the SET oncoprotein, the product of the SET oncogene whose translocation has been implicated in leukemia (Fig. 1C and D) (1, 47). SET is identical to the cellular factor template activating factor-Iβ which stimulates adenoviral core DNA replication (24, 29) and has been shown to be a histone chaperone, which is a factor that can displace and/or assemble nucleosomal histones in an ATP-independent manner (15, 25, 26, 34). While interaction of histone chaperones with histones and their activities to assemble and disassemble nucleosomal histones have been well addressed, their interaction with DNA-binding transcription factors has not been explored. Further, their cellular functions are poorly understood. Thus, characterization of this new interaction would add to our understanding of how histone chaperones may be involved in specific transcription by cooperative interaction with transcription factors as well as their cellular functions.
In vitro and in vivo interactions of KLF5 and SET.
To determine whether SET directly interacts with KLF5 ZF/DBD, a GST pull-down assay was done (Fig. 1E). SET bound KLF5 ZF/DBD (Fig. 1E, lane 3) but not GST alone (Fig. 1E, lane 2), showing that SET and KLF5 ZF/DBD bind directly. To next see whether full-length KLF5 binds SET, a similar GST pull-down assay with full-length KLF5 was done (Fig. 1F), which showed that SET also binds full-length KLF5 (Fig. 1F, lane 3). To further see whether KLF5 and SET interact in the cell, immunoprecipitation was done with specific antibodies against KLF5 and SET (Fig. 1G). Immunoprecipitation of KLF5 followed by immunoblotting with SET antibody showed detection of SET when antibody against KLF5 was used (Fig. 1G, lane 2) but not for control IgG antibody (Fig. 1G, lane 1). SET therefore interacts with KLF5 in vitro and in vivo.
To further examine the expression and cellular localization of SET, immunohistochemistry was done (Fig. 1H). SET (Fig. a) and KLF5 (Fig. b) colocalized to the nucleus in an overlapping pattern (Fig. c), which is supportive of functional interaction.
Functional effects of interaction of KLF5 and SET.
To next address the functional implications of the interaction of SET and KLF5, we first examined the effects on the DNA-binding activity of KLF5 (Fig. 2A). Gel shift analysis under conditions in which KLF5 ZF/DBD showed sequence-specific binding and SET did not bind the probe DNA (Fig. 2A, lanes 2 to 4) showed inhibition of KLF5 DNA-binding activity by the addition of SET (Fig. 2A, lanes 7 and 8). SET did not inhibit the DNA-binding activity of the control NF-κB p50 subunit (Fig. 2B).
FIG. 2.
Effects of SET on KLF5 activity. (A) Effects of SET on KLF5 DNA-binding activity. A gel shift assay with recombinant KLF5 ZF/DBD and SET was performed. Wt and mut represent wild and mutant oligonucleotide competitors (lanes 5 and 6). The amount of recombinant protein is as follows: 150 (+) and 450 (++) ng for SET (lanes 2, 7, and 8) and 10 (+) and 50 (++) ng for KLF5 ZF/DBD (lanes 3 to 8). (B) Gel shift assay of control NF-κB p50 subunit and SET. Gel shift units (0.1) of NF-κB p50 (lanes 2, 4, and 5) and 100 (+) and 300 (++) ng of SET (lanes 3, 4, and 5) were used. (C) Effects of SET on KLF5 transactivation. Cotransfection analysis of effects of SET on KLF5 transcriptional activation.One hundred nanograms of reporter was used in each lane. Effectors are as follows: lanes 2, 3, and 4 through 7, and 12, 13, and 14 through 17 were 83, 250, and 750 ng of KLF5 expression plasmid (pCAG-KLF5), respectively; lanes 5 and 8, 6 and 9, 7 and 10, 15 and 18, 16 and 19, and 17 and 20 were 28, 83, and 250 ng of SET expression plasmid (pCHA-SET/TAF-Iβ). The total amount of effector plasmid was adjusted to 1 μg with the respective control vector. (D) Effects of SET on control NF-κB transactivation. NF-κB transactivation (lanes 2 to 5) was done by transfection of equal amounts (250 ng) of p50 and p65 subunit expression vectors. (E) Effects of SET on KLF5-induced cell growth. SET was transiently transfected into cells stably expressing epitope-tagged (3× FLAG) KLF5 or mock vector in 3T3-3 cells. The cell count on day 5 after transfection compared with mock vector-treated cells is shown. Error bars denote standard errors. (F) BrdU assay showing effects of KLF5 and SET on cell growth by use of adenovirus-mediated transfer (multiplicity of infection, 100) of KLF5 and SET adenoviruses. Empty (lane 1) denotes empty vector alone. Error bars denote standard errors. OD450, optical density at 450 nm. (G) In vitro binding of KLF5 ZF/DBD and SET deletion mutants. Immobilized histidine-tagged SET protein was reacted with GST-KLF5 ZF/DBD fusion protein, separated by SDS-PAGE, and analyzed by immunoblotting with anti-GST antibody. SET deletion mutants are shown by their amino acid numbers in reference to the schematic diagram of functional domains shown above. Lane 1 is GST KLF5 ZF/DBD input, and lane 2 shows that GST KLF5 ZF/DBD does not bind Probond nickel-chelating resin. Lanes 3 to 8 show GST KLF5 ZF/DBD binding to respective resin-bound deletion mutants. Amino acids (aa) 1 to 24 comprise the SET-specific N-terminal region, amino acids 25 to 65 comprise the coiled-coil dimerization domain, amino acids 25 to 119 comprise a region known to inhibit phosphatase PP2A, amino acids 120 to 225 comprise a region with unknown function, and amino acids 226 to 277 comprise the acidic C-terminal region. All experiments were done at least twice with consistent findings.
