Abstract
Ebola viruses (EBOVs) cause rare but highly fatal outbreaks of viral hemorrhagic fever in humans, and approved treatments for these infections are currently lacking. The Ebola VP35 protein is multifunctional, acting as a component of the viral RNA polymerase complex, a viral assembly factor, and an inhibitor of host interferon (IFN) production. Mutation of select basic residues within the C-terminal half of VP35 abrogates its dsRNA-binding activity, impairs VP35-mediated IFN antagonism, and attenuates EBOV growth in vitro and in vivo. Because VP35 contributes to viral escape from host innate immunity and is required for EBOV virulence, understanding the structural basis for VP35 dsRNA binding, which correlates with suppression of IFN activity, is of high importance. Here, we report the structure of the C-terminal VP35 IFN inhibitory domain (IID) solved to a resolution of 1.4 Å and show that VP35 IID forms a unique fold. In the structure, we identify 2 basic residue clusters, one of which is important for dsRNA binding. The dsRNA binding cluster is centered on Arg-312, a highly conserved residue required for IFN inhibition. Mutation of residues within this cluster significantly changes the surface electrostatic potential and diminishes dsRNA binding activity. The high-resolution structure and the identification of the conserved dsRNA binding residue cluster provide opportunities for antiviral therapeutic design. Our results suggest a structure-based model for dsRNA-mediated innate immune antagonism by Ebola VP35 and other similarly constructed viral antagonists.
Keywords: crystal structure, Ebola virus, RNA binding
Ebola viruses (EBOVs) cause severe hemorrhagic fever characterized by fever, shock, coagulation defects, and impaired immunity (1, 2). These manifestations of infection are thought to reflect subversion of the innate immune system coupled with uncontrolled viral replication, particularly in macrophages and dendritic cells (3, 4). EBOV infection of these cells enhances production of proinflammatory cytokines, such as TNF-α and IFN (IFN)-γ, and diminishes stimulation of T cell maturation by dendritic cells (3, 4). Like other negative-strand RNA viruses that impair both innate and adaptive immunity (e.g., influenza, rabies, and measles), EBOV suppresses host IFN activities to replicate, thus resulting in serious disease (5, 6). Only individuals who survive EBOV infection show appreciable amounts of viral-specific antibodies (7), suggesting that EBOV infections lead to shutdown of early immune responses and prevent activation of adaptive immune responses.
Recognition of viral particles and viral RNA, including RNA modifications such as 5′-triphosphate (5′-ppp), by cytosolic pattern recognition receptor helicases RIG-I and MDA-5 leads to activation of transcription factors, including IFN regulatory factor-3 (IRF-3), IRF-7, NF-κB, and AP-1 (8–12). These transcription factors in turn induce expression of a large number of cytokines, such as IFN-α and IFN-β (13). Activated IFN genes operate in both autocrine and paracrine manners to stimulate the activity of additional antiviral genes. Therefore, inhibition of signaling mechanisms that promote IFN responses is a necessary viral countermeasure against the host antiviral system and critical for viral propagation (2).
Ebola viral protein 35 (VP35) is multifunctional, serving as a component of the viral RNA polymerase complex, as a structural/assembly factor, and as a suppressor of host IFN responses (2). Therefore, a functional VP35 is required for efficient viral replication and pathogenesis; knockdown of VP35 leads to reduced viral amplification and reduced lethality in infected mice (14–18). However, limited information is available regarding how VP35 is able to perform multiple functions. VP35 contains an N-terminal coiled-coil domain required for its oligomerization (19, 20) and a C-terminal dsRNA-binding region (15, 16, 21). Oligomerization through the N-terminal oligomerization domain is required for virus replication because oligomerization-defective mutants fail to interact with the viral polymerase (L) protein (15, 22). Similarly, deletion of the N-terminal region or mutation of the coiled-coil domain abrogates IFN suppression by VP35, which can be overcome by tethering a heterologous oligomerization module to the VP35 C terminus or overexpression of the isolated VP35 C terminus (20). The latter observations suggest a model where the coiled-coil domain provides a critical oligomerization function, whereas the C-terminal region of VP35 interacts with host factors to block signaling that triggers IFN responses.
The host factor(s) directly targeted by VP35 have not been definitively identified. Only VP35, and not any other Ebola protein, supports viral growth of a mutant influenza A virus that lacks the IFN suppressor protein NS1A, suggesting that VP35 also inhibits IFN activity (23). A bioinformatics study identified an 8-residue motif in VP35 that has 75% sequence identity to the influenza NS1A protein, including basic residues essential for binding of dsRNA by NS1 (21). Mutation of these residues impaired NS1A inhibition of IFN responses (24). Based on this identity, it was suggested that this motif could also be required for inhibition of IFN responses by VP35 (21). Consistent with these observations, VP35 inhibits phosphorylation, activation, and nuclear localization of IRF-3 and inhibits viral- and dsRNA-induced expression of the IFNβ gene (14, 17, 18, 21, 25, 26). VP35 also inhibits activation of the cellular antiviral kinase RNA-dependent protein kinase (PKR) (16) and activation of the RNAi pathway (27). These pathways are similarly targeted by other virus-encoded dsRNA-binding proteins. Examples include the influenza virus NS1A protein, the reovirus σ3 protein (28), and the vaccinia virus E3L protein (29), which can inhibit host antiviral responses through dsRNA-dependent and -independent mechanisms.
The mechanism by which VP35 inhibits IRF-3 activation and IFNα/β expression is not completely understood, but thus far, VP35 IFN-antagonist function correlates with VP35 dsRNA binding activity. EBOV likely activates the RIG-I pathway (15, 25) and VP35 expression inhibits the RIG-I activated signaling pathway (15). Because coexpression of VP35 can inhibit activation of IFNβ gene expression by overexpressed IKKε or TBK-1, it appears that VP35 may target these kinases to exert its inhibitory effect, although additional mechanisms directed at more upstream points in these pathways cannot be excluded. Indeed, mutation of basic residues such as Arg-312 to alanine (Arg312Ala) severely impairs VP35 dsRNA binding and IFN-antagonist activity, but does not significantly affect VP35 function as part of the viral polymerase complex (15, 17, 21, 25). Viruses containing the Arg312Ala mutation more readily activate IRF-3, compared with a wild-type virus, and induce a much stronger innate immune response than wild-type virus. Taken together, these data correlate dsRNA binding with the inability of the Arg312Ala mutant virus to support viral growth because of aberrant IFN inhibition (22, 25). Strikingly, an Arg312Ala VP35 mutant EBOV is greatly attenuated relative to the wild-type virus in mice (25).
These data suggest that the dsRNA binding activity mediated by the C terminus of VP35 is critical for viral suppression of innate immunity and for virulence (14, 25). Yet, the structure of the VP35 C terminus has not been available, and it is not clear how individual mutations impair dsRNA binding and suppress innate immune signaling. As an initial step toward addressing these questions, we conducted a structural and biochemical analysis of the C-terminal region of Ebola VP35. Here, we report the 3-dimensional structure of the VP35 IFN inhibitory domain (IID) solved by X-ray crystallography and identify residues important for RNA binding. Our structure reveals a unique fold that binds dsRNA. Examination of the VP35 IID structure reveals 2 basic patches that are highly conserved among members of the Filoviridae family (identical among EBOV isolates). Biochemical and NMR-based structural analyses show that one patch contains residues that are required for dsRNA binding and IFN inhibition, whereas the functional significance of the other basic patch remains unknown. Although the structure of VP35 IID is significantly different from that of influenza NS1A RNA-binding domain (RBD), the basic side chains at the dimer interface of the NS1A RBD and those located near Arg-312 in VP35 IID are positioned to form contiguous basic patches, suggesting that NS1A and VP35 may interact with dsRNA in a similar manner. Our results provide structural insights into the role of conserved residues in dsRNA binding that are required for full EBOV virulence and suggest a model for RNA-dependent Ebola VP35 functions.
Results
Crystal Structure of EBOV VP35 Was Solved to High Resolution.
Using NMR-based studies, we identified the minimal structured region from the C terminus of Ebola VP35 IID and the crystal structure of VP35 IID was solved to 1.4-Å resolution (Fig. 1 B and C). In the crystal, there are 2 nearly identical monomers (A and B) of VP35 IID in the asymmetric unit [supporting information (SI) Text, Table S1, and Fig. S1). PROCHECK analysis revealed that 95.2% of the residues are located in the most -favored regions (A, B, L) of the Ramachandran plot, whereas the remaining 4.8% are in the additional allowed regions (a, b, l, p). No electron density is observed for the first 4 residues from monomer A and the first 2 residues from monomer B. Previous studies have shown that full-length VP35 forms oligomers through the N-terminal oligomerization region. However, both dynamic light-scattering and analytical ultracentrifugation experiments demonstrate that VP35 IID is monomeric in solution (Fig. S2). Furthermore, the buried surface area between monomer A and monomer B in the asymmetric unit is ≈490 Å2, indicating that the interaction between the monomers observed in the VP35 IID crystal is weak at best.
Fig. 1.
Crystal structure of VP35 C-terminal IFN-inhibitory domain (IID) reveals a fold that binds dsRNA. (A) Domain organization of VP35. (B) Ribbon representation of VP35 IID. Secondary structural elements that form the α-helical subdomain (orange) and the β-sheet subdomain (yellow). (C) Topology and delimiting sequence markers of VP35 IID.