To further examine the effects of SET on KLF5-dependent transcriptional activation, cotransfection reporter assays were done (Fig. 2C). Using the originally identified KLF5-responsive embryonic vascular smooth muscle (SMemb) promoter (48) and the PDGF-A chain promoter, which is an endogenous target of KLF5 (41), under conditions in which KLF5 showed dose-dependent transactivation of the SMemb and PDGF-A chain promoter activities (Fig. 2C, lanes 2 to 4 and 12 to 14) and SET did not show activation of the respective promoters (Fig. 2C, lanes 8 to 10 and 18 to 20), cotransfection of SET showed dose-dependent inhibition of KLF5-mediated transactivation (Fig. 2C, lanes 5 to 7 and 15 to 17). SET did not inhibit transactivation by NF-κB (Fig. 2D). SET therefore negatively regulates both KLF5 DNA-binding and transactivation activities in a specific manner.
The effects of SET on KLF5 cellular activity were further addressed given that KLF5 is a proto-oncogene which accelerates cell growth (Fig. 2E) (43). Under conditions in which adenovirus-mediated forced expression of KLF5 stimulated the growth of C2/2 cells, expression of SET inhibited cell growth in KLF5-expressing cells to almost basal levels, suggesting coordinated effects of SET and KLF5 on cell growth. To note, SET alone also showed inhibitory effects on cell growth. A BrdU assay further confirmed that SET inhibited cell growth as induced by KLF5 (Fig. 2F).
To determine the region of SET which interacts with KLF5, deletion mutants were made and subjected to a pull-down assay (Fig. 2G). Results collectively showed that a 40-amino-acid stretch (amino acids 25 to 65) known as a coiled-coil dimerization domain of SET (28) did not interact with KLF5, but otherwise, a broad region of SET interacted with KLF5. Initial results with a SET mutant which does not interact with KLF5 did not show effects on cell growth, suggesting that interaction is important for cooperative effects of SET and KLF5 on cell growth (data not shown).
Biological implications of interaction of KLF5 and SET.
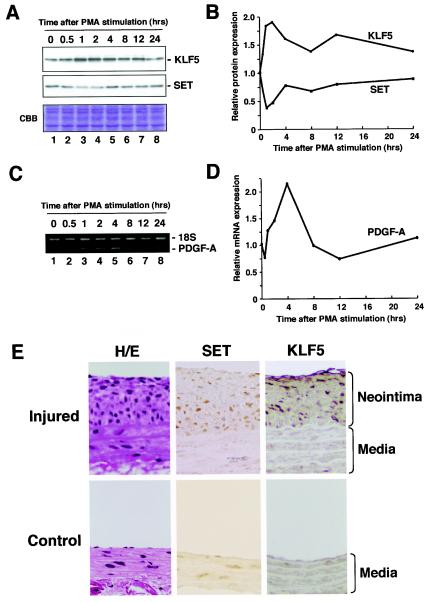
To further examine the biological implications of the interaction of KLF5 and SET, we assessed the effects of SET on KLF5-dependent gene expression. KLF5 and its downstream gene PDGF-A chain are induced by various stimuli (e.g., phorbol ester, angiotensin II, and serum) (14, 48). We examined whether SET expression could also be similarly regulated. Expression levels of KLF5, SET, and PDGF-A chain were assessed at various time intervals after PMA stimulation by Western blotting for KLF5 and SET, and reverse transcription-PCR analysis for the PDGF-A chain (Fig. 3A to D). An induction of KLF5 protein was seen at 2 h with a 1.9-fold increase after PMA stimulation, with a coinciding 2.5-fold decrease in SET protein levels (Fig. 3A and B). PDGF-A mRNA levels showed a 2.2-fold increase at 4 h (Fig. 3C and D). KLF5, SET, and PDGF-A chain levels returned to basal levels at 24 h. Collectively, reciprocal increased KLF5 and coinciding decreased SET correlated with increased PDGF-A chain expression after PMA stimulation.
FIG. 3.
Effects of SET on KLF5 downstream gene expression and pathological states. (A) Induction of KLF5 protein and repression of SET protein after mitogenic stimulation. Cells were starved at the times shown for 24 h, incubated with 100 ng of PMA/ml for the indicated times, and then harvested. Cell lysate was resolved by SDS-PAGE and subjected to Western blotting or Coomassie brilliant blue staining. (B) Quantification of KLF5 and SET protein levels. KLF5 and SET protein levels were normalized by the corresponding Coomassie brilliant blue staining pattern. The relative expression level was shown as the level at 0 h. (C) Induction of PDGF-A chain mRNA expression. Cells were starved at the times shown for 24 h, incubated with 100 ng of PMA/ml for the indicated times, and then harvested. The quantitative reverse transcription-PCR fragmentwas resolved on a 2% agarose gel. (D) Quantification of mRNA expression level for the PDGF-A chain. The expression level of the PDGF-A chain, an endogenous target gene of KLF5, was normalized to that of 18S. (E) KLF5 and SET expression in the pathological neointima. The immunohistochemistry of SET and KLF5 in a balloon injury model of atherosclerosis was examined. The left common carotid artery was denudated by balloon injury, and the neointima was observed 2 weeks after the balloon injury (Injured). The right common carotid artery served as a control (Control). The rat aorta was stained with anti-SET and anti-KLF5 antibodies. Cells in the neointima were clearly positive for SET and KLF5. All experiments were done at least twice with consistent findings. H/E, hematoxylin and eosin staining.
We further examined expression of KLF5 and SET in pathological states by histopathological analysis. An experimental atherosclerosis model (balloon injury) was used in which KLF5 is activated in proliferating neointimal smooth muscle cells after injury (11, 41). Balloon-injured aortas and controls at 2 weeks were examined for expression of KLF5 and SET by immunohistochemistry (Fig. 3E). In contrast to low basal levels of either SET or KLF5 in the nucleus of noninjured control aortic medial cells, proliferating neointimal cells, which form after injury and consist mainly of proliferating smooth muscle cells, showed marked expression of both KLF5 and SET in the nucleus. These findings suggest correlation and colocalization of KLF5 and SET in pathological states and support a functional interaction at the tissue or animal level.
Inhibition of KLF5 acetylation by SET.