VP35 IID Comprises 2 Subdomains.
The VP35 IID structure is organized into 2 subdomains: an N-terminal α-helical subdomain and a C-terminal β-sheet subdomain (Fig. 1 B and C). The α-helical subdomain (residues 221–283) is a 4-helical bundle (α1–α4), arranged in 2 layers, with α1 and α2 helices packed against α3 and α4 helices to form the core of the α-helical subdomain. The α2 and α4 helices pack against the β-sheet subdomain. The β-sheet subdomain (residues 294–340) is composed of a 4-stranded mixed β-sheet (β1–β4) and a short helical segment, α5. The β1 strand connects to α5 helix through a long linker that includes a reverse turn. The α5 helix leads into a hairpin turn, followed by the short β2 strand and the longer β3 and β4 strands. The β2–β4 strands are antiparallel whereas the β1 strand is parallel to the β3 strand (Fig. 1B). Linker 1, which includes Pro-292 and Pro-293 residues, connects the subdomains. Residues between the β2 and β3 strands of VP35 IID (linker 2 and Pro-313 to Pro-318) form a left-handed polyproline Type II helix (PPII), a feature that is not characteristic of canonical dsRNA-binding domains (dsRBDs). The PPII helix packs against the β3 strand and the Type I hairpin loop formed by residues Lys-319 to Gly-323.
VP35 IID Forms a Unique Fold.
The overall structure of VP35 IID represents a unique fold for a dsRBD that is substantially different from the αβββα fold of canonical dsRBDs. Structural comparisons using the DALI server revealed that the α-helical subdomain has a topology similar to many functionally unrelated proteins (DALI z-score >2), including thiocyanate hydrolase and pyruvate carboxylase. However, DALI searches using either the β-sheet subdomain or the complete VP35 IID backbone failed to identify structurally similar or functionally related proteins, suggesting a unique fold for a dsRNA-binding domain. PSI-BLAST analysis of the Ebola VP35 IID sequence against the nonredundant protein sequence database at NCBI/BLAST (http://blast.ncbi.nlm.nih.gov/) revealed that the sequences corresponding to the 2 subdomains of VP35 IID are not found in combination with other domains. The VP35 IID sequence matches with EBOV sequences (E values from 10−70 to 10−62) and Filoviradae family member Marburg VP35 sequences (10−39 to 10−25), whereas the nearest nonfiloviral protein has an E value of 0.88. Our sequence analysis validates our observation that residues within the VP35 IID are likely to form a functionally interdependent folding unit. To further test our observation that the α-helical and β-sheet subdomains are both important for the overall fold and stability of VP35 IID, we generated each subdomain and compared the backbone chemical shifts with the wild-type VP35 IID protein using NMR spectroscopy. Resulting 1H-15N HSQC NMR spectra show significant differences between backbone chemical shifts of VP35 IID and the isolated subdomains. In particular, we observe a collapse in chemical shift dispersion and high-intensity peaks, indicating that the individual subdomains are destabilized and unfolded in solution (Fig. S3). Together, these data support the importance of both subdomains for VP35 IID structure.
Large Hydrophobic Surface Is Buried Between 2 Structurally Interdependent Subdomains in the VP35 IID.
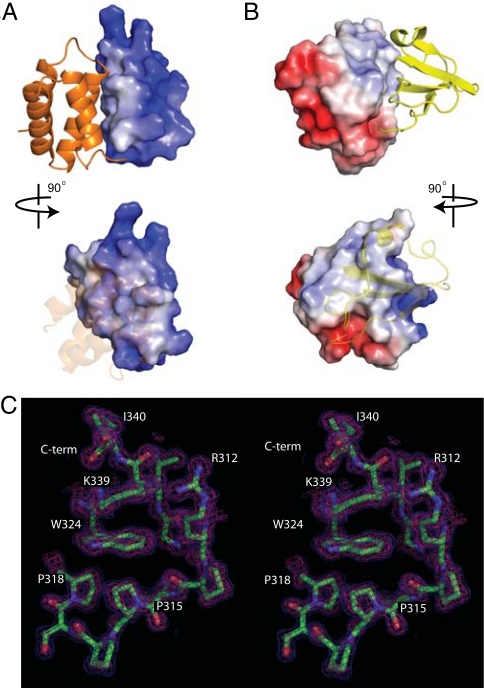
Distribution of surface electrostatic charge across VP35 IID reveals that the α-helical and β-sheet subdomains interact through nonpolar surfaces (Fig. 2 A and B). A total area of 1,405 Å2 is buried between the 2 subdomains, and the interaction surface consists of numerous nonbonded contacts formed by the following residues: Ala-238, Gln-241, Leu-242, Val-245, Ile-246, Leu-249, Ile-278, Ile-280, and Phe-287 from the α-helical subdomain and Pro-293, Ala-306, Cys-307, Pro-315, Pro-318, Ile-320, Asp-321, Gly-323, Trp-324, Val-325, Leu-338, and Ile-340 from the β-sheet subdomain. Interactions between conserved Trp-324 and side chains of Pro-315, Pro-318, and Lys-339 residues are also important to stabilize the β-sheet subdomain structure (Fig. 2C), because mutation of the Trp-324 residue (Trp324Ala) leads to the complete unfolding of VP35 IID (data not shown). Furthermore, the Ile-340 residue in strand β4 is engaged in critical contacts with Phe-239, Leu-242, and Ile-278 that bridge the 2 subdomains. The protein structure is severely destabilized when Ile-340 is deleted or mutated (data not shown). It is interesting to note that all of the residues highlighted here are 100% identical among all known VP35 sequences from Ebola isolates and is highly conserved in the Filoviridae family. Together, these data suggest that Trp-324 and the surrounding hydrophobic core play crucial roles in stabilizing the VP35 IID fold that facilitates VP35 activity.