SET regulates the actions of KLF5 as determined from these studies, but the underlying mechanisms were still not fully understood. One of the functions of SET has been recently shown to be inhibition of histone acetylation (39). It has been shown in the past that the KLF5 family member Sp1 is acetylated by p300 in the ZF/DBD region (45), and as Sp1 and KLF5 have similar ZF/DBDs, and further because SET interacted with the ZF/DBD of KLF5, we reasoned that KLF5 may also be similarly acetylated, and if so, be inhibited by SET. By such, SET may negatively regulate the actions of KLF5 by blocking acetylation.
First, we examined whether KLF5 can be acetylated by pulse-chase experiments (Fig. 4A). Using [3H]acetate and the histone deacetylase inhibitor trichostatin A, cells expressing KLF5 showed a clear uptake, thus showing that KLF5 can be acetylated in vivo. We next examined which region of KLF5 is acetylated by an in vitro acetylation assay by using acetyl [14C]coenzyme A (acetyl-[14C]CoA) (Fig. 4B). Using a catalytic recombinant protein of p300 which acetylates the similar factor Sp1, full-length KLF5 and the KLF5 ZF/DBD region were acetylated but the non-ZF/DBD region was not, showing that the ZF/DBD region is acetylated similar to Sp1 (Fig. 4C). This was specific, as neither acetylase Tip60 (MYST domain containing Tat-interacting protein) nor GCN5 (homologue of mammalian PCAF) acetylated KLF5 ZF/DBD (data not shown).
FIG. 4.
Acetylation of KLF5 and its regulation by SET. (A) Acetylation of KLF5 in vivo. Cells were treated with trichostatin A and labeled with [3H]acetate followed by immunoprecipitation with KLF5 (lane 2) and control normal IgG antibodies (lane 1). (B) Schematic representation of GST-KLF5 fusion mutant constructs. GST-KLF5 comprises full-length KLF5 fused to GST, GST-KLF5-ΔZF/DBD comprises only the N-terminal regulatory domain fused to GST, and GST-KLF5-ZF/DBD comprises only the C-terminal zinc finger DBD fused to GST. (C) Acetylation of KLF5 mutant constructs in vitro by p300. KLF5 proteins (1.2 μg) were incubated with 50 ng of FLAG-p300 HAT domain protein (amino acids 1195 to 1673) in the presence of [14C]acetyl-CoA. Reaction products were separated by SDS-12% PAGE. The difference between pairs (lanes 1 and 2, 3 and 4, and 5 and 6) is the presence of p300 HAT protein in the reaction mixture for the respective KLF5 mutant proteins. The gel was stained with Coomassie brilliant blue (lower panel) and then analyzed with a BAS 1500 phosphorimager (upper panel). (D) Effects of acetylation on KLF5 DNA-binding activity. Acetylation reactions were performed in the presence (+) of acetyl-CoA (AcCoA) and FLAG-p300 HAT domain (lane 3), in the presence of FLAG-p300 HAT domain (lane 2), and in the absence (−) of acetyl-CoA or FLAG-p300 HAT domain (lane 1). Reaction products were resolved by electrophoresis and analyzed with BAS1500. (E) Effects of SET on KLF5 acetylation (lanes 5 to 8). Histone H4 was used as a control (lanes 1 to 4). A schematic diagram of the protocol for order-of-addition experiments is shown. In lanes 3 and 7, the p300 HAT domain was added following the reaction of SET with the substrate (KLF5 ZF/DBD or histone H4) (prior to acetylation), and in lanes 4 and 8, SET was added following the reaction of p300 HAT with the substrate (after acetylation). Acetylation reactions were done essentially as described above. All experiments were done at least twice with consistent findings.
To see whether acetylation affects the DNA-binding activity of KLF5, a gel shift assay was done under acetylation conditions which showed that acetylation of KLF5 has no effect on its DNA-binding activity (Fig. 4D). As p300 acetylated KLF5, we next examined whether SET could inhibit the acetylation of KLF5 (Fig. 4E). As expected, SET inhibited the acetylation of KLF5 ZF/DBD by p300. Order of addition experiments showed that SET was able to inhibit acetylation when reacted with KLF5 prior to the addition of acetylase, but it was unable to react if KLF5 and p300 were reacted beforehand, suggesting that SET inhibits acetylation of KLF5 by masking the protein or inducing a conformational change which does not allow for subsequent acetylation.
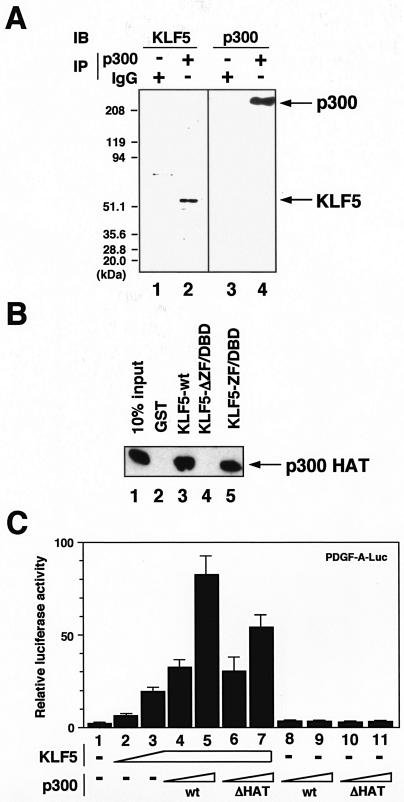
p300 as a transcriptional cofactor of KLF5.
As the coactivator/acetylase p300 acetylated KLF5, we examined whether p300 acts as a coactivator of KLF5, that is, if it interacts with KLF5 and is able to potentiate KLF5-mediated transcriptional activation. First, to examine the interaction between KLF5 and p300, by use of immunoprecipitation with specific antibodies against KLF5 and p300, we show that, under conditions in which p300 is pulled down, immunoblot against KLF5 shows interaction of KLF5 with p300 (Fig. 5A). To next examine whether interaction is direct and by which region interaction is mediated, GST pull-down assays were done with KLF5 deletion mutants and the p300 acetylase catalytic region (Fig. 5B). Results showed that KLF5 ZF/DBD and full-length KLF5, but not non-ZF/DBD region KLF5, interact with p300. Thus, importantly, the ZF/DBD is the interacting domain with p300 in addition to the substrate for acetylation.
FIG. 5.
Interaction and activation of KLF5 by the coactivator and acetylase p300. (A) Interaction of KLF5 and p300 in vivo. p300 was immunoprecipitated from cells followed by immunoblotting against KLF5 (lane 2). Immunoblotting against p300 confirms the immunoprecipitation procedure (lane 4). Normal IgG was used as the control (lane 2). (B) Interaction between KLF5 deletion mutants and p300 HAT domain in vitro. Approximately 2 μg of recombinant GST-KLF5 wild type (wt) (amino acids 1 to 457, lane 3), GST-KLF5-ΔDBD (amino acids 1 to 367, lane 4), and GST-KLF5-DBD (amino acids 368 to 457, lane 5) were used in a GST pull-down assay with 1 μg of the FLAG-p300 HAT domain. The interaction between KLF5 and the p300 HAT domain was analyzed by SDS-10% PAGE of the pull-down reactions and Western blotting with anti-FLAG antibody. The input (lane 1) contains 10% of the FLAG-p300 HAT domain protein. GST protein (lane 2) was used as a control. (C) Effects of p300 on KLF5 transactivation as assessed by reporter cotransfection assay. Cells were transfected with 100 ng of PDGF-A chain luciferase reporter and increasing amounts of KLF5 expression vector plasmids up to 750 ng (lanes 2 to 7). DNA concentrations were maintained constant by ad-dition of the empty vector. Increasing amounts of p300 expression vector plasmids were similarly cotransfected up to 250 ng with 100 ng of PDGF-A-luciferase reporter plasmid in the absence (−) (lanes 8 and 9) or presence of 750 ng of KLF5 expression vector (lanes 4 and 5). Effects of an acetyltransferase region-deleted mutant of p300 (ΔHAT) on KLF5-mediated transcriptional activation were also examined (lanes 6, 7, 10, and 11). All experiments were done at least twice with consistent findings. +, present.
To further see whether p300 potentiates transactivation of KLF5, a cotransfection reporter assay was done (Fig. 5C). Under conditions in which KLF5 activated the PDGF-A chain reporter, the addition of p300 resulted in a dose-dependent increase in transactivation but transactivation did not occur with the addition of p300 alone, thus showing that p300 coactivates KLF5 transcription. p300 is therefore a coactivator of KLF5. A mutant of p300 with the acetylase catalytic region deleted showed reduced activation, suggesting that acetylation and/or interaction through this region with KLF5 is important for transactivation of PDGF-A chain reporter activity by KLF5.
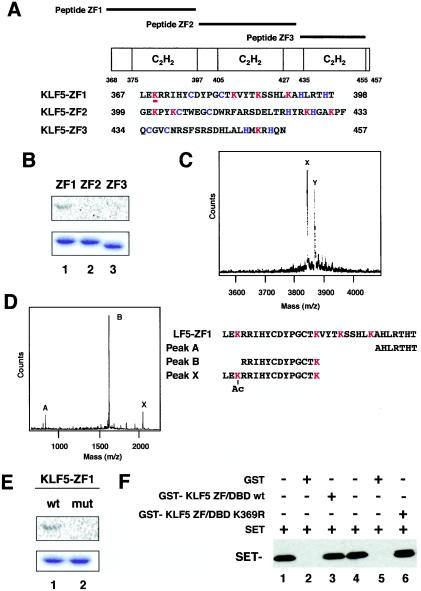
Mapping of the acetylation site of KLF5.
As SET inhibited acetylation of KLF5, we reasoned that understanding the effects of KLF5 acetylation would lead to a better understanding of the actions of SET. For this, we determined the acetylated residue(s) in KLF5 and used nonacetylated point mutants to understand the implications of acetylation in vivo. To determine the acetylation site within the KLF5 ZF/DBD, peptides of each of the three zinc fingers were prepared and subjected to acetylation reactions (Fig. 6A). The first zinc finger was the only peptide acetylated by p300 for both GST proteins and synthetic peptides (Fig. 6B and data not shown). MS of the peptides further showed that the first zinc finger peptide is monoacetylated, as shown by a single shifted peak of 42 m/z (Fig. 6C). The second and third zinc finger peptides did not show a shifted peak and were thus not acetylated in vitro (data not shown).
FIG. 6.
Mapping of the acetylated region and residue of KLF5. (A) Schematic representation of KLF5 zinc finger peptides. ZF1, ZF2, and ZF3 cover each of the zinc fingers, respectively, from the N terminus. (B) Acetylation of KLF5 zinc finger mutants in vitro. Approximately 1.0 μg of purified GST-KLF5 fusion zinc fingers 1, 2, and 3 were incubated with [14C]acetyl-CoA and recombinant FLAG-p300 HAT domain. Reaction products were separated by SDS-10% PAGE. The gel was stained with Coomassie brilliant blue (lower panel) and then analyzed with a BAS 1500 phosphorimager (upper panel). (C) Mass spectrum quantification of acetylated lysines in peptide KLF5-ZF1. A parallel reaction mixture with unlabeled acetyl-CoA was analyzed by MALDI-TOF (MS). The major peak labeled X corresponds to the expected mass of the unmodified peptide KLF5-ZF1. The major peak labeled Y, larger by 42 atomic mass units, represents monoacetylated peptide. (D) Masses of peptides digested withLys-C endopeptidase. The peptide sequences that are suggested from measured masses are shown below. Peak X represents the acetylated fragment. (E) Replacement of acetylated lysine by arginine impairs acetylation of GST-KLF5-zinc finger 1. Approximately 1.0 μg of purified GST-KLF5-zinc finger 1 (lane 1, wild type [wt]) and GST-KLF5-mut zinc finger 1 (K369R) (lane 2, mutant [mut]) were incubated with [14C]acetyl-CoA and 50 ng of FLAG-p300 HAT domain protein. Reaction products were separated by SDS-10% PAGE. The gel was stained with Coomassie brilliant blue (lower panel) and then analyzed with a BAS 1500 phosphorimager (upper panel). (F) Binding assay of K369R and wild-type KLF5 with SET. Wild-type and K369R mutant KLF5 fused to GST were immobilized on GST resin followed by a pull-down assay of SET protein. Lanes 1 and 4 are SET input. Lanes 2 and 5 are GST alone. All experiments were done at least twice with consistent findings. +, present; −, absent.