Fig. 2.
The surface area between the VP35 IID subdomains is hydrophobic. (A and B) Electrostatic representations of the intersubdomain interaction surface for the α-helical subdomain (A) and the β-sheet subdomain (B) reveal hydrophobic surfaces buried between the 2 subdomains. Red, white, and blue represent negative, neutral, and positive electrostatic potentials, respectively (range −5 to +5 kT). (C) Stereographic image showing the Trp-324 side chain making important hydrophobic contacts with residues in β4 strand, α5 helix, and PPII. The 2Fo−Fc map is contoured at 1σ (blue) and 2σ (pink).
Basic Residues Within the VP35 IID Sequence Are Highly Conserved Among Ebola and Marburg Filoviruses.
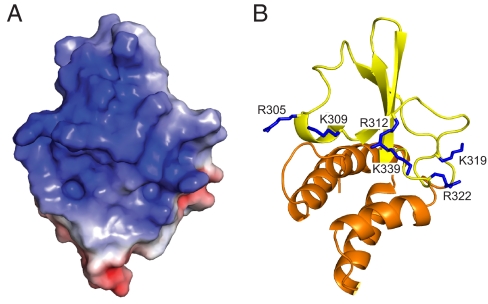
Sequence comparisons among Ebola and Marburg viruses reveal a high level of conservation near the C terminus of VP35 IID, in particular among basic residues (Fig. S4) (21). Examination of the electrostatic surface of the VP35 IID structure shows a large distribution of positive charge and identifies an extended central basic patch in the β-sheet subdomain that connects Arg-305, Lys-309, Arg-312, Lys-319, Arg-322, and Lys-339 residues (Fig. 3). Mutation of Arg-305, Lys-309, or Arg-312 or combinations of these residues to alanine leads to decreased IFN suppression (15, 21, 22). Reduced IFN suppression in virally infected cells results in increased IRF-3 phosphorylation and nuclear localization and leads to reduced viral growth and diminished antagonism of host antiviral signals (15, 22). Similarly, Lys-339 is centrally located in the basic patch near Arg-312 and is likely to impact VP35-mediated functions such as immune inhibition. In contrast, several other basic residues, including Arg-305, Lys-309, Lys-319, and Arg-322, that are in proximity to each other are located on the periphery of the central basic patch in the 3-dimesional structure and may function in a redundant manner (Fig. 4B). Interestingly, a second basic patch consisting of residues Lys-222, Arg-225, Lys-248, and Lys-251 is located on the α-helical subdomain at the face opposite to the central charge surface of VP35 IID (compare Fig. 3A with Fig. S5A). This second basic patch is also 100% identical among members of the Ebola viral isolates.
Fig. 3.
Conserved basic residues in the β-sheet domain form an extended cluster. (A) Electrostatic representation of the solvent-exposed surface of VP35 IID. (B) Conserved basic residues important for IFN antagonism and dsRNA binding are highlighted.
Fig. 4.
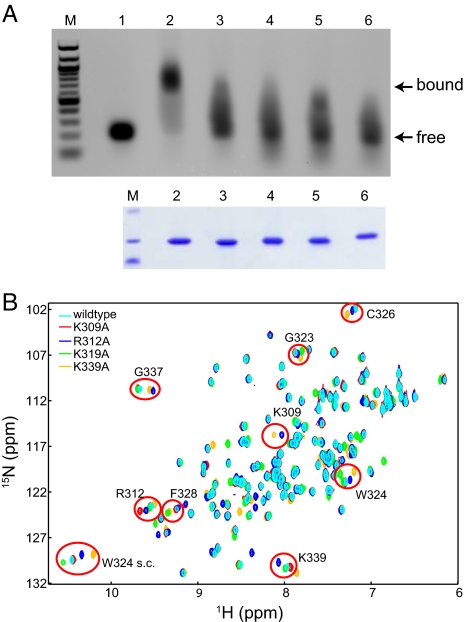
Mutation of conserved basic residues in the β-sheet subdomain diminishes dsRNA binding. (A) Native gel-shift assays show that wild-type VP35 IID and not basic patch mutants bind dsRNA with high affinity. Lanes: M, marker; 1, dsRNA alone; 2, dsRNA plus wild-type VP35 IID; 3, dsRNA plus VP35 IID Lys309Ala; 4, dsRNA plus VP35 IID Arg312Ala; 5, dsRNA plus VP35 IID Lys319Ala; and 6, dsRNA plus VP35 IID Lys339Ala. Coomassie-stained SDS/PAGE gel highlights the purity and the relative amounts of VP35 IID proteins. (B) 1H/15N HSQC NMR-spectra of wild-type and mutant VP35 IID proteins show localized chemical shift changes (red circles).
Conserved Basic Residues Located Near Arg-312 Are Important for dsRNA Binding.
dsRNA has long been recognized as a potent activator of host innate immune signaling that establishes an antiviral state (9, 10), and the ability for Ebola VP35 to block these signals is well documented (3, 15, 16, 23, 26). Therefore, we tested the ability of wild type and central basic patch mutants of VP35 IID to bind heterologous dsRNA. As shown in Fig. 4A, only the wild-type IID and not the central basic patch mutants, including Lys309Ala, Arg312Ala, and Lys339Ala, display dsRNA binding as indicated by a gel shift to a higher molecular mass for the complex. These results demonstrate that the central basic patch is likely to be part of the dsRNA-binding interface of VP35 IID. Interestingly, Lys-319, which is located at the periphery of the central basic patch, displayed reduced, but measurable dsRNA binding, suggesting that Lys-319 may form only a part of the primary dsRNA-binding surface. We further characterized these mutations using NMR-based methods to determine whether the observed differences in dsRNA binding are due to global changes in structure. As shown in Fig. 4B, wild-type and mutant VP35 IID proteins display similar 1H/15N-HSQC NMR resonances, which indicate that the mutant proteins maintain similar overall folds. Minor differences in observed chemical shifts between the wild-type and mutant IID spectra were mapped by sequential assignment of NMR spectra. These results indicate that observed spectral differences are due to subtle changes in the local chemical environment rather than large structural perturbations. Therefore, mutation of residues in the central basic patch that result in diminished dsRNA binding are likely due to the changes in the surface electrostatic potential of VP35 IID (Fig. S6). Sequence-independent dsRNA binding requires specific structural constraints that can position basic residue side chains to make specific contacts with the phosphodiester backbone. The reduced binding we observed upon mutation of select basic residues across the RNA-binding patch suggests that many residues in the central basic patch function in a nonredundant manner.
Second Conserved Basic Patch in VP35 IID Is Not Important for Direct dsRNA Binding.
The structure revealed a second basic patch that is located on the opposite face of the VP35 IID, away from the central RNA-binding residue cluster. We also tested the role of the second basic patch located in the α-helical subdomain toward dsRNA binding. Mutation of either Arg-225 (Arg225Ala) or Lys-251 (Lys251Ala) in VP35 IID did not cause significant changes in the VP35 IID structure (data not shown). Furthermore, native gel-shift assays demonstrate that these VP35 IID mutants retained the ability to bind dsRNA (Fig. S5C). Therefore, it is unlikely that residues contained in the second basic patch directly contribute to dsRNA binding of VP35. However, examination of the surface electrostatic distribution of the α-helical subdomain suggests that this subdomain may play a role in additional interactions, such as those during nucleocapsid formation or replication that require oligomerization of VP35 (30).
Discussion
We have identified residues in Ebola VP35 that are required to form an independently folded unit and identify regions that are important for dsRNA binding. In the structure, we have discovered 2 conserved basic patches located on the surface of VP35 IID, of which only one is important for dsRNA binding and potentially for IFN-inhibition. In the central dsRNA-binding patch, we highlight several conserved basic residues, including Lys-319, Arg-322, and Lys-339, which are critical for dsRNA binding. Location of these residues in the central basic patch together with residues Arg-305, Lys-309, and Arg-312 suggests that mutation of these residues may result in impaired IFN suppression and yield attenuated viruses in vivo because of reduced dsRNA binding.