To next identify the acetylated lysine residue, the acetylated first zinc finger peptide was digested with Lys-C endopeptidase, which cleaves after nonacetylated but not acetylated lysine residues. Analysis of expected fragment masses against actual masses showed that the lysine residue at amino acid number 369 was acetylated (Fig. 6D). To further confirm that this is the only acetylated residue, this lysine was mutagenized to an arginine residue (hereafter referred to as a K369R substitution) to preserve the similar basic charge and then the peptide was subjected to an acetylation assay which showed that the arginine substitution resulted in loss of acetylation (Fig. 6E). SET can bind both K369R and wild-type KLF5, suggesting that interaction is not impaired by this mutation (Fig. 6F).
Functional implications of KLF5 acetylation.
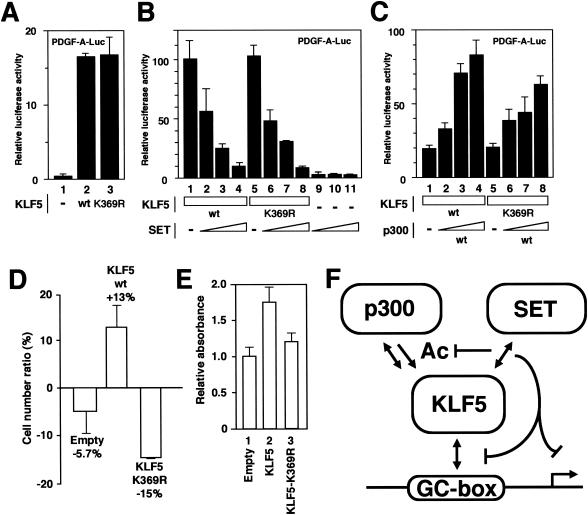
To examine the effects of acetylation on KLF5 function, the transcriptional activity and cellular effects of the K369R mutation were tested. Transcriptional activation examined by cotransfection reporter analysis showed that the K369R mutation showed no effects on PDGF-A chain promoter activity compared to wild-type KLF5 when transfected alone (Fig. 7A). SET also inhibited transactivation by KLF5 similarly for the wild type and the K369R mutant (Fig. 7B), suggesting that interaction is important for the inhibitory effects of SET on KLF5 transactivation. Masking acetylation is a likely result of this interaction. Further, a 20% decrease in PDGF-A chain promoter activity was seen when p300 was cotransfected with the K369R mutant KLF5 construct compared to the KLF5 wild-type construct (Fig. 7C), showing that acetylation is important for transactivation of PDGF-A chain reporter activity by p300 on KLF5.
FIG. 7.
Effects of the KLF5 K369R point mutant. (A) Effects of KLF5 K369R mutant (lane 3) on PDGF-A chain promoter transcriptional activation compared to that of the KLF5 wild type (wt) (lane 2). Seven hundred fifty nanograms of each expression vector was transfected in the presence of 100 ng of the reporter construct (all lanes). −, absent. (B) Effects of SET on KLF5 wild type and K369R mutant. Up to 250 ng of SET expression vector was transfected in the presence of 750 ng of KLF5 expression vector. (C) Effects of the KLF5 K369R mutant on PDGF-A chain promoter transcriptional activation compared to that of the KLF5 wild type in the presence of p300. Increasing amounts of p300 expression vector up to 250 ng were transfected in the presence of 750 ng of KLF5 (wild type, lanes 1 to 4; K369R mutant, lanes 5 to 8) expression vector and 100 ng of the reporter construct. (D) Effects of KLF5 K369R mutant on cell growth. The KLF5 wild type and K369R mutant were transfected with adenovirus and counted on the sixth day in comparison with nontreated cells. Error bars denote standard errors. (E) Effects of the KLF5 K369R mutant on cell growth were similarly assessed by BrdU assay. Error bars denote standard errors. All experiments were done at least twice with consistent findings. (F) Summary of findings. Note that SET negatively regulates DNA binding, transactivation, and acetylation of KLF5. p300 interacts, transactivates, and acetylates KLF5. We envision that this mechanism of interplay between coupled positive regulation by p300 and negative regulation by SET occurs in an inducible setting (e.g., phorbol ester stimulation).
To further examine the effects of acetylation on KLF5 cellular activity, effects on cell growth were investigated. In cells transfected with the wild type or the K369R mutant by adenovirus-mediated transfer, the K369R mutant-transfected cells showed a reduction in cell number (Fig. 7D). A BrdU assay also showed reduced uptake in the K369R mutant-transfected cells, further showing that cell replication is decreased in these cells (Fig. 7E). Interestingly, results of the effects of the nonacetylated K369R mutation of KLF5 closely mimicked the effects of SET on KLF5, suggesting that the effects of SET on KLF5 may at least in part be by inhibition of KLF5 acetylation.
DISCUSSION
Regulatory pathway of coupled interaction and acetylation.
We have described a new regulatory pathway of transcriptional activation and inhibition of a DNA-binding transcription factor through the DBD by coupled interaction and modification (e.g., acetylation) as shown through the opposing actions of p300 and SET on the transcription factor KLF5 (Fig. 7F). SET negatively regulates and p300 positively regulates KLF5 actions, and SET further inhibits acetylation of KLF5 by p300. Importantly, our biochemical and cellular data suggest that the effects of KLF5 acetylation are complementary to the effects of SET on KLF5 on the basis of regulation of transcriptional activation and cell growth. Our findings suggest that the actions of SET are therefore likely mediated at least in part by blocking activation as mediated through this chemical modification. The actions of SET on DNA-binding factors have been hitherto unknown, although past studies had addressed its role in adenoviral DNA replication and as a chaperone for histones (15, 25, 26, 34). The present findings therefore implicate a new role for SET in factor-specific regulation of transcription through a mechanism that has previously only been known for histones. As SET has been shown to similarly inhibit transcription of retinoic acid receptor transcription (39), it is tempting to speculate that this action can be generalized to include at least the nuclear receptor family, which contains a zinc finger-type DBD common to KLF5.
The regulation of acetylation is emerging as a new pathway for the control of transcriptional regulation. Catalytic regulation through actions of deacetylases (17, 33) and coupled regulation with other signaling pathways (i.e., regulation of p53 by coupled acetylation and phosphorylation) (38) and by other interactions has received much attention, but noncatalytic regulation by protein interaction with the substrate and/or enzyme is also a notable regulatory mechanism. SET, as a subunit of the INHAT complex (39), likely masks the substrate protein from acetylation by interaction (e.g., competition) and/or further inducing a conformational change, making it inaccessible to the enzyme. It was also previously shown that DNA binding inhibits interaction and acetylation of Sp1 by the acetylase p300 (45). Further, the human immunodeficiency virus-related tat protein has been shown to modulate acetylation, likely by interaction and by conformational changes to the substrate and/or enzyme, thus modulating acetylation activity and its effects on gene expression (6).
Taken together, modulation of acetylation through noncatalytic actions (e.g., blocking or masking interaction) is a pathway which can confer a new regulatory step to transcriptional regula-tion of gene expression. Note, the combined role of noncatalytic acetylation blocking or masking events with those of deacetylases, which act following acetylation, and further regulation by other interacting regulatory proteins (e.g., complex) is still unknown. Future studies aimed at deciphering the complexity of the regulation of the collective transcription reaction as mediated by acetylation will be necessary to understand the precise role of this regulatory pathway.
Regulatory pathway through the DBD.
Another finding of the present study is that the DBD plays a pivotal role in mediating regulatory interactions. It is well established that the activation and regulatory domain of transcription factors mediates important regulatory interactions (19, 36, 37). In contrast, our findings and those of others suggest that the DBD mediates regulatory interactions particularly for the Sp/KLF family of zinc finger transcription factors. Interaction of the DBD with acetylases has been shown in the past (45), and others have shown interaction with SWI/SNF (12), deacetylase (8), and cell cycle regulatory factor (13, 20) as well as interaction with other zinc finger transcription factors, including Krüppel-like factors (21).
In the present study, we show that there is regulatory interplay by interacting proteins, namely through physical interaction and modification (e.g., acetylation) as mediated by acetylase and its inhibitor (e.g., masking protein) on the DBD. It is tempting to envision that the DBD of at least this subgroup of zinc finger transcription factors mediates a convergence of multiple regulatory pathways contributing to temporospatial regulation of DNA-involved processes ranging from naked DNA to chromatin remodeling to thus affect gene expression. Given that zinc finger transcription factors greatly evolved in genomic complexity in eukaryotes (46), it is likely that they play an important role in specific transcription associated with biological diversification. A better understanding of the actions and regulation of these factors and pathways will add to our understanding of eukaryotic transcriptional regulation.
Functional implications of KLF5-SET interaction.
Our results show a biological setting in which the described mechanisms may contribute to transcriptional regulation of gene expression. The temporospatial biological activity of KLF5 may be dictated by the coordinated regulation of its induced expression and the reduced expression of its negative regulator SET. KLF5 was induced and activated by mitogenic stimulation. In contrast, SET was repressed in response to mitogenic stimulation, and importantly, the repression of SET coincided with the induction of KLF5, which is coupled with expression of a downstream gene. In pathological states, SET and KLF5 were both highly expressed, as examined in an experimental model of atherosclerosis. Expression of SET and KLF5 were both increased in neointimal hyperplasia cells, which are proliferative cells induced in response to a pathological stimulus. Given that their coexpression was seen in late stages of pathogenic evolution, we envision that increased SET expression acted to limit the actions of KLF5 so that uncontrolled inappropriate cell proliferation (e.g., oncogenesis) did not occur.
Interestingly, p300 is also induced by phorbol ester stimulation (23). Together with our findings, our proposed mechanism of coupled positive regulation of KLF5 by p300 and negative regulation by SET may play a role in the biological setting of pathophysiological induction of KLF5 as exemplified by the actions of the model agonist, phorbol ester. Induction of KLF5 activation and of its downstream genes are coupled with up-regulation of its coactivator p300 and down-regulation of its repressor SET, thereby likely resulting in amplification of KLF5 actions. Another interesting issue on the regulation of SET may be that there is possible regulation of SET by its splicing variant and dimerization partner TAF-Iα. We have seen that while SET alone is expressed in KLF5-active smooth muscle cells, TAF-Iα is also expressed in stoichiometric amounts in other cells. Given that TAF-Iα has been suggested to be a negative regulator of SET (28), we believe that the amount of SET relative to TAF-Iα may be critical in regulating SET actions. SET may therefore act only in the absence of TAF-Iα or when SET is found in relative abundance; therefore allowing for dissection of the unique actions of SET as a negative regulator in the TAF-Iα-deficient KLF5-expressing smooth muscle cell. Collectively, our findings suggest a new transcriptional regulatory pathway of a DNA-binding transcription factor by combined use of inducible positive and negative cofactors centered on interaction and acetylation which likely reflects the mechanisms underlying triggered activation by stimuli.
Importantly, the regulatory pathway by p300 and SET allows for bimodal regulation of KLF5, which will efficiently allow for a rapid amplification of coordination and fine tuning of gene regulation such as in gene- and cell type-specific transcriptional regulation (e.g., temporal as well as spatial regulation). Future studies aimed at understanding the regulation and generality of the described mechanisms will further advance our understanding of the biological role of the involved factors and reactions.
Acknowledgments
We thank K. Nagata, Y. Nakatani, V. Ogryzko, and C. Teng for constructs and reagents. We thank Takayoshi Matsumura, Kana Sasaki, Nanae Kada, and Yoshiko Munemasa for assistance.
This study was supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology; New Energy and Industrial Technology Development Organization; Ministry of Health, Labor, and Welfare; Japan Science and Technology Corporation; Sankyo Life Science Foundation; Takeda Medical Research Foundation; Japan Heart Foundation (Zeria grant); and the Applied Enzyme Association.