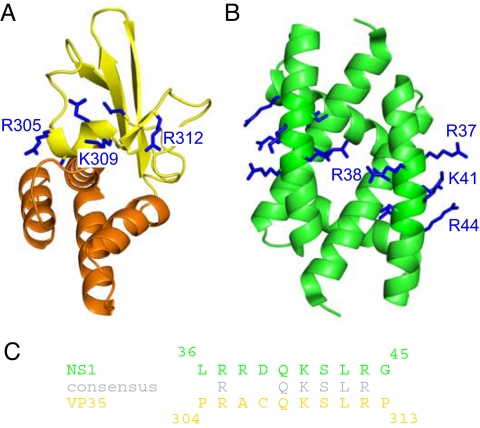
A previous bioinformatics comparison identified a stretch of 8 residues with high sequence similarity with the influenza NS1A protein (21). However, it was not clear whether these residues also share structural similarities simply based on sequence and therefore display mutually overlapping functions. Comparison of the VP35 IID structure from the current study with that of NS1 RNA-binding domain (31, 32) reveals that the overall structures are significantly different (Fig. 5A). Moreover, the short stretch of residues that are highly similar between VP35 and NS1A is incorporated into structurally distinct scaffolds (Fig. 5 A vs. B). In VP35, the 8-residue segment is located within the α5 helix and β2 strand, which in influenza NS1A is incorporated entirely within a single helix (31). NS1 proteins form obligatory dimers, which require contacts from the RNA-binding domain. In contrast, VP35 forms trimers through contacts at the N terminus (ref. 20 and D.W.L. and G.K.A., unpublished observations) that are independent of the dsRNA-binding domain. Moreover, isolated VP35 IID protein is monomeric in solution, even at high concentrations (Fig. S2). Despite these differences, the basic residue side chains, which are important for RNA binding, form similar contiguous patches (Fig. 5 A and B) (21). Altogether, this data along with the ability of influenza NS1 and Ebola VP35 to mutually substitute IFN-suppressive functions suggest that these 2 proteins may target similar components in the host immune system. However, differences we observe between VP35 and NS1 proteins in their overall structures, oligomeric states, and the structural requirements for oligomerization, may reflect additional functionalities performed by these proteins that further contribute to the high virulence displayed by Ebola and influenza viruses.
Fig. 5.
The RNA-binding domains of Ebola VP35 and influenza NS1 proteins are incorporated into distinct scaffolds. (A and B) Structures of the Ebola VP35 IID (this study; left) (A) and the influenza NS1A dsRNA binding domain (PDB ID code 2ZKO) (B). (C) Similar side chains of arginine and lysine residues in the 8-residue alignment between the NS1A and VP35 protein sequences are shown and highlighted in gray (adapted from ref. 21).
Recent studies also show that EBOV can activate antiviral pathways that operate through RIG-I and MDA-5 to activate IFN regulatory factor 3 (IRF-3) (15, 25) and that Ebola VP35 protein can inhibit these antiviral responses (15). Cardenas et al. showed that VP35 may directly interact with host proteins at points proximal to IRF-3, possibly interacting with and preventing the activation or function of the kinases IKKε and TBK-1, which phosphorylate and activate IRF-3 (15). Furthermore, VP35 mutants that are unable to bind dsRNA also show reduced but appreciable suppression of IRF-3 phosphorylation and nuclear localization (15, 22), suggesting that IFN-inhibitory activity of VP35 involves multiple modes. Our structure reveals that residues critical for dsRNA binding and immune inhibition form an extended basic patch. Mutation of these residues results in only minor structural perturbations, yet prevents interactions with dsRNA. Based on these observations, we now propose a model, shown in Fig. 6, where VP35 suppresses IRF-3 activation by dsRNA-dependent activity through direct sequestration of dsRNA and through inhibition of the RIG-I/MDA-5 helicases. Recent structural and biochemical analyses of RIG-I and PKR, an RNA-dependent protein kinase, revealed that additional modifications such as 5′ triphosphate groups can play a role in the RNA recognition by RIG-I to provide additional specificity (11, 12). Alternatively, VP35 can suppress host immune responses through dsRNA-independent activity perhaps by direct binding and inhibition of IRF-3 kinases (15). Our current study suggests that both activities are likely to be mediated through mechanisms that involve interactions with the conserved basic patch centered on Arg-312 and therefore, any changes to residues located on this patch can lead to reduced host immune evasion. Consistent with our model, a recent microarray analysis showed that a recombinant EBOV containing an Arg312Ala mutation leads to activation of antiviral responses, whereas the wild-type virus completely shuts down host innate immune signaling (25). Together, these studies suggest that the conserved basic patch identified in our study can mediate multiple IFN inhibitory mechanisms and is critical for viral replication and pathogenesis.