S. Miyamoto, T. Suzuki, S. Muto, and K. Aizawa contributed equally to this work.
REFERENCES
- 1.Adachi, Y., G. N. Pavlakis, and T. D. Copeland. 1994. Identification and characterization of SET, a nuclear phosphoprotein encoded by the translocation break point in acute undifferentiated leukemia. J. Biol. Chem. 269:2258-2262. [PubMed] [Google Scholar]
- 2.Bieker, J. J. 2001. Krüppel-like factors: three fingers in many pies. J. Biol. Chem. 276:34355-34358. [DOI] [PubMed] [Google Scholar]
- 3.Black, A. R., J. D. Black, and J. Azizkhan-Clifford. 2001. Sp1 and krüppel-like factor family of transcription factors in cell growth regulation and cancer. J. Cell. Physiol. 188:143-160. [DOI] [PubMed] [Google Scholar]
- 4.Chamley-Campbell, J. H., G. R. Campbell, and R. Ross. 1981. Phenotype-dependent response of cultured aortic smooth muscle to serum mitogens. J. Cell Biol. 89:379-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheung, W. L., S. D. Briggs, and C. D. Allis. 2000. Acetylation and chromosomal functions. Curr. Opin. Cell Biol. 12:326-333. [DOI] [PubMed] [Google Scholar]
- 6.Col, E., B. Gilquin, C. Caron, and S. Khochbin. 2002. Tat-controlled protein acetylation. J. Biol. Chem. 277:37955-37960. [DOI] [PubMed] [Google Scholar]
- 7.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doetzlhofer, A., H. Rotheneder, G. Lagger, M. Koranda, V. Kurtev, G. Brosch, E. Wintersberger, and C. Seiser. 1999. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol. Cell. Biol. 19:5504-5511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dynan, W. S., and R. Tjian. 1983. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell 35:79-87. [DOI] [PubMed] [Google Scholar]
- 10.Freiman, R. N., and R. Tjian. 2003. Regulating the regulators: lysine modifications make their mark. Cell 112:11-17. [DOI] [PubMed] [Google Scholar]
- 11.Hoshino, Y., M. Kurabayashi, T. Kanda, A. Hasegawa, H. Sakamoto, E. Okamoto, K. Kowase, N. Watanabe, I. Manabe, T. Suzuki, A. Nakano, S. Takase, J. N. Wilcox, and R. Nagai. 2000. Regulated expression of the BTEB2 transcription factor in vascular smooth muscle cells: analysis of developmental and pathological expression profiles shows implications as a predictive factor for restenosis. Circulation 102:2528-2534. [DOI] [PubMed] [Google Scholar]
- 12.Kadam, S., G. S. McAlpine, M. L. Phelan, R. E. Kingston, K. A. Jones, and B. M. Emerson. 2000. Functional selectivity of recombinant mammalian SWI/SNF subunits. Genes Dev. 14:2441-2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlseder, J., H. Rotheneder, and E. Wintersberger. 1996. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol. Cell. Biol. 16:1659-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawai-Kowase, K., M. Kurabayashi, Y. Hoshino, Y. Ohyama, and R. Nagai. 1999. Transcriptional activation of the zinc finger transcription factor BTEB2 gene by Egr-1 through mitogen-activated protein kinase pathways in vascular smooth muscle cells. Circ. Res. 85:787-795. [DOI] [PubMed] [Google Scholar]
- 15.Kawase, H., M. Okuwaki, M. Miyaji, R. Ohba, H. Handa, Y. Ishimi, T. Fujii-Nakata, A. Kikuchi, and K. Nagata. 1996. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1:1045-1056. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, A., and M. Horikoshi. 1998. Tip60 acetylates six lysines of a specific class in core histones in vitro. Genes Cells 3:789-800. [DOI] [PubMed] [Google Scholar]
- 17.Kochbin, S., A. Verdel, C. Lemercier, and D. Seigneruin-Berny. 2001. Functional significance of histone deacetylase activity. Curr. Opin. Genet. Dev. 11:162-166. [DOI] [PubMed] [Google Scholar]
- 18.Koritschoner, N. P., J. L. Bocco, G. M. Panzetta-Dutari, C. I. Dumur, A. Flury, and L. C. Patrito. 1997. A novel human zinc finger protein that interacts with the core promoter element of a TATA box-less gene. J. Biol. Chem. 272:9573-9580. [DOI] [PubMed] [Google Scholar]
- 19.Lemon, B., and R. Tjian. 2000. Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14:2551-2569. [DOI] [PubMed] [Google Scholar]
- 20.Lin, S. Y., A. R. Black, D. Kostic, S. Pajovic, C. N. Hoover, and J. C. Azizkhan. 1996. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol. Cell. Biol. 16:1668-1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay, J. P., and M. Crossley. 1998. Zinc fingers are sticking together. Trends Biochem. Sci. 23:1-4. [DOI] [PubMed] [Google Scholar]
- 22.Manabe, I., and G. K. Owens. 2001. Recruitment of serum response factor and hyperacetylation of histones at smooth muscle-specific regulatory regions during differentiation of a novel P19-derived in vitro smooth muscle differentiation system. Circ. Res. 88:1127-1134. [DOI] [PubMed] [Google Scholar]
- 23.Masumi, A., I. M. Wang, B. Lefebvre, X. J. Yang, Y. Nakatani, and K. Ozato. 1999. The histone acetylase PCAF is a phorbol-ester-inducible coactivator of the IRF family that confers enhanced interferon responsiveness. Mol. Cell. Biol. 19:1810-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matsumoto, K., K. Nagata, M. Ui, and F. Hanaoka. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582-10587. [PubMed] [Google Scholar]
- 25.Matsumoto, K., M. Okuwaki, H. Kawase, H. Handa, F. Hanaoka, and K. Nagata. 1995. Stimulation of DNA transcription by the replication factor from the adenovirus genome in a chromatin-like structure. J. Biol. Chem. 270:9645-9650. [DOI] [PubMed] [Google Scholar]
- 26.Matsumoto, K., K. Nagata, M. Okuwaki, and M. Tsujimoto. 1999. Histone- and chromatin-binding activity of template activating factor-I. FEBS Lett. 463:285-288. [DOI] [PubMed] [Google Scholar]
- 27.Miller, I. J., and J. J. Bieker. 1993. A novel, erythroid cell-specific murine transcription factor that binds to the CACCC element and is related to the Krüppel family of nuclear proteins. Mol. Cell. Biol. 13:2776-2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyaji-Yamaguchi, M., M. Okuwaki, and K. Nagata. 1999. Coiled-coil structure-mediated dimerization of template activating factor-I is critical for its chromatin remodeling activity. J. Mol. Biol. 290:547-557. [DOI] [PubMed] [Google Scholar]
- 29.Nagata, K., H. Kawase, H. Handa, K. Yano, M. Yamasaki, Y. Ishimi, A. Okuda, A. Kikuchi, and K. Matsumoto. 1995. Replication factor encoded by a putative oncogene, set, associated with myeloid leukemogenesis. Proc. Natl. Acad. Sci. USA 92:4279-4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagata, K., S. Saito, M. Okuwaki, H. Kawase, A. Furuya, A. Kusano, N. Hanai, A. Okuda, and A. Kikuchi. 1998. Cellular localization and expression of template-activating factor I in different cell types. Exp. Cell Res. 240:274-281. [DOI] [PubMed] [Google Scholar]
- 31.Nakatani, Y. 2001. Histone acetylases-versatile players. Genes Cells 6:79-86. [DOI] [PubMed] [Google Scholar]
- 32.Narla, G., K. E. Heath, H. L. Reeves, D. Li, L. E. Giono, A. C. Kimmelman, M. J. Glucksman, J. Narla, F. J. Eng, A. M. Chan, A. C. Ferrari, J. A. Martignetti, and S. Friedman. 2001. KLF6, a candidate tumor suppressor gene mutated in prostate cancer. Science 294:2563-2566. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., and A. Bird. 2000. Histone deacetylases: silencers for hire. Trends Biochem. Sci. 26:1-6. [DOI] [PubMed] [Google Scholar]
- 34.Okuwaki, M., and K. Nagata. 1998. Template activating factor-I remodels the chromatin structure and stimulates transcription from the chromatin template. J. Biol. Chem. 273:34511-34518. [DOI] [PubMed] [Google Scholar]
- 35.Philipsen, S., and G. Suske. 1999. A tale of three fingers: the family of mammalian Sp/XKLF transcription factors. Nucleic Acids Res. 27:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ptashne, M., and A. A. Gann. 1990. Activators and targets. Nature 346:329-331. [DOI] [PubMed] [Google Scholar]
- 37.Roeder, R. G. 1996. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 9:327-335. [PubMed] [Google Scholar]
- 38.Sakaguchi, K., J. E. Herrera, S. Saito, T. Miki, M. Bustin, A. Vassilev, C. W. Anderson, and E. Appella. 1998. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 12:2831-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seo, S. B., P. McNamara, S. Heo, A. Turner, W. S. Lane, and D. Chakravarti. 2001. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell 104:119-130. [DOI] [PubMed] [Google Scholar]
- 40.Shi, H., Z. Zhang, X. Wang, S. Liu, and C. T. Teng. 1999. Isolation and characterization of a gene encoding human Krüppel-like factor 5 (IKLF): binding to the CAAT/GT box of the mouse lactoferrin gene promoter. Nucleic Acids Res. 27:4807-4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shindo, T., I. Manabe, Y. Fukushima, K. Tobe, K. Aizawa, S. Miyamoto, K. Kawai-Kowase, N. Moriyama, Y. Imai, H. Kawakami, H. Nishimatsu, T. Ishikawa, T. Suzuki, H. Morita, K. Maemura, M. Sata, Y. Hirata, M. Komukai, H. Kagechika, T. Kadowaki, M. Kurabayashi, and R. Nagai. 2002. Krüppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat. Med. 8:856-863. [DOI] [PubMed] [Google Scholar]
- 42.Sogawa, K., Y. Kikuchi, H. Imataka, and Y. Fujii-Kuriyama. 1993. Comparison of DNA-binding properties between BTEB and Sp1. J. Biochem. 114:605-609. [DOI] [PubMed] [Google Scholar]
- 43.Sun, R., X. Chen, and V. W. Yang. 2001. Intestinal-enriched Krüppel-like factor (Krüppel-like factor 5) is a positive regulator of cellular proliferation. J. Biol. Chem. 276:6897-6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suzuki, T., T. Yamamoto, M. Kurabayashi, R. Nagai, Y. Yazaki, and M. Horikoshi. 1998. Isolation and initial characterization of GBF, a novel DNA-binding zinc finger protein that binds to the GC-rich binding sites of the HIV-1 promoter. J. Biochem. 124:389-395. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki, T., A. Kimura, R. Nagai, and M. Horikoshi. 2000. Regulation of interaction between the acetyltransferase region of p300 and the DNA-binding domain of Sp1 on and through DNA binding. Genes Cells 5:29-41. [DOI] [PubMed] [Google Scholar]
- 46.Tupler, R., G. Perini, and M. R. Green. 2001. Expressing the human genome. Nature 409:832-833. [DOI] [PubMed] [Google Scholar]
- 47.Von Lindern, M., S. van Baal, J. Wiegant, A. Raap, A. Hagemeijer, and G. Grosveld. 1992. Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′ half to different genes: characterization of the set gene. Mol. Cell. Biol. 12:3346-3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watanabe, N., M. Kurabayashi, Y. Shimomura, K. Kawai-Kowase, Y. Hoshino, I. Manabe, M. Watanabe, M. Aikawa, M. Kuro-o, T. Suzuki, Y. Yazaki, and R. Nagai. 1999. BTEB2, a Krüppel-like transcription factor, regulates expression of the SMemb/Nonmuscle myosin heavy chain B (SMemb/NMHC-B) gene. Circ. Res. 85:182-191. [DOI] [PubMed] [Google Scholar]