Fig. 6.
Model for VP35-mediated IFN inhibition and immune suppression.
Emerging and reemerging viruses such as Ebola are a significant threat to global human health, and the critical role played by Ebola VP35 in host immune suppression is well established. Our crystal structure of VP35 IID now provides a framework to understand structural characteristics that promote interactions between VP35 and dsRNA, which correlates with the ability of Ebola VP35 to antagonize host antiviral signaling pathways. High sequence conservation among filoviruses suggests that VP35 proteins from other family members will likely retain similar architecture. High-resolution structure and solution-state NMR data from this study provide opportunities for targeted antiviral and diagnostic drug design. The unique information afforded by the structure of Ebola VP35 IID will also facilitate future studies to reveal regulatory mechanisms at a key host–viral interface.
Methods
Crystallization and Diffraction Data Collection.
Initial conditions for crystallization were identified by using purified VP35 IID protein (SI Text) and a commercial screen (Hampton Research). In-house optimized native and selenomethionine crystals were grown at 25 °C by the hanging-drop vapor-diffusion method with 17 mg/ml protein solutions diluted with 200 mM sodium citrate (pH 5.8) and 11% (wt/vol) PEG 4000. Crystals were soaked in reservoir solution with 25% glycerol (wt/vol) and frozen in a nitrogen stream. Diffraction data were collected at the Advanced Light Source (beamline 4.2.2) at 100 K (statistics are listed in Table S1).
Structure Determination and Refinement.
Data were processed by using d*TREK (33). Intensities were converted to structure factors by using the CCP4 program TRUNCATE (34). Phases were determined from a multiple-wavelength anomalous dispersion (MAD) experiment performed on a Se-Met substituted crystal. Phasing was performed with the SOLVE/RESOLVE program package (35). ARP/wARP (36) was used to trace the backbone by using the VP35 sequence, which led to an initial model with ≈20% of the residues modeled as alanine. The rest of the model was constructed manually by using XtalView (37) into an electron density map generated from MAD phases to 1.4-Å resolution by using native data. Refinement was performed against the structure factors by using CNS (38) and REFMAC5 (39). Refinement included simulated annealing, followed by conjugate gradient energy minimization. Individual thermal parameters were refined after each cycle of simulated annealing and subject to standard restraints. Water molecules were automatically added by using CNS if a peak >3.0σ was present in Fourier maps with coefficients (Fobs − Fcalc)eiαcalc. The contribution of bulk solvent to structure factors was determined by CNS (default parameters). The model was further refined by using REFMAC5 using MLKF residual function, weight matrix of 0.75, bulk solvent scaling, and individual anisotropic B-factors. The refined model was verified by using a 2Fo−Fc map showing that all residues have continuous electron density. Final refinement statistics are shown in Table S1.
Supplementary Material
Acknowledgments.
We thank Drs. R. DeGuzman, D. Klein, and D. Borek for support and discussions; Dr. M. Shogren-Knaak, Dr. M. Nilsen-Hamilton, Dr. T. Bobik, H. Azzaz, C. Warner, and S. Kakar for helpful discussions; the ISU X-ray Crystallography facility and Dr. J. Hoy for assistance with initial X-ray data collection; Dr. D. Klein for providing the dsRNA; and P. Ramanan, L. Helgeson, D. Peterson, and M. Farahbakhsh for laboratory assistance. This work was supported in part by the Roy J. Carver Charitable Trust Grant 09-3271 (to G.K.A.), a Roy J. Carver Trust Graduate Fellowship (to N.D.G.), and National Institutes of Health Grant AI059536 (to C.F.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.A.W. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID: 3FKE).
This article contains supporting information online at www.pnas.org/cgi/content/full/0807854106/DCSupplemental.
References
- 1.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM, et al., editors. Fields Virology. 5th Ed. Philadelphia: Lippincott Williams & Wilkins; 2006. pp. 1409–1448. [Google Scholar]
- 2.Zampieri CA, Sullivan NJ, Nabel GJ. Immunopathology of highly virulent pathogens: Insights from Ebola virus. Nat Immunol. 2007;8:1159–1164. doi: 10.1038/ni1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosio CM, et al. Ebola and Marburg viruses replicate in monocyte-derived dendritic cells without inducing the production of cytokines and full maturation. J Infect Dis. 2003;188:1630–1638. doi: 10.1086/379199. [DOI] [PubMed] [Google Scholar]
- 4.Mahanty S, et al. Protection from lethal infection is determined by innate immune responses in a mouse model of Ebola virus infection. Virology. 2003;312:415–424. doi: 10.1016/s0042-6822(03)00233-2. [DOI] [PubMed] [Google Scholar]
- 5.Bray M, Geisbert TW. Ebola virus: The role of macrophages and dendritic cells in the pathogenesis of Ebola hemorrhagic fever. Int J Biochem Cell Biol. 2005;37:1560–1566. doi: 10.1016/j.biocel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Bray M, Mahanty S. Ebola hemorrhagic fever and septic shock. J Infect Dis. 2003;188:1613–1617. doi: 10.1086/379727. [DOI] [PubMed] [Google Scholar]
- 7.Baize S, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 8.Saito T, Gale M., Jr Principles of intracellular viral recognition. Curr Opin Immunol. 2007;19:17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007;85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 10.Tortorella D, Gewurz BE, Furman MH, Schust DJ, Ploegh HL. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 11.Cui S, et al. The C-terminal regulatory domain is the RNA 5′-triphosphate sensor of RIG-I. Mol Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 12.Marques JT, et al. A structural basis for discriminating between self and nonself double-stranded RNAs in mammalian cells. Nat Biotechnol. 2006;24:559–565. doi: 10.1038/nbt1205. [DOI] [PubMed] [Google Scholar]
- 13.Basler CF, Garcia-Sastre A. Viruses and the type I interferon antiviral system: Induction and evasion. Int Rev Immunol. 2002;21:305–337. doi: 10.1080/08830180213277. [DOI] [PubMed] [Google Scholar]
- 14.Basler CF, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cardenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartman AL, et al. Inhibition of IRF-3 activation by VP35 is critical for the high level of virulence of Ebola virus. J Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Enterlein S, et al. VP35 knockdown inhibits Ebola virus amplification and protects against lethal infection in mice. Antimicrob Agents Chemother. 2006;50:984–993. doi: 10.1128/AAC.50.3.984-993.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moller P, Pariente N, Klenk HD, Becker S. Homo-oligomerization of Marburgvirus VP35 is essential for its function in replication and transcription. J Virol. 2005;79:14876–14886. doi: 10.1128/JVI.79.23.14876-14886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid SP, Cardenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 22.Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basler CF, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donelan NR, Basler CF, Garcia-Sastre A. A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol. 2003;77:13257–13266. doi: 10.1128/JVI.77.24.13257-13266.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hartman AL, Ling L, Nichol ST, Hibberd ML. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta M, Mahanty S, Ahmed R, Rollin PE. Monocyte-derived human macrophages and peripheral blood mononuclear cells infected with Ebola virus secrete MIP-1alpha and TNF-alpha and inhibit poly-IC-induced IFN-alpha in vitro. Virology. 2001;284:20–25. doi: 10.1006/viro.2001.0836. [DOI] [PubMed] [Google Scholar]
- 27.Haasnoot J, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Imani F, Jacobs BL. Inhibitory activity for the interferon-induced protein kinase is associated with the reovirus serotype 1 sigma 3 protein. Proc Natl Acad Sci USA. 1988;85:7887–7891. doi: 10.1073/pnas.85.21.7887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chang HW, Jacobs BL. Identification of a conserved motif that is necessary for binding of the Vaccinia virus E3L gene products to double-stranded RNA. Virology. 1993;194:537–547. doi: 10.1006/viro.1993.1292. [DOI] [PubMed] [Google Scholar]
- 30.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35–VP40 interaction is sufficient for packaging 3E–5E minigenome RNA into virus-like particles. J Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chien CY, et al. Biophysical characterization of the complex between double-stranded RNA and the N-terminal domain of the NS1 protein from influenza A virus: Evidence for a novel RNA-binding mode. Biochemistry. 2004;43:1950–1962. doi: 10.1021/bi030176o. [DOI] [PubMed] [Google Scholar]
- 32.Yin C, et al. Conserved surface features form the double-stranded RNA binding site of non-structural protein 1 (NS1) from influenza A and B viruses. J Biol Chem. 2007;282:20584–20592. doi: 10.1074/jbc.M611619200. [DOI] [PubMed] [Google Scholar]
- 33.Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D. 1999;55(Pt 10):1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computational Project N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D. 1999;55(Pt 4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrakis A, Sixma TK, Wilson KS, Lamzin VS. wARP: Improvement and extension of crystallographic phases by weighted averaging of multiple-refined dummy atomic models. Acta Crystallogr D. 1997;53(Pt 4):448–455. doi: 10.1107/S0907444997005696. [DOI] [PubMed] [Google Scholar]
- 37.McRee DE. XtalView/Xfit—A versatile program for manipulating atomic coordinates and electron density. J Struct Biol. 1999;125:156–165. doi: 10.1006/jsbi.1999.4094. [DOI] [PubMed] [Google Scholar]
- 38.Brunger AT, et al. Crystallography NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54(Pt 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53(Pt 3):240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.